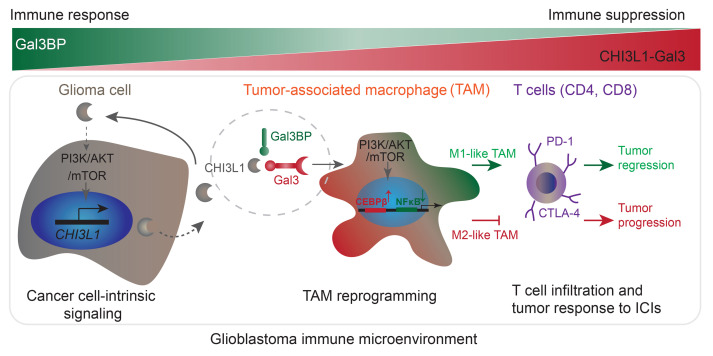
Abstract
Keywords: Oncology
Keywords: Brain cancer, Cancer, Macrophages
Introduction
Glioblastoma (GBM), the most common and lethal primary brain tumor, with a median survival rate of only 15 months, remains incurable despite intensive multimodal treatment of surgical resection, radio-chemotherapy, and antiangiogenic therapy with bevacizumab (1–3). While immunotherapies have been highly effective against some types of cancer, the disappointing results of clinical trials for GBM immunotherapy represent continued challenges (4, 5). Therefore, effective therapies for patients with GBM are urgently needed.
GBM is highly immunosuppressive and resistant to immunotherapy because glioma cells escape effective antitumor immunity by programming the tumor microenvironment (TME) (5, 6). Tumor-associated macrophages (TAMs) and microglia (MG), the major component of the GBM TME, account for up to 30% to 50% of total tumor composition (7). GBM TAMs originate from BM-derived blood monocytes (monocyte-derived macrophages [MDMs]) and brain-resident MG (7, 8). Previous studies reported that MG account for approximately 15% of TAMs and mainly localize in peritumoral areas, whereas MDMs preferentially localize in intratumoral regions and constitute approximately 85% of the total TAM population. MDMs significantly contribute to the immunosuppressive microenvironment of high-grade glioma (9, 10), suggesting different functions of MG and MDMs within the GBM TME. Increasing evidence indicates that protumor M2-like TAMs are frequently accumulated and associated with higher-grade tumors (11, 12). In contrast, repolarization of TAMs toward an antitumor M1-like phenotype results in tumor regression by producing proinflammatory cytokines and key molecules to stimulate T cell antitumor response. This suggests a potential therapeutic strategy of converting M2-like TAMs to M1-like TAMs for the treatment of GBM (7, 13). Therefore, the classification of M1/M2-like TAM phenotypes and the functional plasticity of TAMs regulated through glioma cell–intrinsic mechanisms remain an area of active investigation.
Here, we show that chitinase-3-like 1 (CHI3L1), also known as human homolog YKL-40, predominately modulates the GBM TME using unbiased approaches. CHI3L1 signaling selectively regulates tumor infiltration and cell migration of MDMs and MG by forming distinct protein-binding complexes. CHI3L1 protein complexes further reprogram TAMs to regulate T cell–mediated immune response in GBM progression. Importantly, we developed a peptide to disrupt CHI3L1 protein complexes, which can promote tumor regression in syngeneic mouse GBM models, providing a potential therapeutic strategy to eradicate this devastating brain tumor.
Results
Cancer cell–intrinsic CHI3L1 is regulated by the PI3K/AKT/mTOR pathway in a positive feedback loop.
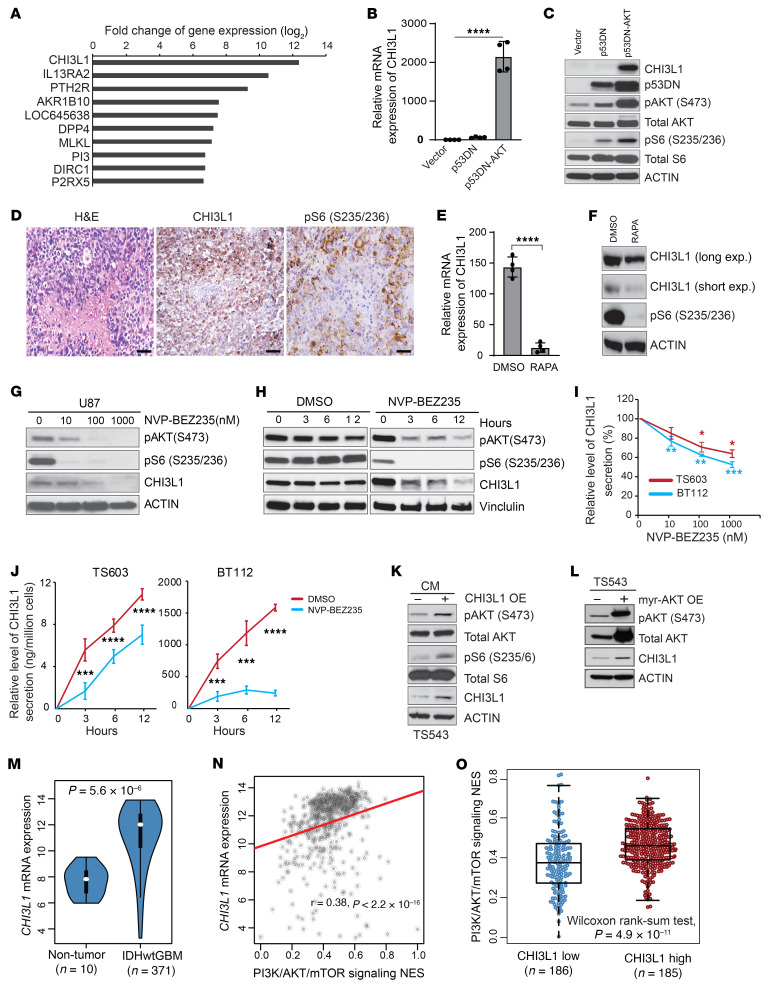
We previously developed a de novo GBM model using a myristoylated form of AKT (myr-AKT) and dominant-negative p53–engineered (p53DN-engineered) human neural stem cells (hNSCs), thereby enabling us to perform precise system-level comparisons between hNSCs and their derived GBM stem cells (GSCs) (14). To identify cancer cell–intrinsic factors for malignant transformation, we performed global analyses of differentially expressed genes between hNSCs and GSCs. CHI3L1 is the most significantly upregulated gene in GSCs derived from hNSCs overexpressing myr-AKT and p53DN (Figure 1A). In vitro and in vivo validations revealed that CHI3L1 is highly expressed in GSCs and their derived tumors with activated AKT signaling (Figure 1, B–D). In contrast, inhibiting AKT/mTOR signaling by rapamycin decreased CHI3L1 mRNA and protein expression in hNSC-p53DN-AKT (Figure 1, E and F).
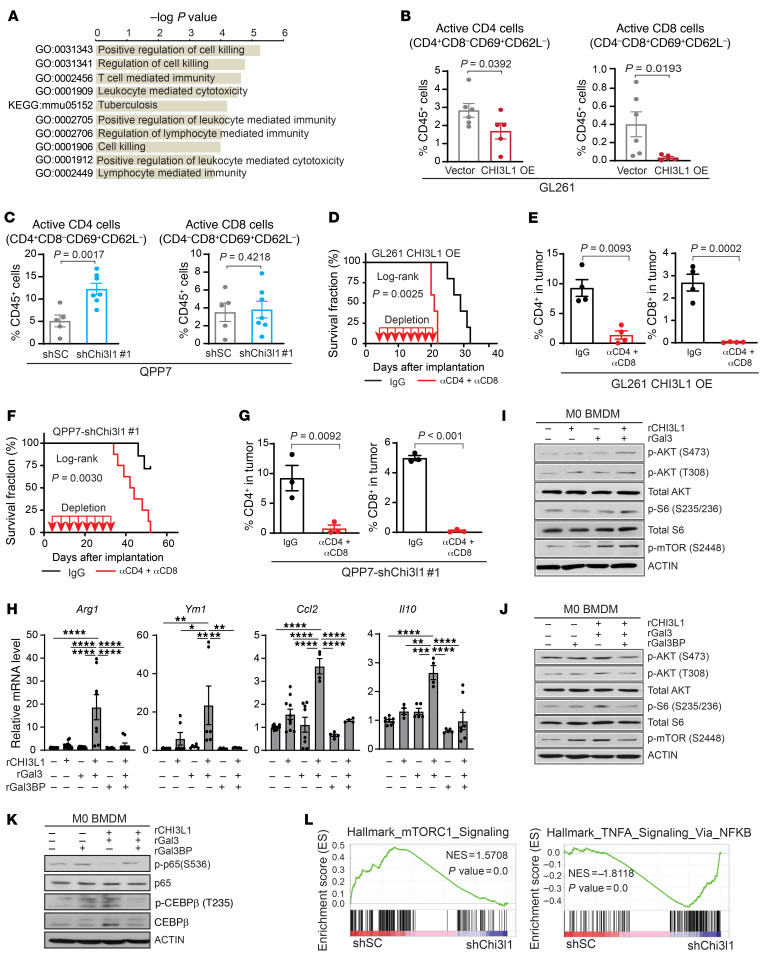
Figure 1. CHI3L1 upregulation is associated with activation of PI3K/AKT/mTOR signaling in GBM.
(A) Top 10 upregulated genes in hNSC versus hNSC-p53DN-AKT ranked by fold change of gene expression. qRT-PCR for CHI3L1 expression (B) and IB analysis of indicated proteins (C) in hNSC expressing p53DN or/and myr-AKT. (D) Representative H&E and IHC images showing indicated proteins in tumors derived from hNSCs-p53DN-AKT. Scale bars: 50 μm. qRT-PCR (E) and IB analysis (F) of indicated genes and proteins in hNSC-p53DN-AKT with rapamycin (RAPA) treatment (100 nM, 24 hours); CHI3L1 signal was shown in both long and short exposure time. IB analysis of indicated proteins in U87 treated with NVP-BEZ235 in a dose- (G) and time-dependent (H) manner. CHI3L1 secretion in the CM was assessed by ELISA from human GBM neurosphere lines treated with NVP-BEZ235 at indicated concentrations after 12 hours of treatment (I) or at a concentration of 1 μM at indicated times (J). (K) IB analysis of indicated proteins in human GBM neurosphere line TS543 treated with CM of TS543 overexpressing (OE) CHI3L1 versus control (K) or overexpressing myr-AKT versus control (L). (M) CHI3L1 mRNA expression in TCGA IDH-WT GBM tumors compared with nontumor brain tissues. Gene expression was normalized by robust multichip average (RMA), and the P values were calculated by Wilcoxon’s rank-sum test. (N) Association between CHI3L1 mRNA expression and the PI3K/AKT/mTOR signature score. Gene expression was normalized by RMA and P values were calculated by Spearman’s rank correlation. (O) Enrichment of the PI3K/AKT/mTOR signature in IDH-WT GBM with high and low levels of CHI3L1 mRNA expression. Data are presented as the mean ± SD (n ≥ 3 replicates). P values were calculated using 1-way ANOVA (B) or 1-tailed, unpaired t test (E, I, and J). *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001.
