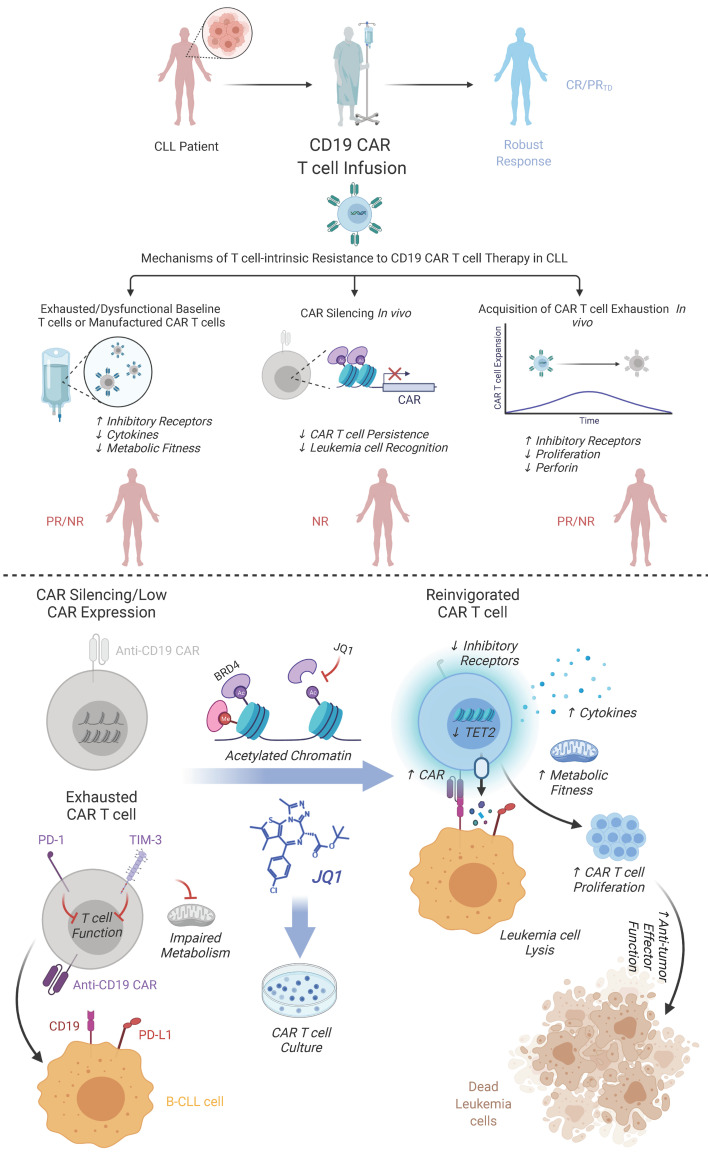
Abstract
Chimeric antigen receptor (CAR) T cells have induced remarkable antitumor responses in B cell malignancies. Some patients do not respond because of T cell deficiencies that hamper the expansion, persistence, and effector function of these cells. We used longitudinal immune profiling to identify phenotypic and pharmacodynamic changes in CD19-directed CAR T cells in patients with chronic lymphocytic leukemia (CLL). CAR expression maintenance was also investigated because this can affect response durability. CAR T cell failure was accompanied by preexisting T cell–intrinsic defects or dysfunction acquired after infusion. In a small subset of patients, CAR silencing was observed coincident with leukemia relapse. Using a small molecule inhibitor, we demonstrated that the bromodomain and extra-terminal (BET) family of chromatin adapters plays a role in downregulating CAR expression. BET protein blockade also ameliorated CAR T cell exhaustion as manifested by inhibitory receptor reduction, enhanced metabolic fitness, increased proliferative capacity, and enriched transcriptomic signatures of T cell reinvigoration. BET inhibition decreased levels of the TET2 methylcytosine dioxygenase, and forced expression of the TET2 catalytic domain eliminated the potency-enhancing effects of BET protein targeting in CAR T cells, providing a mechanism linking BET proteins and T cell dysfunction. Thus, modulating BET epigenetic readers may improve the efficacy of cell-based immunotherapies.
Keywords: Immunology, Oncology
Keywords: Cancer gene therapy, Leukemias, T cells
Introduction
The adoptive transfer of autologous CD19-targeted chimeric antigen receptor (CAR) T cells has shown remarkable activity in patients with B cell malignancies (1–3). Upon treatment with a CAR signaling through 4-1BB/CD3ζ or CD28/CD3ζ, greater than 80% of individuals with relapsed or refractory acute lymphoblastic leukemia (ALL) will achieve complete remission (2–4). Similar therapy can induce durable remissions in relapsed or refractory chronic lymphocytic leukemia (CLL; refs. 1, 5, 6), but only in a small subset of individuals. Many patients with CLL cannot benefit from CAR T cell therapy because of issues with collecting their autologous lymphocytes, especially from patients receiving intensive chemotherapy who possess low numbers of highly proliferative memory T cells (7) and elevated frequencies of exhausted T cells (8). In addition, regardless of sufficient ex vivo proliferation during CAR T cell manufacturing, some individuals exhibit limited in vivo CAR T cell expansion and no persistence. This may be attributed to activation of naturally occurring negative immune checkpoint molecules (e.g., PD-1, TIM-3, LAG-3; ref. 7), a reduction in self-renewal capacity/central memory functions (7), and immunosenescence, all of which can be epigenetically regulated (9–11).
We recently demonstrated that the massive clonal expansion of a single CAR T cell in association with disruption of the methylcytosine dioxygenase gene, TET2, restrained CD8+ T cell differentiation and induced complete remission in a 78-year-old patient with CLL, implicating epigenetic pathway modulation in the amelioration of immune inhibition and improvement of antitumor immunity (12). Another group found that blockade of bromodomain and extra-terminal (BET) proteins in healthy donor T cells endows properties of stem cell–like and central memory T cells with potent effector function in murine T cell receptor (TCR) and CAR gene therapy models (13). BET proteins contain tandem bromodomain signatures that interact with acetylated lysine residues in histones, thereby linking changes in chromatin structure with gene transcription. BET proteins therefore work as “readers” of the histone code and regulate gene activity through the recruitment of transcriptional coactivators or repressors. Thus, optimal differentiation, proliferation, and antitumor potency of CAR T cells depend on an early memory T cell phenotype, and epigenetic programming can enforce this state.
We and others have demonstrated that naive or early memory T cells genetically redirected with CARs have more durable engraftment and antitumor effector function compared with highly differentiated cells (7, 8, 12). However, persistent tumor antigen exposure often leads to exhaustion. T cell exhaustion is an active, tightly regulated process with a controlled and progressive modulation of inhibitory cell-surface molecules. Blockade of these negative signaling pathways (e.g., PD-1/PD-L1) in cancer and chronic viral infections results in enhanced T cell responses as well as prolonged survival and proliferation of antigen-specific CD8+ T cells. In preclinical models, early T cell exhaustion is also a primary factor that hampers the efficacy of CAR-expressing T cells (14). Because exhausted CD8+ T cells occupy a differentiation state that is epigenetically divergent from naive and functional memory CD8+ T cells (9, 10), a critical question is how to identify plasticity of this state and devise reprogramming methods for immunotherapy applications. This is especially relevant in the setting of advanced leukemia, in which autologous CAR T cell quality may strongly influence expansion capacity and antitumor potency upon adoptive transfer (7, 8).
The level and duration of cell-surface CAR expression may also be an important T cell–intrinsic factor influencing clinical efficacy, and receptor extinction as a potential mechanism of resistance has not been widely explored in the context of CAR T cell therapy. The half-life of transgenic antigen receptor expression has been previously implicated as a major determinant of clinical responses to TCR-engineered T cells (15, 16). Transgene shutdown, as defined by an immediate silencing or a gradual decrease in gene expression after differentiation or proliferation of cells transduced with integrating viruses, can occur through multiple epigenetic processes, potentially leading to poor clinical responses to adoptive cell therapies (17).
In this study, we sought to characterize perturbations in phenotypic markers and functional outputs of CAR T cells from patients with CLL as they relate to clinical outcome. We also examined how durability of CAR expression correlates with response. Because epigenetic alterations have been causally associated with transgene extinction, our second goal was to reverse CAR silencing by pharmacologically targeting BET bromodomain “readers” of histone acetylation marks with previously demonstrated roles in gene transcription. Finally, we assessed the impact of BET bromodomain protein antagonists on reversal of dysfunction in exhausted CAR T cells from patients with advanced leukemia who did not respond to therapy, and we investigated the potential mechanisms by which reinvigoration occurs.
Results
Poor responses are associated with T cell exhaustion and CAR extinction in vivo.
We have previously reported the results from 2 clinical studies of CD19 CAR T cells to treat relapsed and highly refractory CLL (ClinicalTrials.gov, NCT01029366 and NCT01747486; refs. 5, 18). Of 46 patients infused, the overall response rate was 51%, with sustained complete responses occurring in only approximately 28% of heavily pretreated patients (5, 18). These individuals were further stratified according to in vivo CAR T cell proliferative capacity. We found that nonresponding (NR) and conventional partially responding (PR) patients exhibited marginal or no expansion of their transferred T cells (7). In contrast, patients who had a complete response (CR) or partial response due to disease transformation (PRTD) possessed CAR T cells with robust proliferative capacity and sustained persistence (7). To investigate the dynamics of circulating CAR T cell antitumor activity, we evaluated the absolute numbers of peripheral blood B-CLL cells (CD19+, kappa or lambda light chain+) in NR and PR patients with detectable but poorly expanded in vivo CAR T cells over time compared with those of CR and PRTD patients (Figure 1A). In these NR and PR patients, there was an early failure of CAR T cells to eradicate disease, whereas the CR and PRTD groups demonstrated a precipitous and sustained decrease in peripheral blood leukemia burden (Figure 1A).
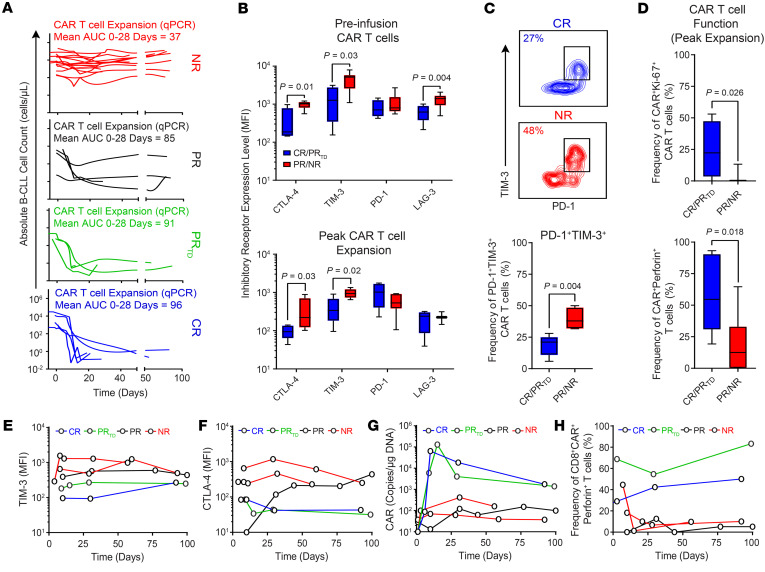
Figure 1. Exhaustion phenotypes are on-treatment biomarkers of resistance to CD19 CAR T cell therapy.
