Abstract
Purpose: Our objective is to identify the predictive factors and predict hospital length of stay (LOS) in dengue patients, for efficient utilization of hospital resources. Methods: We collected 1360 medical patient records of confirmed dengue infection from 2012 to 2017 at Max group of hospitals in India. We applied two different data mining algorithms, logistic regression (LR) with elastic-net, and random forest to extract predictive factors and predict the LOS. We used an area under the curve (AUC), sensitivity, and specificity to evaluate the performance of the classifiers. Results: The classifiers performed well, with logistic regression (LR) with elastic-net providing an AUC score of 0.75 and random forest providing a score of 0.72. Out of 1148 patients, 364 (32%) patients had prolonged length of stay (LOS) (> 5 days) and overall hospitalization mean was 4.03 ± 2.44 days (median ± IQR). The highest number of dengue cases belonged to the age group of 10-20 years (21.1%) with a male predominance. Moreover, the study showed that blood transfusion, emergency admission, assisted ventilation, low haemoglobin, high total leucocyte count (TLC), low or high haematocrit, and low lymphocytes have a significant correlation with prolonged LOS. Conclusion: Our findings demonstrated that the logistic regression with elastic-net was the best fit with an AUC of 0.75 and there is a significant association between LOS greater than five days and identified patient-specific variables. This method can identify the patients at highest risks and help focus time and resources.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10729-021-09571-3.
Keywords: Patient’s length of stay (LOS), Dengue, Predictive models, Healthcare, Elastic-net, Random forest
Highlights
The main objective is to identify the factors which can determine and predict prolonged length of stay for dengue patients.
The findings indicate that essential information available in the first 24 hours of hospitalization for Dengue can be used for predicting prolonged LOS (> 5 days).
The LOS prediction for Dengue patients can be used by clinicians and or hospital managers to design appropriate, case specific, interventions for reduction in length of stay and improved patient outcome.
This system has the potential to help healthcare institutions to improve their decisions about patient management and resource allocation.
Introduction
Dengue is the fastest-growing mosquito-borne disease across the world today [1]. It is a mosquito-borne viral infection that affects infants, young children, and adults. This infection is transmitted by a mosquito bite infected with one of the four serotypes of the dengue virus. Aedes aegypti is the main vector in most of the urban areas of India and Asia. Aedes albopictus is also found as a vector in few areas of southern and eastern India. The World Health Organization (WHO) estimates that nearly 400 million infections occur every year in over 128 countries in Asia, Oceania, America, and Africa. It is evident from the reports that about half of the world’s population is currently at the risk of dengue transmission [1].
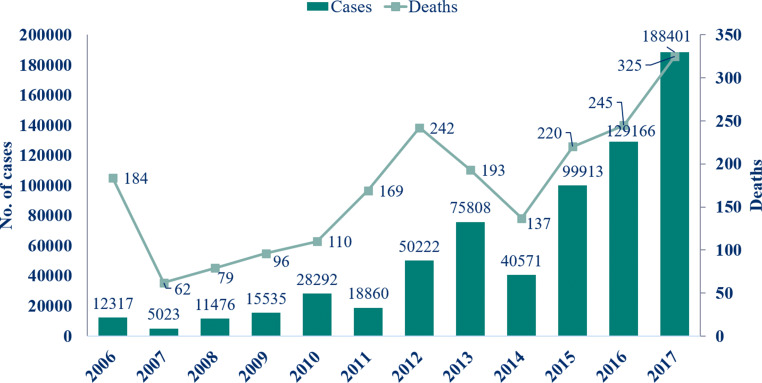
Dengue in India has spread significantly over the past few decades, with rapidly changing epidemiology. According to the data from the Directorate of National Vector Borne Disease Control Programme (NVBDCP) [2] and National Health Profile 2018 [3], in 2017, the spike in cases of dengue was the highest in the last one decade. From less than 60,000 cases in 2009, cases increased to 188,401 in 2017, more than a 300 percent spike. When compared to 75,808 cases in 2013, it is more than a 250 percent spike (Fig 1). The number of outbreaks has risen, and certain states and union territories have become hyperendemic [4]. In 2015, the Indian capital region, Delhi, recorded its worst outbreak since 2006 with over 15,000 confirmed dengue cases [5]. Dengue diseases are characterized by a prolonged length of stay (LOS). Prolonged hospitalization is associated with adverse outcomes for the patients and the hospital, such as high complications, poor outcomes, and high care cost that creates a significant economic burden for the hospital [6]. The overall cost of dengue in 2016 was about US$5.71 billion and US$1.51 billion in 2013 [7]. There has been considerable interest in controlling the use of hospital resources, particularly in dengue diseases; thus, hospitals try to make LOS as short as possible. The LOS can be used as an overall parameter to identify health care resource utilization, healthcare cost, and, severity of illness [8]. Therefore, predicting patients which need the most aggressive early intervention and those who require a moderate amount of intervention to prevent prolonged LOS seems to be crucial. There have been other studies [9–13] that conducted prediction of LOS for other diagnoses with data mining techniques, but very few studies [14–16] have addressed the dengue LOS in hospitals.
Fig. 1.
Incidence of cases and deaths due to dengue in India (Source: Directorate of National Vector Borne Disease Control Programme, Dte. GHS, Ministry of Health and Family Welfare)
We apply data mining techniques to extract useful knowledge and to estimate the LOS for dengue patients. In this paper, we present a system to predict the hospital LOS of patients with confirmed dengue diagnosis. Our contributions are listed as follows:
We collect and examine available information for confirmed dengue patients admitted at Max group of hospitals in the National Capital Region (NCR) of India.
We propose strategies to handle missing values in the collected data.
We predict the LOS of Dengue patients with an encounter at one of Max group healthcare systems at NCR.
We investigate the factors that can be assessed to predict the LOS of dengue patients.
We predict patients which need the most aggressive early interventions, and those patients who require a moderate amount of interventions.
We use logistic regression (LR) with elastic-net and random forest classifiers for the prediction and identification of the important factors associated with the dengue patient data. We internally validate our results with evaluation methods such as recall, precision, and AUC. We seek help from domain experts in the medical domain to validate the results. The experiments show the usefulness of our method in predicting the LOS and identification of predictive factors for classification.
Related work
Dengue mortality and Severity related studies
Acharya et al. [17], have done a prospective cross-sectional study in a total of 364 patients with immunoglobulin m (IgM) dengue serology positive who were admitted to a tertiary care hospital with features of dengue fever. The authors found that the factors such as Age > 40 years, presence of hypotension, platelets < 20,000 cells/mm3, alanine aminotransferase (ALT) > 200U/L, aspartate aminotransferase (AST) > 200U/L, prolonged prothrombin time, presence of renal failure, encephalopathy, multiple organ dysfunction syndrome (MODS), acute respiratory distress syndrome (ARDS) and bleeding tendency (p-value < 0.05) have a significant influence with increased risk of mortality among the dengue patients. In a separate study, Md-Sani et al. [18], built a logistic regression based on a data set of 199 adult patients hospitalization in Kuala Lumpur Hospital, Malaysia. The study identified lethargy, bleeding, pulse rate, serum bicarbonate, and serum lactate to be statistically significant predictors of death. Jain et al. [19], conducted a study to identify the factors that influence dengue-related mortality and disease and found that age, sensorium, and dyspnea have a significant influence on mortality and severity.