CHI3L1 is a secreted glycoprotein with chitin-binding capacity, but lacking chitinase activity (15), that plays a role in tissue remodeling, inflammation, and cancer (16). Although CHI3L1 is highly expressed and associated with a poor clinical outcome in GBM patients (17, 18), CHI3L1 regulation and its molecular mechanism or mechanisms of action remain undefined. To test the hypothesis that CHI3L1 is predominantly upregulated by the PI3K/AKT/mTOR signaling pathway, we treated the GBM neurosphere line TS603 and U87 cells with NVP-BEZ235 (a dual PI3K and mTOR inhibitor). Immunoblot (IB) analysis revealed that CHI3L1 expression was regulated by PI3K/AKT/mTOR signaling in a time- and dose-dependent manner (Figure 1, G and H, and Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI147552DS1). Furthermore, we measured CHI3L1 levels in the conditioned media (CM) of 2 GBM neurosphere lines treated with NVP-BEZ235 or NVP-BKM120 (a pan-PI3K kinase inhibitor). Inhibition of PI3K/AKT/mTOR activation decreased CHI3L1 secretion in a dose- and time-dependent manner (Figure 1, I and J, and Supplemental Figure 1, C and D). Importantly, the CM from GBM neurosphere line TS543 overexpressing CHI3L1 enhanced pAKT, pS6, and CHI3L1 levels over control in TS543 cells (Figure 1K). Conversely, overexpression (OE) of myr-AKT dramatically increased CHI3L1 levels in TS543 (Figure 1L). These results demonstrate a positive feedback loop between CHI3L1 and PI3K/AKT/mTOR signaling.
We analyzed a previously published single-cell GBM transcriptomic patient data set (19), finding that glioma cells express high levels of CHI3L1 and may represent a major source of CHI3L1 in the GBM TME (Supplemental Figure 1, E–G). In The Cancer Genome Atlas (TCGA) GBM data sets, CHI3L1 is highly expressed in tumors versus nontumor tissues and mesenchymal versus proneural and classical subtypes (Figure 1M and Supplemental Figure 1H). Moreover, higher levels of CHI3L1 mRNA expression are significantly associated with PTEN deletions/mutations, PI3K/AKT/mTOR signaling activation, and poor outcome in patients with isocitrate dehydrogenase WT (IDH-WT) GBM (Figure 1, N and O, and Supplemental Figure 1, I and J). Together, these results reinforce that a positive regulatory feedback loop of PI3K/AKT/mTOR-CHI3L1 signaling may play a pivotal role in regulating GBM progression and treatment response.
CHI3L1 plays a predominant role in shaping the landscape of the GBM immune TME.
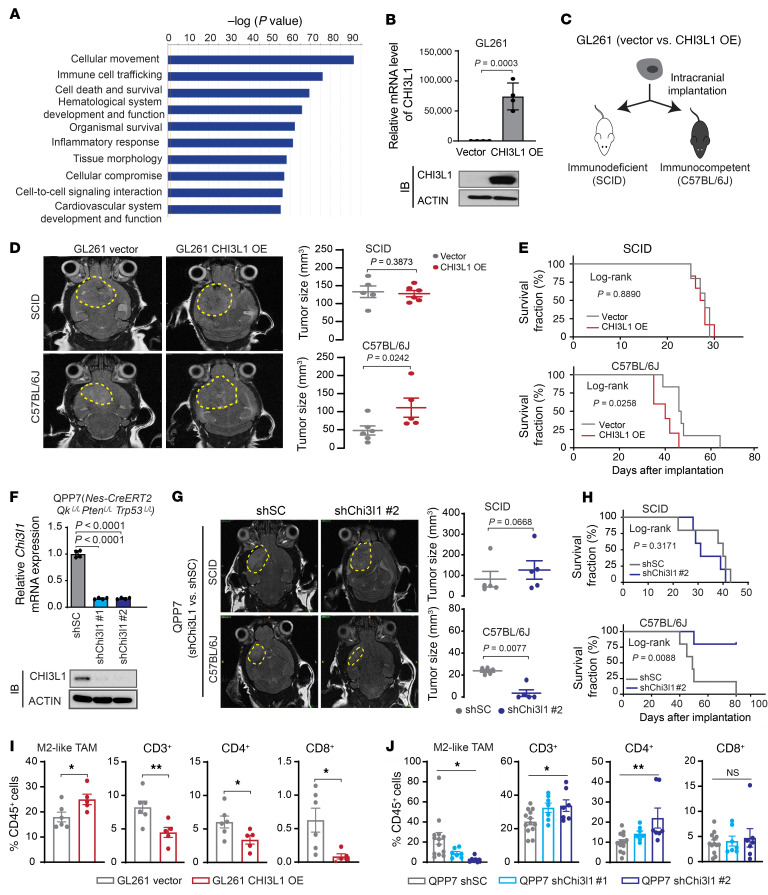
To determine the main function of CHI3L1 in GBM, we performed in vivo CHI3L1 gain-of-function studies in an orthotopic xenograft model of TS543 intracranially implanted in SCID mice. Surprisingly, enforced CHI3L1 expression did not significantly promote tumor progression compared with that in vector controls (Supplemental Figure 2, A–C). In the GBM TCGA data set, CHI3L1-correlated genes (1960 genes) were mainly associated with cellular movement, immune cell trafficking, and cell-to-cell signaling by Ingenuity Pathway Analysis (IPA) (Figure 2A and Supplemental Table 1). These data indicate that CHI3L1 may play a pivotal role in regulating the GBM immune TME. To this end, we compared tumor progression between SCID and immunocompetent (C57BL/6) mice intracranially implanted with murine glioma GL261 cells that have low levels of endogenous CHI3L1, but forced expression of the human CHI3L1 gene (GL261-CHI3L1; Figure 2, B and C). Notably, CHI3L1 OE increased tumor size and decreased survival in C57BL/6 mice, but not in SCID mice (Figure 2, D and E). Conversely, we generated an orthotopic syngeneic mouse model of GBM using QPP7 cells that were derived from a spontaneous murine glioma model, Nes-CreERT2QkL/LPtenL/LTrp53L/L (20), with high levels of endogenous CHI3L1 (Figure 2F and Supplemental Figure 2D). In vivo loss-of-function studies revealed that knockdown (KD) of the mouse Chi3l1 gene (shChi3l1#1 and #2) in QPP7 cells significantly repressed tumor growth in the syngeneic mice, as seen using MRI 4 weeks after implantation (Figure 2F and Supplemental Figure 2, E and F). Importantly, comparative analyses of tumor progression in SCID and C57BL/6 mice bearing QPP7 tumors demonstrated that Chi3l1 KD decreased tumor size and extended the survival of immunocompetent mice, but not immunodeficient mice (Figure 2, G and H). Together, these results suggest that CHI3L1 predominantly regulates the tumor immune TME rather than tumor cells per se.
Figure 2. Tumor progression and the immune microenvironment are implicated in glioma mouse models with altering CHI3L1 expression.
(A) Top 10 biological functional pathways are enriched in CHI3L1-correlated genes in TCGA GBM data sets using IPA. (B) qRT-PCR and IB analyses of the expression levels of CHI3L1 mRNA and protein in GL261 overexpressing vector control or human CHI3L1 gene. Data are presented as mean ± SD. P values were calculated using 1-tailed, unpaired t test. (C) Illustration of 2 orthotopic xenograft models with GL261 CHI3L1 OE or vector control. (D) Representative MRI from mice after intracranial injection of GL261 with CHI3L1 OE or vector. T2 sequences demonstrate infiltrative tumors in the mouse brain (yellow line). Tumor volume was measured by the T2 MRI scan. (E) Kaplan-Meier tumor-free survival analysis of GL261 models. (F) qRT-PCR and IB analyses of the expression levels of CHI3L1 mRNA and protein in QPP7 cells infected with lentivirus carrying shRNA targeting mouse Chi3l1 gene (shChi3l1#1 and #2) versus shRNA scrambled controls (shSC). Data are presented as mean ± SD. P values were calculated by 1-way ANOVA with Dunnett’s multiple comparisons test. (G) Representative MRI from 2 orthotopic xenograft glioma mouse models bearing QPP7 with shChi3l1#2 versus shSC. Tumor volume was measured by the T2 MRI scan. (H) Kaplan-Meier tumor-free survival analysis of QPP7 models. Flow cytometry analyses of the indicated cell populations in GL261 (I) and QPP7(J) syngeneic mouse models with altering CHI3L1 expression. Dots represent mice from the group; data are presented as mean ± SEM. P values were calculated using a 1-tailed, unpaired t test (I) or 1-way ANOVA (J). *P < 0.05; **P < 0.01.
To determine the effect of CHI3L1 on immune cell distribution in the TME of GBM, we analyzed the major cell populations, including TAMs, T cells, NK cells, and myeloid-derived suppressor cells (MDSCs). Flow cytometry of tumors revealed that enforced CHI3L1 expression in GL261 mouse models significantly increased the M2-like TAMs (CD45+CD11b+CD14+MHCII+Ly6C–), but decreased CD3+, CD4+, and CD8+ T cell populations (Figure 2 I and Supplemental Figure 2, G and H). In contrast, Chi3l1 KD in QPP7 syngeneic models significantly decreased the M2-like TAMs, but increased CD3+ and CD4+ T cell populations (Figure 2J and Supplemental Figure 2I). These findings were further validated by performing single-cell mass cytometry (CyTOF) analysis in the QPP7 tumors with Chi3l1 KD versus controls using the additional markers to identify cell populations (e.g., Arg1, iNOS, F4/80, CD90.2, etc.; Supplemental Figure 2, J–L). Notably, QPP7 tumors, compared with GL261 tumors, did not show significant changes of CD8+ T cells, which may support the low mutational load in QPP7, but not in GL261, cells, determining the modest CD8+ T cell infiltration in these syngeneic GBM mouse models (20, 21). Collectively, the gain/loss-of-function studies reveal a predominant role of CHI3L1 in regulating GBM progression by reprogramming the tumor immune microenvironment.
CHI3L1 selectively promotes infiltration of M2-like versus M1-like MDMs and MG.
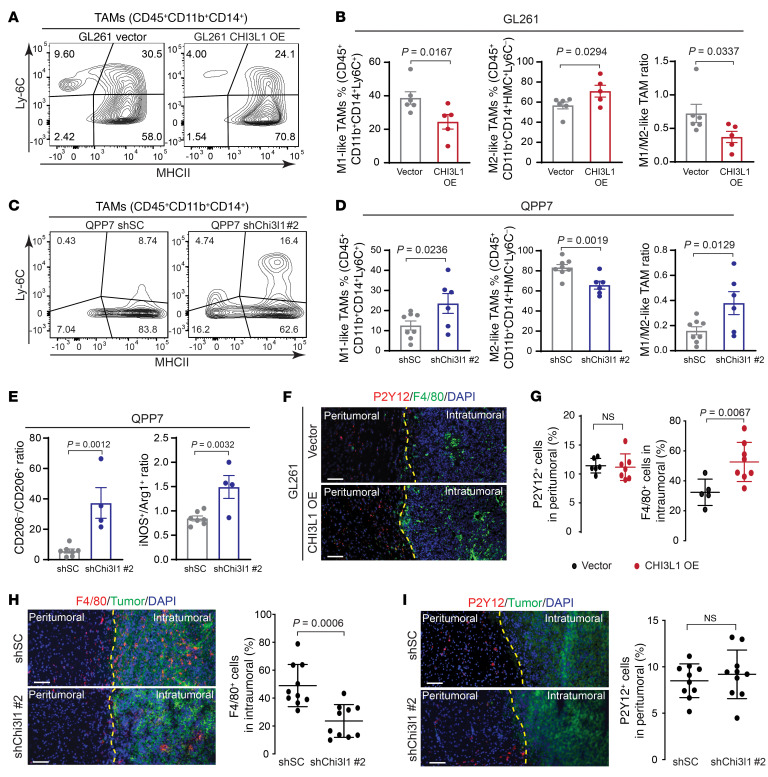
Previous studies reported the involvement of CHI3L1 in macrophage differentiation and recruitment associated with other pathological conditions (22, 23), although the mechanism of its action remains elusive. Furthermore, the M2-like TAMs consistently and significantly changed in response to CHI3L1 expression in our syngeneic mouse models. Therefore, we focused on how CHI3L1 reprograms TAMs in the GBM TME. Further flow cytometry analyses showed that OE of CHI3L1 increased the percentage of M2-like TAMs,but decreased the percentage of M1-like TAMs (CD45+CD11b+CD14+Ly6C+) and the M1-/M2-like TAM ratio in GL261 tumor models (Figure 3, A and B). Conversely, Chi3l1 KD decreased the percentage of M2-like TAMs, but increased the percentage of M1-like TAMs and the M1-/M2-like TAM ratio in QPP7-derived GBM models by flow cytometry (Figure 3, C and D, and Supplemental Figure 3, A and B). Moreover, additional markers used for TAM polarization analyses revealed a significant decrease of the CD45+CD68+CD11b+CD206+ and CD45+Arg1+ populations, but an increase in the ratios of CD206–CD206+ and iNOS+Arg1+ cells in QPP7 tumors (Figure 3E and Supplemental Figure 3C). These results suggest that CHI3L1 regulates TAM polarization toward M2-like phenotype in the GBM TME.
