Abstract
The COVID-19 pandemic brings unprecedented crisis for public health and economics in the world. Detecting specific antibodies to SARS-CoV-2 is a powerful supplement for the diagnosis of COVID-19 and is important for epidemiological studies and vaccine validations. Herein, a rapid and quantitative detection method of anti-SARS-CoV-2 IgG antibody was built based on the optofluidic point-of-care testing fluorescence biosensor. Without complicated steps needed, the portable system is suitable for on-site sensitive determination of anti-SARS-CoV-2 IgG antibody in serum. Under the optimal conditions, the whole detection procedure is about 25 min with a detection limit of 12.5 ng/mL that can well meet the diagnostic requirements. The method was not obviously affected by IgM and serum matrix and demonstrated to have good stability and reliability in real sample analysis. Compared to ELISA test results, the proposed method exhibits several advantages including wider measurement range and easier operation. The method provides a universal platform for rapid and quantitative analysis of other related biomarkers, which is of significance for the prevention and control of COVID-19 pandemic.
Keywords: COVID-19, Evanescent wave fluorescence, Point-of-care testing, Antibody detection
Graphical abstract
1. Introduction
As an astounding pandemic outbreak in the 21st century, the coronavirus disease 2019 (COVID-19) has caused unprecedented public health crisis and social burden all over the world [[1], [2], [3]]. Rapid and accurate diagnostic solutions are particularly important for the prevention and control of the overall epidemic situation [4,5]. At present, the two main forces of diagnostic tests for the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are nucleic acid detection and specific antibody detection [6,7]. Nucleic acid detection plays an irreplaceable role in the diagnosis of COVID-19, and the real-time reverse transcriptase polymerase chain reaction (RT-PCR) assay is regarded as the gold standard. The RT-PCR testing is an effective way for the identification of positive persons in the early stages and the large-scale population screening. However, PCR testing has several limitations, including expensive instrument, high false-negative rates, cumbersome operation, and technical personnel [6,8]. Besides, nucleic acid detection is difficult to be applied for the identification of disease stages, past infection, and immunity situation.
Serological testing of the SARS-CoV-2 specific antibodies is a powerful supplement for the diagnosis of COVID-19 [[9], [10], [11]]. After SARS-CoV-2 infection, immunoglobulin M (IgM) and immunoglobulin G (IgG) is detectable at 3–6 days and 7–14 days, respectively. Therefore, antibody detection is not only useful for finding out the past infection and screening asymptomatic patients, but also for determining different infection stages [12,13]. Compared with nucleic acid detection, antibody detection has higher sensitivity and specificity, shorter turnaround time, lower cost and technical requirement, lower risk of medical staff infection, and is more suitable for the on-site detection. Studies demonstrated that supplementing antibody detection could effectively improve accuracy of the diagnosis [14]. Currently, the serological testing methods of COVID-19 include lateral flow immunoassays (LFIA), chemiluminescence immunoassay (CLIA), and enzyme-linked immunosorbent assay (ELISA) [4,15,16]. LIFA is a cost-effective, fast and simple analytical technique that is quite suitable for pointing-of-care testing (POCT) diagnostics [17,18]. To improve the sensitivity of conventional LIFA, various nanomaterials have been employed as labels for COVID-19 diagnosis [19,20]. Both CLIA and ELISA can be used for qualitative and quantitative serological testing. However, they have the disadvantages of time-consuming and complex detection operation. Thus, it is an urgent need to develop a simple, rapid, and accurate on-site detection method for the serological testing of SARS-CoV-2 specific antibodies.
Evanescent wave fiber optic biosensors have become powerful analytical tools in medical diagnosis, environmental monitoring, and food safety because of their high sensitivity and specificity, easy-to-operation, and potential for point-of-care testing [[21], [22], [23]]. Recently, an optofluidic pointing-of-care testing platform (OPOCT) was developed and the potential in medical testing has been demonstrated from the rapid detection of cholylglycine (CG) in serum [24]. However, the employed indirect competitive immunoassay mechanism is not suitable for the detection of large molecules such as antibodies. Herein, based on sandwich immunoassay principle, the rapid and sensitive on-site detection method of anti-SARS-CoV-2 IgG antibody in serum was built using the OPOCT. The receptor binding domain (RBD) of spike protein (S-protein) of SARS-CoV-2 was immobilized on the fiber biosensor to regarded as the capture probe, which had been widely applied in serological detection of COVID-19 [12,25]. When the sample containing anti-SARS-CoV-2 IgG antibody was introduced over the fiber biosensor, some of them specially bound with the spike protein. The fluorescence-labeled secondary antibody of a certain concentration was added as the fluorescence signal reporter. The higher concentration of anti-SARS-CoV-2 IgG antibody allowed more of them and more fluorescence-labeled secondary antibody bound to the fiber biosensor surface, thus resulting in the higher fluorescence intensity. According to the linear relationship between anti-SARS-CoV-2 IgG antibody concentration and fluorescence intensity, the anti-SARS-CoV-2 IgG antibody in serum can be quantified in about 25 min with simple dilution.
