Probiotics mitigate post-heat stress disorder, preventing coral mortality.
Abstract
Beneficial microorganisms for corals (BMCs) ameliorate environmental stress, but whether they can prevent mortality and the underlying host response mechanisms remains elusive. Here, we conducted omics analyses on the coral Mussismilia hispida exposed to bleaching conditions in a long-term mesocosm experiment and inoculated with a selected BMC consortium or a saline solution placebo. All corals were affected by heat stress, but the observed “post-heat stress disorder” was mitigated by BMCs, signified by patterns of dimethylsulfoniopropionate degradation, lipid maintenance, and coral host transcriptional reprogramming of cellular restructuration, repair, stress protection, and immune genes, concomitant with a 40% survival rate increase and stable photosynthetic performance by the endosymbiotic algae. This study provides insights into the responses that underlie probiotic host manipulation. We demonstrate that BMCs trigger a dynamic microbiome restructuring process that instigates genetic and metabolic alterations in the coral host that eventually mitigate coral bleaching and mortality.
INTRODUCTION
Coral reefs have been undergoing unprecedented mass coral bleaching events in recent decades, fueled by ocean warming (1), heightening the need to devise effective countermeasures to mitigate further declines (2, 3). Increasing sea surface temperatures trigger the disruption of the symbiotic relationship between the coral host and its endosymbiotic algae of the family Symbiodiniaceae (4), resulting in the physical whitening of coral colonies known as “bleaching.” Photosynthetic products from the endosymbiont algae provide more than 90% of the host’s nutritional demands (5). Thus, prolonged periods of heat stress and bleaching lead to coral mortality (6).
Besides endosymbiotic algae, corals are associated with a suite of other organisms (bacteria, protists, fungi, viruses, etc.), collectively referred to as the coral holobiont or metaorganism (7–10). In particular, bacteria are assumed to contribute to coral holobiont biology, notably stress tolerance and adaptation to disparate environments (10–15). The importance of bacteria led to the proposal of the coral probiotic hypothesis (16), which states that microbes support coral biology through selection of the most advantageous holobiont configuration in a given environment. This was later refined by the microbiome flexibility hypothesis to include the notion that the potential or propensity for microbiome change differs among host species (15). The proposal to use these concepts to select and manipulate specific microbes to aid the stress tolerance and resilience of the coral holobiont was dubbed “beneficial microorganisms for corals” (BMCs) (10). Beneficial microorganisms putatively support nitrogen fixation, sulfur cycling, scavenging reactive oxygen species (ROS), and production of antibiotics to thwart pathogens, for example (10, 11, 17).
The proof of concept that manipulating coral microbes improves coral stress tolerance was recently demonstrated in the first experiments to identify the beneficial nature of a selected BMC consortium in ameliorating coral bleaching (18). Nevertheless, exactly “how” these BMCs were associated with functional changes in the host remained unknown. Notably, BMCs do not necessarily need to exert their effect on the coral host directly. Hence, the measured holobiont response does not need to be a perfect reflection of the BMC consortium added. Rather, the BMC consortium may benefit the host indirectly, by means of niche occupation, microbial succession, or the prevention of dysbiosis through pathogen deterrence (10, 11, 18). Furthermore, although the ability of BMCs to ameliorate coral bleaching has been demonstrated (18), it is unknown whether they have the capacity to help corals evade mortality, e.g., through the provisioning of alternate metabolites to compensate for the loss of Symbiodiniaceae.
Despite the diversity of the coral microbiome, which makes it challenging to decipher the contribution of associated microbes to coral holobiont biology, the dynamic nature of the coral microbiome, which can often change markedly—e.g., across sites, species, age, and under stress—further hampers the ability to conduct such studies in the natural environment (15, 19–21). For this reason, manipulation of BMCs in controlled experimental setups, such as mesocosms (22–24), provides an avenue to identify important microbial players and study holobiont responses (and putative underlying mechanisms), while maintaining a quasi-reef environment, to improve and inform the development of biotechnological solutions to promote coral reef resilience.
Here, we used coral mesocosms in combination with multiomics evaluation to assess responses and potentially decipher the mechanisms that underlie the increased stress tolerance and coral mortality evasion, offered by the provisioning of probiotics. In a large-scale effort, fragments of the coral Mussismilia hispida were exposed to thermal stress in a 75-day mesocosm experiment and inoculated with either a M. hispida–tailored BMC consortium or a saline solution placebo. Coral health (measured via Fv/Fm rates and survivorship) (25), microbial activity, and functional responses were assessed through a multiomics approach. Our analysis shows that increased stress tolerance and survivorship of coral holobionts exposed to a BMC consortium coincided with holobiont restructuring and a defined reprogramming of the coral host’s gene expression, targeting cellular reconstruction, immune response, and stress protection during a post-heat stress recovery period.
RESULTS
BMC consortium selection, assembly, and experimental setup
Bacterial strains were isolated from a visually healthy colony of M. hispida. The BMC consortium was assembled with bacterial strains exhibiting (i) at least one of the beneficial traits detailed below, (ii) the absence of antagonist activity against other selected BMCs, and (iii) no previous record of the species/strain being harmful to humans or other marine life. Beneficial traits included nitrogen fixation (nifH), denitrification (nirK), dimethylsulfoniopropionate (DMSP) degradation (dmdA), ROS scavenging potential (measured through catalase activity), and antagonistic activity against two coral pathogens, Vibrio coralliilyticus strain V1 and Vibrio alginolyticus V2 (26, 27).
From an initial 133 obtained isolates, the assembled BMC consortium was composed of the following six bacterial strains: Bacillus lehensis (M20) positive for nifH, nirK, and dmdA; Bacillus oshimensis (M24) positive for dmdA; B. lehensis (M3) positive for nifH and dmdA; Brachybacterium conglomeratum (M1) positive for catalase and nifH; Planococcus rifietoensis (CM29) with antagonistic activity against V1; and Salinivibrio sp. (F2) with antagonistic activity against V1 and V2 (table S1). The experimental BMC consortium consisted of lag phase–grown bacterial strains collected and resuspended in sterile saline solution (0.85% NaCl) at 1 × 108 cells/ml (for details, see fig. S1). The placebo/control consisted of a sterile saline solution (0.85% NaCl), hereafter referred to as the placebo treatment.
BMCs and placebo were applied every 3 days during a simulated heat stress event (maximum temperature of 30°C) and every 5 days for the remainder of the 75-day mesocosm experiment (Fig. 1A), while a control was run in parallel (26°C). We focused on four time points, T0 at the beginning of the experiment, T1 upon reaching peak temperature in the heat stress (30°C), T2 at the end of peak temperature heat stress (30°C for 10 days), and T3 following a 15-day recovery period at 26°C. Microbiome changes associated with BMC treatment were investigated through 16S ribosomal RNA (rRNA) gene metabarcoding (T0, T1, T2, and T3). In addition, patterns and mechanisms underpinning the projected increased stress resilience provided by the BMC treatment were assessed through the evaluation of coral physiology (photosynthetic efficiency of Symbiodiniaceae and visual monitoring of bleaching for T0, T1, T2, and T3) (Fig. 1, B and C), elucidation of metabolic footprints [nuclear magnetic resonance (NMR)/partial least squares-discriminant analysis (PLS-DA) at T0, T1, T2, and T3], and determination of coral transcriptome patterns at the peak of temperature and the end of the experiment (T2 and T3) (see Fig. 1A for experimental design).
Fig. 1. Long-term heat stress experiment and coral bleaching responses to placebo and BMC inoculation.
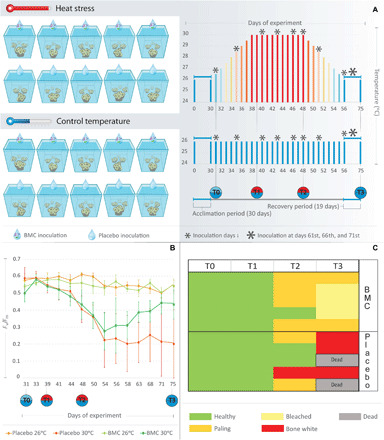
(A) Experimental design and details on temperature, BMC inoculations, and sampling layout. (B) Means of photosynthetic efficiency Fv/Fm ratios (y axis) from coral fragments treated with BMCs or placebo under heat stress temperature regimes (30°C) and control temperature regimes (26°C) during the mesocosm experiment days (x axis). (C) Heatmap based on the bleaching score attributed to coral fragments treated with BMCs or placebo in the heat stress experiment.
Host microbiome shift associated with BMC treatment during heat stress
To confirm the presence of the BMC consortium members in the coral microbiome, the 16S rRNA gene sequences of each of the six BMC members was used to query amplicon sequence variants (ASVs) from both BMC- and placebo-treated coral fragments. We identified three of the six strains throughout various time points of the experiment: CM29 P. rifietoensis (T1 BMC-treated), M24 B. oshimensis (T1 and T2 BMC-treated), and M1 B. conglomeratum (T2 BMC-treated) (Fig. 2A).
Fig. 2. Effects of BMC treatment on coral bacterial community.
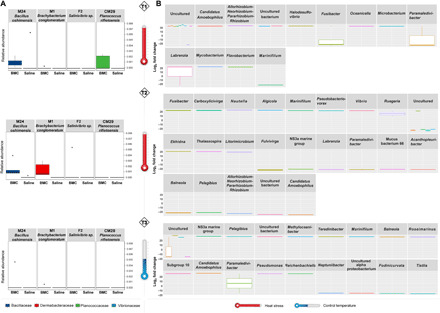
(A) Relative abundance of BMC consortium members in coral fragments treated with BMCs or placebo and exposed to heat stress (T1, P = 0.028; T2, P = 0.0001; T3, P = 0.265; Kruskal-Wallis), where boxes represent the relative mean abundance and stars represent outliers. (B) Boxplot of fold change (FC) of ASVs with differential abundance (P = 0.01) in BMC-treated coral fragments compared with placebo-treated fragments at T1, T2, and T3. Bars with the same color scale belong to the same taxonomic family.
