Abstract
Down syndrome is one of the most common chromosomal anomalies affecting the world’s population, with an estimated frequency of 1 in 700 live births. Despite its relatively high prevalence, diagnostic rates based on clinical features have remained under 70% for most of the developed world and even lower in countries with limited resources. While genetic and cytogenetic confirmation greatly increases the diagnostic rate, such resources are often nonexistent in many low- and middle-income countries, particularly in Sub-Saharan Africa. To address the needs of countries with limited resources, the implementation of mobile, user-friendly and affordable technologies that aid in diagnosis would greatly increase the odds of success for a child born with a genetic condition. Given that the Democratic Republic of the Congo is estimated to have one of the highest rates of birth defects in the world, our team sought to determine if smartphone-based facial analysis technology could accurately detect Down syndrome in individuals of Congolese descent. Prior to technology training, we confirmed the presence of trisomy 21 using low-cost genomic applications that do not need advanced expertise to utilize and are available in many low-resourced countries. Our software technology trained on 132 Congolese subjects had a significantly improved performance (91.67% accuracy, 95.45% sensitivity, 87.88% specificity) when compared to previous technology trained on individuals who are not of Congolese origin (p < 5%). In addition, we provide the list of most discriminative facial features of Down syndrome and their ranges in the Congolese population. Collectively, our technology provides low-cost and accurate diagnosis of Down syndrome in the local population.
Keywords: down syndrome, screening, facial analysis, machine learning, Congo, DRC
Introduction
Down syndrome (DS), caused by a trisomy of chromosome 21, has a prevalence of approximately 1 in 700 live births (1) and is the most common aneuploidy in the population. The phenotype of subjects with this chromosomal disorder has been extensively described in the literature and includes congenital heart defects, respiratory problems, intellectual disability, physical dysmorphology, gastrointestinal tract anomalies, hearing and vision problems, and immune system defects (2–4). Early detection of DS is essential to prevent life-threatening complications in these patients, usually related to their pulmonary and cardiac anomalies. In developed countries, prenatal screening using non-invasive testing is widely available and includes ultrasonography(5). When screening shows an increased risk for DS, more invasive diagnostic tests such as amniocentesis or chorionic villus sampling are used for confirmation(5). However, in regions where access to screening and diagnostic resources is limited, DS is usually identified after birth based on characteristic dysmorphic features observed during physical examination. Although the facial appearance is a key feature used by specialists to identify DS, dysmorphic features are often subtle in young babies and variable among populations with different ancestry. As a consequence, the reported accuracy of identifying DS by family physicians and pediatricians using only physical examination is as low as 64%(6). Hence, there is a need for more accurate alternate methods of diagnosis to improve outcomes through early intervention.
There are still large regions in the world with limited-to-no-access to screening and diagnostic resources for DS(7–9). Not surprisingly, some of those areas have been reported to present high rates of birth defects compared to developed countries. Specifically, the Democratic Republic of the Congo (DRC) has an estimated birth defect rate of 71 per 1,000 children born(10), which is the highest regionally and one of the highest in the world. If affordable and fast screening resources were available, early detection of genetic syndromes in the DRC has the potential to reduce child mortality and morbidity associated with such conditions.
In our previous studies, we showed the potential of one such screening method, facial analysis technology, to identify dysmorphic facial features that are indicative of the presence of genetic syndromes in patients with diverse ancestry with an accuracy of 89% or higher (11–17). Although we found that models trained to identify facial dysmorphology in ethnic- and race-specific groups outperformed models trained in the global population, those former models did not account for phenotypical variations within groups with similar ancestry. For instance, the DS facial model specific to the Asian population was trained with patients from China, Malaysia, Thailand and India, which requires accounting for an important facial phenotype variability. Besides our previous studies, another facial analysis software, Face2Gene (FDNA, Boston, MA) has also been used to report on for the diagnosis of genetic syndromes in specialized (genetic) clinics. Its reported accuracy ranged between 60–69% (18, 19). Lumaka et al (20) evaluated the performance of Face2Gene when trained with a dataset that incorporated photographs of African patients from the DRC, Rwanda and France, similarly concluding that patient ancestry influences the evaluation of facial morphology. However, that study performed differential diagnosis between DS and other syndromes. Hence, no comparisons were made with normative populations, and no interpretable and quantitative reference metrics that could be used at the clinics were provided. In average, DS was identified as the fifth most likely syndrome in African patients with confirmation of DS after training Face2Gene on African populations.
In the current work, we focus on the DRC to implement a screening technology to identify DS in the local population. We use our technology to create a facial model specific to the populations seen in the capital Kinshasa, and to identify the facial features that are most discriminative of DS and their ranges in this population.