Figure 3. CHI3L1 induces M2-like MDM accumulation in vivo.
Representative flow cytometry analyses and quantitation showing the percentages of M1- and M2-like MDMs in tumors derived from GL261 (A and B) and QPP7 (C and D) glioma-bearing mice with altering CHI3L1 expression. (E) Ratio of CD206–:CD206+ cells from the CD45+CD68+CD11b+ cell population and ratio of iNOS+:Arg1+ cells from the CD45+ cell population in QPP7-derived tumors. Each dot represents 1 mouse; data are presented as mean ± SEM. P values were calculated using 1-tailed, unpaired t test. (F) Representative IF images for F4/80+ and P2Y12+ cells in tumor sections from the syngeneic mice bearing GL261-CHI3L1 versus vector control. (G) Quantitation of indicated cells in peritumoral and intratumoral regions, respectively. Representative IF images and quantitation for F4/80+ (H) and P2Y12+ (I) cells in tumor sections from QPP7 glioma-bearing mice with shChi3l1#2 versus shSC. Peritumoral and intratumoral regions are separated using yellow lines. Each dot represents 1 field of the peritumoral or intratumoral region from indicated tumors (n ≥ 3). Data are presented as mean ± SD. P values were calculated using 1-tailed, unpaired t test. Scale bars: 100 μm.
To specifically investigate CHI3L1-regulated MDMs and MG tumoral infiltration, we performed co-immunofluorescence (co-IF) staining to detect F4/80, a mature phagocytic cell marker, and P2Y12, a classic marker for MG (24, 25). Notably, OE of CHI3L1 in GL261-derived glioma models greatly increased F4/80+ cell accumulation in intratumoral regions, but did not significantly change infiltration of P2Y12+ MG, which predominantly reside in peritumoral regions (Figure 3, F and G). In contrast, there was a decreased F4/80+ cell infiltration in QPP7-derived gliomas without a significant change of peritumoral P2Y12+ MG in Chi3l1 KD versus controls (Figure 3, H and I). Furthermore, intratumoral MDMs (M2-like) were significantly accumulated in GL261-CHI3L1 tumors, but reduced in Chi3l1 KD QPP7 tumors, based on IF staining using additional markers (CD206 and CD49D; Supplemental Figure 3, D–G). These data reveal that cancer cell–intrinsic CHI3L1 promotes accumulation of MDMs over MG within the tumor, which provides a mechanistic explanation for the observation of abundant MDM infiltration in tumor lesions, while the preferential occupation of MG is in the periphery (9, 10, 19).
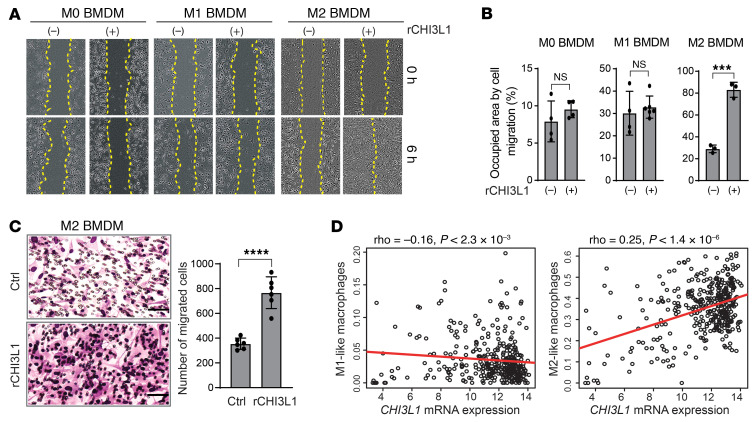
To further verify the effect of CHI3L1 on TAM infiltration, we performed in vitro scratch-wound healing and Transwell assays to examine cell migration of BM-derived macrophages (BMDMs) and MG cells treated with recombinant CHI3L1 protein (rCHI3L1). We generated and confirmed the polarization states of M0, M1, and M2 macrophages using isolated BMDMs from C57BL/6 mice according to a previous standard protocol (ref. 26 and Supplemental Figure 3H). Strikingly, rCHI3L1 treatment promoted M2 BMDM migration, but not M0 and M1 BMDMs (Figure 4, A and B). However, rCHI3L1 treatment did not increase cell migration in a mouse MG cell line (SIM-A9) (Supplemental Figure 3I), supporting our observation that MG tumor infiltration is unaffected by CHI3L1. Moreover, a Transwell assay confirmed that rCHI3L1 promoted cell migration in M2 BMDMs, but not in M0 BMDMs, M1 BMDMs, or MG cells (Figure 4C and Supplemental Figure 3, J–M).
Figure 4. CHI3L1 induces cell migration of M2-like MDM in vitro.
(A) Representative brightfield images showing cell migration at 0 and 6 hours after treatment with CHI3L1 recombinant protein (rCHI3L1) at a concentration of 0.6 μg/mL in M0, M1, and M2 BMDMs by the scratch-wound healing assay. (B) Cell migration was assessed by quantifying occupied areas by migrated cells. (C) Representative brightfield images for cell migration of M2 BMDMs by the Transwell assay. Migration was assessed by determining the number of migrated cells. Data are presented as mean ± SD from at least 3 independent experiments. P values were calculated using 1-tailed, unpaired t test. ***P < 0.001; ****P < 0.0001. (D) Association between CHI3L1 mRNA expression and M1/M2-like macrophage scores in IDH-WT GBM. Gene expression was normalized by RMA, and P values were calculated by Spearman’s rank correlation.
Based on CIBERSORT for gene signatures and correlation analysis in the GBM TCGA data sets (12, 27), CHI3L1 mRNA expression was positively correlated with tumor-promoting M2-like macrophages, but negatively correlated with tumor-killing M1-like macrophages (Figure 4D). Furthermore, analysis of gene set signatures (28) revealed that MDMs, rather than MG, are significantly enriched in tumors with higher levels of CHI3L1 expression in GBM patients (Supplemental Figure 3N). Together, these results demonstrate that CHI3L1 regulates TAM polarization and selectively promotes the migration and accumulation of M2-like MDMs in GBM.
Gal3BP binding to CHI3L1 negatively regulates M2-like macrophage migration.
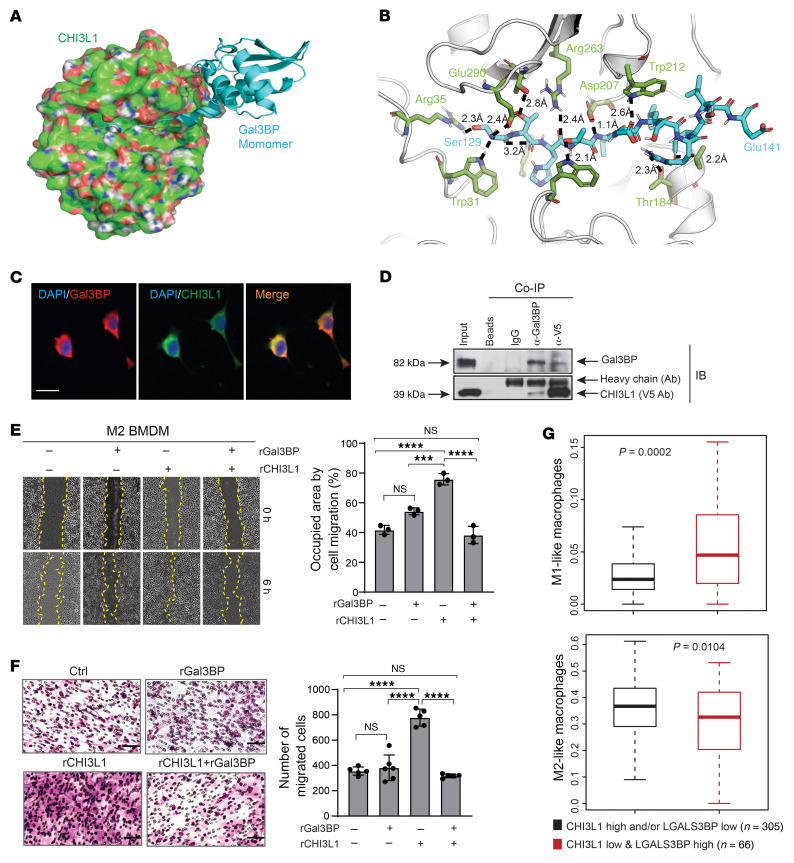
To elucidate how CHI3L1 promotes M2-like MDM migration, we explored CHI3L1 binding proteins using IP coupled to liquid chromatography–mass spectrometry (LC-MS). LC-MS analysis of extracellular or membrane-associated proteins revealed 7 putative binding proteins encoded by the ANXA1, LGALS3BP, GAPDH, PDIA6, BCAP31, ARL6IP5, and MARCKS genes, which are highly associated with CHI3L1 in GBM (Supplemental Figure 4A and Supplemental Table 2). None of these genes have been previously identified as binding partners of CHI3L1. An orthogonal structure-based screening identified galectin 3–binding protein (Gal3BP), encoded by the LGALS3BP gene, as a possible binding partner of CHI3L1 (Figure 5, A and B). Gal3BP, also known as 90K or Mac-2–binding protein, is a secreted glycoprotein upregulated and involved in innate immunity against viral and bacterial infections (29). Interestingly, the domain of Gal3BP predicted to interact with CHI3L1 corresponds to the Gal3BP main dimerization domain (Supplemental Figure 4B), indicating that CHI3L1 can bind monomeric Gal3BP to disrupt its dimerization. Co-IF staining demonstrated strong colocalization of Gal3BP and CHI3L1, which was further supported in live cells by co-IP (Figure 5, C and D).
Figure 5. Gal3BP interacts with CHI3L1 for inhibition of BMDM migration in vitro.
