Abstract
CD8+ T cell responses restricted by MHC-E, a nonclassical MHC molecule, have been associated with protection in an SIV/rhesus macaque model. The biological relevance of HLA-E–restricted CD8+ T cell responses in HIV infection, however, remains unknown. In this study, CD8+ T cells responding to HIV-1 Gag peptides presented by HLA-E were analyzed. Using in vitro assays, we observed HLA-E–restricted T cell responses to what we believe to be a newly identified subdominant Gag-KL9 as well as a well-described immunodominant Gag-KF11 epitope in T cell lines derived from chronically HIV-infected patients and also primed from healthy donors. Blocking of the HLA-E/KF11 binding by the B7 signal peptide resulted in decreased CD8+ T cell responses. KF11 presented via HLA-E in HIV-infected cells was recognized by antigen-specific CD8+ T cells. Importantly, bulk CD8+ T cells obtained from HIV-infected individuals recognized infected cells via HLA-E presentation. Ex vivo analyses at the epitope level showed a higher responder frequency of HLA-E–restricted responses to KF11 compared with KL9. Taken together, our findings of HLA-E–restricted HIV-specific immune responses offer intriguing and possibly paradigm-shifting insights into factors that contribute to the immunodominance of CD8+ T cell responses in HIV infection.
Keywords: Infectious disease
Keywords: T cells
Introduction
There is compelling evidence that effector CD8+ cytotoxic T lymphocytes (CTLs) play a pivotal role in the control of HIV infection (1–3). The paradigm for CTL recognition is via pathogen-derived peptides presented by classical major histocompatibility complex class I (MHC-Ia in general or human leukocyte antigen class I [HLA-Ia] in humans) on the surface of infected cells. The extreme polymorphism of the HLA-Ia genes (4), the propensity of HIV to mutate to escape immune responses, and a remarkably diverse viral peptidome of infected cells together pose a significant hurdle for the design of a universally efficacious HIV vaccine. In contrast, HLA-E, a nonclassical MHC class Ib allele with only 2 major subtypes, 01:01 and 01:03, has a strong preference for monomorphic HLA-I leader sequence peptides (5). HLA-E is ubiquitously expressed at high levels on immune cells in various tissue compartments (5, 6) and plays a dual role in regulating lymphocyte activity due to its potential to inhibit NK and activate αβ CD8+ T cell responses (7). In fact, this allele was first described as the ligand of CD94/NKG2 receptors on NK cells and, through interaction with the inhibitory NKG2A receptor, induced NK-mediated immune tolerance (8).
Although HLA-E is known to be stabilized on the surface of cells by the leader/signal sequences of HLA-Ia alleles (9), under conditions of stress, such as a pathogenic infection (viral and bacterial), it presents a broad array of pathogen-derived peptides that can activate αβ CD8+ T cells (5). Peptide loading onto HLA-Ib molecules, i.e., HLA-E, uses TAP-dependent and -independent mechanisms (10). In a TAP-deficient environment (viral infection or tumor), loading of the canonical leader peptide sequence is hindered (11), leading to a large (>500) HLA-E–bound peptide repertoire. Identification of HLA-E–restricted CD8+ effector T cell responses against cytomegalovirus (CMV) (12–17), hepatitis C virus (18), Epstein-Barr virus (19), and recently hepatitis B virus (20) points to the existence of a subset of human CD8+ T cells that can sense pathogen with their T cell receptors (TCRs) in the context of HLA-E. Recognition of TCRs by peptide (CMV-UL40) presented via HLA-E was first demonstrated in studies using NK-CTL clones (21, 22). In these instances, HLA-E presented a viral immunogen with homology to leader sequence peptide. Peptides presented through this allele can also stimulate αβ TCRs of CD8+ T cells in response to a variety of bacterial pathogens, including Salmonella enterica serovar Typhi and Mycobacterium tuberculosis (5, 23–25). Salmonella-specific responses (26) were long-lasting, suggesting that HLA-E–restricted CD8+ T cells can develop into memory T cells and protect against typhoid fever (27). Recognition of CMV and Salmonella Typhi induced production of IFN-γ and lysis of target cells by granule-dependent pathways (15, 26), whereas CD8+ T cells stimulated in vitro by several M. tuberculosis peptides presented by HLA-E may have regulatory roles (25, 28). Thus HLA-E–restricted CD8+ T cells can exert important immunoregulatory functions.
In the context of simian immunodeficiency virus (SIV), work from Louis Picker’s group highlighted unrecognized flexibility in CD8+ T cell responses and suggested that the current paradigms about immunodominance and epitope selections are not absolute (29, 30). This group showed that vaccinating rhesus macaques with strain 68-1–based RhCMV/SIV vector resulted in stringent control and clearance of a highly pathogenic SIVmac239 challenge in about 50% of the animals (31, 32). This protection correlated with unconventional, broadly targeted Mamu-E–restricted SIV-specific CD8+ T cell responses (30) that largely did not overlap with conventional MHC-Ia–restricted CD8+ T cells. Thus, attenuated CMV strain 68-1 vector inducing MHC-E–restricted SIV-specific CD8+ T cells brings additional effector mechanisms to consider for eliminating virus-infected CD4+ T cells in HIV infection.
The unprecedented protection in macaques vaccinated by persistent RhCMV vectors (33) has prompted interest in whether HIV-1 can be recognized by human CD8+ T cells in the context of HLA-E. Mamu-E and HLA-E are related proteins. Since the Mamu-E gene is highly conserved in both New and Old World primates, it is possible that HLA-E retains the structural and functional potential to be targeted by substantial adaptive T cell responses after HIV-1 infection (34). Furthermore, recent data on surface upregulation of HLA-E in viral infections (20, 30, 35) suggest that HLA-E–restricted HIV-specific CD8+ T cell responses may play an important role in CD8+ T cell surveillance. Despite these potential benefits, targeting of HLA-E–restricted HIV-specific CD8+ T cells in HIV infection and characterization of their functionality have not been addressed before. Furthermore, how such responses compare with those elicited toward HLA-Ia–restricted epitopes remains unknown.
Recent studies aimed to identify potential HLA-E–restricted epitopes in a conserved HIV-1 vaccine immunogen based on binding predictions and assays (36, 37), but HLA-E–restricted HIV-specific CD8+ T cells have, to our knowledge, not yet been reported. In this study, we sought to determine the extent of targeting and functionality of HLA-E–restricted CD8+ T cells during HIV infection and how these responses compare with classic HLA-Ia allele–restricted CD8+ T cells. Both ex vivo and in vitro assays demonstrated that HLA-E–restricted CD8+ T cells have polyfunctional effector profiles including secretion of cytotoxic molecules and an ability to inhibit NL4-3 replication in vitro. HLA-E–restricted CD8+ T cells almost invariably recognize peptides presented by HLA-Ia alleles, and exclusive HLA-E–restricted CD8+ T cell responses were infrequently observed. Thus, HLA-E–restricted CD8+ T cells most likely represent a subpopulation of polyclonal HLA-Ia–restricted cells and highlight a previously unexplored source of adaptive immune responses.
Results
Determining HLA-Ia and HLA-E restriction of 2 HIV-1 Gag epitopes.
In vitro priming of HIV-specific CD8+ T cell responses from HIV-naive individuals is commonly used to identify subdominant responses. By in vitro priming of CD8+ T cells obtained from 2 HIV-seronegative donors with HIV-1 Gag overlapping peptides (OLPs) in long-term cultures, we identified a subdominant effector response for OLPs 7954/7955 (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI148979DS1). Gag OLPs 7954/7955 encompass DCKTILKALGPAATL (aa 329–343)/ILKALGPAATLEEMM (aa 333–347), respectively. Using PBMCs and nested peptides (10 μg/mL) progressively truncated by a single amino acid from the N- or C-termini of OLP 7954 in an IFN-γ ELISPOT assay, we identified a 9-mer, KL9, in individuals who express HLA-A*02 or HLA-B*57. Based on NetMHC predictions of peptide-HLA binding (http://www.cbs.dtu.dk/services/NetMHC/), IC50 for KL9 binding HLA-B*57:01 and -A*02:01 was 226 nM and 4684 nM, respectively, indicating that KL9 could be restricted by more than one class Ia allele.
We next determined whether KL9-specific CD8+ T cells can be elicited in chronic HIV infection and whether these are HLA-B*57:01 and -A*02:01 restricted. Antigen-specific CD8+ T cells were preferentially expanded by long-term culture (7–12 days) using autologous adherent monocytes pulsed with 10 μg/mL KL9. While doing HLA-I restriction studies using parental 721.221 cells (HLA-Ia null) and their derivative transductants expressing single HLA-B*57:01 and -A*02:01 alleles, we were surprised to find peptide-specific reactivities among the 721.221 cells, which suggested an alternative HIV antigen presentation mechanism, perhaps one that is mediated by nonclassical HLA-Ib molecules. We therefore performed peptide binding stabilization assays of HLA-E and KL9, and our data showed that KL9 could bind to HLA-E*01:01 (Supplemental Figure 1A), although not as strongly as B7 signal peptide (B7sp).
Therefore, to determine HLA-Ia/b restriction, the functionality of KL9-specific CD8+ T cell lines was assessed after stimulation of these cells for 6 hours with target antigen-presenting cell (APC) lines (221.B*57:01 or 221.AEH) expressing HLA-B*57:01 or HLA-E*01:01, respectively, and pulsed with 10 μg/mL peptide. Representative data are shown for KL9 response restricted by HLA-B*57 and HLA-E (Figure 1A). Patient CHI-7 (where CHI indicates chronically HIV infected) exhibited a higher CD107a/IFN-γ expression in response to KL9 presented by HLA-E–expressing APCs (221.AEH), compared with when KL9 was presented in the context of HLA-B*57 to CD8+ T cells. These data suggested that KL9 can be dually restricted by HLA class Ia/b alleles.
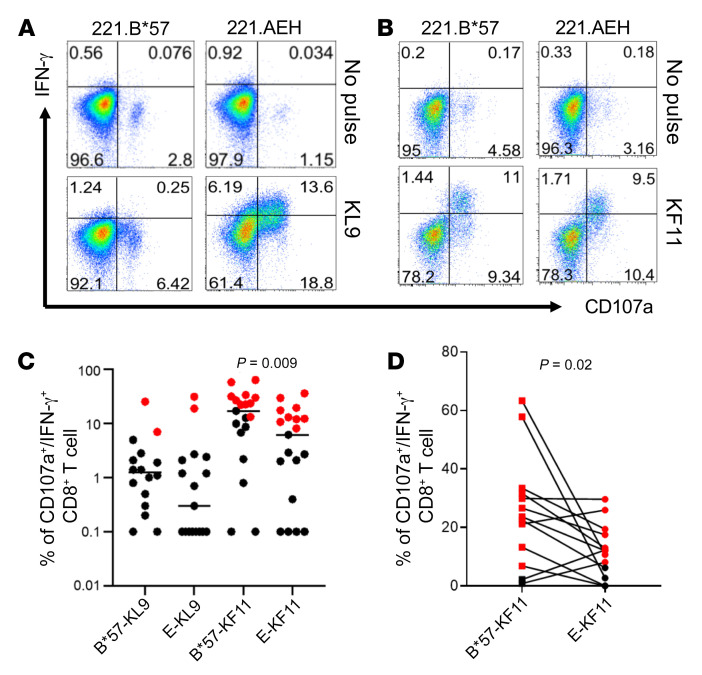
Figure 1. In vitro detection of KL9- and KF11-specific CD8+ T cell responses restricted by HLA-B*57 and HLA-E in chronically HIV-infected individuals.
