Abstract
Introduction
The automated quantitative antigen test (QAT), which detects severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is suitable for mass screening. However, its diagnostic capability differentiated by time from onset and potential contribution to infectivity assessment have not been fully investigated.
Methods
A retrospective, observational study using nasopharyngeal swab specimens from coronavirus disease (COVID-19) inpatients was conducted using LumipulseⓇ SARS-CoV-2 antigen test. Diagnostic accuracy was examined for the early (up to 10 days after onset) and late (over 10 days after onset) stages. Time-course QAT changes and reverse‐transcription quantitative polymerase chain reaction tests results were displayed as locally estimated scatterplot smoothing curve, and receiver operating characteristic curve (ROC) analysis was used to determine the appropriate cutoff value for differentiating the early and late stages.
Results
We obtained 100 specimens from 68 COVID-19 patients, including 51 early-stage and 49 late-stage specimens. QAT sensitivity and specificity were 0.82 (0.72–0.90) and 0.95 (0.75–0.99) for all periods, 0.93 (0.82–0.98) and 1.00 (0.39–1.00) for the early stage, and 0.66 (0.48–0.82) and 0.93 (0.69–0.99) for the late stage, respectively. The ROC analysis indicated an ideal cutoff value of 6.93 pg/mL for distinguishing early-from late-stage specimens. The sensitivity, specificity, positive predictive value, and negative predictive value for predicting the late stage were 0.76 (0.61–0.87), 0.76 (0.63–0.87), 0.76 (0.61–0.87), and 0.76 (0.63–0.87).
Conclusions
QAT has favorable diagnostic accuracy in the early COVID-19 stages. In addition, an appropriate cutoff point can potentially facilitate rapid identification of noncontagious patients.
Keywords: Lumipulse, Quantitative antigen test, Infectivity assessment, Nasopharyngeal swab, COVID-19, SARS-CoV-2
Outbreaks of the novel coronavirus disease 2019 (COVID-19) have become a global concern. The most reliable diagnostic method for COVID-19 in clinical practice is the confirmation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) testing [1]. However, RT-qPCR poses challenges, such as high testing costs, special equipment, and human resource requirements, including, trained laboratory technicians. Consequently, rapid antigen tests (RATs), which are easier to perform, have been developed, although their diagnostic accuracy varies considerably among studies [2].
The newly developed SARS-CoV-2 quantitative antigen test (QAT), Lumipulse® SARS-CoV-2 Ag (Fujirebio, Tokyo, Japan), is an assay based on the chemiluminescence enzyme immunoassay (CLEIA) method [3], and it was approved by the Pharmaceutical Affairs Law as a new COVID-19-antigen test kit on June 19, 2020, in Japan [4].
QAT differs from conventional qualitative antigen tests as it can quantitatively assess antigens in the nasal cavity or saliva with relatively high accuracy. According to a previous report, the concordance of the QAT with RT-qPCR as a reference was reported to be 100% and 85% in specimens with >100 and 10–100 viral copies, respectively, with a 91.4% overall concordance rate [5]. However, its diagnostic capability, when differentiated by time from onset, has not been fully investigated. Moreover, some studies have implied that the RAT is a potentially superior predictor of positive viral culture results to RT-qPCR [6,7], implying that RAT may contribute to the identification of specimens with disease transmissibility. In this study, we aimed to investigate the following: (1) the chronological changes in QAT accuracy in the early and late stages of COVID-19 and (2) the potential contribution of QAT to the estimation of disease transmissibility.
This was a retrospective, observational study of COVID-19 patients admitted to the National Center for Global Health and Medicine (NCGM) with a confirmed RT-qPCR diagnosis prior to admission. Their samples were stored, with written consent having been obtained in a previous prospective study (NCGM-G-003472-02), and ethical consideration was made by way of secondary use with an opt-out option. A universal transport medium (1 mL or 3 mL; COPAN Diagnostics Inc., USA) was used as the viral transport medium (VTM). If the VTM volume was less than 1 mL, it was diluted with 2 mL of saline, of which 500 μL was used for analysis. Patients who had nasopharyngeal swab specimens collected during storage between March 14, 2020, and June 12, 2020, were included.