(A) Absolute numbers of B-CLL cells in the blood of patients treated with CAR T cells are shown (NR with no/poor CAR T cell expansion, n = 15; PR, n = 5; PRTD, n = 3; CR, n = 5). The expansion capacity of CAR T cells in each patient response group as determined by AUC calculations (copies per microgram genomic DNA, AUC 0–28 days) is indicated above each graph. (B) Expression levels of inhibitory receptors (CTLA-4, TIM-3, PD-1, and LAG-3) on preinfusion (top) and postinfusion (bottom) CAR T cells at the peak of in vivo expansion in the blood of the CR/PRTD versus PR/NR patients. (C) Flow cytometric plots of peripheral blood CAR T cells at the peak of in vivo expansion from representative CR and NR patients coexpressing PD-1 and TIM-3 are shown (top). Frequencies of CAR T cells coexpressing these inhibitory receptors in CR/PRTD compared with PR/NR patients are summarized as box plots (bottom). (D) Proportions of peak expansion CAR T cells expressing Ki-67 (top) and perforin (bottom) in CR/PRTD relative to PR/NR patient groups are shown. In all boxplots, boxes extend from the 25th to 75th percentiles; middle line, median; whiskers, minimum, and maximum. P values calculated using a Mann-Whitney test. (E) Longitudinal TIM-3 and (F) CTLA-4 expression on postinfusion CD8+CAR+ T cells at indicated time points. (G) CAR T cell expansion levels and (H) frequencies of perforin-expressing CD8+CAR+ T cells are shown in representative patients from the above response groups.
In order to extend our previous investigations of the determinants of CAR T cell therapeutic efficacy in CLL (5, 7), which were heavily focused on leukapheresis material, the phenotype of CAR+ T cells was analyzed before infusion and at the peak of expansion by multiparameter flow cytometry. CAR+ T cells from NR and PR patients demonstrated significantly higher levels of CTLA-4 and TIM-3 or LAG-3 expression and/or elevated frequencies of inhibitory receptor–expressing T cells compared with those from patients who experienced a CR or PRTD (Figure 1B and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI145459DS1). Because T cell exhaustion is regulated in part by inhibitory receptor coexpression, we next examined frequencies of PD-1+TIM-3+CAR+ T cells and found significantly higher proportions of these cells in NR/PR patients relative to CR/PRTD patients at the time of maximum in vivo expansion (Figure 1C). To further interrogate markers of in vivo CAR T cell potency, we evaluated intracellular expression of Ki-67, a cell-cycle indicator of cycling or recently divided cells. Ki-67 positivity is also indicative of reinvigoration of exhausted T cells in human cancer upon checkpoint blockade (19). Indeed, the frequency of Ki-67+ CAR+ T cells was decreased in NR/PR patients in comparison with those from CR and PRTD patients (Figure 1D). Populations of circulating NR/PR patient CAR T cells expressing cytolytic perforin were also low in frequency after adoptive transfer (Figure 1D). Longitudinal analyses indicated that postinfusion CAR T cells in some patients with CLL with poor outcomes acquired dysfunction or became progressively more exhausted during the initial phase of leukemia clearance, in association with high levels of inhibitory receptor expression (Figure 1, E and F) as well as concomitant proliferative (Figure 1G) and cytolytic defects (Figure 1H).
Transduction efficiency and the level of CD19 CAR expression on the cell surface could affect the proliferation, persistence, and cytotoxicity of transferred cells in patients. We therefore compared the levels of surface CAR expression measured by flow cytometry as MFI for preinfusion and postinfusion CR/PRTD and PR/NR CD8+ T cells. Although no differences between patient groups were observed in preinfusion products, PR/NR patient CD8+ T cells exhibited significantly lower CAR expression levels at peak expansion compared with those of CR/PRTD patients (Figure 2A).
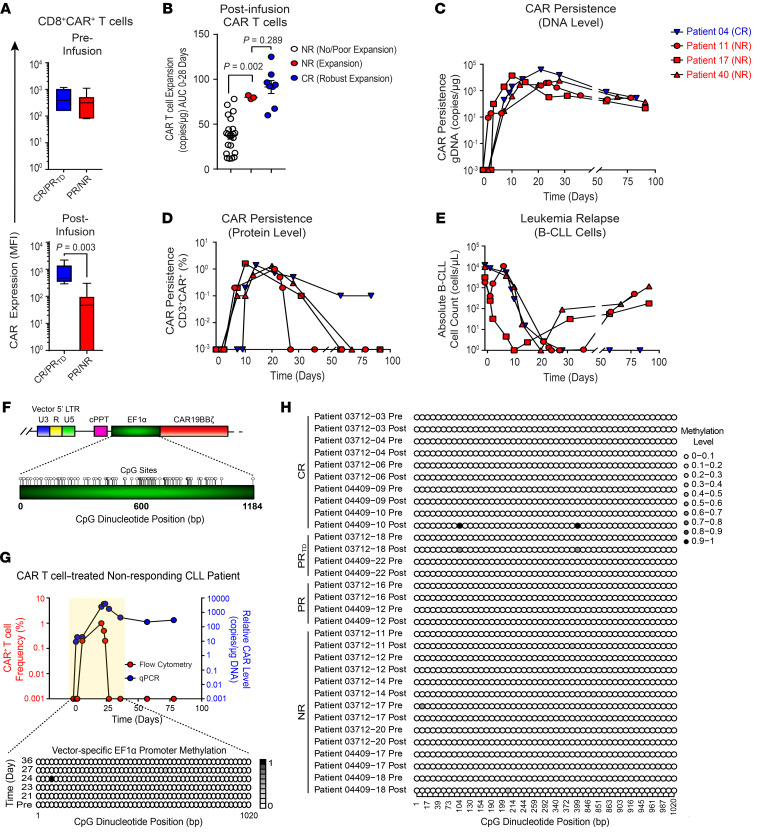
Figure 2. CAR extinction is associated with decreased expression of the transgenic protein despite persistence of the transgenic DNA and occurs independently of promoter methylation.
(A) Boxplots of CD19 CAR expression levels on preinfusion (top) and postinfusion (bottom) CD8+CAR+ T cells in CR/PRTD versus PR/NR groups. (B) CAR T cell expansion capacity as indicated by AUC 0–28-day calculations in NRs with no/poor CAR T cell expansion, NRs with CAR T cell expansion (patients 11, 17, and 40), and CR patients. Data are shown as the mean ± SEM. (C) CAR persistence at the DNA and (D) protein levels in n = 3 NR patients exhibiting CAR T cell expansion compared with a representative CR patient showing similar kinetics. (E) B-CLL burden in the peripheral blood of the same patients above. P values determined with a Mann-Whitney test. (F) CpG dinucleotide clustering within the vector-specific EF1α promoter is shown. Each circle depicts a single CpG site. LTR, long terminal repeat; cPPT, polypurine tract; EF1α, elongation factor 1 alpha promoter; U3, upstream region; U5, downstream region; R, repeat region. (G) Genomic DNA from preinfusion and postinfusion CAR T cells of a representative NR patient who exhibited CAR extinction was isolated followed by bisulfite conversion and targeted sequencing. Longitudinal EF1α promoter methylation analysis is shown, in which each row represents a specific time point, with methylated cytosine residues depicted by shaded circles and nonmethylated residues by unshaded circles. The color indicates the relative methylation level from low (white) to high (black). (H) EF1α promoter bisulfite sequencing results of preinfusion and postinfusion CAR T cells from evaluable patients with CLL with various clinical outcomes (NR, n = 7; PR, n = 2; PRTD, n = 2; CR, n = 5).
A small subset of NR patients (i.e., n = 3 out of 46) had a significantly higher engraftment AUC of CAR-expressing cells compared with the majority of nonresponders (Figure 1A and Figure 2B). This level of CAR T cell engraftment was comparable to that of patients who experienced long-term durable complete remissions (Figure 2B). Levels of proviral DNA were further examined longitudinally to monitor the in vivo persistence of adoptively transferred CAR T cells. The presence of CAR-transduced cells, as determined by the detection of lentiviral integrants, peaked at approximately 1 to 3 weeks after infusion and remained persistent in the peripheral blood of this unusual group of NR patients (Figure 2C). Although the persistence of transferred cells indicated by quantitative PCR (qPCR) was evident at 3 months in these cases, a loss of surface CAR expression measured by flow cytometric antiidiotypic antibody staining was observed soon after adoptive transfer (Figure 2D). This was in contrast to a representative CR patient who displayed similar kinetics of CAR T cell expansion and persistence (Figure 2, C and D). Unlike standard NR patients with CLL whose CAR T cells failed to control leukemia early after infusion (Figure 1A), a transient response to therapy was evident in that the peripheral blood B-CLL cells in these 3 patients were reduced to an undetectable count (Figure 2E). However, the disappearance of CAR expression in this small group of NR patients coincided with leukemic relapse, while continued antitumor activity was observed in the setting of a complete response with sustained CAR expression (Figure 2, D and E). These results suggest that the level and duration of CAR expression may be important for clinical efficacy because patients who experienced complete remissions had ongoing expression, whereas NR patients who originally controlled their leukemia lost cell-surface CAR detection at the time of relapse. Thus, CAR extinction at the cell surface was likely a critical mechanism of resistance to therapy.
Lack of cytosine methylation of the CD19 CAR vector–specific EF1α promoter.
We next sought to understand the mechanism(s) involved in extinction of CAR expression because this could reveal actionable interventions to prevent therapeutic resistance. DNA methylation is a key epigenetic alteration involving addition of a methyl group to cytosine nucleotides. This modification has been implicated in silencing of promoter-driven transgene expression in the context of viral vectors, including lentiviruses (20, 21). We assessed the level of DNA methylation within the vector-specific EF1α promoter used to drive anti-CD19 CAR transgene expression in autologous CLL patient T cells. For these analyses, we focused on promoter regions containing high densities of CpG dinucleotides (Figure 2F). In an NR CLL patient exhibiting rapid CAR extinction kinetics 21 days after adoptive cell transfer, we examined the level of cytosine methylation in vivo and compared that with the level of cytosine methylation in the cellular infusion product. There was a lack of or a minimal level of cytosine methylation within the sequenced regions of the lentiviral vector–specific EF1α promoter, which did not appear to increase with time after infusion (Figure 2G). The data from bisulfite sequencing of 16 patients with CLL with a range of clinical responses at preinfusion (CAR T cell infusion product) and postinfusion (peak of in vivo CAR T cell expansion) time points are displayed in Figure 2H. These experiments demonstrated that lentiviral gene transfer into human T cells did not result in significant DNA methylation of the EF1α promoter either ex vivo during CAR T cell manufacturing or in vivo after adoptive transfer.
Ex vivo BET bromodomain protein inhibition increases CD19 CAR expression and CAR T cell frequency.