2. Materials and methods
2.1. Materials
The recombinant RBD, anti-SARS-CoV-2 S-protein IgG calibrator, anti-SARS-CoV-2 S-protein S IgM calibrator, Alexa Fluor 680 labeled goat anti-human IgG secondary antibody, MERS-CoV-2 antibody, HCoV-HKU1 CoV antibody were purchased from Beijing Biodragon Immunotechnologies CO., Ltd. (Beijing, China). The IgG consists of three human monoclonal antibodies that can specifically recognize SARS‐CoV‐2 spike protein, and the binding affinities (Kd) are 0.06 nM, 0.1 nM, 1.2 nM, respectively. Cardiac troponin I (cTnI) was purchased from Kitgen Biotechnology Co., Ltd (Zhejiang, China). Programmed cell death 1 ligand 1 (PD-L1) was purchased from Cusabio Technology Co., Ltd (Wuhan, China). Bovine serum albumin (BSA), 3-mercaptopropyl-trimethoxysilane (MTS), and N-(4-maleimidobutyryloxy) succinimide (GMBS) were purchased from Sigma-Aldrich (Steinheim, Germany). All other reagents, unless specified, were of analytical grade and supplied by the Beijing Chemical Agents (Beijing, China). The phosphate-buffered saline solution (10 mM PBS, containing 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4, pH = 7.4) and antibody dilution buffer (0.1 % BSA in PBS) were prepared, respectively. The regeneration solution was 0.5 % sodium dodecyl sulfate (SDS, pH = 1.9).
2.2. Preparation of fiber biosensor for detection of S-IgG
To detect the S-IgG antibody, the functional fiber biosensor was prepared according to previous methods [26]. In brief, fiber optic with diameter of 600 μm and length of 5.5 cm (NA = 0.22) was used to prepare biosensor. To improve the excitation efficiency of evanescent wave and collection efficiency of fluorescence, the distal end of fiber optic was tapered through by HF acid tube-etching method to form a taper sensing region with length and diameter of 3.0 cm and 220 μm. The sensing region was initially immersed in piranha solution (H2SO4/H2O2 = 3:1) to activate the hydroxyl groups for 30 min. Then, the thiol groups were introduced onto the fiber biosensor surface by placed it in 2 % MTS solution (v/v in ethanol) for 2 h. After washed three times with ethanol and dried in N2, the fiber biosensor was immersed in GMBS solution for 1 h. Next, the fiber biosensor was placed in RBD solution (0.5 mg/mL) overnight in refrigerator (4 °C). During this process, the RBD was covalently linked to the biosensor surface. Finally, the fiber biosensor was placed in BSA solution (2 mg/mL) for 2 h to blocked up nonspecific adsorption sites. The as-prepared fiber biosensor was placed at 4 °C for use.
2.3. Instrument: OPOCT
A detailed description of the OPOCT can be found in our previous publications [26]. The platform is composed of four parts including optical module, fluidics module, electronic control module and data treatment module (Fig.S1). The system possesses some prominent characteristics including low cost, compact structure, high reliability and portability.
2.4. Immunosensing mechanism of S-IgG
Using OPOCT platform, the immunosensing mechanism for S-IgG based on sandwich assay was illustrated in Fig. 1 a. The RBD functionalized fiber biosensor was placed in optofluidic cell. When the excitation light is introduced into the fiber biosensor, it transmits via total internal reflection (TIR) and the evanescent wave generates on the fiber biosensor surface, which has a limited penetrated depth (typically < 100 nm). First, the PBS solution was pumped over the biosensor surface, and the baseline signal (I b) was recorded by the OPOCT (phase I in Fig. 1b). Second, samples containing various concentrations S-IgG were introduced into the optofluidic cell and incubated for a certain time (phase II in Fig. 1b). During this period, S-IgG specifically bound with RBD immobilized onto fiber optic surface. Third, the fiber biosensor was rinsed with PBS again to remove the excess S-IgG (phase III in Fig. 1b). Fourth, the fluorescently-labeled secondary antibodies were delivered into the cell and incubated for several minutes (phase IV in Fig. 1b). During this process, secondary antibodies bound with S-IgG on the biosensing surface, and the fluorescence was excited by evanescent wave generated on the fiber biosensor surface. Part of the fluorescence coupled back into the fiber biosensor and was real-time detected by the OPOCT. With increasing the S-IgG concentration, the more fluorescence labeled secondary antibodies bound on the fiber biosensor surface and the higher fluorescence intensity (I f) was detected. The net fluorescence signal (I s) of sample was calculated according to Eq. (1)
| Is = If - Ib | (1) |
Fig. 1.
