Abstract
Introduction
Optimal treatment for recurrent glioblastoma isocitrate dehydrogenase 1 and 2 wild-type (rGBM IDH-WT) is not standardized, resulting in multiple therapeutic approaches. A phase III clinical trial showed that tumor treating fields (TTFields) monotherapy provided comparable survival benefits to physician’s chemotherapy choice in rGBM. However, patients did not equally benefit from TTFields, highlighting the importance of identifying predictive biomarkers of TTFields efficacy.
Methods
A retrospective review of an institutional database with 530 patients with infiltrating gliomas was performed. Patients with IDH-WT rGBM receiving TTFields at first recurrence were included. Tumors were evaluated by next-generation sequencing for mutations in 205 cancer-related genes. Post-progression survival (PPS) was examined using the log-rank test and multivariate Cox-regression analysis.
Results
149 rGBM patients were identified of which 29 (19%) were treated with TTFields. No significant difference in median PPS was observed between rGBM patients who received versus did not receive TTFields (13.9 versus 10.9 months, p = 0.068). However, within the TTFields-treated group (n = 29), PPS was improved in PTEN-mutant (n = 14) versus PTEN-WT (n = 15) rGBM, (22.2 versus 11.6 months, p = 0.017). Within the PTEN-mutant group (n = 70, 47%), patients treated with TTFields (n = 14) had longer median PPS (22.2 versus 9.3 months, p = 0.005). No PPS benefit was observed in PTEN-WT patients receiving TTFields (n = 79, 53%).
Conclusions
TTFields therapy conferred a significant PPS benefit in PTEN-mutant rGBM. Understanding the molecular mechanisms underpinning the differences in response to TTFields therapy could help elucidate the mechanism of action of TTFields and identify the rGBM patients most likely to benefit from this therapeutic option.
Keywords: Tumor treating fields (TTFields), PTEN, Recurrent glioblastoma, IDH-wildtype, Predictive biomarker
Introduction
Glioblastomas (GBMs) are malignant brain tumors associated with poor prognosis and a short time to recurrence after initial treatment. The current standard of care involves maximal safe resection followed by concurrent radiotherapy and temozolomide [1]. Treatment at the time of recurrence is variable but commonly involves re-resection, re-irradiation, multiple chemotherapy regimens, and/or clinical trial enrollment. However, no treatments for rGBM have been demonstrated to improve overall survival (OS) in phase III randomized clinical trials (RCTs) [2].
Tumor treating fields (TTFields) are low-intensity electric fields (1–4 V/cm) that alternate at an intermediate frequency (100–300 kHz) to disrupt mitosis and inhibit tumor growth. However, the mechanism of action of TTFields is not entirely understood [3]. TTFields is the only FDA-approved GBM therapy in the past decade to demonstrate prolonged progression-free survival (PFS) and overall survival (OS) when added to the standard of care [4]. In addition, the EF-11 RCT demonstrated that TTFields monotherapy provides comparable survival to physician’s choice chemotherapy in the setting of rGBM [5]. However, as not all GBM patients respond equally to TTFields, understanding which patients will benefit the most from this therapy by identifying predictors of response would have important clinical implications.
In addition, both EF-11 and EF-14 RCTs were performed prior to the routine incorporation of molecular alterations in the diagnosis of infiltrating gliomas [4, 5]. The increased understanding of the clinical implications of mutations in isocitrate dehydrogenase 1 or 2 (IDH1 or IDH2), among other genes, has led to a refinement of the classification of gliomas [6–8]. Therefore, it is important to consider genetic alterations when evaluating the effects of therapeutic interventions, including TTFields.
The goal of this study was (1) to evaluate the survival effects of TTFields in a cohort of patients with IDH wild-type (IDH-WT) recurrent GBM (rGBM) and (2) to investigate possible clinical characteristics or genomic alterations that may predict responsiveness to TTFields. Our results show that mutations in PTEN, a tumor suppressor gene frequently altered in GBM, predict benefit from TTFields in patients with rGBM IDH-WT.
