Abstract
This review provides a brief history of the advances of cellular analysis tools focusing on instrumentation, detection probes, and data analysis tools. The interplay of technological advancement and a deeper understanding of cellular biology are emphasized. The relevance of this topic to drug development is that the evaluation of cellular biomarkers has become a critical component of the development strategy for novel immune therapies, cell therapies, gene therapies, antiviral therapies, and vaccines. Moreover, recent technological advances in single-cell analysis are providing more robust cellular measurements and thus accelerating the advancement of novel therapies.
KEY WORDS: flow cytometry, drug development, single cell
INTRODUCTION
The importance of biomarkers in decision-making during drug development continues to increase; cellular biomarkers, in particular, have become critical components in the strategy for the development of novel immune therapies for oncology, cell therapy, gene therapy, infectious diseases, and vaccines. Flow cytometry, a leading technology in cellular analysis, has been used broadly in the drug development process for over 20 years. In drug discovery, it is used for target identification, to characterize target biology and disease pathways and lead compound characterization and early mechanism of action evaluation (1). It is used preclinically for proof of concept studies, preliminary safety evaluation, and first-in-human dose selection. For chimeric antigen receptor T cell (CAR-T) therapy and other cell-based therapies, flow cytometry is now being used in the manufacturing phases in order to evaluate product quality (purity, viability, and concentration) and post-infusion in order to monitor cellular kinetics (2, 3). Receptor occupancy (RO) assays have become critical measurements in the development of protein-based therapies including bi-specific compounds targeting cell surface receptors such as immune-checkpoint inhibitors (4–6). RO data are often used in pharmacokinetics/pharmacodynamic modeling in order to guide first-in-human dosing ranges (4–6). Nonetheless, the most frequent application of flow cytometry in drug development is likely in the clinical development phase where flow cytometry is used to monitor the cellular composition of the immune system for patient selection, to evaluate the efficacy of cell-depleting therapies and vaccine efficacy, and to assess measurable residual disease in leukemia, along with other numerous applications (7).
The increased importance of cellular biomarkers in the drug development process is coinciding with game-changing advances in high-parameter, single-cell analysis technologies and data reduction tools. Their implementation is expected to have a positive impact on the drug development process and cellular biomarker strategies. By evaluating a greater number of parameters on each individual cell, a deeper understanding of each cell’s function and the overall biology of the immune system is gained. In a drug development setting, increased dimensionality enables a deeper understanding of the impact of novel therapies on the immune system and the targeted disease and from a logistics standpoint, it also increases the efficiency of sample usage which can be very important in clinical trials where minimizing the specimen collection volume per patient visit is critical.
The complex data sets generated from the higher-dimensional instruments create a need for novel data analysis tools. In order to fully explore these content-rich measurements, both supervised and unsupervised data analysis approaches will be required. Although traditional, predefined, hierarchy-based gating approaches continue to be highly valuable, they are time-consuming and can be subjective. Fortunately, there are automated solutions to address these issues. Unsupervised or hypothesis-generating data analysis approaches are valuable to identify cell subsets which were not previously considered and to reveal associations between cell types, between diseased and healthy populations, or between treatment groups, which may not have been included in the initial hypothesis.
EVOLUTION OF TOOLS FOR SINGLE-CELL ANALYSIS
Ever since the earliest microscopes appeared in the 1590s, inventors and scientists continue to make technological improvements which then improves our understanding of biology. Circa 1665 when making observations using a rudimentary compound microscope, Robert Hooke coined the term “cell.” Shortly thereafter using a microscope with improved lenses and magnification capabilities, Anton van Leeuwenhoek provided the first accurate description of cells (red blood cells, bacteria, and sperm).
Critical discoveries which contributed to the early flow cytometer prototypes include Paul Ehrlich’s description of the staining properties of fluorescein in the 1880s, Andrew Moldavan’s system for the photo detection of particles in the 1930s, and Albert Coons’s method for the detection of Streptococcus pneumonia by a UV-excitable fluorophore-conjugated antibody in 1942. A major development arrived in the 1950s with the Coulter Counter developed by Wallace & Joseph Coulter. This instrument consisted of two chambers containing liquid streams separated by a microchannel. As particles move through the microchannel, they displace an equivalent volume of liquid which is detected as a voltage pulse whose height is proportional to the cell number. In the early 1960s while working at IBM’s Watson Laboratory at Columbia University, Kamentsky et al. developed the Rapid Cell Spectrophotometer, which using an arc lamp as a light source could measure cell size and nucleic acid content (8). In 1965, Fulwyler introduced the Los Alamos Flow Microfluorometer, a forerunner of modern flow cytometers based on electronic cell volume and electrostatic deflection for segregation and sorting (9).
Flow Cytometers
The first commercial flow cytometers became available in the late 1960s. Dittrich and Göhde designed a fluorescence-based flow cytometer device (ICP-11) in 1969, which was commercially developed by Partec (10). Next, in the early 1970s, the cytograph/cytofluorograph and the PAS-8000 flow cytometers appeared, and in 1976, Herzenberg et al. at Stanford University designed the first cell sorter (11). Fluorescence-based cytometry initially had numerous names including “impulszytophotometrie” which translates from German to “pulse cytophotometry” in English. Fortunately, it was agreed at the Conference of the American Engineering Foundation in 1976 that the term “flow cytometry” should be used for this type of instrumentation.
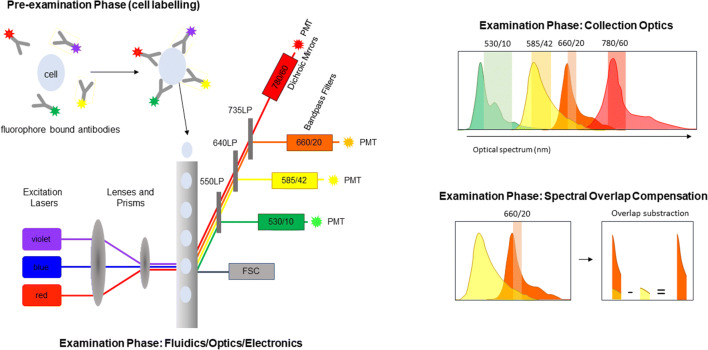
The primary components of these early flow cytometry instruments were the fluidics, optics, and electronics. Fluorescent-labeled particles (typically cells) moving in a fluid stream were interrogated by a laser light source (helium-neon) where the fluorescent probes being excited by the laser light source emitted a signal (Figure 1). These fluorescent signals as well as scattered lighter signals were recorded. The amount of forward-scattered light was being an indication of size and the side-scattered light an indication of “complexity” or granularity. The signature of each particle could be used as the basis for the isolation (sorting) of different populations.
Figure 1.
Conventional flow cytometry. In conventional flow cytometry, cells are labeled with fluorophore-conjugated probes, typically monoclonal antibodies. Within the flow cell, samples then enter the sample stream focused such that a single cell is interrogated by one or more lasers resulting in light dispersion (forward side scatter (FSC)) and excitation of the fluorophores. Emitted fluorescence is then directed to the photodetectors (usually photomultiplier tubes (PMTs)) via a series of long pass (LP), band pass filters and dichroic mirrors. Spectral overlap between fluorophores on the same cell is subtracted by applying spectral overlap compensation (figure adapted from reference (12))
These earliest instruments measured a single fluorescent signal and light scatter and were equipped with water-cooled lasers which required complex facility controls. Moreover, the lasers needed to be manually aligned which required highly skilled operators. Thus, these single-laser instruments with sorting capabilities first introduced by Becton Dickinson in early 1970s were housed in shared core facilities, operated, and maintained by dedicated staff (13). With the introduction of solid-state lasers, benchtop instruments equipped with a single, fixed laser and fixed filters capable of three-color analysis became available in the late 1980s. Sorting was not possible with these instruments, but they were less expensive and could be used directly by research scientists without the need for dedicated operators. As a result of the decreased cost and ease of use, the benchtop flow cytometers became used more broadly and many laboratories purchased their own instruments outside of the core facilities.
When a second laser was added to the benchtop instruments in the mid-1980s, four-color analysis became possible. This advancement was quite important for clinical laboratories given that four-color analysis allowed for the addition of CD45 to standard assays for the detection of the major T lymphocyte subsets (total T cells, CD4, and CD8 T cells) in order to increase the purity of the lymphocyte gate and the overall accuracy of the assay. This class of assay became a critical component in the development of antiviral therapies for HIV/AIDS as the CD4 cell absolute count measurement was a key drug efficacy indicator and a required measurement in the care and treatment of patients receiving these compounds. Additionally, the use of CD45 and SSC to identify cellular populations in blood and bone marrow provided the foundation for characterizing normal and aberrant hematopoiesis, giving rise to the utility of cytometry in diagnosing and monitoring leukemia and lymphomas in clinical laboratories (14). When six-color instruments became available, the assay for the detection of the major lymphocyte subsets (total T cells, CD4 and CD8 T cells, B cells, and NK cells) was reduced from a two-tube assay to a single-tube assay which allowed for greater efficiency and still more accurate measurements.
Leonard Herzenberg and Mario Roederer were leaders in pushing the technology to measure more than four colors. Polychromatic flow cytometry upwards of four colors was first described in 1997, 11-color in 2001, and 17-color in 2004 (15–17). The initial 17-color instrument was configurable and equipped with a variety of lasers, up to 4 lasers, which excited in the green (532 nm), blue (488 nm), violet (407 nm), and red (633 nm) ranges (Table I).
Table I.
Most Commonly Used Lasers in Conventional and Spectral Cytometry
| Laser | Wavelength (nm) |
|---|---|
| UV | 355 |
| Violet | 405 |
| Blue | 488 |
| Yellow green | 561 |
| Red | 633 |
Later, advances in laser technology resulted in the development of a greater range of wavelength choices for solid-state lasers. For example, the introduction yellow-green (561 nm) laser was a notable improvement as it allowed for better excitation of PE and eliminates FITC-PE spillover issues. It should be noted that while the introduction of the yellow-green laser did not necessarily (depending on the instrument) increase the total number of lasers on the instrument or result in greater number of detected parameters, it facilitated the generation of stronger, cleaner signal detection and thus is considered a major advance. This point is something to be aware of when evaluating newer instruments. For example, the 18-color instruments available today can be custom configured with up to 5 lasers and are equipped with digital detection systems which capture a greater amount of the emission signal, thus increasing sensitivity of the measurement (Table I). As a result, these instruments not only have high dimensionality, but they are also more sensitive as a greater portion of the fluorescence emission can now be captured.