Materials
After approval by the ethical committees in the DRC and the US, we collected frontal facial photographs of presumed normative controls and suspected cases of DS using an in-house mobile phone application. Photographs of cases and controls were obtained from Maternité de Binza, Clinique Bondeko, Hôpital Saint Joseph, Hôpital Pédiatrique de Kalembelembe, Centre Mère et Enfant de Bumbu, Pédiatrie de Kimbondo, Village Bondeko network schools, CEIEHMA school and Kikesa School in Kinshasa, DRC. Prior to recruitment at hospital sites, the DRC-based pediatrician visited these establishments and held information sessions with nurses and administrators on the scopes and goals of our study and acquired approval from hospital administrators to conduct the study at these sites. This was followed by short educational sessions with nurses to better train on clinical features to be aware of for newborns and babies that may present with DS. In addition to these sessions, brochures were also given to nurses at our study sites which used text and photographs to highlight some of the facial phenotype characteristics associated with DS, to help identify newborns who may harbor trisomy 21. Once the hospital-based nurses suspected a baby or newborn of having DS, the DRC-based pediatrician was called and consulted with the parents of the child to explain the scope and goals of the study and acquired consent for participation in the study. As this was a pilot study, the pediatrician discussed DS with the parents, however, was unable to offer an official diagnosis, as one had yet to be made at the time of collection.
Sites outside of hospital settings were visited by the local pediatrician on the research team who discussed the scope and goals of this study with center directors and parents for the recruitment of children with DS. The research study was explained in detail to both parents and center administrators prior to obtaining consent for participation. While the participants at these centers were suspected of harboring trisomy 21, genetic confirmation had not been established and children presenting with features of DS at these centers were determined by both the administrators (Joachim Mukau Ebwel) and the DRC-based pediatrician (Kizito B.A. Mosema). Local medical doctors administered and explained the consent to photograph, analyze and publish findings in French or Lingala to the parents/guardians of the subjects. After informed consent, photographs were taken with the mobile phone application and buccal swabs (Zymo Research DNA/RNA Shield Collection tubes, Cat # R1107) were collected for downstream genetic confirmation of suspected cases of Trisomy 21. After genetic confirmations were established downstream, the DRC-based pediatrician contacted/attempted to contact parents of those children in the study to relay such findings (while not an expectation of the study, the DRC-pediatrician determined the best course of action on this matter).
We excluded photographs that were not frontal or had motion/blurring artifacts. In addition, we discarded any photographs with poor illumination or presence of shadows in the face of the patients, as they may affect the quantification of the facial appearance. In total, we included 74 patients (25 females, age 9.60 ± 4.32 years, range 5 days – 18 years) with genetic testing being negative for eight of these patients and trisomy 21 was confirmed for 66 of the suspected cases (25 females, age 9.33 ± 4.46 years, range 5 days – 18 years).
We also acquired facial photographs of 66 normative subjects from the same local population matched to the patients with diagnosis confirmation of DS by sex (age 6.79 ± 4.14 years, range 16 days – 18 years). A flowchart summarizing our data collection approach can be found with our supplementary material (Fig. 1).
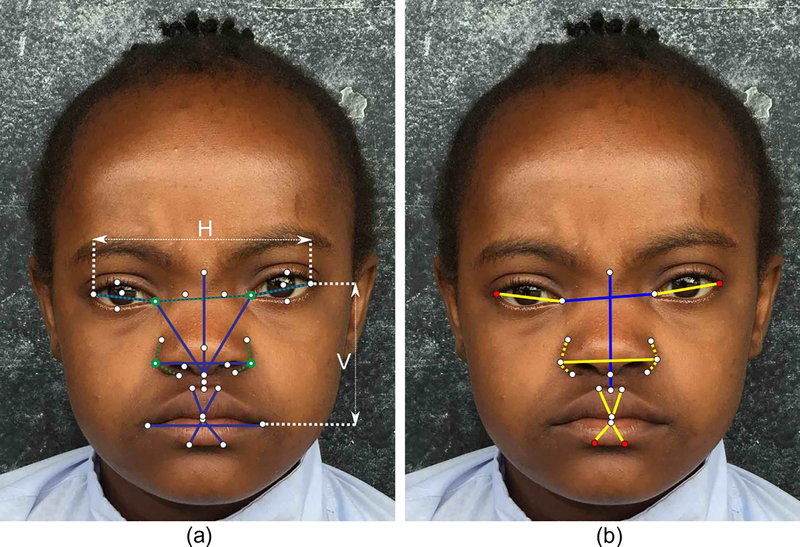
Fig. 1:
(a) Landmarks and metrics used by the facial analysis technology to quantify facial phenotypes. Blue lines represent distances. Horizontal distances are normalized with respect to the distance between the lateral canthi (H), and vertical distances are normalized to the distance between the lateral canthi and the oral commisures (V). Dashed green lines represent angles centered at the landmarks with green circles. Appearance features are calculated at different resolutions around each of the 33 facial landmarks. (b) Metris that are significantly different beteween patients with Down syndrome and normative subjecs in the population of the DRC. Blue lines and landmarks in red represent the distance and appearance metrics, respectively, used by our classifier to identify Down syndrome in the population of the DRC (presented in Tables 2 and 3). The other metrics that are significantly different between patients with Down syndrome and healthy subjects and that are presented in Table 4 are depicted in yellow.