Methods
Patients and tumor samples
We retrospectively searched for patients with infiltrating gliomas using an institutional glioma registry of cases diagnosed between 2010 and 2019. The inclusion criteria for this study were (1) histological diagnosis of IDH-WT GBM; (2) treatment with TTFields at first recurrence; (3) TTFields treatment for more than 4 weeks; and (4) available tumor genomic alteration information (Online Resource 1).
Data for this study were collected from the electronic medical record of Memorial Hermann Hospital (Houston, TX) and compiled utilizing REDCap electronic data capture tools hosted at the University of Texas Health Science Center at Houston (UTHealth) [9, 10]. Data collected included age, sex, Karnofsky performance status score (KPS), diagnosis, tumor location, volumetric extent of resection, initial treatment strategy, treatment strategy at the time of recurrence, use of TTFields, PFS, OS, and post-progression survival (PPS). Tumors were classified by a board-certified neuropathologist following the 2016 WHO Classification of Tumors of the Central Nervous System [6]. Available TTFields data included: average percent usage and therapy start and end dates, which was obtained from the Optune® (Novocure Ltd., Haifa, Israel) usage database. Radiographic extent of resection was classified as gross-total resection (GTR), near-total resection (NTR), or subtotal resection (STR) as previously described [11]. Recurrence and therapeutic strategy were determined by a review of cases by a multidisciplinary tumor board as previously described [12].
A group of IDH-WT rGBM patients without TTFields treatment was utilized as the control cohort. This group was identified using our institutional registry. Patients who met the following criteria were included: (1) diagnosis of IDH-WT GBM; (2) imaging or histological evidence of recurrence; (3) TTFields therapy-naïve; and (4) available tumor genomic alteration information (Online Resource 1).
Ethics declaration
This study was approved by the Institutional Review Board (ID: HSC-MS-17–0917) of UTHealth and Memorial Hermann Hospital, Houston, TX, USA.
Targeted sequencing
Tumor tissue samples were analyzed for genetic alterations by a targeted next-generation sequencing (NGS) panel interrogating 205 genes and 26 gene rearrangements (FoundationOne, Foundation Medicine Inc., Cambridge, MA, USA). The FoundationOne assay was performed in a clinical laboratory improvement amendments (CLIA)-certified laboratory, as previously described [13–15].
Digital droplet PCR (ddPCR)
In a subset of patients (n = 5) TERT promoter mutation was evaluated through ddPCR. FFPE tissue samples were tested using TERT C228T dHsaEXD72405942 and TERT C250T dHsaEXD46675715 (Bio-Rad Laboratories, CA, USA) probes as previously described [16].
Statistical analyses
Descriptive analyses were evaluated by Fisher’s exact test or Mann–Whitney U-test for categorical or continuous variables, respectively. The primary and secondary study outcomes were PPS and OS, respectively. We anticipated that therapeutic strategies at recurrence could bias OS either by including patients without recurrence (sample bias) or taking into consideration the time before recurrence, which was not affected by the treatment initiated after first recurrence (lead-time bias). This lead time bias has been demonstrated in studies of GBM reoperation [17]. The Kaplan–Meier method was used to plot survival curves and the statistical significance was examined by the log-rank test. Multivariate Cox proportional hazard regression models were used to calculate the hazard ratio (HR) estimates with 95% confidence intervals (95% CI), adjusted for the known variables that affect survival. A p-value of < 0.05 was considered statistically significant. All statistical analyses were performed in EZR (v.1.40) [10, 18] and GraphPad Prism (version 9.0, La Jolla, CA, USA).
Results
Cohort characteristics
Five hundred and thirty (530) infiltrating gliomas were identified between 2010 to 2019 from our institutional registry. We selected 29 rGBM patients treated with TTFields that fulfilled the inclusion criteria (Online Resource 1). The median age was 58 years (range 40–70 years). The majority of patients (n = 19, 66%) were male, and 11 (38%) patients had a preoperative KPS ≥ 80. All patients were treated with maximal safe resection with 9 (33%) having gross-total resection (GTR), and 28 (97%) were treated according to the Stupp protocol [1].
