Summary
Microbe-derived acetate activates the Drosophila Immunodeficiency (IMD) pathway in a subset of enteroendocrine cells (EECs) of the anterior midgut. In these cells, the IMD pathway co-regulates expression of antimicrobial and enteroendocrine peptides including tachykinin, a repressor of intestinal lipid synthesis. To determine whether acetate acts on a cell surface pattern recognition receptor or an intracellular target, we asked whether acetate import was essential for IMD signaling. Mutagenesis and RNA interference revealed that the putative monocarboxylic acid transporter Tarag was essential for enhancement of IMD signaling by dietary acetate. Interference with histone deacetylation in EECs augmented transcription of genes regulated by the steroid hormone ecdysone including IMD targets. Reduced expression of the histone acetyltransferase Tip60 decreased IMD signaling and blocked rescue by dietary acetate and other sources of intracellular acetyl-CoA. Thus, microbe-derived acetate induces chromatin remodeling within enteroendocrine cells, co-regulating host metabolism and intestinal innate immunity via a Tip60-steroid hormone axis that is conserved in mammals.
Keywords: monocarboxylic acid transport, acetate, intestinal microbiota, innate immunity, IMD pathway, Tip60, histone acetyltransferase, histone acetylation, ecdysone, ecdysone receptor, Drosophila melanogaster
Graphical Abstract
eTOC blurb
Microbe-derived acetate activates the Drosophila Immunodeficiency (IMD) pathway in a subset of intestinal enteroendocrine cells (EECs), which regulates expression of antimicrobial and enteroendocrine peptides. Jugder et al. reveal that acetate induces chromatin remodeling within EECs by specifically modulating the function of the Tip60 histone acetyltransferase complex via a Tip60-steroid hormone axis that is conserved in mammals.
Introduction
Enteroendocrine cells (EECs) are rare members of the intestinal epithelium with an important role to play in host metabolism. These cells encode enteroendocrine peptides (EEPs) that are packaged in small vesicles at their basolateral surface and control many organismal functions including intestinal contractions, carbohydrate and lipid metabolism, and satiety. By responding to signals in the intestinal lumen via G protein coupled receptors (GPCRs) positioned at their luminal surface, these cells provide a critical link between the host and its intestinal microbiota (Zietek and Daniel, 2015). As further evidence that mammalian EECs respond to microbes, these cells express Toll-like receptors (TLRs) 1, 2, and 4 (Bogunovic et al., 2007). Furthermore, stimulation of a mouse enteroendocrine cell line with LPS leads to secretion of the EEP cholecystokinin and activation of the NF-κB pathway with increased production and secretion of macrophage inhibitory protein 2, tumor necrosis factor (TNF), and transforming growth factor-β. Thus, these cells have functional innate immune signaling pathways that coordinately regulate innate immune effectors and metabolism. This has led some to hypothesize the existence of an immunoendocrine axis in mammals (Bogunovic et al., 2007; Worthington, 2015).
The Drosophila intestine serves as an excellent model in which to uncover EEC signaling pathways that detect and respond to intestinal bacteria. The Drosophila intestine consists of a foregut, midgut, and hindgut (Miguel-Aliaga et al., 2018). The midgut has anterior, middle, and posterior subdivisions with EECs interspersed throughout. Similar to mammals, the varied EEPs packaged along the length of the midgut perform functions appropriate for the region in which they are found (Drucker, 2016; Veenstra, 2009; Veenstra et al., 2008). The Drosophila midgut is transected by the middle midgut, an acidic region of the intestine that prevents the passage of bacteria from the anterior midgut (AMG) to the posterior midgut (PMG) and rectum (Overend et al., 2016). The result is that the preponderance of the commensal microbiota of the fly inhabit the AMG. We have reported evidence of microbial immune surveillance and response roles that are unique to the EECs of this compartment (Kamareddine et al., 2018a; Watnick and Jugder, 2019).
The Drosophila Immunodeficiency (IMD) innate immune signaling pathway, like the tumor necrosis factor (TNF) pathway of mammals, is active in every intestinal cell type. Instead of TLR homologs, the IMD pathway is activated by three peptidoglycan receptors: cell membrane-associated PGRP-LC, cytosolic PGRP-LE, and secreted PGRP-SD (Iatsenko et al., 2016; Leone et al., 2008; Myllymaki et al., 2014). PGRP-LC and PGRP-LE consist of an N-terminal signaling domain fused to a C-terminal peptidoglycan recognition domain. Signaling through these receptors results in phosphorylation and cleavage of Relish, the Drosophila NF-κB homolog, which enters the nucleus to bind to promoters and activate the transcription of many genes including those encoding the antimicrobial peptides (AMPs) Diptericin A (DptA), Cecropin A1 (CecA1), and Defensin (Def) (Myllymaki et al., 2014).
There are three PGRP-LC isoforms: PGRP-LCa, PGRP-LCx, and PGRP-LCy (Werner et al., 2003). The ectodomains of these isoforms are unique, while the transmembrane and intracellular domains are identical. PGRP-LCx activates IMD signaling in response to polymeric peptidoglycan (Kaneko et al., 2004). PGRP-LCa contributes to IMD signaling only when in complex with PGRP-LCx and monomeric peptidoglycan but cannot bind peptidoglycan independently (Chang et al., 2006; Kaneko et al., 2004; Mellroth et al., 2005; Neyen et al., 2012). The function of PGRP-LCy is not yet established. Both peptidoglycan-dependent and independent processes promote IMD signaling via the formation of receptor aggregates or amyloids, which depend on cryptic RIP homotypic interaction domains of PGRP-LC and PGRP-LE (Kleino et al., 2017; Kleino and Silverman, 2014, 2019). Mechanisms of aggregate formation include binding of several PGRP-LCx receptors to one peptidoglycan polymer, increased expression of PGRP-LCx or PGRP-LCa, and peptidoglycan-independent proteolysis of the PGRP-LC ectodomain (Choe et al., 2005; Choe et al., 2002; Gottar et al., 2002; Kleino et al., 2017; Rus et al., 2013; Schmidt et al., 2008).
Our studies center on IMD pathway signaling in EECs of the AMG that express the EEP Tachykinin (Tk+ EECs). Tk acts on enterocytes (ECs) to repress triacylglyceride (TAG) synthesis (Song et al., 2014). Whereas Tk is expressed in EECs throughout the Drosophila intestine (Veenstra et al., 2008), only Tk+ EECs in the AMG coordinate an innate immune response to the intestinal microbiota (Kamareddine et al., 2018a). Interference with IMD pathway signaling in these cells decreases expression of Tk and AMPs. This leads to the accumulation of lipid droplets in ECs of the AMG, depletion of lipid droplets in the adipose tissue known as the fat body (FB), and hyperglycemia. These cells also control the EEP DH31, which activates AMG contractions in response to microbes entering the intestine (Benguettat et al., 2018). Based on these findings, we previously presented a model in which Tk+ EECs of the AMG coordinate a three-pronged response to intestinal microbes involving mechanical, antibacterial, and metabolic branches (Watnick and Jugder, 2019).
