Abstract
Human intestinal enteroids derived from adult stem cells offer a relevant ex vivo system to study biological processes of the human gut. They recreate cellular and functional features of the intestinal epithelium of the small intestine (enteroids) or colon (colonoids) albeit limited by the lack of associated cell types that help maintain tissue homeostasis and respond to external challenges. In the gut, innate immune cells interact with the epithelium, support barrier function, and deploy effector functions. We have established a co-culture system of enteroid/colonoid monolayers and underlying macrophages and polymorphonuclear neutrophils to recapitulate the cellular framework of the human intestinal epithelial niche. Enteroids are generated from biopsies or resected tissue from any segment of the human gut and maintained in long-term cultures as three-dimensional structures through supplementation of stem cell growth factors. Immune cells are isolated from fresh human whole blood or frozen peripheral blood mononuclear cells (PBMC). Monocytes from PBMC are differentiated into macrophages by cytokine stimulation prior to co-culture. The methods are divided into the two main components of the model: (1) generating enteroid/colonoid monolayers and isolating immune cells and (2) assembly of enteroid/colonoid-immune cell co-cultures with separate apical and basolateral compartments. Co-cultures containing macrophages can be maintained for 48 hr while those involving neutrophils, due to their shorter life span, remain viable for 4 hr. Enteroid-immune co-cultures enable multiple outcome measures, including transepithelial resistance, production of cytokines/chemokines, phenotypic analysis of immune cells, tissue immunofluorescence imaging, protein or mRNA expression, antigen or microbe uptake, and other cellular functions.
Basic Protocol 1:
Seeding enteroid fragments onto Transwells for monolayer formation
Alternate Protocol:
Seeding enteroid fragments for monolayer formation using trituration
Basic Protocol 2:
Isolation of monocytes and derivation of immune cells from human peripheral blood
Basic Protocol 3:
Isolation of neutrophils from human peripheral blood
Basic Protocol 4:
Assembly of enteroid/macrophage or enteroid/neutrophil co-culture
Keywords: co-culture, human enteroids, intestinal organoids, macrophages, monolayer, neutrophils, innate immune cells
INTRODUCTION
Intestinal epithelial barrier function depends upon the combined participation of the epithelial cells and underlying immune, nerve, and mesenchymal cells. Traditional in vitro modeling of the human gut mucosa has mainly considered the epithelial cells in the absence of other immune cell populations. The biological processes by which subepithelial cells influence the function of the intestinal epithelium and their coordinated actions have been difficult to explore due to the lack of a relevant in vitro model. Traditional experimental models utilize transformed cell lines, which due to their altered (immortal) nature, do not reflect normal cell behavior. Because these immortalized cell lines are from a specific intestinal region, they fail to recreate the physiology of other intestinal segments. The advent of human intestinal enteroid technology based on adult stem-cell-derived “mini-guts” (Sato et al., 2009; Sato & Clevers, 2013; Jung et al., 2011) has vastly expanded the ability for in-depth study of the human intestinal epithelium. A convenient aspect of the intestinal enteroid system is the ability to generate a single layer of polarized cells (monolayers) with easy access to both apical and basolateral compartments. A downside of the intestinal enteroid model in its current form is the lack of other cell types that contribute to tissue homeostasis and host responses to microbial or pharmacological challenges of the epithelium. With this in mind, we developed a co-culture system using primary human intestinal cells together with immune cells seeking to bridge some of the gaps in modeling the human intestinal epithelium in vitro.
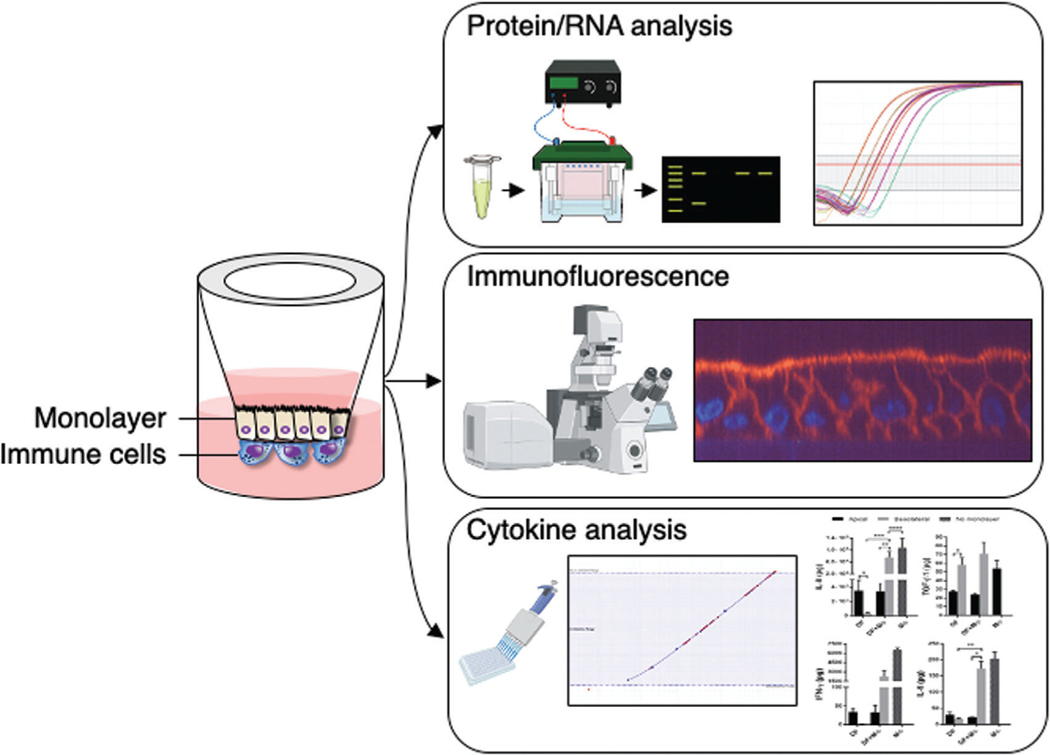
Here, we outline methods for establishing human enteroid monolayers and for their assembly into co-culture with innate immune cells (e.g., monocytes, macrophages, and neutrophils). Long-term, self-renewing enteroid cultures derived from adult intestinal stem cells serve as the source of intestinal epithelial cells. Three-dimensional enteroids are fragmented and subsequently seeded onto the upper chamber of cell culture inserts (e.g., Transwells Corning). Once the fragmented enteroids grow and form a polarized confluent monolayer, immune cells are adhered to the opposite side of the permeable membrane of the cell culture insert or Transwell (TW) facing the basolateral membrane of the epithelial cells to generate the immune cell co-culture (Fig. 1). The monolayer configuration enables the study of both epithelial and immune cells in combination or separately for their contributions to normal physiology or pathology of the human intestine. Basic Protocol 1 presents two methods for generating enteroid fragments and plating them onto permeable membranes or Transwell inserts. One method utilizes an enzymatic treatment to produce enteroid fragments, while the second method achieves enteroid framentation by mechanical means. Once cell culture inserts are seeded with enteroid fragments, it takes 1 to 2 weeks for the monolayer to form and reach confluency. Epithelial cell differentiation into “villus-like” intestinal cells (i.e., absorptive enterocytes, goblet cells, enteroendocrine cells, and Paneth cells) can be induced by incubating monolayers with differentiation medium (DFM) which lacks both Wnt3A and R-Spondin-1 (refer to medium components under Reagents and Solutions at the end of the article) for 5 days (Noel et al., 2017; Yin et al., 2018). Monolayer formation followed by 5 days of differentiation are the rate-limiting steps to co-culture assembly and require strategic experimental planning to coincide with the procurement of monocyte-derived macrophages or fresh neutrophils. The methods described herein are not only applicable to generate an enteroid/macrophage co-culture but also to produce co-cultures of enteroids containing monocytes and neutrophils.