Parallel to the confirmed microbiome incorporation of three of the BMC strains in T1 (CM29 and M24) and T2 (M1and M24) (Fig. 2A), the overall bacterial community structure of BMC-treated corals was significantly different from placebo-treated corals during the heat stress (T2) [permutational multivariate analysis of variance (PERMANOVA), P = 0.05] but became indiscernible during the recovery period (T3) (P = 0.583, stress = 0.15) (fig. S2), where BMC strains were also not detected. The most abundant bacterial phyla identified across all coral fragments were Proteobacteria, followed by Bacteroidetes, throughout the course of the experiment (fig. S3). Despite such consistency at higher taxonomic levels, we found variability over time with regard to bacterial taxa association. Although the relatively most abundant genera associated with corals at T0 were consistently Ruegeria (11.9%), Tistlia (4.6%), and Candidatus Amoebophilus (4.2%), we only found Ruegeria species to be abundant across BMC-inoculated corals (T1: 15.1%; T2: 13.4%; T3: 17.3%), while in placebo-treated coral fragments, Paramaledivibacter spp. were the most prevalent (T1: 21.6%; T2: 13.8%; T3: 3.9%). In addition, ASVs exhibiting significant differences in abundance were identified in BMC-treated coral fragments under thermal stress compared to placebo samples (Fig. 2B). Overall, 13 ASVs were significantly increased [average fold change (FC) = 22.4, P < 0.01] in BMC-treated samples in T1, 23 ASVs in T2 (average FC = 21.8, P < 0.01), and 18 ASVs in T3 (average FC = 21.7, P < 0.01) (Fig. 2B), indicating that BMCs affected the microbiome structure beyond the addition of selected strains. Ruegeria was the most prominent genus found in BMC-inoculated corals and was mainly enriched in T2 samples (FC = 15.3) (Fig. 2B). Despite the observed microbiome structural changes, overall community diversity of BMC- and placebo-treated corals remained similar throughout the course of the experiment [based on Shannon, Chao1, and ASV distribution indexes; analysis of variance (ANOVA), P = 0.8] (fig. S4).
Coral BMC treatment contributes to increased survivorship and recovery from bleaching after acute thermal stress
We compared photosynthetic efficiency (Fig. 1B) and coral holobiont survival (Fig. 1C) to assess the BMC treatment effect. Most notably, survivorship of corals inoculated with the BMC consortium was substantially higher, with 100% of fragments surviving the heat stress treatment (T3) compared to only 60% of the placebo-treated corals (Fig. 1C). Surviving corals in the placebo-treated regime showed a significant decrease in the Fv/Fm average rates (65% decrease, from T0 to T3; P < 0.05) at the end of the experiment compared to the start (Fig. 1B), while photosynthetic efficiencies of BMC-treated corals only decreased at the peak of temperature stress (T2) (P < 0.05) and thereafter returned to the initial average during the recovery period (T3) (P = 0.197).
Post-heat stress disorder and transcriptional reprogramming of BMC-treated coral holobionts
We were further interested in elucidating the coral host transcriptomic response associated with the observed increase in coral survival after heat stress following BMC treatment. RNA sequencing (RNA-seq) was conducted on samples from BMC- and placebo-treated coral fragments at the peak of heat stress (T2; n = 20 samples) and at the end of the experiment (T3; n = 15 samples) from the control and heat stress temperatures. Coral genes were assorted into orthogroups to increase confidence in their annotation and, hence, functional inference. We determined a total of 17,755 orthogroups considered for the gene expression analysis (table S2). As expected, we observed pronounced differences in the response to heat stress when comparing placebo-treated corals at 30° and 26°C at T2 (peak of heat stress) [differential expression of 2294 orthogroups with a false discovery rate (FDR) of <0.05 associated with metabolic disorders, apoptosis, autophagy, and response to stress] (table S2). Significant response differences were also observed after a period of recovery (T3), suggesting heat stress carry-on effects in the transcriptional footprint, which we termed post-heat stress disorder (PHSD) with some signs of recovery (2275 orthogroups with an FDR of <0.05 associated with metabolism, cell death, and oxidative stress) (table S2).
Similar to the physiological and metabolic responses, we did not see significant transcriptomic differences between BMC- and placebo-treated coral samples in T2 in the heat stress treatment, suggesting that both “holobiont systems” react similarly in the peak of heat stress, originally observed in yeast and termed environmental stress response (28, 29). Following this, we focused on differentially expressed orthogroups between BMC- and placebo-treated coral samples subsequent to the heat stress at the recovery time point (T3) to elucidate the transcriptomic footprint associated with BMC-induced recovery (fig. S5). BMCs seemed to exert an overall “healing effect,” as evidenced by increased recovery and stress attenuation processes in coral gene expression. In this regard, a total of 169 orthogroups were differentially expressed because of BMC inoculation, mainly involved in apoptosis, inflammatory response, cytoskeleton, and membrane reorganization (see blue bars for up-regulation and red bars for down-regulation; Fig. 3 and fig. S5). Most of these orthogroups (142 of 169) were down-regulated in BMC-treated corals in comparison to their placebo-treated counterparts, suggesting attenuation of the PHSD due the probiotic treatment.
Fig. 3. Coralhost responses to BMC treatment.
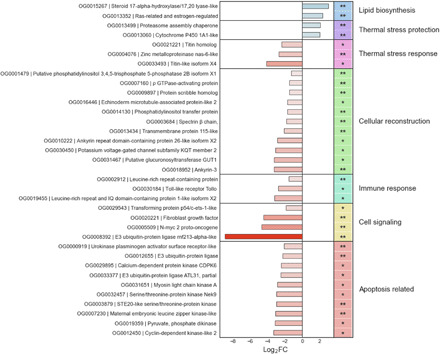
Main orthogroups with significant (FDR < 0.05) differential expression between BMC- and placebo-treated corals from the end of the heat stress temperature experiment (T3). The respective Kyoto Encyclopedia of Genes and Genomes annotation, their FDR value, and log2 FC representing up-regulation (positive values and blue bars) or down-regulation (negative values and red bars) in relation to BMC samples are also shown. Orthogroups marked with ** are conserved among Scleractinia. Orthogroups marked with * are exclusively from M. hispida
Even after the recovery period, the remaining placebo-treated coral survivors were still showing signs of prolonged PHSD, as suggested by the more prominent expression of proteins involved in response to thermal stress, when compared to BMC-treated corals. In addition, orthogroups involved in chromosome condensation (two titin homolog proteins) and DNA methylation (zinc metalloproteinase nas-6-like) were also down-regulated in BMC samples (log2 FC = −4.12, −2.34, and −2.82, respectively). Overall, the ongoing PHSD observed was significantly mitigated in BMC-treated corals. Numerous orthogroups involved in triggering apoptosis were more highly expressed in placebo-treated corals. Following BMC treatment, we observed down-regulation of seven kinases and one kinase receptor, as well as E3 ubiquitins involved in apoptosis signaling (log2 FCs from −8.9 to −2.1; see Fig. 3). Further, mitogen-activated protein kinase signaling orthogroups, such as N-myc 2 proto-oncogene (log2 FC = −4.66) and transforming protein p54/c-ets-1-like (log2 FC = −1.87) involved in kinase-signaling activation, were down-regulated by BMC treatment. PHSD seemed to also trigger inflammatory and innate immune responses, not only potentially through activity of some kinases but also due to the increased expression of Toll-like receptor, leucine-rich repeat protein and domain. By comparison, these orthogroups were all down-regulated in BMC-treated corals, following heat stress (log2 FC = −2.73, −1.55, and −3.17, respectively).
In addition, various orthogroups involved in cytoskeleton organization and anchoring were higher expressed in placebo-treated corals at the end of the recovery period (T3) and, conversely, down-regulated in BMC-treated corals. Orthogroups annotated as echinoderm microtubule-associated protein-like (log2 FC = −1.7), ρ guanosine triphosphatase (GTPase)–activating protein (log2 FC = −1.51), putative phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2B isoform X1 (log2 FC = −1.28), and spectrin β chain (log2 FC = −1.82) were down-regulated in BMC samples. Specifically, the spectrin β chain orthogroup is a complex that is anchored in the cytoplasm via ankyrin proteins, which were down-regulated in BMC-treated corals after the recovery period [ankyrin repeat domain-containing protein 26-like isoform X2 (log2 FC = −2.89) and ankyrin-3 (log2 FC = −3.25)]. In addition, orthogroups associated with the synthesis of membrane or secondary cell wall components, such as phosphatidylinositol transfer protein (log2 FC = −1.28) and glucuronosyltransferase GUT1 (log2 FC = −3.25), were less expressed in BMC-treated corals, as well as cellular adhesion proteins, represented by the transmembrane protein 115-like (log2 FC = −2.12), potassium voltage-gated channel subfamily KQT member 2 (log2 FC = −3.09), and a scribble homolog (log2 FC = −1.51) orthogroups. Conversely, all these orthogroups associated with the cellular response to cope with the prolonged PHSD were more prominently expressed in placebo-treated corals.
On the other hand, following BMC treatment, we found up-regulated expression of 32 orthogroups (table S2), suggesting that BMC treatment resulted in the induction of thermal stress protection and blockage of PHSD through increased expression of the proteasome assembly chaperone (log2 FC = 2.06) and cytochrome P450 1A1-like (log2 FC = 1.99) orthogroups. In addition, the crucial up-regulation of orthogroups associated with biosynthesis of estrogen and steroids (log2 FC = 3.0 and 2.31), critical components of cell membranes, was also observed in BMC-treated corals.
Significant expression differences were also observed when comparing corals inoculated with BMCs or placebo kept at 26°C 17 days after the beginning of the manipulation of their microbiomes (T2), as represented by 2371 orthogroups with FDR < 0.05 mainly associated with the up-regulation of metabolic pathways (specially biosynthesis of fatty acids, cholesterols, and steroids) and cellular signaling and cycle in BMC-treated corals (table S2). However, no long-term BMC reprogramming took place when no thermal stress was applied, as represented by the lack of differential ortholog expression at T3 (i.e., 44 days after the beginning of the microbial therapy at 26°C).