IMD pathway signaling in Tk+ EECs is dependent on PGRP-LC but activated by the short chain fatty acid (SCFA) acetate (Kamareddine et al., 2018a; Shin et al., 2011). Acetate is a fermentation product of the intestinal microbiota with little structural homology to peptidoglycan. Here we examined the mechanism whereby acetate activates IMD signaling. To determine whether acetate acts on a cell surface pattern recognition receptor or an intracellular target, we asked whether acetate import was essential for IMD signaling. We found that the putative monocarboxylic acid transporter Tarag was essential for enhancement of IMD signaling by dietary acetate. Mechanistic studies revealed that acetate is imported into Tk+ EECs, converted into acetyl-CoA, and used to activate expression of ecdysone-regulated genes including PGRP-LC. Acetate-induced increase in PGRP-LC expression was necessary to fully activate IMD signaling both in the presence and absence of infection. This acetate-responsive process was dependent on multiple components of the Tip60 histone acetyltransferase (HAT) complex as well as the variant histone 2A (H2Av). The mammalian nuclear estrogen and androgen receptors interact directly with the Tip60 complex in the presence of their ligand to acetylate the mammalian variant histone 2A, H2A.Z, in nearby nucleosomes, leading to transcriptional activation (Giaimo et al., 2019; Jaiswal and Gupta, 2018). Our data are consistent with a model in which the Drosophila Tip60 complex similarly interacts with the nuclear ecdysone receptor EcR, a homolog of the mammalian sex steroid receptors, to co-regulate host metabolism and innate immunity in a manner responsive to the intestinal microbiota.
Results
Identification of Tarag, a putative acetate transporter that is active in Tk+ EECs.
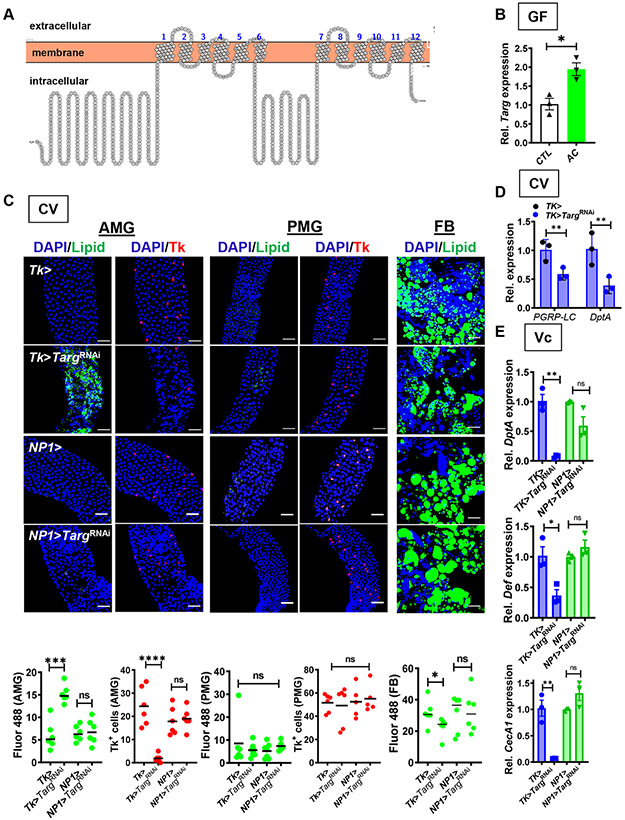
Our previous results show that microbe-derived intestinal acetate activates IMD pathway signaling in Tk+ EECs of the AMG in a PGRP-LC-dependent fashion (Kamareddine et al., 2018a). We reasoned that luminal acetate might act on the ectodomain of PGRP-LC, the cytoplasmic domain of PGRP-LC, or another regulatory protein within the cell. To differentiate between these models, it was essential to know if activation of the IMD pathway depended upon acetate entry into Tk+ EECs. In a previous transcriptomic analysis of whole flies orally infected with wild-type Vibrio cholerae or a mutant that cannot utilize acetate, we observed differential regulation of CG12866, a gene encoding a predicted monocarboxylic acid transporter (MCT) in the SLC16 family (Fig 1A) (Hang et al., 2014; Omasits et al., 2014). While the fly genome encodes 14 MCTs in this family, this particular transporter was more highly expressed in enteroendocrine cells than in other cell types of the intestine (Dutta et al., 2015). To determine if expression of CG12866 was differentially regulated by acetate, we measured its expression in microbe-depleted (GF) flies administered LB broth alone or supplemented with 50 mM acetate. As shown in Fig 1B, intestinal expression of CG12866 increased with acetate supplementation. Based on the hypothesis that this transporter imported products of microbial fermentation, we named it Tarag (Targ) after a fermented Mongolian beverage.
Figure 1: Targ RNAi in Tk+ EECs recapitulates the GF fly phenotype.
(A) Predicted structure of Targ. (B) RT-qPCR quantification of Targ in GF flies alone or supplemented with acetate (AC). (C) Representative micrographs (above) and quantification (below) of Bodipy staining (Lipid) and Tk immunofluorescence in the AMG, PMG, and FB of CV Tk>, NP1>, Tk>TargRNAi, and NP1>TargRNAi flies. (D) Quantification of PGRP-LC and DptA transcription in the intestines of CV flies with the indicated genotype. These data were collected in tandem with Fig S4C and utilize the same control. (E) Intestinal RT-qPCR quantification of DptA, Def, and CecA1 in the setting of oral V. cholerae infection of CV flies with the indicated genotype. The mean measurement is indicated. Error bars represent the standard deviation. A student’s t-test was used to evaluate significance. ns not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. See also Fig S1 and S2.
We reasoned that if Targ were involved in activation of the innate immune response by acetate, mutation, deletion, or RNAi of Targ would replicate the phenotype of a GF fly. To test this, we first examined two available transposon insertion mutants. One of these, TargMI09036, was only available as a heterozygote, suggesting that Targ may be essential. As shown in Fig S1, compared with control yw flies, the TargMB08536 homozygote but not the TargMI09036 heterozygote showed decreased Tk+ EECs and accumulation of lipid in the AMG as well as a decrease in FB lipid stores. In contrast, the abundance of Tk+ EECs and lipid droplets in the PMG remained unchanged. TargMB08536/TargMI09036 heterozygotes as well as heterozygotes carrying a TargMB08536 allele in trans with two independent chromosomal deletions yielded phenotypes similar to that of the TargMB08536 mutant (Fig S1). We hypothesized that, if acetate acts through Targ, its mutation should block rescue of the GF fly phenotype by acetate. In fact, while acetate supplementation rescued the metabolic phenotype of control yw flies, it had no effect on a TargMB08536 mutant (Fig S2).
We questioned whether Targ expression specifically in Tk+ EECs was essential for acetate rescue of the GF fly phenotype. To test this, we knocked down Targ in Tk+ EECs by combining a Tk-specific driver (Tk>) with a Targ-specific RNAi (Tk>TargRNAi). In fact, Tk>TargRNAi flies displayed an intestinal phenotype resembling that of a Targ mutant (Fig 1C). We also observed a small decrease in lipid accumulation in the FB. RNAi of Targ in ECs using the NP1> driver yielded no phenotype, suggesting a cell type-specific role (Fig 1C). We then tested whether Targ RNAi in Tk+ EECs also controlled IMD signaling. As shown in Fig 1D, Targ RNAi decreased expression of both PGRP-LC and DptA. We conclude that Targ participates in the response of Tk+ EECs to microbially-derived acetate in a cell-autonomous fashion.
Our data suggest that Targ controls intestinal IMD signaling in response to the commensal microbiota. We questioned, therefore, whether Targ might also regulate IMD signaling in the setting of intestinal infection. When flies were orally infected with the Gram-negative intestinal pathogen V. cholerae, RNAi of Targ in Tk+ EECs but not ECs decreased expression of IMD-regulated AMPs (Fig 1E). This suggests that Targ plays a role in the innate immune response to both intestinal commensals and pathogens.
Evidence that the cytoplasmic acetyl-CoA pool is critical for modulation of host lipid metabolism by microbially-derived acetate.