Figure 1.
Generation and applications of immune cell co-culture.
Macrophages are derived from monocytes isolated from human PBMC. The separation and recovery of PBMC from human peripheral blood is accomplished by Ficoll-Hypaque density gradient centrifugation (see Current Protocols article: Fuss, Kanof, Smith, & Zola, 2009). PBMC are either used immediately for monocyte isolation or cryopreserved in liquid nitrogen for future use. Monocytes are enriched from PBMC by negative selection using a Pan Monocyte Isolation kit (Miltenyi Biotec) and subsequently differentiated into macrophages by supplementation of macrophage colony-stimulating factor (M-CSF) in the culture medium for 6 days. Monocyte-derived macrophages exhibit a CD14+ CD16low CD64low CX3CR1− phenotype and have the capacity to phagocytose bacteria (Noel et al., 2017). Successful assembly of the co-culture requires careful time coordination to obtain confluent, differentiated enteroid monolayers and fully differentiated macrophages. The required pre-incubation and planning times are built into the protocol described.
Co-cultures can also be assembled with monocytes, macrophages, or neutrophils. Once monocytes are enriched from PBMC (either fresh or frozen), they can be used immediately or can be maintained in culture for 7 to 10 days in the same culture medium used to derive macrophages but omitting M-CSF. Human polymorphonuclear neutrophils (PMN) exhibiting the phenotype CD15+ CD16+ CD14− are used the same day of isolation (Lemme-Dumit, Doucet, Zachos, & Pasetti, 2020). Co-cultures with PMN are short-lived given PMN’s limited lifespan (Kolaczkowska & Kubes, 2013). Details for the isolation and handling of PMN are covered by Kuhns and co-workers (Current Protocols article: Kuhns, Priel, Chu, & Zarember, 2015). Once the PMN are adhered to the bottom of the insert/Transwell, the co-culture may be interrogated for up to 2 hr post assembly.
STRATEGIC PLANNING
It is important to have experience with the propagation and handling of enteroid cultures as these methods (Foulke-Abel et al., 2016; Fujii, Matano, Nanki, & Sato, 2015; Noel et al., 2017; Yin et al., 2018) will not be addressed in detail in this article (also see Current Protocols article: Poole, Rajan, & Maresso, 2018). It is critical to achieve healthy enteroid cultures that need splitting at a minimum ratio of 1:2 every 6 to 7 days prior to use for monolayers. Maintain enteroids in small domes (25 to 30 L of Matrigel; Corning, 356231) in 24-well plates. Enteroid lines recovered from liquid nitrogen storage should be propagated for at least 2 to 3 weeks and observed for sustained growth before monolayer seeding. Careful monitoring of monolayer formation prior to differentiation by measuring transepithelial electrical resistance (TEER; Srinivasan et al., 2015) or visually by low power light microscopy will dictate when to procure a blood donor for monocyte or PMN isolation. Although not ultimately necessary, we highly recommend access to a voltohmmeter to ascertain monolayer formation and maturation and to determine loss or gain of barrier function as an experimental challenge outcome. It is a convenient and non-invasive way to judge monolayer health over time. As for immune cells, macrophages are derived from monocytes after 6 days in medium containing M-CSF and PMN are used fresh the same day of isolation. Monocyte differentiation into macrophages (6 days) should begin a day ahead of the start of monolayer maturation (5 days). The co-culture is assembled on the fifth day of monolayer differentiation. The protocols have incorporated timelines to coincide monolayer differentiation with macrophage derivation from monocytes.
CAUTION:
Universal safety precautions should be followed when handling human blood and tissue samples. Institutional Review Board-approved protocols are required for collection and use of human tissue and for conduct of any research involving human subjects. All solutions and tubes used need to be sterile and proper sterile technique maintained throughout. PBMC and PMN isolation should be performed in a biosafety level 2 (BSL2) cabinet using sterile technique. Enteroids and co-cultures should be prepared and manipulated in a dedicated BSL2 cabinet to avoid contamination.
NOTE:
Co-cultures for bacterial infection require monolayer differentiation in media without antibiotics. All other co-cultures for non-infection experiments are established in media with antibiotics (Lemme-Dumit et al., 2020; Noel et al., 2017).
BASIC PROTOCOL 1
SEEDING ENTEROID FRAGMENTS ONTO TRANSWELLS FOR MONOLAYER FORMATION
Monolayer formation precedes all subsequent steps in the protocol. It is advisable to practice seeding enteroids/colonoids onto inserts/TWs and monitor monolayer formation prior to planning a co-culture experiment (see Strategic Planning and Noel et al., 2017). Practical experience on enteroid/colonoid monolayer formation is important to adequately time blood collection and immune cell isolation. We recommend using one well (from a 24-well plate) with ~100 enteroids to seed two TWs or equivalent inserts (24-well plates, 0.33 cm2 growth area). It is, therefore, important to consider the number of enteroid wells needed for seeding in addition to wells needed for enteroid propagation. Once TWs are seeded, there is a 7 to 14 day growth period until the enteroid fragments grow into monolayers and this is followed by 5 days of differentiation (12 to 19 days in total). Basic Protocol 1 utilizes an enzymatic step to generate enteroid/colonoid fragments. However, some lines will not form monolayers after enzymatic digestion and need to be fragmented by mechanical trituration (Alternate Protocol); this can only be determined empirically. TEER will gradually increase as monolayers become confluent in propagation medium. Alternatively, monolayer confluence may be monitored by light microscopy under low magnification (e.g., inverted light microscope).
Materials
Enteroids/colonoids embedded in Matrigel (phenol red free; Corning, 356231) in non-differentiation medium (NDM) for cell propagation (see recipe; Fujii et al., 2015; Yin et al., 2018)
Collagen IV, from human placenta (MilliporeSigma, C5533; prepare 1 mg/ml in 100 mM or 0.5 M acetic acid; store in single-use aliquots at −20°C)
Sterile PBS
TrypLE Express Enzyme, 1 ×, no phenol red (Thermo Fisher Scientific, 12604013; aliquot and store protected from light at room temperature)
Cultrex Organoid Harvesting solution (biotechne, R&D Systems brand, 3700–100-01)
Enteroid propagation medium: non-differentiation medium (NDM; see recipe) Differentiation medium (DFM; see recipe; Fujii et al., 2015; Yin et al., 2018) Complete medium without growth factors (CMGF-; see recipe)
10 μM Y-27632 (ROCK inhibitor for ileum and colon; see recipe)
10 μM CHIR99021 (GSK-3 inhibitor for ileum and colon; see recipe)
1.0-μm pore Transwell (TW)/culture inserts (PET membrane; MilliporeSigma, MCSP24H48) for 24-well plates for macrophage or monocyte co-cultures
3.0-μm pore Transwell (TW)/culture inserts (PET membrane; Corning, 3472) for 24-well plates for PMN co-cultures (PET membranes are translucent and permit visualization of the underlying immune cells through the monolayer for immunofluorescence)
Sterile mini cell scrapers (United Biosystems, MCS-200) Centrifuge (swinging bucket)
37°C water bath
P200 and P1000 micropipets Sterile Pasteur pipets
Orbital shaker at 4°C
Tissue culture incubator (37°C, 5% CO2)
Voltohmmeter to monitor TEER to establish confluence and differentiation status (World Precision Instruments EVOM2 with STX2 electrode)
Inverted light microscope capable of 5× and 10× magnification
Dilute 1.0 mg/ml collagen IV 1:30 in sterile PBS to obtain a 33 μg/ml solution.