Dengue and other diagnosis LOS studies
Wiratmadja et al. [16], conducted a study to predict hospital LOS of dengue patients using demographic and illness or health-related data set of 370 dengue fever (DF) and dengue haemorrhagic fever (DHF) patients in Bandung, Indonesia. The study identified systolic blood pressure, diastolic blood pressure, haematocrit, leucocytes, lymphocytes, monocytes, and comorbidity score as the most significant predictors. Chakravarty et al. [14], conducted studies with patients admitted with dengue fever in the Paediatric department in Northern India to determine the clinical and laboratory features and found predictive factors for the prolonged hospital admission.
A cross-sectional retrospective study to determine mortality and prolonged hospital stay among patients with confirmed dengue diagnosis based on a data set of 667 hospitalizations was done by Mallhi et al. [15]. The study showed that DHF, elevated alkaline phosphatase (ALP), prolonged prothrombin time (PT), activated partial thromboplastin time (aPTT) and multiple-organ dysfunctions are associated with prolonged hospitalization.
Similarly, studies related to LOS of other diagnoses are, Liu et al. [20], who conducted a comparative analysis to predict LOS which was tested on Geriatric and stroke data sets based on two classification algorithms. Hachesu et al. [12], compared three classification algorithms to predict LOS of heart patients and found SVM was the best fit. Combes et al. [11], explored the prediction of hospital stay in the emergency department using regression models, Blais et al. [10] derived a prediction model as a screening and rating tool using multivariate analysis to quantify variables related to LOS for an acute care medical psychiatric unit, and Azari et al. [9], designed an approach to predict hospital LOS by clustering datasets and using various classifier models such as Bayes net [21], SVM [22], JRIP [23], J48 [24], and Bagging [25]. Most of the prediction studies in dengue disease have attempted to classify DF and DHF or in-hospital mortality [19]. However, very few studies have addressed the LOS prediction problem in dengue patients.
Methods
Data set
The cohort includes patients who have been hospitalized during the study period under the department of Internal Medicine between February 2012 and September 2017. We identified 1360 patients who were admitted to Max group of hospitals in India with dengue disease-related diagnosis. The research study was approved by the institutional Max Healthcare Ethics Committee. We used standard WHO definitions to classify suspected dengue infection [26]. A total of 1148 microbiologically confirmed dengue patients are included in this study. Dengue confirmation is done using two methods,
NS-1 antigen (Panbio Dengue Early ELISA, Standard Diagnostics Inc., Republic of Korea) if the patient presented within 5 days of disease onset.
Dengue serum immunoglobulin M (NIV DEN Immunoglobulin [IgM] Capture ELISA, National Institute of Virology, Pune, India) if the patient presented after 5 days of disease onset.
Patient data are stored in a hospital database management system of Microsoft SQL server database. We extracted data in three phases. In phase 1, demographic and confirmed dengue diagnosis patients were extracted. Information related to administrative and investigations were extracted in phase 2. Then, radiological, procedure, clinical related data were collected in phase 3. We constructed a new data set for hospital LOS of dengue patients from the extracted information. However, 212 patients were removed from the analysis because of the unavailability of platelet count test information, leaving 1148 patients in the final data set.
In the first screening of the features, 40 features were selected, including age, gender, type of admission, blood transfusion, assisted ventilation, lab, and radiological related features of dengue patients, using the data available for 24 hours of hospitalization. Units, value range, and missing percentage of each feature are given in Table 1.
Table 1.
Attributes characteristics of the length of stay prediction dataset of dengue patients (n = 1148)
| Attributes | Unit | Value | Missing data | Method: |
|---|---|---|---|---|
| alternative value | ||||
| Age | Year | 0-87 | 0 | – |
| Gender | – | male; female | 0 | – |
| Length of Stay (LOS) | Days | 1-94 | 0 | – |
| Admission Type | – | emergency; direct | 0 | – |
| Platelet Count | x109/L | 5.0 - 676.0 | 0 | – |
| Haematocrit | % | 18.9 - 58.9 | 10.2 | Median; 40.4 |
| Haemoglobin | g/dL | 4.8 - 19.4 | 15.5 | Median; 13.4 |
| TLC | x109/L | 0.9 - 45.4 | 16.9 | Median; 5.0 |
| Lymphocytes | % | 2.5 - 100.0 | 18.1 | Median; 35 |
| Monocytes | % | 0.0 - 43.7 | 18.1 | Median; 6.0 |
| Neutrophil | % | 7.0 - 95.8 | 18.1 | Median; 56 |
| Eosinophils | % | 0.0 - 36.9 | 18.8 | Median; 1.0 |
| MCH | Pg | 15 – 43 | 18.6 | Median; 29 |
| MCHC | gm/dL | 28.4 - 37.4 | 18.6 | Median; 33.2 |
| MCV | fL | 51.2 - 122.3 | 18.6 | Median; 86.1 |
| RBC Count | x1012/L | 1.8 - 1640.0 | 18.6 | Median; 4.8 |
| RDW | % | 10.6 - 45.5 | 18.6 | Median; 13.7 |
| SGPT (ALT) | IU/L | 8.0 - 6054.0 | 32.4 | Median; 66 |
| SGOT (AST) | IU/L | 13.0 - 18590.0 | 35.1 | Median; 106 |
| Potassium | mmol/L | 2.7 - 6.6 | 34 | Median; 4.1 |
| Sodium | mmol/L | 117.4 - 150.0 | 34.1 | Median; 135.1 |
| Creatinine | mg/dL | 0.1 - 7.6 | 36.4 | Median; 0.7 |
| Albumin | g/dL | 1.3 - 5.3 | 41.5 | Median; 3.6 |
| Bilirubin Total | mg/dL | 0.1 - 17.1 | 44.7 | Median; 0.6 |
| Bilirubin Direct | mg/dL | 0.0 - 7.0 | 44.8 | Median; 0.2 |
| Bilirubin Indirect | mg/dL | 0.0 - 16.6 | 44.8 | Median; 0.4 |
| Globulin | g/dL | 1.0 - 4.8 | 47 | Median; 2.9 |
| Total Protein | g/dL | 2.9 - 8.5 | 46.9 | Median; 6.5 |
| Basophils | % | 0.0 - 3.3 | 49.7 | Removed |
| PT | S | 8.8 - 100.0 | 78.9 | Removed |
| APTT | S | 22.4 - 126.0 | 83 | Removed |
| Left Effusion | – | 1, yes; 0, no | 40 | Regression imputation |
| Right Effusion | – | 1, yes; 0, no | 40 | Regression imputation |
| Bilateral Effusion | – | 1, yes; 0, no | 40 | Regression imputation |
| Abdominal Free Fluid | – | 1, yes; 0, no | 40 | Regression imputation |
| Enlarged Spleen | – | 1, yes; 0, no | 40 | Regression imputation |
| Enlarged Liver | – | 1, yes; 0, no | 0 | – |
| Dialysis | – | 1, yes; 0, no | 0 | – |
| Assisted Ventilation | – | 1, yes; 0, no | 0 | – |
| Blood Component Transfusion | – | 1, yes; 0, no | 0 | – |
Demographic and clinical details are recorded at admission in a predesigned pro-forma, whereas laboratory findings are recorded daily until discharged or dead. The dataset contains demographic, administrative, investigation, and radiological characteristic features with categorical and numerical values. We categorize numerical and categorical data values and derive new fields from existing data in the following features: platelet count, TLC, haematocrit, AST, ALT, haemoglobin, pleural effusion, etc. as shown in Table 2. These features are converted to categorical variables to improve interpretability.