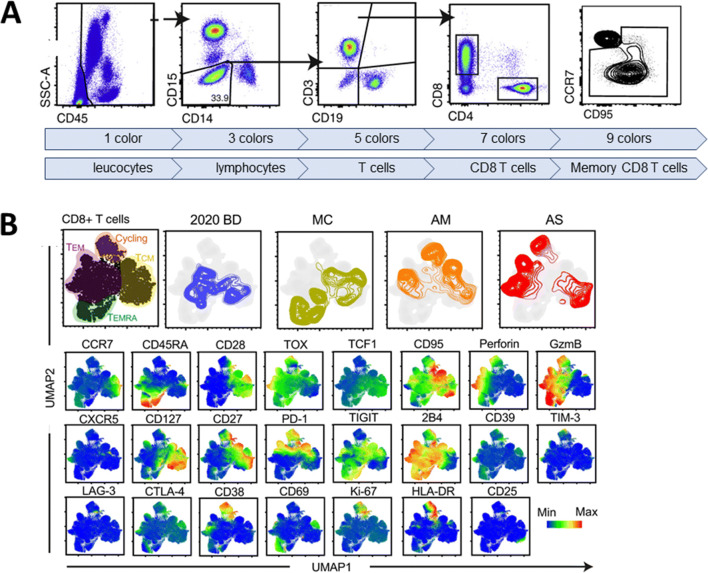
Although it is hard to imagine today, six-color instruments were initially met with some skepticism as some individuals struggled to understand the need for six-color capacity. The Herzenberg lab was the first to address these critiques by sharing data which clearly demonstrated the value of increased dimensionality. Results from the evaluation of an experimental vaccine indicated a lack of response when the entire CD4 T cell population was evaluated; however, when specific CD4 T cell subsets within the parent CD4 T cell population were identified by labeling the cells with additional markers, the data revealed responses to the vaccine (15). This scenario repeats itself often; in that as an increased number of parameters are evaluated, biological responses are revealed which were not observed when less well-defined populations were measured in assays with a lower number of parameters (Figure 2).
Figure 2.
Using high-dimensional cytometry to reveal immune cell populations. A An example of peripheral whole blood labeled with nine fluorophore-conjugated probes. The number of probes influences the type of cells and subsets which can be identified. Hierarchical gating and bivariate plots are used to reveal, leucocytes, lymphocytes, T lymphocytes, CD8+ T lymphocytes, and finally the memory subset of CD8+ T lymphocytes. B Uniform Manifold Approximation and Projection (UMAP) multidimensional analysis of CD8+ T cells from 29 colors flow cytometry assay acquired on BD FACSSymphony. Labeling the samples with a greater number of probes reveals additional cellular subsets and greater information about the biology (figure adapted from reference (18))
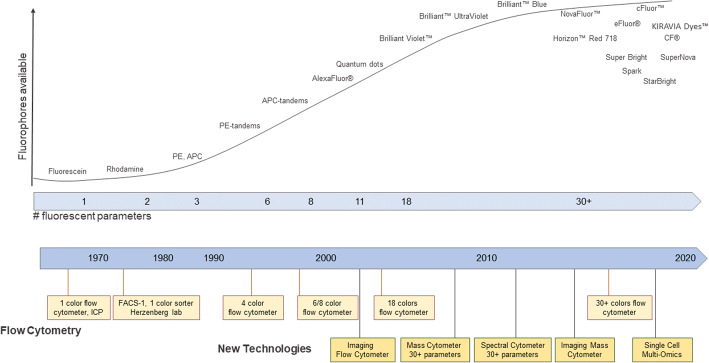
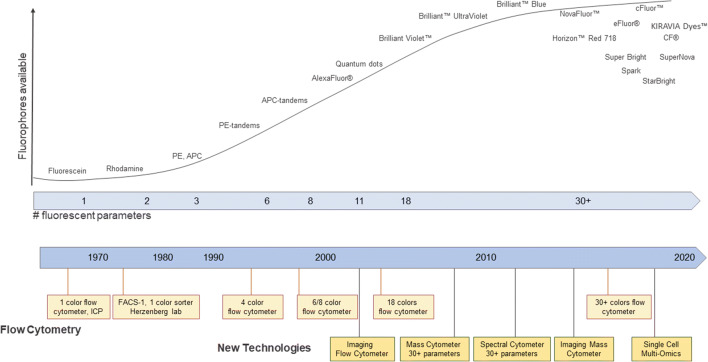
For a period of time in the early 2000s, using commercially available instrumentation polychromatic flow cytometry seemed to have peaked with a maximum of 18-color detection which could be evaluated in a single assay. This limitation was due to the availability of fluorophores as well as limited space within the light spectrum (19). The advancement of new probe chemistry and the design of new types of instruments and solid-state lasers have overcome this limitation (Figure 3 and Table II).
Figure 3.
Evolution of instruments and probes. A schematic representation of the timelines of the emergence of new technologies and probes used in cytometry (figure adapted from reference (20)). ICP Impulscytophotometer, FACS fluorescence-activated cell sorter, PE phycoerythrin, APC allophycocyanin
Table II.
Attributes of Single-Cell Cytometry Technologies (Adapted from reference (21))
| Fluorescence cytometry | Mass cytometry | Omics (Poly-A Seq) cytometry | ||||
|---|---|---|---|---|---|---|
| Flow cytometry | Spectral cytometry | |||||
| Detection method | Lasers with PMT/APD for detection of fluorescent emission spectra | Lasers with PMT/APD for detection of the full spectra | Time-of-flight of ionized heavy elemental isotopes | Nucleic acid sequence | ||
| Type of probe | Fluorophore-conjugated antibodies, fluorescent proteins, or other fluorescent probes | Fluorophore-conjugated antibodies, fluorescent proteins, or other fluorescent probes | Metal-conjugated antibodies or small molecules | Poly-A targeted oligonucleotides, antibody-conjugated oligonucleotide | ||
| Optics | Dichroic mirrors/band pass filters | Dispersive optics: prisms or spectrographs | Ion optics and time of flight mass analyzer | N/A | ||
| Spectral resolution | Captures only a narrow emission bandwidth from a single-laser excitation | Captures the entire fluorophore spectral profile from all lasers | N/A | N/A | ||
| Signal separation | Spectral overlap subtraction (compensation) | Spectral unmixing | N/A, however metal degradation can lead to interference in signal | N/A | ||
| Number of parameters | 8–12 | 18–21 | >30 | >30 | >37 | >100 to 400 |
| Clinical approval* | Yes | No | No | Yes | No | No |
| Sampling efficiency | >95% | >95% | 30–60% | 5 to 65% | ||
| Sample throughput cells/sec | ≈35,000–100,000 | ≈35,000 | Up to 2000 | Up to 10,000 cells total | ||
*Instruments which have, as of July 2021, received IVD clearance as an in vitro diagnostic (IVD) device from the US Food and Drug Administration (FDA) and/or are CE marked under the European Union In Vitro Diagnostic Medical Devices Directive (IVDD) 98/79/EC
PMT photomultiplier tube, APD avalanche photodiodes, N/A not applicable
Mass Cytometers (CyTOF)
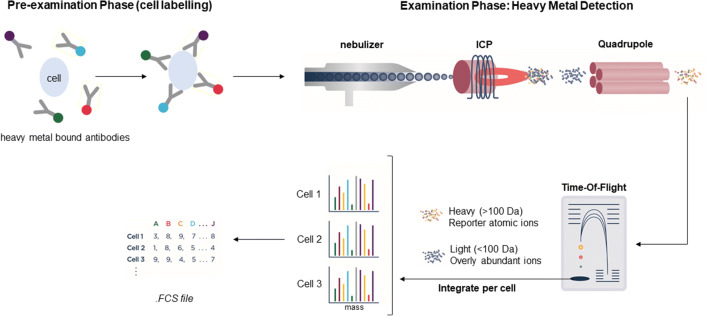
The first solution to the impasse of moving beyond 18-color detection towards higher-dimensional analysis arrived in 2009 with mass cytometry or cytometric time-of-flight (CyTOF®) instruments that combine aspects of both flow cytometry and mass spectrometry (20, 22, 23) (Figure 4). Mass cytometry makes use of heavy metal isotope probes rather than fluorescent probes. An antibody tagged with a specific metal-element binds to a cellular antigen. Then, the metal-tagged cells are quantified in an inductively coupled plasma (ICP) mass spectrometry detection system, i.e., the metal-tagged cells are nebulized into single-cell droplets and introduced into the ICP (24, 25) (Figure 4). The resulting charged atomic ion clouds from cellular components are transferred into the high vacuum of the mass spectrometer along with ions from the argon plasma. To resolve the specific probe ions, the mass cytometer is configured with quadrupole-time-of-flight (qTOF) which acts as a filter and allows only the heavier elemental ions consisting primarily of the masses of the metal-tagged probes, to be quantitated by TOF mass analysis and counted in discrete, time-separated detector channels (Figure 4). The intensity of the signal detected results from the number of specific probe-derived ions striking the detector and is proportional to the amount of probe bound to the target (Figure 4). The CyTOF provides at least three orders of magnitude of resolution between adjacent detection channels, greatly reducing the need for fluorescence spectral overlap compensation and its associated challenges (26, 27).
Figure 4.
Mass cytometry. In mass cytometry, cells are labeled with antibodies tagged with heavy metals. Cells are introduced into the inductively coupled plasma (ICP) by nebulization and are atomized and ionized. After removal of abundant ions, the heavy element composition is determined and signals corresponding to markers are then correlated and data analysis can be performed using any cytometry data analysis software (figure adapted from reference (20))
Mass cytometry allows for profiling about 40-50 parameters on a single cell. The relative ease of designing high-parameter panels is an advantage of mass cytometry. However, low throughput efficiency and lower sensitivity of dimly expressed antigens have been identified as current limitations with the existing instruments. Thus, assays where a large number of events and longer acquisition collection times are required, such as rare event detection, may not be feasible using mass cytometry (28). In addition, these instruments have a large footprint and require facility considerations associated with argon gas storage in high volume labs.
Higher-Dimensional Conventional Flow Cytometers
The progression towards greater than 20-color analysis in conventional flow cytometry has been the outcome of refining the existing elements of flow cytometers rather than the introduction of novel technologies such as is the case with the mass cytometers or spectral cytometers. The high-dimensional, conventional flow cytometers incorporate bandpass and long-pass optical filters and photodetectors for fluorescence detection but also include a variety of state-of-the art enhancements and optimizations which make the higher dimensionality possible. For example, refinements in system fluidics facilitate consistent sample delivery, with high reproducibility and low peak coefficients of variation (CVs). Innovations in the photodetectors result in an increase in the overall detector sensitivity and dynamic range and decreased crosstalk between detectors. To date, assays of up to 28 colors using conventional flow cytometry have been published (29–33).