Methods
Genetic confirmation of suspected DS cases using capillary electrophoresis:
Genetic confirmation for the DS training set was conducted by polymerase chain reaction (PCR) and capillary electrophoresis previously described by Sun et al (21). Conserved regions of segmental duplications of TTC3 on chromosome 21 and KDM2A on chromosome 11 were targeted for amplification. Forward and reverse primers used were designed by Sun et al. (21) that annealed to both TTC3 and KDM2A, enabling amplification of both segments with the same primer pairs. The forward primer was fluorescently labelled with 6-carboxyfluorescein fluorophore on the 5’ end allowing detection during capillary electrophoresis. DNA was isolated from buccal swabs (Zymo Research DNA/RNA Shield Collection tubes, Cat # R1107) iusing the Quick-DNA Magbead Plus Kit (Zymo Research cat # D4081) and 20ng of DNA was used per reaction for each suspected DS case and ran in technical triplicate. The PCR product was treated with ExoSAP-IT PCR Product Cleanup Reagent (ThermoFisher, Catalog Number: 78201.1.ML), following manufacturer’s recommendations, prior to capillary electrophoresis. The PCR products were diluted 1:32 prior to sequencing on the ABI 3500 Genetic Analyzer (Applied Biosystems). The predicted amplicon from TTC3 is 147bp, while the amplicon for KDM2A was predicted at 128bp, allowing fragment analysis to distinguish the two segments.
Area under the curve for the appropriate peaks was used to quantify the proportion of each segment using Microsatellite analysis software (Applied Biosystems). For trisomy 21 confirmation, we expect ~1.5 times more of fragment of size 147bp (from chr. 21) compared to that of fragment of size 128bp (from chr. 11) due to the duplication of chromosome 21. The averages of the ratios were determined for each sample from the triplicates performed. DNA from a trisomy 21 sample from the Coriell Institute’s repository was used as a positive control (Catalog ID: NG06922), while DNA from a non-trisomy 21 male was used a negative control. Statistical analysis of amplicon ratios between confirmed cases, negative controls and positive controls was conducted using the Student t-test.
Facial analysis technology:
The facial analysis technology has been described in detail in our previous works (11–17). In summary, our technology quantifies a set of geometric measurements normalized to the size of the face from 44 anatomical facial landmarks. In addition, it quantifies the appearance around each one of the 33 inner facial landmarks using an improved texture descriptor based on local binary patterns (22) at different levels of resolution to capture patterns at different scales. Fig. 1 illustrates the inner facial landmarks and metric computed from them. From the geometric and appearance features, the most discriminative ones between normative subjects and patients with DS were selected using recursive feature elimination (23) and a support vector machine classifier (24) with a linear kernel was trained to estimate the probability of a subject to present DS.
Experiments:
To characterize quantitatively the facial phenotype of DS in the population of the DRC, we created a computational facial phenotyping model specific to the local population using our facial analysis technology. We trained our classification model using the photographs of the 66 patients with diagnostic confirmation of DS and their matched controls. We evaluated its accuracy, sensitivity and specificity of the technology using cross-validation. Through this process, we trained a facial model using all subjects in our dataset except for one, which was used for testing. We repeated this process iteratively until all patients were tested. To identify the optimal number of discriminative features used for classification, we increased the number of selected features until the area under the receiver operator characteristic curve converged (25). Once the optimal number of features was selected, we calculated the classification threshold as the one that maximized the accuracy of our classifier. For each selected feature, we also estimated its individual discriminative power using the non-parametric Mann-Whitney U test (26).
To show the benefits of a model adapted to the local population of the DRC, we calculated the performance of two other models developed in our previous work(13): a model trained to identify DS in the global population, and a model trained to identify DS in the general African descent population. The latter did not include cases from the DRC. After training the model, we also tested its performance on the 8 patients that were incorrectly diagnosed as DS (based on genetic confirmation) to show the potential of our model in a real setting.