Coat desired number of inserts/TWs with 100 μl diluted human collagen IV (either overnight at 4°C or ≥2 hr at 37°C). Discard unused diluted collagen IV.
Make sure to include TWs for mono-culture controls (no immune cells).
3. Have a minimum of one dense well of enteroids (~100 or more enteroids in a 25–30 μl dome of Matrigel) in a 24-well plate for two TWs.
The seeding enteroids should be as large as possible so that when they are fragmented, the fragments will lay and attach to the TW membrane. Use enteroids that have been in propagation medium for 6–8 days. A ratio higher or lower than one well for two TWs may be more appropriate depending on the well densities.
4. Aspirate medium from the enteroid wells and add 1.0 ml cold Organoid Harvesting solution. Dislodge Matrigel and enteroids from the bottom of the well using a mini cell scraper.
5. Shake plate on an orbital shaker at 4°C as per the Organoid Harvesting solution manufacturer’s recommendations, usually 30–40 min at 250 rpm.
Matrigel removal is critical (see note in step 8).
6. Recover dislodged enteroids using a P1000 micropipet and transfer enteroids to a 15-ml conical tube.
As much as possible, maintain enteroids in an intact state, avoiding small cellular clumps.
7. Add equal volume of CMGF- (see recipe below) and pellet enteroids by centrifugation at 400 × g for 10 min at 4°C.
8. Aspirate and remove as much of the supernatant as possible without disturbing the enteroid pellet; the enteroid pellet should be free of Matrigel.
Any remaining Matrigel will form a translucent layer above the enteroid pellet. To remove residual Matrigel, add 2 ml of Organoid Harvesting solution and gently suspend the pellet using a P1000 micropipet. Place the 15-ml tube on its side on the orbital shaker at 4°C and shake for 10 min. Repeat step 7 to recover the enteroids.
9. Add 50 μl/well of TrypLE Express to the enteroids; mix gently five times by pipetting with a P1000 micropipet.
For example, use four wells plus 200 μl of TrypLE. Once the TrypLE is added to the enteroids, work quickly to limit exposure to the enzyme.
10. Place suspended enteroids in a 37°C water bath for 90 s. Use less time for smaller enteroids (i.e., 75 s). Swirl tube occasionally during the incubation to suspend enteroids.
11. Move tubes to the biosafety cabinet and add 5–8 ml of cold CMGF-. Pellet enteroid fragments by centrifugation as above.
12. Aspirate supernatant and gently suspend enteroid fragments at 100 μl NDM per insert/TW (with 10 μM each Y-27632 and CHIR99021 inhibitors for ileum and colon) using a P1000 micropipet. For example: enteroid fragments from one well will be suspended in 200 μl propagation medium to seed two inserts/TWs. Set aside at room temperature.
13. Aspirate collagen solution from the TWs and wash with 100–200 μl CMGF-. Repeat (two washes in total). Aspirate wash.
14. Gently suspend enteroid fragments using a P1000 micropipet and plate 100 μl per TW using a P200 micropipet.
Gently resuspend the enteroid fragments with a P200 micropipet each time prior to adding 100 μl into TWs. The enteroid fragments will settle and need resuspension between plating.
15. Add 0.6 ml propagation medium (NDM) to the well of the receiver plate (with 10 μM each Y-27632 and CHIR99021 inhibitors for ileum or colon).
16. Incubate at 37°C, 5% CO2.
17. Change medium at 48 hr to NDM without Y-27632 and CHIR99021 inhibitors and continue to incubate at 37°C with medium changes every 2 days.
18. Observe for patch formation and eventual closed monolayers; monolayer confluency is usually reached in 1–2 weeks.
Measure TEER (Srinivasan et al., 2015; refer to voltohmmeter manufacturer for instructions on how to perform TEER measurements) or observe inserts/TWs under an inverted microscope to monitor confluence (TEER range depends on intestinal segment). Raw TEER values of <200 Ω indicate incomplete monolayer coverage of the insert/TW membrane. Newly confluent monolayers will have raw TEER values in the range of 300–500 Ω (100–165 Ω·cm2). TEER values will continue to increase over time while the monolayers are maintained in NDM.
19. Once confluent, change monolayer medium to differentiation medium (DFM) for 5 days. Change DFM on days 2 and 4. Monitor for an increase in TEER as a sign of monolayer maturation. Start monolayer differentiation after 1–3 days of confluency. For experiments interrogating crypt-like epithelia, maintain monolayers in NDM, and proceed to co-culture setup (Basic Protocol 4).
Undifferentiated monolayers in propagation medium will remain viable and confluent for up to 7–10 days; however, it is best to start the differentiation process soon after the monolayers become confluent as older monolayers tend to fall apart. TEER values will increase rapidly over the 5-day period ending at greater than two to three times the initial ohm value after start of monolayer differentiation (Noel et al., 2017).
ALTERNATE PROTOCOL
SEEDING ENTEROID FRAGMENTS FOR MONOLAYER FORMATION USING TRITURATION
Some enteroid or colonoid lines will not form monolayers when fragmented with Try-pLE Express. For these instances, substitute trituration for TrypLE in steps 9–11 of Basic Protocol 1.
Materials
See Basic Protocol 1
After recovery of the enteroids from Matrigel (Basic Protocol 1, step 7), suspend enteroids in NDM (with 10 μM each Y-27632 and CHIR99021 inhibitors for ileum or colon) at 100 μl per TW using a P1000 micropipet.
Using a P200 micropipet, triturate enteroids by vigorously pipetting up and down 25 to 30 times as for fragmenting enteroids for propagation.
Transfer 100 μl of mechanically fragmented enteroids into washed TWs (Basic Protocol 1, step 13).
Proceed with Basic Protocol 1, step 14–19.
BASIC PROTOCOL 2
ISOLATION OF MONOCYTES AND DERIVATION OF IMMUNE CELLS FROM HUMAN PERIPHERAL BLOOD
Monocytes are obtained from human PBMC isolated from whole blood. Refer to Current Protocols article: Fuss et al., 2009 for an in-depth description of PBMC isolation from peripheral blood. The protocol for monocyte isolation using negative selection is abbreviated and the reader is referred to the manufacturer’s recommendations. Monocytes should be plated the same day of isolation to obtain macrophages through M-CSF treatment. Plate monocytes in medium containing M-CSF for 6 days before assembly of the co-culture, 1 day before start of monolayer differentiation.
Materials
Human peripheral blood (~60 ml whole blood is sufficient for isolating the number of monocytes needed to set up more than ten co-cultures; collect blood into ETDA tubes)
Ficoll-Paque PREMIUM (Cytiva, 17544202)
Pan Monocyte Isolation kit (Miltenyi Biotec, 130–096-537)
Macrophage colony-stimulating factor (M-CSF; PreproTech, 300–25)
Monocyte-derived macrophage (MoDM) medium (see recipe)
ETDA tubes (BD Vacutainer™, 366643)
Centrifuge (swinging bucket)
6-well tissue culture plate
15-ml conical tubes
Tissue culture incubator (37°C, 5% CO2)
1. Six days prior to setting up the co-culture, isolate monocytes: Draw 60 ml whole blood into EDTA tubes.
Ideally whole blood should be processed into PBMC on the day of collection.
2. Isolate PBMC by centrifugation gradient over Ficoll-Paque PREMIUM per the manufacturer’s recommendations (also refer Current Protocols article: Fuss et al., 2009).