Two-hundred microliter samples were used to obtain 60 μL of nucleic acid extract using the QIAsymphony DSP virus/Pathogen Mini Kit (QIAGEN, Hilden, Germany). Five microliters of nucleic acid extract was used for RT-qPCR. RT-qPCR was performed using the Applied Biosystems 7500 Real Time PCR System (Applied Biosystems, Foster City, CA, USA) or QuantStudio® 5 (Applied Biosystems). Using the calculated cycle threshold value and PCR amplification efficiency of the N2 primer set, a correlation equation for the Ct value and the number of RNA copies was determined. The Ct value obtained from the reference material (SeraCare, AccuPlex™ SARS-CoV-2 Reference Material Kit) was used to perform a correction to calculate the number of RNA copies. For the samples with negative RT-qPCR results using the National Institute of Infectious Diseases (NIID) method [8], RT-qPCR retesting was performed using a standardized-assay kit (SARS-CoV-2 Direct Detection RT-qPCR Kit; Takara Bio, Japan).
Lumipulse® SARS-CoV-2 Ag testing was performed according to the manufacturer's instructions (Fujirebio Inc.). Briefly, samples were centrifuged at 2,000×g for 10 min, and the supernatant was used for the following test. We used 100 μL of thawed samples to measure antigen levels with a Lumipulse® G1200 automated machine (Fujirebio Inc.), which is based on the CLEIA method. When the antigen level was not measured because it exceeded the detection limit, we tested the diluted sample and calculated the antigen level of the original sample.
In this study, specimens from confirmed patients were used, and the existing cutoff value of 1.34 pg/mL was used to determine QAT positivity. The sensitivity and specificity (95% confidence intervals [CI]) with RT-qPCR results were calculated for the early (up to 10 days after onset) and late (over 10 days after onset) stages. The validity of the results was examined using Cohen's kappa and Gwet's AC1 statistic (AC1). Time-series changes in antigen levels and RT-qPCR results were displayed as the locally estimated scatterplot smoothing (LOESS) curve, and receiver operating characteristic (ROC) curve analysis was conducted with the cutoff value defined as the least distance from the top-left corner of the box. Match-rate and time-series analyses were performed using SAS software, version 9.4 (SAS Institute, Cary, NC). All other analyses were performed using SPSS Statistics (version 26; IBM Corp., Armonk, NY, USA).
This study was conducted in compliance with the guidelines of the Declaration of Helsinki after receiving ethical review approval from the Ethics Committee of the NCGM (NCGM-G-003586-00).
A total of 100 specimens were obtained from confirmed COVID-19 patients, including 51 early-phase and 49 late-stage specimens, with a median time from onset (interquartile range) of 8 (6–9) and 16 (13–19) days, respectively. There were 77 mild cases (no oxygen demand), 14 moderate cases (oxygen in demand except for ventilator), and 9 severe cases (ventilator in demand). The majority of patients were immunocompetent except for one case of metastatic cancer and three cases of human immunodeficiency virus infection in mild disease and two cases of solid organ transplantation in severe disease. The antiviral agents used were Hydroxychloroquine (32 cases; 18 mild, 6 moderate, and 8 severe), inhaled Ciclesonide (11 cases; 10 mild and 1 moderate), Favipiravir (6 cases; 3 mild and 3 moderate), and Lopinavir-Ritonavir (1 moderate case). Among the collected VTM specimens, 70 of 100 specimens were positive, while 30 were negative, as determined by the NIID assay. Of these negative specimens, 10 were confirmed positive by the Takara assay, resulting in 80 positive and 20 negative specimens by RT-qPCR. For all specimens, QAT sensitivity and specificity for all periods were 0.82 (0.72–0.90) and 0.95 (0.75–0.99), respectively (Table 1 ). QAT sensitivity and specificity for the early phase were 0.93 (0.82–0.98) and 1.00 (0.39–1.00) and those for the late stage were 0.66 (0.48–0.82) and 0.93 (0.69–0.99), respectively.
Table 1.