Positively isolated CD8+ T cells were cultured with KL9- or KF11-pulsed (10 μg/mL) autologous adherent monocytes for 7–12 days. Polyfunctional activation was assessed after 5 hours of stimulation with target cells (221.B*57 or 221.AEH) pulsed with the peptide of interest. No peptide pulse was used as a negative control. 221.B*57 and 221.AEH are antigen-presenting 721.221 cell lines expressing single HLA-B*57:01 and HLA-E*01:01 alleles, respectively. Representative data from 2 HIV-infected individuals are shown in A and B. (A) KL9-specific reactivity restricted by B*57:01 or E*01:01 in patient CHI-7. (B) KF11-specific reactivity restricted by B*57:01 or E*01:01 in patient CHI-6. (C) Summary of KL9- and KF11-specific reactivity in HIV-infected individuals (n = 20) based on CD107a/IFN-γ expression. (D) Net frequency (CD107a/IFN-γ) of paired HLA-E and HLA-B*57 responses specific for KF11 is shown for HIV-infected individuals tested (n = 13) who mounted an HLA-E–restricted CD8+ T cell response. Black and red dots are negative and positive responses, respectively. Error bars represent median value. Wilcoxon matched-pairs signed rank test was used in C and D to determine statistical significance.
Since KL9, a subdominant epitope, could be presented in the context of both HLA-E and HLA-B*57 allele, we next sought to determine whether a well-described B*57-restricted epitope could also be presented by HLA-E. We selected KF11 as a model epitope owing to its high immunogenicity in chronic HIV infection and its ability to bind to HLA-E*01:01 (Supplemental Figure 1B). We analyzed the functionality of CD8+ T cell responses specific for KL9 and KF11 presented by HLA-Ia/b alleles in 20 PBMC samples from CHI individuals. HLA-Ia/b genotype and clinical characteristics of the study cohort are shown in Supplemental Table 2. Representative data are shown for KF11 response restricted by HLA-B*57 and HLA-E (Figure 1B). In this individual, KF11 response restricted by both HLA-Ia (B*57:01) and HLA-Ib (E*01:01) alleles showed CD107a/IFN-γ production as a functional response. Figure 1C summarizes data from all 20 CHI individuals tested, indicating percentage CD8+ T cells in response to both KL9 and KF11 peptides when presented by either HLA-B*57 or HLA-E allele. These data show that both KL9 and KF11 peptides can be dually presented by HLA-Ia and -Ib alleles. For either epitope, the frequency of positive responders was similar regardless of HLA-I allele restriction. However, a higher frequency was observed when KF11 was presented by HLA-B*57 compared with HLA-E presentation (P = 0.009; Figure 1C). When responses that were positive to either or both HLA-E– and HLA-B*57–mediated presentation of KF11 were compared in a pairwise manner within each individual, HLA-B*57–restricted responses were still higher than those specific for HLA-E (P = 0.02; Figure 1D).
Details of responder frequency and magnitude of both epitope-specific responses are shown in Supplemental Table 3. For KL9, we observed very few responses. They were unique to either HLA-B*57 allele (patients CHI-2 and CHI-21) or HLA-E*01 (patient CHI-7) except for one dually restricted response seen in patient CHI-10. In contrast, B*57-KF11 responses were observed in 11 of 19 samples (58%) tested, and nearly two-thirds of these were dually HLA-Ia/b–restricted (7/11, 64%; Supplemental Table 3). Overall, unique HLA-E–restricted CD8+ T cell responses were observed only in patients CHI-17 and CHI-18. In summary, unique HLA-E–restricted responses were seen in only 3 of 20 (15%) whereas dually restricted responses were observed in 8 of 20 (40%) individuals tested.
PBMCs stimulated with KL9 or KF11 (10 μg/mL) elicited CD8+ T cells with multifunctional profile as manifested by dual production of CD107a/b with IFN-γ, IL-2, TNF-α, and MIP-1β (Supplemental Figure 2). As compared with the KF11 stimulation, KL9 responses were either lower (Supplemental Figure 2, A and B) or similar in frequency (Supplemental Figure 2C).
Generation of a panel of cell lines to discriminate between classical HLA class Ia– and E–restricted CD8+ T cell responses.
A prior study showed that monomorphic leader peptides from HLA class I molecules are perfectly suited for tight and deep binding into the HLA-E groove (11). In contrast, pathogen-derived antigens presented by HLA-E in a TAP-deficient environment adopt different conformation, docking more superficially into the groove. Our data from binding stabilization assays show that both KL9 and KF11 can bind HLA-E*01:01 (Supplemental Figure 1). However, because of the low HLA binding affinities of the KL9 or KF11 peptides for HLA-E and/or peptide stabilization, we were unable to construct stable HIV-1 peptide–specific HLA-E tetramers for either peptide. To circumvent this issue and to discriminate between classical HLA class Ia– and HLA-E–restricted T cells, we first disrupted HLA-E expression in the 721.221 cell line using a CRISPR/Cas9 system. We obtained seven HLA-E–knockout clones that were verified by genotyping, and based on these data, clone 41A3 was used in follow-up studies. A heterozygous deletion in the HLA-E allele in this clone resulted in a loss of function, which was confirmed by a lack of surface HLA-E expression as assessed by staining with HLA-E mAbs 3D12 and 4D12 (which bind to empty HLA-E). Next, individual class Ia (HLA-A*02:01, HLA-B*57:01, HLA-B*57:03), HLA-E*01:01, or HLA-E*01:03 alleles were singly transduced into the 41A3 clone, generating a total of 5 target cell lines, each expressing only a single aforementioned HLA class I allele. This approach facilitated the discrimination between classical and nonclassical class I–restricted responses. Supplemental Figure 3 shows the surface expression of HLA-Ia (A*02:01 and B*57) or HLA-Ib (E*01 and F) transductants in the 41A3 clone.
HLA-E*01–restricted KF11-specific responses are mediated by CD94– CD8+ T cells.
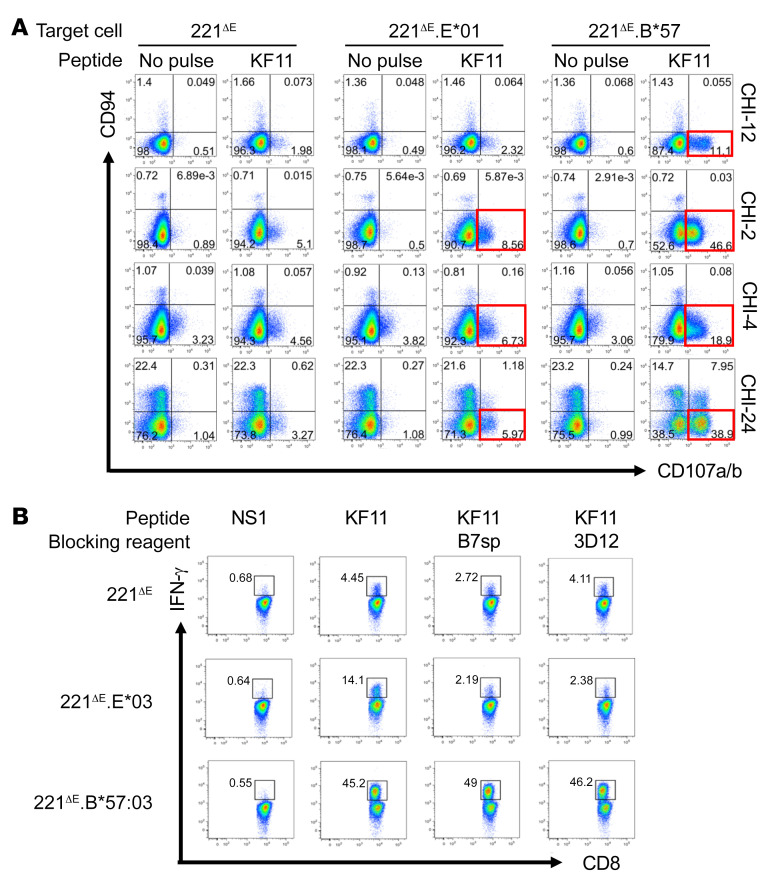
It is well known that NK/NKT cells recognize HLA-E–bound peptide via their CD94/NKG2 receptors (38). To confirm that the responses observed in our assays were due to CD8+ T cells, we performed the following assay. In vitro–cultured CD8+ T cells from 4 HIV-infected individuals (CHI-2, CHI-4, CHI-12, and CHI-24) were tested for KF11-specific responses with 221ΔE.E*01:01, 221ΔE.E*01:03, and 221ΔE.B*57:03 target cells pulsed with 10 μg/mL peptide. In these 4 separate patients, KF11 was recognized in the context of HLA-E*01:01 or HLA-B*57:03 by CD94– non-NK/NKT CD8+ T cells (Figure 2A). HLA-E restriction was validated in 1 patient (CHI-24) by 2 different blocking strategies (Figure 2B), i.e., using B7sp and the anti–HLA-E mAb 3D12. KF11-specific activation (IFN-γ production) in the context of HLA-E*01:03 but not HLA-B*57:03 was blocked using both approaches.
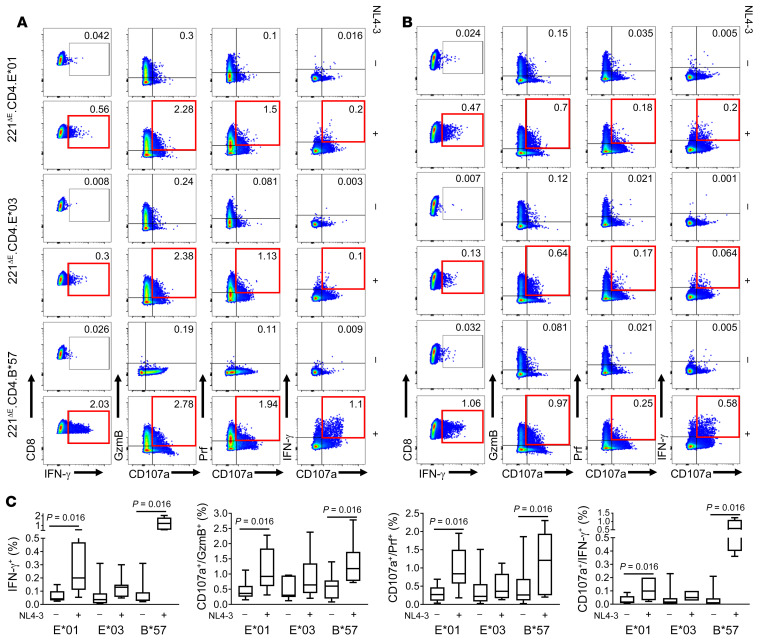
Figure 2. Recognition of KF11 in the context of HLA-E*01 by non-NK/NKT CD8+ T cells from HIV-infected individuals, using the 221ΔE cell panel and in vitro–generated CD8+ T cell lines.