3. Isolate monocytes from PBMC by using Pan Monocyte Isolation kit, according to the manufacturer’s instructions.
The PBMC and monocyte isolation will take 4–5 hr.
4. Plate monocytes in MoDM medium with 50 ng/ml M-CSF into 6-well plates at 1 × 106cells/ml. Plate 2 ml per well.
For monocytes, omit M-CSF from medium.
5. Differentiate monocytes into macrophages for 6 days replacing culture medium (MoDM + M-CSF) every other day.
Observe for cell adherence to the plate and production of cell extensions. Rounding and loss of adhesion are signs of cell death.
6. Five days before assembly of the co-culture (refer to step 19 in Basic Protocol 1), change propagation medium (NDM) of the confluent enteroid/colonoid monolayers to differentiation medium (DFM; no Wtn3A, Rspo1, and SB 202190) if necessary, for the experiment. Change DFM on day 2 and 4. The co-culture is assembled on day 5 of enteroid/colonoid monolayer differentiation.
BASIC PROTOCOL 3
ISOLATION OF NEUTROPHILS FROM HUMAN PERIPHERAL BLOOD
Human neutrophils are sensitive cells which can be easily activated after isolation or by contaminants and have a relatively short lifespan. All reagents and material that come in contact with PMN should be pyrogen-free. The reader is referred to Current Protocols article: Kuhns et al., 2015 (and Alternate Protocol 1 of that article) for Ficoll-Paque detailed isolation of PMN and removal of erythrocytes. The PMN are isolated the same day of co-culture assembly. The isolation process takes ~4 hr and should be performed using strict sterile technique.
Materials
Human peripheral blood (~30 ml whole blood is sufficient for isolating the number of PMN needed to set up more than ten co-cultures; collect blood into ETDA tubes; start PMN isolation early in the day of co-culture that represents day 5 of differentiated enteroid/colonoid monolayers)
Ficoll-Paque PREMIUM (Cytiva, 17544202)
6% dextran (Alfa Aesar, J63702; see Current Protocols article: Kuhns et al., 2015 for preparation)
PBS (1×), pH 7.4 (Quality Biological, 114–058-101)
PBS (10×), pH 7.4 (Quality Biological, 119–069-131)
Cold sterile MilliQ water
ETDA tubes (BD Vacutainer™ 366643)
50-ml conical tubes
Centrifuge
Hemacytometer (or other means for counting cells) Inverted light microscope
1. In a 50-ml conical tube, transfer 15 ml anticoagulated blood and add PBS (1 ) toa final volume of 50 ml.
If starting with 30 ml of whole blood, set up duplicate samples.
2. Centrifuge for 10 min at 400 × g, 21°C with the acceleration/deacceleration brake set at 5/5.
3. Aspirate supernatant (mix of PBS and plasma) without disturbing the pellet and suspend cells in PBS (1×) to a final volume of 35 ml.
4. Dispense 15 ml Ficoll-Paque PREMIUM in a new 50-ml conical tube and carefully layer the 35 ml of washed blood cells from step 3.
5. Centrifuge for 35 min at 300 × g, 21°C without brake.
It is important to avoid the use of the brake during spin-down process to maintain the gradient.
6. Collect PBMC layer and remove Ficoll-Paque without disturbing the PMN/erythrocytes pellet.
7. Suspend PMN/erythrocytes pellet in remaining liquid and transfer to a new 50-ml conical tube.
It is important to transfer to a fresh conical tube to avoid PBMC contamination that can remain on the tube walls.
8. Add PBS (1×) to a final volume of 22.5 ml.
9. Add 7.5 ml of 6% dextran (dilution 1:4). Mix tube contents by gentle inversion, ten times.
10. Set aside to allow erythrocytes to sediment by gravity for 15–20 min at room temperature.
11. Transfer supernatant to a new 50-ml conical tube and bring volume to 50 ml with PBS (1×).
12. Centrifuge for 10 min at 300 × g, 21°C with the acceleration/deacceleration brake set at 5/5.
13. Aspirate supernatant and suspend pellet in the remaining liquid.
14. Add 18 ml cold sterile water to lyse erythrocytes. Mix cell suspension by inversion for 20 s, and immediately add 2 ml cold 10× PBS and 20 ml cold 1× PBS.
15. Centrifuge for 10 min at 300 × g at 4°C with the acceleration/deacceleration brake set at 5/5.
16. Repeat steps 10 and 11 if necessary, to lyse any remaining erythrocytes.
17. Suspend PMN in 1–2 ml DFM without antibiotics and count using a hemacytometer (see Current Protocols article: Strober, 2015) or an automatic cell counter.
Keep PMN suspension at 4°C while counting.
18. Adjust PMN concentration to 1 × 107 viable cells/ml in DFM without antibiotics. Immediately proceed to co-culture set up.
BASIC PROTOCOL 4
ASSEMBLY OF ENTEROID/MACROPHAGE OR ENTEROID/NEUTROPHIL CO-CULTURE
Co-culture of monolayers and macrophages is useful to model intestinal homeostasis (resident cells) and under activating conditions, immunity to pathogens. PMN in enteroid co-cultures model inflammation. The day of co-culture assembly will coincide with 5 days of monolayer maturation and 6 days of monocyte-derived macrophage differentiation. PMN are recovered from whole blood the same day the co-culture is established. Perform all steps under a biosafety cabinet.
Materials
Immune cells (e.g., macrophages, monocytes, polymorphonuclear neutrophils, neutrophils; see Basic Protocols 2 and 3)
Sterile PBS (1×), pH 7.4
NDM or DFM (without antibiotics for bacterial infections; see recipe)
15-ml sterile conical tubes
Sterile cell scrapers (United Biosystems, MCS-200)
12-well flat bottom tissue culture plate(s)
Small metal forceps
Sterile Pasteur pipets
P200 micropipet
FIREBOY Safety Bunsen burner (Integra Biosciences Corp, 144010) Centrifuge
Hemacytometer (or other means to count cells)
Light microscope
Tissue culture incubator (37°C, 5% CO2)
1. The morning of the experiment, measure TEER if this is a read-out parameter of the assay. Return plate to the tissue culture incubator until macrophages are ready or when PMN isolation is complete (~4 hr). Once PMN are in DFM without antibiotics, proceed to step 7 without delay.
2. Using a sterile cell scraper, gently remove attached macrophages in a 6-well plate and collect in a 15-ml centrifuge tube.
3. Pellet macrophages by centrifugation (swinging bucket; 400 × g for 5 min room temperature).
4. Suspend macrophages in sterile PBS and count using trypan blue to determine viability (see Current Protocols article: Strober, 2015).
5. Determine number of cells needed (1 × 105 per monolayer) and transfer volume to a new 15-ml centrifuge tube.
6. Pellet macrophages by centrifugation and suspend in enteroid medium (propagation medium or DFM) with 10 ng/ml M-CSF at a concentration of 2 × 106 viable cells/ml. Set aside at room temperature.
7. Using metal forceps (flame to sterilize), pick up a monolayer TW and gently invert into an empty 12-well plate. Retain the 12-well plate lid.
Some of the apical medium will drain; however, the monolayer will retain some medium. Set up the number of inverted monolayers needed.
8. Gently aspirate any remaining medium on the bottom (now facing up) of culture inserts using a sterile Pasteur pipet attached to an aspirator.
Angle the Pasteur pipet 90° to the insert to aspirate any medium.
9. Using a P200 micropipet, gently add 50 μl of the macrophage suspension (1 105 cells) in enteroid medium (propagation or DFM) plus M-CSF onto the bottom of the insert. For PMN, add 50 μl (5 × 105 cells) in DFM without antibiotics.