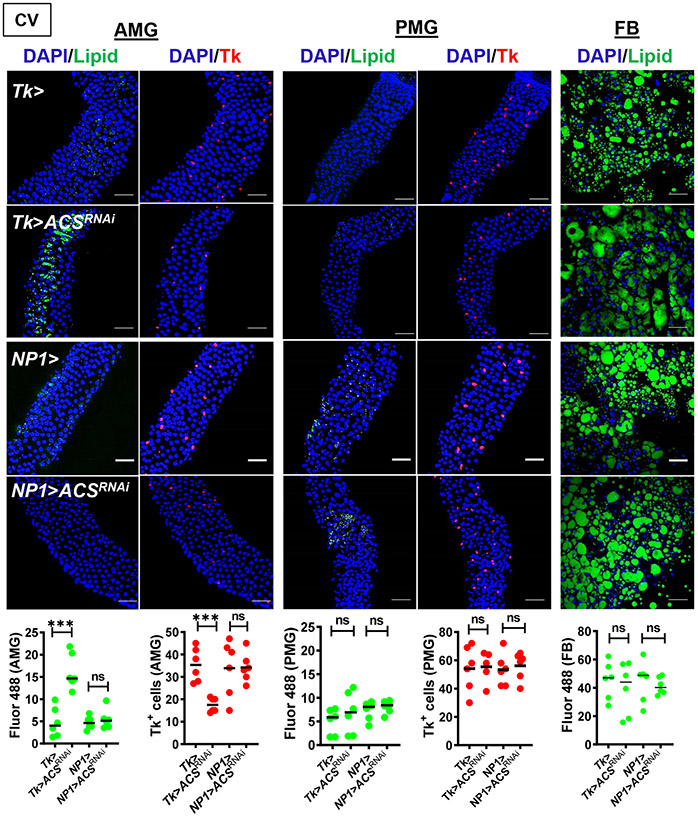
After import into cells, acetate is converted into acetyl-CoA by acetyl-CoA synthase (ACS). To further bolster our hypothesis that acetate imported by Targ acts inside the cell to mediate metabolic homeostasis, we assessed the impact of ACS RNAi. RNAi of ACS in Tk+ EECs but not ECs largely reproduced the intestinal Targ RNAi phenotype (Fig 2) but did not decrease lipid stores in the FB. In RNAi experiments, in general, we found that the FB phenotype was less pronounced. We hypothesize that this is related to the efficiency of RNAi.
Figure 2: ACS RNAi in Tk+ EECs but not ECs phenocopies the GF fly phenotype.
Representative micrographs (above) and quantification (below) of Bodipy staining (Lipid) and Tk immunofluorescence in the AMG, PMG, and FB of CV Tk>, NP1>, Tk>ACSRNAi, and NP1>ACSRNAi flies. The mean measurement is indicated. Error bars represent the standard deviation. A student’s t-test was used to evaluate significance. ns not significant, ***p<0.001.
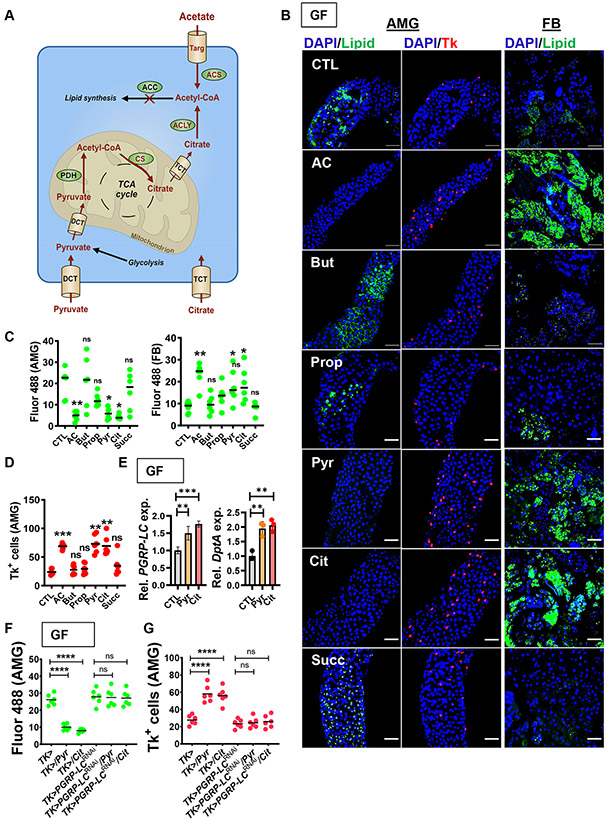
In addition to acetate, pyruvate and citrate increase intracellular acetyl-CoA pools through well-conserved metabolic pathways (Fig 3A). In mammals and presumably flies, these molecules enter cells through specific transporters. Pyruvate is then imported into mitochondria and metabolized to acetyl-CoA by the action of pyruvate dehydrogenase. Mitochondrial acetyl-CoA is converted to citrate by the first enzyme in the tricarboxylic acid (TCA) cycle, citrate synthase (CS). Citrate that enters the cytoplasm either through the cell membrane or from the mitochondria is converted to acetyl-CoA by cytoplasmic ATP-citrate lyase (ACLY).
Fig 3: Augmentation of acetyl-CoA pools by supplementation with acetate, pyruvate, or citrate rescues the GF fly phenotype.
(A) The metabolic pathways that contribute to the cytoplasmic acetyl-CoA pool. Dicarboxylic transport (DCT); Tricarboxylic transporter (TCT); Acetyl CoA synthase (ACS); Pyruvate dehydrogenase (PDH); Acetyl CoA carboxylase (ACC), Citrate synthase (CS), ATP-citrate lyase (ACLY); Aconitase (Acon). (B) Representative micrographs and (C-D) quantification of Bodipy staining (Lipid) in the AMG and FB and Tk immunofluorescence in the AMG of GF yw flies alone (CTL) or treated with acetate (AC), butyrate (But), propionate (Prop), pyruvate (Pyr), citrate (Cit), or succinate (Succ). (E) RT-qPCR quantification of PGRP-LC and DptA in the intestines of GF yw flies fed LB alone (CTL) or supplemented with pyruvate (Pyr) or citrate (Cit). Quantification of (F) Bodipy staining (Lipid) and (G) Tk immunofluorescence in the AMG of GF Tk> or Tk>PGRP-LCRNAi flies fed LB alone or supplemented with pyruvate (Pyr) or citrate (Cit). The mean measurement is indicated. Error bars represent the standard deviation. For total fluorescence and Tk+ cell quantification, a Brown-Forsythe ANOVA with a Dunnett’s T3 multiple comparisons test (C-AMG, D) or ordinary one-way ANOVA with Dunnett’s multiple comparisons test (C-FB, E, F) was used to evaluate significance. ns not significant, *p<0.05, **p<0.01, *** p<0.001. See also Fig S3-4.
We predicted that, if acetyl-CoA were a critical intermediate, we should be able to rescue the phenotype of GF hosts by dietary supplementation with pyruvate and citrate but not other carbon sources. As shown in Fig 3B-D, supplementation with pyruvate and citrate as well as acetate rescued lipid metabolism and Tk expression in the GF intestine. However, supplementation with other SCFAs, such as propionate and butyrate, and intermediates in the TCA cycle such as succinate had no effect. To further establish the parallels between rescue of the GF fly phenotype by acetate, citrate, and pyruvate, we showed that citrate and pyruvate supplementation increased transcription of PGRP-LC and DptA (Fig 3E) and that, as was previously shown for acetate, phenotype rescue by these molecules was absolutely dependent on expression of PGRP-LC (Fig 3F-G) (Kamareddine et al., 2018a).