(A) Immunosensing mechanism of S-IgG detection using OPOCT platform; (b) Typical fluorescence signal trace for S-IgG detection.
The proportional relationship between I s and the S-IgG concentration in the samples was used for determination of dose-response curve. Finally, the fiber biosensor was regenerated with 0.5 % SDS solution (pH = 1.9) to remove the S-IgG and secondary antibodies. The signal trace returned to the baseline after washed with PBS solution, and the fiber biosensor could be used for next detection.
2.5. Detection of IgG using OPOCT biosensor
Under optimized conditions, a whole process of IgG detection was carried out as follows. First, the RBD modified biosensor was embedded into sample cell as biorecognition element. Then, sample (100 μL) containing various concentrations of IgG was introduced into sample cell and incubated for 7 min. During this period, IgG binds to the optical fiber surface through specific antigen-antibody reaction. After were rinsed to remove excess IgG with PBS buffer, 100 μL fluorescent labeled secondary antibody was introduced and reacted for 7 min. In this stage, secondary antibody bound with IgG and the fluorescence intensity was real time recorded. Finally, the biosensor surface was regenerated by SDS solution (0.5 %, pH = 1.9) for 5 min. After washing with PBS buffer, the biosensor can be reused for the next test. The typical signal traces were demonstrated in Fig. 1b, and the net fluorescence signal was calculated by subtracting baseline value. The higher concentration of IgG in the sample resulted in higher fluorescence signal. Every concentration of IgG was measured three times. To obtain dose-response relationship, a four-parameter logistic equation was employed to fit the signal plotted against the concentration of IgG.
3. Results and discussion
3.1. Feasibility of S-IgG detection using the OPOCT
To evaluate the feasibility of IgG detection using the OPOCT biosensor, two control experiments were carried out. As can be seen from Fig. 2 a, when 125 ng/mL S-IgG and fluorescence labeled secondary antibody was sequentially introduced into the biosensor, the fluorescence intensity was by far higher than that of only fluorescence labeled secondary antibody. The measured fluorescence intensities were displayed as Fig.S2a. To further verify these results, the fiber biosensors were also characterized by the confocal microscope (Nikon A1, Japan, 638 nm) (Fig. 2b). Results showed that no fluorescence was observed for the original biosensor modified with RBD (Left in Fig. 2b). When only secondary antibody was introduced over the fiber biosensor surface, a little fluorescence was observed due to non-specific adsorption (Middle in Fig. 2b). With 125 ng/mL IgG, a clear fluorescence image with high brightness was observed after the S-IgG and the fluorescence-labeled secondary antibody were sequentially incubated with biosensor (Right in Fig. 2b). The intensities of the confocal microscope images were consistent with the results of OPOCT sensor (Fig.S2b). All of these results confirmed that the S-IgG was successfully immobilized on the fiber biosensor surface, and the fluorescence signal detected by the OPOCT originated from the specific binding between RBD and S-IgG. Based on the sandwich immunoassay mechanism, the S-IgG could be quantitatively detected by the OPOCT.
Fig. 2.
(A)Typical curves for baseline, secondary antibody, 125 ng/mL IgG and secondary antibody using the proposed biosensor (b)Laser confocal microscope observation of original biosensor, secondary antibody, 125 ng/mL IgG and secondary antibody.
3.2. Optimization of detection conditions
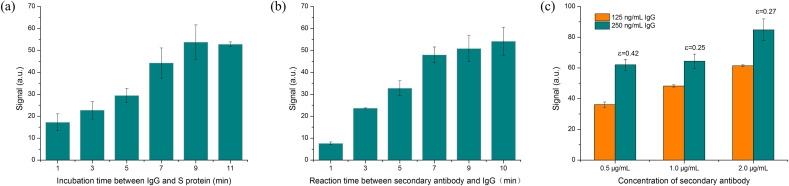
To improve the detection performance of S-IgG, several detection conditions, including the incubation time between S-IgG antibodies and RBD, the reaction time between secondary antibodies and S-IgG, and the secondary antibodies concentration, were optimized. First, the effect of the incubation time between S-IgG antibodies and RBD was investigated. As shown in Fig. 3 a, the fluorescence intensity increased over the incubation time when the incubation time was less than 9 min. This should contribute to the more S-IgG bound with RBD immobilized on the fiber biosensor, thus allowing the more fluorescence labeled secondary antibody to bind with S-IgG and the more fluorescence molecules to be excited. However, the longer incubation time did not further increase the fluorescence intensity. Next, the reaction time between secondary antibodies and S-IgG was optimized. Fig. 3b shown that the fluorescence intensity rapidly increased and gradually reached a plateau. To consider the fluorescence intensity and detection time, the incubation time of both secondary antibodies and S-IgG was chosen as 7 min.