Stratedigm’s S1000EON is configurable with up to six lasers and 30-color detection. The BioRad ZE5 Cell Analyzer is another innovative new flow cytometer with flexible configurations and makes possible up to 27-color analysis with up to five lasers. The NovoCyte Quanteon™ instrument is equipped with 25 fluorescence channels and four lasers. The CytoFLEX from Beckman Coulter is equipped with six lasers and detectors for 21-color analysis. The BD FACSymphony™ instruments can be configured with a variable number of detectors and up to nine lasers allowing up to a theoretical 50-color detection capabilities. Mostly due to the limitations of available spectrally distinct fluorophores, the reality of conventional flow cytometry panels with higher than 28 colors remains technically challenging.
Spectral Cytometers
The next game-changing technology was spectral cytometry, which allows for increased number of fluorophores, improved detection sensitivity, and reduced requirements for fluorescence spectral overlap compensation (19, 34–36). The concept of measuring fluorescent spectrum by flow cytometry emerged in the 1970s (37) and was refined in the 2000s by Paul Robinson at Purdue University (38–40). A couple of years later, the first commercial spectral cytometer was launched by Sony Biotechnology in 2012. These spectral cytometers were initially using prisms along with many photomultiplier tube (PMT) photodetectors to collect fluorescence signals.
In 2017, Cytek Biosciences released the Aurora spectral cytometer, which has since gained popularity in the cytometry community. Currently, this instrument can be configured with up to five lasers and three scatter detectors and 64 fluorescent detectors (41). This instrument uses avalanche photodiodes (APD) for light detection rather than PMTs. ADPs yield superior signal-to-noise ratio in the red and near infrared spectral regions compared to PMTs (41, 42).
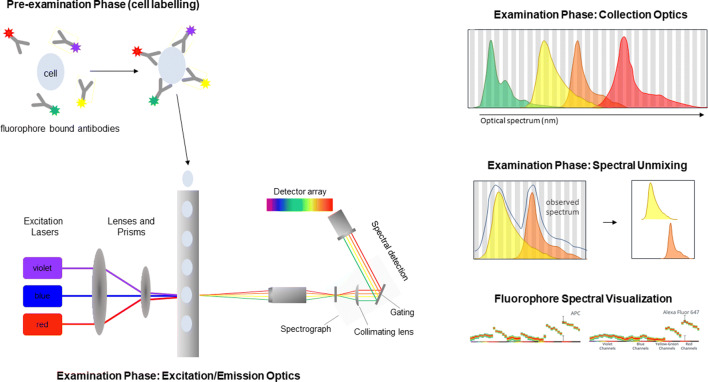
A unique feature of spectral instruments is that they do not have the typical filters (long pass and band pass) and dichroic mirrors used in the optical configurations of conventional flow cytometers and thus can measure the fluorescence emission of any available fluorophore excited by the on-board lasers (12). By using multiple detectors spanning the entire optical spectrum rather than in dedicated individual detectors, spectral cytometers collect the full emission spectrum of individual fluorophores (Figure 5) (12). Given that fluorophores are unique in their spectral signatures, fluorophores which could not previously be used together, due to large spectral overlap, can now be used in combination and accurately distinguished from one another (12). Overall, spectral instruments allow for greater flexibility in panel design (7).
Figure 5.
Spectral Cytometry. Sample preparation is similar to that of conventional flow cytometry; detection antibodies conjugated with fluorophores are used for labeling. Within the flow cell, sample then enters the sample stream focused such that a single cell is interrogated by one or more lasers resulting in light dispersion and activation of the fluorophores. Emitted fluorescence is then collected by a detector array. Spectral overlap between multiple fluorophores on the same cell then separated using spectral unmixing, enabling the simultaneous detection of fluorophores with overlapping emission spectra (figure adapted from reference (12); emission spectra from www.cytekbio.com)
In spectral cytometers, collected light signals from individual fluorophores are separated by spectral unmixing rather than compensation/spectral overlap subtraction (12). This results in better conservation of signal resolution compared to compensation, easing some of the challenges cytometrists have encountered while optimizing compensation matrices for high-parameter flow cytometry panels (7, 19). In addition, most spectral cytometers can treat autofluorescence as a parameter and thus can extract it from fluorescent signal, further enhancing signal resolution (7). This may significantly reduce the background, particularly when analyzing highly autoflorescent cell lines and certain tissues such as the brain (35). An additional advantage of spectral flow cytometry is it follows a similar workflow to conventional flow cytometry, using the same reagents and similar sample staining procedures.
Despite its greater flexibility, spectral flow cytometry does not remove the need for careful fluorophores/antigen selection, with general rules for panel design still applying (43). Development of high parameter panels remains complex and requires careful upfront development (43). Although numerous new dyes have been developed in the past couple of years, the availability of “spectrally unique” fluorophores remains a limitation for panel design (41). Finally, the parallel development of data analysis tools is also critical to enable analysis of the complex data output generated, an aspect discussed later in this review.
Since the release of the first commercial spectral cytometer, numerous papers have been published which demonstrated their applications including a 40-color OMIP-069 in 2020 (44–49). More recently, its applications in the context of cancer immunotherapy studies have also been discussed (7, 41). Spectral flow cytometry applications in academic research and drug discovery have already been reported, and standard implementation of this technology in multicentric clinical studies will likely occur in the near future.
Single-Cell Imaging
Single-cell imaging is useful when it is important to understand cellular morphology, subcellular localization, internalization, colocalization and trafficking of molecules, or cell binding characteristics (50). Depending on the system, the cells will be in suspension or positioned on a slide or microplate.
Imaging Flow Cytometry
The imaging flow cytometry system from Amnis, first described in 1979, combines flow cytometry with an imaging system (51). The newest instrument, ImageStream-X MarkII, is equipped with five-laser and can process 60,000 cells per second. In addition, up to 12 images (brightfield image, a dark field image, and multiple fluorescence images) can be captured per cell using a charge-coupled device (CCD) camera, positioned behind the cell stream and 20×, 40×, or 60× objectives. These features enable colocalization analysis of fluorescent proteins at a resolution down to 500 nm within cellular environments (52). This is the ability to determine not only the position of the signal (“spots”) within a cell, but also the ability to analyze whether spot location from 2 channels overlap. The technology can be used to screen rare events at the subcellular level for a range of molecular diagnostic applications such as morphology-based diagnosis of rare cells in blood diseases or circulating tumor cells (53), in the detection of protein aggregation associated with a range of neurodegenerative diseases, in CRISPR-based genetic screening for the identification of protein candidates that affect stress granule assembly (54). These capabilities allow the quantitation of cellular morphology and the intensity and location of fluorescent probes on, in, or between cells, even in rare subpopulations and highly heterogeneous samples (55–59).
Imaging Mass Cytometry
Imaging mass cytometry (Hyperion Imaging System, Fluidigm Corporation) creates dimensional multiplexed images with subcellular resolution of up to 37 markers by coupling a laser ablation system to a mass cytometer (60–62). Suitable samples include tissue sections stained with metal-tagged antibodies such as standard formalin-fixed paraffin-embedded, frozen tissue sections, and fixed cells deposited on glass microscope slides (63–65). Advantage of this type of technology includes the ability to evaluate tissue architectures in high-dimensional space in situ (66–68). Another approach, called multiplexed ion beam imaging (MIBI), uses an ion beam to liberate metal ion reporters, which are quantified by mass spectrometry (61).
EVOLUTION OF PROBES
In addition to new instrumentation, novel probes and chemistries were required for flow cytometry to expand first up to 18-color detection and more recently towards 50-color detection (Figure 3). Since 2012, the fluorescent probes commonly used in flow cytometry for conjugation to monoclonal antibodies and antigen detection could be grouped into five general categories: small organic dyes, phycobiliproteins, tandem dyes, inorganic fluorescent nanocrystals, and organic polymer dyes (69). More recently, additional novel probe technologies have become available.
As with any detection probe, the ideal fluorescent probe for high-dimensional cytometry would generate a strong signal but display very low background. In flow cytometry, the brightness of the dye signal is dictated by its absorbance cross-section and quantum efficiency. Several independent factors influence the background of a fluorescent probe, and one is its autofluorescence in the same channel detector as the cells. Other contributors to background include signal spillover into other detectors as well as the ability to be excited by more than one laser. New probes continue to appear on the market; some were created with novel chemistries such as Phiton™ technology, and others were created with modifications of existing chemistries. Advantages of the new probes include additional choices of emission spectra (i.e., more colors), increased signal strength, and decreased background contributions. Depending on the context, there may be independent value to having different spectral overlaps compared to having improved signal-to-noise ratios. For example, a narrow emission spectrum will help to maximize signal-to-noise and minimize compensation and cross-laser excitation.
Small organic dyes such as fluorescein isothiocyanate (FITC) and Texas Red were the first fluorophores used in flow cytometry. The advantages of this category of probes are that they deliver consistent, discreet excitation profiles, are relatively stable, and are easy to conjugate to antibodies. In addition, organic dyes are soluble and thus less susceptible to aggregation and precipitation. They are not sensitive to organic solvents; this property along with their small size makes them well-suited for intracellular and intranuclear staining as well as phospho-flow applications. Texas Red which is susceptible to hydrolysis has largely been replaced by molecules with similar spectral properties but better solubility and photostability properties. Despite its continued wide usage, FITC is known to be pH sensitive and has a high rate of photobleaching and a broad emission spectrum. Other disadvantages of the initial organic dyes are that the difference between excitation wavelength and emission wavelength, named the Stokes shift, is small (50–100 nm); thus, the signals are not among the brightest. A wide variety of commercially available next-generation, small organic dyes which address many of the limitations of the initial organic dyes have become available.
The Alexa Fluor® (AF) family of dyes which includes AF350, AF430, AF488, AF532, AF546, AF568, AF594, and AF647 is a unique type of small organic dye which is synthesized through sulfonation of other dyes such as fluorescein, cyanine, coumarin, and rhodamine (70). The AF dyes are named according to their excitation wavelengths, e.g., AF488 is optimally excited at 488 nm. AF dyes are compatible with standard optical filters on most existing instruments with advantages including brightness, photostability, increased pH tolerance, and increased stability in aqueous formulation and compatibility with common fixatives. More recently, other new families of small organic dyes include eFluors® from eBioscience, Spark from BioLegend and Horizon™ Red 718 from Becton Dickinson have been introduced.
Phycobiliproteins are large proteins such as phycoerythrin (PE) and allophycocyanin (APC) which have large Stokes shifts (75–200 nm) and thus generate very bright signals. Until organic polymer dyes became available in 2012, PE and APC were the brightest dyes available and the best choice for detecting dim antigens. Moreover, due to their large size, this class of molecule most often results in a 1:1 or 1:2 molar fluorophore to protein ratio. When purified to eliminate aggregates and 1:2 molar conjugates, the phycobiliprotein conjugates are well-suited for quantitative measurements (71). Another protein-based dye, PerCP, a peridinin-chlorophyll protein complex, is derived from photosynthetic dinoflagellates.