Together, we found that inoculation with BMCs instigated restructuring of the transcriptional network and cellular homeostasis, up-regulating key orthogroups associated with PHSD mitigation, such as steroids biosynthesis and stress protection proteins, although the general pattern suggested dampening PHSD through down-regulation of stress-related downstream pathways, e.g., apoptosis (Fig. 3). The proximate cause of the transcriptional reprogramming was the BMC treatment that resulted in a restructured host microbiome, which, in turn, suggests a signal cascade from the microbes to the coral host, during the recovery period, corroborating the notion that the holobiont is the functional biological unit.
Metabolic restructuring of BMC-treated coral holobionts after heat stress
We obtained metabolic profiles from the thermal stress experiment using NMR to identify metabolic mechanisms associated with the microbial and genomic restructuring underpinning the increased thermal tolerance of BMC-treated corals. Sample complexity led to strong overlapping 1H resonances, challenging the elucidation of metabolic patterns (Fig. 4A). Nevertheless, the characterized peaks at 2.88 parts per million (ppm) (singlet from the S-Methyl groups) for DMSP and at 2.58 ppm (singlet from the S-Methyl groups) for dimethyl sulfoxide (DMSO) were found to be separated and well defined, as well as correlated with different treatments (Fig. 4B). DMSP variation was found to be positively correlated to the BMC-treated corals, and DMSO variation was found to show a negative correlation to the BMC-treated corals in the PLS-DA loading plot (Fig. 4B). To explore the correlations in more detail, comparing metabolic variations overtime (i.e., T1, T2, and T3), the area under the curve representing the direct quantitative ratio for the selected DMSP and DMSO peaks was integrated and represented as boxplots, which indicated significant decreases in the DMSP and DMSO levels observed at T2 and T3 in BMC-treated samples exposed to thermal stress (P < 0.05; Fig. 4Cfor DMSP and Fig. 4D for DMSO). We therefore used both DMSP and DMSO as important proxies for the metabolic assessment because of their clear separation from overlapped profiles, as well as their importance in sulfur cycling and microbial structuring (i.e., role of DMSP-related chemotaxis of V. coralliilyticus and antimicrobial activity of DMSO). In addition, lipids, despite their predominant presence in every sample, were also positively correlated to the BMC-treated corals as indicated by the PLS-DA loading plot (Fig. 4B). Nevertheless, the strong overlapping signals (at ~5.26, ~2.74, 1.19, and 0.87 ppm), representing positive and negative trends of a unique compound, prevented the possibility of annotating specific compounds. Future efforts should include the analysis of broader molecular spectra by using liquid chromatography–mass spectrometry.
Fig. 4. Metabolic restructuring due to BMC treatment and heat stress.
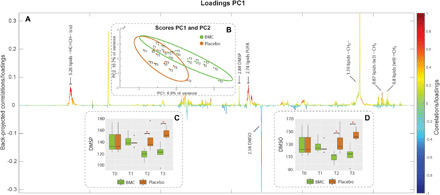
Color-coded loading plot (A) (in which colors indicate variation intensity) and score plot with 95% confidence ellipses showing sample clustering by PLS-DA (B) from PLS-DA of the 1H NMR dataset comparing the metabolic patterns from coral fragments treated with BMCs and placebo during the thermal stress experiment. Peaks from the loading plot (resonances from annotated as lipids, DMSP, and DMSO) pointing upward are correlated with BMC-treated samples (grouped in the positive quadrant of PC1 (in the score plot), and those pointing downward are correlated with the placebo samples [grouped in the negative quadrant of PC1 (Principal Components 1) in the score plot]. Boxplots are provided to access semiquantitative evaluations of the characteristic DMSP peak at 2.88 ppm (C) and DMSO peak at 2.58 ppm (D) across sampling time and treatment independently.
DISCUSSION
The promise of coral probiotics to increase the stress tolerance of corals has been very recently shown (11, 18, 30), although the effect that BMCs exert on the holobiont or whether BMCs can increase survivability of corals under stress remained elusive. Here, we show that the inoculation of coral fragments with a native BMC consortium instigated holobiont changes at the level of the microbiome, host gene expression, and metabolism, which coincide with an increase in coral survival rates (Fig. 5). Hence, our results provide a first insight into the putative mechanistic underpinnings of how the coral (host) responds to BMC inoculation, although the detailed functional changes that cause the altered phenotype await further elucidation. Our results argue for PHSD recovery improvement of the metaorganism by the BMC consortium, as indicated by changes at the coral host, Symbiodiniaceae, and bacterial compartment level. From the results obtained, a number of key findings emerge that we discuss in the following.
Fig. 5. Probiotics-mediated mitigation of coral PHSD.
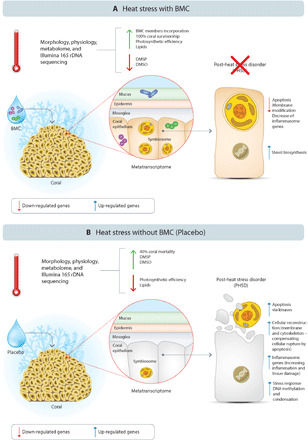
Summary of the overall differential recovery mechanisms observed at the end of the 75-day mesocosm experiment, comparing the process in BMC-treated (A) and placebo-treated (B) M. hispida fragments.
We observed major changes in microbial community structure observed during heat stress (12, 31, 32) in conjunction with the dynamic microbiome restructuring following the recovery period, indicating that M. hispida exhibits microbiome adaptation. Thus, it may fit into the “microbiome conformer” type previously suggested (14, 15) and observed for this coral species regarding other impacts (33–35). Following this notion, the level of microbiome flexibility may be considered as a factor to identify corals with high(er) manipulative potential. Corals that naturally alter their microbial composition and potentially uptake microbes from the environment are more likely to “accept” inoculants (14, 15, 36). Notably, shifts in metaorganism microbial composition are, potentially, rapid and versatile means of adaptation to environmental change (12–15).
It is important to consider that the host’s ability to take up microorganisms from the environment is hypothesized to increase when under stress, a conclusion based on the finding that many host microbiomes appear less ordered when stressed (14, 21, 37). Inoculation with high numbers of different BMC cells (i.e., a consortium) may therefore ensure (and improve) uptake of at least some microorganisms exhibiting beneficial characteristics, which may, at the same time, preclude colonization by pathogens considering that “space is limited.” The use of bacterial consortia provides a combination of beneficial mechanisms to increase stress tolerance, even if not all members of the BMC successfully associate with the coral holobiont (18, 31, 38–40). Here, we show that the use of a bacterial consortium resulted in incorporation of some of the selected BMCs, which were found in the microbiome of BMC-treated corals during the thermal stress, i.e., at T1 and T2 (see Fig. 2A). Notably, members of the BMC consortium were not detected after the 15-day period (T3; i.e., recovery). This suggests three things: first, a dynamic restructuring of the microbiome can happen on a relatively small time scale (12, 14, 41); second, incorporation of BMCs might be facilitated under stress (in this experiment, during the peak of heat stress) because coral defense is compromised or selection for beneficial microbes is supported; and third, it is currently unclear how long the beneficial effect of BMCs is lasting. From our results, it appears that BMC members colonized coral fragments during stress and instigated significant changes in the coral holobiont but reverted to the original microbiome structure after ceasing (or the absence) of stress [sensu Ziegler et al. (14) who used the term “microbiome recovery”]. Accordingly, the duration of the presence of the stressor might determine the longevity of the BMC effect, which suggests that repeated addition of BMCs might be needed to ensure a long-lasting effect under natural conditions (11).
The early and detectable incorporation of some of the BMC consortium members into the coral microbiome and the subsequent microbial restructuring were correlated with significant improvements in coral recovery after thermal stress, as most convincingly demonstrated by mortality evasion. Heat stress–driven mortality and/or decrease in Fv/Fm rates observed in fragments that were not treated with BMCs suggest damage to the temperature-related photosystem II electron transport of the Symbiodiniaceae through chronic photoinhibition (42), which ultimately leads to a breakdown in symbiosis and results in loss/expelling of the Symbiodiniaceae, i.e., bleaching (43). Notably, bleaching is a symptomatic phenotype, i.e., loss of Symbiodiniaceae can occur through multiple processes, including host cellular apoptosis (44) or necrosis, and eventually death from starvation (6, 45), which was corroborated by the up-regulation of different kinases directly involved in triggering apoptosis in placebo-treated corals (46, 47). Our transcriptome results indicate that BMCs did not buffer the immediate heat stress response in M. hispida but exerted its effect during recovery, supported by the gene expression patterns and coral physiology. Most notably, we observed low Fv/Fm rates for both BMC and placebo treatments at T2 (during heat stress), but only the BMC treatment promoted recovery at T3, as indicated by the “return-to-normal” Fv/Fm rates. Our interpretation is that BMCs exert their effect through mitigation of the effects from what we term PHSD. The molecular evidence for this condition includes not only apoptosis activity, which may be triggering inflammatory responses, but also membrane and cellular reconstruction due to tissue loss caused by recent-past heat stress. In this regard, the remaining placebo-treated surviving corals seemed to be still struggling from the effects of recent heat stress, even 19 days after the end of the heat stress period, while all BMC-treated corals seem to have recovered.
As an analogy to posttraumatic stress disorder (48), coral PHSD is characterized by the contrast of the coral response and its attempts to recover from a heat stress event while still fading due to the cellular, immune, and metabolic consequences of such stress. The significant up-regulation of numerous kinases and receptors, as well as signaling molecules, by the remaining placebo-treated survivor corals at T3 suggests ongoing apoptosis (47). In addition, the oxidative stress increased by thermal photodamage to the photosynthetic apparatus of Symbiodiniaceae might be further contributing to trigger inflammatory responses (49). Previous studies have also found expression of immune-related and apoptosis genes in corals affected by heat stress for extended periods of time (50, 51), suggesting a persistent bleaching effect on the coral transcriptome of susceptible corals (52). We hypothesize that down-regulation of orthogroups involved in apoptosis and the concomitant up-regulation of thermal stress protection proteins, such as chaperones, promoted by BMC inoculation, protected corals from tissue damage and Symbiodiniaceae loss, with consequences for coral survival. Such prominent recovery promoted by coral probiotics indicates that if the selection of BMCs based on the hypothetical framework proposed by Peixoto et al. (10, 11) already provides measurable benefit and survivorship improvement, more careful selection of BMCs could result in even larger improvements. It is worthwhile and interesting to also highlight that such reprogramming was also observed when no stress was applied (at 26°C), but only at an early stage. While it seems that inoculation of BMCs rapidly trigger change of response norms from the host, long-term BMC reprogramming and exerted effects are only manifested under and subsequent to stress.