Fig. 3.
(A) Incubation time between IgG and RBD (IgG concentration was 125 ng/mL, secondary antibody concentration was 0.5 μg/mL); (b)Optimization of reaction time between second antibodies and IgG (IgG concentration was 125 ng/mL, secondary antibody concentration was 0.5 μg/mL); (c) Optimization of secondary antibody concentration (The concentration of IgG was 125 and 250 ng/mL, both the incubation time and reaction time were 7 min).
Finally, the optimization of secondary antibody concentration was performed, and 0.5, 1.0, and 2.0 μg/mL secondary antibody were tested (Fig. 3b). Results showed that higher concentration of secondary antibodies indeed improved the fluorescence signal of S-IgG detection, whose concentrations were 125 and 250 ng/mL. A sensitivity index (ε) was introduced to determine the optimal concentration. The sensitivity index was defined as ε=(S2–S1)/S2, where S1 and S2 are the fluorescence signal of 125 ng/mL and 250 ng/mL IgG, respectively. High value of ε is conducive to sensitive determination of analyte and is favorable to achieve 0.30. For these three concentrations, the sensitivity indexes were evaluated to be 0.42, 0.25 and 0.27 when the secondary antibody concentrations were 0.5, 1, and 2 μg/mL, respectively. To select the most suitable concentration, the ε should be as high as possible to realize a better sensitivity. Therefore, 0.5 μg/mL AF680-labeled secondary antibody were chosen as the optimal concentration for subsequent IgG measurements. In addition, the lower concentration secondary antibody can also reduce the reagent consumption thus reducing the testing cost.
3.3. Determination of IgG using the OPOCT biosensor
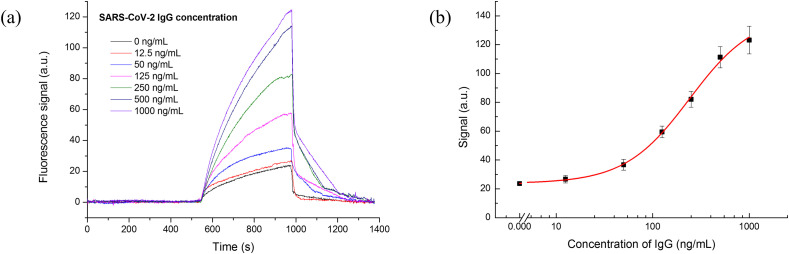
The quantification detection of S-IgG was carried out under optimal conditions using the OPOCT. Fig. 4 a showed the typical fluorescence intensity curves for different concentrations of S-IgG. With increasing concentrations of S-IgG, the fluorescence intensity increased faster and the higher fluorescence signal was obtained, which contributed to the more fluorescence labeled secondary antibody bound to the fiber biosensor surface based on sandwich immunoreaction principle. To reuse the fiber biosensor, the SDS solution (0.5 %, pH = 1.9) was pumped into the cell, both S-IgG and fluorescence labeled secondary antibody bound onto the biosensor surface could be simultaneously removed. The signal traces returned to the baseline after washed by the PBS. and the whole detection process was less than 25 min. The dose-response curve of S-IgG was plotted against the logarithm of S-IgG concentration using a four-parameter logistic equation (Fig. 4b). The error bars corresponded to the standard deviation of the data points in triple experiments. The linear response of IgG ranges from 82.89 to 702.91 ng/mL. The limit of detection (LOD) was 12.5 ng/mL using three times standard deviation of the mean blank values. Several studies showed that the IgG concentration ranged between 1.4 and 4200 μg/mL in COVID-19 patients' serum, and the neutralization antibodies concentration should be higher than 1.0 μg/mL to ensure the effective protection [[27], [28], [29], [30]]. Therefore, the sensitivity of the OPOCT is adequate for detecting S-IgG in COVID-19 patients’ serum, and was also comparable to those of other immunoassays [12,20,27,[31], [32], [33], [34], [35]] (Table S1). Compared with the reported works, our method can achieve quantitative detection of IgG with high sensitivity (LOD = 12.5 ng/mL) and relatively short assay time (about 25 min). Moreover, the device is portable and cheaper, and the reusability of fiber biosensor can effectively reduce the cost of testing. Our method is expected to provide a powerful tool for the existing COVID-19 serological assays.
Fig. 4.
(A) Typical signal curves of IgG detection using OPOCT system; (B) dose-response curve of IgG detection (0.5 μg/mL anti-IgG antibody, incubation and reaction time were both 7 min).