Results
Genetic Confirmation:
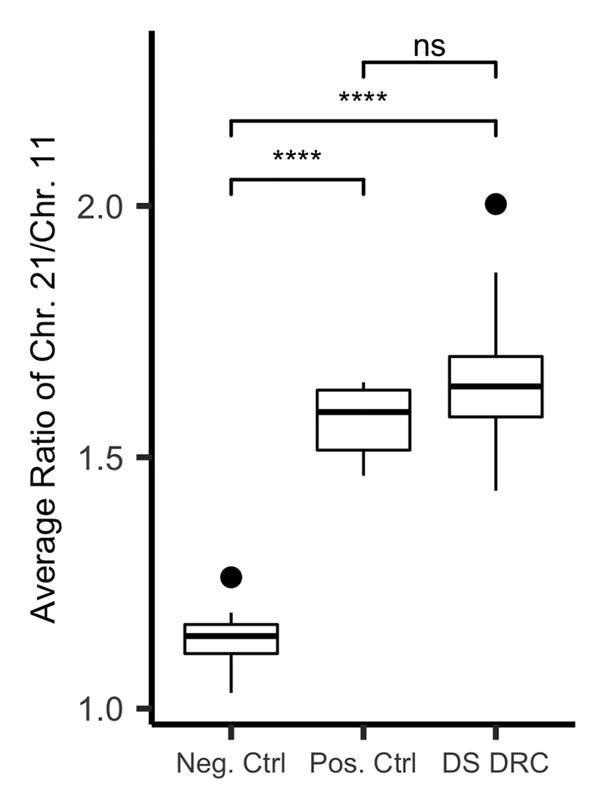
To genetically confirm an aneuploidy of chromosome 21 in the DRC patients suspected of having DS, we utilized qRT-PCR that targeted segmental duplications on chromosome 21 and 11. When the ratio of both fragments were plotted (Chr. 21/ Chr. 11), as shown in Fig. 2, we observed no overlap between negative and positive samples, enabling accurate confirmation. Negative controls had a minimum ratio of 1.03, a maximum ratio of 1.26 and a mean of 1.14 ± 0.06, whereas positive DS controls had a minimum ratio of 1.46, a maximum ratio of 1.64 and a mean of 1.572 ± 0.07. Our confirmed cases of DS from DRC had a minimum ratio of 1.43, a maximum ratio of 2.00 with the mean being 1.65 ± 0.10. The ratio of fragments from chromosome 21 to chromosome 11 for both positive controls and confirmed cases of DS from our DRC cohort were significantly different than the ratios observed for the negative control (p<=0.001). Collectively, using a low-cost and user-friendly qRT-PCR our approach confirmed 66 cases of DS out of 74 clinically suspected subjects based on ratios of segmental duplications of chromosomes 21 and 11.
Fig. 2:
Distribution of the ratios of segmental duplications of chromosome 11 versus chromosome 21 for negative controls, independent positive controls and cases of Down syndrome identified in the DRC. **** indicates a p value < 0.001 as measured by the t-test.
Facial Analysis:
The performance of our three models identifying DS in the local population of the DRC is summarized in Table 1. Cross-validation of the model trained for the local population of the DRC showed a classification accuracy of 91.67% (95.45% sensitivity and 87.88% specificity) for an optimal threshold of 45%. The list of the five optimal discriminative features selected by our technology is provided in Table 2 together with their ranges in the control population and in the subjects with DS.
Table 1.
Performance of the models trained on the DRC, African descent and global populations when tested on the local population of the DRC. P-values were calculated to compare the performance of the models trained on the African and Global populations with the one trained with the local population of the DRC using the McNemar’s test. PPV and NPV stand for positive predictive value and negative predictive value, respectively.
| Accuracy | Sensitivity | Specificity | PPV (precision) | NPV | p-value | |
|---|---|---|---|---|---|---|
| DRC | 91.67% | 95.45% | 87.88% | 88.73% | 95.08% | - |
| African | 77.27% | 89.39% | 65.15% | 71.95% | 86.00% | 0.037 |
| Global | 75.76% | 95.45% | 56.06% | 68.48% | 92.50% | 0.003 |
Table 2.
Features selected by the facial analysis technology to identify DS in the DRC population. Horizontal features (H) are normalized to the distance between the lateral canthi. Vertical features (V) are normalized to the distance between the oral commissures and the lateral canthi.
| Feature | Normative | DS | p-value |
|---|---|---|---|
| Distance between medial canthi (H) | 0.41± 0.03 | 0.46 ± 0.03 | <0.001 |
| Nose length (V) | 0.76 ± 0.06 | 0.65 ± 0.06 | <0.001 |
| Philtrum length (V) | 0.24 ± 0.04 | 0.22 ± 0.04 | 0.023 |
| Average texture at lateral canthi | N/A | N/A | 0.0127 |
| Texture at lower border of upper lip | N/A | N/A | 0.502 |
The model trained on the general African descent population provided an accuracy of 77.27% (89.39% sensitivity and 65.15% specificity). On the other hand, the model trained with the general population provided an accuracy of 75.76% in the DRC population (95.45% sensitivity and 56.06% specificity). The contingency tables used to calculate the performance of our classifiers are available in our supplementary material (Table 1). Table 3 also shows the ranges reported in the global population and in the population with African ancestry. Finally, our model discarded correctly the presence of DS in 5 of the 8 patients that were diagnosed incorrectly by experts, representing a 62% reduction in the false positive rate.
Table 3.