It is tempting to speculate that the increased host survivorship observed in this study is a direct consequence of the transcriptomic changes discussed above, which arguably will result in altered metabolic profiles. For instance, the observed changes in the metabolomic profile of corals treated with BMC supports the hypothesis that the selected microbes play a direct role in increasing coral stress tolerance as evidenced by correspondence between selected traits of BMC bacteria (i.e., DMSP degradation) and observed metabolic changes. Shifts in BMC-treated metabolomic profiles were signified by a decrease in the DMSP concentration and lipidic reservoir maintenance. This connects directly to the presence of M24 in the 16S rDNA data: M24 was found exclusively in BMC-treated samples at T1 and T2, indicating its incorporation into the coral microbiome, and was selected because of its ability to degrade DMSP (table S1). Notably, DMSP is mostly produced by algae (Symbiodiniaceae), and its degradation generates antimicrobial compounds, helping to control pathogens (53–55). Peixoto et al. (10, 11) suggested that this is a desirable BMC trait. In parallel to the DMSP degradation as one of the direct mechanisms provided by the BMC consortium to ameliorate heat stress, the BMC treatment may have also indirectly influenced DMSP metabolism, through the enrichment of bacteria able to assimilate DMSP, such as Ruegeria (56), the most abundant genus found in BMC-treated coral samples during the course of the experiment. This genus has been previously observed to inhibit and control the growth or pathogenicity of V. coralliilyticus (57). These observed traits may have triggered the molecular responses observed by the host. These results highlight the importance of microbiome restructuring to coral resilience (12, 15) and the additional potential role of the M. hispida BMC consortium in modulating the microbial colonization and succession of inoculated coral fragments. This parallel colonization/succession/enrichment of beneficial microbes has also been observed in other hosts, including humans, as a result of the use of pre- or probiotics (58, 59).
The increasing frequency and severity of ocean warming events has caused coral die-offs worldwide in the last few years (1, 60–62). The development and better understanding of novel interventions to mitigate large-scale coral mortality is one of the climate priorities for the coming decades (63, 64). The results of this study provide three completely novel insights that can aid the development of tools to promote human-accelerated environmental adaptation of corals: (i) BMC treatment and heat stress are both necessary conditions to trigger a long-term BMC thermal protection effect, whereas neither on its own is sufficient; (ii) the BMC thermal protection effect manifests after the heat stress and affects recovery; and (iii) such BMC-promoted protection mitigates coral PHSD, preventing mortality. Our results support the potential of microbiome restructuring to aid in the environmental adaptation of the coral metaorganism to global change (15) and identify a suite of microbial-mediated host responses underlying coral survival and recovery to thermal bleaching provided through BMCs. This is most prominently highlighted by the marked increase of 40% in coral survival rates following thermal stress and prior BMC treatment. This was accompanied by overall shifts in the coral microbiome that suggest a dynamic restructuring of the microbiome, due partially to the incorporation of BMC members and the relative increase of other bacteria. We further show that such microbiome restructuring directly affects the host, exerting beneficial effects and PHSD mitigation, as evidenced by transcriptional reprogramming (i.e., down-regulating apoptosis and inflammatory triggering molecules and up-regulating thermal stress protection proteins). In this light, our results reinforce the promise and potential of coral probiotics as an effective tool to rehabilitate coral reefs, particularly because the ability to “recover” is what eventually makes the difference in the real world, i.e., not only the difference in responding to heat stress but also in surviving the heat stress. In this regard, our data also suggest that prophylactic inoculation of BMCs, a few weeks before thermal events, can be advantageous for corals to sustain heat stress as it supposedly allows them to more rapidly and readily recover from thermal stress.
MATERIALS AND METHODS
Ethics approval and consent to participate
Permission for sampling was obtained from the System of Authorization and Information on Biodiversity. The microbial survey permits were obtained from CNPq (National Council for Scientific and Technological Development, Brazil) and SISGEN (National System for the Management of Genetic Heritage and Associated Traditional Knowledge) (number A620FE5).
Sampling procedures
M. hispida colonies were collected by SCUBA diving at the Coroa Vermelha reefs, Santa Cruz de Cabrália County, Bahia, Brazil. Coral colonies were collected at three sites along the reef: site A (16°20′57.99″ S; 038°58′45.00″ W), site B (16°20′39.30″ S; 038°58′38.10″ W), and site C (16°22′02.20″ S; 039°0′15.63″ W), at depths between 1.5 and 10 m on 26 to 29 January 2017. Corals were transported in sterile plastic bags and then packed in Styrofoam boxes containing 800 g of ice and were sent by air cargo to Rio de Janeiro. Upon arrival at the research station, around 13 hours after sampling, coral colonies were transferred to 1500-liter tanks with constant sea water flow and air bubbling for a 3-day preliminary acclimatization period. After that, coral colonies were fragmented using a diamond-based band saw (Gryphon Corp., CA, USA) in ~7-cm fragments with at least three polyps each, sawn in the coenosarc, and placed in the experimental system for acclimatization and healing. About 4 days after sawing, the coral fragments showed the first signs of healing and were kept in acclimation conditions (26°C) until all fragments reached Fv/Fm rates of around 0.6.
Isolation of bacterial strains from coral
Three previously tagged colonies (5 to 15 cm) of the thermally resistant coral M. hispida collected from Maraú, Bahia, Brazil (13°56′10.9″ S; 38°55′38.71″ W) were used as a source to isolate BMCs. Two different approaches were used for bacterial isolation. First, 0.5 g of each coral macerate was resuspended in 45 ml of sterile saline solution (0.85% NaCl) and then shaken for 16 hours. After incubation, triplicate subsamples (100 μl) of 10−3, 10−4, and 10−5 dilutions were inoculated into petri dishes containing 20 ml of marine agar medium (Marine Agar Zobell 2216, HiMedia Laboratories, Mumbai, India), diluted marine agar medium (Marine Agar Medium 2× diluted with 2.5% NaCl and agar adjusted), 2.5% NaCl Luria-Bertani medium (10 g of tryptone, 5 g of yeast extract, 25 g of NaCl, and 15 g of agar to 1000 ml of distilled water) or marine water medium (1000 ml of sea water and 13 g of agar). In addition, coral fragments of ~0.5 mm were placed directly onto dishes with these culture media. All the plates were incubated at 26°C for 48 hours. A total of 133 bacterial colonies were isolated, based on bacterial colony morphology, with 67 derived from macerated slurries and 52 derived from mini fragments. Each different morphological colony was stored in an ultrafreezer with a final concentration of 20% glycerol and removed when necessary for functional screening.
Functional screening for probiotic and bacterial 16S rRNA gene sequencing
Each morphologically different bacterial isolate was screened for beneficial traits for corals, as proposed by Peixoto et al. (10). Sixty-seven morphologically distinct bacterial strains were recovered from macerated slurries and 52 from microfragments of the coral placed directly onto the agar medium. The isolates were then screened for beneficial traits, as previously outlined by Peixoto et al. (9), and tested via a proof-of-concept study (18). Antagonistic activity against V. coralliilyticus YB strain (DSM19607) (V1) and V. alginolyticus (BAA450) (V2) was tested by the agar diffusion method (65). First, 20 μl of each bacterial strain was spot-inoculated onto 2.5% NaCl LB medium, placing three spots for each strain (representing replicates). The plates were incubated at 26°C for as long as necessary for the strain to grow. The strains were inactivated by chloroform volatilization, followed by pouring 3 ml of semisolid 2.5% NaCl LB medium (0.7% agar) containing the Vibrio indicators over the inactivated spots. These plates were then incubated at 28°C for 16 hours, and the antagonistic activity was indicated by inhibition halos around or no detection of Vibrio growth over the colony spot. The same procedure was repeated for both V1 and V2 in separate plates. Among the remaining candidates, one strain, identified as P. rifietoensis (CM29), was an antagonist against V. coralliilyticus YB (DSM19607) (V1), while another strain, identified as Salinivibrio sp. (F2), showed antagonistic activity against both V1 and V. alginolyticus (BAA450) (V2). The strains were screened for ROS scavenger enzyme activity, based on qualitative (production or no production) and quantitative (bubble amount) catalase production when 50 ml of their liquid culture was mixed with 50 μl of 3% (v/v) hydrogen peroxide.
Nitrogen-cycling genes, as nitrogenase subunits (nifH) and denitrification (nirK), as well as the DMSP degradation (dmdA) gene were screened by polymerase chain reaction (PCR) from the genomic DNA samples (for additional information about primer and PCR cycling, see table S3). From the initial 133 isolated strains, 33 (25%) isolates demonstrated high catalase activity and positive results for the amplification (PCR) of the genes nifH (12 strains, i.e., 9%), nirK (5 strains, 11%), and dmdA (11 strains, 8%). Almost half of the isolates (49% or 65 of 133) were identified as belonging to the genus Vibrio and were excluded from the following steps, considering that they are regularly postulated to be coral pathogens (66, 67). A total of 38 strains positive for at least one screened trait described above had their nearly full-length 16S rRNA gene PCR-amplified and sequenced (table S3). The sequencing electropherograms were processed using the Ribosomal Database Project II (68) to remove low-quality bases. Sequences of each isolate were assembled into contigs using Bioedit 7.0.5.3 (69). The bacterial 16S rRNA gene sequences were aligned with sequences from the National Center for Biotechnology Information (NCBI) database (70). All sequences were deposited in the NCBI database under an individual accession number, given below (see table S4). Bacteria strains identified as potential human or marine pathogens as well as those with antagonistic activity against any member of the selected BMC consortium (assessed by the agar diffusion method cited above) were excluded.