The median time to progression from initial diagnosis was 4.7 months. All 29 rGBM patients received TTFields therapy for first GBM recurrence, with a median TTFields start time of 51 days (range 2–161 days) from the diagnosis of recurrence. TTFields was used for a median of 176 days (range 41–961 days), while the median percentage of daily usage was 59% (range 3–88%). The 29 rGBM patients were concurrently treated with reoperation (n = 12, 41%), temozolomide re-challenge (n = 19, 66%), bevacizumab (n = 24, 83%), and/or irinotecan (n = 15, 52%). Table 1 summarizes the demographic and clinical characteristics of the TTFields-treated rGBM cohort.
Table 1.
Characteristics of tumor treating fields (TTFields)-treated patients with recurrent glioblastoma (rGBM) IDH-WT and PTEN mutation status (n = 29)
| Characteristics, N (%) | TTFields-treated patients N = (29) | PTEN wild-type N = 15 | PTEN mutant N = 14 | p-value |
|---|---|---|---|---|
|
| ||||
| Age, median (IQR) | 58 (52–63) | 61 (54–64) | 58 (51–60) | 0.457 |
| Male | 19 (66) | 10 (67) | 9 (64) | 1.000 |
| White/Caucasian | 26 (90) | 12 (80) | 14 (100) | 0.224 |
| Pre-operative KPS ≥ 80 | 11 (38) | 7 (47) | 4 (29) | 0.450 |
| Gross total resection | 9 (31) | 5 (33) | 4 (29) | 1.000 |
| Chemoradiotherapy with TMZ | 28 (97) | 14 (93) | 14 (100) | 1.000 |
| Second resection | 12 (41) | 6 (40) | 6 (43) | 1.000 |
| Salvage TMZ | 19 (66) | 10 (67) | 9 (64) | 1.000 |
| Salvage bevacizumab | 24 (83) | 15 (100) | 9 (64) | 0.017 |
| Salvage SRS | 8 (28) | 3 (20) | 5 (36) | 0.427 |
| TTFields usage percentage, median (IQR)* | 59 (39–76) | 59 (37–74) | 64 (44–78) | 0.600 |
Significant p-values are bolded
TMZ temozolomide, KPS Karnofsky Performance Status, SRS stereotactic radiosurgery, WT wildtype, IQR interquartile range, N number
10 patients did not have available information (4 PTEN wildtype and 6 PTEN mutant)
The most common genomic alterations identified in the tumors of TTFields-treated patients were in TERT promoter (76%), CDKN2A/B (72%), EGFR (55%), PTEN (48%), TP53 (28%), PIK3CA (21%), NF1 (14%), PIK3R1 (14%), RB1 (14%), CDK4 (10%), MDM2 (10%), MDM4 (10%), BCOR (7%), GLI1 (7%), MYC (7%), PDGFRA (7%), and BRAF (7%), which were similar to the frequencies reported in larger series of GBM [19–22]. Detailed information on the genomic alterations in the TTFields-treated patients is included in Online Resource 2.