For monocytes, add 50 μl (1 × 105 cells) in enteroid medium (propagation or DFM).
10. Repeat until macrophages/immune cells have been deposited onto the inverted inserts.
11. Gently place the lid onto the 12-well plate.
The lid will contact the 50-μl bubble containing the macrophages/immune cells and form a bevel.
12. Return plate to the 37°C, 5% CO2 incubator for 2 hr.
Longer incubations of the inverted inserts do not improve adherence of the immune cells. Keep the original insert/TW receiver 24-well plate with medium at 37°C, 5% CO2. Alternatively, a new 24-well plate can be used to keep upright the co-culture insert/TWs.
13. Move plate from the incubator with the inverted TWs and place under the biosafety cabinet.
14. Using metal forceps (flame to sterilize), pick up the inverted TWs and place upright into the original or a new 24-well plate.
15. Add 100 μl enteroid medium (propagation or DFM) to the apical side and 600 μl to the well of the receiver plate plus M-CSF at 10 ng/ml for macrophages (omit for PMN or monocytes), if using a new 24-well plate.
If the original 24-well receiver plate is used, add M-CSF to the remaining medium to 10 ng/ml if co-culturing monolayers with macrophages.
16. Return plate to the incubator and run a co-culture time course. The standard co-culture time is 24 hr at the time of seeding macrophages onto the bottom of the inserts (22 hr after placing the upright inserts/TWs at 37°C, 5% CO2).
PMN co-cultures run for up to 2 hr after returning the inserts/TWs to their upright position. These experiments run for a total of 4 hr. Monocyte co-cultures can be extended up to 48 hr, as for macrophage co-cultures; see Background Information section below.
17. Measure TEER at 24 hr of co-culture and again at the end of any treatment (i.e., bacterial infection) if this is a read-out.
Some of the macrophages will drop from the bottom of the insert/TW onto the receiver plate at >48 hr. Any treatment is for up to 24 hr after co-culture establishment (e.g., 24 hr co-culture is treated or interrogated for up to 24 hr, or 48 hr total).
REAGENTS AND SOLUTIONS
CHIR99021
CHIR99021 (biotechne, R&D Systems brand, 4423)
Prepare 10 mM in DMSO.
Store aliquots at −20°C for up to 1 month.
Complete medium without growth factors (CMGF-)
Advanced DMEM/F12 (Gibco brand, Thermo Fisher Scientific, 12634028)
10 mM HEPES buffer (Gibco brand, Thermo Fisher Scientific, 15630080)
1% GlutaMAX (Thermo Fisher Scientific, 35050061)
1% penicillin-streptomycin (Quality Biological, 120-095-721)
Store at 4°C for up to 1 month.
Differentiation medium (DFM)
CMGF- (without pen/strep for bacterial infection of co-cultures)
50 ng/ml EGF (biotechne, R&D Systems brand, 236-GMP)
2% B27 Supplement (50×; Gibco brand, Thermo Fisher Scientific, 17504044)
10 nM gastrin (AnaSpec, AS-64149)
500 nM A 83–01 (biotechne, Tocris brand, 2939)
125 μg/ml Primocin (InvivoGen ant-pm-2; antimicrobial reagent for primary cells; omit for bacterial infections of co-culture)
10% Noggin-conditioned medium (HEK293T cells stably expressing murine Noggin-Fc from Dr. Gijs R. van den Brink, Tytgat Institute for Liver and Intestinal Research and Department of Gastroenterology and Hepatology, Academic Medical Center, Amsterdam, The Netherlands)
Store at 4°C for1 week.
Monocyte-derived macrophages (MoDM) complete medium
RPMI (Thermo Fisher Scientific, 11875093)
10% FBS, heat inactivated (MilliporeSigma, F4135)
1 × MEM non-essential amino acids (MilliporeSigma, M7145)
1 mM sodium pyruvate (100 mM solution; MilliporeSigma, S8636)
55 μM 2-mercaptoethanol (Gibco brand, Thermo Fisher Scientific, 21985023)
1% penicillin-streptomycin (Quality Biological, 120-095-721)
50 ng/ml macrophage colony-stimulating factor (M-CSF; PreproTech, 300-25)
Store at 4°C for up to 2 weeks. Omit M-CSF for culturing monocytes.
Non-differentiation medium (NDM)
CMGF- (see recipe)
50 ng/ml EGF (biotechne, R&D Systems brand,236-GMP)
2% B27 Supplement (50; Gibco brand, Thermo Fisher Scientific, 17504044)
10 nM Gastrin (AnaSpec, AS-64149)
500 nM A 83-01 (biotechne, Tocris brand, 2939)
10 μM SB 202190 (MilliporeSigma, S7067; prepare stock solution in DMSO)
125 μg/ml Primocin (InvivoGen ant-pm-2; antimicrobial reagent for primary cells)
50% Wnt3A-conditioned medium (L Wnt3A from ATCC, CRL-2647)
15% R-Spondin 1-conditioned medium (HEK293T cells stably expressing mouse Rspo1-Fc from Dr. Calvin Kuo, Stanford University).
10% Noggin-conditioned medium (HEK293T cells stably expressing murine Noggin-Fc from Dr. Gijs R. van den Brink, Tytgat Institute for Liver and Intestinal Research and Department of Gastroenterology and Hepatology, Academic Medical Center, Amsterdam, The Netherlands).
Store at 4°C for1 week.
Y-27632
Y-27632 dihydrochloride (biotechne, R&D Systems brand, 1254)
Prepare 10 mM in sterile double-distilled H2O.
Store aliquots at −20°C for up to 1 month.
COMMENTARY
Background Information
Human intestinal enteroids/colonoids offer a relevant and practical experimental ex vivo model to study physiology, developmental biology, pathophysiology, or host-pathogen interactions of the intestinal epithelium (Blutt et al., 2019; Costantini et al., 2018; Foulke-Abel et al., 2014; Foulke-Abel et al., 2016; Koestler et al., 2019; Noel et al., 2017; Current Protocols article: Poole et al., 2018; Ranganathan et al., 2019; Saxena et al., 2016; Yoo & Donowitz, 2019; Zachos et al., 2016; Zou et al., 2019, Lemme-Dumit et al., 2020). Because enteroids recapitulate normal and diseased intestinal physiology, e.g., chloride ion (Cl−) secretion defects in cystic fibrosis (Cil et al., 2017; de Winter-de Groot et al., 2018; Duan et al., 2019; Liu, Walker, Cook, Ootani, & Clarke, 2012), and are readily established from biopsies or surgical tissues, the use and applications of this model continue to expand. The use of human enteroids/colonoids as a surrogate for the small intestinal or colonic epithelium, respectively, overcomes multiple major drawbacks of immortalized cell lines. Traditional human intestinal culture lines do not recapitulate normal intestinal physiology, often vary phenotypically between subclones of the same line or after certain passage numbers and many lines are aneuploid. Enteroids derived from different intestinal segments retain the physiological characteristics of the tissue of origin including microbe infectivity tropisms (Blutt, Crawford, Ramani, Zou, & Estes, 2018; Chan et al., 2019; Haga et al., 2020; Rajan et al., 2018). Long-term culture of enteroids (years) do not introduce phenotypic or genetic changes (Sato & Clevers, 2013); supplementation with Wnt3A maintains the indefinite epithelial self-renewal capacity without the need for transformation. While the spheroidal three-dimensional enteroid growth forms have been employed in many studies, the ability to generate enteroid monolayers offers a practical conveniency by facilitating tissue manipulation and by expanding the information (i.e., outcome readouts) that can be derived from this model (In et al., 2016; In, Foulke-Abel, Clarke, & Kovbasnjuk, 2019).