(A) In vitro–cultured CD8+ T cells from 4 chronically HIV-infected individuals (CHI-12, CHI-2, CHI-4, and CHI-24) were tested for upregulation of CD107a/b in CD94-expressing CD8+ T cells in response to KF11 (10 μg/mL). APCs or target cells included 221ΔE.E*01, 221ΔE.B*57, and 221ΔE, the latter serving as a negative control. In addition, within each APC, KF11 stimulation was assessed in comparison with no-peptide-pulse control (red boxes). (B) Two different blocking strategies in patient CHI-24 were used to confirm that KF11 was presented in the context of HLA-E. KF11-specific activation of CD8+ T cells (IFN-γ production) in the context of HLA-E*01:03 but not HLA-B*57:03 was blocked by excess leader sequence B7sp peptide or by the HLA-E–specific 3D12 mAb, both at 10 μg/mL.
HLA-E–mediated KF11 peptide presentation in NL4-3–infected cells.
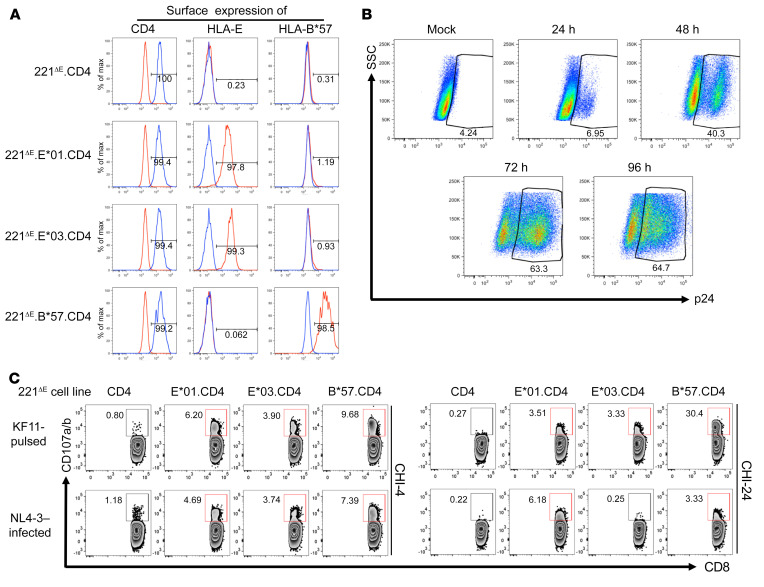
Although data from the prior experiments demonstrated that HIV peptides could be presented exogenously by HLA-E, the question remained of whether this peptide could be loaded onto nascent HLA-E within an infected cell. To address this question, we first transduced CD4 coreceptor into each of the 6 cell lines generated (Supplemental Figure 3) to render them HIV permissive. The surface expression of CD4 was confirmed on 221ΔE.CD4, 221ΔE.E*01:01.CD4, 221ΔE.E*01:03.CD4, and 221ΔE.B*57:01.CD4 cell lines (Figure 3A). We next determined whether these cell lines support permissive HIV-1 infection. We observed an increased frequency of intracellular Gag-p24 staining in a representative NL4-3–infected cell line (221ΔE.CD4) over 96 hours after infection (Figure 3B). We then determined whether CD8+ T cells could recognize the KF11 peptide presented by infecting target cells with NL4-3 virus for 48 hours and coculturing them with CD8+ T cells at 1:1 effector/target cell ratio. Target cells pulsed with 10 μg/mL KF11 were used as positive controls, while 221ΔE.CD4 cells (KF11-pulse or NL4-3–infected) alone served as negative controls. Cultured CD8+ T cells from 2 HIV-infected individuals showed degranulation after incubation with NL4-3–infected 221ΔE.E*01:01.CD4, 221ΔE.E*01:03.CD4, or 221ΔE.B*57:01.CD4 target cells compared with the negative control (221ΔE.CD4 target cells; Figure 3C). These data demonstrate that KF11 could be processed endogenously and loaded onto HLA-E.
Figure 3. Activation of KF11-specific CD8+ T cells by HIV-1–infected target cells expressing HLA-E and CD4.
(A) Confirmation of surface expression of CD4 by 221ΔE.CD4, 221ΔE.E*01.CD4, 221ΔE.E*03.CD4, and 221ΔE.B*57.CD4 cell lines. Staining by isotype-matched negative control mAbs is also depicted in each case. (B) Percentages of p24+ cells and increasing intensity of cytoplasmic p24 staining (mAb Kc57) in a representative NL4-3–infected cell line (221ΔE.CD4) over 96 hours. (C) Degranulation of CD94–CD8hi T cells from patients CHI-4 and CHI-24 after incubation with NL4-3–infected 221ΔE.CD4, 221ΔE.E*01.CD4, 221ΔE.E*03.CD4, or 221ΔE.B*57.CD4 target cells. Effector cells were generated by in vitro stimulation with 10 μg/mL KF11 peptide. Target cells were assessed 48 hours after infection with 18% of the cells staining for p24. KF11-pulsed target cells were included as positive controls, while 221ΔE.CD4 cells (KF11-pulsed or NL4-3–infected) served as negative controls.
HLA-E–restricted responses are readily detected using PBMCs taken ex vivo from HIV-infected individuals.
In vitro assays indicated that HLA-E–specific responses can be detected after long-term enrichment or expansion of antigen-specific CD8+ T cells. However, as long-term culturing can impact T cell phenotype and function (39), we wanted to more closely mimic an in vivo setting. Thus, we performed ex vivo assays to determine whether HLA-E responses can be readily identified.
To identify KL9 and KF11 responders, we used PBMCs from HIV-infected individuals who expressed HLA-A*02 (n = 36), B*57 (n = 13), or both alleles (n = 7) and stimulated them with 10 μg/mL KL9 and KF11 peptides in an overnight IFN-γ ELISPOT assay. Individuals who were either A*02, B*57, or both responded to the KL9 peptide with a frequency of 11%, 69%, and 57%, respectively. KL9 responses are more frequently targeted by individuals expressing HLA-B*57 than -A*02 allele. In comparison, the immunodominant B*57-restricted KF11 elicited 92% and 71% responder frequency in individuals with B*57 or both A*02 and B*57 allele carriage, respectively. Among B*57-expressing individuals who mounted both KL9 and KF11 responses, we observed a higher response magnitude of KF11 (Figure 4A).
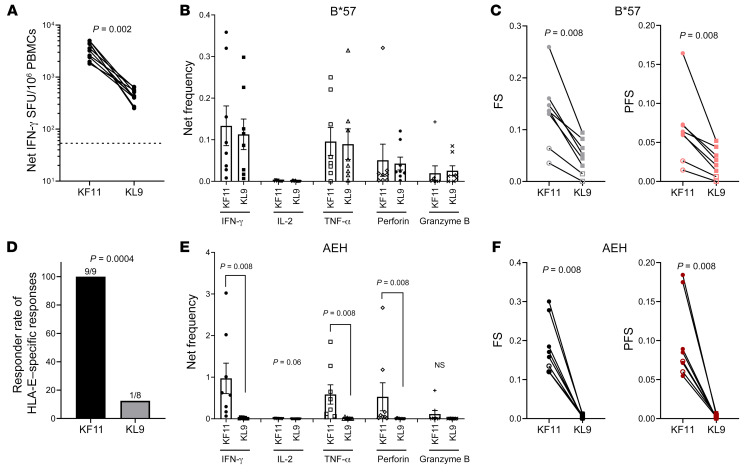
Figure 4. Ex vivo detection of KL9- and KF11-specific CD8+ T cell responses restricted by either HLA-B*57 or HLA-E in chronically HIV-infected individuals.
Positive isolated CD8+ T cells from PBMCs were cultured with target cells pulsed with 10 μg/mL peptide without long-term expansion. (A) Comparison between KF11- and KL9-induced IFN-γ response using ex vivo ELISPOT (n = 9). The dashed line represents the positive cutoff value of 55 SFU/106 PBMCs. (B and C) Net frequency of production of cytokine/effector molecules by CD8+ T cells responding to KF11/KL9 presentation by HLA-B*57 using the K562.B*57 cell line as APCs (B); and functional score (FS) and polyfunctional score (PFS) (C) (n = 8). Net frequency was calculated by subtraction of the no-peptide-pulse control of the identical APCs. (D) Comparison of responder rates between KF11-induced (n = 9) and KL9-induced (n = 8) CD8+ T cell response using HLA-E–expressing AEH cell line as APCs. (E and F) Data similar to those in B and C but for HLA-E–restricted responses: net frequency of production of cytokine/effector molecules (E); and functional score and polyfunctional score (F) (n = 8). Error bars represent mean ± SEM. Wilcoxon matched-pairs signed rank test was used in A–C, E, and F, and Fisher’s exact test in D, to determine statistical significance.
To determine HLA-I restriction of these 2 epitopes, we then performed intracellular cytokine staining assays in which APCs expressing either B*57 (K562.B*57 cell line) or HLA-E allele (721.221.AEH cell line) and pulsed with 10 μg/mL peptide were used as targets. Representative data are shown in Supplemental Figure 4. At an inter-epitope level a similar magnitude of response was elicited when KF11 and KL9 were presented by HLA-B*57 as assessed by dual expression of CD107a with IFN-γ, IL-2, TNF-α, perforin, and granzyme B (Figure 4B). However, the functional and polyfunctional scores of KF11 were higher than those of KL9 as calculated by Combinatorial Polyfunctionality Analysis of Antigen Specific T cell subsets (http://rglab.github.io/COMPASS/) (Figure 4C). In the context of peptide presentation by HLA-E–expressing APCs, we observed a higher responder rate for KF11 compared with KL9 (Figure 4D). In addition, CD8+ T cell responses for KF11 were higher when dual expression of CD107a with IFN-γ, TNF-α, perforin, and granzyme B was assessed (Figure 4E) and when the functional and polyfunctional scores were compared with those of KL9 (Figure 4F). These data suggest that the observed immunodominance of KF11 over KL9 responses in the ex vivo assays could be attributed to HLA-E restriction.
Next, we determined whether, at an intra-epitope level, the functionality of CD8+ T cells responding to the same peptide but presented by APCs expressing HLA-B*57 or HLA-E differed. Cumulative data show that KF11 elicited robust CD8+ T cell responses with comparable responder frequencies when presented by both APCs (n = 9; patient 1–patient 9, Supplemental Table 4 and Figure 5A). However, the magnitude of HLA-E–restricted responses was higher when assessed for dual function (CD107a plus other markers; Figure 5B) and when the functional and polyfunctional scores were determined using COMPASS (40) analysis (Figure 5, C–E). In contrast, ex vivo KL9-specific CD8+ T cell responses were largely restricted by B*57 and were polyfunctional in nature (Supplemental Figure 5).