TTFields therapy in recurrent IDH-WT GBM
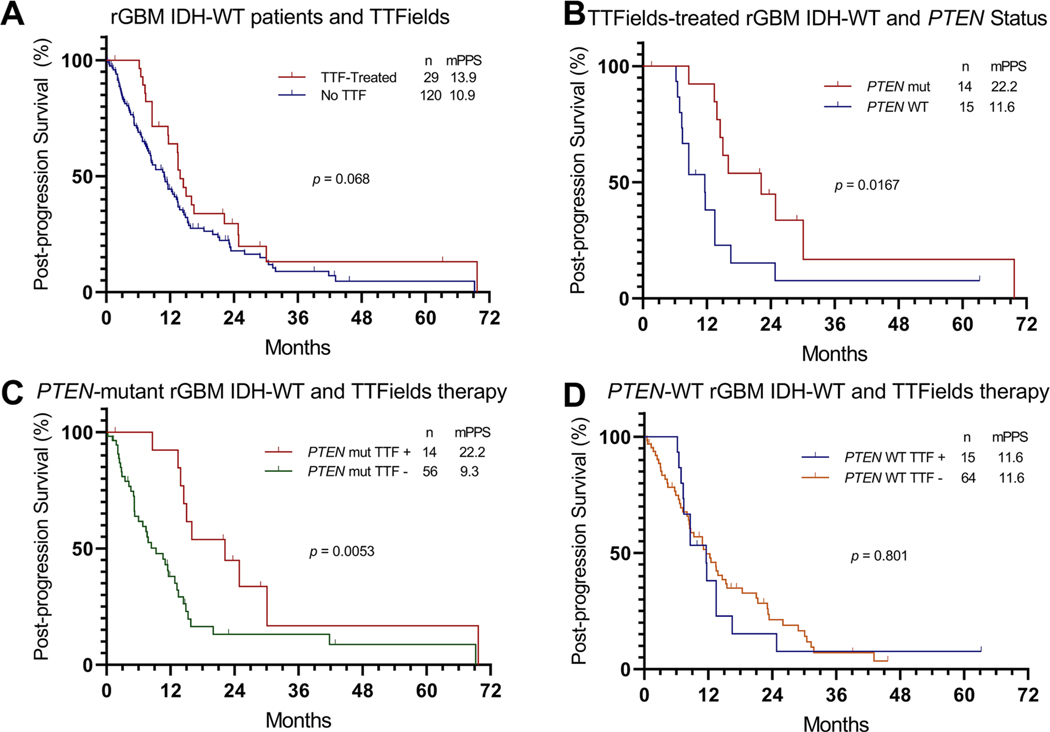
The rGBM patients treated with TTFields (n = 29) did not show a statistically significant increase in PPS compared to rGBM patients that did not receive TTFields (n = 120), (13.9 months VS. 10.9 months, p = 0.068), Fig. 1a. Similarly, no significant increase in OS was observed in rGBM patients treated with TTFields (Online Resource 3A).
Fig. 1.
Post-progression survival (PPS) differences in response to tumor treating fields (TTFields) treatment of recurrent glioblastoma (rGBM) isocitrate dehydrogenase wild-type (IDH-WT). a PPS of recurrent GBM IDH-WT by TTFields therapy b PPS of recurrent GBM IDH-WT patients treated with TTFields by PTEN status. c PPS of PTEN-mutant rGBM IDH-WT by TTFields therapy. d PPS of PTEN-WT rGBM IDH-WT by TTFields therapy. TTF TTFields.
Predictors of survival in rGBM patients treated with TTFields
Univariate analysis did not show a significant correlation between PPS and demographic or clinical characteristics in TTFields-treated patients (Online Resource 4). However, analysis of genetic alterations in rGBM patients treated with TTFields revealed a significantly longer PPS in patients with PTEN-mutant tumors (n = 14) compared to patients with PTEN-WT (n = 15) tumors (22.2 months vs. 11.6 months, p = 0.0167), Fig. 1b. Multivariate Cox-regression analysis adjusting for age and preoperative KPS demonstrated that preoperative KPS ≥ 80 (HR 0.30, p = 0.026) and PTEN mutation (HR 0.23, p = 0.003) independently correlated with improved PPS in TTFields-treated patients (Table 2). In addition to prolonged median PPS, there was an increased median OS in TTFields-treated patients with PTEN-mutant compared to PTEN-WT rGBM (30.8 vs. 16.6 months, p = 0.007), Online Resource 3B.
Table 2.
Multivariate analysis of post-progression survival of patients with recurrent glioblastoma (rGBM) IDH-WT treated with tumor treating fields (TTFields) (n = 29)
| Variables | HR (CI 95%) | p-value |
|---|---|---|
|
| ||
| Age (years) | 0.95 (0.90–1.01) | 0.128 |
| Preoperative KPS ≥ 80 | 0.30 (0.11–0.87) | 0.026 |
| PTEN mutation | 0.23 (0.09–0.63) | 0.003 |
Significant p-values are bolded
Multivariate analysis was performed using a Cox regression model HR hazard ratio, CI confidence interval, KPS Karnofsky Performance Status, WT wildtype
TTFields therapy improves survival of patients with PTEN-mutant rGBM
To confirm our results, we evaluated the effects of TTFields treatment in patients with PTEN-mutant rGBM. Among the 149 patients with GBM included in this study, 70 (47%) harbored a PTEN mutation, while 79 (53%) were PTEN-WT.