We further hypothesized that, if acetyl-CoA pools were critical for microbe-mediated modulation of host lipid metabolism, RNAi of the genes encoding CS and ACLY might also interfere with this process. In fact, when we knocked these genes down in Tk+ EECs but not ECs, lipid accumulated, and Tk+ EECs were depleted in the AMG of conventionally-raised (CV) flies (Fig S3). These observations support our claim that increased acetyl-CoA pools in Tk+ EECs are essential for rescue of the GF phenotype.
Targ is specific for monocarboxylic acids.
We show that dietary supplementation with the dicarboxylate pyruvate and the tricarboxylate citrate rescues the GF state. We reasoned that if pyruvate and citrate entered the cell via transporters other than Targ, supplementation with these carbon sources should rescue the metabolic phenotype of the TargMB08536 mutant. As shown in Fig S4A, supplementation with pyruvate and citrate but not acetate rescued the TargMB08536 mutant phenotype. The similarities between acetate, citrate, and pyruvate rescue of the GF fly, the involvement in this rescue of intracellular enzymes that convert these three molecules to acetyl-CoA, the homology of Targ to MCTs, and the observation that Targ blocks the action of acetate but not pyruvate or citrate strongly support our contention that Targ functions as an acetate transporter in the pathway under study.
Increased acetyl-CoA pools activate IMD signaling.
Luminal acetate amplifies PGRP-LC expression and activates IMD signaling in a PGRP-LC-dependent fashion (Kamareddine et al., 2018a). To demonstrate that ACS and ACLY, proteins that are predicted to increase acetyl-CoA pools, also modulate intestinal PGRP-LC expression and IMD pathway signaling, we measured PGRP-LC and DptA transcription in flies with ACS and ACLY knocked down in Tk+ EECs. As shown in Fig S4B, RNAi of ACS and ACLY in these cells decreased transcription of DptA, CecA1, and PGRP-LC in the intestine. ACLY RNAi also led to a decrease in AMP expression in the setting of V. cholerae infection (Fig S4C).
IMD-regulated AMPs are principally transcribed in enterocytes of the AMG (Dutta et al., 2015) but controlled by a signaling cascade in Tk+ EECs (Kamareddine et al., 2018a). Because Tk is a secreted, diffusible signal, we questioned whether Tk itself might control AMP expression in the intestine. To test this, we knocked down Tk in Tk+ EECs and measured expression of DptA. As shown in Fig S4D, RNAi of Tk in Tk+ EECs reduced expression of DptA approximately 5-fold. This suggests the hypothesis that Tk functions as a cytokine in the intestinal innate immune response.
Histone deacetylases (HDACs) are involved in microbial modulation of host lipid metabolism.
Our observations support a model in which the cytoplasmic acetyl-CoA pool in Tk+ EECs is critical for microbial modulation of IMD pathway signaling and host lipid metabolism. Two cellular functions that utilize acetyl-CoA are fatty acid synthesis by acetyl-CoA carboxylase (ACC) and post-translational protein acetylation. We first ruled out a role for ACC in the metabolic response to acetate by RNAi of ACC in Tk+ EECs and ECs of CV flies (Fig S5A). As predicted, RNAi of ACC in either of these cell types had no effect on AMG lipid accumulation or Tk expression. In contrast, RNAi of ACC in the ECs of GF flies prevented lipid accumulation but had no effect on Tk expression (Fig S5B). This is consistent with the previous observation that the intestinal lipid accumulation that accompanies Tk depletion is the result of deregulated lipid synthesis (Song et al., 2014).
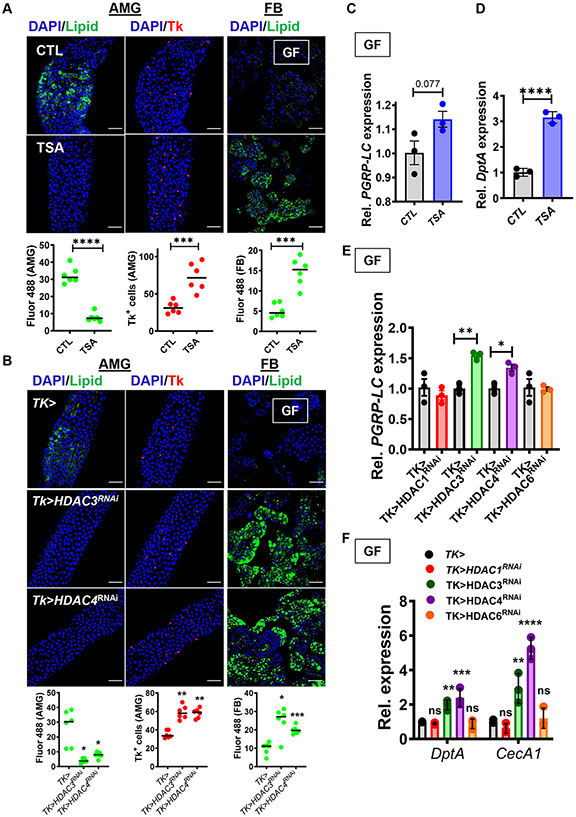
Acetyl-CoA pools have previously been linked to histone acetylation in yeast and mammalian intestinal cells (Takahashi et al., 2006; Wellen et al., 2009). To evaluate the role of histone acetylation in intestinal lipid homeostasis, we first administered the HDAC inhibitor trichostatin A (TSA) to GF flies as previously described (Tao et al., 2004). As shown in Fig 4A, TSA rescued the intestinal phenotype of GF flies and replenished FB stores. This suggests that histone acetylation promotes lipid homeostasis in a GF environment. We hypothesized that, if histone acetylation were important specifically in Tk+ EECs, we could also restore intestinal lipid homeostasis in GF flies by knocking down HDAC expression in these cells. We pursued EEC-specific RNAi of the Drosophila HDACs 1,3,4, and 6. RNAi of HDACs 3 and 4 but not 1 and 6 reduced intestinal lipid stores and increased lipid accumulation in the FB (Fig 4B and Fig S5C). Furthermore, overexpression of HDAC3 in CV flies recapitulated the metabolic phenotype of the GF fly (Fig S5D). This strongly suggests that the acetyl-CoA pool in Tk+ EECs acts in concert with histone acetylation to modulate intestinal lipid metabolism.
Fig 4: Inhibition of protein deacetylation inhibits accumulation of TAG and activates IMD pathway signaling in the GF fly intestine.
Representative micrographs (above) and quantification (below) of Bodipy staining (Lipid) and Tk immunofluorescence in the AMG and FB of (A) GF yw flies fed LB broth alone (CTL) or supplemented with trichostatin A (TSA), and (B) GF Tk> and Tk>HDAC3RNAi and Tk>HDAC4RNAi flies. This data was acquired in tandem with the data presented in Fig S5C and uses the same driver control. RT-qPCR quantification of (C) PGRP-LC and (D) DptA in GF yw flies fed LB broth alone (CTL) or supplemented with trichostatin A (TSA). RT-qPCR quantification of (E) PGRP-LC and (F) DptA and CecA1 in GF Tk> flies or with Tk> specific HDAC RNAi as noted. The mean measurement is indicated. Error bars represent the standard deviation. A student’s t-test (A, C, D), a Brown-Forsythe ANOVA with a Dunnett’s T3 multiple comparisons test (B) or an ordinary one-way ANOVA with Dunnett’s multiple comparisons test (E) was used to evaluate significance. ns not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. See also Fig S5.
Histone deacetylases decrease IMD pathway signaling.