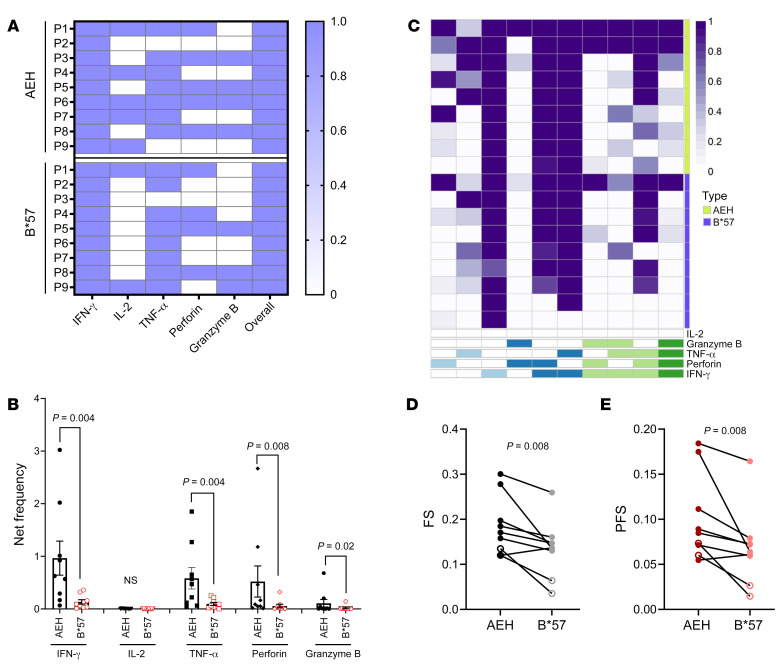
Figure 5. Ex vivo intracellular cytokine staining–based polyfunctional profile comparison of KF11-specific responses restricted by HLA-B*57 versus HLA-E in chronically HIV-infected individuals.
(A) Heatmap showing the KF11 response restricted by HLA-E or B*57 in each of the 9 individuals tested (positive response was assigned a value of 1 and negative response a value of 0). (B) Net frequency of production of cytokine/effector molecules by CD8+ T cells responding to KF11 presented by the AEH cell line and the K562.B*57 cell line. Net frequency was calculated by subtraction of the no-peptide-pulse control of the identical APCs. (C) Heatmap of COMPASS functional analysis. (D and E) Data for functional score (FS) and polyfunctional score (PFS). Error bars represent mean ± SEM. Wilcoxon matched-pairs signed rank test was used in B, D, and E to determine statistical significance.
We next determined the antigen sensitivity of HLA-E–restricted KF11 CD8+ T cell responses in comparison with the same peptide presented by HLA-B*57. Using log fold KF11 peptide dilutions (10–0.1 μg/mL), our data showed a dose-dependent response for HLA-E–restricted KF11 CD8+ T cells, while no significant decrease in responses was seen for HLA-B*57–restricted CD8+ T cells (n = 5; Supplemental Figure 6). These results were expected considering that HLA-E has a lower predicted peptide binding affinity for KF11 compared with B*57. Nevertheless, in 2 of the 5 individuals tested, E-restricted responses were still detectable at 0.1 μg/mL.
CD8+ T cells are known to cross-present antigens, so it is possible that epitopes are presented via the HLA-Ia of these cells. To rule out that cross-presentation plays a role in the observed antigen-specific CD8+ T cell response, we cultured CD8+ T cells in the presence of 10 μg/mL peptide without APCs. No significant CD8+ T cell response was detected in CD8+ T cell–only control as compared with those cocultured with peptide-pulsed AEH cells, demonstrating that cross-presentation did not account for the observed findings and indicating that the majority of the HLA-E–restricted specific CD8+ T cell responses aforementioned were stimulated by peptide presentation via APCs expressing single alleles (Supplemental Figure 7). HLA-E restriction was further validated in 1 patient by use of a B7sp blocking strategy ex vivo (Supplemental Figure 8). KF11-specific CD8+ T cell responses (CD107a/IFN-γ and CD107a/perforin) in the context of HLA-E but not HLA-B*57 were significantly diminished.
HIV-infected target cells expressing HLA-E and CD4 activate bulk CD8+ T cells ex vivo.
To determine whether HIV-infected targets expressing HLA-E only were capable of activating ex vivo–derived CD8+ T cells, we performed similar infectivity assays (Figure 3) using bulk CD8+ T cells rather than in vitro–expanded effector cell lines. In this assay, CD8+ T cells were activated as measured by upregulation of CD107a with IFN-γ, perforin, and granzyme B (n = 7; Figure 6, A and B). Although the frequency of activated T cells induced by HLA-E*01–expressing targets (E*03-induced responses did not achieve statistical significance as compared with the uninfected background) was lower in comparison with those responding to HLA-B*57–expressing targets, these data are in line with our findings using expanded CD8+ T cells that HLA-E–bound HIV peptides, loaded endogenously, induced T cell activation in ex vivo CD8+ T cells obtained from HIV-infected patients (Figure 6C).
Figure 6. Ex vivo–based bulk CD8+ T cell activation by HIV-1–infected target cells expressing HLA-E and CD4.
221ΔE cell lines expressing CD4 and either HLA-E*01:01, HLA-E*03:01, or HLA-B*57:01 were infected with NL4-3 virus (MOI = 0.5) for 12 hours, cultured for 48 hours, then cocultured with freshly isolated bulk CD8+ T cells from HIV-infected individuals (n = 7). CD8+ T cell production of IFN-γ alone or CD107a in combination with granzyme B, perforin, or IFN-γ was assessed after 12 hours of coculture with APCs. (A and B) HLA-E*01:01–, HLA-E*03:01–, and HLA-B*57:01–restricted CD8+ T cell responses from 2 representative individuals (patient 5 and patient 9; Supplemental Table 4). Red boxes indicate positive responses compared with uninfected controls. (C) Cumulative data from 7 individuals with both HLA-E*01:01– and HLA-B*57:01–restricted responses. Median values are shown, and error bars represent interquartile range. Data within groups were compared using Wilcoxon signed rank test.
Single-cell TCR sequencing shows distinct TCR-αβ clones restricted by HLA-E and HLA-B*57.
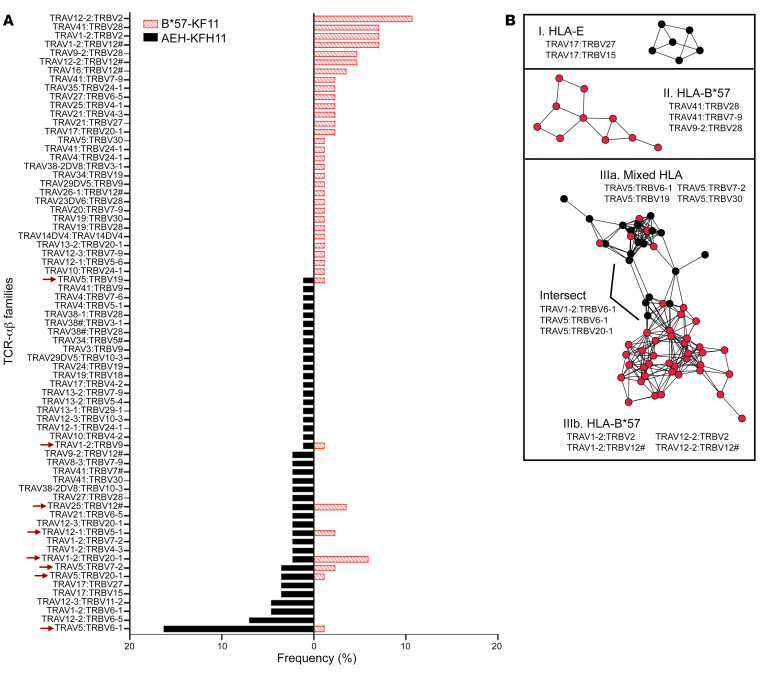
To determine whether the same CD8+ T cell responds to KF11 presented by either HLA-E or B*57, we evaluated the TCR usage from activated CD8+ T cells in the context of AEH or K562.B*57 cells pulsed with 10 μg/mL KF11 using CD8+ T cells isolated from a chronically infected individual. Epitope-specific CD8+ T cells were single-cell-sorted based on the expression of T cell–specific activation-induced markers, CD69 and CD137 (41), in an ex vivo assay. We next performed single-cell RNA sequencing to obtain paired TCR-αβ profiles. Our TCR sequencing results identified 79 TCR-αβ clones from CD8+ T cells activated by B*57 and AEH cells but with different frequencies (Figure 7A). Interestingly, only a small proportion of TCR clones (8 of 79) was shared between these 2 groups. To further investigate differences in TCR repertoire between HLA-E– and B*57-restricted CD8+ T cells, we examined similarity in TCR-αβ CDR3 sequences obtained from KF11-specific T cells using a bioinformatics tool, TCRdist2 (42), and visualized the results as a network graph (Figure 7B). Of 151 TCR-αβ sequences analyzed, 120 clones (79%) were linked with at least 1 partner in our data set, generating a total of 329 edges. TCR nodes were color-coded according to their observed HLA restriction (HLA-E, black; B*57, red), and the 3 largest TCR clusters, containing a total of 77 clones, are presented. These included one HLA-E–restricted cluster (I; n = 6 clones), one B*57-restricted cluster (II; n = 10 clones), and one extended cluster displaying a mixture of both HLA-E and B*57 restriction (III; n = 61 clones). Seventeen additional clusters containing 4 or fewer clones were not assessed further. Notably, clusters I and II were enriched for TCR variable genes identified as being mono-HLA-restricted in Figure 7A, including TRAV17-containing pairs for HLA-E and TRAV41-containing pairs for B*57. Cluster III formed a more extended network architecture, with 2 distinct subclusters displaying either a mixture of HLA-E– and B*57-restricted clones (IIIa) or predominantly B*57-restricted clones (IIIb), which were linked by edges between a relatively small number of clones (referred to as Intersect in Figure 7B). Each subcluster encoded distinct TCR variable genes, with IIIa dominated by TRAV15-containing clones and IIIb dominated by clones containing TRAV1-2 or TRAV12-2 and TRBV2 or TRBV12. Intersect clones encoded a mixture of these genetic features, but often contained alternative αβ variable gene pairings or distinct CDR3s. The genetic similarity of TCR sequences located in subcluster IIIa suggests that most of these clones may be cross-reactive for HLA-E and B*57; indeed, one identical TCR-αβ sequence that was found in both HLA-E– and B*57-stimulated T cells is located in subcluster IIIa, while a second identical sequence is located in the intersect of IIIa and IIIb. Taken together, our data suggest distinct TCR-αβ profiles as restricted by HLA-E and B*57 with a subset of clones that harbor the potential to cross-recognize KF11 presented by both HLA-I alleles.
Figure 7. Ex vivo single-cell-based TCR-αβ profiles of CD8+ T cells activated by KF11-pulsed AEH or B*57 target cells.
(A) The frequencies of TCR-αβ variable gene pairs used to recognize KF11 presented by either HLA-E or B*57. Arrows indicate cases where the same variable gene pair was isolated from both HLA-E– and B*57-restricted T cells. (B) Network graphs depict clusters of similar KF11-specific TCR-αβ sequences restricted by HLA-E only (black nodes; cluster I, top), B*57 only (red nodes; cluster II, middle), or both HLAs (cluster III, bottom), identified using TCRdist with a Hamming distance threshold of 150. Cluster III included subclusters of TCRs restricted by both HLAs (Mixed, IIIa) or B*57 only (IIIb), and a small number of clones that linked these subclusters (labeled Intersect). The predominant variable gene pairs present within each cluster and subcluster are indicated.
HLA-E–restricted KL9- and KF11-specific responses can be generated by in vitro priming of CD8+ T cells obtained from HIV-seronegative donors.