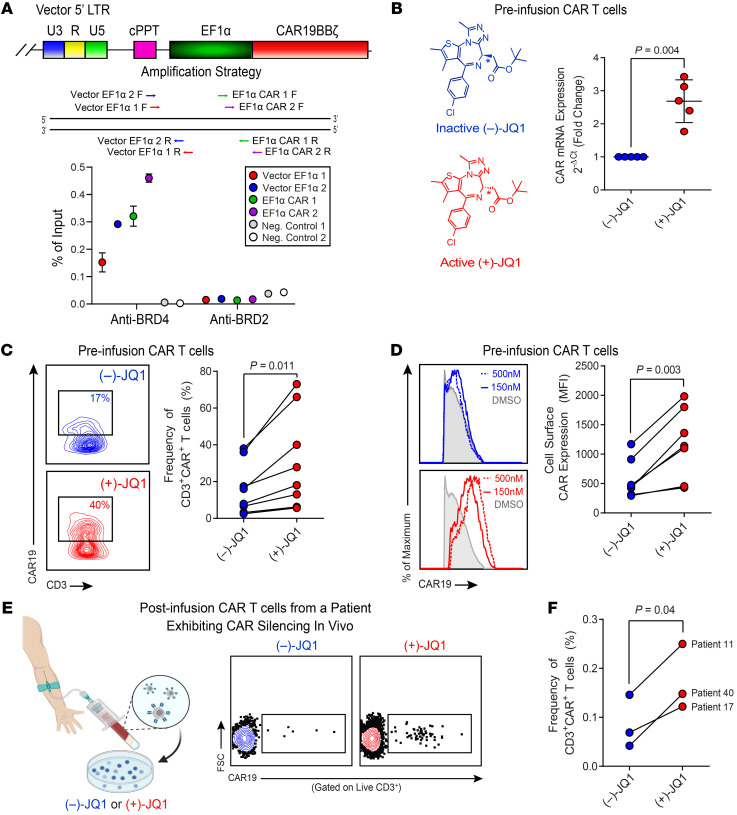
Because our data supported the premise that cytosine methylation does not appear to play a significant role in the downregulation of CD19 CAR expression, we explored alternative molecular mechanisms underlying transgene extinction. We recently reported the relationship between integrated CD19 CAR vectors and genomic features, bound proteins, and sites of epigenetic modification in the patient cohorts described herein (22). Proximity of lentiviral integration sites to BRD4 and BRD2 promoters were among the influential negative variables predictive of clinical outcome (22). Localization of BRD4 and BRD2 at promoters and other regions important for transcription has been shown to repress gene expression, including virally integrated genes (23–25). To investigate whether BRD4 and/or BRD2 bound at the CD19 CAR vector–specific EF1α promoter, we performed ChIP assays on CAR T cell samples from patients with CLL using anti-BRD4 and anti-BRD2 antibodies. BRD4 but not BRD2 was highly enriched at various regions of the lentiviral EF1α promoter (Figure 3A), suggesting that BRD4 recruitment may contribute to regulation of CD19 CAR expression.
Figure 3. JQ1, a BET bromodomain inhibitor, rescues CAR expression.
(A) ChIP analysis of the CAR vector–specific EF1α promoter for BET bromodomain protein localization. A schematic of the PCR amplification for ChIP is shown (top). CAR T cells from nonresponding patients with CLL (n = 3) were subjected to ChIP with anti-BRD4 and anti-BRD2 antibodies. Enrichment of BRD4 and BRD2 at the vector-specific EF1α promoter was measured by qPCR. Data are shown as the mean ± SEM (bottom). (B) CAR T cells from nonresponders were treated with (–)-JQ1 (left panel) or (+)-JQ1 for 4 days, and real-time RT-PCR was subsequently performed to determine expression levels of the CAR transgene relative to CD3 epsilon, which served as a loading/normalization control. Fold expression levels of CAR in active JQ1-treated patient T cells compared with those in T cells treated with an inactive control are shown (n = 5, paired t test). Data are shown as the mean ± SEM. (C) Changes in the frequencies of nonresponding CLL patient CD3+CAR+ T cells (representative contour plots, left; graphical data summary, right) and (D) CAR expression levels as denoted by MFI of an antiidiotypic antibody (representative histograms, left; summarized data, right) after a 4-day incubation with 150–500 nM (–)-JQ1 or (+)-JQ1 are shown (n = 7, paired t test). (E) PBMCs were isolated from the peripheral blood of patients who exhibited CAR silencing in vivo (left). Cells were subsequently treated with (–)-JQ1 or (+)-JQ1 and analyzed for CAR expression (representative flow cytometric plots, middle). (F) Frequencies of CD3+CAR+ T cells from n = 3 patients were quantified (paired t test).
Given the role of BET proteins in transcriptional modulation, we hypothesized that the BRD4 chromatin adaptor may act as a repressor of CAR transgene expression. The impact of this potential epigenetic silencing mechanism was therefore assessed in a short-term culture model in which cryopreserved CLL patient CAR+ T cells were thawed and incubated in the presence of JQ1, a potent and selective BET small molecule inhibitor. During the course of 4 days, treatment of NR patient CAR T cells with the pharmacologically active form of JQ1 augmented CAR mRNA levels in preinfusion CAR T cells (Figure 3B). We next assessed protein-level changes via flow cytometric analysis of CAR positivity and found that JQ1 increased the frequency of CAR+ T cells (Figure 3C) as well as cell-surface expression of the anti-CD19 synthetic receptor (Figure 3D) in infusion products.
Because histone deacetylases (HDACs) are enzymes that remove acetyl groups from proteins (e.g., histones) and BET proteins are readers of acetylated chromatin status, they share many common targets and affect similar cellular processes. Thus, we tested whether treatment of CAR T cells from patients with CLL with pan-HDAC inhibitors led to upregulation of CAR expression. Unlike JQ1, HDAC inhibitors did not augment CAR expression (Supplemental Figure 2). Furthermore, JQ1 treatment did not affect CAR expression when a retroviral construct possessing a murine stem cell virus promoter was used to drive transgene expression (Supplemental Figure 3). Active JQ1 treatment markedly increased CAR expression only when transduction was carried out with the EF1α promoter–containing lentiviral vector, which further supports the above mechanism of BRD4-mediated transgene silencing.
We next sought to reactivate CAR expression in gene-modified T cells. There were sufficient numbers of engineered lymphocytes from 1 NR patient whose infusion product was confirmed to be lentiviral vector–positive by qPCR (data not shown), but CAR negative by flow cytometry. To evaluate the effect of BRD4 inhibition on potential reversal of CAR silencing, cells were treated with JQ1 as described above. Treatment of patient-derived T cells with active JQ1 was able to upregulate CAR expression at the mRNA and protein levels (Supplemental Figure 4, A and B). Patient T cell function was assessed using a cytotoxicity assay. JQ1-treated CAR+ T cells demonstrated CD19-specific lysis in a dose-dependent manner (Supplemental Figure 4C). Finally, we treated post–CAR T cell infusion PBMCs from patients who exhibited CAR silencing in vivo with (–)-JQ1 or (+)-JQ1 and effectively recused CAR T cell loss (Figure 3, E and F). Taken together, these data suggest that downregulation or extinction of CAR expression may be reversed by ex vivo BRD4 protein inhibition.
JQ1 ameliorates PD-1–mediated inhibition of CD19 CAR T cells.
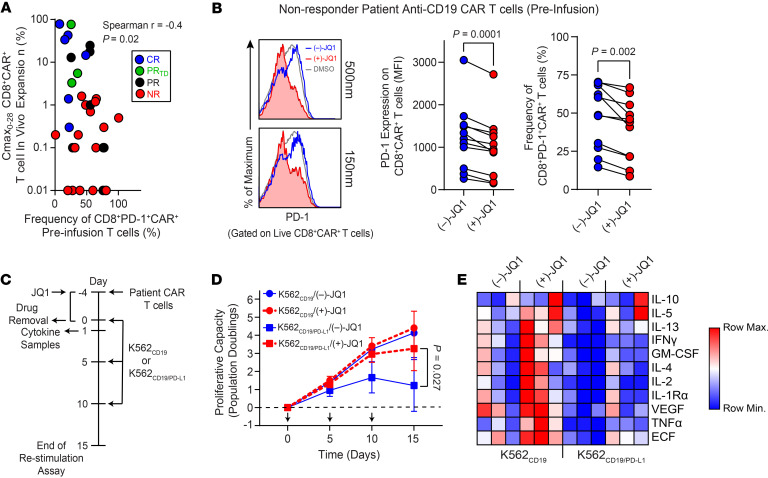
We recently found that anti-CD19 CAR T cells from PR or NR patients with CLL contained higher frequencies of PD-1–expressing CD8+ T cells in the infusion product when compared with CRs or patients who had a PRTD (7). Thus, we reasoned that PD-1 upregulation on manufactured preinfusion CAR T cells may not only serve as a predictive indicator of poor in vivo CAR T cell activity and clinical outcomes but also represent a tractable point of intervention with checkpoint blockade or related modulation strategies. Accordingly, we found that there was a significant negative correlation between the frequency of CD8+PD-1+CAR+ T cells in the infusion product and the maximum degree of in vivo CAR T cell proliferation in the first 28 days of therapy (Figure 4A). Although CLL cells from patients enrolled in our trials of CAR T cell therapy had detectable levels of a cognate ligand for PD-1, there was no association with expression of PD-L1 nor other B cell surface markers (HLA-DR, CD80, CD10, CD86, CD40, and CD70) and likelihood of response (Supplemental Figure 5). This further supports the notion that T cell–intrinsic fitness is likely the major determinant of clinical outcome in the setting of CD19-directed CAR T cell therapy of CLL.
Figure 4. JQ1 reinvigorates dysfunctional CAR T cells from nonresponding patients with CLL.
(A) Spearman’s Rho correlation performed between the frequency of preinfusion CD8+PD-1+CAR+ T cells and the maximum proportion (Cmax) of in vivo expanded CD8+CAR+ T cells. Each dot represents an individual patient (n = 36) differentially colored according to therapeutic response. (B) CD8+CAR+ T cells were treated with 150–500 nM (–)-JQ1 or (+)-JQ1 for 4 days followed by evaluation of PD-1 levels. Flow cytometry histograms of PD-1 expression on CLL patient CAR+CD8+ T cells (left) are shown. PD-1 expression levels (middle) and proportions of CD8+CAR+ T cells after a 4-day incubation with 150 nM (–)-JQ1 or (+)-JQ1 are depicted (n = 11, paired t test). (C) CAR+ T cells were treated with (–)-JQ1 or (+)-JQ1 followed by stimulation with K562CD19 or K562CD19/PD-L1 cells. Functional properties of CAR T cells were then analyzed. (D) Proliferation of CAR T cells restimulated with K562 cells as indicated above (n = 3, paired t test). Arrows indicate stimulation time points. (E) Heatmap of cytokine profiles for CLL patient CAR T cells (n = 3) treated with (–)-JQ1 or (+)-JQ1 and stimulated with irradiated K562CD19 or K562CD19/PD-L1 cells is presented. Colors represent scaled cytokine data.