Tandem dyes consist of two covalently linked fluorophores selected such that when the first fluorophore (the donor) is excited, its emission, being the same as the excitation spectrum for the second (the acceptor), will excite the acceptor. This process is known as Förster or fluorescence, resonance energy transfer (FRET). The development of tandem dye technology was a major advancement in flow cytometry as it expanded the pool of available options which could be used with existing lasers and detectors (Table I). For this reason, tandem dyes were quickly adopted and continue to be broadly used. The first available tandem dyes were composed of a large protein donor, such as PE or APC, attached to a small molecule (cyanine) acceptor (e.g., PE-Cy7, PE-Cy5.5, APC-Cy5.5, and APC-Cy7). More recently, tandem dyes are being generated with the newer dyes being used as donor such as synthetic organic polymers or acceptor molecules such as eFluors®. The tandems with synthetic organic polymers do not require the emission of the donor to overlap with the excitation of the acceptor which allows for tandem with high Stokes shifts, emitting as far out as 785 nm with a 405-nm excitation. Advantages of tandem dyes include their brightness, with large Stokes’s shift values (150–300 nm). The major disadvantages of tandem dyes include lot-to-lot variability and stability issues (72, 73). Next-generation tandem products such as Fire™ and Dazzle™ from BioLegend reduce some of the issues associated with tandem dyes.
Multilayered Quantum Dot (QD) particles or Qdot nanocrystals are made of a semiconductor core (typically made of cadmium, selenium, and tellurium) covered by an outer shell of zinc sulfate with a diameter of 2–15 nm (74–76). QDs are further coated with polymers which facilitate the attachment of biomolecules such as proteins, monoclonal antibodies, oligonucleotides, or streptavidin. Unlike fluorescent probes, their emission is not measured in Stokes shifts; laser excitation results in the creation of excitons proportional to the size of the QD and to the wavelength of the emitted fluorescence. Varying the amounts of cadmium and selenium results in QDs which can emit energy in the form of light over a range of wavelengths generating very bright signals and offering the advantages photostability and long shelf life (77, 78). The invention of QDs was instrumental in the progress of flow cytometry towards 18-color analysis. Disadvantages include their size preventing the diffusion through cell membranes (79, 80). Moreover, nonspecific protein adsorption to the QD particle surface may prevent QD-conjugated ligands from interacting with their respective cellular receptors further decreasing signal in the readouts (81). Although QDs are optimally excited with UV or violet lasers, they can also be minimally excited by multiple lasers, thus leading to compensation challenges. Thus, these reagents have now largely been replaced with the polymer dyes.
Polymer dyes consist of fluorescent monomeric subunits which can be modified to absorb and emit light at specific wavelengths based on the length of the polymer chain and the attached molecular subunits. Introduced in 2012, polymers are as bright as phycobiliproteins and Qdots and are available in a wide range of emission wavelengths which, like organic dyes, retain discreet excitation profiles. Brilliant Violet™ (BV421), the first dye available within this category, truly revolutionized multiparameter flow cytometry as it was the first bright dye which could be excited off the violet (405 nm) laser (82). At the time of its release, most instruments including clinical instruments were already equipped with violet lasers; thus, BV421 increased panel design options for many users. Given that polymer dyes are only excited at specific wavelengths, there are no issues with multiple laser excitation. Other advantages include photostability and reliable conjugation to antibodies. Currently, there are numerous subcategories of commercially available polymer dyes such as multiple Brilliant Violet™ (BV), Brilliant™ Ultraviolet (BUV), and Brilliant™ Blue (BB) and Super Bright dyes from eBioscience. Beckman Coulter has introduced a proprietary line of polymer dyes, SuperNova. In addition, polymer-dye tandem structures are also available.
More recently, novel types of probes have become available. The Phiton™ technology from Phitonex, Inc. was introduced to the flow cytometry community at CYTO, the annual meeting of congress of the International Society for the Advancement of Cytometry in 2019. This technology is used to create NovaFluor™ dyes which share high-level principles in common with the polymer dye technology. The Phiton™ technology uses synthetic DNA oligonucleotides that self-assemble to form a stable DNA macrostructure on which fluorophores can be placed at optimal distances. The brightness of the NovaFluor™ probes can be increased by additional fluor-loading on the Phiton™ arms or by controlling how many Phiton™-based fluorescent labels are conjugated to the antibody. The resulting NovaFluor™, like the polymer dyes, can be tuned to span the UV, visible, and near infrared (NIR) spectra, with enhanced fluorescence transfer efficiencies, minimized cross-laser excitation, and reduced spectral spill over.
In March 2020, Sony introduced the KIRAVIA Dyes™. This class of dyes employs an organic backbone that separates fluorophores to minimize quenching effects. This structure facilitates for high fluorophore-to-protein (F:P) ratios. In June 2020, Cytek Biosciences introduced cFluor™ probes at the CYTO 2020 congress. cFluor™ probes consist of cyanine-based fluorescence. Their excitation and emission properties can be adjusted by altering the length of the polymethine chain. To date, there are over 30 cFluor™ available spanning the blue, visible, far-red, and NIR spectra.
Biotium currently offers over 30 CF® novel dyes spanning the visible, far-red, and NIR spectra. These dyes are the result chemical engineering aimed to reduce dye aggregation and increase solubility and photostability. Newer fluorescent nanoparticle-based dyes include StarBright Dyes from Bio-Rad.
Other probes used in flow cytometry such as fluorescent proteins, nucleic acid probes, calcium indicators, cell proliferation, and viability dyes are not directly related to the focus of this review but are discussed in detail elsewhere (69).
EVOLUTION OF TOOLS FOR DATA ANALYSIS
Data analysis tools have advanced considerably since the early days of the cytofluorograph and flow cytometers where data was visualized from printed analog histograms and scatter plots. A major advancement came in 1984 when the concept of the Flow Cytometry Standard file was first introduced (83). This standardized data output format allowed users to collect data on any flow cytometer and read the file using other software on another computer. With the commercialization and clinical adoption of flow cytometry, instrument vendors integrated software into the computer workstations which allowed users to use the tools to automate daily quality control processes, visualize data in histograms or bivariate plots, and obtain various statistical outputs such as population frequency and fluorescence intensity values. By the 1990s, 3rd party software packages became available such as FlowJo and FCS Express. The availability of independent analysis software gave the user more flexible and sophisticated data analysis tools. Some of these software platforms also had the capability to integrate into Laboratory Information Management Systems (LIMS), thus creating an end-to-end workflow for clinical labs.
Foundational work on minimum data standards (84–87) and the birth of computational approaches and collaborative data analysis initiatives showed the importance of standardizing data analysis approaches (88, 89). Through the pioneering work of The Flow Cytometry: Critical Assessment of Population Identification Methods (FlowCAP), a consortium of immunologists, bioinformaticians, statisticians, and clinical scientists facilitated the evaluation and comparison of the performance of various computational methods for identifying cell populations across multiple studies. The FlowCAP initiative demonstrated the value and ability of algorithm-based approaches for identification of cell types associated with clinical outcomes efficiently and reproducibly, while also shedding light on some of the limitations of these techniques. The FlowCAP challenges demonstrated that depending on the task at hand (e.g., rare cells and heterogeneous populations), some algorithms performed better or worse. This effort propelled new commercial companies to offer cytometry informatics services (90).
Since the mid-2000s, a continuous maturation of computational cytometry science and algorithm-based approaches has emerged for cell-type identification. These approaches fall into three categories: (1) supervised, (2) unsupervised, and (3) mixed machine learning. Supervised algorithms, such as flowDensity, flowLearn, and OpenCyto, are designed to recapitulate manual, predefined gating of known cell populations and markers. The advantages of these approaches include reducing subjectivity in gate placement resulting in lower variability and decreased time for gating and improved overall efficiency, yet still closely following the principles of manual gating, which remains the gold standard for gating of cytometry data. Unsupervised algorithms largely fall into 2 categories: (1) dimensionality reduction and (2) clustering algorithms. Principal Component Analysis (PCA), T-distributed Stochastic Neighbor Embedding (tSNE), and Uniform Manifold Approximation and Projection (uMAPS) are among the tools currently being used with high-dimensional flow cytometry data sets for mapping and visualization of data into a 2D space. Clustering algorithms that group the cells together based on similarities such Phenograph and FlowSOM are also widely used. Unsupervised approaches can potentially reveal multidimensional relationships between cells not observed by prescribed gating strategies. Machine learning is a broader approach that utilizes a combination of algorithm-based tools to fin signals at cellular level in large datasets and identify biomarkers of disease severity, progression, or clinical response to therapeutic interventions. Computational approaches have also provided tools to improve the preprocessing steps of analysis such as sample quality assessment, normalization, and transformation (91). Many of the algorithms have been reviewed in recent publications to compare performance and speed across a wide variety of patient samples in immune, infectious, and oncology disease areas (92–96). More recently, computational multi-omics approaches have elucidated immune heterogeneity in COVID-19 patients infected with SARS-CoV-2 virus (97). This is an important step forward in understanding the biology of this disease and advancing the development of effective therapies to combat this devastating pandemic.
Moving into this new phase of high complexity cytometry and cellular biomarkers, a different model of considering cellular biomarkers will be warranted in order to best leverage the content-rich data sets. Hypothesis-driven biomarkers (current state) in many cases will be combined with results generated from unsupervised data analysis pipelines. Moreover, the drug development teams tasked with brining forward new scientific insights will look different. A partnership with computational scientists will be essential to fully interrogate all biomarker data and clinical outcomes to generate insights that will facilitate the development of life changing therapies.
MULTI-OMICS
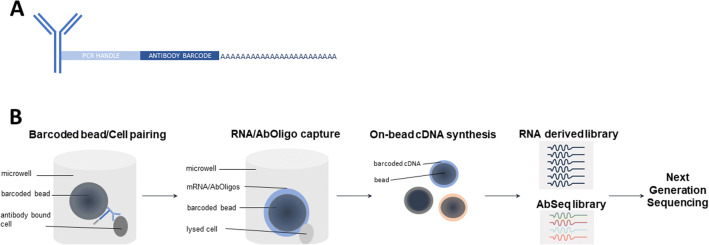
One of newest innovation in single-cell analysis is Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-Seq), developed at the New York Genome Center, and the related technology RNA Expression and Protein Sequencing (REAP-seq) developed at Merck (98, 99) (Figure 6). These technologies combine proteomics and transcriptomics at the single-cell level with next-generation sequencing (NGS) in order to provide a high resolution of cellular differences. The cells are labeled with antibody-oligonucleotide conjugates or antibody-derived tags (ADTs) (Figure 6). DNA-barcoded antibodies convert the detection of proteins into quantitative, sequenceable readouts. A potentially limitless number of markers could be used for immunophenotyping and unbiased transcriptome analysis given that the issues related to spectral overlap or isotopes availability are not relevant.