(A) A binding model of Gal3BP monomer (cyan from PDB 6GFB) and CHI3L1 (green surface with red/blue/white shades corresponding to O/N/H atoms from PDB 1HJV_A). (B) Detailed view from A of the binding mode of Ser129-Glu141 of Gal3BP (cyan) and CHI3L1 (green). The 10 hydrogen bonds are indicated with dashed lines and distances. Several hydrophobic contacts are also shown in the protein-binding complex. (C) Representative IF images showing colocalization of proteins in TS603 cells. Scale bar: 20 μm. (D) IB analysis of protein-binding complexes after co-IP with indicated antibodies in TS603 overexpressing V5-tagged CHI3L1. (E) Representative brightfield images from the scratch-wound healing assay showing M2 BMDM cell migration at 0 and 6 hours after treatment with rCHI3L1 (2.5 μg/mL) and/or rGal3BP (5.0 μg/mL). Cell migration was assessed by quantifying occupied areas by migrated cells. (F) Representative brightfield images from the Transwell assay for M2 BMDM cell migration under treatment with rCHI3L1 (2.5 μg/mL) and/or rGal3BP (5.0 μg/mL). Migration was assessed by determining the number of migrated cells. In E and F, data are presented as mean ± SD from at least 3 independent experiments. P values were calculated using 1-way ANOVA with Tukey’s multiple comparisons test. ***P < 0.001; ****P < 0.0001. Scale bar: 50 μm. (G) Boxplots showing enrichment of M1/M2-like macrophage signature in 2 indicated groups of TCGA GBMs. P values were calculated by Wilcoxon’s rank-sum test.
To determine whether Gal3BP binding to CHI3L1 promotes MDM migration, we treated M2 BMDMs with rCHI3L1 and recombinant Gal3BP protein (rGal3BP). Surprisingly, rGal3BP significantly attenuated rCHI3L1-induced M2 BMDM migration, as found by scratch-wound healing assay (Figure 5E) and Transwell migration assay (Figure 5F). Analyzing IF staining of tumors derived from GL261-CHI3L1–bearing syngeneic mice, we observed the mutually exclusive expression patterns of Gal3BP and F4/80 (Supplemental Figure 4C), indicating a negative correlation between Gal3BP expression levels and MDM distribution in glioma. Assessment of correlation between Gal3BP expression and gene signatures of M1/M2-like macrophages revealed a significant positive correlation between LGALS3BP mRNA expression and M1-like macrophages compared with M2-like macrophages in the GBM TCGA database (Supplemental Figure 4D). Furthermore, the M1-like macrophage signature was found to be highly enriched in GBMs with low levels of CHI3L1 expression, but high levels of LGALS3BP expression, whereas a decrease of M2-like macrophage signature was shown in these tumors (Figure 5G). These results indicate that Gal3BP binding to CHI3L1 could negatively regulate CHI3L1-induced M2-like MDM infiltration in the GBM TME.
CHI3L1 binds to Gal3, resulting in selective migration of MDMs, which is negatively regulated by Gal3BP.
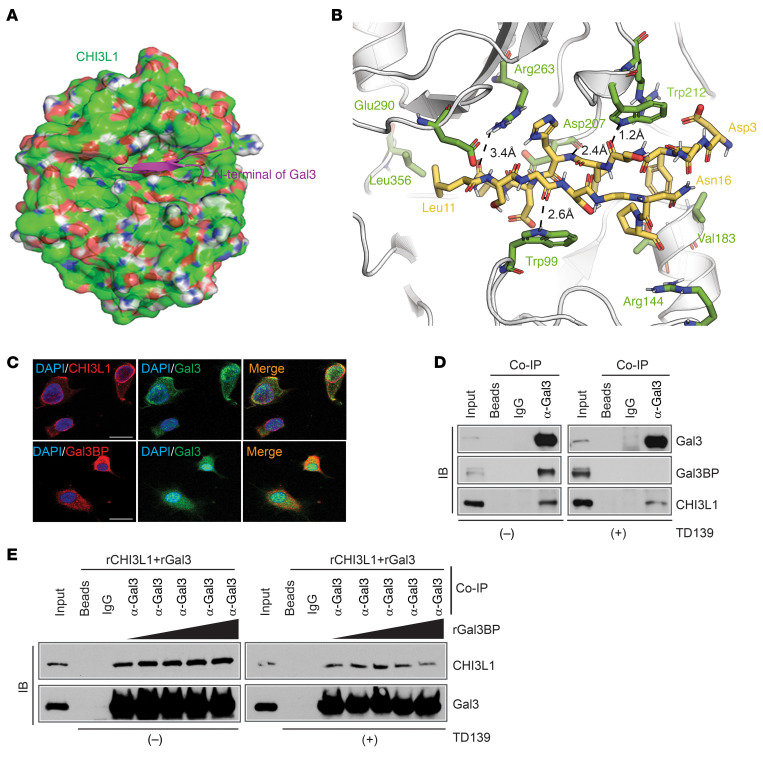
Galectin 3 (Gal3), encoded by the LGALS3 gene as a binding partner of Gal3BP, plays a critical role in macrophage migration and activation (30–32). Therefore, we hypothesized that Gal3 is also involved in CHI3L1-mediated MDM migration. In silico docking of the N-terminal domain of Gal3 and CHI3L1 suggests that Gal3 interacts with CHI3L1 in the same binding pocket as Gal3BP (Figure 6, A and B). Consistent with this prediction, we verified Gal3-Gal3BP binding as well as Gal3-CHI3L1 binding by co-IF and co-IP assays (Figure 6, C and D). Gal3BP was shown to bind with a conserved carbohydrate-recognition domain (CRD) at the C-terminal domain of Gal3 (32). Interestingly, the co-IP assay demonstrated that TD139, a high-affinity and potent inhibitor of Gal3, completely disrupted Gal3-Gal3BP, but not Gal3-CHI3L1, interactions, indicating what we believe to be a novel binding mechanism of Gal3 and CHI3L1 (Figure 6D). Importantly, Gal3 and Gal3BP are predicted to compete for the same binding site in CHI3L1, which was validated by a co-IP assay by adding an increasing amount of rGal3BP into the mixture of rCHI3L1 plus recombinant Gal3 protein (rGal3) with or without TD139 treatment in vitro (Figure 6E). Computational estimates of the interaction-free energy between these proteins using the FastContact server (33) suggested that Gal3BP binds to CHI3L1 more strongly than Gal3 (ΔGbinding = –9.9 kcal/mol and –4.3 kcal/mol, respectively). Of note, sequence alignments of the binding domains in CHI3L1, Gal3, and Gal3BP showed high conservation between the human and mouse, indicating the evolutionarily conserved functions of these genes (Supplemental Figure 5A).
Figure 6. Gal3BP competes with Gal3 for binding with CHI3L1.
(A) A binding model of N-terminal Gal3 (magenta Asp3-Pro17 from PDB 6FOF) and CHI3L1 (green surface with red/blue/white shades corresponding to O/N/H atoms from PDB 1HJV_A). (B) Detailed view from A of the binding mode of Asp3-Asn16 of Gal3 (yellow) and CHI3L1 (green). (C) Representative IF images showing colocalization of proteins in TS603 cells. Scale bar: 20 μm. (D) IB analysis of protein-binding complexes using co-IP with Gal3 antibody in TS603-V5-CHI3L1 cells treated with DMSO or TD139 (10 μM for 24 hours). (E) IB analysis of Gal3 and CHI3L1 protein binding in the mixture of rGal3 and CHI3L1 (200 ng rGal3 + 200 ng rCHI3L1) by adding different amounts of rGal3BP (0, 100, 200, 400, 800 ng/sample) with or without TD139 (10 μM for 1 hour).
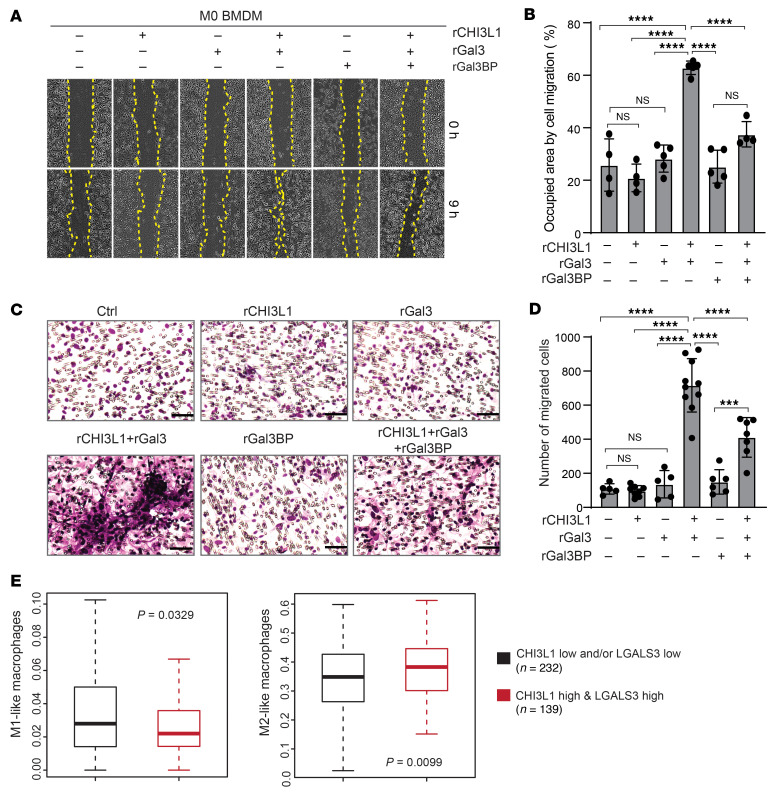
To determine whether Gal3 and Gal3BP are associated with selective migration of M1/M2-like MDMs, we detected expression levels of Gal3 and Gal3BP in polarized BMDMs. Quantitative real-time PCR (qRT-PCR) and IB analyses revealed highly expressed Gal3 in M2 BMDMs compared with M0 and M1 BMDMs (Supplemental Figure 5, B and C). Interestingly, we observed that an MG cell line (SIM-A9) expressed higher levels of Gal3BP compared with polarized BMDMs and the murine macrophage cell line RAW264.7 (Supplemental Figure 5, C and D), indicating a potential mechanism by which CHI3L1 effectively induces cell migration in M2-like MDMs, but not MG. We, therefore, hypothesized that CHI3L1 cooperates with Gal3 to selectively promote M2 BMDM migration. To this end, scratch-wound healing and Transwell assays revealed that treatment of M0 BMDMs (lower endogenous levels of Gal3 expression) with rCHI3L1+ rGal3 significantly increased cell migration (Figure 7, A–D). To test whether Gal3BP inhibits CHI3L1-Gal3 induced MDM migration, M0 BMDMs were treated with rCHI3L1+rGal3+rGal3BP, resulting in a significant decrease in cell migration compared with those treated with rCHI3L1+rGal3 (Figure 7, A–D). These data suggest that Gal3BP competes with Gal3 to bind CHI3L1, leading to inhibition of MDM migration by disrupting the CHI3L1-Gal3 protein complex. We analyzed the enrichment of M1/M2-like macrophage signatures associated with the expression of these genes in TCGA GBM samples and observed increased M2- and decreased M1-like macrophage signatures in tumors with high mRNA expression levels of CHI3L1 and LGALS3 (Figure 7E). Furthermore, a significant increase of M1-like macrophage signatures was shown in the GBMs with low levels of CHI3L1 and LGALS3 expression, but high levels of LGALS3BP expression (Supplemental Figure 5E), indicating a predominant association between Gal3BP and a proinflammatory M1-like phenotype. Collectively, these results suggest that CHI3L1 binds with Gal3, forming a protein complex, promoting infiltration of M2-like MDMs, which is negatively regulated by Gal3BP via a competitive interaction.
Figure 7. The CHI3L1-Gal3-Gal3BP binding complex regulates BMDM migration.