Ranges of the geometric features selected to identify DS in the DRC population compared with the global population and the African descent population. Horizontal features (H) are normalized to the distance between the lateral canthi. Vertical features (V) are normalized to the distance between the oral commissures and the lateral canthi.
| Normative | Down syndrome | |||||
|---|---|---|---|---|---|---|
| Features | DRC | African | Global | DRC | African | Global |
| Distance between medial canthi (H) | 0.41± 0.03 | 0.42 ± 0.04 | 0.41 ± 0.04 | 0.46 ± 0.03 | 0.44 ± 0.03 | 0.44 ± 0.04 |
| Nose length (V) | 0.76 ± 0.06 | 0.79 ± 0.13 | 0.77 ± 0.12 | 0.65 ± 0.06 | 0.69 ± 0.10 | 0.69 ± 0.11 |
| Philtrum length (V) | 0.24 ± 0.04 | 0.25 ± 0.05 | 0.26 ± 0.05 | 0.22 ± 0.04 | 0.22 ± 0.05 | 0.24 ± 0.05 |
Discussion
Down syndrome is one of the most common chromosomal anomalies in the world’s population, resulting from an aneuploidy of fragments or the whole of chromosome 21 (1). Clinical features associated with DS enable diagnosis to be made early in life in most of the developed world, however despite the well-known features associated with this condition, diagnostic success rates based on clinician assessments are estimated to be below 70% (6, 27). Accuracy rates from clinicians are significantly reduced in the developing world, particularly in Sub-Saharan Africa, resulting in limited prevalence data and long-term patient success in those countries (6, 28, 29). While genetic testing greatly increases the odds of accurate diagnosis of DS, the methods come with high financial and technical burdens, preventing countries with limited resources from utilizing such technologies. To address these deficiencies in clinical diagnosis and limited resources, low-cost and user-friendly technologies must be implemented to improve clinical diagnostic accuracy and reduce mortality and morbidity of those living with a genetic condition. Given that the DRC is estimated to have one of the highest rates of birth defects in the world, coupled with limited clinical and genetic resources, we sought to determine if facial recognition software was capable of accurately identifying cases of DS (10).
Technology training on data from population of local ethnicity increases accuracy:
When assessing the performance of our facial recognition application using study populations acquired from Kinshasa, the urban capital of DRC, we see that the performance of the software is highly variable depending on the source/ethnicity of the subjects individuals used for the training of the facial analysis technology. If the software is trained using data from a global general population, the application can distinguish those with DS from the DRC with 75.76% accuracy (95.45% sensitivity and 56.06% specificity). These performance indices are marginally improved if the software is trained on the general population of African ancestry (including African Americans) resulting in 77.27% accuracy (89.39% sensitivity and 65.15% specificity). However, the performance of the technology improved significantly (p<5% using a McNemar’s test) when it was trained on data from subjects of Congolese origin enabling 91.67% accuracy (95.45% sensitivity and importantly 87.88% specificity). These findings highlight the importance of utilizing local subjects, including controls, for training prior to using the application for diagnostic approaches. If trained with global or African descent controls, we see a low specificity in performance, indicating that the application misidentifies a large percentage of individuals of Congolese descent as being dysmorphic who otherwise are not. These results likely stem from the fact that Africa has the highest level of genetic human diversity on the planet, which has subsequently resulted in unique characteristics and physical features of the thousands of ethno-linguistic subgroups of individuals on the continent. Due to this level of diversity an “African” training model is generally unlikely to prove successful, however these challenges can be overcome if technology training is performed on a targeted population of interest, as our findings demonstrate.
Facial analysis identifies discriminative facial features specific to Down syndrome in the DRC:
Our technology utilized geometric and appearance measures derived from facial landmarks to effectively distinguish normative controls from those individuals harboring a condition that results in a facial phenotype, as illustrated with DS. The classifier identified significant differences in three geometric facial features including the distance between medial canthi, nose length and philtrum length that were most important for discriminating cases of DS from unaffected individuals of Congolese descent. Other geometric facial features such as the distance between medial and lateral canthi, the distance between nose alas, upper lip width, lower lip width and the angle at the alas of the nose were also found to be significantly different between cases of DS and normative DRC controls, however they were not needed for accurate distinction.
Study limitations:
Although our facial analysis provided 91.67% accuracy identifying DS in the local population from Kinshasa (DRC), its accuracy may be affected by the imperfect matching in our dataset. Our patients with DS were matched with controls by sex and race, but they were not perfectly matched by age because of data unavailability. Moreover, since our technology was designed to distinguish patients with DS from normative subjects, it could potentially indicate in the future that patients harboring other syndromes have a similar phenotype to DS, if not genetically confirmed as it was done for this study. Although the current study focuses on the quantitative characterization of the Congolese facial phenotype of DS, our technology is designed for screening purposes and its goal is to identify patients at risk for referral to preventive and specialized care. In this scenario, the identification of a patient at risk, even if the cause is not DS, may be beneficial to initiate preventive care. Finally, although our study covered the full pediatric age range, we believe that the highest potential of this technology is the screening of newborns and babies. Future work will focus on these populations, to make this technology more impactful.