Acetate rescue of lipid metabolism depends on activation of IMD pathway signaling through PGRP-LC (Kamareddine et al., 2018a). We hypothesized that if acetate rescue of the GF fly phenotype depended on histone acetylation, IMD pathway activation by acetate should also depend on histone acetylation. In fact, we found that the histone deacetylase inhibitor TSA activated transcription of PGRP-LC and DptA in the intestines of GF flies, although the effect on PGRP-LC did not reach the significance threshold (Fig 4C and D). Similarly, RNAi of HDAC3 and HDAC4 but not HDAC1 and HDAC6 in Tk+ EECs of GF flies resulted in increased transcription of PGRP-LC, DptA, and CecA1 (Fig 4E and F), and overexpression of HDAC3 in CV flies decreased DptA and CecA1 transcription (Fig S5E). This is consistent with a model in which acetate activates IMD pathway signaling through histone acetylation.
Overexpression of PGRP-LCx or PGRP-LCa rescues the metabolic phenotype of GF flies.
Acetate activation of IMD signaling in the GF Drosophila intestine and metabolic rescue is dependent on the presence of the peptidoglycan receptor PGRP-LC, and acetate ingestion leads to increased PGRP-LC expression (Kamareddine et al., 2018a). Previous investigators have shown that increased PGRP-LCx and PGRP-LCa but not PGRP-LCy expression can result in activation of IMD signaling in the absence of peptidoglycan (Choe et al., 2005; Choe et al., 2002). To determine if this was also true in Tk+ EECs, we overexpressed PGRP-LCx and PGRP-LCa using the Tk> driver in GF flies and assessed AMP expression, lipid accumulation in ECs, and numbers of Tk+ EECs. Overexpression of PGRP-LCx and PGRP-LCa led to an increase in DptA and CecA1 transcription (Fig S5 F-G), a reduction in lipid accumulation in ECs, and an increase in Tk+ EECs (Fig S5H). To determine whether expression of the PGRP-LCa isoform alone, which cannot bind peptidoglycan, was sufficient to activate this pathway, we tested previously generated strains expressing all three isoforms (PGRP-LCaxy), PGRP-LCa alone, or PGRP-LCy alone in a PGRP-LCE12 background (Neyen et al., 2012). As expected, PGRP-LCaxy was sufficient to rescue IMD signaling and intestinal homeostasis in the PGRP-LCE12 background, while no rescue was observed with PGRP-LCa or PGRP-LCy alone (Fig S5I-J). We conclude that, in uninfected CV and GF flies, PGRP-LCx and PGRP-LCa have roles to play in intestinal IMD signaling and lipid metabolism. PGRP-LCx and PGRP-LCa also function together in the context of peptidoglycan recognition, and elimination of peptidoglycan contamination is challenging (Chang et al., 2005). Therefore, while acetate is essential for activation of IMD signaling in the AMG, these experiments do not rule out an accessory role for peptidoglycan.
Acetate, histone acetylation, and ecdysone signaling collaborate in intestinal IMD pathway signaling and maintenance of metabolic homeostasis.
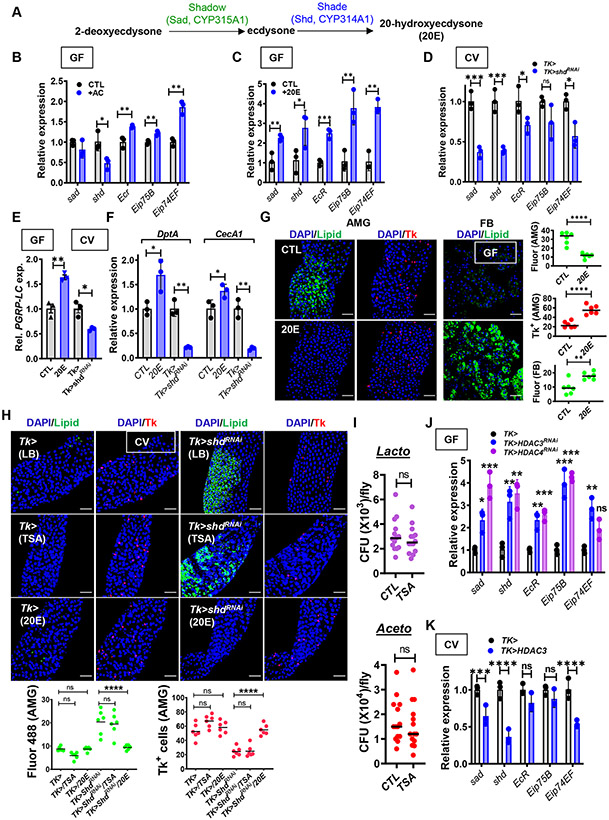
Synthesis of the steroid hormone ecdysone from cholesterol is a multi-step process in which the final four steps are carried out by a series of mitochondrial cytochrome P450 proteins encoded by the Halloween genes (Gilbert, 2004). The final two steps are carried out by Shadow (Sad), which converts 2-deoxyecdysone to ecdysone, and Shade (Shd), which converts ecdysone to its most active form, 20-hydroxyecdysone (20E) (Fig 5A). The ecdysone receptor EcR and ultraspiracle (USP) are transcription factors located in the nucleus that bind to ecdysone response elements (Ec-REs) most commonly through heterodimer formation (Koelle et al., 1991; Thomas et al., 1993). While ecdysone binding is not necessary for association of EcR and USP with DNA at every Ec-RE, it may stabilize the interaction of the complex with DNA (Thomas et al., 1993). Upon binding of ecdysone, this heterodimer activates transcription of many genes, some of which are best known for their involvement in development including ecdysone-induced proteins 75B (Eip75B) and 74EF (Eip74EF) (Xu et al., 2020). Like acetate, 20E also increases PGRP-LC transcription (Rus et al., 2013; Zheng et al., 2018). Therefore, we questioned whether acetate supplementation might increase ecdysone-dependent regulation in the gut (Kamareddine et al., 2018a). To test this, we supplemented GF flies with acetate or 20E and measured the change in transcription of 20E biosynthetic and ecdysone-regulated genes. Supplementation with acetate and 20E resulted in overlapping but distinct patterns of expression. Both acetate and 20E supplementation increased transcription of EcR, Eip75B, and Eip74EF in GF flies (Fig 5B and C). In contrast, while 20E increased transcription of the 20E synthesis genes sad and shd, acetate did not have a significant effect on sad and decreased transcription of the Halloween gene shd. RNAi of shd in Tk+ EECs of CV flies decreased transcription of the same set of genes activated by 20E supplementation (Fig 5D). Based on these data, we hypothesize that acetate increases transcription of 20E-regulated genes by a mechanism other than activation of 20E synthesis in Tk+ EECs. One possibility is that acetate acts downstream of 20E synthesis to increase transcription of 20E-dependent genes and, therefore, results in negative feedback on 20E synthesis via transcriptional repression of shd.
Fig 5: Acetate, histone acetylation, and ecdysone-dependent regulation are essential for IMD pathway signaling and maintenance of metabolic homeostasis.