BET bromodomain protein antagonism has previously been shown to downregulate PD-L1 on tumors in association with an increase in the activity of cytotoxic T cells (26). We thus hypothesized that BET inhibition may affect CAR T cell activity at least partially through the PD-1/PD-L1 axis. To determine whether BET blockade could also potentially ameliorate immune suppression in a T cell–intrinsic manner, we incubated dysfunctional CLL patient preinfusion CAR T cells with JQ1 and assessed expression levels of PD-1 after a 4-day culture. Incubation of CAR T cells from a CLL patient with active JQ1 resulted in a significant decrease in both the expression level of PD-1 and frequency of CD8+PD-1+CAR+ T cells (Figure 4B). The effect of JQ1 treatment on PD-1 signaling occurred independently of any changes in CAR expression; CRISPR/Cas9-mediated knockout of PDCD1 (encodes PD-1) had no effect on surface or intracellular CAR levels (Supplemental Figure 6). In addition, PD-1 ligation did not result in a significant change in CAR expression as determined upon stimulation with leukemia targets expressing CD19/PD-L1 versus CD19 alone (Supplemental Figure 6). In agreement with other studies (27, 28), JQ1 treatment did not affect the expression levels of T cell activation markers (Supplemental Figure 7A). In addition, JQ1 had no impact on the apoptosis sensitivity of these gene-modified cells, as indicated by annexin V staining (Supplemental Figure 7B).
We next investigated the influence of JQ1 treatment on CLL patient CAR T cell activity. Samples of expanded CAR T cells were thawed and treated with JQ1 or an inactive stereoisomer for 4 days followed by inhibitor washout and functional assessments, including proliferative capacity (i.e., one of the best in vitro predictive correlates of clinical outcome with CAR T cell therapy of CLL; refs. 5, 7, 12, 29) and cytokine/chemokine production (Figure 4C). We initially tested CAR T cells by repeatedly stimulating these lymphocytes with CD19-expressing K562 leukemia cells and confirmed that these conditions promoted T cell expansion in an antigen-dependent manner as observed in patients (Supplemental Figure 8). Repeated stimulation of NR patient infusion product CAR T cells with irradiated CD19+/PD-L1+ K562 targets resulted in a marked decrease in T cell proliferation compared with serial exposure to CD19+ K562 cells. Suppression of expansion during this in vitro “stress test” was largely rescued by pretreatment of dysfunctional CAR T cells with active JQ1 (Figure 4D). In contrast, CAR T cells expanded with (+)-JQ1 versus (–)-JQ1 showed similar in vitro cytotoxic activity against the CD19+ ALL cell lines NALM-6 and NALM-6PD-L1 after short-term coculture (Supplemental Figure 9A). After repeated antigen stimulation (Supplemental Figure 9B), CAR T cells generated in the presence of (+)-JQ1 over (–)-JQ1 demonstrated marginally higher cytolytic capacity over time at high effector to target cell ratios (Supplemental Figure 9C).
JQ1 treatment also increased expression levels of various immunoregulatory cytokines and chemokines previously reported to be produced by CAR T cells in CLL during successful therapy (5, 6, 30), and this was observed in cells stimulated with CD19+ K562 targets with and without PD-1 ligation (Figure 4E and Supplemental Figure 10). JQ1 treatment of PBMCs isolated from the blood of NR patients after CAR T cell treatment also resulted in reduced frequencies of CD8+PD-1+CAR+ T cells in ex vivo culture (Supplemental Figure 11A). When these PBMCs pretreated with (+)-JQ1 were stimulated with phorbol 12-myristate 13-acetate/ionomycin to bypass antigen receptor signaling, increased proportions of CD8+CAR+ T cells were found to coexpress effector cytokines, relative to the matched (–)-JQ1 control (Supplemental Figure 11B). These findings suggest that pathways in addition to PD-1 could inhibit cytokine production via BET bromodomain proteins. BET blockade may therefore have the effect of increasing cytokine production not only by relieving PD-1–mediated inhibition but also by attenuating other negative feedback pathways. Thus, reducing the association of BET epigenetic readers with histone acetylation signatures can reinvigorate exhausted CLL patient CAR T cell antitumor function beyond what may be seen by checkpoint blockade of the PD-1/PD-L1 axis.
JQ1 treatment reduces suppression of glycolysis and oxidative phosphorylation in exhausted CAR T cells from patients with CLL.
We recently reported that CAR T cells derived from NR patients with CLL exhibit impaired mitochondrial biogenesis (31), which is a key determinant of optimal CAR T cell antitumor function (32). Furthermore, there is a significant positive correlation between mitochondrial mass and the presence of CD27+ CAR T cells that do not express inhibitory receptors, including PD-1 (31). To extend these findings, we measured the mitochondrial respiration of CD8+CAR+ T cells from responding and NR patients. In line with the bioenergetic deficiencies observed in exhausted CD8+ T cells in chronic viral infection (33), NR CD8+CAR+ T cells exhibited a significantly lower mitochondrial spare respiratory capacity (SRC) compared with those derived from responding patients (Supplemental Figure 12A). SRC enhances the survival and function of memory T cells by providing a contingency source of ATP under conditions of increased cellular activity or for cells encountering metabolic stress, including nutrient depletion and oxygen deprivation.
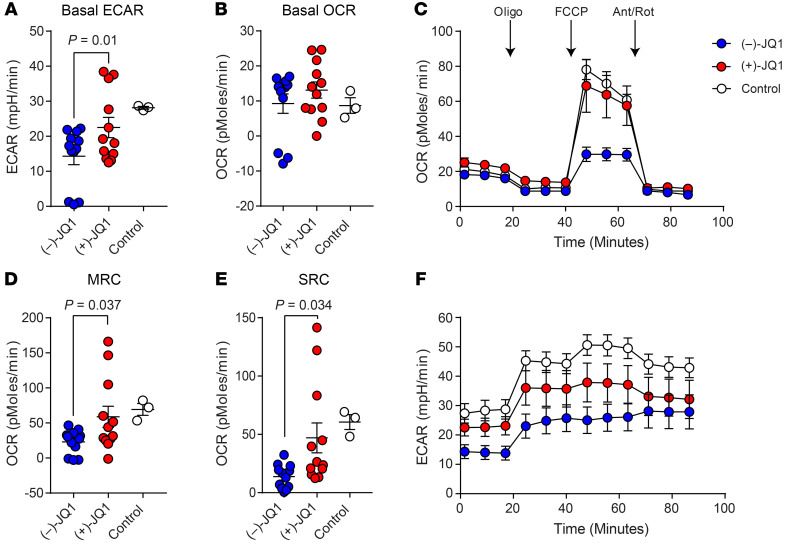
Because CAR T cells from PR/NR patients are exhausted (7) and mitochondrial respiration is also significantly reduced in exhausted CD8+ T cells in chronic viral infection (33), we next asked whether JQ1-mediated reinvigoration of these lymphocytes is associated with enhancement of metabolic fitness. Retrospective CAR+ T cell samples from patients with CLL confirmed to be exhausted based on RNA-Seq data and various functional assessments (7) were treated with JQ1 or an inactive enantiomer for 4 days. CD4+ and CD8+ T cell populations were subsequently isolated from the CAR-transduced patient samples and subjected to Seahorse mitochondrial flux analysis. JQ1-treated CD8+CAR+ T cells had a higher basal extracellular acidification rate (ECAR) (Figure 5A) but not a significantly different oxygen consumption rate (OCR) (Figure 5B) compared with matched samples incubated with the inactive control. Introduction of the mitochondrial decoupler FCCP permitted assessment of the maximal respiratory capacity (MRC) and SRC (Figure 5C). Pharmacological BET inhibition resulted in increased MRC (Figure 5D) and SRC (Figure 5E) in dysfunctional CD8+CAR+ T cells. Overall ECAR and the change in ECAR in response to oligomycin injection were augmented in the group treated with (+)-JQ1 (Figure 5F). The effects of JQ1 on enhancement of mitochondrial respiration were specific to CD8+CAR+ T cells: CD4+CAR+ T cell metabolic features were indistinguishable between (+)-JQ1 and (–)-JQ1 treatments (Supplemental Figure 12, B–G). CAR expression levels did not correlate with parameters of metabolic activity, including the basal OCR (Supplemental Figure 12H) and SRC (Supplemental Figure 12I). These results suggest that suppressed glycolysis and diminished oxidative phosphorylation, hallmarks of exhausted CD8+ T cells (33), can be ameliorated in dysfunctional CLL patient CD8+CAR+ T cells via BET bromodomain protein inhibition.
Figure 5. JQ1 ameliorates suppression of mitochondrial respiration and glycolysis in dysfunctional CD8+ CAR T cells from patients with CLL.
Metabolic flux profiling was performed on purified, dysfunctional CD8+ T cells from CLL patient infusion products after a 4-day treatment with 150 nM (–)-JQ1 (blue) or (+)-JQ1 (red). The metabolic profile of functional CD8+ CAR T cells from CR patients is shown as a control. (A) The extracellular acidification rate (ECAR) and (B) oxygen consumption rate (OCR) were measured at baseline. (C) Longitudinal OCR was quantified during a mitochondrial stress test performed by injection of oligomycin (Oligo), a mitochondrial decoupler (FCCP), followed by electron transport chain inhibitors, antimycin A/rotenone (Ant/Rot). (D) Maximal respiratory capacity (MRC) and (E) spare respiratory capacity (SRC) of CD8+ CAR T cells treated with (–)-JQ1 or (+)-JQ1 were determined after FCCP injection. (A–E) n = 12, paired t test. Data are shown as the mean ± SEM. (F) ECAR was also measured over time during the same mitochondrial stress test. Data are shown as the mean ± SEM.
BET protein blockade improves the production of CD19 CAR T cells from poor-quality lymphocytes in patients with CLL.