Table 2.
The demographic, investigation, clinical and procedure characteristics of the length of stay data set (n = 1148)
| Variable | Value | ||
|---|---|---|---|
| LOS (Days) | 1, > 5; 0,≤ 5 | ||
| Gender | 1, male; 0, female | ||
| Age | 1, 0-10 else 0; 1, 10-20 else 0; 1, 20-30 else 0; 1, 30-40 else 0; 1, 40-50 else 0; 1, 50-60 else 0, 1, > 60 else 0 | ||
| Marital Status | 1, single else 0; 1, married else 0; 1, unknown else 0 | ||
| Address | 1, NCR ; nonNCR, 0 | ||
| Channel | 1, cash else 0; 1, corporate else 0; 1, PSU else 0; 1, TPA else 0 | ||
| Type of admission | 1, emergency else 0; 1, direct else 0 | ||
| Previous admission | 1, yes; 0, no | ||
| Investigations | Normal | Low | High |
| Heamatocrit (%) | if((men & hct≥ 40 & hct≤ 50) or (women & hct≥ 36 & hct≤ 46)), 1 else 0 | if((men & hct< 40) or (women & hct< 36)), 1 else 0 | if((men & hct> 50) or (women & hct> 46)), 1 else 0 |
| Haemoglobin (g/dL) | if((men & age≥ 13yr & hgb≥ 13 & hgb≤ 17) or (women & age≥ 13yr & hgb≥ 12 & hgb≤ 15) or (age< 13yr &hgb≥ 11 & hgb≤ 15)), 1 else 0 | if((men & age≥ 13yr & hgb< 13) or (women & age≥ 13yr & hgb< 12) or (age< 13yr & hgb< 11)), 1 else 0 | if((men & age≥ 13yr & hgb> 15) or (women & age≥ 13yr & hgb< 17) or (age< 13yr & hgb> 15)), 1 else 0 |
| TLC (x109/L) | if((tlc≥ 4) & (age≥ 17yr & tlc≤ 10) or (age< 17yr & tlc≤ 15)), 1 else 0 | if(tlc< 4), 1 else 0 | if((age≥ 17yr & tlc> 10) or (age< 17yr & tlc> 15)), 1 else 0 |
| Lymphocytes (%) | if((age≥ 8yr & lym≥ 20 & lym≤ 40) or (age< 8yr & lym≥ 40 & lym≤ 75)), 1 else 0 | if((age≥ 8yr & lym< 20) or (age< 8yr & lym< 40)), 1 else 0 | if((age≥ 8yr & lym> 40) or (age< 8yr & lym> 75)), 1 else 0 |
| Monocytes (%) | if(mono≥ 2 & mono≤ 10), 1 else 0 | if(mono< 2), 1 else 0 | if(mono> 10), 1 else 0 |
| Neutrophils (%) | if(neutro≥ 20 & neutro≤ 45), 1 else 0 | if(neutro< 20), 1 else 0 | if(neutro> 45), 1 else 0 |
| Eosinophils (%) | if(eos≥ 1 & eos≤ 6), 1 else 0 | if(eos< 1), 1 else 0 | if(eos> 6), 1 else 0 |
| MCH (Pg) | if(mch≥ 26 & mch≤ 34), 1 else 0 | if(mch< 26), 1 else 0 | if(mch> 34), 1 else 0 |
| MCHC (gm/dL) | if(mchc≥ 32 & mchc≤ 36), 1 else 0 | if(mchc< 32), 1 else 0 | if(mchc> 36), 1 else 0 |
| MCV (fL) | if(mcv≥ 80 & mcv≤ 100), 1 else 0 | if(mcv< 80), 1 else 0 | if(mcv> 100), 1 else 0 |
| RBC Count (x1012/L) | if(rbc≥ 4.5 & rbc≤ 5.5), 1 else 0 | if(rbc< 4.5), 1 else 0 | if(rbc> 5.5), 1 else 0 |
| RDW (%) | if(rdw≥ 11.5 & rdw≤ 14.5), 1 else 0 | if(rdw< 11.5), 1 else 0 | if(rdw> 14.5), 1 else 0 |
| ALT (IU/L) | if((men & alt≥ 17 & alt≤ 63) or (women & alt≥ 14 & alt≤ 54)), 1 else 0 | if((men & alt< 17) or (women & alt< 14)), 1 else 0 | if((men & alt> 63) or (women & alt> 54)), 1 else 0 |
| AST (IU/L) | if(ast≥ 15 & ast≤ 41), 1 else 0 | if(ast< 15), 1 else 0 | if(ast> 41), 1 else 0 |
| Potassium (mmol/L) | if(k ≥ 3.6 & k ≤ 5.1), 1 else 0 | if(k < 3.6), 1 else 0 | if(k > 5.1), 1 else 0 |
| Sodium (mmol/L) | if(na≥ 136 & na≤ 144), 1 else 0 | if(na< 136), 1 else 0 | if(na> 144), 1 else 0 |
| Creatinine (mg/dL) | if((men & cr≥ 0.6 & cr≤ 1.2) or (women & cr≥ 0.4 & cr≤ 1)), 1 else 0 | if((men & cr< 0.6) or (women & cr< 1.4)), 1 else 0 | if((men & cr> 1.2) or (women & cr> 1)), 1 else 0 |
| Albumin (g/dL) | if(albm≥ 3.5 & albm≤ 5), 1 else 0 | if(albm< 3.5), 1 else 0 | if(albm> 5), 1 else 0 |
| Bilirubin Total (mg/dL) | if(tbil≥ 0.3 & tbil≤ 1.2), 1 else 0 | if(tbil< 0.3), 1 else 0 | if(tbil> 1.2), 1 else 0 |
| Bilirubin Direct (mg/dL) | if(dbil≥ 0.1 & dbil≤ 0.5), 1 else 0 | if (dbil< 0.1), 1 else 0 | if(dbil> 0.5), 1 else 0 |
| Bilirubin Indirect (mg/dL) | if(ibil≥ 0.1 & ibil≤ 1), 1 else 0 | if(ibil< 0.1), 1 else 0 | if(ibil> 1), 1 else 0 |
| Globulin (g/dL) | if(glb≥ 2.9 & glb≤ 3.3), 1 else 0 | if(glb< 2.9), 1 else 0 | if(glb> 3.3), 1 else 0 |
| Total Protein (g/dL) | if(tp≥ 6.5 & tp≤ 8.1), 1 else 0 | if(tp< 6.5), 1 else 0 | if(tp> 8.1), 1 else 0 |
| Platelet Count (x109/L) | Severe Thrombocytopenia | Moderate Thrombocytopenia | Mild Thrombocytopenia |
| if(plt< 20), 1 else 0 | if(plt> 20 & plt≤ 50), 1 else 0 | if(plt> 50 & plt≤ 100), 1 else 0 | |
1 There are two types of admission (emergency & direct) and we have created two binary independent variables for this and the binary variables only reflect the presence of the label as ’1’ or ’0’ for the individual patients. For example, if the patient is admitted to an emergency then the value is 1, if not then 0. Similarly, for the channel, there are 4 channels (cash, corporate, PSU, TPA) and we have created 4 independent binary variables for this and converted values as 0/1 similar to what we have done for the type of admission, 2 TLC: total leucocyte count; ALT: alanine aminotransferase; AST: aspartate aminotransferase; RBC: red blood cell count; RDW: red cell distribution width; hct: Heamatocrit; hgb:Haemoglobin, lym:Lymphocytes; mono:Monocytes; neutro:Neutrophils; eos:Eosinophils; k:Potassium; na:Sodium; cr:Creatinine; alb:Albumin; tbil:Bilirubin Total; dbil:Bilirubin Direct; ibil:Bilirubin Indirect; glb:Globulin; tp:Total Protein; plt:Platelet Count
The target feature, LOS in the initial data set could take 28 different values. Figure 2 illustrates the distribution of length of stays in terms of count and percentage from February 2012 to September 2017. The most frequent LOS is 4 days (293, 25.5%) and the least LOS is 1 day (14, 1.2%). In this experiment, we binned the LOS values into two classes to build robust predictive models. Usually, patients with dengue infection have an average hospital stay between 3 and 5 days [14, 27–31]. We used > 5 days as a cut-off point for prolonged hospitalization (median LOS in the present study is 4.03 ± 2.44 (median ± IQR)). We divided LOS into two different functional groups: First, we merged LOS of 1 to 5 days into one bin labeled ’≤ 5 days’ and is coded as a 0 representing those patients for whom the moderate amount [9] of intervention is required to reduce LOS. Second, we pull all the length of stays longer than 5 days into the second bin labelled ’> 5 days’ and is coded as a 1. The patients in the second group are the most in need of early aggressive intervention [9] to prevent long - term hospital admissions.