Probiotic preparation
A total of six bacterial strains—M20 B. lehensis (NCBI access number MK308622), M24 B. oshimensis (MK308624), M3 B. lehensis (MK308617), M1 B. conglomeratum (MK308603), CM29 P. rifietoensis (MK308593), and F2 Salinivibrio sp. (MK308616)—were selected to compose the M. hispida BMC consortium, based on the beneficial traits cited above and described in table S1. The probiotic consortium suspension contained a total of 108 cells/ml. The cell number of each individual BMC strain was estimated by an optical density spectrophotometer [optical density at 600 nm (OD600)] [UV-1800 spectrophotometer (Agilent Cary 60, Agilent Technologies)], measurements for cultures grown at 26°C in 100 ml of LB medium for 8, 16, 22, 30, 42, 48, and 54 hours and correlated directly with the number of colony-forming units (CFUs) of each strain at each time point. The CFUs were assessed by subsampling (100 μl) each serial dilution of each strain at each time point, plating on LB agar medium, and incubating under the same conditions. The results were normalized to 1 ml of medium to estimate the cell number at the sampling points (fig. S1). As the probiotic consortium is composed of a diverse combination of bacteria, each strain was collected proportionally at the peak of its growth curve to compose a consortium with a final concentration of 108 cells/ml. The cultures were centrifuged at 5000g for 2 min, and the cell pellets washed three times with saline solution (0.85% NaCl), followed by centrifugation and resuspension in 50 ml of saline solution.
Mesocosm experimental design
The experimental mesocosm used for this experiment consisted of two water baths (100 cm by 50 cm by 10 cm) per temperature (total of four water baths), where five individual aquariums (each with 1.3 liters of capacity; 15 cm by 11 cm by 12 cm) from each treatment were randomly distributed in the mesocosm. Each completely individualized aquarium was an independent true biological replica, with its own individual sump (8.7 liters) and circulation pump; the sump and aquarium assembly together contained a total of 10 liters of seawater. The water flow between the sump and the aquariums was driven by a water pump (Mini A, Sarlo Better, São Caetano do Sul, Brazil) at a flow rate of 250 ml min−1, providing a 10-fold recirculation of the experimental aquarium volume per hour. Every 2 days, 10% of the sump water was changed and the salinity adjusted to 34 practical salinity unit with deionized water if necessary. The aquariums were supplied with natural seawater from the Marine Aquarium of Rio de Janeiro research station where the experiment was performed. The replicates received individual continuous air-bubbling circulation through air pumps (HG-370, Sunsun) connected to silicone air hoses and flow controllers. The water in the water bath was homogenized by two aquarium pumps (SB 1000A, Sarlo Better) to maintain homogeneous temperatures, and there was no water exchange between the aquariums and the water baths. Thermostat controls MT-518ri (Full Gauge, Canoas, Brazil) measured and controlled the temperature of each water bath, activating the cooling system or heaters as needed. The water baths were connected to a 1000-liter freshwater reservoir at 18°C to provide cooling water through water pumps (Better 2000, Sarlo Better) when the temperature-controlled thermostat activated the corresponding pump. The heating system consisted of two 100-W heaters (Atman, China) in the water bath. Physical-chemical parameters of the water, including pH, salinity, and dissolved oxygen (OD), were measured on the sampling days, using a multiparameter probe (Model HI 9,828, Hanna Instruments, Barueri, São Paulo). The experiments followed artificial day/night cycles (12 hours/12 hours) with 150 μmol of photons m−2 s−1 from 06:00 to 10:00 and from 14:00 to 18:00 hours, and 250 μmol of photons m−2 s−1 from 10:00 to 14:00 hours, modulated with light dimmers and a shade cloth. Each replicate individual aquarium had its own lighting system, consisting of six 3W of blue-light and three 3W white-light light-emitting diodes (LEDs), each controlled by a potentiometer. Four coral fragments (~7 cm) of M. hispida were placed randomly in each aquarium, and a single fragment was randomly used as a sampling unit for each treatment and sampling time.
Mesocosm experiment
A total of 80 coral fragments of M. hispida were exposed to two temperature regimes, 30°C (heat stress temperature regime) and 26°C (control temperature regime), and two treatments, placebo or BMCs. A total of four coral fragments (~7 cm) were placed randomly in each aquarium, consisting of a completely independent replica, and each treatment used five of these aquaria. One fragment was randomly used as a sampling unit for each treatment and sampling time. All coral fragments were first maintained under the same conditions at 26°C for 30 days to allow them to heal and acclimate to the experimental conditions. For the heat stress temperature experiment, the temperature was increased, from day 0 to day 8 by 0.5°C per day up to 30°C, which was maintained for 10 days. Then, the temperature was decreased to 26° by 1°C per day, followed by 23 days of recovery. All control experiment aquariums were maintained at 26°C during the 75 experimental days. Sampling points were before heat stress (T0), at the peak of temperature (T1), at the last day of high temperature (T2), and after the recovery period (T3). Samples from the control temperature experiment were also taken in parallel at the same time points. The placebo and BMCs were inoculated on the first day of the experiment and every 5 days thereafter; during the 10 days at the temperature peak, inoculations were performed every 3 days. A detailed schematic view of the experimental design is shown in Fig. 1A. Inoculations were performed by removing the coral fragments from the aquarium and placing them in a sterile petri dish to inoculate 1 ml of the respective treatment above the fragments. After the inoculation, the fragments were immediately returned to their respective aquariums, and the individual petri dishes were rinsed into the aquarium water. Raw data generated in this work are available in the NCBI Sequence Read Archive under the BioProject accession number PRJNA649484.
Assessment of coral health and microbiome
Coral health was assessed during the experiment using different proxies, including visual monitoring of bleaching and algal photosynthetic parameters. The coral visual response was assessed by color score based on the tissue appearance: (i) white (>80% of colony white, with no visible pigmentation), (ii) pale (>10% colony affected by pigment loss), or (iii) fully pigmented (<10% colony with pale coloration). Coral mortality was scored as 0. Each replicate was photographed at each sampling time, with a Canon T3i digital camera, under the same conditions, and the color was scored on the basis of the photographic assessment.
The photochemical efficiency of the Symbiodiniaceae was assessed using pulse amplitude–modulated (PAM) fluorometry. We used a submersible diving-PAM system (Walz GmbH, Effeltrich, Germany) fitted with a red-emitting diode (LED; peak at 650 nm). To avoid nonphotochemical processes of dissipation of PSII excitation energy, measurements were taken after sunset, after at least 30 min of darkness, to ensure full photochemical dissipation of the reaction centers. The maximum quantum yield of PSII photochemistry was determined as Fv/Fm. The diving PAM was configured as follows: measuring light intensity = 5; saturation pulse intensity = 8; saturation pulse width = 0.8; gain = 2; and damping = 2. The same coral fragment from each replicate (= 4 fragments) was used to measure chlorophyll fluorescence at different sampling times during the experiment. The statistical significance of the results was analyzed in Paleontological Statistics software (PAST3).
Assessment of coral microbiome through 16S rRNA gene amplicon sequencing
The coral microbiome was assessed by 16S rRNA gene amplicon sequencing analysis. Samples of the mucus layer, tissue, and skeleton of the coral were collected with sterile clippers at the sampling time. Samples from each sampling time point were macerated with a mortar and pestle under dry conditions. Total DNA was extracted from 0.5 g of the macerated mucus, tissue, or skeleton using the PowerBiofilm DNA Isolation Kit (MO BIO Laboratories Inc.), following the manufacturer’s instructions. The DNA concentration was determined using the Qubit 2.0 Fluorometer High Sensitivity DNA Kit (Invitrogen, USA).
To amplify the hypervariable regions V5 and V6 of the bacterial 16S rRNA gene, the primers 784 forward (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGGATTAGATACCCTGGTA-3′) and 1061 reverse (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCRRCACGAGCTGACG AC-3′) (71) were used (Illumina adapter sequences underlined). Triplicate PCRs (using 1 μl of input DNA) were performed with the QIAGEN Multiplex PCR kit, with a final primer concentration of 0.3 μM in a final reaction volume of 10 μl. In addition to samples, null template PCRs were run (no template DNA input) to account for putative kit contaminants. Thermal cycler conditions were as follows: initial denaturation at 95°C for 15 min, 27 cycles of 95°C for 30 s, 55°C for 90 s, and 72°C for 30 s, followed by a final extension at 72°C for 10 min. Then, 5 μl of each PCR product was run on a 1% agarose gel to confirm successful amplification. Triplicate PCRs for each sample were pooled and samples cleaned using the ExoProStar 1-Step (GE Healthcare, UK). Samples were indexed using the Nextera XT Index Kit v2 (dual indices and Illumina sequencing adaptors added). Successful addition of indexes was confirmed by comparing the length of the initial PCR product to the corresponding indexed sample on a 1% agarose gel. Samples were cleaned and normalized using the SequalPrep Normalization Plate Kit (Invitrogen, Carlsbad, CA, USA). The samples were then pooled in an Eppendorf tube (4 μl per sample) and concentrated using the CentriVap Benchtop Vacuum Concentrator (Labnoco, USA). The quality of the library was assessed using the Agilent High Sensitivity DNA Kit in the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and quantified using Qubit (Qubit dsDNA High Sensitivity Assay Kit, Invitrogen). Library sequencing was performed at 5 pM with 20% phiX on the Illumina MiSeq Illumina platform at the King Abdullah University of Science and Technology (KAUST) Bioscience Core Lab at 2 × 301 bp paired-end V3 chemistry, according to the manufacturer’s specifications.