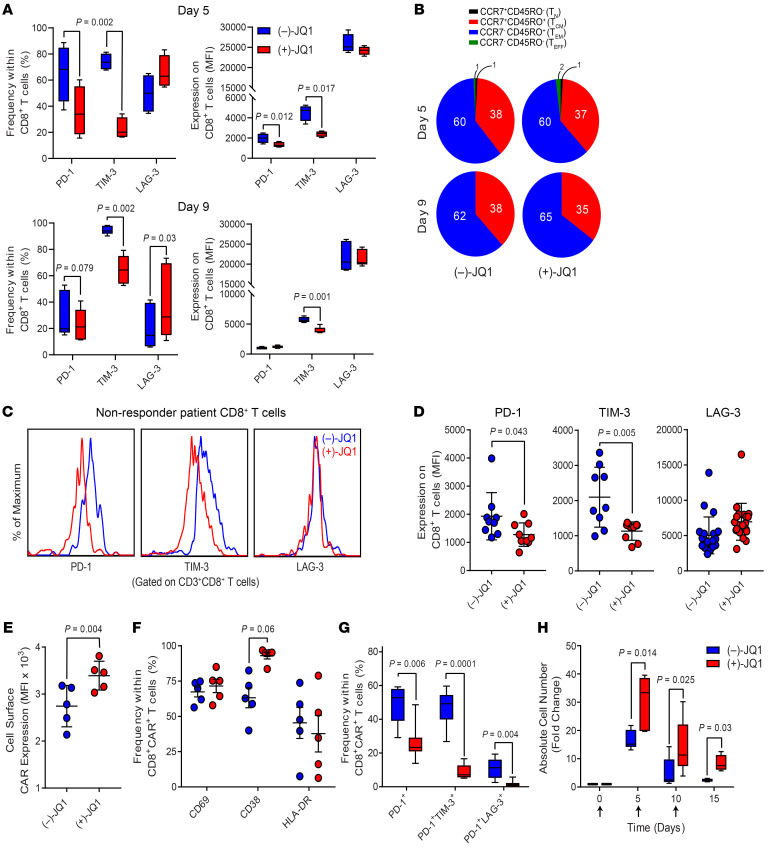
A number of patients with CLL and other cancers are unable to receive CAR therapy because of inadequate T cell numbers or because of manufacturing failures likely related to intrinsic T cell dysfunction, often associated with intensive prior therapies. Thus, actionable autologous CAR T cell manufacturing improvements have the potential to increase the number of patients eligible for therapy, treat patients sooner, and improve response rates. Given the observed reinvigoration of dysfunctional CAR T cells from patients with CLL mediated by short-term BET inhibition, we postulated that incorporating JQ1 into cellular engineering/expansion processes would lead to generation of less defective and more potent final CAR T cell products. We recently reported that reducing the duration of ex vivo culture limits differentiation and enhances the antitumor potency of CAR T cells (34). Based on these findings, we evaluated the inhibitory phenotypes of CAR T cells generated from bulk CLL patient CD3+ T cells continuously exposed to (+)-JQ1 or (–)-JQ1 and harvested from cultures at early (day 5) compared with later (day 9) time points. Frequencies of CD8+ T cells expressing PD-1 and TIM-3 but not LAG-3 and the expression levels of these inhibitory receptors were significantly reduced during both 5- and 9-day CAR T cell manufacturing processes incorporating active JQ1 (Figure 6A). Although JQ1 has been shown to maintain healthy donor CD8+ T cells with functional properties of early memory T cells (13), we did not see such an effect after CAR T cell expansion (Figure 6B). We next proceeded to test JQ1 in small-scale CAR T cell test expansions using retrospective leukapheresis material from heavily pretreated and relapsed patients who did not respond to CD19 CAR T cell therapy. During 5-day CAR T cell cultures, BET inhibition significantly reduced the inhibitory phenotype of CD8+ T cells (i.e., characterized initially by inhibitory receptor levels), even in these highly refractory patients (Figure 6, C and D). Accordingly, CLL patient CAR T cell manufacturing in the presence of (+)-JQ1 over (–)-JQ1 resulted in the production of products with higher CAR expression (Figure 6E), independently of significant changes in CD8+CAR+ T cell activation (Figure 6F). Incorporation of active JQ1 into CAR T cell expansion had similar effects on CD4+ T cell inhibitory, differentiation, and activation phenotypes (Supplemental Figure 13, A–C). Furthermore, CAR T cells expanded with (+)-JQ1 relative to (–)-JQ1 possessed lower frequencies of CD8+ T lymphocytes expressing PD-1 and coexpressing multiple inhibitory receptors (Figure 6G), signifying a reduction in conventional exhaustion phenotypes. Finally, the resultant CAR T cells produced in the presence of (+)-JQ1 compared with (–)-JQ1 exhibited much more robust proliferative capacity after repeated exposure to CD19 (Figure 6H). The beneficial effect of active JQ1 on proliferative potency was not observed during the primary CAR T cell expansion (Supplemental Figure 13D). These results indicate that BET inhibition during small-scale manufacturing can produce CD4+ and CD8+ CAR T cells with diminished exhaustion and improved function.
Figure 6. JQ1 treatment improves production of CAR T cells from dysfunctional autologous T cells.
Bulk CD3+ T cells were purified from PBMCs of patients with CLL and stimulated through CD3 and CD28 in the presence of 150 nM (–)-JQ1 or (+)-JQ1. Early (day 5) and late (day 9) harvested CD8+ T cells were evaluated for (A) the frequencies and MFI of inhibitory molecules (PD-1, TIM-3, and LAG-3) and (B) T cell differentiation phenotype. TN, naive-like; TCM, central memory; TEM, effector memory; TEFF, effector. (C) CD3+ T cells were isolated from leukapheresis material from highly relapsed/refractory patients with CLL treated with CD19 CAR T cells. After activation and expansion in the presence of (–)-JQ1 or (+)-JQ1, cells were analyzed for an inhibitory phenotype defined by expression of PD-1, TIM-3, and LAG-3. Representative flow cytometric histograms of inhibitory receptor expression on CD8+ T cells are shown. (D) Summarized inhibitory receptor expression data on (–)-JQ1– or (+)-JQ1–treated and expanded T cells from multiple relapsed/refractory patients with CLL are shown (statistical analyses were performed using paired t tests). After transduction of the above patient T cells with a lentiviral vector encoding an anti-CD19 4-1BBζ CAR and subsequent expansion in the presence of 150 nM (–)-JQ1 or (+)-JQ1, CAR T cells were assessed for (E) CAR expression, (F) activation status, (G) proportions of CAR T cells expressing PD-1 or coexpressing multiple inhibitory receptors, and (H) proliferative potency after repetitive stimulation with irradiated K562CD19 cells (n = 7, paired t tests). Data are shown as the mean ± SEM.
BET inhibition in dysfunctional CLL patient CD8+CAR+ T cells is associated with a unique transcriptomic reinvigoration profile.
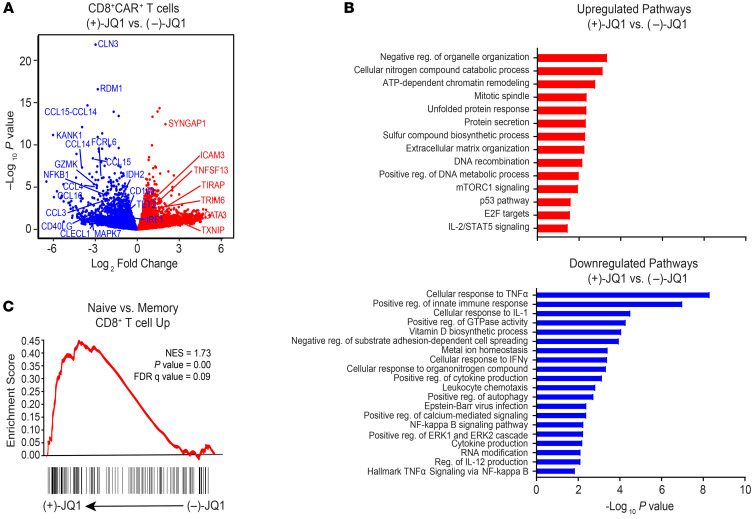
We next explored the molecular pathways that contribute to JQ1-mediated amelioration of CD8+CAR+ T cell dysfunction. The transcriptomes of dysfunctional CAR T cells from patients with CLL (n = 4) were analyzed after 4 days of (+)-JQ1 or (–)-JQ1 treatment (Supplemental Table 1). Principal component analysis revealed that despite patient variability, transcriptional profiles clustered based on BET inhibitor condition (Supplemental Figure 14). Several genes enriched in dysfunctional CD8+ tumor-infiltrating lymphocytes (e.g., CCL3, CCL4, NFKB1, GZMK, CD160, IRF1; refs. 35–38) and those encoding chromatin modulators implicated in T cell exhaustion (e.g., TET2, IDH2; refs. 11, 12, 39–41) were downregulated after incubation with (+)-JQ1 (Figure 7A). In contrast, defective CAR T cells treated with (+)-JQ1 exhibited enrichment of genes associated with robust T cell–mediated antitumor immunity and effector function (e.g., GATA3, ICAM3, TIRAP; refs. 42–44) as well as mitochondrial metabolism (e.g., TXNIP; ref. 45 and Figure 7A).
Figure 7. JQ1 treatment of dysfunctional CAR T cells from patients with CLL is associated with a unique transcriptional profile.
(A) Volcano plot showing differentially expressed genes identified through RNA-Seq of relapsed/refractory CLL patient CAR+CD8+ T cells treated with (–)-JQ1 or (+)-JQ1 for 4 days (n = 4). Downregulated genes are colored in blue and upregulated genes are depicted in red. Unadjusted P values are shown. (B) Gene ontology terms associated with genes that were significantly upregulated (red) and downregulated in the above JQ1-treated patient CD8+CAR+ T cells. (C) Representative GSEA results from running the unfiltered CD8+CAR+ T cell (+)-JQ1 versus (–)-JQ1 rank list against the Molecular Signatures Database (MSigDB) gene ontology collections.
Using gene set enrichment analysis (GSEA) to conduct unbiased comparisons of (+)-JQ1 versus (–)-JQ1 CAR T cells (Supplemental Table 2), we identified enrichment of gene expression profiles involved in chromatin remodeling and positive regulation of metabolic processes, including mTORC1 activation, which promotes the development of effector CD8+ T cells (46), and IL-2/STAT5 signaling (Figure 7B). JQ1 treatment also resulted in downregulation of pathways enriched in exhausted CD8+ T cells, such as metal ion homeostasis, positive regulation of cytokine production, and organonitrogen compound metabolism (10). TNF cellular response and signaling pathways, which were downregulated in clonally expanded TET2-deficient CD8+ CAR T cells with robust proliferative and antitumor capacity in a patient with advanced leukemia (12), were also diminished in response to BET bromodomain protein blockade (Figure 7B). Notably, significant enrichment of naive over memory-related genes was observed after exposure of CAR T cells of patients with CLL to active JQ1 (Figure 7C). Together, our results suggest quantitative alterations in genes and pathways known to drive CD8+ T cell exhaustion/dysfunction and to restrain differentiation, leading to augmentation and/or persistence of antitumor T cell efficacy. These changes may collectively lead to the reinvigoration and improved functionality of defective CAR T cells after (+)-JQ1 treatment.
JQ1 promotes CAR T cell reinvigoration in patients with CLL by suppressing TET2.