Fig. 2.
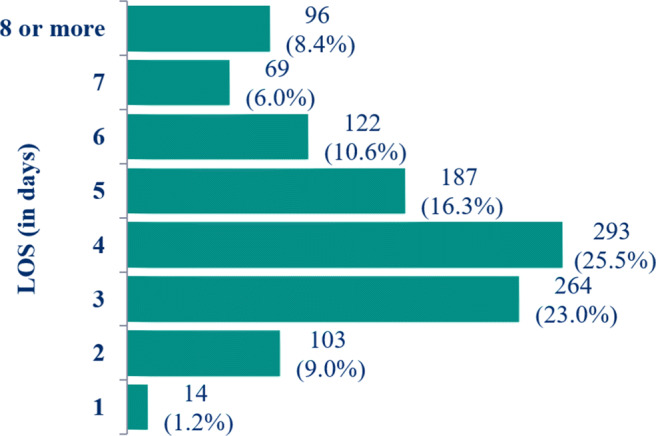
Distribution of the Length of Stay (in days) in Dengue data from February 2012 to September 2017
Case-inclusion criteria:
All patients (children, and adults) for whom there was a serologically confirmed dengue infection were included.
Case-exclusion criteria:
A case was excluded if dengue serology was negative for febrile illness patients.
Data pre-processing
As a step of data cleaning, we removed duplicate records and fields with more than 50% missing data.
Missing values handling
Secondary use of Electronic Health Record (EHR) data can be challenging, because the patient records within the EHR may be inconsistent and incomplete. The presence or absence of information, the timing, and other characteristics of the collected data may vary considerably from patient to patient. EHR data, especially for laboratory measurements, often contain missing values due to various reasons such as time and cost constraints [32]. While hospital systems are capable of capturing the entirety of data measurements, some patient data are still found missing from databases [33]. The rates of missing data in the EHR have previously been reported from 20% to 80% [34, 35]. In this study, extracted data have many laboratory measurements that are missing at any given hour during the first 24 hours of a patient’s hospital admission (Table 1) and in this case, data may be missing not at random because measurements are taken at different schedules and frequencies. These shortcomings make it harder for algorithms to capture patterns in medical data sets. One approach to handle incomplete data is to discard all cases consisting of missing values; however, this can potentially remove a significant portion of training data and is generally not desirable. Alternatively, a more common approach is to apply data imputation. We followed three steps to handle the missing values which are as follows,
Feature removal: If a feature has more than 50% of records with missing values, such as PT, APTT, and basophils then they were determined not to be an effective feature in the analysis and as a result, were removed (Table 1).
Median imputation: If a continuous feature has less than 50% missing values, then the median values of records were replaced instead of missing values (Table 1) because non-normality and some outliers were detected in these features.
Regression imputation: Regression imputation using R software was applied to those features that were in nominal or ordinal type (Table 1). We imputed the missing values of these features using the regression imputation model with the test AUC score shown in Fig. 3. The high values of AUC provide good confidence in the imputed values.
Fig. 3.
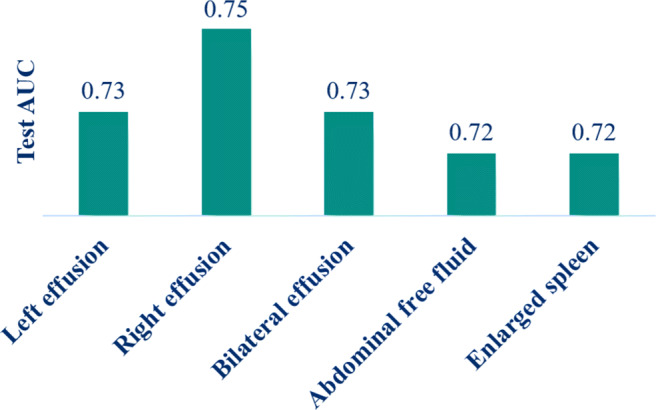
Features with 40% missing data values and their test AUC using regression imputation
Impact of missing data imputation
The statistical impact of missing data is evaluated and the details and statistical results of imputed variables are available in the Supplemental Material (see Tables 1, 2 and 3; Figs. 1 and 2 of the Supplemental Material).
Table 3.