Figure 6.
Multi-omics. A schematic representation of the simultaneous measurement of gene and protein expression using a combination of oligo-conjugated antibodies (Abseq) and capture beads. A Samples are labeled with antibodies conjugated with oligonucleotides. Structure of oligo-conjugated antibody (PCR polymerase chain reaction). B Workflow. Labeled samples are mixed with barcoded beads and lysed. mRNA and antibody oligos bound to beads are then converted to cDNA prior to proceeding to generation of mRNA derived library and AbSeq libraries and next-generation sequencing (NGS).
ADT-labeled cells are encapsulated at random as single cells along with a single bead (100) (Figure 6). The RT primers are anchored to the bead and are made up several components including an oligo dT to prime the RT, a unique molecular identifier (UMI) to uniquely tag each molecule, and a barcode unique to each bead to distinguish one cell from all other cells. Next, the cells are lysed, allowing the mRNA molecules and the antibody oligos to hybridize to the RT primers by their poly A tails (Figure 6). The cDNA is then synthesized from both ADTs and cellular mRNAs. The cDNAs are amplified, purified, and finally pooled for sequencing in one scRNA-seq run using existing methods such as 10x Genomics, Drop-seq, and ddSeq (101).
Even more so than with other high-content systems, data analysis for multi-omics is such a critical and complex aspect of the overall workflow that there is an almost obligatory need for bioinformatic expertise (102). A variety of analysis tools are required. Data visualization tools described above can be used for the identification of novel cell clusters, whereas determining nucleic acid sequences requires the same type of data analysis tools as would be used in any scRNA-seq experiment. Recommendations and latest developments can be found on the developer’s website https://cite-seq.com/computational-tools/.
This technology, first described in 2017, is rapidly evolving and now includes the ability to multiplex samples through a process coined “Cell Hashing” and the ability to detect intracellular proteins. Current applications include the characterization of tumor heterogeneity for tumor classification and a deeper characterization of cells and host-pathogen interactions (103–105).
The multitude of uses of multi-omics makes it an appealing and exciting new technology for single cell profiling; however, the high cost, limited throughput, and comparatively slow data turnaround time are some of the current limitations.
DISCUSSION
The recent advances in instrumentation, reagents, and software are propelling the technology for single-cell analysis forward. Given the recent rapid pace of growth, it is highly likely that this progress will continue at a similarly vigorous rate.
The most evident outcome of these new developments has been in the ability to detect a greater number of parameters per cell which is driven by the combination of instruments with a greater number of lasers and detectors and an increase in the types of probes available. The new probes display a variety of features such as increased signal to noise characteristics, greater stability, and resistance to environmental fluctuations. The availability of more wavelength choices for solid-state lasers combined with the ability to include a greater number of lasers on the instruments has broadened the available wavelengths for excitation and opened up the possibility of using newer fluorophores while decreasing some of the existing compensation issues. An equally important but perhaps less obvious contributor to increased dimensionality is the quality of the signals generated. The newer instruments are not only capable of collecting signals from a greater number and more diverse array of probes, but they are also able to capture a greater portion of the fluorescent signals. This functionality originally introduced in the digital instruments has matured in its presentation in the spectral instruments and other higher-dimensional instruments.
The principal significance of the new technologies is that they are facilitating a deeper understanding of basic immunology, hematopoiesis, and hematopathology as well as other areas of cellular investigation. In addition, their implementation in drug development is providing a deeper and truer understanding of the impact of novel therapeutic interventions. However, given the aggressive timelines during drug development and the likely necessity of quickly addressing the next global pandemic, significant challenges remain before greater than 30-parameter analysis become routine practice in drug development and in particular, in clinical trials. Not only are additional fluorophores needed, there is also a critical need to decrease the assay development and data analysis timelines for high-content assays. These requirements could be satisfied with integrated automation solutions, better tools for panel design, and improved data analysis software packages (106). As discussed above, data analysis has been identified as a rate-limiting step in cytometry and major contributor to data variability. Not only are new data analysis tools needed, but a pathway to implement them in regulated environments is critical. Too often a lack of clarity from regulators regarding implementation of new software packages impedes their implementation (107). Additional requirements for successful adoption of these new higher-content solutions to single-cell analysis in the context of multicenter clinical trials include more robust strategies for instrument setup and cross-instrument standardization procedures.
The National Institute of Standards and Technology (NIST) has recently established the Flow Cytometry Standards Consortium (https://www.nist.gov/programs-projects/nist-flow-cytometry-standards-consortium). This project is part of broader advanced therapy program at NIST. The high-level goals of this initiative are to develop best practices for flow cytometry measurement and new standards in order to facilitate more accurate quantitation and improve reproducibility. The consortium’s specific aims are geared towards addressing the many of concerns mentioned above.
In order to select and implement new tools for single-cell analysis at a given stage of drug development, one must consider the needs of the program and the biological questions to be answered (Table II). In choosing one approach over another, one should consider the context of the biology first and foremost, in other words the selection of the instrument based on the purpose. For direction regarding the validation of new instruments and assays for use in drug development, the new CLSI guidance document, H62-Validation of Assays Performed by Flow Cytometry, should be consulted (108). The 37-member document development committee (DDC) for this consensus document included representatives from the biopharma community, instrument, and reagent vendors, as well as regulatory agencies including the Food and Drug Administration and the National Institute of Standards and Technology.
The growth and merging of high-dimensional cytometry instrumentation, advanced analysis tools, and single-cell RNA/DNA methods will position the field of drug development to pioneer new insights in preclinical and human biology. Success depends on the integration of biological, clinical, and computational scientists to leverage traditional workflows with advanced analytics and machine learning approaches to gain actionable insights from “big data.”
AUTHOR CONTRIBUTION
Shibani Mitra-Kaushik wrote and designed sections on the history of flow cytometry and fluorescent dyes. Anita Mehta-Damani wrote and designed sections on mass cytometry, imaging mass cytometry, and imaging flow cytometry. Jennifer J. Stewart wrote and designed section on single-cell multi-omics. Cherie Green wrote and designed section on automated data analysis. Virginia Litwin designed the overall flow and edited the manuscript. Christèle Gonneau wrote and designed sections on spectral cytometry, tables and figures, and overall formatting and editing of the manuscript.
DECLARATIONS
CONFLICT OF INTEREST
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
REFERENCES
- 1.Raventos-Suarez C, Long B. A multiparameter approach to cell cycle analysis as a standard tool in oncology drug discovery. In: Litwin V, Marder P, editors. Flow Cytometry in Drug Discovery and Development. Wiley-Blackwell, John Wiley & Sons, Inc.: New Jersey; 2011. pp. 99–122. [Google Scholar]
- 2.Sarikonda G, Mathieu M, Natalia M, Pahuja A, Xue Q, Pierog PL, et al. Best practices for the development, analytical validation and clinical implementation of flow cytometric methods for chimeric antigen receptor T cell analyses. Cytom Part B - Clin Cytom [Internet]. 2021 Jan 1 [cited 2021 Mar 19];100(1):79–91. Available from: https://pubmed.ncbi.nlm.nih.gov/33373096/ [DOI] [PubMed]
- 3.Sarikonda G, Pahuja A, Kalfoglou C, Burns K, Nguyen K, Ch’en IL, et al. Monitoring CAR-T cell kinetics in clinical trials by multiparametric flow cytometry: benefits and challenges [Internet]. Vol. 100, Cytometry Part B - Clinical Cytometry. John Wiley and Sons Inc; 2021 [cited 2021 Mar 19]. p. 72–8. Available from: https://pubmed.ncbi.nlm.nih.gov/32573972/ [DOI] [PubMed]