Representative brightfield images and quantitation for cell migration of M0 BMDMs treated with rCHI3L1 (2.5 μg/mL), rGal3 (2.5 μg/mL), rGal3BP (5.0 μg/mL), and combinations in the scratch-wound healing assay (A and B) and the Transwell assay (C and D). Cell migration was assessed by quantifying the occupied area or by counting the number of migrating cells, respectively. Data are presented as the mean ± SD from at least 3 independent experiments. P values were calculated using 1-way ANOVA with Tukey’s multiple comparisons test. ***P < 0.001; ****P < 0.0001. Scale bars: 50 μm. (E) Boxplots showing enrichment of M1/M2-like macrophage signature in 2 indicated groups of TCGA GBMs. P value was calculated by Wilcoxon’s rank-sum test.
CHI3L1-Gal3 protein complex induces tumor immunosuppression by reprogramming MDMs.
To delineate molecular mechanisms of CHI3L1 protein complex-induced tumor progression, we performed RNA-Seq analysis on TAMs isolated from orthotopic xenograft glioma models in C57BL/6 mice bearing the isogenic line QPP7 with shChi3l1, relative to QPP7 with shSC. Gene Ontology (GO) analysis showed that signaling pathways regulating cell killing, leukocyte-mediated cytotoxicity, and lymphocyte-mediated immunity were enriched in TAMs derived from Chi3l1 KD tumors (Figure 8A). We hypothesized that CHI3L1 signaling reprograms TAMs toward a protumor phenotype, leading to dysregulation of the tumor-infiltrating lymphocyte–mediated (TIL-mediated) antitumor immune responses. To this end, we observed that CHI3L1 OE significantly decreased the active CD4+ (CD69+CD62L–) and CD8+ (CD69+CD62L–) TILs in GL261 tumors (Figure 8B). Conversely, the active CD4+ T cells were significantly increased in QPP7 Chi3l1 KD tumors, while enrichment of active CD8+ T cells did not reach significance (Figure 8C). These results suggest that dysregulation of T cell–mediated antitumor immune response contributes to CHI3L1-induced tumor progression.
Figure 8. CHI3L1 protein complexes regulate MDM reprogramming in immune suppression and stimulation.
(A) Enrichment of top 10 GO biological pathways in TAMs derived from C57BL/6 mice bearing QPP7 with shChi3l1 compared with shSC. (B) Flow cytometry analysis showing active CD4+ and CD8+ cells in GL261 tumors with CHI3L1 OE compared with vector controls. (C) Flow cytometry analysis showing active CD4+ and CD8+ cells in QPP7 tumors with Chi3l1 KD compared with shSC. Depletion antibodies against CD4 and CD8 (10 mg/kg) were injected intraperitoneally every 3 days for a total of 8 times after tumor implantation. Kaplan-Meier tumor-free survival analysis of mice bearing GL261 overexpressing CHI3L1 (D) and mice bearing QPP7 with Chi3l1 KD (F). Flow cytometry analysis showing CD4+ and CD8+ cell populations within the tumors from GL261 models (E) and QPP7 models (G) with antibody depletion. Each dot represents 1 mouse; data are presented as mean ± SEM. P values were calculated using a 1-tailed, unpaired t test. (H) qRT-PCR for indicated gene expression in M0 BMDMs treated with rCHI3L1 (2.5 μg/mL), rGal3 (2.5 μg/mL), rGal3BP (5.0 μg/mL), and combinations for 24 hours. Data are presented as mean ± SEM from at least 2 independent experiments. P values were calculated using 1-way ANOVA with Tukey’s multiple comparisons test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (I–K) IB analysis of indicated protein levels in M0 BMDMs treated with rCHI3L1 (2.5 μg/mL), rGal3 (2.5 μg/mL), rGal3BP (5.0 μg/mL), and combinations for 30 minutes or 4 hours (p-p65 and p65). (L) GSEA plots depicting mTOR1 and TNFA/NFKB signaling pathways in TAMs derived from C57BL/6 mice bearing QPP7 with shChi3l1 compared with shSC. NES, normalized enrichment score.
Previous studies revealed that TAM depolarization, rather than depletion, profoundly affects cancer progression by changing gene expression and switching between phenotypes of immune suppression and immune stimulation (34, 35). We further demonstrated that depleting peripheral and intratumoral MDMs, but not MG, by systemic delivery of clodronate liposomes (36, 37) did not repress tumor progression in the syngeneic orthotopic glioma model of C57BL/6 mice bearing GL261-CHI3L1 (Supplemental Figure 6, A–E). Gene set enrichment analysis (GSEA) showed that hallmark pathways in immune stimulation, including IFN-α, IFN-γ, IL6/JAK/STAT3 signaling, and inflammatory response, were enriched in TAMs derived from tumors with Chi3l1 KD compared with controls (Supplemental Figure 6, F and G). Moreover, genes associated with immune suppression (Arg1, Ym1, Ccl2, Il10) were downregulated in TAMs from shChi3l1 tumors; however, genes associated with immune stimulation (Nos2, Il6, Il12b, Ifng) were upregulated in these TAMs (Supplemental Figure 6H). To further examine the involvement of T cells in CHI3L1-mediated tumor progression, we performed antibody depletion studies of CD4+ and CD8+ cells in GL261-CHI3L1 and QPP7-shChi3l1#1 derived syngeneic tumor models, respectively. Treatment with anti-CD4 and anti-CD8 antibodies significantly enhanced CHI3L1 OE–induced tumor progression in GL261 models (Figure 8, D and E), but attenuated Chi3l1 KD–mediated tumor regression in QPP7 models (Figure 8, F and G, and Supplemental Figure 6I). Together, these results reveal that silencing CHI3L1 reprograms a TAM switch from immune suppression to stimulation, which is required for T cell–mediated antitumor response.
To elucidate the downstream signaling pathways of CHI3L1 protein complex–regulated TAM reprogramming, we treated M0 BMDMs with rCHI3L1+rGal3. Interestingly, genes related to antiinflammation (Arg1, Ym1, Ccl2, Il10) were increased compared with any single-agent treatment by qRT-PCR assessment. Notably, upregulation of these genes by rCHI3L1+rGal3 treatment was significantly inhibited in M0 BMDMs treated with rGal3BP (Figure 8H). These data indicate that the CHI3L1-Gal3 protein complex reprograms TAMs toward an M2-like phenotype by regulating gene expression associated with immune suppression, which is negatively regulated by Gal3BP. Given the positive feedback loop of the CHI3L1-PI3K/AKT/mTOR signaling pathway (Figure 1), we hypothesized that the CHI3L1-Gal3 protein complex activates the PI3K/AKT/mTOR pathway, which significantly controls a macrophage switch between immune stimulation and suppression by regulating NF-κB and CEBPβ activation (35). IB analysis showed that rCHI3L1+rGal3 treatment increased the levels of p-AKT (T473 and S308), p-S6, and p-mTOR compared with either agent alone (Figure 8I), while those were inhibited by the addition of rGal3BP in M0 BMDMs (Figure 8J). To evaluate the activation of PI3K/AKT/mTOR downstream transcription factors, we found that rCHI3L1 plus rGal3 stimulated C/EBPβ expression and simultaneously inhibited p65-RelA phosphorylation in M0 BMDMs, which were also reversed by treating with rGal3BP (Figure 8K). GSEA showed enrichment of mTOR1 signaling in TAMs derived from tumors with shSC and enrichment of TNFA signaling via the NF-κB pathway in TAMs derived from tumors with shChi3l1, further supporting the involvement of these transcription factors in MDM reprogramming by CHI3L1 protein complexes (Figure 8L). Collectively, these results indicate that CHI3L1 protein complexes reprogram MDMs toward an immunosuppression or immunostimulation phenotype by controlling a transcriptional regulatory program of PI3K/AKT/mTOR–NF-κB/CEBPβ (Supplemental Figure 6J).
Gal3BP mimetic peptide inhibits CHI3L1-Gal3 complex–induced tumor progression.
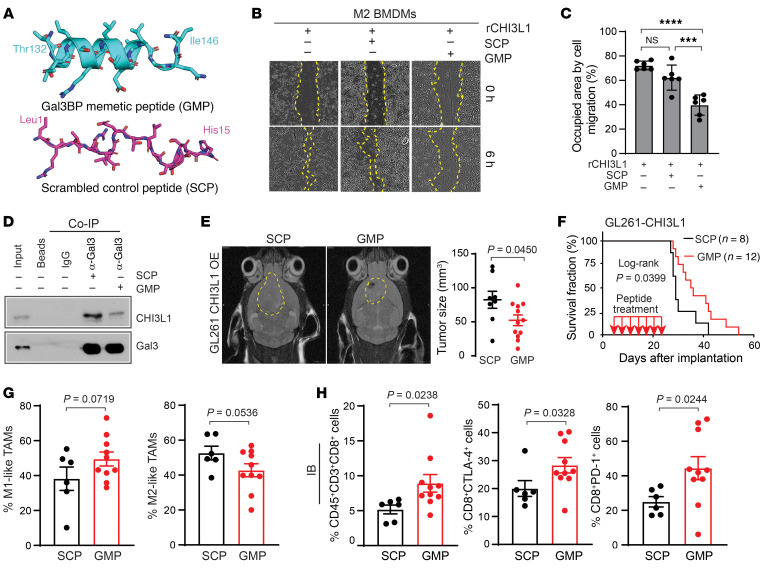
To investigate whether disruption of CHI31L-Gal3 protein–binding complex can reverse MDM-mediated immune suppression and thereby attenuate glioma progression, we designed a Gal3BP mimetic peptide (GMP) to disrupt the interaction between Gal3 and CHI3L1. Molecular dynamics (MD) showed that GMP (T132LDLSRELSEALGQI146), rather than scrambled control peptide (SCP) (L1RTRLEETLSSDTSH15), behaves as a linear peptide capable of recapitulating interaction with CHI3L1 (Figure 9A). To test GMP inhibiting CHI3L1-Gal3–induced macrophage migration in vitro, M2 BMDMs were treated with rCHI3L1 in combination with GMP and SCP, respectively. Of note, scratch-wound healing assay analysis revealed that rCHI3L1-promoted M2 BMDM migration was significantly inhibited by GMP compared with SCP treatment (Figure 9, B and C). In contrast with SCP, GMP also attenuated rCHI3L1+rGal3-induced M0 BMDM migration (Supplemental Figure 7, A and B). To verify that GMP recapitulates Gal3BP to compete with Gal3 binding with CHI3L1, we found that GMP treatment resulted in decreasing binding of CHI3L1 and Gal3 in THP-1 cells by the co-IP assay (Figure 9D). Moreover, GMP inhibited rCHI3L1-induced BMDM migration in a dose-dependent manner (Supplemental Figure 7C). These data demonstrate that this peptide can mimic Gal3BP to disrupt CHI31L-Gal3 protein interaction and BMDM migration.
Figure 9. A peptide-mimicking Gal3BP attenuates BMDM migration and CHI3L1-induced tumor progression.
(A) Snapshot from MD of GMP and SCP. (B) Representative brightfield images for cell migration of M2 BMDMs treated with rCHI3L1 (0.6 μg/mL) with or without GMP or SCP at a concentration of 30 μM in the scratch-wound healing assay. (C) Cell migration was assessed by quantifying the occupied area of migrated cells. Data are presented as mean ± SD from at least 3 independent experiments. P values were calculated using 1-way ANOVA with Tukey’s multiple comparisons test. ***P < 0.001; ****P < 0.0001. (D) IB analysis of protein-binding complexes using co-IP with Gal3 antibody in THP-1 cells treated with SCP or GMP (20 μM for 24 hours). (E) Representative MRI from mice after intracranial injection of GL261-CHI3L1 cells after the treatment of SCP and GMP, respectively. Tumor volume was measured by T2 sequences for infiltrative tumors in the mouse brain (yellow line). (F) Kaplan-Meier tumor-free survival analysis of mice bearing GL261-CHI3L1 tumors treated with indicated peptides. Frequency of M1/M2-like MDMs (G) and CD8+ T cells with expression of PD-1 and CTLA-4 (H) in tumors derived from syngeneic mice bearing GL261-CHI3L1 under treatment with GMP versus SCP. Each dot represents 1 mouse; data are presented as the mean ± SEM. P values were calculated using a 1-tailed, unpaired t test.