Some important features of all 1148 patients in the study population comparing those LOS≤ 5 days with those LOS> 5 days
| Parameter | LOS ≤ 5 days | LOS > 5 days | Overall | p value* | |
|---|---|---|---|---|---|
| (n = 784) | (n = 364) | (n = 1148) | |||
| LOS | n (%) | 784 (68%) | 364 (32%) | 1148 | |
| Age | Median (IQR) | 27 (1-87) | 30 (1-85) | 28 (1-87) | 0.00000 |
| Male Gender | 474 (60.5%) | 218 (59.9%) | 692 (60.3%) | 0.90560 | |
| Admission Type | Emergency | 529 (67.5%) | 274 (75.3%) | 803 (69.9%) | 0.00897 |
| Direct | 255 (32.5%) | 90 (24.7%) | 345 (30.1%) | 0.00934 | |
| Platelet Count (x109/L) | Median (x109/L) | 100 (8-569) | 135 (5-676) | 105 (5-676) | 0.00095 |
| Thrombocytopenia n(%) | 405 (71.6%) | 161 (28.4%) | 566 (49.3%) | 0.02267 | |
| TLC (x109/L) | Mean (x109/L) | 5.5 (1.3-43.6) | 5.5 (0.9-45.4) | 5.5 (0.9-45.4) | 0.00095 |
| Low (%) | 175 (22.3%) | 84 (23.1%) | 259 (22.6%) | 0.03935 | |
| High (%) | 62 (7.9%) | 49 (13.5%) | 111 (9.7%) | 0.00430 | |
| Lymphocytes (%) | Mean | 34 (3.0-92.0) | 30 (2.5-78.8) | 34 (2.5-92.0) | 0.00000 |
| Low | 90 (11.5%) | 102 (28%) | 192 (16.7%) | 0.00000 | |
| High | 212 (27.0%) | 33 (9.1%) | 245 (21.3%) | 0.00000 | |
| Neutrophils (%) | Mean (%) | 55 (7.0-91.9) | 60 (14.8-95.8) | 55 (7.0-95.8) | 0.00000 |
| High | 585 (74.6%) | 326 (89.6%) | 911 (79.4%) | 0.00000 | |
| ALT (IU/L) | Mean (IU/L) | 62.5 (11-6054) | 62.5 (8-3650) | 62.5 (8-6054) | 0.00062 |
| Low | 14 (1.8%) | 16 (4.4%) | 30 (2.6%) | 0.01728 | |
| High | 403 (51.4%) | 159 (43.7%) | 562 (49%) | 0.01769 | |
| AST (IU/L) | Mean (IU/L) | 108 (13-18590) | 108 (14-12559) | 108 (13-18590) | 0.00052 |
| High | 728 (92.9%) | 313 (86%) | 1041 (90.7%) | 0.00030 | |
| Haematocrit (%) | Low | 41.1 ± 5.7 | 39.4 ± 6.1 | 40.6 ± 5.9 | 0.00000 |
| Haemoglobin (g/dL) | Low | 13.3 ± 1.9 | 12.6 ± 2.2 | 13.1 ± 2 | 0.00000 |
| Eosinophils (%) | Low | 1 (0.0-36.9) | 1 (0.0-18.9) | 1 (0.0-36.9) | 0.00003 |
| RBC Count (x1012/L) | Low | 4.67 (1.76-7.7) | 4.67 (2.48-8.6) | 4.67 (1.76-8.6) | 0.00002 |
| RDW (%) | High | 13.8 (10.8-45.5) | 13.8 (10.8-31.2) | 13.8 (10.8-45.5) | 0.00042 |
| Creatinine (mg/dL) | High | 0.7 (6.1-6.3) | 0.7 (0.1-7.6) | 0.7 (0.1-7.6) | 0.00029 |
| Albumin (g/dL) | High | 3.6 (2.0-4.8) | 3.6 (1.3-5.0) | 3.6 (1.3-5.0) | 0.00000 |
| Radiological Findings | Bilateral effusion | 59 (7.5%) | 40 (11%) | 99 (8.6%) | 0.06690 |
| Right effusion | 151 (19.3%) | 92 (25.3%) | 243 (21.2%) | 0.02485 | |
| Left effusion | 66 (8.4%) | 46 (12.6%) | 112 (9.8%) | 0.03277 | |
| Assisted Ventilation | 8 (1.0%) | 21 (5.8%) | 29 (2.5%) | 0.00000 | |
| Blood Transfusion | 130 (16.6%) | 145 (39.8%) | 275 (24%) | 0.00000 |
TLC: total leucocyte count; ALT: alanine aminotransferase; AST: aspartate aminotransferase; RBC: red blood cell count; RDW: red cell distribution width. Categorical variables are summarized as n (%), Continuous variables are presented as mean±SD or median (range) if SD> 50% of the mean. *Pearson chi-square/Fisher exact test applied
Feature engineering
Feature engineering is a key task in data preparation but a work-intensive component of machine learning applications [36]. Initially, we collected 40 predictors using the available data for 24 hours of hospitalization, which included age, admission type, several predictors related to the investigation data, blood transfusion, assisted ventilation, and radiological related predictors of dengue patients and then we generated new features from these predictors. For feature generation of lab data, we used the standard lab reference range provided by Max healthcare system and coded them based on whether it is below within or above the reference range (Table 2) and for other features, we categorized them into binary variables. After completion of the feature generation process, we prepared a list of 389 independent variables and one dependent variable including administrative, demographics, pathology, radiology, and procedure.
After pre-processing the data, the final dataset was randomly split into two subsets, a training set (70%) and a testing set (30%).
Feature selection
The selection of relevant attributes may also benefit from domain knowledge. Based on studies conducted in [15, 20, 37, 38], factors that often appear in dengue-cases have been selected as initial attributes then these attributes were validated by clinicians to ensure that no unrelated factors are used as predictors.
We use different techniques such as information value [39], variable importance using random forest [40], recursive feature elimination using logistic regression [39], chi-square [39], and L1 [39] feature selection methods to select variables and then finally each technique voted whether they selected the variable. As a final step, the vote was counted and the variables with higher votes were used in the modelling process. We removed sensitive patient information such as episode and location ID from the dataset.
Predictive models and algorithms
Logistic regression with elastic-net
We use LR with elastic-net [41] model that utilized EHR data to predict the LOS. It is a regularized regression technique that linearly combines the L1 and L2 penalties of lasso [42] and ridge methods. L1 regularization helps in sparsifying the weight vectors, while L2 regularization limits the weight value to protect against outliers. Together, elastic-net can find a stable and sparse weight vector for logistic regression [41]. The elastic-net estimator is defined as,
| 1 |
N is the number of observations
yi is the binary response at observation i
Xi is data, a vector of d values at observation i
λ1 and λ2 are positive regularization parameter which interpolates between L1 and L2 norm β
The parameter β is a coefficient of features
The LOS probability for Dengue hospitalization can be formulated as:
| 2 |
where Xi are independent variables and P is the probability of prolonged LOS (> 5 days) following dengue infection.
We did a grid search of 100 values for different values of λ1 and λ2 and selected the best with the lowest cross-validation error.
Random forest
Random forest [40] is an ensemble of multiple decision trees. In a decision tree each internal node (non-leaf node) denotes a test on an attribute, each branch represents an outcome of the test, and each leaf node (or terminal node) holds a class label [43]. The process of building a random forest involves constructing individual decision trees from bootstrap samples of the data using only a subset of predictors in each node of each tree.
In our experiments, the random forest model is tuned via 10-fold cross-validation over 10 combinations of hyperparameter values (number of decision trees, number of features). We set the default values for the algorithm e.g. 100 for the number of trees, Gini index for splitting and computing variable importance, five observations are set as the minimal number of observations required for forming terminal nodes, and the square root of the number of variables is used to split each node.
The algorithms were executed using R, an open-source software application for statistical computing and data mining [23]. Glmnet and caret libraries were used for LR with elastic-net and random forest algorithms respectively.
We also compared other methods like support vector machine (SVM) and extraTrees to predict LOS but these were excluded because of unsatisfactory predictive performance.
Evaluation measures
We used 70% of the data for training and the remaining 30% for testing. For fine-tuning of the model parameters, we used 10-fold cross-validation on the training dataset; the training data was first divided into ten folds, nine folds were used to train the model, and the remaining fold was used to assess the model performance/generalizability.