Coral microbiome data analysis
The microbiota associated with M. hispida fragments was investigated by sequencing the V5 to V6 variable region of the 16S rRNA gene. A library of 3,501,072 good-quality reads with a mean length of 283.64 bp was generated. Demultiplexed raw sequences were imported into QIIME2 2019.4 for analysis. Sequences were merged, denoised, dereplicated, clustered, and trimmed using the DADA2 (“dada2 denoise-paired”) plugin with the following parameters: -p-trim-left-f 5 --p-trim-left-r 5 --p-trunc-len-f 250 --p-trunc-len-r 250, and 4775 ASVs were obtained. The ASVs were classified taxonomically using the naive Bayes machine-learning classifier (72) with the q2-feature-classifier parameter, using the SILVA132 (73) trained classifier clustered at 99% identity as the reference database. A rooted phylogenetic tree was created for downstream analyses, using the programs MAFFT2 (74) and FastTree with CAT-like rate approximation category through Q2-alignment and Q2-phylogeny plugins. The microbial QIIME2 output (qza files) were imported to R programming language version 3.6.0 with the function qiime2R and analyzed with the Phyloseq (75), parsed with the dplyr package (76), and the barplots, boxplots, and statistical test (Kruskal-Wallis and ANOVA) were generated with ggplot2 (77). The data were tested for differential abundance using DESeq2 (78) on a model of negative binomial distribution (NEB); a Wald test with parametric fitting of dispersions to the mean intensity was used for differential abundance estimation, using a cutoff value of P = 0.01. For nonmetric multidimensional scaling analyses, the data were log2-transformed (log x + 1), and ordination was performed with the Bray-Curtis distance matrix until a solution was reached (i.e., stress). The results were plotted with function plot_ordination using the samples, treatments, or environment metadata for features ordination. The significance of the results was evaluated with PERMANOVA, using 999 random permutation tests with pseudo F-ratios through the Adonis function of the Vegan package (79) in R. The community structure of the microbiome (represented by diversity and richness measures) was evaluated (from 0 to 3000 reads) using classic ecological indexes of α diversity (observed ASVs, Chao1, and Shannon) on the rarefaction curve plateau using Phyloseq package.
Gene expression analysis
The coral host gene expression response from T2 and T3 samples were explored to elucidate patterns and genes associated with the physiological differences observed between the BMC and placebo treatments. Total RNA was extracted from both the BMC and placebo treatment samples from the heat stress temperature (30°C) and control temperature (26°C) experiment for sequence analysis, from T0 (five representative replicas), T2 (20 samples, five replicas of each treatment and temperature), and T3 (15 samples, five and three replicas from BMC- and placebo-treated corals of heat stress temperature; four and three replicas from BMC- and placebo-treated corals of control temperature; some of the samples from control temperature had not enough material for RNA extraction). RNA-seq was performed using the Illumina HiSeq 6000 platform (Illumina Inc., San Diego, CA, USA). Sequence reads were quality-controlled using KneadData v0.7.4 with the GRCh38.p13 human genome as a reference for potential decontamination, yielding between 14,372,271 and 74,483,759 paired-end reads at 2 × 150 bp per sample after quality trimming and filtering. De novo transcriptomes were coassembled using rnaSPAdes v3.13.1 (80), producing 520,555 representative transcripts from 496,603 putative genes; genes were estimated by rnaSPAdes.
We used TransDecoder v5.5.0 (81) for gene modeling in a multistep process to minimize false positives. In particular, we used the following procedure: (i) TransDecoder.LongOrfs, with transcript-to-gene mappings assigned by rnaSPAdes, to generate putative open reading frames (ORFs); (ii) hmmscan (hmmer v3.3.1 suite) (82) to identify protein domains using the PFAM v33.1 and TIGRFAM v15.0 databases; (iii) Diamond v0.9.30.131 (83) blastp against all Scleractinia (stony corals) proteomes available in NCBI (GCA_002571385.1, GCF_002042975.1, GCA_003704095.1, GCF_004143615.1, GCF_002571385.1, GCF_003704095.1, and GCF_000222465.1); and (iv) TransDecoder.Predict with the putative ORFs from (i), the protein domains from (ii), and the alignments from (iii) using the --single_best_only argument. This procedure generated a single ORF per transcript to yield 130,183 ORFs from 114,118 genes.
High-quality genes were annotated by using Diamond’s blastp against NCBI’s nr database (v2020.04.01), and taxonomic lineages were extrapolated from NCBItaxid using the get_taxonomy_lineage_from_identifier function from soothsayer v2020.08.24 (https://github.com/jolespin/soothsayer) with ete3 backend (84). PhyloDB v1.076 was used for additional annotations such as Kyoto Encyclopedia of Genes and Genomes ortholog assignments.
Orthogroups were identified using OrthoFinder v2.4.0 (85) with the high-quality proteins generated from our TransDecoder procedure and all of the Scleractinia proteomes listed previously. Annotations for orthogroups were assigned by using the most common organism-agnostic annotation within the grouping.
We assessed differential expression through a comparative genomics perspective for increased ecological interpretability. We aggregated the counts for each orthogroup to generate an orthogroup expression table with 27,140 orthogroup features. We filtered the expression table to include only orthogroups that were in at least 95% of the samples to yield a filtered count table with 17,755 orthogroup features.
For differential expression analysis, we used the 17,755 orthogroup set with the glmFIT and glmLRT models from edgeR v3.28.0 (86) and visualized the distributions and differentially expressed groups (DEGs) using plot_volcano from the soothsayer Python package (https://github.com/jolespin/soothsayer) (fig. S6). To build our design matrix for the generalized linear models, we use a global categorical approach where we used each (time point, treatment, and temperature) grouping as a category and built a binary matrix. We estimated dispersion using the global dataset (not including T0 samples as they were not applicable) using estimateGLMCommonDisp and estimateGLMTagwiseDisp. This model structure allowed us to address variance between conditions while also providing a means to do time-specific contrasts.
Each condition had at least three biological replicates. The conditions investigated were the following using a threshold of FDR < 0.05: (i) placebo (26°C) versus placebo (30°C) (T2: 2294 DEGs; T3: 2795 DEGs); (ii) BMC (26°C) versus BMC (30°C) (T2: 35 DEGs; T3: 1426 DEGs); (iii) placebo (30°C) versus BMC (30°C) (T2: 0 DEGs; T3: 169 DEGs); and (iv) placebo (26°C) versus BMC (26°C) (T2: 2371 DEGs; T3: 0 DEGs) (table S2). As only minor differences were seen between BMC 30°C and placebo 30°C in T2, we focused on T3 transcriptome responses. For this, a graphic showing the DEGs with significant difference (FDR P < 0.05) between the condition BMC 30°C and placebo 30°C at T3 highlighted in the discussion was generated using the log2 FC and presented in Fig. 3, highlighting the mechanisms that might be involved in coral survivorship given by BMC treatment. The log2 FC and FDR values for each orthogroups and condition investigated (described in the paragraph above) can be found on table S2.
Metabolomic analysis
Fragments from each coral sample produced (300.00 mg) were homogenized with 80% methanol (1.50 ml) using zirconia bids and sonicated for 8 min at room temperature. The extraction mixtures were centrifuged at 10,000g for 10 min at 4°C, and the supernatants were concentrated to dryness under vacuum. This procedure was repeated three times for maximum recovery. The residues were resuspended in methanol-d4 (200.00 ml) for NMR data acquisition, using 3-mm tubes and a 600-MHz Bruker Avance III equipped with a 5-mm TCI H-C/N-D cryoprobe and a SampleJet autosampler cooled samples to 6°C while waiting in the queue. The one-dimensional (1D) spectra (noesypr1D) experiment was used to assess the metabolomic profile of the dataset, and 2D experiments heteronuclear single-quantum coherence and heteronuclear multiple-bond correlation (hsqcedetgpsisp2.2 and hmbcetgpl3nd, respectively) were used to confirm the identity of key compounds. Quality control samples were included, and they have shown to be according to the expected. The spectra were processed using NMRPipe and imported into MATLAB for normalization, scaling, and multivariate analysis, using an in-house toolbox [developed in the Edison laboratory (70)]. The PLS-DA analysis was done using the 1D NMR spectra in full resolution, and the boxplot was constructed using the area under the curve of the peaks related to DMSP (2.88 ppm) and DMSO (2.58 ppm). The statistical difference of DMSP and DMSO between sampling times and treatments was assessed with an independent samples t test.