3.4. Effects of IgM, other proteins, and serum on IgG detection
IgM, a major active component in the human serum after infection with COVID-19, is considered as another effective analyte for serological diagnosis. Although IgM can be positive from the 3–6 days, high levels of IgM and IgG can be simultaneously detected from the second week of symptom's onset. Since both of them have high affinity to RBD, IgM may affect the binding between IgG and RBD, thus resulting in the inaccurate results. To evaluate the effect of IgM on quantitative detection of IgG, IgM was introduced into the cell individually. When 100 ng/mL IgM was introduced and incubated, a little fluorescence intensity was detected after secondary antibody was added, which was similar as that of the non-specific adsorption (Fig. 5 a). This indicated that the cross-reactivity between fluorescence labeled secondary antibody and IgM was negligible. Then, the mixture of various concentrations of IgG and IgM was added and incubated, the detected fluorescence intensity of the mixture increased with increasing the concentration of IgG and IgM, which was slightly lower than that of the IgG solution in the absence of IgM. This might contribute to the site-hindrance effect originated from the binding between IgM and RBD, and allow the less IgG bound to the RBD immobilized on the fiber bio-probe.
Fig. 5.
(A) Effect of IgM with different concentration (10, 50, 100 ng/mL) for nonspecific adsorption and IgG detection; (b) Matrix effect of serum on the nonspecific adsorption and specific detection of IgG using biosensor (IgG was added to PBS, total serum, 5 times, 10 times, and 20 times diluted serum, respectively).
To further evaluate the sensor selectivity, the signal response of five different analytes including BSA, cTnI, PD-L, MERS-CoV-2 antibody, HCoV-HKU1 CoV antibody was detected. Fig. S3 demonstrates that even the concentration of other proteins is much higher than IgG, these analytes cause little signal response compared with IgG, which is similar with the non-specific adsorption. The slight rise for MERS-CoV-2 antibody and HCoV-HKU1 CoV antibody may be because both of them belongs to coronavirus family. The mixture of MERS-CoV-2 antibody, HCoV-HKU1 CoV antibody and IgG was also measured, and the fluorescence signal was quite close to the IgG detection value. These results demonstrated that the detected fluorescence signal originated from highly specific binding reaction between antigen and antibody, which had little influence from the interfering substances. The selectivity of the proposed biosensor derived from two aspects. On the one hand, both the immobilized RBD and fluorescent labeled second antibody had high specificity for SARS-CoV-2 IgG. On the other hand, the cleaning stage during the detection process could effectively eliminate the interference.
Due to the complication of serum components (e.g., albumins, polypeptides and globulins), they may affect the interaction between antibody and antigen [28]. Dilution has become an effective method to reduce the matrix effect of the serum on the immunoassay of the clinical samples [28]. We investigated the matrix effect of serum on the S-IgG detection based on sandwich immunoassay principle. 50 ng/mL and 100 ng/mL S-IgG were spiked in PBS solution, original serum, and various diluted serums, respectively. These samples were detected using the OPOCT according to above described method. As shown in Fig. 5b, the fluorescence intensities of all samples were similar when the S-IgG concentration was 50 ng/mL or 100 ng/mL, respectively. The serum matrix had no significant influence on the binding between S-IgG and RBD, even for the original serum samples. Therefore, the serum sample can be directly detected using the OPOCT and RBD modified bio-probe without dilution, which benefits to simplify the detection procedure and save the detection time.
3.5. Regeneration performance of S-IgG biosensor
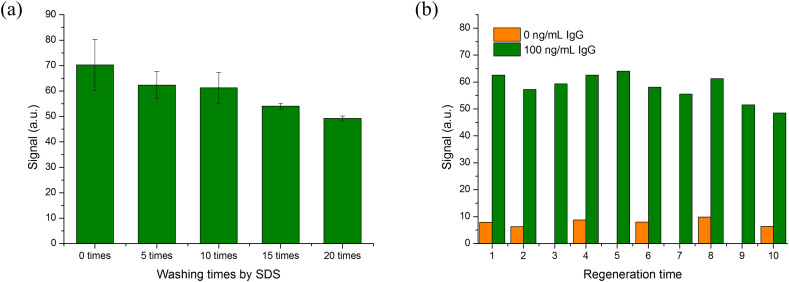
Most of the existing S-IgG rapid on-site immunoassay methods (e.g. LFIA) are disposable. For clinical biomedical analyses, the parallel detection of samples is important to improve the accuracy. Thus, compared with disposable biosensing method, using the same sensor to complete parallel analysis of samples can reduce the difference between batches, ensure the repeatability of the results and also save the detection cost. Herein, we investigated reusability of RBD modified fiber bio-probe. Because the regeneration solution is generally harsh (strong acid or base), this may damage the activity of biorecognition molecules immobilized onto the biosensor surface. The SDS solution (0.5 %, pH = 1.9) was employed to reuse the fiber bio-probe. To evaluate the reusability of fiber bio-probe, the effect of SDS solution on the activity of RBD was initially investigated. The RBD modified fiber bio-probe was repeatedly washed by the SDS solution according to the S-IgG detection procedure. The RBD activity was tested using the OPOCT after washed 5, 10, 15, and 20 times, respectively. As shown in Fig. 6 a, after the fiber bio-probe was washed 10 times, the detectable fluorescence intensity had no significant decrease, indicating that the RBD immobilized on the fiber bio-probe surface still kept a high binding affinity with the S-IgG and a good tolerability for SDS regeneration solution. However, more washed times could reduce the binding affinity.