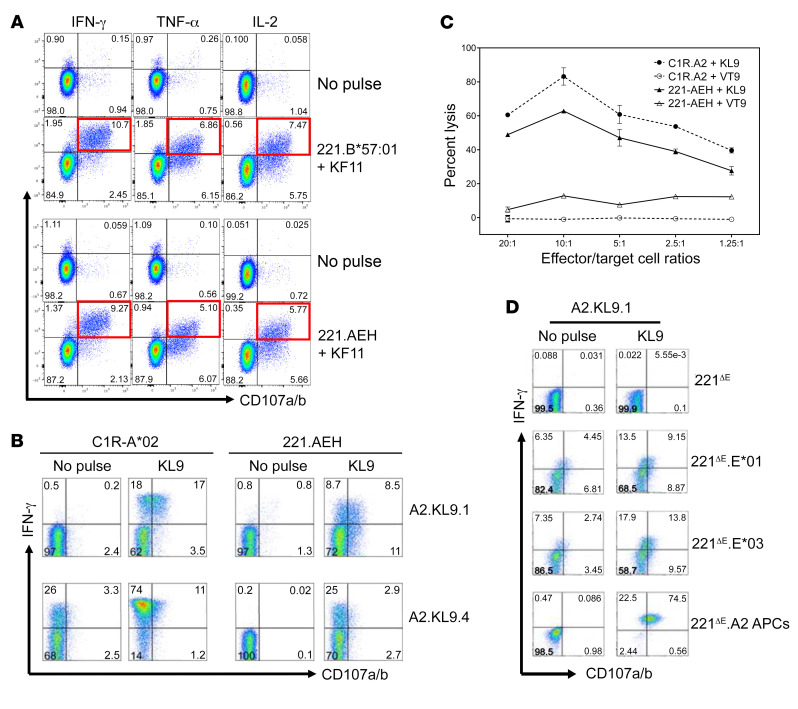
Finally, as HLA-E–restricted CD8+ T cells may have important implications for HIV-1 vaccines (30, 43–45), we next determined whether such responses can be primed in HIV-seronegative donors as shown in a recent study (44). We generated HLA-E–restricted KF11-specific CD8+ T cell lines by in vitro priming using PBMCs obtained from 3 HIV-seronegative healthy donors. HLA-E*01–restricted responses were detected in 1 of 3 donors. Data in Figure 8A show functional characterization of KF11-specific polyfunctional responses of CD8+ T cell lines that were restricted by peptide (10 μg/mL) presented by both HLA-Ia and -Ib allomorphs. Similarly, HLA-E*01–restricted KL9-specific CD8+ T cell lines were successfully generated from 2 HIV-seronegative donors (Figure 8B). HLA-A*02:01– and HLA-E*01:01–restricted KL9-specific responses demonstrated degranulation and IFN-γ production in A*02.KL9.1 and A*02.KL9.4 CD8+ T cell lines. KL9-specific lysis of cognate peptide–pulsed C1R-A*02 and 221.AEH target cells by A*02.KL9.1 T cells at multiple effector/target cell ratios was observed as determined by the chromium release assay (Figure 8C). Coculture of peptide-pulsed 221ΔE, 221ΔE.E*01:01, 221ΔE.E*01:03, or 221ΔE.A*02 cells with A*02-restricted KL9-specific CD8+ T cells resulted in upregulation of surface CD107a/b expression and intracellular IFN-γ production (Figure 8D).
Figure 8. In vitro priming and functional characterization of HLA-E*01–restricted KF11- and KL9-specific CD8+ T cells obtained from 2 HIV-seronegative donors.
(A) KF11-specific polyfunctional responses of CD8+ T cell lines from 2 donors that are restricted by both HLA-Ia (221.B*57:01) and HLA-Ib (221.AEH) allomorphs. (B) HLA-A*02:01–restricted (C1R-A*02 used as APCs) and HLA-E–restricted (221.AEH used as APCs) KL9-specific degranulation and IFN-γ production by A*02:01-restricted KL9-specific CD8+ T cell clones (A2.KL9.1 and A2.KL9.4.) from different donors. (C) KL9-specific lysis of cognate peptide–pulsed (10 μg/mL) C1R.A2 and 221.AEH target cells by A2.KL9.1 T cells as determined by the chromium release assay. Error bars represent mean ± SEM. (D) Upregulation of surface CD107a/b expression and intracellular IFN-γ production in the A*02-restricted KL9-specific CD8+ T cells after stimulation by peptide-pulsed 221ΔE, 221ΔE.E*01, 221ΔE.E*03, or 221ΔE.A2 APCs. No response or low-level responses were observed with the no-peptide-pulse negative control.
Discussion
The role of HLA-E–restricted CD8+ T cell responses has not been explored despite the potential advantages that this arm of adaptive immunity can offer in the setting of HIV infection and vaccination. For example, HLA-E–restricted CD8+ T cell responses can enhance response breadth, a qualitative feature that has been associated with HIV control (46, 47); circumvent interindividual heterogeneity of CD8+ T cell responses associated with the highly polymorphic HLA-Ia alleles (36, 48); and mitigate immune evasion strategies commonly associated with HLA-Ia–restricted T cell responses. Thus, targeting HLA-E–restricted CD8+ T cells could be beneficial for the development of a globally effective HIV vaccine, as the dimorphic HLA-E allele can perhaps induce similar antigenic responses across most vaccinees regardless of their HLA-Ia genotype.
In this study, we describe HLA-E–restricted HIV-specific CD8+ T cell responses toward 2 Gag epitopes, a subdominant KL9 and an immunodominant KF11 epitope, that can be detected in chronically HIV-infected individuals (both ex vivo and in vitro) and also be primed in vitro from HIV-seronegative individuals. The latter has important implications for the induction of HLA-E–restricted CD8+ T cells in HIV vaccines. The HLA-E–restricted CD8+ T cell responses observed in our study are non-NKT CD8+ T cell responses that are multifunctional with effector characteristics. In addition, HLA-E–restricted CD8+ T cells can recognize KF11 not only when presented exogenously but also when loaded endogenously within an infected cell. Prior work showed that eluted HLA-E peptidome encompassed a large assortment of peptides, ranging from short (8- to 10-mer) to longer peptides (11- to 17-mer) (5, 11, 23, 49). Moreover, eluted peptides had hydrophobic amino acids on p2 and p9 consistent with a binding motif that is similar to HLA-A*02 (11), and prior work showed that structural recognition of HLA-E overlaps with that of HLA-Ia (50). Based on the above, it is not surprising that KL9 and KF11 can bind both HLA-E and HLA-Ia molecules. Although we only examined 2 Gag epitopes for HLA-E–restricted responses, based on findings from other infectious pathogens, it is likely that HLA-E allele presents a yet-to-be-characterized broad repertoire of epitopes in Gag as well as other HIV-1 protein–derived epitopes in HIV infection. Recent work by Hannoun et al. (36) identified 2 subdominant epitopes, derived from Vif and Pol, encoded in the conserved HIV vaccine immunogen with an inferred HLA-E restriction in addition to its documented presentation via HLA-Ia allele, although the former was not validated experimentally. HLA-E–specific reagents, as described here, will be key for such future studies to evaluate the protein-specific targeting of HLA-E–restricted responses and determine their biological role.
Several in silico HLA-I binding prediction tools have been commonly used to identify potential peptides capable of eliciting immune responses. Experimental validation is important as binding does not always equate with immunogenicity. In fact, prior macaque studies showed that only a minor fraction of MHC-E–restricted peptides exhibited canonical MHC-E binding motifs (30). For HIV, 5 peptides (3 Gag and 1 each in Pol and Vif) have been reported to bind HLA-E (35, 36, 51). However, no study has determined the recognition of such peptide/HLA-E complexes by antiviral CD8+ T cells. One possible reason for this lack of immune data could be the use of PBMCs rather than enriched CD8+ T cells in coculture assays. Intriguingly, recent work showed that epitopes against which HLA-E–restricted CD8+ T cell responses were identified did not, however, exhibit HLA-E binding or showed lower binding affinity (37, 52, 53). This was observed for M. tuberculosis and SIV, and in the latter case these responses were, surprisingly, those associated with protection. Thus, binding of peptides to HLA-E may not be the sole determinant of the elicitation, nor of the strength, of an HLA-E–restricted response. In our study, we show that KL9 and KF11 peptides bind HLA-E at levels similar to or slightly lower than B7 signal peptide (Supplemental Figure 1). Future work will delineate the mechanisms that facilitate the binding and stabilization of low- to medium-affinity HIV-1 peptides to HLA-E to induce a CD8+ T cell response.
Studies in CMV (12) showed the presence of CMV-UL40–derived HLA-E–restricted CD8+ T cell responses, some of which could be as frequently targeted as those restricted by HLA-Ia alleles. In our ex vivo analyses, KF11-specific responses exhibited higher magnitude and functional scores than those specific for KL9. Among KF11-specific responses, those restricted by HLA-E were higher in magnitude and showed enhanced functionality/polyfunctionality compared with those restricted by HLA-B*57. On the contrary, KL9, although recognized by HLA-E, was mainly presented by B*57 allele. These intriguing data suggest that immunodominance of KF11-specific CD8+ T cell responses could perhaps be due to a synergistic contribution of both HLA-Ia– and HLA-Ib–mediated peptide presentations. Interestingly, even though ex vivo HLA-E–restricted KF11 specific responses were higher than those restricted by HLA-B*57 (at 10 μg/mL peptide concentration), an opposite trend was seen in in vitro assays. These data suggest that although effector HLA-E–restricted CD8+ T cells are readily detected and could play a role in viral control, they do not appear to proliferate to the same extent as those specific for HLA-Ia. One plausible reason for this observation may be the relatively shallow versus tight binding of KF11 in the peptide binding groove of HLA-E and B*57, respectively. Nevertheless, HLA-E–restricted CD8+ T cells can serve as an additional source of antiviral effectors against a pliable pathogen that readily escapes innate and adaptive immune recognition. A caveat regarding our ex vivo and in vitro assays is the use of 10 μg/mL peptide for presentation by both HLA-E– and B*57-expressing targets. Although this peptide or an even higher concentration is commonly used (44, 54–57), a prior study indicated that even 1 μg/mL peptide is superphysiological (58). Nevertheless, our data from this assay show that HLA-E–restricted T cell responses were detectable at peptide concentration as low as 0.1 μg/mL. Our observation of CD8 activation induced by HLA-E–expressing HIV-infected targets overcomes the biases associated with exogenous peptide loading and provides compelling evidence that HLA-E–restricted CD8+ T cells are induced during HIV infection. Specifically, our data show that endogenously HLA-E–loaded HIV-1 peptides can activate antigen-specific and bulk CD8+ T cells to induce multiple effector molecules.
Prior work from Hansen et al. in a nonhuman primate model (30) suggested that MHC-E–restricted CD8+ T cell responses are poorly primed in SIV infection. Based on our data on 2 Gag epitopes in HIV, we detected very few CD8+ T cell responses restricted only by HLA-E; most responses were also restricted by HLA-Ia alleles. In contrast to KF11, KL9 responses were more readily detected after in vitro expansion, perhaps owing to the subdominant nature of the epitope and also to their low frequency ex vivo. In support of the latter, Hannoun et al. (36) reported infrequent and low-magnitude response to an in vitro–expanded culture specific for a subdominant epitope with a possible HLA-E restriction that was not unequivocally established. In comparison with HIV, data from SIV/macaque studies (30) showed low dual recognition (25%) of both MHC-Ia– and MHC-E–restricted CD8+ T cell responses.