Acknowledgments
We thank D. Pimenta and R. Grilo for logistical and sampling support in the Maraú peninsula and A. O’rourke for technical support and discussions. We also thank the CCRC NMR Facility, especially A. S. Edison, for instrumentation for NMR data collection. Funding: This research project won the Great Barrier Reef Foundation’s Out of the Blue Box Reef Innovation Challenge People’s Choice Award supported by The Tiffany & Co. Foundation. C.R.V. was supported through the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) Project Numbers 433042944 and 458901010. R.S.P. was supported through KAUST grant number BAS/1/1095-01-01 and the Rio de Janeiro Marine Aquarium Research Center. E.P.S. received support from the Graduate Programs of Science (Microbiology) and Plant Biotechnology and Bioprocess Engineering (PBV)/Federal University of Rio de Janeiro, the Brazilian Government Research Agency CAPES, and the CAPES PRINT international mobility grant. Author contributions: E.P.S., H.D.M.V., G.A.S.D., and R.S.P conceived and designed the study; G.A.S.D. and E.P.S. collected the coral samples; E.P.S., C.S.M.A.M., H.D.M.V., C.L.S.V., J.G.R., P.M.C., P.M.R., J.M.A., and G.A.S.D. performed the BMC selection and mesocosm experiment; R.M.B. performed the metabolomic analysis; J.L.E., C.L.D., K.E.N., L.M.P., C.R.V., and G.P. were involved with DNA- or RNA-seq, bioinformatics, and statistical analyses; E.P.S., J.L.E., C.L.D., C.R.V., L.M.P., M.J.S., A.S.R., A.M., R.M.B., and R.S.P. analyzed and interpreted the data; E.P.S., C.R.V., and R.S.P. wrote the manuscript; J.L.E., C.L.D., M.J.S., H.D.M.V., A.S.R., A.M., and R.M.B. were involved in the critical revision of the manuscript; and R.S.P., K.E.N., and C.R.V. provided financial support. All author(s) read and approved the final manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. M. hispida transcripts, gene models, and annotations are available at https://doi.org/10.6084/m9.figshare.13344758.v1.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/33/eabg3088/DC1
REFERENCES AND NOTES
- 1.Hughes T. P., Anderson K. D., Connolly S. R., Heron S. F., Kerry J. T., Lough J. M., Baird A. H., Baum J. K., Berumen M. L., Bridge T. C., Claar D. C., Eakin C. M., Gilmour J. P., Graham N. A. J., Harrison H., Hobbs J. A., Hoey A. S., Hoogenboom M., Lowe R. J., McCulloch M. T., Pandolfi J. M., Pratchett M., Schoepf V., Torda G., Wilson S. K., Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359, 80–83 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Van Oppen M. J. H., Gates R. D., Blackall L. L., Cantin N., Chakravarti L. J., Chan W. Y., Cormick C., Crean A., Damjanovic K., Epstein H., Harrison P. L., Jones T. A., Miller M., Pears R. J., Peplow L. M., Raftos D. A., Schaffelke B., Stewart K., Torda G., Wachenfeld D., Weeks A. R., Putnam H. M., Shifting paradigms in restoration of the world’ s coral reefs. Glob. Chang. Biol. 23, 1–12 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Peixoto R. S., Sweet M., Bourne D. G., Customized medicine for corals. Front. Mar. Sci. 6, 686 (2019). [Google Scholar]
- 4.LaJeunesse T. C., Parkinson J. E., Gabrielson P. W., Jeong H. J., Reimer J. D., Voolstra C. R., Santos S. R., Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Muscatine L., Porter J. W., Reef corals: Mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27, 454–460 (1977). [Google Scholar]
- 6.Rädecker N., Pogoreutz C., Gegner H. M., Cárdenas A., Roth F., Bougoure J., Guagliardo P., Wild C., Pernice M., Raina J., Meibom A., Voolstra C. R., Heat stress destabilizes symbiotic nutrient cycling in corals. Proc. Natl. Acad. Sci. U.S.A. 118, e2022653118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohwer F., Seguritan V., Azam F., Knowlton N., Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10 (2002). [Google Scholar]
- 8.Littman R., Willis B. L., Bourne D. G., Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ. Microbiol. Rep. 3, 651–660 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Jaspers C., Fraune S., Arnold A. E., Miller D. J., Bosch T. C. G., Voolstra C. R., Resolving structure and function of metaorganisms through a holistic framework combining reductionist and integrative approaches. Fortschr. Zool. 133, 81–87 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Peixoto R. S., Rosado P. M., Leite D. C. A., Rosado A. S., Bourne D. G., Beneficial microorganisms for corals (BMC): Proposed mechanisms for coral health and resilience. Front. Microbiol. 8, 341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peixoto R. S., Sweet M., Villela H. D. M., Cardoso P., Thomas T., Voolstra C. R., Høj L., Bourne D. G., Coral probiotics: Premise, promise, prospects. Annu. Rev. Anim. Biosci. 9, 265–288 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Ziegler M., Seneca F. O., Yum L. K., Palumbi S. R., Voolstra C. R., Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat. Commun. 8, 14213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bang C., Dagan T., Deines P., Dubilier N., Duschl W. J., Fraune S., Hentschel U., Hirt H., Hülter N., Lachnit T., Picazo D., Pita L., Pogoreutz C., Rädecker N., Saad M. M., Schmitz R. A., Schulenburg H., Voolstra C. R., Weiland-Bräuer N., Ziegler M., Bosch T. C. G., Metaorganisms in extreme environments: Do microbes play a role in organismal adaptation? Fortschr. Zool. 127, 1–19 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Ziegler M., Grupstra C. G. B., Barreto M. M., Eaton M., BaOmar J., Zubier K., Al-Sofyani A., Turki A. J., Ormond R., Voolstra C. R., Coral bacterial community structure responds to environmental change in a host-specific manner. Nat. Commun. 10, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voolstra C. R., Ziegler M., Adapting with microbial help: Microbiome flexibility facilitates rapid responses to environmental change. Bioessays 42, 2000004 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Reshef L., Koren O., Loya Y., Zilber-Rosenberg I., Rosenberg E., The coral probiotic hypothesis. Environ. Microbiol. 8, 2068–2073 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Robbins S. J., Singleton C. M., Chan C. X., Messer L. F., Geers A. U., Ying H., Baker A., Bell S. C., Morrow K. M., Ragan M. A., Miller D. J., Forêt S.; ReFuGe2020 Consortium, Voolstra C. R., Tyson G. W., Bourne D. G., A genomic view of the reef-building coral Porites lutea and its microbial symbionts. Nat. Microbiol. 4, 2090–2100 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Rosado P. M., Leite D. C. A., Duarte G. A. S., Chaloub R. M., Jospin G., Rocha U. N., Saraiva J. P., Dini-Andreote F., Eisen J. A., Bourne D. G., Peixoto R. S., Marine probiotics: Increasing coral resistance to bleaching through microbiome manipulation. ISME J. 13, 921–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams A. D., Brown B. E., Putchim L., Sweet M. J., Age-related shifts in bacterial diversity in a reef coral. PLOS ONE 10, e0144902 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster N. S., Reusch T. B. H., Microbial contributions to the persistence of coral reefs. ISME J. 11, 2167–2174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaneveld J. R., Mcminds R., Thurber R. V., Stress and stability: Applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2, 1–8 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Silva D. P., Duarte G., Villela H. D. M., Santos H. F., Rosado P. M., Rosado J. G., Rosado A. S., Ferreira E. M., Soriano A. U., Peixoto R. S., Adaptable mesocosm facility to study oil spill impacts on corals. Ecol. Evol. 9, 5172–5185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva D. P., Villela H. D. M., Santos H. F., Duarte G. A. S., Ribeiro J. R., Ghizelini A. M., Vilela C. L. S., Rosado P. M., Fazolato C. S., Santoro E. P., Carmo F. L., Ximenes D. S., Soriano A. U., Rachid C. T. C. C., Thurber R. L. V., Peixoto R. S., Multi-domain probiotic consortium as an alternative to chemical remediation of oil spills at coral reef and adjacent sites. Microbiome 9, 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Yang Q., Ling J., Long L., Huang H., Yin J., Wu M., Tang X., Lin X., Zhang Y., Dong J., Shifting the microbiome of a coral holobiont and improving host physiology by inoculation with a potentially beneficial bacterial consortium. BMC Microbiol. 21, 130 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voolstra C. R., López C. B., Perna G., Cárdenas A., Hume B. C. C., Rädecker N., Barshis D. J., Standardized short-term acute heat stress assays resolve historical differences in coral thermotolerance across microhabitat reef sites. Glob. Change Biol 26, 4328–4343 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Ben-Haim Y., Thompson F. L., Thompson C. C., Cnockaert M. C., Hoste B., Swings J., Rosenberg E., Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int. J. Syst. Evol. Microbiol. 53, 309–315 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Alves N. J., Neto O. S. M., Silva B. S. O., De Moura R. L., Francini-Filho R. B., Castro C. B., Paranhos R., Bitner-Mathé B. C., Kruger R. H., Vicente A. C. P., Thompson C. C., Thompson F. L., Diversity and pathogenic potential of Vibrios isolated from Abrolhos Bank corals. Environ. Microbiol. Rep. 2, 90–95 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Gasch A. P., Spellman P. T., Kao C. M., Harel O. C., Eisen M. B., Storz G., Botstein D., Brown P. O., Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dixon G., Abbott E., Matz M., Meta-analysis of the coral environmental stress response: Acropora corals show opposing responses depending on stress intensity. Mol. Ecol. 29, 2855–2870 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Morgans C. A., Hung J. Y., Bourne D. G., Quigley K. M., Symbiodiniaceae probiotics for use in bleaching recovery. Restor. Ecol. 28, 282–288 (2020). [Google Scholar]
- 31.Santos H. F., Duarte G. A. S., Rachid C. T. C. C., Chaloub R. M., Calderon E. N., Marangoni L. F. B., Bianchini A., Nudi A. H., Carmo F. L., van Elsas J. D., Rosado A. S., Castro C. B., Peixoto R. S., Impact of oil spills on coral reefs can be reduced by bioremediation using probiotic microbiota. Sci. Rep. 5, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pootakham W., Pootakham W., Mhuantong W., Yoocha T., Putchim L., Jomchai N., Sonthirod C., Naktang C., Kongkachana W., Tangphatsornruang S., Heat-induced shift in coral microbiome reveals several members of the Rhodobacteraceae family as indicator species for thermal stress in Porites lutea. Microbiol. Open 12, 935 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leite D. C., Leão P., Garrido A. G., Lins U., Santos H. F., Pires D. O., Castro C. B., van Elsas J. D., Zilberberg C., Rosado A. S., Peixoto R. S., Broadcast spawning coral Mussismilia hispida can vertically transfer its associated bacterial core. Front. Microbiol. 8, 176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leite D. C., Salles J. F., Calderon E. N., Castro C. B., Bianchini A., Marques J. A., van Elsas J. D., Peixoto R. S., Coral bacterial-core abundance and network complexity as proxies for anthropogenic pollution. Front. Microbiol. 9, 833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leite D. C., Salles J. F., Calderon E. N., van Elsas J. D., Peixoto R. S., Specific plasmid patterns and high rates of bacterial co-occurrence within the coral holobiont. Ecol. Evol. 8, 1818–1832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villela H., Microbiome flexibility provides new perspectives in coral research. Bioessays 42, e2000088 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Sweet M., Burian A., Fifer J., Bulling M., Elliott D., Raymundo L., Compositional homogeneity in the pathobiome of a new, slow-spreading coral disease. Microbiome 7, 139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos H. F., Carmo F. L., Paes J. E. S., Rosado A. S., Peixoto R. S., Bioremediation of mangroves impacted by petroleum. Water Air Soil Pollut. 216, 329–350 (2011). [Google Scholar]