We compared the PPS of patients with PTEN-mutant rGBM between those treated with TTFields and those who did not receive TTFields therapy. Our results show an increased PPS in patients with PTEN-mutant rGBM that received TTFields (n = 14, 22.2 months) compared to those who did not receive TTFields therapy (n = 56, 9.3 months), p = 0.0053, Fig. 1c. Multivariate analysis adjusting for the most established covariates of survival in GBM (age and KPS), as well as other therapies used at recurrence (bevacizumab, temozolomide, and TTFields), showed that TTFields therapy was independently associated with improved PPS in patients with PTEN-mutant IDH-WT rGBM, p = 0.003 (Table 3). Although patients with PTEN-mutant rGBM that received TTFields had an approximate doubling of the median OS (30.8 months) compared to those who did not receive TTFields therapy (16.8 months), the difference was not statically significant (p = 0.054, Online Resource 3C).
Table 3.
Multivariable analysis of post-progression survival of patients with PTEN-mutant recurrent glioblastoma (rGBM) IDH-WT (N = 70)
| Variables | HR (CI 95%) | p-value |
|---|---|---|
|
| ||
| Age (years) | 1.00 (0.98–1.03) | 0.778 |
| Male | 0.77 (0.42–1.42) | 0.404 |
| Preoperative KPS ≥ 80 | 0.59 (0.30–1.17) | 0.130 |
| Non-GTR | 1.72 (0.85–3.48) | 0.134 |
| Salvage TMZ | 0.84 (0.43–1.65) | 0.610 |
| Salvage bevacizumab | 0.91 (0.48–1.73) | 0.775 |
| Salvage TTFields | 0.29 (0.12–0.66) | 0.003 |
Significant p-values are bolded
Multivariate analysis was performed using a Cox regression model
TTFields tumor treating fields, KPS Karnofsky Performance Status, GTR Gr oss-total resection TMZ temozolomide, HR hazard ratio, CI confidence interval, WT wildtype
Importantly, the improved PPS and OS observed with TTFields treatment in patients with PTEN-mutant rGBM was not observed in the n = 79 patients with PTEN-WT rGBM. There was no significant difference in median PPS between patients with PTEN-WT rGBM who received TTFields (n = 15, 11.6 months) and those who did not receive TTFields therapy (n = 64, 11.6 months), p = 0.801 (Fig. 1d). There was no significant difference in median OS between patients with PTEN-WT rGBM who received TTFields (n = 15, 16.6 months) and those who did not receive TTFields therapy (n = 64, 21.5 months), p = 0.078 (Online Resource 3D).
We also examined demographic and clinical characteristics that might explain the survival differences between patients with PTEN-mutant and PTEN-WT tumors treated with TTFields. One hundred percent (100%, n = 15) of patients with PTEN-WT rGBM treated with TTFields were simultaneously treated with salvage bevacizumab, which is higher than the 64% (n = 14) of patients with PTEN-mutant rGBM treated with TTFields (p = 0.017) that also received bevacizumab. However, no significant differences in TTFields usage percentage or any other clinical characteristics were observed between TTFields-treated patients with PTEN-mutant or PTEN-WT rGBM (Table 1).
The type of PTEN mutations and its biological effect were subsequently evaluated demonstrating 6 missense mutations, 4 nonsense mutations, 2 frameshift mutations, and 2 loss copy number alterations. From the 14 mutations; 7 are known to cause loss-of-function of the gene and 7 are likely to cause loss-of-function of the gene. Moreover, PTEN mutations reported in this study are considered pathogenic or likely pathogenic according to ClinVar and OncokB databases (Online Resource 5) [23, 24].