(A) Positions of Shadow (Sad) and Shade (Shd) in the 20E synthesis pathway. (B-D) RT-qPCR quantification of the 20E-regulated genes sad, shd, EcR, Eip75B, and Eip74F in (B) GF yw flies alone (CTL) or supplemented with acetate (+AC), (C) GF yw flies alone (CTL) or supplemented with 10 μM 20E (+20E) or (D) CV Tk> and Tk>shdRNAi flies. (E) RT-qPCR quantification of PGRP-LC in GF yw flies alone (CTL) or supplemented with 10 μM 20E, and in Tk> and Tk>shdRNAi CV flies. (F) RT-qPCR quantification of DptA and CecA1 in GF yw flies alone (CTL) or supplemented with 10 μM 20E (+20E) and in CV Tk> and Tk>shdRNAi flies. (G) Representative micrographs (left) and quantification (right) of Bodipy staining (Lipid) and Tk immunofluorescence in the AMG and Bodipy staining (Lipid) of the FB of GF flies alone (CTL) or supplemented with 10 μM 20-hydroxyecdysone (20E). (H) Representative micrographs and quantification of Bodipy staining (Lipid) and Tk immunofluorescence in the AMG of CV Tk> and Tk>shdRNAi flies alone or supplemented with 10 μM trichostatin A (TSA) or 20E. (I) Quantification of colony-forming units (CFU) of Lactobacillus and Acetobacter species in the intestines of CV yw flies alone (CTL) or supplemented with 10 μM trichostatin A (TSA). RT-qPCR quantification of sad, shd, EcR, Eip75B, and Eip74F in (J) GF Tk>, Tk>HDAC3RNAi, and Tk>HDAC4RNAi flies and (K) CV Tk> and Tk>HDAC3 flies. The mean measurement is indicated. Error bars represent the standard deviation. Significance was assessed using a student’s t-test in all measurements except those in (H-I) where a one-way ANOVA with Dunnett’s multiple comparisons test was used. ns not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
In S2 cells, 20E administration potentiates the IMD pathway response to peptidoglycan by increasing expression of PGRP-LC (Flatt et al., 2008; Kleino and Silverman, 2014; Rus et al., 2013). Furthermore, in control flies, increased 20E synthesis in the setting of desiccation is sufficient to activate PGRP-LC transcription and IMD signaling in Malpighian tubules (Zheng et al., 2018). To demonstrate that a similar process is at work in the uninfected intestine and sufficient to activate IMD signaling, we measured the change in PGRP-LC, DptA, and CecA1 expression with 20E supplementation of GF flies or RNAi of shd in Tk+ EECs of CV flies. As shown in Fig 5 E and F, 20E supplementation of GF flies increased and shd RNAi in CV flies decreased transcription of these genes. This suggests that ecdysone-dependent transcriptional regulation in Tk+ EECs increases intestinal IMD signaling via activation of PGRP-LC transcription.
Because 20E activates IMD pathway signaling in the intestine, we questioned whether it might also rescue the metabolic phenotype of GF flies. In fact, when we supplemented GF flies with 20E, lipid stores in the gut were decreased, numbers of Tk+ EECs increased, and lipid stores in the FB were replenished (Fig 5G). Furthermore, the metabolic phenotype of Tk>shdRNAi CV flies recapitulated that of GF flies and could be rescued by 20E supplementation but not TSA (Fig 5H). Importantly, TSA supplementation had no effect on the abundance of the commensal microbiota of CV flies (Fig 5I).
Histone acetylation increases transcription of 20E synthesis and effector genes.
Because TSA did not rescue the metabolic phenotype of a shd RNAi fly, we hypothesized that histone acetylation might act in tandem with 20E. Therefore, we measured the impact of HDAC3 and 4 RNAi or HDAC3 overexpression on transcription of 20E synthesis and effector genes. As shown in Fig 5J and K, RNAi of these HDACs in GF flies increased transcription and HDAC3 overexpression in CV flies decreased transcription of 20E synthesis and target genes. These results suggest that histone deacetylases alter expression of 20E synthesis genes and 20E regulatory targets in the Drosophila intestine. Because our data also showed that histone deacetylation modulates IMD pathway signaling, we hypothesized that histone acetylation and 20E might function in the same pathways to modulate IMD pathway signaling.
Ecdysone regulation and histone acetylation are upstream of IMD pathway signaling.
We showed that acetate uptake, histone deacetylation, and ecdysone synthesis activate transcription of IMD pathway-regulated genes and reduce intestinal lipid accumulation but not how these processes might be positioned in a common regulatory pathway. To test this, we performed an epistasis experiment in which we compared the phenotypes of a driver-only control fly (Tk>) and flies with EEC-specific RNAi of Targ, PGRP-LC or PGRP-LE alone or with dietary supplementation with TSA or 20E. A Rel RNAi fly was included as a negative control. As expected, the AMGs of all untreated RNAi flies demonstrated accumulation of lipid droplets and a decrease in Tk+ cells (Fig S6). Interference with histone deacetylation by TSA rescued PGRP-LERNAi RNAi flies, confirming that PGRP-LE is not in the regulatory pathway under study (Kamareddine et al., 2018a). TSA supplementation only partially rescued CV Tk>TargRNAi flies. This likely represents a low level of acetylation in the absence of Targ. 20E supplementation completely rescued Tk>TargRNAi and Tk>PGRP-LERNAi but not Tk>PGRP-LCRNAi or Tk>RelRNAi flies. These results show that 20E supplementation can compensate for a paucity of intracellular acetate and suggest that histone deacetylation and 20E-dependent transcriptional activation are upstream of PGRP-LC in IMD pathway signaling.
The dATAC complex activates transcription of 20E synthesis genes but does not participate in the intestinal response to acetate.
Because 20E synthesis and EcR-dependent transcription are extensively regulated by chromatin remodeling complexes including histone acetyltransferases (HATs) (Bodai et al., 2012; Borsos et al., 2015; Pahi et al., 2015; Pankotai et al., 2010; Yamanaka et al., 2013), we hypothesized the involvement of a HAT in activation of ecdysone signaling in response to microbially-derived acetate.
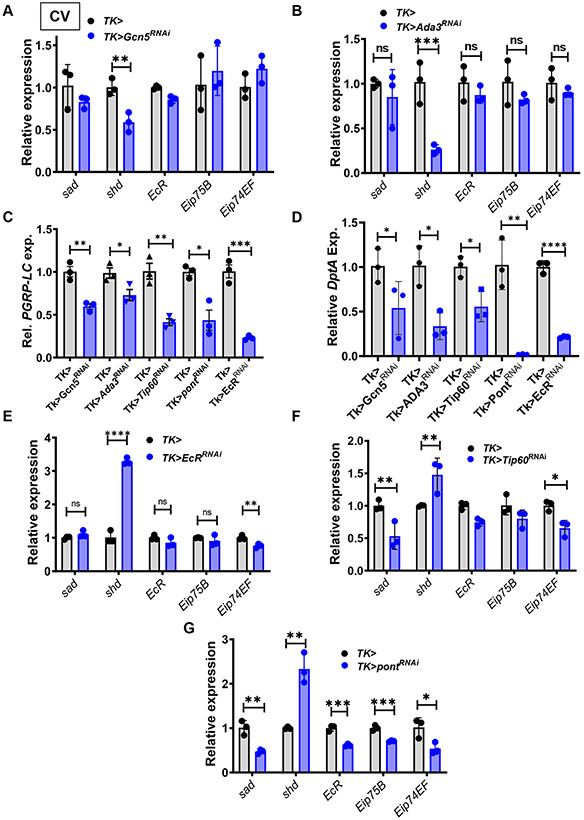
The ATAC complex, which includes the HAT Gcn5 and the adaptor protein Ada3, has previously been implicated in regulation of the Halloween genes in the prothoracic gland during development (Pankotai et al., 2010). To determine whether this could be involved in the intestinal response to acetate, we first knocked down Gcn5 and Ada3 in Tk+ EECs of CV flies and measured transcription of 20E synthesis and effector genes. RNAi of Gcn5 and Ada3 in Tk+ EECs decreased shd transcription but did not decrease overall transcription of Eip75B or Eip74EF (Fig 6A and B). One explanation for this observation is that synthesis of 20E in Tk+ cells does not contribute to overall intestinal Eip75B and Eip74EF transcriptional regulation. In addition, a small decrease in PGRP-LC transcription and DptA transcription was observed (Fig 6C and D). RNAi of Ada3 also resulted in lipid accumulation in enterocytes and a reduction in Tk+ EECs (Fig S7A). To determine whether this complex was responsive to acetate, we treated GF Tk>Ada3RNAi RNAi flies with acetate or 20E. As shown in Fig S7B, RNAi of Ada3 in Tk+ EECs did not block rescue of intestinal lipid accumulation or Tk expression by acetate or 20E. We conclude that, while the ATAC HAT complex regulates 20E synthesis in the intestine, it does not mediate the communication that occurs between the microbiota and the intestine by way of microbial acetate production. We then tested RNAi of the CREB-binding protein or CBP encoded by nejire and CG1894, a proven and a putative HAT, respectively. However, neither one of these reproduced the GF fly phenotype (Fig S7C).