Fig. 6.
(A) Signal for the OPOCT sensor with multiple washing times by SDS solution; (b) Signal for the OPOCT sensor with multiple regeneration times in the absence and presence of IgG (100 ng/mL). (Detection conditions: 0.5 μg/mL secondary antibody, incubation time 7 min, reaction time 7 min, each SDS washing time 5 min).
To reuse the fiber bio-probe, the S-IgG and fluorescence labeled secondary antibody bound to the fiber bio-probe surface should completely be removed by the SDS solution. Therefore, multiple immunoassay cycles of S-IgG were performed to verify regeneration performance. As illustrated in Fig. 6b, the RBD could maintain high binding capability with the S-IgG and the fluorescence intensity had no significant decrease after 8th immunoassay cycles, and the regenerated fiber bio-probe had still few nonspecific adsorptions after 10th immunoassay cycles. However, with continually increasing the immunoassay times, the detectable fluorescence intensity gradually decreased. The regenerated biosensor surface was also observed using confocal microscope (Nikon A1, Japan) after 5, 10, 15, and 20 times regenerations (Fig.S4). The amount of the residual fluorescence labeled secondary antibodies on the fiber bio-probe surface increased with increasing immunoassay cycles. Therefore, the prepared fiber bio-probe could be reused about eight immunoassay cycles. For more immunoassay cycles, the activity loss of RBD and incomplete elution of S-IgG/second antibody complex might result in the poor performance. We also tested the regeneration performance for SARS-CoV-2 IgG antibody detection in total serum samples. The results were shown as Fig.S5. The fiber biosensor also maintained a good detective performance after repeated use for 7 times, which was similar with the detection results in PBS buffer. This result further proved that our method was not interfered by the serum matrix. Increasing the reusability of fiber bio-probe will be the focus of our next research because it can effectively increase the accuracy of detection and reduce the detection cost.
3.6. Analysis of serum samples
To evaluate reliability of the proposed method, a commercial ELISA kit was obtained and a series of S-IgG concentration were measured from 0 to 2000 ng/mL (Fig.S6). The ELISA exhibited a linear detection range from 3.66 to 29.38 ng/mL, which is lower but narrower than this study. The difference mainly came from the shorter incubation times compared to ELISA. For ELISA, the incubation time for IgG to RBD and HRP-conjugated second antibodies to IgG were 45 and 30 min respectively, while the time was both 7 min in our assay. With shorter incubation time, the antibodies could not bind so sufficiently with the immobilized RBD in the binding process, so the detection concentration was higher for our assay. Both methods were used to detect the actual samples. For OPOCT biosensor, the recovery was from 67.78% to 125.59 % with RSD value within 18.37 % when the spiked concentrations were of 100, 250, and 500 ng/mL (Table S2). For ELISA, when the spiked concentrations of 5, 25 and 50 ng/mL, the recovery was in the range of 71.10–100.5 % and the RSD value was within 6.61 % (Table S3). As can be seen, the two methods showed good reliability in their respective linear range, and our method has wider detection range and shorter detection cycle.
To further verify the performance of serum antibody tests, the proposed method had been applied successfully in the IgG antibodies detection after two doses vaccine (Sinopharm, COVID-19). Finger-prick blood samples were collected before and after vaccination from volunteers. Considering the small volume of finger-prick blood samples, the subsequent detection was carried out with 20 times dilution. Two vaccine recipients were selected and the blood sample from 0 to 5 weeks were analyzed (Fig.S7). Before vaccination, no antibody was detected. After 7 days, a very low amount of IgG antibody was detected in vaccinated 2, and then the antibody concentration gradually increased. After the second injection, the antibody concentration significantly increased, which was similar with those of previous studies [30]. Antibody concentrations were different, but the relative trend was consistent, which might originate from the individual differences. Therefore, this work can be used to examine the level of SARS-Cov-2 IgG antibody, including COVID-19 patients and vaccinator.