Immortalized cells lines consist of a single cell type while enteroid epithelial cells can give rise to multiple cell types found in normal intestinal epithelia: stem cells, Paneth cells, enteroendocrine cells, goblet cells, microfold cells, and Tuft cells (Ding et al., 2020; Fasciano, Blutt, Estes, & Mecsas, 2019; Howitt et al., 2016; Noel et al., 2017; Sato et al., 2009). Medium manipulation also permits modeling of the enteroids to reflect a more crypt-like or villus-like cellular compartment that is lacking with traditional cell lines. A crypt-like epithelium is induced by maintaining enteroids in propagation medium (NDM) and removal of Wnt signaling (DFM) promotes epithelial differentiation to mimic a villus-like epithelium of the small intestine or surface cells of the colon. Other medium manipulations can further drive enrichment of specific epithelial cell types (Beumer et al., 2018; Beumer et al., 2020) to suit experimental needs. For example, the rare M cell type found in the follicle-associated epithelium of Peyer’s patches or isolated lymphoid follicles can be induced in ileal enteroids grown as spheroids or monolayers by addition of tumor necrosis factor alpha (TNF-α) and receptor activator of NF-κB ligand (RANKL; Fasciano et al., 2019; Ranganathan et al., 2019; Wood, Rios, & Williams, 2016) or retinoic acid, lymphotoxin, and RANKL (Ding et al., 2020). Small intestinal enteroids were recently engineered to increase the differentiation and numbers of hormone-secreting enteroendocrine cells by over expression of neurogenin 3 (Chang-Graham et al., 2019). Medium manipulation or over expression of a targeted transcription factor as a means to influence intestinal epithelial developmental physiology is often incomplete when using transformed cell lines. In all, human enteroids have become the choice physiologically relevant experimental model to study the intestinal epithelium. Because it employs primary human cells, this model has high translational value for interrogation of disease- and health-associated cellular pathways and the evaluation of preventive and therapeutic tools.
It is important to bear in mind, however, that although useful for ex vivo studies, enteroid monolayers represent a reductionist model of the intestinal epithelium. The physiology of the intestinal epithelium is the result of the functional collaboration of epithelial and underlying immune, nerve, and mesenchymal cells. In the enteroid model, growth factors secreted by mesenchymal and other underlying cells are provided in the medium. Exposing enteroid/colonoid monolayers to purified cytokines, chemokines, or proteases may mimic immune cell simulation; however, the outcome may reflect a narrow and perhaps incomplete epithelial response. The contributions of immune cells to epithelial physiology or host defenses are lacking unless these are added to the enteroids in a co-culture system. The co-culture system described here overcomes the limits of monoculture models by incorporating, in the same tissue culture framework, intestinal epithelial monolayers in close proximity to immune cells. All intestinal segments are amenable to monolayer formation and co-culture (Noel et al., 2017). Macrophage co-cultures with ileal enteroids and colonoids have also been established by our group (Noel et al., 2017 and J. F. Staab and N. C. Zachos, unpublished results) and in principle any segment, including stomach (Sebrell et al., 2019), could be combined with innate immune cells to address specific experimental questions. The interchangeability of the enteroids and immune cells in this co-culture model also supports personalized medicine and preclinical translational studies.
Critical Parameters and Troubleshooting
A key variable for generating monolayers is to have well-established, healthy enteroid cultures propagating at a rate that requires splitting every 6–7 days. When planning the culture of monolayers, it is important to consider the number of wells needed for seeding in addition to those needed to maintain enteroid propagation and maintenance. Often, it takes 2–3 weeks of enteroid expansion to generate enough wells for seeding monolayers and continued propagation. Monolayer formation across culture inserts/Transwells (TWs) is not always complete. It is, therefore, advisable to seed at least two additional monolayers per experiment. Ultimately, the type of experimental inquiry will dictate the number of co-cultures needed. For endpoints that require recovery and lysis of the monolayer for RNA or protein isolation, pooling duplicate or triplicate samples is recommended. We have found that the RNA or protein yield from one monolayer is only adequate for molecular analysis of only a few genes or proteins. For interrogation immune cells, a combination of three to four co-cultures will yield ample material for RNA or protein analysis, or fluorescence-activated cell sorting (FACS). Ideally, replicate experiments should include monolayers derived from two or more enteroid lines and two or more healthy blood donors to account for biological variability.
Monolayer formation in culture inserts/TWs of larger pore membranes (3.0 μm) usually takes longer but these inserts are needed for basolateral bacterial cell invasion assays (Koestler et al., 2019; Ranganathan et al., 2019). Here we use the 3.0-μm pore culture inserts/TWs for PMN co-cultures to allow PMN migration through the monolayer (Lemme-Dumit et al., 2020). We have not noticed differences in phenotypic features of the monolayers (e.g., TEER, differentiation) grown on 3.0-, 1.0-, or 0.4-μm pore culture inserts/TWs (Noel et al., 2017 and J. F. Staab, J. M. Lemme-Dumit, R. Latanich, and N. C. Zachos, unpublished data).
It is important to monitor confluence by light microscopy and/or TEER to plan for the start of differentiation, which will determine the day of co-culture assembly. Monolayers in NDM with TEER values >70 Ω·cm2 have reached confluence and are ready for differentiation. Try to start the differentiation process 1 to 3 days upon monolayer confluence because older monolayers sometimes lose integrity after culture in DFM for 5 days. It is common to find that some monolayers lag in reaching confluence when it is necessary to begin differentiation in order to keep with immune cell maturation or isolation plans. Under these circumstances, we have found that monolayers can complete their closing in DFM if they are near confluence (~90% closed). This process takes 1 to 2 days in DFM and thus day 5 of differentiation should now be considered as the fifth day after changing the medium to DFM. There is no appreciable difference in day 5 TEER values or other physiological parameters between monolayers differentiated from fully closed or nearly closed monolayers. Medium changes affect TEER values; therefore, it is recommended that measurements are performed before replacing the medium. The times to measure TEER should be during monolayer growth, before assembly of the co-cultures, 24 hr later (pre-treatment), and 0 to 24 hr post treatment.
A key step that affects enteroid fragment adhesion to culture inserts/TWs is the human collagen IV coating. The stock 1 mg/ml solution in acetic acid is prepared in the shipping vial (5 mg) and allowed to reconstitute at 4°C for several hours with occasional mixing (refer to MilliporeSigma recommendations). The collagen must be completely dissolved to allow for the enteroid fragments to properly adhere and spread onto the culture insert/TW. The stock collagen suspension should be stored in single-use aliquots at −20°C and can be used for several months. Replace the collagen IV stock solution at the first sign that enteroid fragments are failing to adhere to culture inserts/TWs. We find that the culture inserts/TWs need to be coated with a saturating concentration of collagen IV (10 μg/cm2) for efficient adhesion and spreading of enteroid fragments. Collagen coating can be done the day before seeding by placing the culture inserts/TWs with 0.33 μg/ml collagen IV in a Parafilm-sealed receiver plate at 4°C. On the day of seeding, move the plate from 4°C storage and place it under the biosafety cabinet at the start of the protocol to allow the plate to come to room temperature. Culture inserts/TWs with 0.33 μg/ml collagen IV can be left at 4°C for up to 2 days without detriment. This is a convenient method to save time (2 hr) on the day of seeding.