Fig 6: Acetate supplementation and Tip60 RNAi in Tk+ cells result in inverse patterns of 20E and IMD-regulated gene expression in the Drosophila intestine.
RT-qPCR quantification of sad, shd, EcR, Eip75B, and Eip74F in the intestines of CV Tk>, Tk>Gcn5RNAi, and (A) Ada3RNAi (B) flies. RT-qPCR quantification of (C) PGRP-LC and (D) DptA in the intestines of CV Tk>, Tk>Gcn5RNAi, Tk>Ada3RNAi, Tk>Tip60RNAi, Tk>pontRNAi and Tk>EcRRNAi flies. RT-qPCR quantification of ecdysone-regulated genes in the intestines of CV Tk> and (E) Tk>EcRRNAi, (F) Tk>Tip60RNAi, and (G) Tk>pontRNAi flies. All experiments were performed in CV flies. The mean measurement is indicated. Error bars represent the standard deviation. Significance was assessed using a student’s t-test. ns not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
The Tip60 HAT complex activates transcription of ecdysone-regulated genes in the intestine in response to acetate.
We noted that 20E as well as the ATAC complex increased transcription of shd, while acetate supplementation repressed shd transcription. These distinct patterns of regulation support our contention that the ATAC complex does not mediate the effect of acetate and, further, suggested that acetate does not increase 20E synthesis. Many chromatin remodeling complexes interact with EcR. For instance, the lysine methyltransferase Trr interacts directly with EcR to trimethylate lysine 4 of histone 3 at Ec-RE’s (Sedkov et al., 2003). Similarly, the ATP-dependent nucleosome remodeling factor complex (NURF) interacts with EcR when ecdysone is bound and is thought to cause nucleosome sliding (Badenhorst et al., 2005). We predicted that, if acetate decreased transcription of shd and increased transcription of 20E-regulated genes through its action on EcR, RNAi of EcR in Tk+ EECs should have an effect on shd opposite to that of acetate. As shown in Fig 6C-E, shd transcription increased upon EcR RNAi, while transcription of Eip74EF, DptA, and PGRP-LC decreased. No change in overall transcription of EcR in the intestine was noted. This is likely due to the fact that EcR is highly expressed in most cells of the intestine, and Tk+ cells are only a small proportion of these (Dutta et al., 2015). The regulatory pattern associated with EcR RNAi was the inverse of that observed with acetate supplementation. Therefore, we hypothesized that acetate might activate a HAT that interacts directly with EcR.
In mammals, the Tip60 HAT complex is a well-established directly-interacting co-activator of steroid nuclear receptors (Jaiswal and Gupta, 2018). To determine if Tip60 might be essential for activation of 20E-dependent transcriptional regulation, we knocked down both Tip60 and Pontin, the ATPase in the Tip60 complex, in Tk+ EECs. We observed a similar pattern of expression for both components of the Tip60 complex, namely a decrease in sad transcription, an increase in shd transcription, and small decreases in EcR-regulated genes (Fig 6F and G). Furthermore, we noted decreases in PGRP-LC and DptA (Fig 6C-D). We conclude that the Tip60 complex increases transcription of EcR-regulated genes in Tk+ EECs, increases IMD pathway signaling, and, therefore, may act at Ec-REs to potentiate transcription.
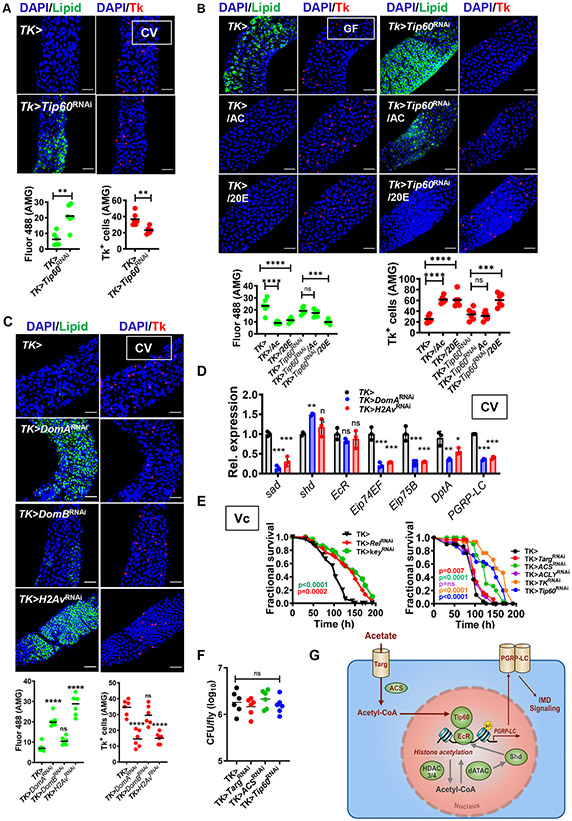
The Tip60 complex regulates lipid metabolism in the AMG.
We hypothesized that RNAi of Tip60 complex components should also recapitulate the GF lipid storage phenotype. Therefore, we assessed lipid accumulation in the intestines of Tip60 and Pontin RNAi flies. RNAi of Tip60 (Fig 7A) and pontin (Fig S7C) resulted in accumulation of lipid droplets in the AMG and a paucity of Tk+ EECs. We hypothesized that if the Tip60 complex were directly responsive to acetate, the phenotype of GF Tk>Tip60RNAi flies should not be rescued by acetate. In fact, that is what was observed (Fig 7B). We previously noted that supplementation with 20E overrides a paucity of transported acetate (Fig S6). 20E similarly rescued the phenotype of GF Tip60 RNAi flies (Fig 7B).
Fig 7: Evidence that intestinal acetate acts in concert with the Tip60 HAT complex to activate IMD signaling.
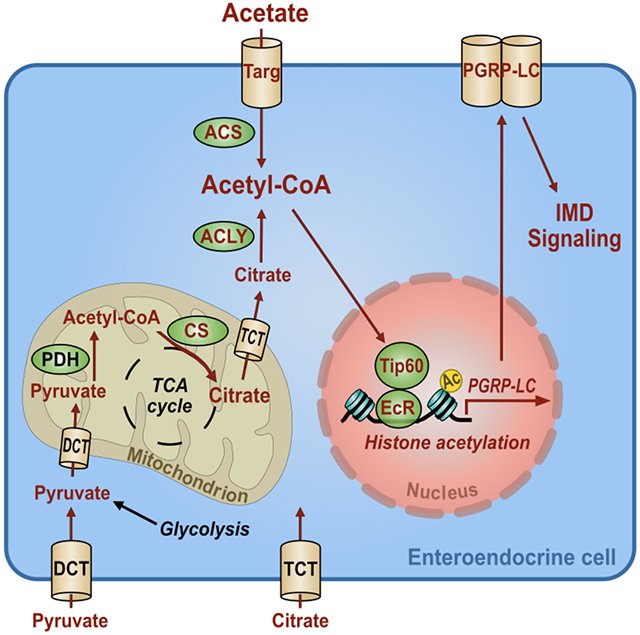
Representative micrographs and quantification of Bodipy staining (Lipid) and Tk immunofluorescence in the AMG of (A) CV Tk> and Tk>Tip60RNAi flies, (B) GF Tk> and Tk>Tip60RNAi flies fed LB alone or supplemented with 50 mM acetate (AC) or 10 μM 20E (These data were collected in tandem with that shown in Fig S7B and utilize the same control), or (C) CV Tk>, Tk>DomARNAi, Tk>DomBRNAi, and Tk>H2AvRNAi flies. (D) RT-qPCR quantification of the indicated transcripts in CV Tk>, Tk>DomARNAi, and Tk>H2AvRNAi flies. (E) Fractional survival of flies with the indicated genotypes continuously fed V. cholerae. (F) A model of the proposed signal transduction pathway required for activation of innate immune signaling by microbially-derived acetate in the intestinal lumen. Acetate enters the enteroendocrine cell (EEC) through the MCT Tarag. It is then converted into acetyl-CoA by ACS. Acetyl-CoA acts in concert with the histone acetylase complex Tip60 and the ecdysone receptor EcR to activate transcription of PGRP-LC through acetylation of H2Av. This increases signaling through the IMD pathway. For A-D and F, the mean measurement is indicated. Error bars represent the standard deviation. Significance was assessed using either a student’s t-test (A) or a one-way ANOVA with Dunnett’s multiple comparisons test (B-D and F). ns not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. For survival assays, statistically significant differences with respect to a driver-only control were calculated by log-rank analysis. p values are color-coded. See also Fig S7.