4. Conclusions
In this study, a rapid and quantitative detection method for anti-SARS-CoV-2 S1 IgG for COVID-19 was developed using OPOCT biosensor. The all-fiber evanescent wave fluorescence platform was employed which exhibits the advantages of compact structure, small size, high sensitivity, and convenient operation. This method realized sensitive quantitative detection of IgG over the range of 12.5–1000 ng/mL. The LOD of 12.5 ng/mL is comparable to other immunoassay for IgG detection, which well satisfies the diagnostic requirements in POC testing. The IgM and other components in serum have no significant effects on IgG detection, and the total serum can be tested directly by the proposed method without dilution. These qualities make it possible to achieve early diagnosis and timely clinical decision-making for COVID-19 with good stability and accuracy. The evaluation of the regeneration performance indicated that the biosensor can be reused for about 8 times. The characterization results demonstrated that the activity loss mainly arose from the incomplete elution and destruction of surface RBD. The reliability of our method was proved by analyzing serum samples and compared with a commercial ELISA kit. Additionally, the platform can readily be applied to the determination of other biomarkers by using respective specific antibodies. With the above attractive properties, especially high sensitivity and reusability, this method is very suitable and favors a great potential for rapid quantitative POCT for serological analysis of COVID-19.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (21675171) and the Outstanding Innovative Talents Cultivation Funded Programs 2020 of Renmin University of China.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2021.122800.
CRediT author statement
Dan Song, Jiayao Liu, Wenjuan Xu, Xiangzhi Han: conducted SARS-CoV-2 IgG antibody detection experiments and analyzed data. Hongliang Wang, Yuan Chen, Yuxin Zhuo: produced fiber bio-probe and characterized it. Feng Long: carried out the statistical analysis and were responsible for writing and editing the manuscript. All authors have read and proof edited the manuscript.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bchetnia M., Girard C., Duchaine C., Laprise C. The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a review of the current global status. J. Infect. Public Health. 2020;13(11):1601–1610. doi: 10.1016/j.jiph.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acter T., Uddin N., Das J., Akhter A., Kim S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: a global health emergency. Sci. Total Environ. 2020;730:138996. doi: 10.1016/j.scitotenv.2020.138996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakraborty I., Maity P. COVID-19 outbreak: migration, effects on society, global environment and prevention. Ence Total Environ. 2020;728:138882. doi: 10.1016/j.scitotenv.2020.138882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilic T., Weissleder R., Lee H. Molecular and immunological diagnostic tests of COVID-19: current status and challenges. iScience. 2020;23(8):101406. doi: 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji T., Liu Z., Wang G., Guo X., Akbar khan S., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q. Detection of COVID-19: a review of the current literature and future perspectives. Biosens. Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J. Adv. Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui F., Zhou H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020;165:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morales-Narváez E., Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 2020;163:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sidiq Z., Hanif M., Kumardwivedi K., Chopra K.K. Benefits and limitations of serological assays in COVID-19 infection. Indian J. Tubercul. 2020;67:S163–S166. doi: 10.1016/j.ijtb.2020.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long Q.X., Tang X.J., Shi Q.L., Li Q., Huang A.L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 11.Winter A., Hegde S. The important role of serology for COVID-19 control. Lancet Infect. Dis. 2020;20(7):758–759. doi: 10.1016/S1473-3099(20)30322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z.T., Yi Y.X., Luo X.M., Xiong N., Liu Y., Li S.Q., Sun R.L., Wang Y.Q., Hu B.C., Chen W., Zhang Y.C., Wang J., Huang B.F., Lin Y., Yang J.S., Cai W.S., Wang X.F., Cheng J., Chen Z.Q., Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng L., Li Y., Liu J., Guo L., Wang Z., Xu X.X., Song S., Hao C.L., Liu L., Xin M., Xu C.L. Rapid, ultrasensitive and highly specific biosensor for the diagnosis of SARS-CoV-2 in clinical blood samples. Mater. Chem. Front. 2020;4:2000–2005. [Google Scholar]
- 14.Ravi N., Cortade D., Ng E., Wang S. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter L., Garner L., Smoot J., Li Y., Zhou Q., Saveson C., Sasso J., Gregg A., Soares D., Beskid T., Jervey S., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan X., Yang C.M., He Q., Chen J.H., Yu D.M., Jie L., Zhai S.Y., Qin Z.F., Du K., Chu Z.H., Qin P.W. Current and perspective diagnostic techniques for COVID-19. ACS Infect. Dis. 2020;6(8):1998–2016. doi: 10.1021/acsinfecdis.0c00365. [DOI] [PubMed] [Google Scholar]