Understanding Results
TEER measurements are key in co-culture experiments because they quickly inform on the monolayer barrier integrity and fate of the experiment. After the initial 24 hr of co-culture (2 hr for PMN co-cultures), the monolayer can be treated apically with bacteria, virus, cytokines, or compound of choice for up to 24 hr (2 hr for PMN co-culture). Treatments that destroy the barrier function of the monolayers are likely toxic and should be scaled back in time or application (drug concentration, bacteria numbers). Alternatively, loss of barrier function may be the intended or expected result of the experiment. A starting point for cytokine or drug concentrations can be based on experiments in neoplastic intestinal epithelium cell lines (e.g., T-84 or CaCo-2).
Bacterial infection should be based on multiplicity of infection (MOI) relative to the number of immune cells. A range of MOI from 10 to 100 should inform on immune cell function and activity without overwhelming and destroying the monolayers.
Cytokine concentrations can be determined from apical and basolateral medium at the end of the co-culture experiment. These are best performed on a multiplex platform due to the small apical volumes (recovery is usually <100 μl) in the inserts/TWs. A confounding variable is discerning the cytokine contribution from the monolayer versus the immune cells. Comparison to monocultures is helpful but may be limited unless there is a significant change in cytokine levels upon treatment.
Replicate experiments with multiple enteroid lines derived from the small intestine (e.g., duodenum, jejunum, ileum) or colon (e.g., ascending, transverse, descending) should be considered when planning experiments. Certain experiments may only be performed with a given intestinal segment because of bacterial or viral tropism (Ranganathan, Smith, Foulke-Abel, & Barry, 2020); therefore, multiple same-segment enteroid lines should be incorporated into the experimental plan. In general, small intestine enteroid lines have similar phenotypes (e.g., TEER, cytokine expression) that are different from colon-derived lines.
Time Considerations
Propagation, seeding of culture inserts/TWs, and monolayer confluence can take up to 3 to 4 weeks. Another 5 days are required for differentiation of the monolayers, followed by an initial 24 hr of co-culture and another 24 hr of treatment (for macrophages or monocytes). In all, a co-culture experiment from enteroid seeding into culture inserts/TWs to co-culture harvest may take 5 to 6 weeks. It is advisable to use a stepwise approach and become comfortable with seeding and culturing monolayers prior to attempting immune cell co-cultures. Adding immune cells to monolayers significantly increases the time commitment and complexity of the experimental method. Large experiments with >24 co-cultures may require two investigators working separately to establish monolayers, and isolate and culture immune cells. Harvesting the components of the co-culture for analysis is often the most time-intensive part of the procedure. Preparation of monolayer lysates for bacterial plating is best performed with two investigators working in tandem: one investigator preparing monolayer lysates while a second scientist performs serial dilutions and plating for colony-forming units. Other harvest procedures can be performed by a single researcher. Collected apical and basolateral medium are either pooled or stored separately at −20°C until sampled for cytokine or other biomolecule levels. Co-cultures destined for immunofluorescence can be fixed and stored at 4°C for several weeks prior to antibody staining. Monolayer or immune cell lysates can be stored at −80°C for protein or RNA isolation purification at a later date. Many of the downstream assays can be conveniently performed on banked frozen samples from replicate experiments. Ultimately, co-culture experiments are at best a low to medium throughput platform that require strong resource and time commitments. Human enteroid and immune cell co-cultures can be established in a reliable and reproducible manner. This refined ex vivo model is suitable to unravel mechanistic biological processes relevant to the human gut.
Acknowledgments
The protocols developed for co-culture studies were funded by the National Institutes of Health (NIAID P01-AI125181 to MP and NZ). The authors also wish to acknowledge the Integrated Physiology and Imaging Cores of the Hopkins Conte Digestive Disease Basic and Translational Research Core Center (NIH NIDDK P30-DK089502) for resources used to develop the human immune-enteroid co-culture models.
Footnotes
Conflicts of Interest Statement
The authors report no conflicts of interest.
Literature Cited
- Beumer J, Artegiani B, Post Y, Reimann F, Gribble F, Nguyen TN, … Clevers H. (2018). Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nature Cell Biology, 20(8), 909–916. doi: 10.1038/s41556-018-0143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer J, Puschhof J, Bauza-Martinez J, Martinez-Silgado A, Elmentaite R, James KR, … Clevers H. (2020). High-resolution mRNA and secretome atlas of human enteroendocrine cells. Cell, 181(6), 1291–1306 e1219. doi: 10.1016/j.cell.2020.04.036. [DOI] [PubMed] [Google Scholar]
- Blutt SE, Crawford SE, Ramani S, Zou WY, & Estes MK (2018). Engineered human gastrointestinal cultures to study the microbiome and infectious diseases. Cellular and Molecular Gastroenterology and Hepatology, 5(3), 241–251. doi: 10.1016/j.jcmgh.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blutt SE, Klein OD, Donowitz M, Shroyer N, Guha C, & Estes MK (2019). Use of organoids to study regenerative responses to intestinal damage. American Journal of Physiology. Gastrointestinal and Liver Physiology, 317(6), G845–G852. doi: 10.1152/ajpgi.00346.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MC, Cheung SKC, Mohammad KN, Chan JCM, Estes MK, & Chan PKS (2019). Use of human intestinal enteroids to detect human norovirus infectivity. Emerging Infectious Diseases, 25(9), 1730–1735. doi: 10.3201/eid2509.190205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang-Graham AL, Danhof HA, Engevik MA, Tomaro-Duchesneau C, Karandikar UC, Estes MK, … Hyser JM (2019). Human intestinal enteroids with inducible neurogenin-3 expression as a novel model of gut hormone secretion. Cellular and Molecular Gastroenterology and Hepatology, 8(2), 209–229. doi: 10.1016/j.jcmgh.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cil O, Phuan PW, Gillespie AM, Lee S, Tradtrantip L, Yin J, … Verkman AS (2017). Benzopyrimido-pyrrolo-oxazine-dione CFTR inhibitor (R)-BPO-27 for antisecretory therapy of diarrheas caused by bacterial enterotoxins. FASEB Journal, 31(2), 751–760. doi: 10.1096/fj.201600891R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini V, Morantz EK, Browne H, Ettayebi K, Zeng XL, Atmar RL, … Vinje J. (2018). Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerging Infectious Diseases, 24(8), 1453–1464. doi: 10.3201/eid2408.180126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter-de Groot KM, Janssens HM, van Uum RT, Dekkers JF, Berkers G, Vonk A, … Beekman JM (2018). Stratifying infants with cystic fibrosis for disease severity using intestinal organoid swelling as a biomarker of CFTR function. European Respiratory Journal, 52(3), 1702529. doi: 10.1183/13993003.02529-2017. [DOI] [PubMed] [Google Scholar]