Based on prior work, mainly in M. tuberculosis, HLA-E–restricted CD8+ T cell responses are likely to be pleiotropic (helper, cytotoxic, and/or regulatory), with functional features that are unique and/or shared with responses restricted by HLA-Ia alleles. Specifically, in M. tuberculosis–infected individuals, HLA-E–restricted CD8+ T cells displayed a unique functional profile compared with HLA-A*02–restricted CD8+ T cells (25). However, functional and phenotypic characterization of HLA-E–restricted CD8+ T cells in other infectious diseases is largely limited (5, 14). In the current study, in both ex vivo and in vitro assays, we found that HLA-B*57– and HLA-E–restricted KF11 responses elicited similar response rates of effector functions such as IFN-γ, IL-2, TNF-α, perforin, granzyme B, and CD107a, but the magnitude of such responses and the polyfunctional profile varied. Future work to comprehensively study the characteristics of HIV-specific CD8+ T cell response induced by HLA-E would provide important insights into the biological relevance of this cell subset and be useful for T cell–based vaccine as well as therapeutic strategies.
While M. tuberculosis–specific HLA-E tetramers were used in 2 prior studies (23, 25), our attempts to synthesize HIV-specific HLA-E tetramers in this work failed as a result of instability issues. Nevertheless, we used a CRISPR knockout approach to discriminate between HLA-Ia– and HLA-Ib–restricted responses. In both ex vivo and in vitro assays, enriched CD8+ T cells were stimulated with peptide-pulsed target cells overexpressing HLA-E, i.e., 721.221.AEH. It should be noted, however, that 721.221.AEH cells also express a variety of other nonclassical MHC class I antigens, such as HLA-F and -G (59, 60), which can present peptides and stimulate CD8+ T cells. While most cultured tumor cell lines express HLA-E, including the parental 721.221 cell line, our HLA-E–knockout and single-allele transductant cell lines mitigated this caveat and will pave the way for the future conduct of comprehensive studies to determine the full extent of HLA-E–restricted HIV-specific CD8+ T cell repertoire targeting.
In summary, dually HLA class Ia– and class Ib–restricted, i.e., HLA-E–restricted responses elicited against 2 HIV-1–derived Gag epitopes were frequently observed both ex vivo and in vitro. The HLA-E–restricted responses were not mediated by NK/NKT CD8+ T cells and were polyfunctional. Importantly, these cells were capable of recognizing endogenously loaded HIV-1 peptide and were activated by the latter targets to produce multiple effector molecules. Future studies should examine the extent of HLA-E targeting across the HIV proteome and focus on mechanisms involved in the elicitation of these cells in the context of HIV infection. Furthermore, it is important to understand what role HLA-E–restricted CD8+ T cells are playing in preventing and controlling HIV infection and whether HIV-1 vaccines, including the CMV vector–based vaccine, can induce these responses. Further development of HLA-E–specific reagents promises to enhance our understanding of the biological relevance of HLA-E–restricted CD8+ T cells in infection and vaccination.
Methods
Further information can be found in Supplemental Methods.
Study cohort and HLA-I genotyping
PBMCs were collected from 8 HIV-seronegative volunteers (University of Texas at El Paso and University of Alabama at Birmingham) and 20 chronically HIV-1–infected individuals (University of Alabama at Birmingham). HLA class I alleles were resolved by bidirectional sequencing of exons 2–4 at each locus. Supplemental Table 2 lists the clinical parameters and HLA class Ia and Ib alleles of the HIV-infected individuals.
Cell lines
Human B lymphoblastoid cell line 721.221 (abbreviated here as 221) is devoid of functional classical HLA class Ia genes owing to mutations in the HLA complex. However, 221 cell lines can be readily transfected to induce cell surface HLA-A/B/C expression (61). HLA-E is expressed at the mRNA level, but the surface expression is lacking, and the latter was induced by transfection of a DNA construct (HLA-E*01:01 fused to the HLA-A*02:01 leader peptide) with the resulting cell line termed 721.221.AEH or AEH (9). Both 221 and AEH cell lines were provided by Dan Geraghty (Fred Hutchinson Cancer Research Center, Seattle, Washington, USA). For HLA-E studies, in addition to 221-derived AEH, other HLA-Ia–null cell lines such as K562 (62) were used in which HLA-Ia/b alleles (63) were selectively transfected. 221 cells were cultured in R20, whereas 221.AEH cells and K562 cells were cultured in R20 supplemented with 200 mU/mL hygromycin B (Sigma-Aldrich) and 0.2 mg/mL Geneticin (Corning). Other cell lines used in this study for assessing T cell specificity included peptide-pulsed T2 cells (ATCC CRL-1992) and C1R cells expressing a full-length HLA-A*02:01 (C1R.A2) (64) maintained in RPMI 1640 medium supplemented with 10% heat-inactivated FBS (R-10).
Peptides
The peptides KF11 (KAFSPEVIPMF, Gag [162–172]), KL9 (KALGPAATL, Gag [335–343]), IW9 (ISPRTLNAW, Gag [147–155]), HLA-B*57–binding NS1 (HTWTEQYKF, dengue virus NS1 [26–34]) (65), HLA-A*02:01–binding VT9 (VLSEWLPVT, Rift Valley fever virus nucleocapsid protein N [121–129]) (64), and B7sp (VMAPRTVLL, HLA-B*07:01 [3–11]) (63, 66) were purchased from Genemed Synthesis. HIV-1 consensus B Gag peptide set was obtained from the NIH AIDS Reagent Program (8117, lot 130047). All single peptides were used at 10 μg/mL unless otherwise specified.
In vitro expansion of KL9- and KF11-specific CD8+ T cells from chronically HIV-infected individuals
CD8+ T cells were positively selected from freshly thawed PBMCs obtained from HIV-infected individuals (Dynabeads CD8 Positive Isolation Kit, Thermo Fisher Scientific). The CD8-negative fraction, used as a source for autologous APCs, was irradiated (3000 cGy), plated at 1.25 × 106 cells per well of 48-well cluster plates, and pulsed for 3 hours at 37°C in serum-free RPMI containing 10 μg/mL of the KL9 and/or KF11 peptide, 3 μg/mL of human β2-microglobulin (MP Biomedicals), and 1% human albumin (Sigma-Aldrich). Exogenous β2-microglobulin was added as it governs the stability of the MHC complex (67, 68) to impact T cell response. After removal of nonadherent cells, half a million purified CD8+ T cells were added to each well 1, 4, 7, and 10 days later in complete medium (RPMI 1640 with 10% autologous serum, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 2 mM l‑glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin) supplemented with IL-7 (10 ng/mL; Genzyme), IL-2 (20 U/mL; Proleukin, Prometheus Laboratories Inc.), and IL-4 (250 U/mL). CD8+ T cells were assessed for antigen specificity on days 7–12 after expansion.
In vitro priming of KL9- and KF11-specific CD8+ T cells from HIV-1–seronegative donors
Using PBMCs, peptide-specific CD8+ T cells were primed as described previously (64, 69). Briefly, monocyte-derived DCs were cultured for 7 days in a complete medium supplemented with GM-CSF (1000 U/mL; Leukine [sargramostim], Bayer HealthCare Pharmaceuticals) and IL-4 (500 U/mL; PeproTech). Irradiated DCs (3000 cGy) were pulsed with 10 μg/mL peptide in RPMI containing β2-microglobulin at 100 μg/mL and 1% human albumin for 3 hours and used to prime positively selected autologous CD8+ T cells (purity >98%; Dynabeads, Invitrogen) at a T cell/DC ratio of 5:1. CD8+ T cells were restimulated every 7 days thereafter with cognate peptide–pulsed autologous monocytes, and IL-7 (10 ng/mL) was added on the day of priming and restimulation; IL-2 (20 U/mL) was added on day 1 and every 3–4 days later.
Ex vivo IFN-γ ELISPOT
ELISPOT assay was performed as previously described (70). Nitrocellulose plates (MilliporeSigma) coated overnight with anti–IFN-γ antibody were blocked with R-10 medium for 2 hours. PBMCs were washed and rested overnight at 37°C and 5% CO2. Cells (105 cells per well) were plated in triplicate and stimulated with peptide (10 μg/mL) for 22–24 hours. Cells cultured in medium without peptide and in medium with phytohemagglutinin (PHA) (5 μg/mL) were used as negative and positive controls, respectively. Plates were washed and treated with biotinylated anti–IFN-γ antibody for 2 hours followed by streptavidin–alkaline phosphatase for 1 hour, and finally developed with the NBT/BCIP substrate for 5–10 minutes. Counts were determined by CTL ImmunoSpot analyzer (version 5). Data were normalized to spot-forming units per 106 cells (SFU/106). A positive response (71, 72) was defined as at least 55 SFU/106 cells and at least 4 times background (negative control).
Detection of HLA-Ia/b–restricted CD8+ T cell responses
In vitro intracellular cytokine staining.
Briefly, expanded CD8+ T cell effector function was assessed by coculturing of these cells with peptide-pulsed (10 μg/mL) target cells expressing single HLA-I alleles for 6 hours in the presence of costimulatory molecules and anti-CD107a–FITC as well as monensin and brefeldin A. For in vitro HLA-E blocking assay, APCs were pretreated with either HLA-E–specific peptide (mAb 3D12, 10 μg/mL) or B7sp (20 μg/mL) for 2 hours before peptide pulse as mentioned above. Cells were surface-stained at 4°C for 30 minutes and washed before fixation and permeabilization using Cytofix/Cytoperm (BD Biosciences). Cells were stained intracellularly with anti–IFN-γ, anti–IL-2, anti–TNF-α, and anti–MIP-1β at 4°C for 30 minutes. Cells were acquired on an LSR II flow cytometer (BD Immunocytometry Systems) and data analyzed by FlowJo (Tree Star Inc.). Gating for CD8+ T cells was performed on small lymphocytes, singlets, and viable CD8+ T cells. Between 50,000 and 100,000 and between 40,000 and 60,000 live CD8+ events were collected per sample of cell lines and cultured T cells, respectively, from infected donors. Criteria for a positive response using an intracellular cytokine staining assay were determined using χ2 with Yates’s correction, P < 0.0001.
Ex vivo intracellular cytokine staining.
Untouched CD8+ T cells were isolated from PBMCs of chronically HIV-infected individuals with detectable antigen-specific CD8+ T cell response as measured by ex vivo ELISPOT assay. Cell lines 721.221.AEH (HLA-E) and K562.B*57:01 (HLA-B*57) were used as APCs. For ex vivo HLA-E blocking assay, APCs were pretreated with B7sp (10, 20, 50 μg/mL) for 2 hours. APCs were pulsed with peptide (10 μg/mL) for 2 hours and washed 3 times before being added to the CD8+ T cells at a 1:1 ratio. Costimulatory molecules (anti-CD28 and anti-CD49d), anti-CD107a–FITC antibodies, and monensin and brefeldin A (5 μg/mL; eBioscience) were added and the cells cocultured for 12 hours. Cell surface and intracellular cytokine/effector molecule production was assessed as described in Supplemental Methods.