Conclusion:
Due to the limited scientific data on the topic of Down syndrome in the DRC, prevalence rates and outcomes for those affected with this condition are largely unknown. However, reports based on medical expert opinions suggest that the vast majority of those living with DS in the Congo frequently do not live past 2 years of age, due to lack of diagnosis, treatment options and associated costs (30). While traditional cytogenic testing serves as a gold standard to diagnose cases of trisomy 21, other technologies can effectively accomplish accurate genetic confirmations at a fraction of the cost and expertise. Here, we demonstrated that DS can be successfully diagnosed using smart phone-based applications with high sensitivity and specificity, coupled with a much simpler genetic technique of qRT-PCR, should genetic confirmations be warranted. Based on local medical opinions, the cost of diagnosing DS in the DRC is estimated to cost $500 USD (30), a price that is largely outside of the financial capability of a population that resides in one of the most poverty-stricken countries in the world (31). Utilizing the user-friendly and accurate applications presented here could effectively offer a diagnostic solution for less than $10 USD, as well as greatly expanding much needed surveillance data on inborn genetic conditions in countries where medical expertise and infrastructure are lacking, as is the case in the DRC.
Supplementary Material
Table 4.
Geometric features with significantly different ranges between the normative population and the patients with DS in the DRC that were not selected for classification. Horizontal features (H) are normalized to the distance between the lateral canthi. Vertical features (V) are normalized to the distance between the oral commissures and the lateral canthi.
| Feature | Mean: Normal | Std: Normal | Mean: Syndromic | Std: Syndromic | p-value |
|---|---|---|---|---|---|
| Distance between medial and lateral canthi (H) | 0.30 | 0.01 | 0.27 | 0.01 | <0.001 |
| Distance between nose alas (H) | 0.43 | 0.03 | 0.46 | 0.04 | <0.001 |
| Upper lip width (V) | 0.16 | 0.03 | 0.18 | 0.03 | <0.001 |
| Lower lip width (V) | 0.18 | 0.03 | 0.17 | 0.04 | 0.001 |
| Angle at the alas of the nose (degrees) | 54.86 | 5.36 | 57.56 | 6.20 | 0.011 |
Acknowledgements
This work was partially supported by the National Center for Advancing Translational Sciences under grant UL1 TR001876. This work was also supported by the A. James Clark Distinguished Professor of Molecular Genetics Endowment to Professor Eric Vilain, MD, PhD, A. James Clark Professor, Children’s National Hospital. M. S. B. was supported by the Fogarty International Center of the National Institutes of Health (NIH) under Award Number D43TW009343 and the University of California Global Health Institute (UCGHI); The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or UCGHI.
Footnotes
This technology is protected by the following US patents: US patent no. 9,443,132 (Linguraru) and US patent no. 10,204,260 (Linguraru).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability
The facial photographs collected for this study are not publicly available to preserve patient privacy. The facial analysis technology is not yet publicly available. Interested individuals should contact Dr. Linguraru for details (mlingura@childrensnational.org).
References
- 1.Mai CT, Isenburg JL, Canfield MA, Meyer RE, Correa A, Alverson CJ, Lupo PJ, Riehle-Colarusso T, Cho SJ, Aggarwal D, Kirby RS, Network NBDP, National population-based estimates for major birth defects, 2010–2014, Birth Defects Res. 111, 1420–1435 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan KD, Evans D, Pandey A, Hraha TH, Smith KP, Markham N, Rachubinski AL, Wolter-Warmerdam K, Hickey F, Espinosa JM, Blumenthal T, Trisomy 21 causes changes in the circulating proteome indicative of chronic autoinflammation, Sci. Rep. 7, 14818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asim A, Kumar A, Muthuswamy S, Jain S, Agarwal S, “Down syndrome: an insight of the disease,” J. Biomed. Sci. 22, 41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park AH, Wilson MA, Stevens PT, Harward R, Hohler N, Identification of Hearing Loss in Pediatric Patients with Down Syndrome, Otolaryngol. Neck Surg. 146, 135–140 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Kazemi M, Salehi M, Kheirollahi M, Down syndrome: Current status, challenges and future perspectives, Int. J. Mol. Cell. Med. 5, 125–133 (2016). [PMC free article] [PubMed] [Google Scholar]
- 6.Sivakumar S, Larkins S, Accuracy of clinical diagnosis in Down’s syndrome, Arch. Dis. Child. 89, 691 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christianson A, Modell B, Medical Genetics in Developing Countries, Annu. Rev. Genomics Hum. Genet. 5, 219–265 (2004).Pre-proof [DOI] [PubMed] [Google Scholar]
- 8.Ballantyne A GIPA., Medical Genetic Service in Developing Countries. The Ethical, Legal and Social Implications of Genetic Testing and Screening (2006).