- 39.P. Pandey, S. Bisht, A. Sood, A. Aeron, G. D. Sharma, D. K. Maheshwari, Consortium of plant growth-promoting-bacteria: Future perspectives in agriculture, in Bacteria in Agrobiology: Plant Probiotics, D. K. Maheshwari, Ed. (Springer, 2012), pp 185–200. [Google Scholar]
- 40.de Souza R. S. C., Armanhi J. S. L., Arruda P., From microbiome to traits: Designing synthetic microbial communities for improved crop resiliency. Front. Plant Sci. 11, 1179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sweet M. J., Brown B. E., Dunne R. P., Singleton I., Bulling M., Evidence for rapid, tide-related shifts in the microbiome of the coral Coelastrea aspera. Coral Reefs 36, 815–828 (2017). [Google Scholar]
- 42.Takahashi S., Bauwe H., Badger M. R., Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol. 144, 487–494 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lesser M. P., Stochaj W. R., Tapley D. W., Shick J. M., Bleaching in coral reef anthozoans: Effects of irradiance, ultraviolet radiation, and temperature on the activities of protective enzymes against active oxygen. Coral Reefs 8, 225–232 (1990). [Google Scholar]
- 44.Bieri T., Onishi M., Xiang T., Grossman A. R., Pringle J. R., Relative contributions of various cellular mechanisms to loss of algae during cnidarian bleaching. PLOS ONE 11, e0152693 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunn S. R., Pernice M., Green K., Hoegh-guldberg O., Dove S. G., Thermal stress promotes host mitochondrial degradation in symbiotic cnidarians: Are the batteries of the reef going to run out? PLOS ONE 7, e39024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cross T. G., Toellner D. S., Henriquez N. V., Deacon E., Salmon M., Lord J. M., Serine/threonine protein kinases and apoptosis. Exp. Cell Res. 256, 34–41 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Kurokawa M., Kornbluth S., Caspases and kinases in a death grip. Cell 138, 838–854 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yehuda R., Hoge C. W., McFarlane A. C., Vermetten E., Lanius R. A., Nievergelt C. M., Hobfoll S. E., Koenen K. C., Neylan T. C., Hyman S. E., Post-traumatic stress disorder. Nat. Rev. Dis. Primers. 1, 15057 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Weiss V. M., Cellular mechanisms of Cnidarian bleaching: Stress causes the collapse of symbiosis. J. Experim. Biol. 211, 3059–3066 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Thomas L., Palumbi S. R., The genomics of recovery from coral bleaching. Proc. R. Soc. B 284, 20171790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinzón J. H., Kamel B., Burge C. A., Harvell C. D., Medina M., Weil E., Mydlarz L. D., Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral. R. Soc. Open Sci. 21, 40214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savary R., Barshis D. J., Voolstra C. R., Cárdenas A., Evensen N. R., Prandi G. B., Fine M., Meibom A., Fast and pervasive transcriptomic resilience and acclimation of extremely heat-tolerant coral holobionts from the northern Red Sea. Proc. Natl. Acad. Sci. U.S.A. 118, e2023298118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garren M., Son K., Raina J.-B., Rusconi R., Menolascina F., Shapiro O. H., Tout J., Bourne D. G., Seymour J. R., Stocker R., A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raina J. B., Tapiola D., Motti C. A., Foret S., Seemann T., Tebben J., Willis B. L., Bourne D. G., Isolation of an antimicrobial compound produced by bacteria associated with reef-building corals. PeerJ 4, e2275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raina J. B., Clode P. L., Cheong S., Bougoure J., Kilburn M. R., Reeder A., Forêt S., Stat M., Beltran V., Hall P. T., Tapiolas D., Motti C. M., Gong B., Pernice M., Marjo C. E., Seymour J. R., Willis B. L., Bourne D. G., Subcellular tracking reveals the location of dimethylsulfoniopropionate in microalgae and visualises its uptake by marine bacteria. eLife 6, e23008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wirth J. S., Wang T., Huang Q., White R. H., Whitman W. B., Dimethylsulfoniopropionate sulfur and methyl carbon assimilation in Ruegeria species. MBio 11, 2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miura N., Motone K., Takagi T., Aburaya S., Watanabe S., Aoki W., Ueda M., Ruegeria sp. strains isolated from the reef-building coral Galaxea fascicularis inhibit growth of the temperature-dependent pathogen Vibrio coralliilyticus. J. Mar. Biotechnol. 21, 1–8 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Dahiya D. K., Malik R., Puniya M., Shandilya U. K., Dhewa T., Kumar N., Kumar S., Puniya A. K., Shukla P., Gut microbiota modulation and its relationship with obesity using prebiotic fibers and probiotics: A review. Front. Microbiol. 8, 563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shin D., Chang S. Y., Bogere P., Won K. H., Choi J. Y., Choi Y. J., Lee H. K., Hur J., Park B. Y., Kim Y., Heo J., Beneficial roles of probiotics on the modulation of gut microbiota and immune response in pigs. PLOS ONE 14, e0220843 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eakin C. M., Morgan J. A., Heron S. F., Smith T. B., Liu G., Filip L. A., Baca B., Bartels E., Bastidas C., Bouchon C., Brandt M., Bruckner A. W., Williams L. B., Cameron A., Causey B. D., Chiappone M., Christensen T. R. L., Crabbe M. J. C., Day O., de la Guardia E., Pulido G. D., DiResta D., Agudelo D. L. G., Gilliam D. S., Ginsburg R. N., Gore S., Guzmán H. M., Hendee J. C., Delgado E. A. H., Husain E., Jeffrey C. F. G., Jones R. J., Dahlgren E. J., Kaufman L. S., Kline D. I., Kramer P. A., Lang J. C., Lirman D., Mallela J., Manfrino C., Maréchal J. P., Marks K., Mihaly J., Miller W. J., Mueller E. M., Muller E. M., Toro C. A. O., Oxenford H. A., Taylor D. P., Quinn N., Ritchie K. B., Rodríguez S., Ramírez A. R., Romano S., Samhouri J. F., Sánchez J. A., Schmahl G. P., Shank B. V., Skirving W. J., Steiner S. C. C., Villamizar E., Walsh S. M., Walter C., Weil E., Williams E. H., Roberson K. W., Yusuf Y., Caribbean corals in crisis: Record thermal stress, bleaching, and mortality in 2005. PLOS ONE 5, e13969 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bahr K. D., Rodgers K. S., Jokiel P. L., Impact of three bleaching events on the reef resiliency of Kāne’ohe Bay, Hawai’i. Front. Microbiol. 4, 398 (2017). [Google Scholar]
- 62.Duarte G. A. S., Villela H. D. M., Deocleciano M., Silva D., Barno A., Cardoso P. M., Vilela C. L. S., Rosado P., Messias C. S. M. A., Chacon M. A., Santoro E. P., Olmedo D. B., Szpilman M., Rocha L. A., Sweet M., Peixoto R. S., Heat waves are a major threat to turbid coral reefs in Brazil. Front. Mar. Sci. 7, 179 (2020). [Google Scholar]
- 63.National Academies of Science, Engineering and Medicine, A Decision Framework for Interventions to Increase the Persistence and Resilience of Coral Reefs (National Academies Press, 2019). [Google Scholar]
- 64.Kleypas J., Son K., Raina J.-B., Rusconi R., Menolascina F., Shapiro O. H., Tout J., Bourne D. G., Seymour J. R., Stocker R., Designing a blueprint for coral reef survival. Biol. Conserv. 257, 109107 (2021). [Google Scholar]
- 65.Giambiagi-Marval M., Mafra M. A., Penido E. G. C., Bastos M. C. F., Distinct groups of plasmids correlated with bacteriocin production in Staphylococcus aureus. J. Gen. Microbiol. 136, 1591–1599 (1990). [DOI] [PubMed] [Google Scholar]
- 66.Vidal-Dupiol J., Ladrière O., Meistertzheim A. L., Fouré L., Adjeroud M., Mitta G., Physiological responses of the scleractinian coral Pocillopora damicornis to bacterial stress from Vibrio coralliilyticus. J. Experim. Biol. 214, 1533–1545 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Vidal-Dupiol J., Ladrière O., Garzón D. D., Sautière P. E., Meistertzheim A. L., Tambutté E., Tambutté S., Duval D., Fouré L., Adjeroud M., Mitta G., Innate immune responses of a scleractinian coral to vibriosis. J. Biol. Chem. 286, 22688–22698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cole S. E., Lariviere F. J., Merrikh C. N., Moore M. J., A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol. Cell 34, 40–50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hall T., BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999). [Google Scholar]
- 70.Altschul S., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 71.Andersson A. F., Lindberg M., Jakobsson H., Nyrén P., Engstrand L., Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLOS ONE 3, 2836 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bokulich N. A., Kaehler B. D., Rideout J. R., Dillon M., Bolyen E., Knight R., Huttley G. A., Caporaso J. G., Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Henderson G., Yilmaz P., Kumar S., Forster R. J., Kelly W. J., Leahy S. C., Guan L. L., Janssen P. H., Improved taxonomic assignment of rumen bacterial 16S rRNA sequences using a revised SILVA taxonomic framework. PeerJ 7, e6496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katoh K., Kuma K. I. H., Miyata T., Mafft version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mcmurdie P. J., Holmes S., Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE 8, 61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.H. Wickham, R. Francois, L. Henry, K. Müller, Dplyr: A Grammar of Data Manipulation, R Package Version 0.7.3 (2017).
- 77.Wickham H., ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 3, 180–185 (2011). [Google Scholar]
- 78.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.J. Oksanen, F. G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, P. R. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, E. Szoecs, H. Wagne, Vegan: Community ecology package. R package version 2.0-10 (2019).
- 80.Bushmanova E., Antipov D., Lapidus A., Prjibelski A. D., rnaSPAdes: A de novo transcriptome assembler and its application to RNA-Seq data. Gigascience 8, giz100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Couger M. B., Eccles D., Li B., Lieber M., Manes M. D. M., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C. N., Henschel R., Le Duc R. D., Friedman N., Regev A., De novo transcript sequence reconstruction from RNA-Seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eddy S. R., Accelerated profile HMM searches. PLOS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 84.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Emms D. M., Kelly S., OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robinson M. D., Mccarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/33/eabg3088/DC1