To assess the antitumor effect of GMP in vivo, GMP and SCP were administered directly into brain tumors by an implantable guide-screw system in C57BL/6 mice bearing GL261-CHI3L1 orthotopic tumors. Notably, GMP treatment reduced tumor growth and extended animal survival (median survival of 36 days) compared with SCP (median survival of 29 days; Figure 9, E and F). To validate the antitumor effect of GMP on tumors with the endogenous level of CHI3L1, the orthotopic syngeneic mice bearing QPP7 glioma were treated by local delivery of GMP and SCP into the brain, respectively. Consistently, we found that the treatment of GMP decreased tumor size and increased mouse survival in the QPP7 model (Supplemental Figure 7, D and E).
To evaluate the changes of immune cell populations following peptide treatment, we performed flow cytometry of cells harvested from syngeneic C57BL/6 mice bearing GL261-CHI3L1 glioma. Following GMP treatment, an increase of M1-like TAMs (49.5% ± 4.0% vs. 38.2% ± 6.7%, P = 0.0719) and decrease of M2-like TAMs (42.8% ± 3.8% vs. 52.7% ± 3.9%, P = 0.0536) were observed compared with what was seen with SCP treatment (Figure 9G). In contrast with SCP treatment, GMP treatment significantly increased T cells (CD3+), particularly CD8+ cells (Figure 9H and Supplemental Figure 7F). The CD4+ cell population increased under GMP versus SCP treatment (41.9% ± 4.5% vs. 39.8% ± 4.0%), indicating that Tregs, a subset of CD4+ cells, could influence total CD4+ cell composition, proliferation, and recruitment (Supplemental Figure 7G). These results suggest that GMP could reprogram TAMs from protumor to antitumor phenotype, which would indirectly promote CD8+ T cell–mediated antitumor immune response.
Despite increasing the tumor-infiltrating T cells after GMP treatment, T cell exhaustion is a hallmark of GBM local immune dysfunction due to the upregulation of multiple immune checkpoints, such as PD-1 and CTLA-4 (38, 39). Therefore, we assessed expression levels of these immune checkpoints in CD4+ and CD8+ T cells and found that both PD-1 and CTLA-4 were significantly upregulated in CD8+ T cells from GMP-treated tumors compared with those in the SCP-treated tumors in the GL261-CHI3L1 model (Figure 9H). The CD4+ T cells from tumor-bearing mice displayed a trend of elevated levels of PD-1 and CTLA-4 following the treatment with GMP versus SCP, respectively (Supplemental Figure 7G). We also evaluated the expression of PD-L1, a ligand of PD-1, which is upregulated in activated leukocytes and cancer cells (40). Interestingly, GMP treatment significantly decreased PD-L1 expression in CD45+, CD8+, and glioma cells, which suggests that disrupting CHI3L1-Gal3 interaction may lead to the reduction of T cell exhaustion (Supplemental Figure 7, H and I).
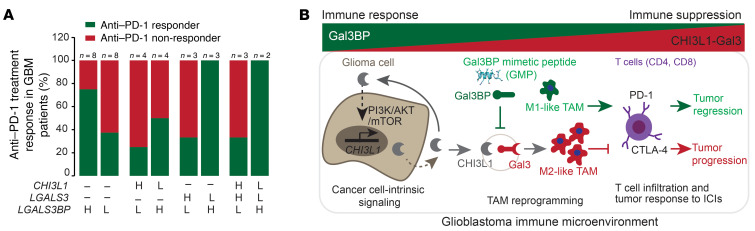
Together, our results indicate that CHI3L1 protein–binding complexes with Gal3 or Gal3BP modulate TAM-mediated immune suppression and stimulation, leading to resistance or response to immune-checkpoint therapy. To evaluate whether gene expression of CHI3L1, LGALS3, and LGALS3BP is associated with patient response to immunotherapy with immune-checkpoint inhibitors (ICIs), we analyzed bulk RNA-Seq profiles of GBM from 16 GBM patients with treatment of PD-1 inhibitors (nivolumab or pembrolizumab; ref. 41). Consistently, higher levels of LGALS3BP expression were associated with anti–PD-1 responders, whereas lower levels of LGALS3BP expression were associated with anti–PD-1 nonresponders (Figure 10A and Supplemental Figure 7J). Moreover, higher levels of LGALS3BP combined with lower levels of LGALS3 and/or lower levels of CHI3L1 were associated with anti–PD-1 responders, while lower levels of LGALS3BP combined with higher levels of LGALS3 and/or higher levels of CHI3L1 were associated with anti–PD-1 nonresponders (Figure 10A). Collectively, these data suggest that the CHI3L1 protein–binding complex modulates TAM–mediated T cell immunity, which underlies these proteins as the key determinants of the response to immune-checkpoint therapy. Therapeutically, disrupting the CHI3L1-Gal3 protein complex using GMP may synergize with ICIs to effectively promote tumor regression for GBM patients (Figure 10B).
Figure 10. The levels of CHI3L1, LGALS3, and LGALS3BP mRNA expression predict anti–PD-1 response in GBM patients.
(A) Histogram analysis of the distribution of anti–PD-1 treatment responders and nonresponders in GBM patients following anti–PD-1 treatment from a previous data set (41). The n represents the number of patients characterized with indicated gene expression. (B) Schematic drawing indicates that glioma cell–intrinsic CHI3L1 binding with Gal3 forms a protein-binding complex modulating the TAM-mediated immune microenvironment for tumor progression, which is negatively regulated by Gal3BP or GMP.
Discussion
Although the GBM TME plays a crucial role in regulating tumor progression and is increasingly recognized as a therapeutic target, understanding the underlying cellular and molecular mechanisms governing glioma cells and their surrounding components remains challenging. In this study, we discovered that cancer cell–intrinsic CHI3L1 plays a predominant role in modulating the GBM TME by forming a protein complex with Gal3 or Gal3BP to promote macrophage-mediated immune suppression. Our efforts to understand the mechanisms governing GBM immune suppression resulted in a peptide as an immunostimulatory drug candidate and pharmacological modifications of CHI3L1-Gal3/Gal3BP protein complexes as potential therapeutics for patients with GBM.
Increasing evidence suggests that tumor-intrinsic mechanisms dictate various noncancerous cells (e.g., regulating tumor infiltration of myeloid cells and lymphocytes) within the TME, which exert multifaceted functions, ranging from antitumor to protumor activities (12, 13, 42). The findings in this study demonstrate that cancer cell–intrinsic CHI3L1 is upregulated by the PI3K/AKT/mTOR signaling axis in a positive feedback loop, which plays a predominant role in modulating the GBM immune microenvironment by inducing M2-like MDM infiltration and repolarization in a paracrine mechanism. Genetically, CHI3L1 gene expression is significantly associated with loss of chromosome 10q encompassing PTEN in GBM (18). Our work reinforces the positive correlation between CHI3L1 gene expression and PTEN deletions/mutations or other mechanisms leading to PI3K/AKT/mTOR activation (e.g., NF1 mutations). These findings deepen our understanding of tumor-intrinsic signaling pathways driven by genetic alterations in the regulation of the GBM immune microenvironment for tumor progression and treatment response.
In exploring the role of CHI3L1 for regulating the GBM immune microenvironment, we discovered that CHI3L1 binding with Gal3 promotes MDM infiltration and reprograms MDMs toward a tumor-promoting M2-like phenotype, which is negatively regulated by Gal3BP. Increased expression and secretion of Gal3 were observed in both human and mouse M2-polarized macrophages compared with monocytes and M1-polarized macrophages (30, 43). However, increased levels of Gal3BP and a proinflammatory phenotype were observed in human monocyte–derived M1 macrophages in vitro and plasma from patients with cardiovascular disease or hepatitis C infection (44, 45). In this study, our finding of CHI3L1-Gal3 protein complex–induced selective migration of M2-polarized BMDMs provides a mechanistic explanation for a long-standing observation, namely that highly infiltrating M2-like MDMs associated with both human and mouse GBM (9, 10, 12, 19). In addition to promoting M2-like MDM accumulation, the present study also provides the mechanisms for immunosuppression that enable GBM to escape immune surveillance by which the CHI3L1-Gal3 protein complex activates the AKT/mTOR-mediated transcriptional regulatory network (NF-κB and CEBPβ), leading to a macrophage switch toward immune suppression from immune stimulation (35).
Reducing immunosuppression and overcoming immunotherapy resistance are becoming therapeutic areas of great interest for the treatment of GBM as well as other solid tumors (46, 47). Our findings provide a rationale for disrupting the CHI3L1-Gal3 protein complex by the addition of Gal3BP to reduce the degree of tumor immunosuppression and improve antitumor immune response in the GBM TME. Interestingly, a previous study showed that local and systemic increases in Gal3BP levels inhibited tumor growth by stimulation of the residual cell–mediated immune defense of the nude mouse (48). Although the function of Gal3BP is controversial in physiologic and pathologic conditions, elevated levels of Gal3BP in bacterial and viral infections and the neoplastic context suggest its crucial role in immune response as an immunostimulatory molecule (29, 49, 50). In our study, GMP being locally delivered into brain tumor led to tumor regression in the treated animals combined with reduced M2-like MDMs and increased M1-like MDMs and CD8+ T cells in the TME, indicating that this peptide can modify CHI3L1 protein complexes and thereby reprogram the immune microenvironment. Although CD8+ T cells were significantly increased by GMP treatment, we observed elevated levels of CTLA-4 and PD-1 expression in these T cells, a hallmark feature of T cell exhaustion, suggesting that GMP may synergize with ICIs to form more effective immunotherapy for GBM treatment. Based on analyzing a publicly available clinical data set (41), the higher and lower levels of LGALS3BP combined with LGALS3 or CHI3L1 gene expression are associated with response to anti–PD-1 immunotherapy, reinforcing the mechanism of CHI3L1-Gal3/Gal3BP protein complexes in regulating protumor or antitumor immunity in GBM. In summary, the findings in this study shed light on a crucial molecular mechanism of macrophage-mediated immunosuppression in GBM, indicating the development of a more effective treatment for patients with this devastating brain cancer.
Methods
Detailed methods can be found in the Supplemental Methods.
Cell lines.
GBM patient–derived neurosphere lines (TS543, TS603, BT112) and hNSC lines were used and cultured as described previously (14). Mouse glioma cell line QPP7, provided by Jian Hu (MD Anderson Cancer Center, Houston, Texas, USA), was cultured in the serum-free NSC medium. U87, GL261, RAW246.7, and 293T from ATCC were cultured in DMEM supplemented with 10% FBS (Sigma-Aldrich) and penicillin/streptomycin (P/S) (Gibco, Thermo Fisher Scientific). SIM-A9 from ATCC was cultured in DMEM/F12 supplemented with 10% FBS, 5% horse serum, and P/S. The THP-1 cell line was purchased from ATCC and cultured in RPMI-1640 supplemented with 10% FBS, 0.05 mM 2-mercaptoethanol, and P/S. All cell lines were verified to be mycoplasma free using the MyCoAlert PLUS Mycoplasma Detection Kit (Lonza, catalog LT07-710), and cultured at 37°C with 5% CO2.