HIV-1 infectivity assay
HIV-1 infectivity assays were conducted using in vitro–generated antigen-specific CD8+ T cell lines and bulk ex vivo–isolated CD8+ T cells. In vitro–cultured CD8+ T cells were stimulated with peptide-pulsed (10 μg/mL) or NL4-3–infected target cells at a T cell/target cell ratio of 1:1 in the presence of brefeldin A (5 μg/mL) solution for 6 hours. Target cells were spinoculated (1065g for 2 hours at 32°C) with NL4-3 virus containing supernatants using a volume that infected 90% of T1 cells in 24 hours. T1 (174 × CEM.T1) (ATCC CRL-1991 Homo sapiens) is the parental line for the T2 line (174 × CEM.T2) (ATCC CRL-1992). Cells were washed twice 12 hours later and cultured for up to 4 days. Productive infection was monitored by intracellular p24 staining with the mAb phycoerythrin (Kc57) (Beckman Coulter).
For the ex vivo–based assay, each target cell was resuspended in 10% R-10 medium (RPMI plus 10% FBS), and 200,000 cells per tube in a total volume of 100 μL were spinfected, washed twice 12 hours later, transferred to a 96-well V-bottom plate, and cultured for an additional 48 hours. Freshly isolated CD8+ T cells were added to washed targets at an effector/target cell ratio of 1:1 and cocultures incubated for 12 hours at 37°C, 5% CO2 in the presence of anti-CD28, anti-CD49d, CD107a (FITC), GolgiStop, and GolgiPlug (BD Biosciences). Cells were washed 2 times with FACS wash (PBS plus 2% FBS). Surface staining (anti-CD3, anti-CD8, anti-CD19, and dead cell dye) was performed for 30 minutes at 4°C. After washing once with FACS wash buffer, cells were permeabilized with Cytofix/Cytoperm. Next, cells were washed 2 times with Perm/Wash buffer (BD Biosciences), and intracellular cytokine staining was performed using antibodies against IFN-γ, TNF-α, perforin, and granzyme B. Cells were fixed in 4% formalin and analyzed on a FACSymphony (BD Biosciences), and at least 300,000 events were acquired.
Cytotoxicity assay
Target cells were labeled with sodium chromate (51Cr; PerkinElmer), pulsed with KL9 (10 μg/mL) for 1 hour at 37°C, washed, and admixed with cultured T cells at various effector/target cell ratios for 4 hours. Released 51Cr in the supernatants was determined with a MicroBeta counter (PerkinElmer). An irrelevant peptide was used as the control for spontaneous release. Specific percentage lysis was calculated as [(cpm experimental – cpm spontaneous)/(cpm total – cpm spontaneous)] × 100.
Ex vivo activation-induced marker–based single-cell sorting and TCR sequencing
Activation-induced marker–based (AIM-based) single-cell TCR sequencing was performed as previously described (73). PBMCs were peptide-pulsed (KF11, 10 μg/mL) in the presence of anti-CD28 and anti-CD49d for 18 hours. Cells were surface-stained for 30 minutes at 4°C with LIVE/DEAD Aqua stain (Thermo Fisher Scientific), anti-CD3–Alexa Fluor 780, anti-CD8–FITC, anti-CD19–PerCP/Cy5.5, anti-CD137–phycoerythrin, and anti-CD69–allophycocyanin CD69+CD137+ CD8+ T cells were identified as activated CD8+ T cells (41), and single cells were sorted directly into 96-well plates containing 3 μL lysis buffer (RNaseOUT [Thermo Fisher Scientific], dNTPs, and X-100) using a FACS sorter (BD Immunocytometry Systems).
TCR sequencing was performed at the Institute for Immunology and Infectious Diseases in Perth, Australia. Single-cell lysates underwent oligo-dT–primed reverse transcription. The assay uses uniquely tagged primers for reverse transcription and template switching with a preamplification step to increase the yield and transcript length of the single-cell cDNA library. Samples were sequenced on an Illumina MiSeq using a 2 × 300 bp paired-end chemistry kit (Illumina Inc.). Reads were quality-filtered and passed through a demultiplexing tool to assign reads to individual wells and mapped to the TCRB and TCRA loci. TCR clonotypes were assigned using MiXCR software before analysis.
TCR clustering analysis
TCR-αβ sequences were examined using the bioinformatics tool TCRdist2 (https://github.com/kmayerb/tcrdist2) (42), implemented in Python version 3.9. TCRdist calculates a Hamming distance between pairs of TCR sequences based on amino acid similarity in complementarity-determining regions (CDRs) 1, 2, 2a, and 3 of the α and β chains, with penalties for gaps and additional weight given to both CDR3 motifs. The resulting matrix of pairwise TCR distance values was imported into Cytoscape version 3.8 (74) using the aMatReader plug-in and visualized as a network graph (yFiles Organic Layout, default parameters) with edges filtered at an upper threshold of 150 distance units.
Statistics
Data were analyzed using Fisher’s exact test, χ2 with Yates’s correction, and Wilcoxon ranked test (2-tailed) for paired comparison. GraphPad Prism version 8.0 software was used to perform these analyses. Significance was determined as P value less than 0.05 unless otherwise stated.
Study approval
All subjects enrolled for this study provided written informed consent, and the study procedures were approved by the Institutional Review Boards at the University of Texas at El Paso and the University of Alabama at Birmingham.
Author contributions
AB, JKM, and PAG conceptualized and designed the study. MNG, KQ, SS, AA, YD, SA, MCC, LAV, JT, and NM conducted experiments and/or curated data. AB, KQ, OOY, JKM, PAG, and MB conducted formal data analysis. JKM, PAG, and AB acquired funding. JKM, PAG, and AB supervised the study. AB, MNG, KQ, JKM, and PAG wrote the manuscript. SS, AA, YD, SA, MCC, LAV, JT, NM, OOY, and MB reviewed and edited the manuscript. The order of the 3 co–first authors is based on contribution to study conception and design, experimental conduct, data analysis, and/or manuscript preparation.
Supplementary Material
Acknowledgments
We thank Spyros Kalams and Simon Mallal (University of Vanderbilt) for providing us with the K562.B*57-expressing cell line. We also thank Stephen Migueles and Mark Conners for providing patient samples, and Dana Indihar for her help with the ELISPOT assays. This work was funded by NIH grants R01-AI102663 (to JKM) and R01-AI162168 (to AB and PAG).
Version 1. 07/06/2021
In-Press Preview
Version 2. 08/16/2021
Electronic publication
Funding Statement
AB and PAG
JKM
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(16):e148979.https://doi.org/10.1172/JCI148979.
Contributor Information
Anju Bansal, Email: anjubansal@uabmc.edu.
Mika N. Gehre, Email: Mika.gehre@gmail.com.
Kai Qin, Email: qinkai@uab.edu.
Sarah Sterrett, Email: sarahsterrett@uabmc.edu.
Ayub Ali, Email: aali@mednet.ucla.edu.
Ying Dang, Email: dangying@kanglinbio.com.
Sojan Abraham, Email: sojan.abraham@fda.hhs.gov.
Margaret C. Costanzo, Email: mcostanzo@hivresearch.org.
Leon A. Venegas, Email: leon.venegas@astrazeneca.com.
Jianming Tang, Email: jtang@uab.edu.
N. Manjunath, Email: pariswamy100@gmail.com.
Mark A. Brockman, Email: mark_brockman@sfu.ca.
Otto O. Yang, Email: oyang@mednet.ucla.edu.
June Kan-Mitchell, Email: jkanmitchell@utep.edu.
Paul A. Goepfert, Email: paulg@uab.edu.
References
- 1.Jin X, et al. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189(6):991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ndhlovu ZM, et al. Magnitude and kinetics of CD8+ T cell activation during hyperacute HIV infection impact viral set point. Immunity. 2015;43(3):591–604. doi: 10.1016/j.immuni.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitz JE, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283(5403):857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 4.Robinson J, et al. IPD — the immuno polymorphism database. Nucleic Acids Res. 2013;41(D1):D1234–D1240. doi: 10.1093/nar/gks1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joosten SA, et al. Characteristics of HLA-E restricted T-cell responses and their role in infectious diseases. J Immunol Res. 2016;2016:2695396. doi: 10.1155/2016/2695396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez J, et al. Expression of HLA-E molecules in the placental tissue of women infected with HIV-1 and uninfected women. Placenta. 2017;49:33–36. doi: 10.1016/j.placenta.2016.08.082. [DOI] [PubMed] [Google Scholar]
- 7.Pietra G, et al. HLA-E and HLA-E-bound peptides: recognition by subsets of NK and T cells. Curr Pharm Des. 2009;15(28):3336–3344. doi: 10.2174/138161209789105207. [DOI] [PubMed] [Google Scholar]
- 8.Petrie EJ, et al. CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence. J Exp Med. 2008;205(3):725–735. doi: 10.1084/jem.20072525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee N, et al. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol. 1998;160(10):4951–4960. [PubMed] [Google Scholar]
- 10.Abdulhaqq SA, et al. Vaccine-mediated inhibition of the transporter associated with antigen processing is insufficient to induce major histocompatibility complex E-restricted CD8+ T cells in nonhuman primates. J Virol. 2019;93(19):e00592-19. doi: 10.1128/JVI.00592-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lampen MH, et al. Alternative peptide repertoire of HLA-E reveals a binding motif that is strikingly similar to HLA-A2. Mol Immunol. 2013;53(1–2):126–131. doi: 10.1016/j.molimm.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Jouand N, et al. HCMV triggers frequent and persistent UL40-specific unconventional HLA-E-restricted CD8 T-cell responses with potential autologous and allogeneic peptide recognition. PLoS Pathog. 2018;14(4):e1007041. doi: 10.1371/journal.ppat.1007041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allard M, et al. HLA-E-restricted cross-recognition of allogeneic endothelial cells by CMV-associated CD8 T cells: a potential risk factor following transplantation. PLoS One. 2012;7(11):e50951. doi: 10.1371/journal.pone.0050951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazzarino P, et al. Identification of effector-memory CMV-specific T lymphocytes that kill CMV-infected target cells in an HLA-E-restricted fashion. Eur J Immunol. 2005;35(11):3240–3247. doi: 10.1002/eji.200535343. [DOI] [PubMed] [Google Scholar]
- 15.Pietra G, et al. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc Natl Acad Sci U S A. 2003;100(19):10896–10901. doi: 10.1073/pnas.1834449100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romagnani C, et al. HLA-E-restricted recognition of human cytomegalovirus by a subset of cytolytic T lymphocytes. Hum Immunol. 2004;65(5):437–445. doi: 10.1016/j.humimm.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan LC, et al. The presence of HLA-E-restricted, CMV-specific CD8+ T cells in the blood of lung transplant recipients correlates with chronic allograft rejection. PLoS One. 2015;10(8):e0135972. doi: 10.1371/journal.pone.0135972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulte D, et al. The HLA-E(R)/HLA-E(R) genotype affects the natural course of hepatitis C virus (HCV) infection and is associated with HLA-E-restricted recognition of an HCV-derived peptide by interferon-gamma-secreting human CD8(+) T cells. J Infect Dis. 2009;200(9):1397–1401. doi: 10.1086/605889. [DOI] [PubMed] [Google Scholar]