- 4.Stewart JJ, Green CL, Jones N, Liang M, Xu Y, Wilkins DEC, et al. Role of receptor occupancy assays by flow cytometry in drug development [Internet]. Vol. 90, Cytometry. Part B, Clinical cytometry. Cytometry B Clin Cytom; 2016 [cited 2021 Mar 19]. p. 110–6. Available from: https://pubmed.ncbi.nlm.nih.gov/26704557/ [DOI] [PubMed]
- 5.Green CL, Stewart JJ, Högerkorp CM, Lackey A, Jones N, Liang M, et al. Recommendations for the development and validation of flow cytometry-based receptor occupancy assays [Internet]. Vol. 90, Cytometry. Part B, Clinical cytometry. Cytometry B Clin Cytom; 2016 [cited 2021 Mar 19]. p. 141–9. Available from: https://pubmed.ncbi.nlm.nih.gov/26566147/ [DOI] [PubMed]
- 6.Hilt E, Sun YS, McCloskey TW, Eck S, McIntosh T, Grugan KD, et al. Best practices for optimization and validation of flow cytometry-based receptor occupancy assays. Cytom Part B - Clin Cytom [Internet]. 2021 Jan 1 [cited 2021 Mar 19];100(1):63–71. Available from: https://pubmed.ncbi.nlm.nih.gov/33259706/ [DOI] [PubMed]
- 7.Sanjabi S, Lear S. New cytometry tools for immune monitoring during cancer immunotherapy [Internet]. Vol. 100, Cytometry Part B - Clinical Cytometry. John Wiley and Sons Inc; 2021 [cited 2021 Mar 19]. p. 10–8. Available from: https://pubmed.ncbi.nlm.nih.gov/33432667/ [DOI] [PubMed]
- 8.Kamentsky LA, Melamed MR, Derman H. Spectrophotometer: new instrument for ultrarapid cell analysis. Science (80- ) [Internet]. 1965 [cited 2021 Mar 19];150(3696):630–1. Available from: https://pubmed.ncbi.nlm.nih.gov/5837105/ [DOI] [PubMed]
- 9.Fulwyler MJ. Electronic separation of biological cells by volume. Science (80- ) [Internet]. 1965 [cited 2021 Mar 19];150(3698):910–1. Available from: https://pubmed.ncbi.nlm.nih.gov/5891056/ [DOI] [PubMed]
- 10.Dittrich W, Göhde W. Impulsfluorometrie bei Einzelzellen in Suspensionen [Internet]. Vol. 24, Zeitschrift fur Naturforschung - Section B Journal of Chemical Sciences. Z Naturforsch B; 1969 [cited 2021 Mar 19]. p. 360–1. Available from: https://pubmed.ncbi.nlm.nih.gov/4389205/ [PubMed]
- 11.Herzenberg LA, Sweet RG, Herzenberg LA. Fluorescence-activated cell sorting. Sci Am [Internet]. 1976 [cited 2020 Sep 21];234(3):108–17. Available from: https://pubmed.ncbi.nlm.nih.gov/1251180/ [DOI] [PubMed]
- 12.Nolan JP, Condello D. Spectral flow cytometry. Curr Protoc Cytom. 2013;SUPPL.63:1–18. doi: 10.1002/0471142956.cy0127s63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapiro HM. The evolution of cytometers. Cytometry [Internet]. 2004 Mar 1 [cited 2021 Apr 25];58A(1):13–20. Available from: 10.1002/cyto.a.10111 [DOI] [PubMed]
- 14.Stelzer GT, Shults KE, Loken MR. CD45 gating for routine flow cytometric analysis of human bone marrow specimens. Ann N Y Acad Sci [Internet]. 1993 [cited 2021 Apr 20];677(1):265–80. Available from: https://pubmed.ncbi.nlm.nih.gov/8494214/ [DOI] [PubMed]
- 15.De Rosa SC, Herzenberg LA, Herzenberg LA, Roederer M. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med [Internet]. 2001 [cited 2021 Mar 19];7(2):245–8. Available from: https://pubmed.ncbi.nlm.nih.gov/11175858/ [DOI] [PubMed]
- 16.Roederer M, De Rosa S, Gerstein R, Anderson M, Bigos M, Stovel R, et al. 8 color, 10-parameter flow cytometry to elucidate complex leukocyte heterogeneity. Cytometry [Internet]. 1997 Dec 1 [cited 2021 Mar 19];29(4):328–39. Available from: https://pubmed.ncbi.nlm.nih.gov/9415416/ [DOI] [PubMed]
- 17.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system [Internet]. Vol. 4, Nature Reviews Immunology. Nature Publishing Group; 2004 [cited 2021 Mar 19]. p. 648–55. Available from: https://pubmed.ncbi.nlm.nih.gov/15286731/ [DOI] [PubMed]
- 18.Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin JB, Olsson A, et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell [Internet]. 2020 Oct 1 [cited 2021 Apr 25];183(1):158-168.e14. Available from: https://pubmed.ncbi.nlm.nih.gov/32979941/ [DOI] [PMC free article] [PubMed]
- 19.Robinson JP. Spectral flow cytometry—Quo vadimus? Cytom Part A. 2019;95(8):823–824. doi: 10.1002/cyto.a.23779. [DOI] [PubMed] [Google Scholar]
- 20.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry [Internet]. Vol. 33, Trends in Immunology. Trends Immunol; 2012 [cited 2020 Sep 21]. p. 323–32. Available from: https://pubmed.ncbi.nlm.nih.gov/22476049/ [DOI] [PMC free article] [PubMed]
- 21.Mistry AM, Greenplate AR, Ihrie RA, Irish JM. Beyond the message: advantages of snapshot proteomics with single-cell mass cytometry in solid tumors [Internet]. Vol. 286, FEBS Journal. Blackwell Publishing Ltd; 2019 [cited 2021 Apr 25]. p. 1523–39. Available from: https://pubmed.ncbi.nlm.nih.gov/30549207/ [DOI] [PMC free article] [PubMed]
- 22.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem [Internet]. 2009 Aug 15 [cited 2020 Oct 15];81(16):6813–22. Available from: https://pubmed.ncbi.nlm.nih.gov/19601617/ [DOI] [PubMed]
- 23.Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. Highly multiparametric analysis by mass cytometry [Internet]. Vol. 361, Journal of Immunological Methods. J Immunol Methods; 2010 [cited 2020 Oct 15]. p. 1–20. Available from: https://pubmed.ncbi.nlm.nih.gov/20655312/ [DOI] [PubMed]
- 24.Jiménez-Lamana J, Szpunar J, Łobinski R. New frontiers of metallomics: elemental and species-specific analysis and imaging of single cells. In: Advances in Experimental Medicine and Biology [Internet]. Springer New York LLC; 2018 [cited 2020 Oct 12]. p. 245–70. Available from: https://pubmed.ncbi.nlm.nih.gov/29884968/ [DOI] [PubMed]
- 25.Shinko D, Ashhurst TM, McGuire HM, Charles KA. Staining of phosphorylated signalling markers protocol for mass cytometry. In: Methods in Molecular Biology [Internet]. Humana Press Inc.; 2019 [cited 2021 Mar 21]. p. 139–46. Available from: https://pubmed.ncbi.nlm.nih.gov/31077104/ [DOI] [PubMed]
- 26.Lee BH, Rahman AH. Acquisition, processing, and quality control of mass cytometry data. In: Methods in Molecular Biology [Internet]. Humana Press Inc.; 2019 [cited 2020 Oct 12]. p. 13–31. Available from: https://pubmed.ncbi.nlm.nih.gov/31077096/ [DOI] [PubMed]
- 27.Spitzer MH, Nolan GP. Mass cytometry: single cells, many features [Internet]. Vol. 165, Cell. Cell Press; 2016 [cited 2020 Oct 12]. p. 780–91. Available from: https://pubmed.ncbi.nlm.nih.gov/27153492/ [DOI] [PMC free article] [PubMed]
- 28.Sommer U, Eck S, Marszalek L, Stewart JJ, Bradford J, McCloskey TW, et al. High-sensitivity flow cytometric assays: considerations for design control and analytical validation for identification of Rare events [Internet]. Vol. 100, Cytometry Part B - Clinical Cytometry. John Wiley and Sons Inc; 2021 [cited 2021 Mar 19]. p. 42–51. Available from: https://pubmed.ncbi.nlm.nih.gov/32940947/ [DOI] [PubMed]
- 29.Swanson PA, Seder RA. <scp>OMIP-067</scp> : 28-color flow cytometry panel to evaluate human T-cell phenotype and function. Cytom Part A [Internet]. 2020 Aug 23 [cited 2020 Sep 20];cyto.a.24189. Available from: 10.1002/cyto.a.24189 [DOI] [PubMed]
- 30.Payne K, Li W, Salomon R, Ma CS. OMIP-063: 28-color flow cytometry panel for broad human immunophenotyping. Cytom Part A [Internet]. 2020 Aug 1 [cited 2020 Sep 20];97(8):777–81. Available from: https://pubmed.ncbi.nlm.nih.gov/32298042/ [DOI] [PubMed]
- 31.Liechti T, Roederer M. OMIP-058: 30-parameter flow cytometry panel to characterize iNKT, NK, unconventional and conventional T cells. Cytom Part A [Internet]. 2019 Sep 23 [cited 2020 Sep 20];95(9):946–51. Available from: 10.1002/cyto.a.23850 [DOI] [PubMed]
- 32.Liechti T, Roederer M. OMIP-060: 30-parameter flow cytometry panel to assess T cell effector functions and regulatory T cells. Cytom Part A [Internet]. 2019 Nov 1 [cited 2020 Sep 20];95(11):1129–34. Available from: https://pubmed.ncbi.nlm.nih.gov/31334913/ [DOI] [PubMed]
- 33.Mair F, Prlic M. OMIP-044: 28-color immunophenotyping of the human dendritic cell compartment. Cytom Part A. 2018;93(4):402–405. doi: 10.1002/cyto.a.23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feher K, von Volkmann K, Kirsch J, Radbruch A, Popien J, Kaiser T. Multispectral flow cytometry: the consequences of increased light collection. Cytom Part A. 2016;89(7):681–689. doi: 10.1002/cyto.a.22888. [DOI] [PubMed] [Google Scholar]
- 35.Niewold P, Ashhurst TM, Smith AL, King NJC. Evaluating spectral cytometry for immune profiling in viral disease. Cytom Part A [Internet]. 2020 Sep 17 [cited 2020 Sep 20];cyto.a.24211. Available from: 10.1002/cyto.a.24211 [DOI] [PubMed]
- 36.Sanders CK, Mourant JR. Advantages of full spectrum flow cytometry. J Biomed Opt. 2013;18(3):037004. doi: 10.1117/1.JBO.18.3.037004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wade CG, Rhyne RH, Woodruff WH, Bloch DP, Bartholomew JC. Spectra of cells in flow cytometry using a vidicon detector. J Histochem Cytochem [Internet]. 1979 [cited 2020 Sep 20];27(6):1049–52. Available from: https://pubmed.ncbi.nlm.nih.gov/110874/ [DOI] [PubMed]
- 38.Robinson J, Rajwa B, Gregori G, Jones J, Patsekine V. Collection hardware for high speed multispectral single particle analysis. In: ISAC. 2004.
- 39.Robinson JP. Multispectral cytometry: the next generation. Biophotonics Int. 2004:36–40.
- 40.Robinson JP, Gregori G, Rajwa B, Jones J, Patsekin V. Multispectral detector and analysis system. USA; Patent US7280204B2, 2007.