Concordance of the QAT with the RT-qPCR in different time periods.
| (A) QAT vs RT-qPCR: all periods | ||||
|---|---|---|---|---|
| RT-qPCR |
||||
| Positive | Negative | Total | ||
| Antigen test | Positive | 66 | 1 | 67 |
| Negative | 14 | 19 | 33 | |
| Positive coincidence rate | 0.82 (0.72–0.90) | |||
| Negative coincidence rate | 0.95 (0.75–0.99) | |||
| Cohen's kappa (SD) | 0.75 (0.62–0.87) | |||
| Gwet's AC1 statistic (SD) | 0.62 (0.45–0.78) | |||
| (B) QAT vs RT-qPCR: from day 1 to day 10 after onset (the early stage) | ||||
| RT-qPCR | ||||
| Positive | Negative | Total | ||
| Antigen test | Positive | 44 | 0 | 44 |
| Negative | 3 | 4 | 7 | |
| Positive coincidence rate | 0.93 (0.82–0.98) | |||
| Negative coincidence rate | 1.00 (0.39–1.00) | |||
| Cohen's kappa (SD) | 0.92 (0.84–1.00) | |||
| Gwet's AC1 statistic (SD) | 0.69 (0.38–1.00) | |||
| (C) QAT vs RT-qPCR: over 10 days after onset (the late stage) | ||||
| RT-qPCR | ||||
| Positive | Negative | Total | ||
| Antigen test | Positive | 22 | 1 | 23 |
| Negative | 11 | 15 | 26 | |
| Positive coincidence rate | 0.66 (0.48–0.82) | |||
| Negative coincidence rate | 0.93 (0.69–0.99) | |||
| Cohen's kappa (SD) | 0.52 (0.27–0.76) | |||
| Gwet's AC1 statistic (SD) | 0.52 (0.30–0.73) | |||
QAT, SARS-CoV-2 quantitative antigen test; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; SD, standard deviation.
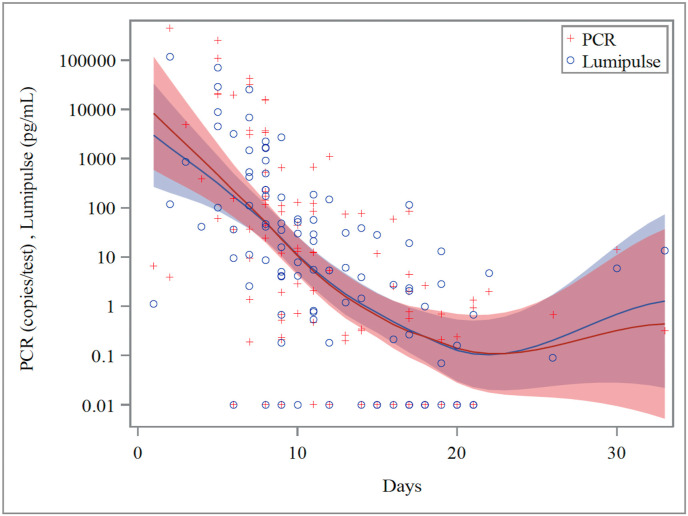
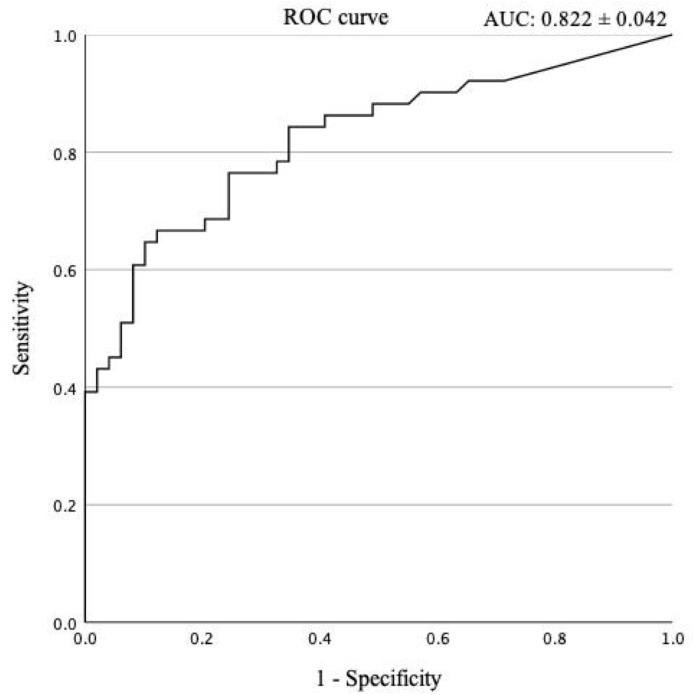
LOESS curve was used to compare QAT and RT-qPCR along the time series (Fig. 1 ). The correlation coefficients between QAT and SARS-CoV-2 RNA in all periods, in the early stage, and in the late stage were 0.883, 0.882, and 0.794, respectively (Supplementary Figure 1). The ROC analysis for samples in all periods indicated that the most appropriate cutoff value to distinguish early-phase from late-stage specimens was 6.93 pg/mL (Fig. 2 ). Using new cutoff values, the sensitivity, specificity, positive predictive value, and negative predictive value for predicting the late stage was 0.75 (0.61–0.86), 0.76 (0.62–0.87), 0.75 (0.61–0.86), and 0.76 (0.62–0.87), respectively.