- Ding S, Song Y, Brulois KF, Pan J, Co JY, Ren L, … Greenberg HB (2020). Retinoic acid and lymphotoxin signaling promote differentiation of human intestinal M cells. Gastroenterology, 159, 214–226.e1. doi: 10.1053/j.gastro.2020.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan T, Cil O, Tse CM, Sarker R, Lin R, Donowitz M, & Verkman AS (2019). Inhibition of CFTR-mediated intestinal chloride secretion as potential therapy for bile acid diarrhea. FASEB Journal, 33(10), 10924–10934. doi: 10.1096/fj.201901166R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasciano AC, Blutt SE, Estes MK, & Mecsas J. (2019). Induced differentiation of M cell-like cells in human stem cell-derived ileal enteroid monolayers. Journal of Visualized Experiments,149, e59894. doi: 10.3791/59894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Ettayebi K, Blutt SE, … Donowitz M. (2014). Human enteroids as an ex-vivo model of host-pathogen interactions in the gastrointestinal tract. Experimental Biology and Medicine, 239(9), 1124–1134. doi: 10.1177/1535370214529398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulke-Abel J, In J, Yin J, Zachos NC, Kovbasnjuk O, Estes MK, … Donowitz M. (2016). Human enteroids as a model of upper small intestinal ion transport physiology and pathophysiology. Gastroenterology, 150(3), 638–649.e638. doi: 10.1053/j.gastro.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Matano M, Nanki K, & Sato T. (2015). Efficient genetic engineering of human intestinal organoids using electroporation. Nature Protocols, 10(10), 1474–1485. doi: 10.1038/nprot.2015.088. [DOI] [PubMed] [Google Scholar]
- Fuss IJ, Kanof ME, Smith PD, & Zola H. (2009). Isolation of whole mononuclear cells from peripheral blood and cord blood. Current Protocols in Immunology, 85(1), 7–1. doi: 10.1002/0471142735.im0701s85. [DOI] [PubMed] [Google Scholar]
- Haga K., Ettayebi K., Tenge VR., Karandikar UC., Lewis MA., Lin SC., … Estes MK. (2020). Genetic manipulation of human intestinal enteroids demonstrates the necessity of a functional fucosyltransferase 2 gene for secretor-dependent human norovirus infection. mBio, 11(2). doi: 10.1128/mBio.00251-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, … Garrett WS (2016). Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science, 351(6279), 1329–1333. doi: 10.1126/science.aaf1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- In J, Foulke-Abel J, Zachos NC, Hansen AM, Kaper JB, Bernstein HD, … Kovbasnjuk O. (2016). Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cellular and Molecular Gastroenterology and Hepatology, 2(1), 48–62.e3. doi: 10.1016/j.jcmgh.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- In JG, Foulke-Abel J, Clarke E, & Kovbasnjuk O. (2019). Human colonoid monolayers to study interactions between pathogens, commensals, and host intestinal epithelium. Journal of Visualized Experiments, 9(146), e59357. doi: 10.3791/59357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung P, Sato T, Merlos-Suárez A, Barriga FM, Iglesias M, Rossell D, … Batlle E. (2011). Isolation and in vitro expansion of human colonic stem cells. Nature Medicine, 17(10), 1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- Koestler BJ, Ward CM, Fisher CR, Rajan A, Maresso AW, & Payne SM (2019). Human intestinal enteroids as a model system of shigella pathogenesis. Infection and Immunity, 87(4), e00733–18. doi: 10.1128/IAI.00733-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowska E, & Kubes P. (2013). Neutrophil recruitment and function in health and inflammation. Nature Reviews Immunology, 13(3), 159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- Kuhns DB, Priel DAL, Chu J, & Zarember KA (2015). Isolation and functional analysis of human neutrophils. Current Protocols in Immunology, 111(1), 7–23. doi: 10.1002/0471142735.im0723s111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemme-Dumit JM, Doucet M, Zachos NC, & Pasetti MF (2020). Host-cell interactions and innate immune response to an enteric pathogen in a human intestinal enteroid-neutrophil co-culture. bioRxiv, 2020.09.03.281535. doi: 10.1101/2020.09.03.281535. [DOI] [Google Scholar]
- Liu J, Walker NM, Cook MT, Ootani A, & Clarke LL (2012). Functional Cftr in crypt epithelium of organotypic enteroid cultures from murine small intestine. American Journal of Physiology. Cell Physiology, 302(10), C1492–1503. doi: 10.1152/ajpcell.00392.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel G, Baetz NW, Staab JF, Donowitz M, Kovbasnjuk O, Pasetti MF, & Zachos NC (2017). A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Scientific Reports, 7, 45270. doi: 10.1038/srep45270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole NM, Rajan A, & Maresso AW (2018). Human intestinal enteroids for the study of bacterial adherence, invasion, and translocation. Current Protocols in Microbiology, 50(1), e55. doi: 10.1002/cpmc.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A, Vela L, Zeng XL, Yu X, Shroyer N, Blutt SE, … Maresso AW (2018). Novel segment- and host-specific patterns of enteroaggregative escherichia coli adherence to human intestinal enteroids. mBio, 9(1), e02419–17. doi: 10.1128/mBio.02419-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan S, Doucet M, Grassel CL, Delaine-Elias B, Zachos NC, & Barry EM (2019). Evaluating Shigella flexneri pathogenesis in the human enteroid model. Infection and Immunity, 87(4). doi: 10.1128/IAI.00740-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan S, Smith EM, Foulke-Abel JD, & Barry EM (2020). Research in a time of enteroids and organoids: How the human gut model has transformed the study of enteric bacterial pathogens. Gut Microbes, 12(1), 1795492. doi: 10.1080/19490976.2020.1795389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, & Clevers H. (2013). Growing self-organizing mini-guts from a single intestinal stem cell: Mechanism and applications. Science, 340(6137), 1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, … Clevers H. (2009). Single Lgr5 stem cells build cryptvillus structures in vitro without a mesenchymal niche. Nature, 459(7244), 262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Saxena K, Blutt SE, Ettayebi K, Zeng XL, Broughman JR, Crawford SE, … Estes MK (2016). Human intestinal enteroids: A new model to study human rotavirus infection, host restriction, and pathophysiology. Journal of Virology, 90(1), 43–56. doi: 10.1128/JVI.01930-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebrell TA, Hashimi M, Sidar B, Wilkinson RA, Kirpotina L, Quinn MT, … Bimczok D. (2019). A novel gastric spheroid co-culture model reveals chemokine-dependent recruitment of human dendritic cells to the gastric epithelium. Cellular and Molecular Gastroenterology and Hepatology, 8(1), 157–171.e153. doi: 10.1016/j.jcmgh.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan B, Kolli AR, Esch MB, Abaci HE, Shuler ML, & Hickman JJ (2015). TEER measurement techniques for in vitro barrier model systems. Journal of Laboratory Automation, 20(2), 107–126. doi: 10.1177/2211068214561025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strober W. (2015). Trypan blue exclusion test of cell viability. Current Protocols in Immunology, 111(1), A.3B.1–A.3B.3. doi: 10.1002/0471142735.ima03bs111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MB, Rios D, & Williams IR (2016). TNF-α augments RANKL-dependent intestinal M cell differentiation in enteroid cultures. American Journal of Physiology. Cell Physiology, 311(3), C498–507. doi: 10.1152/ajpcell.00108.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Tse CM, Avula LR, Singh V, Foulke-Abel J, de Jonge HR, & Donowitz M. (2018). Molecular basis and differentiation-associated alterations of anion secretion in human duodenal enteroid monolayers. Cellular and Molecular Gastroenterology and Hepatology, 5(4), 591–609. doi: 10.1016/j.jcmgh.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo JH, & Donowitz M. (2019). Intestinal enteroids/organoids: A novel platform for drug discovery in inflammatory bowel diseases. World Journal of Gastroenterology, 25(30), 4125–4147. doi: 10.3748/wjg.v25.i30.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos NC, Kovbasnjuk O, Foulke-Abel J, In J, Blutt SE, de Jonge HR, … Donowitz M. (2016). Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. Journal of Biological Chemistry, 291(8), 3759–3766. doi: 10.1074/jbc.R114.635995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou WY, Blutt SE, Crawford SE, Ettayebi K, Zeng XL, Saxena K, … Estes MK (2019). Human intestinal enteroids: New models to study gastrointestinal virus infections. Methods in Molecular Biology, 1576, 229–247. doi: 10.1007/7651_2017_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Key Reference
- Noel et al. 2017. See above.
- Describes the first phenotypic study of a human enteroid/macrophage co-culture model and its applicability to study intestinal physiology as well as innate host and macrophage responses to pathogenic bacteria.