The Kruskal-Wallis H test is used to check the statistically significant differences between two or more groups of an independent variable on a continuous dependent variable [44]. For checking the association between two categorical varables, the Chi-square test [45] is used when the expected frequencies are higher than 5, whereas Fisher’s exact test [46] is performed when the expected table values are smaller than 5. Statistically significant differences are determined by p-value < 0.05 (Table 3). The results have been summarized in terms of means ± standard deviation and median ± IQR for continuous features.
Performance measures: We assess a set of performance measures including sensitivity or recall, precision or positive predictive value (PPV), and AUC for each model. We use traditional performance measures for classification that are based on the four values of the confusion table: true positive (TP), false positive (FP), true negatives (TN), and false negatives (FN). We use these values to compute a positive predictive value (PPV) or precision, negative predictive value (NPV), sensitivity or recall, and specificity as in Eq. 3, 4, 5, and 6.
| 3 |
| 4 |
| 5 |
| 6 |
In addition, the Receiver Operating Characteristic Curve (ROC) is graphed and the areas under the ROC (AUC) [43] are analyzed.
Results
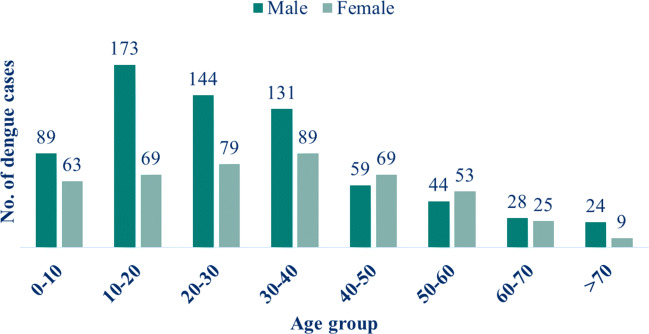
Of the 1148 dengue confirmed cases, 974 (84.8%) belonged to the adult’s age group (> 12 years) and 174 (15.2%) to the paediatric age group (≤ 12 years) in this study. Larger proportions of positive cases were observed among adult cases. The majority of the dengue cases were noted in the age group of 10-20 years (21.1%), where there was a male predominance. The next majority of cases were reported among 20-30 years (19.4%) followed by 30-40 years (19.2%). The least number of cases were seen in the age group of > 70 years (Fig. 4). The distribution of males and females across the different age groups was statistically the same (p > 0.05).
Fig. 4.
Age and gender-wise distribution of dengue confirmed cases
The mean age of 1,148 patients was 58.2 ± 13.0 years (SD) (aged > 0 to ≤ 87), the majority (60.3%) males (Table 3). The mean LOS was 4.03 ± 2.44 (median ± IQR). Prolonged hospitalization (> 5 days) was seen in 32% (364/1148) of patients while LOS was ≤ 5 days among 68% (784/1148) of patients. The characteristics of patients with or without prolonged LOS were compared and shown in Table 3.
Laboratory and radiological investigations
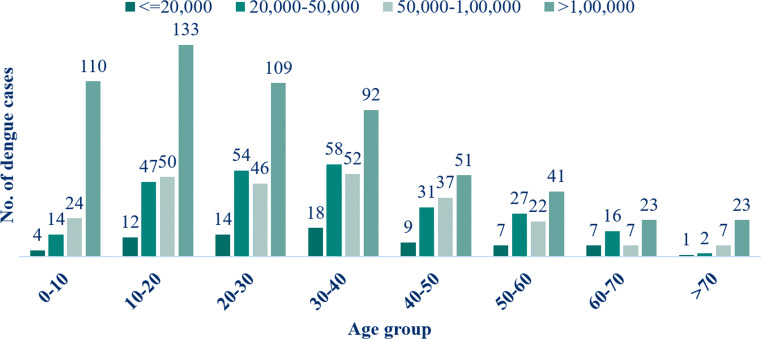
Considering > 5 days as ”prolonged LOS”, serum creatinine, platelet count, total leucocyte count, alanine aminotransferase (ALT), aspartate aminotransferase (AST), haematocrit (%), haemoglobin (g/dL), assisted ventilation, blood transfusion, and admission type emergency were identified as highly statistical significant independent predictors of LOS (p < 0.05) (Table 3). Platelet count was done in all the confirmed cases out of which 72 patients (6.3%) had platelet count ≤ 20,000 (severe thrombocytopenia), 249 patients (21.7%) ranged from 20,000 – 50,000 (moderate thrombocytopenia), 245 patients (21.3%) ranged from 50,000 – 1,00,000 (mild thrombocytopenia) while the remaining 582 patients (50.7%) were above 1,00,000 (Fig. 5). Of these cases, 49.3% had thrombocytopenia (< 1,00,000) while the remaining 50.7% had normal platelet counts (Table 3). A significant association was observed between the thrombocytopenia and the age groups. Thrombocytopenia was found to be more severe in age groups of 30-40 years than in the older age group and this difference was significant (p < 0.05).
Fig. 5.
Platelet counts and age-wise distribution of dengue cases
Important factors
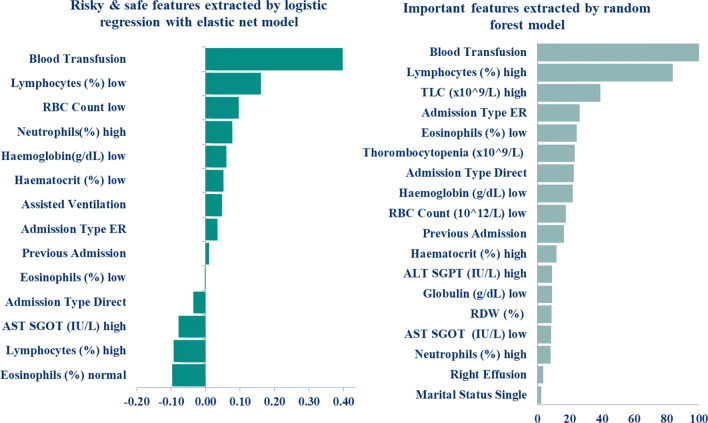
The relative importance of each variable in the model evaluation is linked to the importance of each feature in making a prediction and it does not relate to model accuracy [43]. Based on the model performance, we have extracted the topmost risky and protective features for a longer LOS. We have called features associated with a longer LOS (positive correlation) as risky features, whereas safe features are those which demonstrate an inverse relationship with longer LOS. Top features based on LR with elastic-net are reported in Fig. 6. The most significant factors for a longer stay are lymphocytes, total leucocyte count, alanine aminotransferase (ALT), aspartate aminotransferase (AST), red blood cell count (RBC), red cell distribution width (RDW), haematocrit, neutrophils as well as platelet count. Admission type emergency, blood transfusion, marital status being single and, right effusion were also significant in predicting prolonged LOS. On the other hand, eosinophils and AST and high lymphocyte are the safe features which contribute to a shorter LOS.
Fig. 6.
Top risky and safe features extracted by logistic regression with elastic-net model. Positive and negative values represent the risky and safe features respectively and top 18 important variables extracted by the random forest model. A higher relative weight value represents the higher importance of the features
The random forest model, with earlier parameter setting, was used to extract important factors in Fig. 6, features with a great impact on LOS are listed in order of variable importance. The random forest also identified some top features such as lymphocytes, eosinophils, haemoglobin, and marital status which agree with elastic-net.