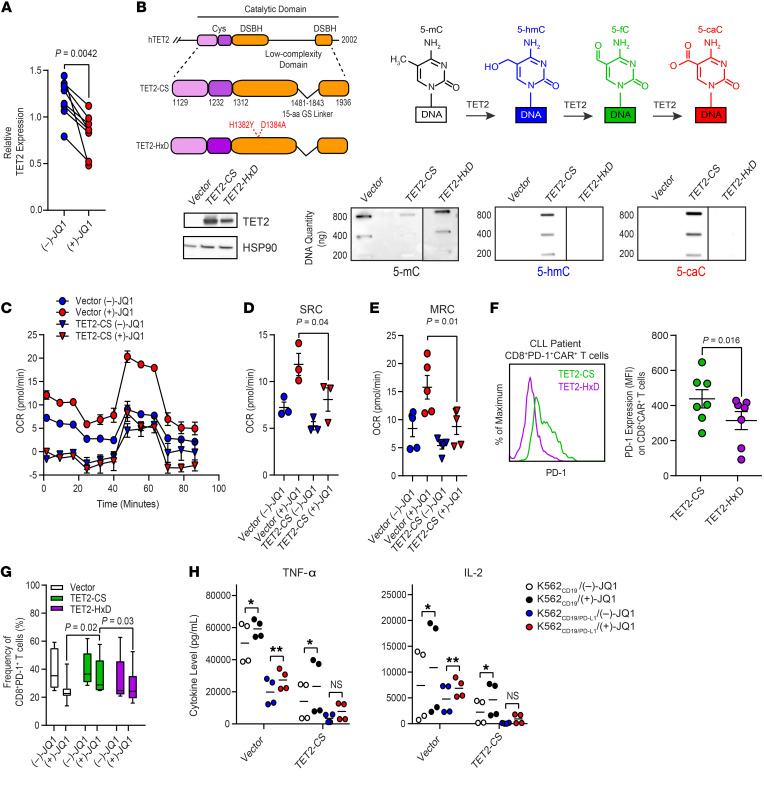
We next investigated the mechanism of how JQ1 ameliorates CAR T cell dysfunction. Our RNA-Seq data revealed that TET2 expression was downregulated in defective CAR T cells from patients with CLL after (+)-JQ1 treatment (Figure 7A). We therefore focused on investigating BET bromodomain protein–mediated regulation of TET2 expression as a potential determinant of CAR T cell reinvigoration. The generation of CD19 CAR T cells from leukapheresis products from patients with CLL expanded in the presence of (+)-JQ1 relative to an inactive (–)-JQ1 enantiomer, resulting in a significant downregulation of TET2 expression (Figure 8A). This finding suggested that pharmacological inhibition of BET proteins during cell manufacturing has the potential to modulate TET2, which is a major epigenetic regulator of CAR T cell differentiation, proliferation, persistence, and antitumor function (12).
Figure 8. Downregulation of TET2 expression by JQ1 treatment contributes to reinvigoration of CAR T cells from patients with CLL.
(A) TET2 mRNA reduction in CD19 CAR T cells transduced and expanded in the presence of 150 nM (–)-JQ1 or (+)-JQ1 (n = 8; paired t test). (B) Depiction of the organization of the human (h)TET2 catalytic domain and structures of FLAG-tagged TET2-CS and TET2-HxD with highlighted targets for mutagenesis in red (top left panel). Schematic of the sequential oxidations of 5-mC to 5-hmC and to 5-fC and to 5-caC catalyzed by TET2 is shown (top right panel). Immunoblot of TET2 protein levels in HEK293T cells is depicted. HSP90 was used as a loading control (bottom left panel). Dot blots for 5-mC, 5-hmC, and 5-caC in genomic DNA isolated from the above HEK293T cells transfected with an empty vector, TET2-CS, and TET2-HxD are shown (bottom right panel, blots are representative of 3 independent experiments). (C) OCR, (D) SRC, and (E) MRC of expanded CLL patient CAR T cells transduced with vector alone or TET2-CS and subsequently treated with (–)-JQ1 or (+)-JQ1 for 4 days (n = 3–4; paired t test). (F) Levels of PD-1 expression on CLL patient CD8+ CAR+ T cells transduced with TET2-CS or TET2-HxD (representative histograms, left panel; graphical data summary, right panel; n = 7, paired t test). (G) Frequency of CD8+ PD-1+ CLL patient T cells transduced with vector alone or lentiviral vectors encoding TET2-CS and TET2-HxD followed by treatment with (–)-JQ1 or (+)-JQ1 (n = 6–12, 2-tailed t test). Data are shown as the mean ± SEM. (H) Levels of TNF-α and IL-2 elaborated by CD8+CAR+ T cells transduced with vector alone or TET2-CS and subsequently treated with (–)-JQ1 or (+)-JQ1 and stimulated with irradiated K562CD19 or K562CD19/PD-L1 cells (n = 4, paired t test). *P ≤ 0.05, **P ≤ 0.01, NS, not significant.
Because of the complexity of TET2-regulated gene expression (47) and the possibility that TET2 may modify T cell function through a noncatalytic property (48) as observed with other epigenetic modifiers (49, 50), we first examined whether CAR T cell proliferative potency was affected, at least partially, by a catalytic activity–dependent function of TET2. To address this, we transduced cells with lentiviruses expressing the enzyme truncated to the minimal regions necessary for catalytic activity (CS) or a dead version of the catalytic domain (HxD) as a control. TET2 overexpression for both constructs was found to be uniform by Western blot of cell lysates (Figure 8B). TET2-CS catalyzed stepwise oxidation of 5-mC to 5-hmC, and 5-caC, while no oxidation of 5-mC was observed with TET2-HxD (Figure 8B), confirming the predicted activity of these constructs.
Overexpression of TET2-CS in CAR T cells significantly diminished proliferative capacity after repeated stimulation with CD19-expressing leukemia targets (Supplemental Figure 15A). In addition, forced expression of the TET2 catalytic domain compared with TET2-HxD in CD8+ T cells resulted in decreased metabolic fitness, as manifested by reduced OCR (Supplemental Figure 15B), SRC (Supplemental Figure 15C), MRC (Supplemental Figure 15D), and ECAR (Supplemental Figure 15E) during a mitochondrial stress test. When CD8+ T cells from patients with CLL were ectopically transduced with TET2-CS, JQ1-mediated metabolic reinvigoration was abrogated (Figure 8, C–E). Based on the observation that JQ1 treatment of exhausted CAR T cells from NR patients with CLL significantly diminished PD-1–mediated proliferative CAR T cell suppression, we reasoned that introduction of TET2-CS has the potential to hamper CAR T cell function through upregulation of PD-1 expression. Indeed, PD-1 levels were significantly elevated on CAR T cells transduced with TET2-CS compared with TET2-HxD (Figure 8F). Consistent with the potential for JQ1 to ameliorate CAR T cell dysfunction through a TET2-dependent mechanism, we also observed reversal of the JQ1-induced reduction in CD8+PD-1+ T cell frequency with TET2-CS overexpression (Figure 8G). The JQ1-treated TET2-CS expressing anti-CD19 CAR T cells exhibited decreased TNF-α and IL-2 production compared with those transduced with a control vector. The partial reversal of PD-1/PD-L1–triggered suppression of CAR T cell cytokine elaboration was diminished by forced TET2-CS expression (Figure 8H). Collectively, these findings demonstrated that TET2 may contribute to the reversal of CAR T cell dysfunction after BET bromodomain protein blockade through PD-1/PD-L1–dependent and –independent mechanisms.
Discussion
Identification of the determinants underlying failure of CAR T cells and further discovery of predictors of response and resistance are essential in order to ultimately improve the efficacy of this therapy. To date, few reports have described on-treatment prognostic markers of CAR T cell potency and clinical efficacy in advanced B cell malignancies. Sheih and colleagues recently reported on a small subset of patients with B-cell ALL, non-Hodgkin’s lymphoma, and CLL in which transcriptomic signatures of CAR T cell activation and proliferative expansion were progressively decreased without enrichment of exhaustion-related genes or an increase in coexpression of inhibitory receptors after infusion (51). In another investigation, patients with ALL who either did not achieve remission or relapsed within a few weeks of their initial response exhibited a LAG-3hi/TNF-αlo phenotype in peripheral blood CD8+ T cells at the time of leukapheresis. The presence of this cellular phenotype prior to cellular manufacturing was associated with poorly proliferative CAR T cells after adoptive transfer (8). Although we report high proportions of CD8+PD-1+ T cells in the infusion products of patients with CLL who had poor responses, neither PD-1 expression levels nor frequencies of this cell population alone at the height of in vivo CAR T cell expansion segregated PR/NR from CR/PRTD patients in our present study. However, we found that PR/NR patients with CLL with detectable postinfusion CAR T cells exhibited phenotypic and functional attributes consistent with T cell exhaustion/dysfunction at the time of peak in vivo proliferation, independently of baseline tumor burden/CD19 antigen load. Potential prospective biomarkers associated with failure of CAR T cell therapy in CLL include high levels of CTLA-4 and TIM-3 on the surface CD8+CAR+ T cells as well as increased frequencies of CAR T cells coexpressing PD-1 and TIM-3 enriched in PR/NR relative to CR/PRTD patients. These rapidly acquired markers of exhaustion were accompanied by functional attributes consistent with poor proliferation and cytotoxic capacity. Such on-treatment biomarkers are not only useful in helping to characterize clinical responses early during the course of cellular immunotherapy but also in informing the type of immunological failure and adapting subsequent treatment options. Thus, our findings indicate a multifaceted but potentially predictable interplay between patient-intrinsic and CAR T cell–intrinsic factors that could significantly affect clinical outcome following CD19 CAR T cell therapy in relapsed or refractory CLL.
Importantly, we demonstrated that progressive loss of cell-surface CAR expression can be seen in clinical trial participants undergoing CD19 CAR T cell therapy. These findings revealed that CAR silencing limited the therapeutic efficacy of CAR T cells targeting CLL, given that leukemia relapse in a small subset of NR patients coincided with the disappearance of CAR at the protein but not DNA level. In the context of immunotherapeutic strategies, T cells can be modified using integrating vectors to permanently express therapeutic transgenes to potentiate recognition of a tumor antigen, increase proliferative potential, prolong in vivo persistence, and/or improve migratory capacity to tissues. However, loss of vector-driven transcriptional activity in cells containing proviral DNA has been observed early after transduction (52), which drastically limits the efficacy of vector-based gene delivery platforms. Although attempts to optimize vector design, particularly with γ-retroviruses, have augmented transgene expression, these systems remain susceptible to promoter shutdown (16, 53).
Bisulfite sequencing analysis of CpG sites within the vector-specific EF1α promoter in preinfusion and postinfusion CAR T cells of patients with CLL revealed either the absence or only low levels of DNA methylation. Further, ex vivo use of HDAC inhibitors failed to augment expression of the anti-CD19 CAR transgene, which had decreased expression in vivo in PR/NR patients or progressed to full extinction in 3 NR patients who initially exhibited sufficient therapeutic levels of CAR T cell expansion to effect complete remission. These results were somewhat surprising given reports that these epigenetic mechanisms potentiate transgene silencing of integrating vectors, particularly in embryonic stem cells (ESCs; refs. 54, 55). This may be attributed to inherent differences in transduced mature human T lymphocytes relative to ESCs and other cell types.