Fig. 1.
Variation of SARS-CoV-2 Ag concentration and RT-qPCR results over time. Relationship between days after onset and SARS-CoV-2 Ag concentration or RT-qPCR results according to the locally estimated scatterplot smoothing curve. The plus sign and circle sign correspond to the results of RT-qPCR and Lumipulse® SARS-CoV-2 Ag testing, respectively. The red and blue lines show the LOESS curves and the 95% confidence intervals for RT-qPCR and Lumipulse® SARS-CoV-2 Ag testing, respectively. Negative test results were represented by a Y-axis value of 0.01 for convenience. Ag, antigen; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2
Fig. 2.
ROC analysis for predicting the late stage after onset. Receiver operating characteristic (ROC) analysis was performed to determine the cutoff value of the SARS-CoV-2 Ag concentration to differentiate between the early (up to 10 days after the onset) and late (over 10 days after the onset) stages. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2
In this study, we analyzed the diagnostic accuracy of QAT against RT-qPCR in the early and late stages of COVID-19. QAT and RT-qPCR were well correlated in a time series using the LOESS curve, and QAT results showed high concordance with those of RT-qPCR, suggesting minimal concern for missing patients in the early stage. However, QAT sensitivity in the late stage was lower than in the early stage, with a declined correlation coefficient. Thus, it may be inadvisable to exclude COVID-19 in the late stages when using QAT. However, it is more important to reliably distinguish patients in the early stages of the disease when the virus is more contagious.
Aoki et al. reported on the positive cutoff for QAT using SARS-CoV-2 infected and uninfected patients, and set the cutoff at 1.03 pg/mL [9]. This was a positive criterion that did not consider the time of onset. In the current study, we have presented a cutoff to determine whether the patient is within 10 days or later after onset, and it seems reasonable that our cutoff is higher than that for the positivity criterion. In addition, in the time-course of antigen levels shown by Aoki et al. only one sample fell below our cutoff within 10 days from onset, and this sample had also been judged negative for QAT by the authors. Given these results, we believe that our findings, which suggest that a high cutoff for QAT may distinguish between early- and late–stage COVID-19 from onset, are consistent with those of previous studies.
Regarding the duration of infectious viral shedding, the presence of a viable virus should be confirmed. Although a viral culture could be a surrogate marker for the presence of viable viruses, this cannot be readily performed [10]. According to Singanayagam et al. in 133 SARS-CoV-2 culture-positive samples obtained from 253 patients, the culture-positive period for almost all samples was within 10 days, whereas only two samples were positive until days 11 and 12 [11]. The authors estimated a 6.0% (95% CI: 0.2%–31.2%) probability of culturing the virus in samples 10 days after disease onset. Arons et al. reported that among 53 samples collected from 43 patients, viable virus was isolated for up to 9 days in COVID-19 patients with typical symptoms [12]. Only one specimen from a patient with atypical symptoms was reportedly culture positive on day 13. Based on these studies, several institutions, including those in Japan, have now established exemptions from infection control measures 10 days after disease onset [13,14].