- 9.Akinmoladun J, Anumba D, Fetal imaging and diagnosis services in developing countries – A call to action, Trop. J. Obstet. Gynaecol. 36, 1 (2019). [Google Scholar]
- 10.Christianson A, Howson CP, Modell B, March of Dimes gloal report on birth defects (2006; https://www.marchofdimes.org/materials/global-report-on-birth-defects-the-hidden-toll-of--d2unzZI5_VWOaLZnw6iHcx7hbpMWtWzTuIOU3DabcVY.pdf).
- 11.Zhao Q, Okada K, Rosenbaum K, Zand DJ, Sze R, Summar M, Linguraru MG, Hierarchical constrained local model using ICA and its application to Down syndrome detection., Med. Image Comput. Comput. Assist. Interv. 16, 222–229 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Cerrolaza JJ, Porras AR, Mansoor A, Zhao Q, Summar M, Linguraru MG, Identification of dysmorphic syndromes using landmark-specific local texture descriptors, 2016 IEEE 13th Int. Symp. Biomed. Imaging, 1080–1083 (2016). [Google Scholar]
- 13.Kruszka P, Porras AR, Sobering AK, Ikolo FA, La Qua S, Shotelersuk V, Chung BHY, Mok GTK, Uwineza A, Mutesa L, Moresco A, Obregon MG, Sokunbi OJ, Kalu N, Joseph DA, Ikebudu D, Ugwu CE, Okoromah CAN, Addissie YA, Pardo KL, Brough JJ, Lee N-C, Girisha KM, Patil SJ, Ng ISL, Min BCW, Jamuar SS, Tibrewal S, Wallang B, Ganesh S, Sirisena ND, Dissanayake VHW, Paththinige CS, Prabodha LBL, Richieri-Costa A, Muthukumarasamy P, Thong M-K, Jones KL, Abdul-Rahman OA, Ekure EN, Adeyemo AA, Summar M, Linguraru MG, Muenke M, Down syndrome in diverse populations, Am. J. Med. Genet. Part A 173, 42–53 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Kruszka P, Addissie YA, McGinn DE, Porras AR, Biggs E, Share M, Crowley TB, Chung BHY, Mok GTK, Mak CCY, Muthukumarasamy P, Thong M-K, Sirisena ND, Dissanayake VHW, Paththinige CS, Prabodha LBL, Mishra R, Shotelersuk V, Ekure EN, Sokunbi OJ, Kalu N, Ferreira CR, Duncan J-M, Patil SJ, Jones KL, Kaplan JD, Abdul-Rahman OA, Uwineza A, Mutesa L, Moresco A, Obregon MG, Richieri-Costa A, Gil-da-Silva-Lopes VL, Adeyemo AA, Summar M, Zackai EH, McDonald-McGinn DM, Linguraru MG, Muenke M, 22q11.2 deletion syndrome in diverse populations., Am. J. Med. Genet. A 173, 879–888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruszka P, Porras AR, Addissie YA, Moresco A, Medrano S, Mok GTK, Leung GKC, Tekendo-Ngongang C, Uwineza A, Thong M-K, Muthukumarasamy P, Honey E, Ekure EN, Sokunbi OJ, Kalu N, Jones KL, Kaplan JD, Abdul-Rahman OA, Vincent LM, Love A, Belhassan K, Ouldim K, El Bouchikhi I, Shukla A, Girisha KM, Patil SJ, Sirisena ND, Dissanayake VHW, Paththinige CS, Mishra R, Klein-Zighelboim E, Gallardo Jugo BE, Chávez Pastor M, Abarca-Barriga HH, Skinner SA, Prijoles EJ, Badoe E, Gill AD, Shotelersuk V, Smpokou P, Kisling MS, Ferreira CR, Mutesa L, Megarbane A, Kline AD, Kimball A, Okello E, Lwabi P, Aliku T, Tenywa E, Boonchooduang N, Tanpaiboon P, Richieri-Costa A, Wonkam A, Chung BHY, Stevenson RE, Summar M, Mandal K, Phadke SR, Obregon MG, Linguraru MG, Muenke M, Noonan syndrome in diverse populations, Am. J. Med. Genet. Part A 173, 2323–2334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruszka P, Porras AR, de Souza DH, Moresco A, Huckstadt V, Gill AD, Boyle AP, Hu T, Addissie YA, Mok GTK, Tekendo-Ngongang C, Fieggen K, Prijoles EJ, Tanpaiboon P, Honey E, Luk H-M, Lo IFM, Thong M-K, Muthukumarasamy P, Jones KL, Belhassan K, Ouldim K, El Bouchikhi I, Bouguenouch L, Shukla A, Girisha KM, Sirisena ND, Dissanayake VHW, Paththinige CS, Mishra R, Kisling MS, Ferreira CR, de Herreros MB, Lee N-C, Jamuar SS, Lai A, Tan ES, Ying Lim J, Wen-Min CB, Gupta N, Lotz-Esquivel S, Badilla-Porras R, Hussen DF, El Ruby MO, Ashaat EA, Patil SJ, Dowsett L, Eaton A, Innes AM, Shotelersuk V, Badoe Ë, Wonkam A, Obregon MG, Chung BHY, Trubnykova M, La Serna J, Gallardo Jugo BE, Chávez Pastor M, Abarca Barriga HH, Megarbane A, Kozel BA, van Haelst MM, Stevenson RE, Summar M, Adeyemo AA, Morris CA, Moretti-Ferreira D, Linguraru MG, Muenke M, Williams-Beuren syndrome in diverse