- 17.Guo J.C., Chen S.Q., Guo J.H., Ma X. Nanomaterial labels in lateral flow immunoassays for point-of-care-testing. J. Mater. Sci. Technol. 2021;60:90–104. [Google Scholar]
- 18.Guo J.C., Chen S.Q., Tian S.L., Liu K., Ma X., Guo J.H. A sensitive and quantitative prognosis of C-reactive protein at picogram level using mesoporous silica encapsulated core-shell up-conversion nanoparticle based lateral flow strip assay. Talanta. 2021;230:122335. doi: 10.1016/j.talanta.2021.122335. [DOI] [PubMed] [Google Scholar]
- 19.Guo J.C., Chen S.Q., Tian S.L., Liu K., Ni J., Zhao M., Kang Y.J., Ma X., Guo J.H. 5G-enabled ultra-sensitive fluorescence sensor for proactive prognosis of COVID-19. Biosens. Bioelectron. 2021;181:113160. doi: 10.1016/j.bios.2021.113160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C.W., Shi D.W., Wan N., Yang X.S., Liu H.F.F., Gao H.X., Zhang M.L., Bai Z.K., Li D.C., Dai E.H., Rong Z., Wang S.Q. Development of spike protein-based fluorescence lateral flow assay for the simultaneous detection of SARS-CoV-2 specific IgM and IgG. Analyst. 2021;146:3908–3917. doi: 10.1039/d1an00304f. [DOI] [PubMed] [Google Scholar]
- 21.Yin M.J., Gu B., An Q.F., Yang C., Guan Y.L., Yong K.T. Recent development of fiber-optic chemical sensors and biosensors: mechanisms, materials, micro/nano-fabrications and applications. Coord. Chem. Rev. 2018;376:348–392. [Google Scholar]
- 22.Jiao L.Z., Zhong N.B., Zhao X.D., Ma S.X., Fu X.L., Dong D.M. Recent advances in fiber-optic evanescent wave sensors for monitoring organic and inorganic pollutants in water. TrAC Trends Anal. Chem. (Reference Ed.) 2020;127:115892. [Google Scholar]
- 23.Tang Y., Long F., Gu C., Wang C., Han S., He M. Reusable split-aptamer-based biosensor for rapid detection of cocaine in serum by using an all-fiber evanescent wave optical biosensing platform. Anal. Chim. Acta. 2016;933:182–188. doi: 10.1016/j.aca.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 24.Liu J., Xu W., Zhu A., Kang H., Long F. Reusable optofluidic point-of-care testing platform with lyophilized specific antibody for fluorescence detection of cholylglycine in serum. Microchim. Acta. 2020;187:439. doi: 10.1007/s00604-020-04424-2. [DOI] [PubMed] [Google Scholar]
- 25.To K.W., Tsang T.Y., Leung W.S., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long F., Li W., Song D., Han X., Zhu A. Portable and automated fluorescence microarray biosensing platform for on-site parallel detection and early-warning of multiple pollutants. Talanta. 2019;210:120650. doi: 10.1016/j.talanta.2019.120650. [DOI] [PubMed] [Google Scholar]
- 27.Tan X., Krel M., Dolgov E., Park S., Fan X. Rapid and quantitative detection of SARS-CoV-2 specific IgG for convalescent serum evaluation. Biosens. Bioelectron. 2020;169:112572. doi: 10.1016/j.bios.2020.112572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’B Cells. Cell. 2020;182(1):73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ju B., Zhang Q., Ge J.W., Wang R., Sun J., Ge X.Y., Yu J.Z., Shan S.S., Zhou B., Song S., Tang X., Yu J.F., Lan J., Yuan J., Wang H.Y., Zhao J.J., Zhang S.Y., Wang Y.C., Shi X.L., Zhang L.Q. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020:1–8. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 30.Ma H., Zeng W., He H., Zhao D., Yang Y., Jiang D., Zhou P., Qi Y., He W., Zhao C., Yi R., Wang X., Wang B., Xu Y., Yang Y., Kombe A., Ding C., Xie J., Gao Y., Cheng L., Li Y., Ma X., Jin T. 2020. COVID-19 Diagnosis and Study of Serum SARS-CoV-2 Specific IgA, IgM and IgG by a Quantitative and Sensitive Immunoassay. [Google Scholar]
- 31.Rashed M., Kopechek J., Priddy M., Hamorsky K., Palmer K., Mittal N., Valdez J., Flynn J., Williams S. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2020;171:112709. doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elledge S.K., Zhou X.X., Byrnes J.R., et al. Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection. Nat. Biotechnol. 2021;39:928–935. doi: 10.1038/s41587-021-00878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Funari R., Chu K.Y., Shen A.Q. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens. Bioelectron. 2020;169:112578. doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z.H., Wang X.Q., Wei X.J., Zheng S.W., Lenhart B.J., Xu P.S., Li J., Pan J., Albrecht H., Liu C. Multiplex quantitative detection of SARS-CoV-2 specific IgG and IgM antibodies based on DNA-assisted nanopore sensing. Biosens. Bioelectron. 2021;181:113134. doi: 10.1016/j.bios.2021.113134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu J.X., Chenier M., Zhang Y.S., Xu C.Q. A microflow cytometry-based agglutination immunoassay for point-of-care quantitative detection of SARS-CoV-2 IgM and IgG. Micromachines. 2021;12:433. doi: 10.3390/mi12040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.