Another major epigenetic mechanism of transcriptional regulation involves BET bromodomain proteins. BRD4 has been proposed as a positive regulator of transcription that links histone acetylation with transcriptional regulators, such as the Mediator complex and P-TEFb (56). Paradoxically, BRD4 has been shown to be a transcriptional repressor, but the mechanisms behind this effect are largely unknown (23, 25). Because BRD4 was localized at the vector-specific EF1α promoter in CLL patient T cells with low levels of CAR on the surface, we assessed the impact of BET inhibition on transgene expression. BET bromodomain inhibitors exert biologic effects by displacing the acetylated histone readers BRD2, BRD3, and BRD4 from chromatin. We previously reported that the most influential variables predictive of negative clinical outcome in CD19 CAR T cell–treated PR/NR patients included proximity to the epigenetic modifications H4K16ac, H4K8ac, H3K14ac, and H4K12ac (22). Acetylation of histone H3 lysine 14 (H3K14ac) and H4 dual lysines — either K5 and K12 or K8 and K16 — is crucial for BRD4 binding to acetylated histone peptides (57), and JQ1-mediated displacement of BRD4 at the EF1α promoter via JQ1 rescues CAR expression in dysfunctional CLL patient T cells. This may explain, at least in part, why BRD4-mediated repression of lentiviral EF1α promoter–driven transgene expression was observed after CAR T cell infusion into PR/NR patients. An alternative explanation is that the BRD4-mediated repression of the CAR transgene is linked to the 3D genome reorganization of CAR T cells. Accordingly, it is likely that JQ1 treatment can reorganize long-range interactions such that the CAR transgene can be transcribed. This enhancement of CAR transgene expression, however, needs to be clinically evaluated to determine whether it is sufficient to improve objective responses.
In an attempt to elucidate the potential mechanisms responsible for mediating CAR T cell exhaustion in CLL, we examined whether chromatin signaling was associated with expression of inhibitory molecules and suppressed antitumor effector and metabolic functions. Preincubation with JQ1 of PBMCs from patients with CLL during CAR T cell expansion resulted in a significant decrease in the expression of PD-1 and TIM-3, which are well-characterized markers of T cell exhaustion. We observed the same effect of JQ1 in manufactured NR CLL patient CAR T cell infusion products. The profound decrease in CAR T cell proliferative capacity and cytokine elaboration that was seen upon engagement of the PD-1/PD-L1 axis was ameliorated during continuous culture with active JQ1, implicating PD-1 as a major control pathway of reinvigoration in the setting of BET bromodomain protein blockade. BET bromodomain protein inhibition also reversed several metabolic defects in dysfunctional CLL patient CD8+ CAR T cells, suggesting that dysregulated metabolism is a major driver of CD8+ T cell exhaustion in CAR T cell therapy of CLL and that metabolic inhibition of exhaustion through epigenetic programming may maximize the efficacy of cellular immunotherapy.
To reveal how BET protein inhibition can ameliorate CAR T cell exhaustion, we compared the transcriptional profiles of poorly functional CAR T cells treated with active versus inactive JQ1. In addition to diminished expression of genes and pathways that are uniquely enriched in exhausted T cells, we identified several pathways that are upregulated in CAR T cells treated with (+)-JQ1: chromatin remodeling, positive regulation of metabolic processes, and IL-2/STAT5 signaling. Notably, T cells engineered with CARs incorporating a truncated cytoplasmic domain of IL-2Rβ for STAT5 recruitment exhibit superior proliferation and reduced terminal differentiation in vitro and in vivo (58). In contrast to a study showing that JQ1 prevented effector differentiation and maintained CD8+ T cells with functional properties of stem cell–like and central memory T cells, we did not observe effects of JQ1 on memory CD8+ T cell differentiation in CLL. This is likely attributed to the effector or terminally differentiated state of lymphocytes dominating the T cell compartment in CLL (59) that were used in this study and in most clinical scenarios as the “seed” populations for CAR T cell production. However, we provide the unexpected observation that JQ1 treatment led to the significant enrichment of naive over memory-related T cell gene programs. Naive T cell deficits at baseline and after chemotherapy impair the potential for efficacious CAR T cell therapy in cancer (60), and strategies that prevent a decline in desirable features of naive differentiation states have the potential to positively influence CAR T cell expansion quality and antitumor function. To what extent the above pathways contribute to the exhaustion-ameliorating effects of JQ1 in CAR T cells is a focus of our ongoing investigations.
The potency-enhancing effects of JQ1 resulted at least in part from the transcriptional modulation of TET2. TET2 deficiency is known to potentiate the antitumor activity of antigen-specific T cells, including CAR T cells (12, 41), and even minor changes in TET2 expression can lead to pronounced functional consequences in the context of malignant hematopoiesis and stem cell self-renewal (61). Interestingly, treatment of Nanog (i.e., a pluripotency transcription factor) overexpressing ESCs with cobalt chloride, which inhibits iron-dependent dioxygenases, including TET2, has been shown to induce a dose-dependent loss of response to JQ1. Further, the ability of Nanog overexpression to maintain adequate levels of pluripotency-associated transcription factors during JQ1 treatment is blunted in TET1/2 double-mutant ESCs (62). Although we have reported that TET2 deficiency in CAR T cells maintains a central memory state and prevents effector differentiation (12), the contribution of TET2 in driving T cell exhaustion has not been well characterized. In this regard, we found that overexpression of the TET2 catalytic domain suppressed antigen-dependent CAR T cell proliferation and hampered metabolic function. In addition, forced expression of TET2-CS led to upregulation of PD-1 and suppressed effector cytokine production. Notably, PD-1/PDL-1 axis-dependent and -independent reinvigoration of exhausted CAR T cells mediated by JQ1 was reversed by TET2-CS overexpression. This suggests a critical role for methylcytosine dioxygenases in bridging “reading” of the histone code to lineage-specific chromatin accessibility and gene expression profiles that program T cell dysfunction. Pharmacological intervention through BET bromodomain protein blockade, which can transiently suppress TET2 expression, is potentially safe and suitable for this purpose. In addition, several other factors affect important steps in CAR T cell fate determination. Further exploration of JQ1-induced gene expression alterations will be crucial to achieving a comprehensive understanding of the reinvigoration mechanisms.
In summary, we showed that BET inhibition has potential use for adoptive cellular therapies hampered by synthetic antigen receptor silencing and the preexisting or acquired dysfunctional state of T cells. This report also provides a basis for differential CAR T cell persistence and antitumor function in clinical trials and provides insights into the mechanisms underlying human T cell exhaustion. These findings are applicable to a broad range of adoptive immunotherapy platforms and identify actionable cellular manufacturing improvement pathways to generate more potent products or rational in vivo combination strategies involving epigenetic drugs and engineered T cells.
Methods
Complete methods are in the Supplemental Methods.
Accession information.
The RNA-Seq data reported in this paper have been deposited in NCBI’s Gene Expression Omnibus (GSE175981).
Study approval.
This research was conducted according to University of Pennsylvania IRB-approved protocols. Written informed consent was provided by participants.
Author contributions
JAF designed the project and supervised the research. WK and JAF wrote the manuscript. WK, AD, IJ, CJO, MF, YW, IK, MG JED, EW, JX, JS, TJR, VGB, and KH performed the experiments. WK, WW, CJO, TY, JKE, RMJ, JKJ, KTM, MMD, SFL, GV, and JAF analyzed the results. JJM, SFL, and DLP provided patient samples. MVM, JEB, BLL, DLP, FDB, RMK, CHJ, MMD, SFL, and GV provided critical feedback and edited the manuscript.
Supplementary Material
Acknowledgments
We acknowledge Roddy O’Connor, Omkar Kawalekar, Steven Avino, Stefan Lundh, McKensie Collins, Theresa Colligon, Gabriela Plesa, Yolanda Mahnke, Felipe Bedoya, Joseph Cohen, Fang Chen, Natalka Koterba, and Farzana Nazimuddin for technical assistance; Vanessa Gonzalez, Christopher Nobles, and Edward Pequignot for help with patient sample analysis; and Anne Chew, Brian Keith, Jianxin You, Elizabeth Hexner, Donald Siegel, James Riley, and Michael Milone (University of Pennsylvania) for their helpful suggestions. We thank the Human Immunology Core for providing leukocytes, the Clinical Cell and Vaccine Production Facility for GMP cell manufacturing, and the Hospital of the University of Pennsylvania Apheresis Unit for PBMC collections.
This work was supported by the Bob Levis Funding Group (to BLL, CHJ, and JAF). AD receives funding from the Training in HIV Pathogenesis Program (T32 AI007632) from the National Institute of Allergy and Infectious Diseases. Additional funding comes from P01 CA214278 (to RMY, DLP, CHJ, SFL, and JAF), R01 CA241762 (to JJM, FDB, SFL, and JAF), U54 CA244711 (to RMY, CHJ, and JAF), P30 CA016520-44S3 (to SFL and JAF), and P30 CA016520-44S4 (to SFL and JAF) grants from the National Cancer Institute; a U01 AG066100 (to RMY, CHJ, and JAF) grant from the National Institute on Aging; an R01 GM118501 (to RMK) grant from the National Institute of General Medical Sciences; an Emerging Cancer Informatics Center of Excellence award from the Penn Institute for Biomedical Informatics and Abramson Cancer Center (to GV and JAF); the Gabrielle’s Angel Foundation (to JAF); an Alliance for Cancer Gene Therapy Investigator Award in Cell and Gene Therapy for Cancer (to RMY and JAF); and Novartis (to JJM, RMY, BLL, DLP, FDB, CHJ, SFL, and JAF).
D. Campana, C. Imai, and others at St. Jude Children’s Research Hospital designed, developed, and provided, under material transfer agreements, the CAR used in this study.
The graphical abstract was created using BioRender.
Version 1. 08/16/2021
Electronic publication
Funding Statement
Center of Excellence Award
Investigator Award in Cell and Gene Therapy for Cancer
Footnotes
Conflict of interest: JS, MVM, JJM, RMY, BLL, DLP, FDB, CHJ, MMD, SFL, and JAF are inventors of intellectual property licensed by the University of Pennsylvania to Novartis and have received patent royalties (USA Patent Number WO/2015/157252; Argentina Patent Number AR099974A1; USA Patent Numbers WO/2016/164580, WO/2017/049166, US20200087376A1, and US20200370012A1; European Patent Office Patent Number EP3788369A1). JJM receives funding from Novartis, Incyte, IASO Biotherapeutics, and Kite/Gilead; is a consultant for Simcere of America, Shanghai Unicar Therapy, Johnson & Johnson, Poseida Therapeutics, and IASO Biotherapeutics; and serves on the medical advisory board and scientific advisory board (SAB) for IASO Biotherapeutics. BLL and CHJ are scientific founders of Tmunity Therapeutics, for which they have founder’s stock. BLL, MMD, CHJ, and JAF have received royalties from Tmunity Therapeutics. CHJ and JAF are founders of DeCART Therapeutics. BLL is a consultant for Novartis and Terumo and SAB member for Avectas, Patheon/Thermo Fisher Scientific Viral Vector Services, Immuneel, In8bio, Ori Biotech, and Vycellix. MMD is a consultant and member of the SAB for Cellares Corporation. SFL is a consultant for Gilead/Kite. JAF is a consultant for Guidepoint and LEK Consulting.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(16):e145459.https://doi.org/10.1172/JCI145459.
Contributor Information
Weimin Kong, Email: kongw@mail.med.upenn.edu.
Alexander Dimitri, Email: Alexander.Dimitri@pennmedicine.upenn.edu.
Wenliang Wang, Email: wangwl.me@gmail.com.
In-Young Jung, Email: Inyoung.Jung@pennmedicine.upenn.edu.
Christopher J. Ott, Email: christopher_ott@dfci.harvard.edu.
Maria Fasolino, Email: maria.d.fasolino@gmail.com.