From both the methods, the most significant factors were blood transfusion, admission type emergency, assisted ventilation, and thorombocytopenia. haemoglobin low, TLC high are also strong predictors of prolonged LOS of dengue patients. haematocrit (low and high) played a notable role as well since analysis revealed that patients < 40% and > 50% haematocrit value for men and patients < 36% and > 46% haematocrit value for women statistically had increased mean LOS. A low (critical value< 15-20%) haematocrit may cause cardiac failure or death [47–49] and a high (critical value> 60%) may cause spontaneous blood clotting [47–49]. The low value of lymphocytes signifies that the patients are more likely to have prolonged LOS. Furthermore, the previous admission also increases risk for a prolonged LOS.
Thus, the most remarkable features influencing LOS for dengue patients obtained by algorithms are blood transfusion, admission type emergency, assisted ventilation, haemoglobin low, TLC high, haematocrit low and high, lymphocytes low and, previous admission.
Table 4 shows the performance measures of the LR with elastic-net and random forest classifiers. The LR with elastic-net model achieved an AUC of 0.75, whereas the random forest model exhibited an AUC of 0.72. ROC curve analysis with area under the curve (AUC) of 0.75 demonstrated that LR with elastic-net model has good ability to predict prolonged hospitalisation among patients with dengue (Fig. 7). Confusion matrices for both the models are available in Fig. 3 of the Supplemental Material.
Table 4.
Comparison of the performance measures for the predictive models on test data
| Model | AUC | Sens | Spec | PPV | NPV |
|---|---|---|---|---|---|
| LR with elastic-net | 0.75 | 0.24 | 0.97 | 0.82 | 0.72 |
| Random forest | 0.72 | 0.31 | 0.91 | 0.64 | 0.73 |
This performance result for each model on 345 records which is 30% of the data (n = 1148)
Fig. 7.
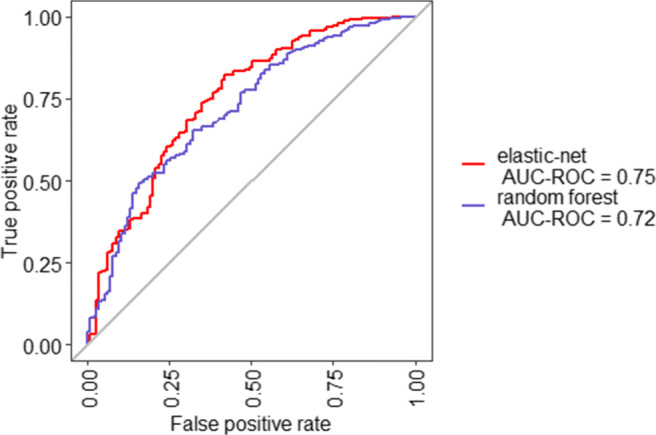
Receiver-operating characteristics curve analysis of both the models on test data predicting prolonged hospitalisation among dengue patients
Discussion
This research investigated the determinants of hospital LOS in the patient’s representative of confirmed dengue diagnosis within our group of healthcare centers. Previous studies have predicted in-hospital mortality [19] of dengue patients. Our findings indicated that there is a significant association between LOS greater than 5 days and amount of lymphocytes, leucocyte, alanine aminotransferase (ALT), aspartate aminotransferase (AST), red blood cell count (RBC), red cell distribution width (RDW), haematocrit, neutrophils as well as platelet count. Admission type being emergency, blood transfusion, marital status being single, and right effusion were also significant in predicting prolonged LOS.
In general, we found that LR with elastic-net model trained on data from Max group healthcare systems, was able to predict the prolonged LOS better than random forest. We plan to use this predictive model as a screening tool to proactively identify high-risk patients to receive a care coordination intervention to reduce prolonged LOS of Dengue patients. This is in direct contrast to the department care, which is reactive, requiring patient’s symptoms to be present, to receive care by clinicians and care coordinators. We believe that transforming care coordination from reactive to a proactive activity carries great potential to reduce LOS. As enrolment in the hospital is still in progress, it remains to be seen whether this model can be translated into the real world. If the model is successful, there are potential implications for LOS and cost reduction. Long stay and care coordination activities are often expensive and aligning care coordination resources with patients responsible for large costs to the health system may optimize resource allocation.
We found that both models have good overall performance. The alignment of important variables between these two models provides more confidence in the prediction. Implementing any of these models can enable efficient management of hospital resources and plan for preventive interventions for patients with intense conditions. As a result, this study provides better insight into the underlying factors that influence the LOS.
The operating point selection in the ROC curve is for operational reason. Our aim is to predict dengue patients with a risk of higher length of stay, so as to direct limited hospital resources to the high risk group. Therefore, we have chosen to trade-off low sensitivity for higher specificity. More specifically, to reduce the Type I error where the false positives are minimized, and we should be able to identify few patients with higher specificity for an early intervention and optimized resource allocation.
As we know, healthcare data is generally not fully structured, it is distributed across various locations. We are aware that our current study has several limitations, which could be addressed in subsequent works. First, while our model is likely generalizable to Max group healthcare systems from which the data is collected, however, obtained information is not clinically exhaustive, as present work has fully relied on the demographic, administrative, investigation, and radiological characteristic data retrieved from hospital electronic databases. It may not generalize other parts of the Indian healthcare system with different demographics, practice patterns. The model may need to be developed for each community, using the process we describe. Second, we did not collect data from outside of our healthcare environment, where we may miss earlier predictors of prolonged LOS in our model.