Positive-RT-qPCR persistency has been reported even after meeting the existing quarantine-release requirement of 10 days from disease onset. In this study, 33/49 samples remained RT-qPCR-positive after 10 days of illness. A similar challenge existed with the Lumipulse® SARS-CoV-2 Ag, where 23/49 specimens remained positive. However, using a cutoff of 6.93 pg/mL, which is higher than the existing cutoff of 1.34 pg/mL, might have contributed to the prediction that the specimens were no longer infectious. RT-qPCR provides a diagnosis based on the cycle threshold value. The problem with this diagnostic method is that the cycle threshold cutoff value is not validated among diagnostic reagents [15]. The same 10-day quarantine is prescribed for asymptomatic pathogen carriers and those with slightly mild disease, although the time from onset is sometimes ambiguous [13]. However, a uniform response potentially overestimates quarantining large numbers of people who are no longer a public health threat.
Previous studies have suggested that RAT is a potentially superior predictor of viral culture results to RT-qPCR. Pekosz et al. found concordance between SARS-CoV-2 VeroE6 TMPRSS2 culture, RT-qPCR, and rapid antigen testing and revealed that RAT had a higher positive predictive value than RT-qPCR (90% vs. 70%) [6]. Kohmer et al. reported that the antigen-detecting rapid diagnostic test correlated better with cell culture infectivity in vitro than RT-qPCR (61.8%–82.4% vs 51.6%) [7]. Our previous study also implied the utility of qualitative antigen testing in predicting the appropriate duration of quarantine [16]. Qualitative antigen tests generally tend to be less sensitive, and false-negative results in contagious patients have been a challenge. In contrast, Lumipulse® SARS-CoV-2 Ag allows quantitative evaluation of antigens, which may minimize the problem of false negatives in the early stages of infection. In addition, this test may be used to differentiate contagious patients by setting a value other than the original cutoff for a positive result.
Finally, the optimal strategy for preventing the spread of infection could depend on the disease prevalence in the community. Even testing with suboptimal accuracy, such as rapid antigen testing, can be useful in situations where a pandemic is spreading rapidly. Mathematical models showed that frequent testing and strict isolation of patients using RATs could contribute to pandemic control at justifiable cost by reducing local transmission of the virus and its mortality [17,18]. Although RT-PCR is a diagnostic method with high sensitivity and specificity, it requires more financial and human resources. Other convenient diagnostic methods could ensure efficient use of limited resources. QAT, with its relatively high diagnostic accuracy, might contribute to the control of the current pandemic.
There are some limitations in our study. First, we set 10 days from the COVID-19 onset as the criterion for relaxation of isolation. However, long-term shedding of the infectious virus has been reported in severe disease and immunocompromised cases [19,20]. These specific cases should be considered separately from our study. Second, although viral culture serves as a surrogate marker of viral infectivity, we did not perform viral culture in our study. Therefore, we lack a strict determination of infectivity for individual cases. Discontinuation of isolation using our cutoff requires caution, and its use may be limited to cases with mild-to-moderate disease.
In conclusion, Lumipulse® SARS-CoV-2 Ag is a rapid test with favorable diagnostic accuracy in the early stages of COVID-19. The use of an appropriate cutoff value potentially facilitates the rapid identification of noncontagious patients.
Authorship statement
Concept and design: H.N, K.Y.
Acquisition, analysis, or interpretation of data: H.N, K.Y.
Data collection: M.S, N.K.
Drafting of the manuscript: H.N.
Statistical analysis: H.N, G.Y.
Preparation and handling of specimens: A.M.
Critical revision of the manuscript for important intellectual content: All authors.
All the authors meet the ICMJE authorship criteria.
Funding
This work was supported by Fujirebio Inc., Japan, whose funds were used to conduct the current research and prepare the article.
Declaration of competing interest
K.Y. received research grants from Fujirebio Inc. For their submitted work, and received research grants from Mizuho Medy, Co., Ltd., and VisGene Inc., outside the submitted work. N.O. received grants from Sanofi K.K., Japan, and Eiken Chemical Co. Ltd., Japan, outside the submitted work.