Discussion
The results of this study suggest that the effects of TTFields are influenced by tumor-related factors, particularly, the PTEN mutation status. Our study reveals, for the first time, a molecular biology predictor of responsiveness to TTFields therapy, i.e., that compared to PTEN-WT, PTEN-mutant GBM IDH-WT patients have an almost-doubling of median PPS due to TTFields at the time of recurrence (11.6 months vs. 22.2 months, respectively, p = 0.0167). More importantly, we did not observe differences in TTFields compliance, which is a known factor that influences TTFields efficacy [25], between patients with PTEN-mutant and PTEN-WT rGBM. Additionally, we found that TTFields-treated patients with KPS ≥ 80 derived a PPS benefit, which has been demonstrated in several GBM studies prior to the advent of TTFields therapy [26–28]. Importantly, large studies have demonstrated that PTEN mutations do not confer an outcome benefit in GBM IDH-WT [19–21]. Accordingly, we did not observed survival differences in either OS (PTEN-WT 21.5 vs. PTEN-mutant 16.8-months, p = 0.062) or PPS (PTEN-WT 11.6 vs. PTEN-mutant 9.3-months, p = 0.296) in patients not treated with TTFields.
The improved survival observed in patients with PTEN-mutant GBM might indicate a relationship between the mechanism of action of TTFields and PTEN’s cellular function. PTEN, a gene found in chromosome 10, is commonly mutated in human cancers including ~ 50% of GBM IDH-WT tumors [19–21]. Given the frequent loss-of-function of PTEN in cancers and its function (inhibition of PI3K pathway), PTEN is recognized as a bona fide tumor suppressor gene [29]. PTEN is involved in maintaining mitotic spindle architecture and promoting chromosome alignment and segregation [29]. These functions overlap with the postulated mechanism of action of TTFields, which involves induction of abnormal spindle formation and subsequent mitotic arrest or delay [30, 31]. Even though the mechanism is not yet fully elaborated, it is believed that TTFields cause an improper attachment of chromosomes to the spindle fibers [3, 32, 33]. Loss of PTEN function causes disruption of proper spindle assembly and chromosome segregation, which results in mitotic catastrophe [29, 34, 35]. Therefore, it is possible that the effects of TTFields therapy, which can inhibit the polymerization of microtubules and the assembly of the mitotic spindle apparatus, would be enhanced by loss-of-function mutations in PTEN.
Future directions
In vitro experiments using PTEN-mutant and PTEN-WT cell lines will facilitate the evaluation of TTFields’ effects on the mitotic spindle and mitotic division. Similarly, studies with animal models using PTEN-WT and PTEN-mutant tumor models could shed light into the mechanism of action of TTFields and the possible sensitizing effects of PTEN mutations. Finally, a prospective study evaluating the effects of TTFields on GBM patient survival in the context of the tumor’s genomic alterations (including PTEN mutation status) will be critical to confirm our results.
Limitations
Some limitations of our study include its retrospective nature and the potential for selection bias, as not all patients with rGBM in our institution had information available on the mutations present in the tumor. Also, the effects of concurrent systemic chemotherapies cannot be entirely segregated from the effects of TTFields. However, as Table 1 shows, the only significant difference in concurrent therapy use with TTFields at recurrence was more frequent salvage bevacizumab use in the PTEN-WT compared to the PTEN-mutant cohort. Other limitations of our study are the relatively small sample size of TTFields-treated patients and unknown MGMT promoter methylation status for most patients. Lastly, mutation analysis was performed in tissue from the initial resection. However, recent studies have shown that GBM may evolve stochastically from early driver events that are shared both at presentation and recurrence. Therefore, differences in genetic alterations between initial and recurrent tumors are not expected in critical oncogenic drivers like PTEN [36].
Conclusions
Compared to patients with PTEN-WT GBM, those with PTEN-mutant GBM derived a significantly improved survival benefit when treated with TTFields at recurrence. Understanding the molecular mechanisms underpinning and predicting the differences in response to TTFields therapy could help elucidate the mechanisms of action of TTFields, thereby identifying those patients that will benefit the most from this therapeutic option.
Supplementary Material
Acknowledgments
Funding Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number K08CA241651 (LYB). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s11060-021-03755-1.