- 19.Jorgensen PB, et al. Epstein-Barr virus peptide presented by HLA-E is predominantly recognized by CD8(bright) cells in multiple sclerosis patients. PLoS One. 2012;7(9):e46120. doi: 10.1371/journal.pone.0046120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burwitz BJ, et al. MHC-E-restricted CD8+ T cells target hepatitis B virus-infected human hepatocytes. J Immunol. 2020;204(8):2169–2176. doi: 10.4049/jimmunol.1900795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia P, et al. Human T cell receptor-mediated recognition of HLA-E. Eur J Immunol. 2002;32(4):936–944. doi: 10.1002/1521-4141(200204)32:4<936::AID-IMMU936>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.Pietra G, et al. The analysis of the natural killer-like activity of human cytolytic T lymphocytes revealed HLA-E as a novel target for TCR alpha/beta-mediated recognition. Eur J Immunol. 2001;31(12):3687–3693. doi: 10.1002/1521-4141(200112)31:12<3687::AID-IMMU3687>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 23.McMurtrey C, et al. T cell recognition of Mycobacterium tuberculosis peptides presented by HLA-E derived from infected human cells. PLoS One. 2017;12(11):e0188288. doi: 10.1371/journal.pone.0188288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prezzemolo T, et al. Functional signatures of human CD4 and CD8 T cell responses to mycobacterium tuberculosis. Front Immunol. 2014;5:180. doi: 10.3389/fimmu.2014.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prezzemolo T, et al. Detailed characterization of human Mycobacterium tuberculosis specific HLA-E restricted CD8+ T cells. Eur J Immunol. 2018;48(2):293–305. doi: 10.1002/eji.201747184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salerno-Goncalves R, et al. Ex vivo kinetics of early and long-term multifunctional human leukocyte antigen E-specific CD8+ cells in volunteers immunized with the Ty21a typhoid vaccine. Clin Vaccine Immunol. 2010;17(9):1305–1314. doi: 10.1128/CVI.00234-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fresnay S, et al. Salmonella Typhi-specific multifunctional CD8+ T cells play a dominant role in protection from typhoid fever in humans. J Transl Med. 2016;14:62. doi: 10.1186/s12967-016-0819-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Meijgaarden KE, et al. Human CD8+ T-cells recognizing peptides from Mycobacterium tuberculosis (Mtb) presented by HLA-E have an unorthodox Th2-like, multifunctional, Mtb inhibitory phenotype and represent a novel human T-cell subset. PLoS Pathog. 2015;11(3):e1004671. doi: 10.1371/journal.ppat.1004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen SG, et al. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science. 2013;340(6135):1237874. doi: 10.1126/science.1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen SG, et al. Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science. 2016;351(6274):714–720. doi: 10.1126/science.aac9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen SG, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473(7348):523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen SG, et al. Immune clearance of highly pathogenic SIV infection. Nature. 2013;502(7469):100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fruh K, Picker L. CD8+ T cell programming by cytomegalovirus vectors: applications in prophylactic and therapeutic vaccination. Curr Opin Immunol. 2017;47:52–56. doi: 10.1016/j.coi.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu HL, et al. The role of MHC-E in T cell immunity is conserved among humans, rhesus macaques, and cynomolgus macaques. J Immunol. 2018;200(1):49–60. doi: 10.4049/jimmunol.1700841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nattermann J, et al. HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antivir Ther. 2005;10(1):95–107. doi: 10.1177/135965350501000107. [DOI] [PubMed] [Google Scholar]
- 36.Hannoun Z, et al. Identification of novel HIV-1-derived HLA-E-binding peptides. Immunol Lett. 2018;202:65–72. doi: 10.1016/j.imlet.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walters LC, et al. Detailed and atypical HLA-E peptide binding motifs revealed by a novel peptide exchange binding assay. Eur J Immunol. 2020;50(12):2075–2091. doi: 10.1002/eji.202048719. [DOI] [PubMed] [Google Scholar]
- 38.Lee N, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci U S A. 1998;95(9):5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akinsiku OT, et al. Interleukin-2 production by polyfunctional HIV-1-specific CD8 T cells is associated with enhanced viral suppression. J Acquir Immune Defic Syndr. 2011;58(2):132–140. doi: 10.1097/QAI.0b013e318224d2e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boppana S, et al. Cross-reactive CD8 T-cell responses elicited by adenovirus type 5-based HIV-1 vaccines contributed to early viral evolution in vaccine recipients who became infected. J Virol. 2020;94(2):e01632-19. doi: 10.1128/JVI.01632-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolfl M, et al. Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood. 2007;110(1):201–210. doi: 10.1182/blood-2006-11-056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dash P, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547(7661):89–93. doi: 10.1038/nature22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malouli D, et al. Cytomegaloviral determinants of CD8+ T cell programming and RhCMV/SIV vaccine efficacy. Sci Immunol. 2021;6(57):eabg5413. doi: 10.1126/sciimmunol.abg5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang H, et al. HLA-E-restricted, Gag-specific CD8+ T cells can suppress HIV-1 infection, offering vaccine opportunities. Sci Immunol. 2021;6(57):eabg1703. doi: 10.1126/sciimmunol.abg1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verweij M, et al. Modulation of MHC-E transport by viral decoy ligands is required for RhCMV/SIV vaccine efficacy. Science. 2021;372(6541):eabe9233. doi: 10.1126/science.abe9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geldmacher C, et al. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J Virol. 2007;81(5):2440–2448. doi: 10.1128/JVI.01847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murakoshi H, et al. CD8+ T cells specific for conserved, cross-reactive Gag epitopes with strong ability to suppress HIV-1 replication. Retrovirology. 2018;15(1):46. doi: 10.1186/s12977-018-0429-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grant EJ, et al. The unconventional role of HLA-E: the road less traveled. Mol Immunol. 2020;120:101–112. doi: 10.1016/j.molimm.2020.02.011. [DOI] [PubMed] [Google Scholar]
- 49.Celik AA, et al. The diversity of the HLA-E-restricted peptide repertoire explains the immunological impact of the Arg107Gly mismatch. Immunogenetics. 2016;68(1):29–41. doi: 10.1007/s00251-015-0880-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoare HL, et al. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat Immunol. 2006;7(3):256–264. doi: 10.1038/ni1312. [DOI] [PubMed] [Google Scholar]
- 51.Davis ZB, et al. A conserved HIV-1-derived peptide presented by HLA-E renders infected T-cells highly susceptible to attack by NKG2A/CD94-bearing natural killer cells. PLoS Pathog. 2016;12(2):e1005421. doi: 10.1371/journal.ppat.1005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruibal P, et al. Peptide binding to HLA-E molecules in humans, nonhuman primates, and mice reveals unique binding peptides but remarkably conserved anchor residues. J Immunol. 2020;205(10):2861–2872. doi: 10.4049/jimmunol.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walters LC, et al. Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding. Nat Commun. 2018;9(1):3137. doi: 10.1038/s41467-018-05459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pohlmeyer CW, et al. CD8+ T cells from HLA-B*57 elite suppressors effectively suppress replication of HIV-1 escape mutants. Retrovirology. 2013;10:152. doi: 10.1186/1742-4690-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu XG, et al. Important contribution of p15 Gag-specific responses to the total Gag-specific CTL responses. AIDS. 2002;16(3):321–328. doi: 10.1097/00002030-200202150-00002. [DOI] [PubMed] [Google Scholar]
- 56.Qin K, et al. CD8 T cells targeting adapted epitopes in chronic HIV infection promote dendritic cell maturation and CD4 T cell trans-infection. PLoS Pathog. 2019;15(8):e1007970. doi: 10.1371/journal.ppat.1007970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schaubert KL, et al. Generation of robust CD8+ T-cell responses against subdominant epitopes in conserved regions of HIV-1 by repertoire mining with mimotopes. Eur J Immunol. 2010;40(7):1950–1962. doi: 10.1002/eji.200940079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kloverpris HN, et al. Early antigen presentation of protective HIV-1 KF11Gag and KK10Gag epitopes from incoming viral particles facilitates rapid recognition of infected cells by specific CD8+ T cells. J Virol. 2013;87(5):2628–2638. doi: 10.1128/JVI.02131-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiani Z, et al. HLA-F on HLA-Null 721.221 cells activates primary NK cells expressing the activating killer Ig-like receptor KIR3DS1. J Immunol. 2018;201(1):113–123. doi: 10.4049/jimmunol.1701370. [DOI] [PubMed] [Google Scholar]
- 60.Mandelboim O, et al. Multiple receptors for HLA-G on human natural killer cells. Proc Natl Acad Sci U S A. 1997;94(26):14666–14670. doi: 10.1073/pnas.94.26.14666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimizu Y, DeMars R. Production of human cells expressing individual transferred HLA-A,-B,-C genes using an HLA-A,-B,-C null human cell line. J Immunol. 1989;142(9):3320–3328. [PubMed] [Google Scholar]
- 62.Lisovsky I, et al. Functional analysis of NK cell subsets activated by 721.221 and K562 HLA-null cells. J Leukoc Biol. 2015;97(4):761–767. doi: 10.1189/jlb.4AB1014-499R. [DOI] [PubMed] [Google Scholar]
- 63.Michaelsson J, et al. A signal peptide derived from hsp60 binds HLA-E and interferes with CD94/NKG2A recognition. J Exp Med. 2002;196(11):1403–1414. doi: 10.1084/jem.20020797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu W, et al. The nucleocapsid protein of Rift Valley fever virus is a potent human CD8+ T cell antigen and elicits memory responses. PLoS One. 2013;8(3):e59210. doi: 10.1371/journal.pone.0059210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Townsley E, et al. Distinct activation phenotype of a highly conserved novel HLA-B57-restricted epitope during dengue virus infection. Immunology. 2014;141(1):27–38. doi: 10.1111/imm.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strong RK, et al. HLA-E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J Biol Chem. 2003;278(7):5082–5090. doi: 10.1074/jbc.M208268200. [DOI] [PubMed] [Google Scholar]
- 67.Berko D, et al. Membrane-anchored β2-microglobulin stabilizes a highly receptive state of MHC class I molecules. J Immunol. 2005;174(4):2116–2123. doi: 10.4049/jimmunol.174.4.2116. [DOI] [PubMed] [Google Scholar]
- 68.Obermann S, et al. Peptide-beta2-microglobulin-major histocompatibility complex expressing cells are potent antigen-presenting cells that can generate specific T cells. Immunology. 2007;122(1):90–97. doi: 10.1111/j.1365-2567.2007.02616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kan-Mitchell J, et al. Degeneracy and repertoire of the human HIV-1 Gag p17(77-85) CTL response. J Immunol. 2006;176(11):6690–6701. doi: 10.4049/jimmunol.176.11.6690. [DOI] [PubMed] [Google Scholar]
- 70.Boppana S, et al. HLA-I associated adaptation dampens CD8 T-cell responses in HIV Ad5-vectored vaccine recipients. J Infect Dis. 2019;220(10):1620–1628. doi: 10.1093/infdis/jiz368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fiore-Gartland A, et al. Pooled-peptide epitope mapping strategies are efficient and highly sensitive: an evaluation of methods for identifying human T cell epitope specificities in large-scale HIV vaccine efficacy trials. PLoS One. 2016;11(2):e0147812. doi: 10.1371/journal.pone.0147812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boaz MJ, et al. Concordant proficiency in measurement of T-cell immunity in human immunodeficiency virus vaccine clinical trials by peripheral blood mononuclear cell and enzyme-linked immunospot assays in laboratories from three continents. Clin Vaccine Immunol. 2009;16(2):147–155. doi: 10.1128/CVI.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qin K, et al. Elevated HIV infection of CD4 T cells in MRKAd5 vaccine recipients due to CD8 T cells targeting of adapted epitopes. J Virol. doi: 10.1128/jvi.00160-21. [published online June 2, 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.