Acknowledgements
We thank the staff from the Disease Control and Prevention Center, Department of Respirology, National Center for Global Health and Medicine, as well as those from the AIDS Clinical Center, National Center for Global Health and Medicine, for collecting the clinical samples.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jiac.2021.08.015.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.WHO Diagnostic testing for SARS-CoV-2. Interim guidance. https://www.who.int/publications/i/item/diagnostic-testing-for-sars-cov-2
- 2.Dinnes J., Deeks J.J., Adriano A., Berhane S., Davenport C., Dittrich S., et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2020;8:CD013705. doi: 10.1002/14651858.CD013705. PMID: 32845525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arakawa H. Development of highly sensitive analytical methods for biologically relevant materials and their pharmaceutical applications. Chem Pharm Bull (Tokyo) 2017;65(12):1099–1112. doi: 10.1248/cpb.c17-00787. PMID: 29199216. [DOI] [PubMed] [Google Scholar]
- 4.Ministry of health, labour and welfare. https://www.pmda.go.jp/files/000235412.pdf
- 5.Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K., et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int J Infect Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. PMID: 32800855, PMCID: PMC7422837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pekosz A., Parvu V., Li M., Andrews J.C., Manabe Y.C., Kodsi S., et al. Antigen-based testing but not real-time polymerase chain reaction correlates with severe acute respiratory syndrome coronavirus 2 viral culture. Clin Infect Dis. 2021 doi: 10.1093/cid/ciaa1706. ciaa1706. Epub 2021 Jan 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohmer N., Toptan T., Pallas C., Karaca O., Pfeiffer A., Westhaus S., et al. The comparative clinical performance of four SARS-CoV-2 rapid antigen tests and their correlation to infectivity in vitro. J Clin Med. 2021;10(2):328. doi: 10.3390/jcm10020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Institute of Infectious Diseases Japan: manual for the detection of Pathogen 2019-nCoV ver.2.6. https://www.niid.go.jp/niid/images/epi/corona/2019-nCoVmanual20200217-en.pdf
- 9.Aoki K., Nagasawa T., Ishii Y., Yagi S., Okuma S., Kashiwagi K., et al. Clinical validation of quantitative SARS-CoV-2 antigen assays to estimate SARS-CoV-2 viral loads in nasopharyngeal swabs. J Infect Chemother. 2021;27(4):613–616. doi: 10.1016/j.jiac.2020.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leland D.S., Ginocchio C.C. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev. 2007;20(1):49–78. doi: 10.1128/CMR.00002-06. PMID: 17223623, PMCID: PMC1797634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J., et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32):2001483. doi: 10.2807/1560-7917.ES.2020.25.32.2001483. PMID: 32794447, PMCID: PMC7427302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., et al. Public Health – seattle & King County and CDC. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382(22):2081–2090. doi: 10.1056/NEJMoa2008457. PMCID: PMC7200056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ministry of health, labour and welfare. https://www.mhlw.go.jp/content/000745527.pdf
- 14.Centers for Disease Control and Prevention Interim guidance on ending isolation and precautions for adults with COVID-19. https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html
- 15.Chang M.C., Hur J., Park D. Interpreting the COVID-19 test results: a guide for physiatrists. Am J Phys Med Rehabil. 2020;99(7):583–585. doi: 10.1097/PHM.0000000000001471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto K., Suzuki M., Yamada G., Sudo T., Nomoto H., Kinoshita N., et al. Utility of the antigen test for coronavirus disease 2019: factors influencing the prediction of the possibility of disease transmission. Int J Infect Dis. 2021;104:65–72. doi: 10.1016/j.ijid.2020.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Z., Pandey A., Bai Y., Fitzpatrick M.C., Chinazzi M., y Piontti A.P., et al. Comparative cost-effectiveness of SARS-CoV-2 testing strategies in the USA: a modelling study. Lancet Public Health. 2021;6(3):e184–e191. doi: 10.1016/S2468-2667(21)00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paltiel A.D., Zheng A., Sax P.E. Clinical and economic effects of widespread rapid testing to decrease SARS-CoV-2 transmission. Ann Intern Med. 2021;174(6):803–810. doi: 10.7326/M21-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarhini H., Recoing A., Bridier-Nahmias A., Rahi M., Lambert C., Martres P., et al. Long-term severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectiousness among three immunocompromised patients: from prolonged viral shedding to SARS-CoV-2 superinfection. J Infect Dis. 2021;223(9):1522–1527. doi: 10.1093/infdis/jiab075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Kampen J.J., van de Vijver D.A., Fraaij P.L., Haagmans B.L., Lamers M.M., Okba N., et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat Commun. 2021;12(1):267. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.