- 41.Bonilla DL, Reinin G, Chua E. Full spectrum flow cytometry as a powerful technology for cancer immunotherapy research. Front Mol Biosci [Internet]. 2020 Jan 29 [cited 2021 Jul 12];7. Available from: /pmc/articles/PMC7878389/ [DOI] [PMC free article] [PubMed]
- 42.Lawrence WG, Varadi G, Entine G, Podniesinski E, Wallace PK. A comparison of avalanche photodiode and photomultiplier tube detectors for flow cytometry. Imaging, Manip Anal Biomol Cells, Tissues VI. 2008;6859:68590M. [Google Scholar]
- 43.Ferrer-Font L, Pellefigues C, Mayer JU, Small SJ, Jaimes MC, Price KM. Panel design and optimization for high-dimensional immunophenotyping assays using spectral flow cytometry. Curr Protoc Cytom [Internet]. 2020 Mar 1 [cited 2021 Jul 12];92(1):e70. Available from: 10.1002/cpcy.70 [DOI] [PubMed]
- 44.Futamura K, Sekino M, Hata A, Ikebuchi R, Nakanishi Y, Egawa G, Kabashima K, Watanabe T, Furuki M, Tomura M. Novel full-spectral flow cytometry with multiple spectrally-adjacent fluorescent proteins and fluorochromes and visualization of in vivo cellular movement. Cytom Part A. 2015;87(9):830–842. doi: 10.1002/cyto.a.22725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmutz S, Valente M, Cumano A, Novault S. Spectral cytometry has unique properties allowing multicolor analysis of cell suspensions isolated from solid tissues. PLoS One [Internet]. 2016 Aug 1 [cited 2020 Sep 20];11(8). Available from: https://pubmed.ncbi.nlm.nih.gov/27500930/ [DOI] [PMC free article] [PubMed]
- 46.Park LM, Lannigan J, Jaimes MC. <scp>OMIP-069</scp> : forty-color full spectrum flow cytometry panel for deep immunophenotyping of major cell subsets in human peripheral blood. Cytom Part A [Internet]. 2020 Aug 31 [cited 2020 Sep 20];cyto.a.24213. Available from: 10.1002/cyto.a.24213 [DOI] [PMC free article] [PubMed]
- 47.Silvin A, Chapuis N, Dunsmore G, Goubet AG, Dubuisson A, Derosa L, et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020; Available from:. 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed]
- 48.Sahir F, Mateo JM, Steinhoff M, Siveen KS. Development of a 43 color panel for the characterization of conventional and unconventional T-cell subsets, B cells, NK cells, monocytes, dendritic cells, and innate lymphoid cells using spectral flow cytometry. Cytom Part A [Internet]. 2020 [cited 2021 Apr 25]; Available from: https://pubmed.ncbi.nlm.nih.gov/33336868/ [DOI] [PMC free article] [PubMed]
- 49.Liu C, Martins AJ, Lau WW, Rachmaninoff N, Chen J, Imberti L, et al. Time-resolved systems immunology reveals a late juncture linked to fatal COVID-19. Cell. 2021;184(7):1836–1857.e22. doi: 10.1016/j.cell.2021.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vorobjev IA, Barteneva NS. Imaging flow cytometry methods and protocols. Methods Mol Biol [Internet]. 2016 [cited 2020 Oct 15];1389(November):2–12. Available from: https://link.springer.com/content/pdf/10.1007%2F978-1-4939-3302-0.pdf
- 51.Kay DB, Cambier JL, Wheeless LL. Imaging in flow. J Histochem Cytochem [Internet]. 1979 [cited 2020 Oct 12];27(1):329–34. Available from: https://pubmed.ncbi.nlm.nih.gov/374597/ [DOI] [PubMed]
- 52.Holzner G, Mateescu B, van Leeuwen D, Cereghetti G, Dechant R, Stavrakis S, et al. High-throughput multiparametric imaging flow cytometry: toward diffraction-limited sub-cellular detection and monitoring of sub-cellular processes. Cell Rep [Internet]. 2021 Mar 9 [cited 2021 Jul 21];34(10). Available from: https://pubmed.ncbi.nlm.nih.gov/33691119/ [DOI] [PubMed]
- 53.Bigorra L, Larriba I, Gutiérrez-Gallego R. Machine learning algorithms for accurate differential diagnosis of lymphocytosis based on cell population data. Br J Haematol [Internet]. 2019 Mar 1 [cited 2021 Jul 21];184(6):1035–7. Available from: https://pubmed.ncbi.nlm.nih.gov/29790152/ [DOI] [PubMed]
- 54.Wheeler EC, Vu AQ, Einstein JM, DiSalvo M, Ahmed N, Van Nostrand EL, et al. Pooled CRISPR screens with imaging on microraft arrays reveals stress granule-regulatory factors. Nat Methods [Internet]. 2020 Jun 1 [cited 2021 Jul 21];17(6):636–42. Available from: https://pubmed.ncbi.nlm.nih.gov/32393832/ [DOI] [PMC free article] [PubMed]
- 55.Barteneva NS, Fasler-Kan E, Vorobjev IA. Imaging flow cytometry: coping with heterogeneity in biological systems [Internet]. Vol. 60, Journal of Histochemistry and Cytochemistry. J Histochem Cytochem; 2012 [cited 2020 Oct 15]. p. 723–33. Available from: https://pubmed.ncbi.nlm.nih.gov/22740345/ [DOI] [PMC free article] [PubMed]
- 56.Basiji DA. Principles of Amnis imaging flow cytometry. Methods Mol Biol [Internet]. 2016 [cited 2020 Oct 15];1389:13–21. Available from: https://pubmed.ncbi.nlm.nih.gov/27460235/ [DOI] [PubMed]
- 57.Han Y, Gu Y, Zhang AC, Lo YH. Review: imaging technologies for flow cytometry [Internet]. Vol. 16, Lab on a Chip. Royal Society of Chemistry; 2016 [cited 2020 Oct 15]. p. 4639–47. Available from: https://pubmed.ncbi.nlm.nih.gov/27830849/ [DOI] [PMC free article] [PubMed]
- 58.Mikami H, Lei C, Nitta N, Sugimura T, Ito T, Ozeki Y, et al. High-speed imaging meets single-cell analysis. Chem. Elsevier Inc. 2018;4:2278–300.
- 59.Lau AKS, Wong TTW, Shum HC, Wong KKY, Tsia KK. Ultrafast microfluidic cellular imaging by optical time-stretch. Methods Mol Biol. 2016;1389:23–45. doi: 10.1007/978-1-4939-3302-0_3. [DOI] [PubMed] [Google Scholar]
- 60.Bodenmiller B. Multiplexed epitope-based tissue imaging for discovery and healthcare applications [Internet]. Vol. 2, Cell Systems. Cell Press; 2016 [cited 2020 Oct 12]. p. 225–38. Available from: https://pubmed.ncbi.nlm.nih.gov/27135535/ [DOI] [PubMed]
- 61.Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med [Internet]. 2014 [cited 2020 Oct 12];20(4):436–42. Available from: https://pubmed.ncbi.nlm.nih.gov/24584119/ [DOI] [PMC free article] [PubMed]
- 62.Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods [Internet]. 2014 [cited 2020 Oct 12];11(4):417–22. Available from: https://pubmed.ncbi.nlm.nih.gov/24584193/ [DOI] [PubMed]
- 63.Ijsselsteijn ME, van der Breggen R, Farina Sarasqueta A, Koning F, de Miranda NFCC. A 40-marker panel for high dimensional characterization of cancer immune microenvironments by imaging mass cytometry. Front Immunol [Internet]. 2019 Oct 29 [cited 2020 Oct 15];10(OCT):2534. Available from: 10.3389/fimmu.2019.02534/full [DOI] [PMC free article] [PubMed]
- 64.Jackson HW, Fischer JR, Zanotelli VRT, Ali HR, Mechera R, Soysal SD, et al. The single-cell pathology landscape of breast cancer. Nature [Internet]. 2020 Feb 27 [cited 2020 Oct 15];578(7796):615–20. Available from: https://www.nature.com/articles/s41586-019-1876-x [DOI] [PubMed]
- 65.Schapiro D, Jackson HW, Raghuraman S, Fischer JR, Zanotelli VRT, Schulz D, et al. HistoCAT: Analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods [Internet]. 2017 Aug 31 [cited 2020 Oct 15];14(9):873–6. Available from: https://pubmed.ncbi.nlm.nih.gov/28783155/ [DOI] [PMC free article] [PubMed]
- 66.Damond N, Engler S, Zanotelli VRT, Schapiro D, Wasserfall CH, Kusmartseva I, et al. A map of human type 1 diabetes progression by imaging mass cytometry. Cell Metab [Internet]. 2019 Mar 5 [cited 2020 Oct 12];29(3):755-768.e5. Available from: https://pubmed.ncbi.nlm.nih.gov/30713109/ [DOI] [PMC free article] [PubMed]
- 67.Warren C, McDonald D, Capaldi R, Deehan D, Taylor RW, Filby A, et al. Decoding mitochondrial heterogeneity in single muscle fibres by imaging mass cytometry. Sci Rep [Internet]. 2020 Dec 1 [cited 2020 Oct 12];10(1). Available from: https://pubmed.ncbi.nlm.nih.gov/32948797/ [DOI] [PMC free article] [PubMed]
- 68.Schulz D, Zanotelli VRT, Fischer JR, Schapiro D, Engler S, Lun XK, et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst [Internet]. 2018 Jan 24 [cited 2020 Oct 12];6(1):25-36.e5. Available from: /pmc/articles/PMC5791659/?report=abstract [DOI] [PMC free article] [PubMed]
- 69.McKinnon KM. Flow cytometry: an overview. Curr Protoc Immunol [Internet]. 2018 Feb 1 [cited 2021 Mar 20];2018:5.1.1-5.1.11. Available from: https://pubmed.ncbi.nlm.nih.gov/29512141/ [DOI] [PMC free article] [PubMed]
- 70.Panchuk-Voloshina N, Haugland RP, Bishop-Stewart J, Bhalgat MK, Millard PJ, Mao F, et al. Alexa dyes, a series of new fluorescent dyes that yield exceptionally bright, photostable conjugates. J Histochem Cytochem [Internet]. 1999 [cited 2021 Mar 20];47(9):1179–88. Available from: https://pubmed.ncbi.nlm.nih.gov/10449539/ [DOI] [PubMed]
- 71.Kantor AB, Moore WA, Meehan S, Parks DR. A quantitative method for comparing the brightness of antibody-dye reagents and estimating antibodies bound per cell. Curr Protoc Cytom [Internet]. 2016 Jul 1 [cited 2021 Jul 19];77(1):1.30.1-1.30.23. Available from: 10.1002/cpcy.6 [DOI] [PubMed]
- 72.Carter A. Tandem dye breakdown. eICCS Newsletter Vol5 No 2 [Internet]. 2014; Available from: https://www.cytometry.org/public/newsletters/eICCS-5-2/article4.php
- 73.Hulspas R, Dombkowski D, Preffer F, Douglas D, Kildew-Shah B, Gilbert J. Flow cytometry and the stability of phycoerythrin-tandem dye conjugates. Cytom Part A [Internet]. 2009 Nov [cited 2021 Apr 20];75(11):966–72. Available from: https://pubmed.ncbi.nlm.nih.gov/19777547/ [DOI] [PubMed]
- 74.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science (80- ) [Internet]. 1998 Sep 25 [cited 2021 Mar 20];281(5385):2013–6. Available from: https://pubmed.ncbi.nlm.nih.gov/9748157/ [DOI] [PubMed]
- 75.Ibáñez-Peral R, Bergquist PL, Walter MR, Gibbs M, Goldys EM, Ferrari B. Potential use of quantum dots in flow cytometry. Int J Mol Sci [Internet]. 2008 Dec [cited 2021 Mar 21];9(12):2622–38. Available from: https://pubmed.ncbi.nlm.nih.gov/19330097/ [DOI] [PMC free article] [PubMed]