Availability of data and material The datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Declarations
Ethical approval This study was approved by the institutional review board of The University of Texas Health Science Center at Houston and Memorial Hermann Hospital, Houston, TX and it was in accordance with the 1964 Helsinki Declaration and its later amendments.
Conflict of interest CBP declares patent applications with Novocure, Ltd. and support from the AACR-Novocure Career Development Award for TTFields Research.
References
- 1.Stupp R, Mason WP, van den Bent MJ et al. (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996. 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 2.Wen PY, Weller M, Lee EQ et al. (2020) Glioblastoma in adults: a society for neuro-oncology (sno) and european society of neuro-oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 10.1093/neuonc/noaa106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hottinger AF, Pacheco P, Stupp R (2016) Tumor treating fields: A novel treatment modality and its use in brain tumors. Neuro Oncol 18:1338–1349. 10.1093/neuonc/now182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupp R, Taillibert S, Kanner A et al. (2017) Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma a randomized clinical trial. JAMA - J Am Med Assoc 318:2306–2316. 10.1001/jama.2017.18718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupp R, Wong ET, Kanner AA et al. (2012) NovoTTF-100A versus physician’s choice chemotherapy in recurrent glioblastoma: A randomised phase III trial of a novel treatment modality. Eur J Cancer 48:2192–2202. 10.1016/j.ejca.2012.04.011 [DOI] [PubMed] [Google Scholar]
- 6.Louis DN, Perry A, Reifenberger G et al. (2016) The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131:803–820. 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 7.Brat DJ, Aldape K, Colman H et al. (2018) cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV.” Acta Neuropathol 136:805–810. 10.1007/s00401-018-1913-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brat DJ, Aldape K, Colman H et al. (2020) cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol 139:603–608. 10.1007/s00401-020-02127-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris PA, Taylor R, Minor BL et al. (2019) The REDCap consortium: building an international community of software platform partners. J Biomed Inform 95:103208. 10.1016/j.jbi.2019.103208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dono A, Wang E, Lopez V et al. (2020) Molecular characteristics and clinical features of multifocal glioblastoma. J Neurooncol. 10.1007/s11060-020-03539-z [DOI] [PubMed] [Google Scholar]
- 11.Esquenazi Y, Friedman E, Liu Z et al. (2017) The survival advantage of “supratotal” resection of glioblastoma using selective cortical mapping and the subpial technique. Neurosurgery 81:275–288. 10.1093/neuros/nyw174 [DOI] [PubMed] [Google Scholar]
- 12.Morris SAL, Zhu P, Rao M et al. (2019) Gamma knife stereotactic radiosurgery in combination with bevacizumab for recurrent glioblastoma. World Neurosurg 127:e523–e533. 10.1016/j.wneu.2019.03.193 [DOI] [PubMed] [Google Scholar]
- 13.Frampton GM, Fichtenholtz A, Otto GA et al. (2013) Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31:1023–1031. 10.1038/nbt.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwaederle M, Krishnamurthy N, Daniels GA et al. (2018) Telomerase reverse transcriptase promoter alterations across cancer types as detected by next-generation sequencing: a clinical and molecular analysis of 423 patients. Cancer 124:1288–1296. 10.1002/cncr.31175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dono A, Wang E, Lopez-Rivera V et al. (2020) Molecular characteristics and clinical features of multifocal glioblastoma. J Neurooncol. 10.1007/s11060-020-03539-z [DOI] [PubMed] [Google Scholar]