BMDM culture and polarization.
BMDMs were isolated from male and female C57BL/6 mice as previously described (26). Briefly, femur bones were isolated from mice, and IMDM (ATCC) supplemented with 10% FBS and P/S was used to flush the BM into a petri dish. After 4- to 6-hour incubation, floating cells were collected and resuspended in the medium with 20 ng/mL M-CSF (PeproTech). On day 6, the fully differentiated cells were designated as in an M0 state. To induce BMDM polarization toward an M1 state, 100 ng/mL LPS (Invitrogen) and 50 ng/mL IFN-γ (PeproTech) were added to the M0 cells for 24 hours. To induce BMDM polarization toward an M2 state, 20 ng/mL IL-4 (PrproTech) was added to the M0 cells for 72 hours.
Intracranial xenograft tumor models, macrophage depletion, T cell depletion, and peptide treatment.
Male and female ICR SCID and C57BL/6 mice (4 to 6 weeks of age) were purchased from Taconic Biosciences and The Jackson Laboratory, respectively. The intracranial xenograft tumor models were established as previously described (14). Cells in 5 μL DPBS were injected in the following amounts: TS543 vector control or CHI3L1 OE, 1 × 104 cells; QPP7 scrambled control or CHI3L1 KD, 1 × 105 cells; and GL261 vector control or CHI3L1 OE, 1 × 105 cells. For tumor models with macrophage depletion, chodrosome or control liposome (Clodrosome, catalog CLD-8901, Encapsula Nano Sciences) was injected into animals through the tail vein. For tumor models with T cell depletion, IgG (catalog BE0090, BioXCell) or anti-CD4 (catalog BE0003-1, BioXCell) and anti-CD8 (catalog BE0061, BioXCell) antibodies were given to animals through intraperitoneal injection. For mice treated with peptides, 5 μL of 20 μM SCP or GMP was delivered into the mouse brain every 4 days for a total of 7 times.
ELISA.
Secreted CHI3L1 protein in the cell culture supernatant was measured by using the Quantikine Human CHI3L1 Immunoassay (catalog DC3L10, R&D Systems). CHI3L1 content in the CM was quantified per million cells, and no CHI3L1 was detected in DMEM or NSC medium supplemented with EGF and bFGF.
Co-IP and MS.
TS603 cells overexpressing CHI3L1 with V5 tag (TS603 CHI3L1_V5_OE) or THP-1 cells treated with the peptides were collected, and protein-protein interaction was crosslinked with 2 mM dithiobis(succinimidyl propionate) (DSP). Membrane proteins were extracted with the Membrane Protein Extraction Kit (catalog 89842, Thermo Fisher), and approximately 500 μg of protein was used for co-IP assay using the Co-Immunoprecipitation Kit (catalog 26149, Thermo Fisher). For MS, 10 μg of each of the TS603 CHI3L1_V5_OE Co-IP samples were separated in a 12% SDS-PAGE gel and analyzed by MS at the University of Pittsburgh Biomedical MS Center.
IB, IHC, and IF.
Cells were lysed on ice using RIPA buffer (Millipore) supplemented with protease and phosphatase inhibitors (Roche). The protein concentration was determined by the BCA method, and approximately 15 to 30 μg of total proteins were loaded and analyzed by Western blotting with indicated antibodies. For IHC staining, brain tissues were fixed in 10% formalin overnight and embedded in paraffin. For IF staining, fresh brain tissues were immediately frozen in OCT on dry ice. IHC and IF staining were performed as we described previously (14). Additional information about antibodies is provided in the Supplemental Methods.
Scratch-wound healing assay.
BMDMs were polarized to the indicated status (M0, M1, or M2) and seeded in 12-well plates at 80%–90% confluency. The cells were switched to the medium without FBS for 6 hours of starvation. Scratches were made using pipette tips, and fresh IMDM with indicated recombinant proteins and/or peptides was added. Images of the scratches were captured at indicated times. For the SIM-A9 scratch-wound healing assay, 12-well plates were coated with 10 μg/mL fibronectin (Sigma-Aldrich) at 37°C overnight before seeding cells.
Transwell migration assay.
Polarized BMDMs were starved by removing FBS for 6 hours. Cells were collected and 2 × 105 cells in 200 μL of IMDM were added into each Transwell insert (catalog MCMP24H48, Millipore); 700 μL of IMDM containing 2% FBS and the indicated recombinant proteins was added to the bottom of the plates. After 14-hour incubation, Transwell inserts were stained with the HEMA 3 Stain Set (Fisher Scientific). Insert membranes were separated and mounted on glass slides with CYTOSEAL XYL (Thermo Fisher), and images were taken by an inverted microscope (Leica DM 2500). For the SIM-A9 cells, the inserts were coated with 10 μg/mL fibronectin overnight in advance of seeding cells.
RNA isolation, qRT-PCR, and RNA-Seq.
RNA was extracted and cDNA was synthesized as described previously (14). qRT-PCR was performed using PowerSYBR Green PCR Master Mix (Applied Biosystems) and detected with a StepOnePlus Real-Time PCR System (Applied Biosystems). Primers are listed in Supplemental Table 3. Each reaction was performed in duplicate or triplicate. The relative expression of genes was normalized to human RPL39 or mouse 18S ribosomal RNA. For RNA-Seq experiments, cells from intracranial xenograft tumors were isolated and incubated with antibodies for immune cell types. Macrophages were isolated by FACS and RNA was then isolated and sent to the Health Sciences Sequencing Core at UPMC Children’s Hospital of Pittsburgh for RNA-Seq. RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE174177).
MRI and bioluminescent imaging.
MRI and bioluminescent imaging of mice were performed at Rangos Research Center Animal Imaging Core. The tumor size of mice detected by MRI was analyzed with ITK-SNAP. For bioluminescent imaging, mice were intraperitoneally injected with d-luciferin (150 mg/Kg; GoldBio), and images were captured by the IVIS Lumina S5 system (PerkinElmer).
Brain tumor cell isolation.
Mice with neurological deficits or moribund appearance were sacrificed. Tumors were separated and homogenized for 15 minutes at 37°C in collagenase IV cocktail (3.2 mg/mL collagenase type IV, 2 mg/mL soybean trypsin inhibitor, and 1.0 mg/mL deoxyribonuclease I; Worthington Biochemical). Red blood cells were lysed using ACK lysing buffer (Gibco, Thermo Fisher Scientific). Cell suspensions were filtered through 70 μm strainers, centrifuged at 400g for 5 minutes, and resuspended in cold FACS buffer (DPBS with 1% BSA) for further analysis.
Flow cytometry and CyTOF.
About 3 × 106 cells were used for each staining panel. Cells were incubated with 1.0 μg of TruStain fcX (BioLegend) for 10 minutes on ice to block Fc receptors, followed by staining with the combination of indicated antibodies. After staining, cells were washed with FACS buffer 3 times and incubated with Fixation Buffer (BioLegend) at room temperature for 20 minutes. Cells were washed with FACS buffer, resuspended in Cyto-Last Buffer (BioLegend), and analyzed by either a BD LSRFortessa or BD FACSAria II SORP. For CyTOF, samples were prepared as described above for flow cytometry. Three pairs of samples (scrambled shRNA versus Chi3l1 KD) with similar tumor sizes were chosen and the staining procedure was followed as previously described (51). The samples were analyzed on a Helios2 CyTOF system (Fluidigm) at the Longwood Medical Area CyTOF Core (Dana-Farber Cancer Institute, Boston, Massachusetts, USA). Additional information about flow cytometry antibodies is provided in the Supplemental Methods.
Structure analysis of protein-protein interaction.
Prediction of protein-protein interaction was based on available human protein structures for binding of CHI3L1 and other putative protein candidates. Protein structures of CHI3L1 (PDB 1HJV), Gal3 (PDB 6FOF), and Gal3BP monomer (PDB 6GFB) were used for protein-protein interaction analyses. Docked poses of CHI3L1 with Gal3BP (monomer of dimerization domain) and CHI3L1 with Gal3 were predicted using ClusPro (52) and further analyzed with FastContact for energetic complementarity (33).
Peptide design.
GMP was designed using MD simulations in AMBER18 on the GPU-accelerated code with the AMBER ff14SB force field (53, 54). The tLeap binary was used to solve structures in an octahedral TIP3P water box with a 15 Å distance from the peptide surface to the box edges and a closeness parameter of 0.75 Å. The system was neutralized and solvated in 150 mM NaCl. The nonbonded interaction cutoff was set to 8 Å. Hydrogen bonds were constrained using the SHAKE algorithm and an integration time step of 2 fs. Simulations were carried out by equilibrating the system for 5 ns at NPT, using a Berendsen thermostat to maintain a constant pressure of 1 atm followed by 300 ns NVT production at 300 K.
Statistics.
Data were calculated with GraphPad Prism and presented as mean ± SD or ± SEM. P < 0.05 was considered statistically significant, and it was determined by the indicated tests in figure legends. Scratch-wound healing areas of cell migration, Transwell migration assay cell number, and IF staining positive cells were analyzed by using Fiji software (ImageJ, NIH). Flow cytometry data were gated, analyzed, and visualized using FlowJo software (BD). CyTOF data were analyzed with Cytobank (Cytobank Inc.). TCGA GBM data sets were used for clinical GBM analysis, and RNA-Seq data (41) were used for correlation analysis between gene expression (CHI3L1, LGALS3, and LGALS3BP) and GBM patient response to anti–PD-1 treatment.
Study approval.
All animal experiments were performed with the approval of the University of Pittsburgh’s Institutional Animal Care and Use Committee (IACUC) (protocols 18031339, 21049271).
Author contributions
AC and BH contributed to the conception and design of the study. AC, YJ, ZL, VS, YG, and J Ma performed experiments. US and CJC contributed to protein-protein interaction prediction and peptide design. LW, HZ, CC, SL, XL, and QW contributed to bioinformatics analysis for RNA-Seq, TCGA data sets, and clinical data sets. AC and BH analyzed and interpreted data and wrote the original manuscript. J Michel, YLW, YS, DY, LK, NMA, POZ, GK, SA, DS, GKG, and XX provided resources and technical support. AC, LHM, CJC, DY, IFP, and BH reviewed and edited the manuscript.
Supplementary Material
Acknowledgments
The authors would like to thank Ronald A. DePinho for critically evaluating the manuscript; Jiangong Ren and Jian Hu from UT Texas MD Anderson Cancer Center for providing the mouse glioma cell line QPP7; Junfei Zhao and Raul Rabadan from Columbia University for providing detailed information about their published RNA-Seq data sets; Samuel K. Wyman and Jackson E. Landman from Rangos Research Center Animal Imaging Core for assistance with MRI imaging; Nathan A. Yates and Xuemei Zeng from BioMS Center for assistance with MS experiments; William A. MacDonald from Health Sciences Sequencing Core for assistance with RNA-Seq; Krishna Prasadan from Rangos Research Center Cell Imaging Core for assistance with the confocal image; and Esther P. Jane and Daniel R. Premkumar for sharing antibodies and reagents. This work was supported by the Scientific Program Fund from the Children’s Hospital of Pittsburgh (to BH and IFP), the Translational Brain Tumor Research Fund (to IFP), the Walter L. Copeland Fund of the Pittsburgh Foundation (to BH), the Connor’s Cure Fund from the V Foundation (to BH and IFP), and NIH grant 2GM097082 (to CJC and US).