Conclusion
The study indicates that routinely collected hospital data can be used to identify the prolonged LOS of dengue patients and may also provide insight into the factors influencing hospital LOS of dengue patients that can easily be interpreted by the clinician. Our model results show that LR with elastic-net and a random forest model can predict dengue patient’s LOS, but still, LR with elastic-net is the best fit with an AUC of 0.75. We intend to implement the derived model in our information systems for real-time feedback to the clinician to reduce the long LOS of dengue confirmed patients during the admission. This could potentially help clinicians in planning for preventive interventions, thereby leading to improvement in health services and to manage the hospital resources more efficiently. Also, we intend to conduct a follow-up study to measure and potentially improve the predictive performance of the model, after system implementation is rolled out.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.W.H. Organization. Dengue. http://www.searo.who.int/india/topics/dengue/en/
- 2.N.V.B.D.C. Programme. Dengue/dhf situation in india. http://nvbdcp.gov.in/index4.php?lang=1&level=0&linkid=431&lid=3715
- 3.C.B. of Health Intelligence. National health profile 2018. http://www.cbhidghs.nic.in/Ebook/National%20Health%20Profile-2018%20(e-Book)/files/assets/common/downloads/files/NHP%202018.pdf
- 4.Chakravarti A, Matlani M, Kashyap B, Kumar A, et al. Ind J Med Microbiol. 2012;30(2):222. doi: 10.4103/0255-0857.96699. [DOI] [PubMed] [Google Scholar]
- 5.Organization WH Dengue and severe dengue. fact sheet; 2018. https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue/
- 6.Ruangkriengsin D, Phisalprapa P. J Med Assoc Thai. 2014;97(Suppl 3):S206. [PubMed] [Google Scholar]
- 7.Hariharan D, Das MK, Shepard DS, Arora NK. Int J Infect Dis. 2019;84:S68. doi: 10.1016/j.ijid.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Gómez V, Abásolo JE. Paradigma. 2009;3(1):1. [Google Scholar]
- 9.Azari A, Janeja VP, Mohseni A. Int J Knowl Discov Bioinf (IJKDB) 2012;3(3):44. doi: 10.4018/jkdb.2012070103. [DOI] [Google Scholar]
- 10.Blais MA, Matthews J, Lipkis-Orlando R, Lechner E, Jacobo M, Lincoln R, Gulliver C, Herman JB, Goodman AF. Administr Policy Mental Health Mental Health Serv Res. 2003;31(1):15. doi: 10.1023/A:1026044106172. [DOI] [PubMed] [Google Scholar]
- 11.Combes C, Kadri F, Chaabane S (2014) In: 10èMe conférence francophone de modélisation, optimisation et simulation-MOSIM’14
- 12.Hachesu PR, Ahmadi M, Alizadeh S, Sadoughi F. Healthcare Inf Res. 2013;19(2):121. doi: 10.4258/hir.2013.19.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lella L, Di Giorgio A, Dragoni AF (2015) In: AI-AM/NetMed@ AIME, pp 11–21
- 14.Chakravarty A, Krishnan A (2018) IJTDH
- 15.Mallhi TH, Khan AH, Sarriff A, Adnan AS, Khan YH (2017) BMJ Open 7(7):e016805 [DOI] [PMC free article] [PubMed]
- 16.Wiratmadja II, Salamah SY, Govindaraju R (2018) J Eng Technol Sci 50(1)
- 17.Acharya V, Khan MF, Kosuru S, Mallya SD. Int J Res Med Sci. 2018;6(5):1605. doi: 10.18203/2320-6012.ijrms20181495. [DOI] [Google Scholar]
- 18.Md-Sani SS, Md-Noor J, Han WH, Gan SP, Rani NS, Tan HL, Rathakrishnan K, A-Shariffuddin MA, Abd-Rahman M. BMC Infect Diseases. 2018;18(1):1. doi: 10.1186/s12879-018-3141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain S, Mittal A, Sharma SK, Upadhyay AD, Pandey RM, Sinha S, Soneja M, Biswas A, Jadon RS, Kakade MB et al (2017) In: Open forum infectious diseases, vol 4. Oxford University Press US, pp ofx056 [DOI] [PMC free article] [PubMed]
- 20.Liu P, El-Darzi E, Vasilakis C, Chountas P, Huang W, Lei L (2004) In: pacific asia conference on information systems 2004
- 21.Bouckaert RR. Artif Intell Tools. 2008;11(3):369. [Google Scholar]
- 22.Saravanan N, Siddabattuni VK, Ramachandran K. Expert Syst Appl. 2008;35(3):1351. doi: 10.1016/j.eswa.2007.08.026. [DOI] [Google Scholar]
- 23.Cohen WW (1995) In: Machine learning proceedings 1995. Elsevier, pp 115–123
- 24.Salzberg SL (1994) C4. 5: Programs for machine learning by j. ross quinlan. Morgan Kaufmann Publishers, Inc. 1993
- 25.Breiman L. Mach Learn. 1996;24(2):123. [Google Scholar]
- 26.W.H. Organization, S.P. for Research, T. in Tropical Diseases, W.H.O.D. of Control of Neglected Tropical Diseases, W.H.O. Epidemic, P. Alert, Dengue: guidelines for diagnosis, treatment prevention and control World Health Organization (2009)
- 27.Agarwal R, Kapoor S, Nagar R, Misra A, Tandon R, Mathur A, Misra A, Srivastava K, Chaturvedi U. South Asian J Trop Med Public Health. 1999;30(4):735. [PubMed] [Google Scholar]
- 28.Goh K, Ng S, Chan Y, Lim S, Chua E. South Asian J Trop Med Public Health. 1987;18(3):295. [PubMed] [Google Scholar]
- 29.Khalil MAM, Tan J, Khalil MAU, Awan S, Rangasami M. BMC Res Notes. 2014;7(1):1. doi: 10.1186/1756-0500-7-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lye D, Chan M, Lee V, Leo Y. Singapore Med J. 2008;49(6):476. [PubMed] [Google Scholar]
- 31.Shahin W, Aly M. Afro-Egyptian J Infect Endem Diseases. 2013;3(4):127. doi: 10.21608/aeji.2013.18255. [DOI] [Google Scholar]
- 32.Chen D, Liu S, Kingsbury P, Sohn S, Storlie CB, Habermann EB, Naessens JM, Larson DW, Liu H. NPJ Digit Med. 2019;2(1):1. doi: 10.1038/s41746-019-0122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adibuzzaman M, DeLaurentis P, Hill J, Benneyworth BD (2017) In: AMIA Annual Symposium Proceedings, vol 2017. American Medical Informatics Association, pp 384 [PMC free article] [PubMed]
- 34.Chan KS, Fowles JB, Weiner JP. Med Care Res Rev. 2010;67(5):503. doi: 10.1177/1077558709359007. [DOI] [PubMed] [Google Scholar]
- 35.Kharrazi H, Wang C, Scharfstein D (2014) Prospective ehr-based clinical trials: the challenge of missing data [DOI] [PMC free article] [PubMed]
- 36.Bengio Y, Courville A, Vincent P. IEEE Trans Pattern Anal Mach Intell. 2013;35(8):1798. doi: 10.1109/TPAMI.2013.50. [DOI] [PubMed] [Google Scholar]
- 37.Pongpan S, Wisitwong A, Tawichasri C, Patumanond J. Int J Clin Pediatr. 2013;2(1):12. doi: 10.1155/2013/845876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suwarto S, Ulhaq S, Widjaja B. Univ Med. 2017;36(1):19. [Google Scholar]
- 39.Li J, Cheng K, Wang S, Morstatter F, Trevino RP, Tang J, Liu H. ACM Comput Surv (CSUR) 2017;50(6):1. doi: 10.1145/3136625. [DOI] [Google Scholar]
- 40.Breiman L. Mach Learn. 2001;45(1):5. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 41.Zou H, Hastie T. J R Stat Soc Ser B (Stat Methodol) 2005;67(2):301. doi: 10.1111/j.1467-9868.2005.00503.x. [DOI] [Google Scholar]
- 42.Tibshirani R. J R Stat Soc Ser B (Stat Methodol) 1996;58(1):267. [Google Scholar]
- 43.Han J, Pei J, Kamber M (2011) Data Mining: Concepts and Techniques. The Morgan Kaufmann Series in Data Management Systems. Elsevier Science. https://books.google.co.in/books?id=pQws07tdpjoC
- 44.Kruskal WH, Wallis WA. J Amer Stat Assoc. 1952;47(260):583. doi: 10.1080/01621459.1952.10483441. [DOI] [Google Scholar]
- 45.Cochran WG (1952) Ann Math Stat:315–345
- 46.Fisher RA. J R Stat Soc. 1922;85(1):87. doi: 10.2307/2340521. [DOI] [Google Scholar]
- 47.Boissonnault WG (2011) Elsevier Saunders, St Louis
- 48.DeVita VT, Lawrence TS et al (2008) Wolters Kluwer, pp 1897–1898
- 49.Fischbach FT, Dunning MB (2009) A manual of laboratory and diagnostic tests. Lippincott Williams & Wilkins
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.