populations, Am. J. Med. Genet. Part A 176, 1128–1136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowsett L, Porras AR, Kruszka P, Davis B, Hu T, Honey E, Badoe E, Thong M-K, Leon E, Girisha KM, Shukla A, Nayak SS, Shotelersuk V, Megarbane A, Phadke S, Sirisena ND, Dissanayake VHW, Ferreira CR, Kisling MS, Tanpaiboon P, Uwineza A, Mutesa L, Tekendo-Ngongang C, Wonkam A, Fieggen K, Batista LC, Moretti-Ferreira D, Stevenson RE, Prijoles EJ, Everman D, Clarkson K, Worthington J, Kimonis V, Hisama F, Crowe C, Wong P, Johnson K, Clark RD, Bird L, Masser-Frye D, McDonald M, Willems P, Roeder E, Saitta S, Anyane-Yeoba K, Demmer L, Hamajima N, Stark Z, Gillies G, Hudgins L, Dave U, Shalev S, Siu V, Gupta N, Kabra M, Ades A, Dubbs H, Raible S, Kaur M, Salzano E, Jackson L, Deardorff M, Kline A, Summar M, Muenke M, Linguraru MG, Krantz ID, Cornelia de Lange syndrome in diverse populations, Am. J. Med. Genet. Part A 179, 150–158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurovich Y, Hanani Y, Bar O, Nadav G, Fleischer N, Gelbman D, Basel-Salmon L, Krawitz PM, Kamphausen SB, Zenker M, Bird LM, Gripp KW, Identifying facial phenotypes of genetic disorders using deep learning, Nat. Med. 25, 60–64 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Gurovich Y, Hanani Y, Bar O, Fleischer N, Gelbman D, Basel-Salmon L, Krawitz P, Kamphausen SB, Zenker M, Bird LM, Gripp KW, DeepGestalt - Identifying Rare Genetic Syndromes Using Deep Learning, (2018) (available at http://arxiv.org/abs/1801.07637). [DOI] [PubMed]
- 20.Lumaka A, Cosemans N, Lulebo Mampasi A, Mubungu G, Mvuama N, Lubala T, Mbuyi-Musanzayi S, Breckpot J, Holvoet M, de Ravel T, Van Buggenhout G, Peeters H, Donnai D, Mutesa L, Verloes A, Lukusa Tshilobo P, Devriendt K, Facial dysmorphism is influenced by ethnic background of the patient and of the evaluator, Clin. Genet. 92, 166–171 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Sun L, Fan Z, Weng X, Ye X, Long J, Fu K, Yan S, Wang B, Zhuo Y, Liu X, Lao K, Rapid detection of Down’s syndrome using quantitative real-time PCR (qPCR) targeting segmental duplications on chromosomes 21 and 11, Gene 552, 272–276 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Ye J, Janardan R, Li Q, in Advances in Neural Information Processing Systems 17, Saul LK, Weiss Y, Bottou L, Eds. (MIT Press, 2005), pp. 1569–1576. [Google Scholar]
- 23.Guyon I, Weston J, Barnhill S, Vapnik V, Gene Selection for Cancer Classification using Support Vector Machines, Mach. Learn. 46, 389–422 (2002). [Google Scholar]
- 24.Cortes C, Vapnik V, Support-vector networks, Mach. Learn. 20, 273–297 (1995). [Google Scholar]
- 25.Bradley AP, The use of the area under the ROC curve in the evaluation of machine learning algorithms, Pattern Recognit. 30, 1145–1159 (1997). [Google Scholar]
- 26.Mann H, Whitney D, On a test of wether one of two random variables is stochastically larger than the other, Ann. Math. Stat. 18, 50–60 (1947). [Google Scholar]
- 27.Christianson AL, Down syndrome in sub-Saharan Africa, J. Med. Genet. 33, 89–92 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lebese V, Aldous C, Malherbe HL, South African congenital disorders data, 2006 – 2014, South African Med. J. 106, 992–995 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Malherbe HL, Christianson AL, Aldous C, Need for services for the care and prevention of congenital disorders in South Africa as the country’s epidemiological transition evolves South African Med. J. 105, 186–188 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Living with Down’s syndrome in Kinshasa, DRC | Africa: | Al Jazeera. [Google Scholar]
- 31.Democratic Republic of Congo Overview.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The facial photographs collected for this study are not publicly available to preserve patient privacy. The facial analysis technology is not yet publicly available. Interested individuals should contact Dr. Linguraru for details (mlingura@childrensnational.org).