- 16.Corless BC, Chang GA, Cooper S et al. (2019) Development of novel mutation-specific droplet digital pcr assays detecting TERT promoter mutations in tumor and plasma samples. J Mol Diagn 21:274–285. 10.1016/j.jmoldx.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldman DA, Hovinga K, Reiner AS et al. (2018) The relationship between repeat resection and overall survival in patients with glioblastoma: A time-dependent analysis. J Neurosurg 129:1231–1239. 10.3171/2017.6.JNS17393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanda Y (2013) Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant 48:452–458. 10.1038/bmt.2012.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan Y, Takayasu T, Hines G, Dono A, Hsu SH, Zhu JJ, Riascos-Castaneda RF, Kamali A, Bhattacharjee MB, Blanco AI, Tandon N (2020) Landscape of genomic alterations in IDH wild-type glioblastoma identifies PI3K as a favorable Prognostic factor. JCO Precis Oncol 4:575–584 [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Lichtenberg T, Hoadley KA et al. (2018) An integrated tcga pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173:400–416.e11. 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U et al. (2012) The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:1–20. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zehir A, Benayed R, Shah RH et al. (2017) Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23:703–713. 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landrum MJ, Lee JM, Benson M et al. (2018) ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46:D1062–D1067. 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ballo MT, Urman N, Lavy-Shahaf G et al. (2019) Correlation of tumor treating fields dosimetry to survival outcomes in newly diagnosed glioblastoma: a large-scale numerical simulation-based analysis of data from the phase 3 ef-14 randomized trial. Int J Radiat Oncol Biol Phys 104:1106–1113. 10.1016/j.ijrobp.2019.04.008 [DOI] [PubMed] [Google Scholar]
- 26.Mirimanoff RO, Gorlia T, Mason W et al. (2006) Radiotherapy and temozolomide for newly diagnosed glioblastoma: recursive partitioning analysis of the EORTC 26981/22981-NCIC CE3 phase III randomized trial. J Clin Oncol 24:2563–2569. 10.1200/JCO.2005.04.5963 [DOI] [PubMed] [Google Scholar]
- 27.Stark AM, Stepper W, Mehdorn HM (2010) Outcome evaluation in glioblastoma patients using different ranking scores: KPS, GOS, mRS and MRC. Eur J Cancer Care (Engl) 19:39–44. 10.1111/j.1365-2354.2008.00956.x [DOI] [PubMed] [Google Scholar]
- 28.Chambless LB, Kistka HM, Parker SL et al. (2015) The relative value of postoperative versus preoperative Karnofsky Performance Scale scores as a predictor of survival after surgical resection of glioblastoma multiforme. J Neurooncol 121:359–364. 10.1007/s11060-014-1640-x [DOI] [PubMed] [Google Scholar]
- 29.Hou SQ, Ouyang M, Brandmaier A et al. (2017) PTEN in the maintenance of genome integrity: from DNA replication to chromosome segregation. BioEssays 39:1–9. 10.1002/bies.201700082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessler AF, Frömbling GE, Gross F et al. (2018) Effects of tumor treating fields (TTFields) on glioblastoma cells are augmented by mitotic checkpoint inhibition. Cell Death Discov. 10.1038/s41420-018-0079-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giladi M, Schneiderman RS, Voloshin T et al. (2015) Mitotic spindle disruption by alternating electric fields leads to improper chromosome segregation and mitotic catastrophe in cancer cells. Sci Rep 5:1–16. 10.1038/srep18046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giladi M, Schneiderman RS, Porat Y et al. (2014) Mitotic disruption and reduced clonogenicity of pancreatic cancer cells in vitro and in vivo by tumor treating fields. Pancreatology 14:54–63. 10.1016/j.pan.2013.11.009 [DOI] [PubMed] [Google Scholar]
- 33.Kirson ED, Dbalý V, Tovaryš F et al. (2007) Alternating electric fields arrest cell proliferation in animal tumor models and human brain tumors. Proc Natl Acad Sci USA 104:10152–10157. 10.1073/pnas.0702916104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He J, Zhang Z, Ouyang M et al. (2016) PTEN regulates EG5 to control spindle architecture and chromosome congression during mitosis. Nat Commun. 10.1038/ncomms12355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, Hou SQ, He J et al. (2016) PTEN regulates PLK1 and controls chromosomal stability during cell division. Cell Cycle 15:2476–2485. 10.1080/15384101.2016.1203493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barthel FP, Johnson KC, Varn FS et al. (2019) Longitudinal molecular trajectories of diffuse glioma in adults. Nature 576:112–120. 10.1038/s41586-019-1775-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.