Yan Wang, Email: wangyan0816@gmail.com.
Irina Kulikovskaya, Email: ikulikov@pennmedicine.upenn.edu.
Minnal Gupta, Email: minnalg@pennmedicine.upenn.edu.
Todd Yoder, Email: Todd.Yoder@pennmedicine.upenn.edu.
Jamie E. DeNizio, Email: jamiedenizio@gmail.com.
John K. Everett, Email: everj@pennmedicine.upenn.edu.
Erik F. Williams, Email: Erik.Williams@Pennmedicine.upenn.edu.
Jun Xu, Email: junpeggy@hotmail.com.
John Scholler, Email: schoj@pennmedicine.upenn.edu.
Tyler J. Reich, Email: treich001@gmail.com.
Vijay G. Bhoj, Email: vbhoj@pennmedicine.upenn.edu.
Kathleen M. Haines, Email: haineskm@pennmedicine.upenn.edu.
Marcela V. Maus, Email: mvmaus@mgh.harvard.edu.
J. Joseph Melenhorst, Email: mej@upenn.edu.
Regina M. Young, Email: ryoung@upenn.edu.
Julie K. Jadlowsky, Email: juliejad@pennmedicine.upenn.edu.
Katherine T. Marcucci, Email: katie.marcucci@gmail.com.
James E. Bradner, Email: james.bradner@novartis.com.
Bruce L. Levine, Email: levinebl@mail.med.upenn.edu.
David L. Porter, Email: David.Porter@pennmedicine.upenn.edu.
Frederic D. Bushman, Email: bushman@mail.med.upenn.edu.
Rahul M. Kohli, Email: rkohli@upenn.edu.
Carl H. June, Email: cjune@upenn.edu.
Megan M. Davis, Email: suhoski@pennmedicine.upenn.edu.
Simon F. Lacey, Email: simon.lacey@uphs.upenn.edu.
Golnaz Vahedi, Email: vahedi@pennmedicine.upenn.edu.
Joseph A. Fraietta, Email: jfrai@upenn.edu.
References
- 1.Porter DL, et al. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maude SL, et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med. 2018;378(5):439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park JH, et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N Engl J Med. 2018;378(5):449–459. doi: 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee DW, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385(9967):517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porter DL, et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci Transl Med. 2015;7(303):303ra139. doi: 10.1126/scitranslmed.aac5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geyer MB, et al. Safety and tolerability of conditioning chemotherapy followed by CD19-targeted CAR T cells for relapsed/refractory CLL. JCI Insight. 2019;5:e122627. doi: 10.1172/jci.insight.122627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraietta JA, et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med. 2018;24(5):563–571. doi: 10.1038/s41591-018-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finney OC, et al. CD19 CAR T cell product and disease attributes predict leukemia remission durability. J Clin Invest. 2019;129(5):2123–2132. doi: 10.1172/JCI125423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sen DR, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354(6316):1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pauken KE, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354(6316):1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghoneim HE, et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell. 2017;170(1):142–157. doi: 10.1016/j.cell.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraietta JA, et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature. 2018;558(7709):307–312. doi: 10.1038/s41586-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kagoya Y, et al. BET bromodomain inhibition enhances T cell persistence and function in adoptive immunotherapy models. J Clin Invest. 2016;126(9):3479–3494. doi: 10.1172/JCI86437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long AH, et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat Med. 2015;21(6):581–590. doi: 10.1038/nm.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan RA, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314(5796):126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowicki TS, et al. Epigenetic suppression of transgenic T-cell receptor expression via gamma-retroviral vector methylation in adoptive cell transfer therapy. Cancer Discov. 2020;10(11):1645–1653. doi: 10.1158/2159-8290.CD-20-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitsuyasu RT, et al. Prolonged survival and tissue trafficking following adoptive transfer of CD4zeta gene-modified autologous CD4(+) and CD8(+) T cells in human immunodeficiency virus-infected subjects. Blood. 2000;96(3):785–793. [PubMed] [Google Scholar]
- 18.Frey NV, et al. Long-term outcomes from a randomized dose optimization study of chimeric antigen receptor modified T cells in relapsed chronic lymphocytic leukemia. J Clin Oncol. 2020;38(25):2862–2871. doi: 10.1200/JCO.19.03237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang AC, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545(7652):60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aranyi T, et al. Systemic epigenetic response to recombinant lentiviral vectors independent of proviral integration. Epigenetics Chromatin. 2016;9:29. doi: 10.1186/s13072-016-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann A, et al. Epigenetic regulation of lentiviral transgene vectors in a large animal model. Mol Ther. 2006;13(1):59–66. doi: 10.1016/j.ymthe.2005.07.685. [DOI] [PubMed] [Google Scholar]
- 22.Nobles CL, et al. CD19-targeting CAR T cell immunotherapy outcomes correlate with genomic modification by vector integration. J Clin Invest. 2020;130(2):673–685. doi: 10.1172/JCI130144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakamaki JI, et al. Bromodomain protein BRD4 is a transcriptional repressor of autophagy and lysosomal function. Mol Cell. 2017;66(4):517–532. doi: 10.1016/j.molcel.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boehm D, et al. BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism. Cell Cycle. 2013;12(3):452–462. doi: 10.4161/cc.23309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu SY, et al. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20(17):2383–2396. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu H, et al. BET bromodomain inhibition promotes anti-tumor immunity by suppressing PD-L1 expression. Cell Rep. 2016;16(11):2829–2837. doi: 10.1016/j.celrep.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mele DA, et al. BET bromodomain inhibition suppresses TH17-mediated pathology. J Exp Med. 2013;210(11):2181–2190. doi: 10.1084/jem.20130376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banerjee C, et al. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J Leukoc Biol. 2012;92(6):1147–1154. doi: 10.1189/jlb.0312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraietta JA, et al. Ibrutinib enhances chimeric antigen receptor T-cell engraftment and efficacy in leukemia. Blood. 2016;127(9):1117–1127. doi: 10.1182/blood-2015-11-679134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turtle CJ, et al. Durable molecular remissions in chronic lymphocytic leukemia treated with CD19-specific chimeric antigen receptor-modified T cells after failure of ibrutinib. J Clin Oncol. 2017;35(26):3010–3020. doi: 10.1200/JCO.2017.72.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Bruggen JAC, et al. Chronic lymphocytic leukemia cells impair mitochondrial fitness in CD8(+) T cells and impede CAR T-cell efficacy. Blood. 2019;134(1):44–58. doi: 10.1182/blood.2018885863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawalekar OU, et al. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity. 2016;44(3):712. doi: 10.1016/j.immuni.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 33.Bengsch B, et al. Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8(+) T cell exhaustion. Immunity. 2016;45(2):358–373. doi: 10.1016/j.immuni.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghassemi S, et al. Reducing ex vivo culture improves the antileukemic activity of Chimeric Antigen Receptor (CAR) T cells. Cancer Immunol Res. 2018;6(9):1100–1109. doi: 10.1158/2326-6066.CIR-17-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller BC, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20(3):326–336. doi: 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satpathy AT, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol. 2019;37(8):925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alvisi G, et al. IRF4 instructs effector Treg differentiation and immune suppression in human cancer. J Clin Invest. 2020;130(6):3137–3150. doi: 10.1172/JCI130426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woroniecka K, et al. T-cell exhaustion signatures vary with tumor type and are severe in glioblastoma. Clin Cancer Res. 2018;24(17):4175–4186. doi: 10.1158/1078-0432.CCR-17-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carty SA, et al. The loss of TET2 promotes CD8+ T cell memory differentiation. J Immunol. 2018;200(1):82–91. doi: 10.4049/jimmunol.1700559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bunse L, et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat Med. 2018;24(8):1192–1203. doi: 10.1038/s41591-018-0095-6. [DOI] [PubMed] [Google Scholar]
- 41.Tyrakis PA, et al. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature. 2016;540(7632):236–241. doi: 10.1038/nature20165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tai TS, et al. GATA-3 regulates the homeostasis and activation of CD8+ T cells. J Immunol. 2013;190(1):428–437. doi: 10.4049/jimmunol.1201361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koneru M, et al. Defective adhesion in tumor infiltrating CD8+ T cells. J Immunol. 2006;176(10):6103–6111. doi: 10.4049/jimmunol.176.10.6103. [DOI] [PubMed] [Google Scholar]
- 44.Imanishi T, et al. mTORC1 signaling controls TLR2-mediated T-cell activation by inducing TIRAP expression. Cell Rep. 2020;32(3):107911. doi: 10.1016/j.celrep.2020.107911. [DOI] [PubMed] [Google Scholar]
- 45.Klein Geltink RI, et al. Mitochondrial priming by CD28. Cell. 2017;171(2):385–397. doi: 10.1016/j.cell.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollizzi KN, et al. mTORC1 and mTORC2 selectively regulate CD8+ T cell differentiation. J Clin Invest. 2015;125(5):2090–2108. doi: 10.1172/JCI77746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hon GC, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell. 2014;56(2):286–297. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ito K, et al. Non-catalytic roles of Tet2 are essential to regulate hematopoietic stem and progenitor cell homeostasis. Cell Rep. 2019;28(10):2480–2490. doi: 10.1016/j.celrep.2019.07.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jani A, et al. A novel genetic strategy reveals unexpected roles of the Swi-Snf-like chromatin-remodeling BAF complex in thymocyte development. J Exp Med. 2008;205(12):2813–2825. doi: 10.1084/jem.20080938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaiyachati BH, et al. BRG1-mediated immune tolerance: facilitation of Treg activation and partial independence of chromatin remodelling. EMBO J. 2013;32(3):395–408. doi: 10.1038/emboj.2012.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheih A, et al. Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nat Commun. 2020;11(1):219. doi: 10.1038/s41467-019-13880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart CL, et al. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1982;79(13):4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krall WJ, et al. Increased levels of spliced RNA account for augmented expression from the MFG retroviral vector in hematopoietic cells. Gene Ther. 1996;3(1):37–48. [PubMed] [Google Scholar]
- 54.Knight S, et al. Safer, silencing-resistant lentiviral vectors: optimization of the ubiquitous chromatin-opening element through elimination of aberrant splicing. J Virol. 2012;86(17):9088–9095. doi: 10.1128/JVI.00485-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther. 2005;16(11):1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 56.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54(5):728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dey A, et al. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A. 2003;100(15):8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kagoya Y, et al. A novel chimeric antigen receptor containing a JAK-STAT signaling domain mediates superior antitumor effects. Nat Med. 2018;24(3):352–359. doi: 10.1038/nm.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riches JC, et al. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood. 2013;121(9):1612–1621. doi: 10.1182/blood-2012-09-457531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das RK, et al. Naïve T-cell deficits at diagnosis and after chemotherapy impair cell therapy potential in pediatric cancers. Cancer Discov. 2019;9(4):492–499. doi: 10.1158/2159-8290.CD-18-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finley LWS, et al. Pluripotency transcription factors and Tet1/2 maintain Brd4-independent stem cell identity. Nat Cell Biol. 2018;20(5):565–574. doi: 10.1038/s41556-018-0086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.