- 76.Petryayeva E, Algar WR, Medintz IL. Quantum dots in bioanalysis: a review of applications across various platforms for fluorescence spectroscopy and imaging [Internet]. Vol. 67, Applied Spectroscopy. SAGE PublicationsSage UK: London, England; 2013 [cited 2021 Apr 25]. p. 215–52. Available from: 10.1366/12-06948 [DOI] [PubMed]
- 77.Chattopadhyay PK, Perfetto SP, Yu J, Roederer M. The use of quantum dot nanocrystals in multicolor flow cytometry [Internet]. Vol. 2, Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. Wiley Interdiscip Rev Nanomed Nanobiotechnol; 2010 [cited 2021 Mar 21]. p. 334–48. Available from: https://pubmed.ncbi.nlm.nih.gov/20101649/ [DOI] [PubMed]
- 78.Chattopadhyay PK. Quantum dot technology in flow cytometry [Internet]. Vol. 102, Methods in Cell Biology. Methods Cell Biol; 2011 [cited 2021 Mar 20]. 463–477 p. Available from: https://pubmed.ncbi.nlm.nih.gov/21704850/ [DOI] [PubMed]
- 79.Valizadeh A, Mikaeili H, Samiei M, Farkhani SM, Zarghami N, Kouhi M, et al. Quantum dots: synthesis, bioapplications, and toxicity [Internet]. Vol. 7, Nanoscale Research Letters. Nanoscale Res Lett; 2012 [cited 2021 Apr 20]. Available from: https://pubmed.ncbi.nlm.nih.gov/22929008/ [DOI] [PMC free article] [PubMed]
- 80.Hu L, Zhong H, He Z. Toxicity evaluation of cadmium-containing quantum dots: a review of optimizing physicochemical properties to diminish toxicity. Colloids Surfaces B Biointerfaces [Internet]. 2021 Apr [cited 2021 Mar 20];200:111609. Available from: https://pubmed.ncbi.nlm.nih.gov/33588242/ [DOI] [PubMed]
- 81.Barroso MM. Quantum dots in cell biology [Internet]. Vol. 59, Journal of Histochemistry and Cytochemistry. SAGE PublicationsSage CA: Los Angeles, CA; 2011 [cited 2021 Apr 25]. p. 237–51. Available from: 10.1369/0022155411398487 [DOI] [PMC free article] [PubMed]
- 82.Chattopadhyay PK, Gaylord B, Palmer A, Jiang N, Raven MA, Lewis G, et al. Brilliant violet fluorophores: a new class of ultrabright fluorescent compounds for immunofluorescence experiments. Cytom Part A [Internet]. 2012 Jun [cited 2021 Mar 21];81 A(6):456–66. Available from: https://pubmed.ncbi.nlm.nih.gov/22489009/ [DOI] [PubMed]
- 83.Dean P, Bagwell C, Lindmo T, Murphy R, Salzman G. Introduction to flow cytometry data file standard. Cytometry [Internet]. 1990 [cited 2021 Jul 25];11(3):321–2. Available from: https://pubmed.ncbi.nlm.nih.gov/2340768/ [DOI] [PubMed]
- 84.Lee JA, Spidlen J, Boyce K, Cai J, Crosbie N, Dalphin M, et al. MIFlowCyt: the minimum information about a flow cytometry experiment. Cytom Part A [Internet]. 2008 Oct [cited 2021 Apr 20];73(10):926–30. Available from: https://pubmed.ncbi.nlm.nih.gov/18752282/ [DOI] [PMC free article] [PubMed]
- 85.Spidlen J, Moore W, Parks D, Goldberg M, Blenman K, Cavenaugh JS, et al. Data file standard for flow cytometry, Version FCS 3.2. Cytom Part A [Internet]. 2021 Jan 1 [cited 2021 Apr 20];99(1):100–2. Available from: https://pubmed.ncbi.nlm.nih.gov/32881398/ [DOI] [PMC free article] [PubMed]
- 86.Spidlen J, Gentleman RC, Haaland PD, Langille M, Le Meur N, Ochs MF, et al. Data standards for flow cytometry [Internet]. Vol. 10, OMICS A Journal of Integrative Biology. OMICS; 2006 [cited 2021 Apr 20]. p. 209–14. Available from: https://pubmed.ncbi.nlm.nih.gov/16901228/ [DOI] [PMC free article] [PubMed]
- 87.Spidlen J, Breuer K, Brinkman R. Preparing a minimum information about a flow cytometry experiment (MIFlowCyt) compliant manuscript using the international society for advancement of cytometry (ISAC) FCS file repository (FlowRepository.org). Curr Protoc Cytom [Internet]. 2012 Jul [cited 2021 Apr 20];Chapter 10(SUPPL.61). Available from: https://pubmed.ncbi.nlm.nih.gov/22752950/ [DOI] [PubMed]
- 88.Finak G, Langweiler M, Jaimes M, Malek M, Taghiyar J, Korin Y, et al. Standardizing flow cytometry immunophenotyping analysis from the human immunophenotyping consortium. Sci Rep [Internet]. 2016 Feb 10 [cited 2021 Apr 20];6. Available from: https://pubmed.ncbi.nlm.nih.gov/26861911/ [DOI] [PMC free article] [PubMed]
- 89.Brinkman RR. Improving the rigor and reproducibility of flow cytometry-based clinical research and trials through automated data analysis [Internet]. Vol. 97, Cytometry Part A. Wiley-Liss Inc.; 2020 [cited 2021 Apr 20]. p. 107–12. Available from: https://pubmed.ncbi.nlm.nih.gov/31515945/ [DOI] [PMC free article] [PubMed]
- 90.Aghaeepour N, Finak G, Hoos H, Mosmann TR, Brinkman R, Gottardo R, et al. Critical assessment of automated flow cytometry data analysis techniques. Nat Methods [Internet]. 2013 Mar [cited 2021 Apr 20];10(3):228–38. Available from: https://pubmed.ncbi.nlm.nih.gov/23396282/ [DOI] [PMC free article] [PubMed]
- 91.Fletez-Brant K, Špidlen J, Brinkman RR, Roederer M, Chattopadhyay PK. flowClean: automated identification and removal of fluorescence anomalies in flow cytometry data. Cytom Part A [Internet]. 2016 May 1 [cited 2021 Apr 20];89(5):461–71. Available from: https://pubmed.ncbi.nlm.nih.gov/26990501/ [DOI] [PMC free article] [PubMed]
- 92.Shannon CP, Blimkie TM, Ben-Othman R, Gladish N, Amenyogbe N, Drissler S, et al. Multi-omic data integration allows baseline immune signatures to predict hepatitis B vaccine response in a small cohort. Front Immunol [Internet]. 2020 Nov 30 [cited 2021 Apr 20];11. Available from: https://pubmed.ncbi.nlm.nih.gov/33329547/ [DOI] [PMC free article] [PubMed]
- 93.Duetz C, Bachas C, Westers TM, Van De Loosdrecht AA. Computational analysis of flow cytometry data in hematological malignancies: future clinical practice? [Internet]. Vol. 32, Current Opinion in Oncology. Lippincott Williams and Wilkins; 2020 [cited 2021 Apr 20]. p. 162–9. Available from: https://pubmed.ncbi.nlm.nih.gov/31876546/ [DOI] [PubMed]
- 94.Mair F, Hartmann FJ, Mrdjen D, Tosevski V, Krieg C, Becher B. The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur J Immunol [Internet]. 2016 Jan 1 [cited 2021 Apr 20];46(1):34–43. Available from: https://pubmed.ncbi.nlm.nih.gov/26548301/ [DOI] [PubMed]
- 95.Montante S, Brinkman RR. Flow cytometry data analysis: recent tools and algorithms. Int J Lab Hematol [Internet]. 2019 May 1 [cited 2020 Oct 12];41(S1):56–62. Available from: https://pubmed.ncbi.nlm.nih.gov/31069980/ [DOI] [PubMed]
- 96.Weber LM, Robinson MD. Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytom Part A [Internet]. 2016 Dec 1 [cited 2020 Oct 12];89(12):1084–96. Available from: https://pubmed.ncbi.nlm.nih.gov/27992111/ [DOI] [PubMed]
- 97.Su Y, Chen D, Yuan D, Lausted C, Choi J, Dai CL, et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell [Internet]. 2020 Dec 10 [cited 2021 Apr 20];183(6):1479-1495.e20. Available from: https://pubmed.ncbi.nlm.nih.gov/33171100/ [DOI] [PMC free article] [PubMed]
- 98.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods [Internet]. 2017 Aug 31 [cited 2020 Oct 15];14(9):865–8. Available from: https://www.nature.com/articles/nmeth.4380 [DOI] [PMC free article] [PubMed]
- 99.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, et al. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol [Internet]. 2017 Oct 1 [cited 2021 Mar 19];35(10):936–9. Available from: https://pubmed.ncbi.nlm.nih.gov/28854175/ [DOI] [PubMed]
- 100.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell [Internet]. 2015 May 30 [cited 2021 Mar 19];161(5):1202–14. Available from: https://pubmed.ncbi.nlm.nih.gov/26000488/ [DOI] [PMC free article] [PubMed]
- 101.Yamawaki TM, Lu DR, Ellwanger DC, Bhatt D, Manzanillo P, Arias V, et al. Systematic comparison of high-throughput single-cell RNA-seq methods for immune cell profiling. BMC Genomics [Internet]. 2021 Dec 1 [cited 2021 Apr 20];22(1). Available from: https://pubmed.ncbi.nlm.nih.gov/33472597/ [DOI] [PMC free article] [PubMed]
- 102.Gao S. Data analysis in single-cell transcriptome sequencing. In: Methods in Molecular Biology [Internet]. Humana Press Inc.; 2018 [cited 2021 Mar 19]. p. 311–26. Available from: https://pubmed.ncbi.nlm.nih.gov/29536451/ [DOI] [PubMed]
- 103.Tirosh I, Suvà ML. Deciphering human tumor biology by single-cell expression profiling [Internet]. Vol. 3, Annual Review of Cancer Biology. Annual Reviews Inc.; 2019 [cited 2021 Mar 19]. p. 151–66. Available from: 10.1146/annurev-cancerbio-030518-
- 104.Gutierrez-Arcelus M, Teslovich N, Mola AR, Polidoro RB, Nathan A, Kim H, et al. Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions. Nat Commun [Internet]. 2019 Dec 1 [cited 2021 Mar 19];10(1). Available from: https://pubmed.ncbi.nlm.nih.gov/30737409/ [DOI] [PMC free article] [PubMed]
- 105.Chattopadhyay PK, Roederer M, Bolton DL. A deadly dance: the choreography of host–pathogen interactions, as revealed by single-cell technologies [Internet]. Vol. 9, Nature Communications. Nature Publishing Group; 2018 [cited 2021 Mar 19]. Available from: https://pubmed.ncbi.nlm.nih.gov/30401874/ [DOI] [PMC free article] [PubMed]
- 106.Brummelman J, Haftmann C, Núñez NG, Alvisi G, Mazza EMC, Becher B, et al. Development, application and computational analysis of high-dimensional fluorescent antibody panels for single-cell flow cytometry. Nat Protoc [Internet]. 2019 Jul 1 [cited 2020 Sep 20];14(7):1946–69. Available from: https://www.nature.com/articles/s41596-019-0166-2 [DOI] [PubMed]
- 107.Gonneau C, Wang L, Mitra-Kaushik S, Trampont PC, Litwin V. Recent global standardization for quantitative flow cytometry. Bioanalysis. 2021;in press. [DOI] [PubMed]
- 108.Validation of assays performed by flow cytometry. 1st ed. CLSI document H62. Wayne, PA: Clinical Laboratory Standards Institute; 2021.