Version 1. 07/06/2021
In-Press Preview
Version 2. 08/16/2021
Electronic publication
Footnotes
Glioblastoma is a highly malignant and incurable brain tumor characterized by intrinsic and adaptive resistance to immunotherapies. However, how glioma cells induce tumor immunosuppression and escape immunosurveillance remains poorly understood. Here, we find upregulation of cancer-intrinsic chitinase-3-like 1 (CHI3L1) signaling modulating an immunosuppressive microenvironment by reprogramming tumor-associated macrophages (TAMs). Mechanistically, CHI3L1 binding with galectin 3 (Gal3) selectively promotes TAM migration and infiltration with a protumor M2-like, but not an antitumor M1-like, phenotype in vitro and in vivo, governed by a transcriptional program of NF-κB/CEBPβ in the CHI3L1/Gal3-PI3K/AKT/mTOR axis. Conversely, galectin 3–binding protein (Gal3BP) negatively regulates this process by competing with Gal3 to bind CHI3L1. Administration of a Gal3BP mimetic peptide in syngeneic glioblastoma mouse models reverses immune suppression and attenuates tumor progression. These results shed light on the role of CHI3L1 protein complexes in immune evasion by glioblastoma and as a potential immunotherapeutic target for this devastating disease.
Contributor Information
Apeng Chen, Email: apeng.chen@pitt.edu.
Yinan Jiang, Email: yinan.jiang12@chp.edu.
Zhengwei Li, Email: lizhengwei1006@163.com.
Lingxiang Wu, Email: woolingxiang@gmail.com.
Ulises Santiago, Email: ulises.osa@gmail.com.
Han Zou, Email: HANZOU@pitt.edu.
Chunhui Cai, Email: chunhuic@pitt.edu.
Vaibhav Sharma, Email: VSharma@som.geisinger.edu.
Yongchang Guan, Email: gyc821129@163.com.
Lauren H. McCarl, Email: lam287@pitt.edu.
Jie Ma, Email: maj5@upmc.edu.
Yijen L. Wu, Email: yijenwu@pitt.edu.
Joshua Michel, Email: Joshua.Michel@chp.edu.
Yi Shi, Email: yi.shi@pitt.edu.
Liza Konnikova, Email: liza.konnikova@yale.edu.
Nduka M. Amankulor, Email: amankulornm@upmc.edu.
Pascal O. Zinn, Email: zinnpo@upmc.edu.
Gary Kohanbash, Email: gary.kohanbash2@chp.edu.
Sameer Agnihotri, Email: sameer.agnihotri@pitt.edu.
Songjian Lu, Email: songjian@pitt.edu.
Xinghua Lu, Email: xinghua@pitt.edu.
Dandan Sun, Email: sund@upmc.edu.
George K. Gittes, Email: gittesgk@upmc.edu.
Qianghu Wang, Email: wangqh@njmu.edu.cn.
Xiangwei Xiao, Email: xiangwei.xiao@chp.edu.
Dean Yimlamai, Email: dean.yimlamai@yale.edu.
Ian F. Pollack, Email: POLLACI@upmc.edu.
Carlos J. Camacho, Email: ccamacho@pitt.edu.
Baoli Hu, Email: baolihu@pitt.edu.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Desjardins A. Neuro-oncology: what is the optimal use of bevacizumab in glioblastoma? Nat Rev Neurol. 2015;11(8):429–430. doi: 10.1038/nrneurol.2015.127. [DOI] [PubMed] [Google Scholar]
- 3.Furnari FB, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 4.Buerki RA, et al. Immunotherapy of primary brain tumors: facts and hopes. Clin Cancer Res. 2018;24(21):5198–5205. doi: 10.1158/1078-0432.CCR-17-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim M, et al. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15(7):422–442. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 6.Sampson JH, et al. Brain immunology and immunotherapy in brain tumours. Nat Rev Cancer. 2020;20(1):12–25. doi: 10.1038/s41568-019-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hambardzumyan D, et al. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330(6005):841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, et al. Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Res. 2017;77(9):2266–2278. doi: 10.1158/0008-5472.CAN-16-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinton L, et al. The immune suppressive microenvironment of human gliomas depends on the accumulation of bone marrow-derived macrophages in the center of the lesion. J Immunother Cancer. 2019;7(1):58. doi: 10.1186/s40425-019-0536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komohara Y, et al. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J Pathol. 2008;216(1):15–24. doi: 10.1002/path.2370. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32(1):42–56. doi: 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quail DF, Joyce JA. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31(3):326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu B, et al. Epigenetic activation of WNT5A drives glioblastoma stem cell differentiation and invasive growth. Cell. 2016;167(5):1281–1295. doi: 10.1016/j.cell.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fusetti F, et al. Crystal structure and carbohydrate-binding properties of the human cartilage glycoprotein-39. J Biol Chem. 2003;278(39):37753–37760. doi: 10.1074/jbc.M303137200. [DOI] [PubMed] [Google Scholar]
- 16.Lee CG, et al. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol. 2011;73:479–501. doi: 10.1146/annurev-physiol-012110-142250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwamoto FM, et al. Serum YKL-40 is a marker of prognosis and disease status in high-grade gliomas. Neuro Oncol. 2011;13(11):1244–1251. doi: 10.1093/neuonc/nor117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelloski CE, et al. YKL-40 expression is associated with poorer response to radiation and shorter overall survival in glioblastoma. Clin Cancer Res. 2005;11(9):3326–3334. doi: 10.1158/1078-0432.CCR-04-1765. [DOI] [PubMed] [Google Scholar]
- 19.Darmanis S, et al. Single-cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21(5):1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shingu T, et al. Qki deficiency maintains stemness of glioma stem cells in suboptimal environment by downregulating endolysosomal degradation. Nat Genet. 2017;49(1):75–86. doi: 10.1038/ng.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genoud V, et al. Responsiveness to anti-PD-1 and anti-CTLA-4 immune checkpoint blockade in SB28 and GL261 mouse glioma models. Oncoimmunology. 2018;7(12):e1501137. doi: 10.1080/2162402X.2018.1501137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawada M, et al. Chitinase 3-like 1 promotes macrophage recruitment and angiogenesis in colorectal cancer. Oncogene. 2012;31(26):3111–3123. doi: 10.1038/onc.2011.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu N, et al. Chitinase-3-Like-1 promotes M2 macrophage differentiation and induces choroidal neovascularization in neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2019;60(14):4596–4605. doi: 10.1167/iovs.19-27493. [DOI] [PubMed] [Google Scholar]
- 24.Haynes SE, et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci. 2006;9(12):1512–1519. doi: 10.1038/nn1805. [DOI] [PubMed] [Google Scholar]
- 25.Butovsky O, et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17(1):131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ying W, et al. Investigation of macrophage polarization using bone marrow derived macrophages. J Vis Exp. 2013;(76):50323. doi: 10.3791/50323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen B, et al. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller S, et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 2017;18(1):234. doi: 10.1186/s13059-017-1362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loimaranta V, et al. Galectin-3-binding protein: a multitask glycoprotein with innate immunity functions in viral and bacterial infections. J Leukoc Biol. 2018;104(4):777–786. doi: 10.1002/JLB.3VMR0118-036R. [DOI] [PubMed] [Google Scholar]
- 30.MacKinnon AC, et al. Regulation of alternative macrophage activation by galectin-3. J Immunol. 2008;180(4):2650–2658. doi: 10.4049/jimmunol.180.4.2650. [DOI] [PubMed] [Google Scholar]
- 31.Sano H, et al. Human galectin-3 is a novel chemoattractant for monocytes and macrophages. J Immunol. 2000;165(4):2156–2164. doi: 10.4049/jimmunol.165.4.2156. [DOI] [PubMed] [Google Scholar]
- 32.Inohara H, et al. Interactions between galectin-3 and Mac-2-binding protein mediate cell-cell adhesion. Cancer Res. 1996;56(19):4530–4534. [PubMed] [Google Scholar]
- 33.Champ PC, Camacho CJ. FastContact: a free energy scoring tool for protein-protein complex structures. Nucleic Acids Res. 2007;35(Suppl_2):W556–W560. doi: 10.1093/nar/gkm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pyonteck SM, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19(10):1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneda MM, et al. PI3Kgamma is a molecular switch that controls immune suppression. Nature. 2016;539(7629):437–442. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Rooijen N, et al. In vitro and in vivo elimination of macrophage tumor cells using liposome-encapsulated dichloromethylene diphosphonate. Virchows Arch B Cell Pathol Incl Mol Pathol. 1988;54(4):241–245. doi: 10.1007/BF02899217. [DOI] [PubMed] [Google Scholar]
- 37.Fulci G, et al. Depletion of peripheral macrophages and brain microglia increases brain tumor titers of oncolytic viruses. Cancer Res. 2007;67(19):9398–9406. doi: 10.1158/0008-5472.CAN-07-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woroniecka K, et al. T-cell exhaustion signatures vary with tumor type and are severe in glioblastoma. Clin Cancer Res. 2018;24(17):4175–4186. doi: 10.1158/1078-0432.CCR-17-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medikonda R, et al. A review of glioblastoma immunotherapy. J Neurooncol. 2021;151(1):41–53. doi: 10.1007/s11060-020-03448-1. [DOI] [PubMed] [Google Scholar]
- 40.Topalian SL, et al. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao J, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25(3):462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wellenstein MD, de Visser KE. Cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity. 2018;48(3):399–416. doi: 10.1016/j.immuni.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 43.Novak R, et al. Galectin-1 and galectin-3 expression profiles in classically and alternatively activated human macrophages. Biochim Biophys Acta. 2012;1820(9):1383–1390. doi: 10.1016/j.bbagen.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 44.Gleissner CA, et al. Galectin-3 binding protein, coronary artery disease and cardiovascular mortality: Insights from the LURIC study. Atherosclerosis. 2017;260:121–129. doi: 10.1016/j.atherosclerosis.2017.03.031. [DOI] [PubMed] [Google Scholar]
- 45.Shaked I, et al. Macrophage inflammatory markers are associated with subclinical carotid artery disease in women with human immunodeficiency virus or hepatitis C virus infection. Arterioscler Thromb Vasc Biol. 2014;34(5):1085–1092. doi: 10.1161/ATVBAHA.113.303153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomaszewski W, et al. Brain tumor microenvironment and host state: implications for immunotherapy. Clin Cancer Res. 2019;25(14):4202–4210. doi: 10.1158/1078-0432.CCR-18-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson CM, et al. Mechanisms of immunotherapy resistance: lessons from glioblastoma. Nat Immunol. 2019;20(9):1100–1109. doi: 10.1038/s41590-019-0433-y. [DOI] [PubMed] [Google Scholar]
- 48.Jallal B, et al. Suppression of tumor growth in vivo by local and systemic 90K level increase. Cancer Res. 1995;55(15):3223–3227. [PubMed] [Google Scholar]
- 49.Ullrich A, et al. The secreted tumor-associated antigen 90K is a potent immune stimulator. J Biol Chem. 1994;269(28):18401–18407. doi: 10.1016/S0021-9258(17)32322-0. [DOI] [PubMed] [Google Scholar]
- 50.Kalayci O, et al. Role of 90K protein in asthma and TH2-type cytokine expression. Ann Allergy Asthma Immunol. 2004;93(5):485–492. doi: 10.1016/S1081-1206(10)61417-2. [DOI] [PubMed] [Google Scholar]
- 51.Mitsialis V, et al. Single-cell analyses of colon and blood reveal distinct immune cell signatures of ulcerative colitis and Crohn’s disease. Gastroenterology. 2020;159(2):591–608. doi: 10.1053/j.gastro.2020.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Comeau SR, et al. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32(Suppl_2):W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salomon-Ferrer R, et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald. J Chem Theory Comput. 2013;9(9):3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 54.Maier JA, et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.