The metabolic pathways of acetate, pyruvate, and citrate intersect at acetyl-CoA production, and we have shown that pyruvate and citrate but not acetate can rescue the phenotype of a GF Targ mutant because these carboxylic acids enter the cell through distinct transporters (Fig 3A and S4A). Neither acetate, citrate nor pyruvate could rescue GF Tip60 RNAi flies (Fig S7D). We, therefore, conclude that acetyl-CoA acts in concert with the Tip60 complex to potentiate 20E-dependent transcription at Ec-REs. This leads to PGRP-LC-dependent IMD pathway activation, Tk expression, and establishment of lipid homeostasis.
Evidence that the Tip60 complex functions in chromatin remodeling.
In order to further investigate the role of chromatin remodeling in activation of 20E-dependent genes via the action of Tip60, we examined the role of Domino. Domino (Dom) is a component of the Tip60 complex with homology to the SWI/SNF family of proteins, which remodel chromatin by means of a DNA-dependent ATPase domain (Ruhf et al., 2001). There are two Dom isoforms A and B. DomB participates in a complex with a chaperone function similar to that of the yeast SWR1 complex, which loads dimers comprised of H2A.Z and H2B onto histone 3 (H3)/histone 4 (H4) tetrasomes (Luk et al., 2010; Scacchetti et al., 2020). In contrast, DomA associates with the Tip60 complex, which is similar to the Nu4 complex of yeast (Kusch et al., 2004; Scacchetti et al., 2020). Nu4 acetylates the tails of H2A and H4 to accelerate loading of H2A.Z onto nucleosomes and also acetylates H2A.Z in nucleosomes to activate transcription (Giaimo et al., 2019). In Drosophila, H2Av immunoprecipitates with the Tip60 complex, and DomA has been implicated in acetylation of both H2Av and H4 (Kusch et al., 2004; Scacchetti et al., 2020). We, therefore, hypothesized that, along with Tip60 and Pontin, DomA might participate in acetylation of H2Av to activate a subset of ecdysone-regulated genes. To test this, we knocked down DomA and DomB in Tk+ EECs. As shown in Fig 7C, DomA but not DomB RNAi recapitulated the metabolic phenotype of GF flies. Furthermore, RNAi of H2Av also blocked repression of intestinal lipid synthesis by the microbiota (Fig 7C). To confirm that DomA and H2Av act similarly to other members of the Tip60 complex, we then measured the effect of DomA and H2Av RNAi on transcription of 20E synthesis and target genes as well as IMD pathway genes. In fact, RNAi of DomA and H2Av decreased sad, Eip74EF, Eip75B, PGRP-LC, and DptA transcription (Fig 7D). DomA RNAi increased transcription of shd, similar to what was observed for Tip60 and EcR RNAi. We also compared the impact of DomA and H2Av RNAi with that of other components of this pathway on TAG accumulation in whole flies and intestinal Tk transcription. As shown in Fig S7E and F, there were comparable increases in TAG stores and decreases in Tk transcription for all RNAi lines. Taken together, these observations support a model in which acetyl-CoA derived from microbial acetate is used by the Tip60 complex to acetylate H2Av. This, in turn, promotes transcription of ecdysone-regulated genes leading to activation of IMD signaling.
Acetate-activated IMD signaling modulates intestinal infection.
Our data suggest that the pathway of microbial acetate uptake and metabolism that we have uncovered activates IMD signaling in the presence of both intestinal commensals and the intestinal pathogen V. cholerae. We and others have previously demonstrated that IMD signaling increases Drosophila susceptibility to V. cholerae infection (Berkey et al., 2009; Fast et al., 2018; Fast et al., 2020; Wang et al., 2013). This effect is multi-factorial. We, therefore, questioned whether acetate signaling might also play a role in susceptibility to V. cholerae infection. Because we had not previously tested this, we first documented that RNAi of Rel and Key in Tk+ EECs decreased susceptibility to infection (Fig 7E). We then knocked down each component of the acetate response pathway using the TK> driver and appropriate RNAi and tested susceptibility of these flies to ingestion of V. cholerae. RNAi of each component of the pathway except ACLY decreased susceptibility to infection. As we have previously reported for IMD RNAi in the gut, decreased susceptibility to infection did not correlate with decreased pathogen burden (Fig 7F) (Wang et al., 2013). Rather, our previous findings suggest that lethality reflects the wasting of chronic infection and that genetic manipulations that promote lipid and carbohydrate storage or block wasting delay death (Hang et al., 2014; Kamareddine et al., 2018b). These results show that the pathway under investigation participates in the intestinal innate immune response to both commensals and pathogens and that, similar to RNAi of IMD pathway components, RNAi of components that block the intestinal response to acetate modulates susceptibility to infection.
Discussion
There are likely multiple lines of communication between the host intestine and its resident microbiota. However, the precise dialog is difficult to discern in higher organisms. Here we use the invertebrate model Drosophila melanogaster to trace the message from bacterium to host. We previously showed that microbe-derived acetate activates IMD pathway signaling in a subset of enteroendocrine cells in the AMG (Kamareddine et al., 2018a). These cells express AMPs, Tk, and DH31 in response to this activation. We hypothesize that the bactericidal activity of AMPs, increased intestinal contractions caused by DH31 signaling, and sequestration of lipids in the FB by Tk collaborate to decrease the burden of luminal and intracellular intestinal bacteria (Watnick and Jugder, 2019).
Here we provide evidence for the following mechanism underlying activation of IMD pathway signaling by intestinal acetate (Fig 7G). Acetate produced by intestinal microbes is transported into EECs by the putative MCT Targ and then converted into acetyl-CoA by ACS. While associated with the nuclear steroid receptor EcR, the Tip60 complex uses the augmented acetyl-CoA pool to acetylate H2Av at EcR-regulated promoters resulting in increased transcription of PGRP-LC and IMD signaling.
Our data suggest that metabolic sensing of acetate plays a role in the intestinal response to both commensal and pathogenic microbes. The dependence of AMG innate immune activation on products of microbial fermentation in addition to peptidoglycan may ensure immune activation only by metabolically active microbes. Furthermore, low-level immune activation by commensal metabolites may provide the host intestine with resistance against pathogen colonization. Last, we hypothesize that the synergistic impact of acetate and peptidoglycan on IMD signaling produces a robust response to florid bacterial infection.
In Drosophila, Tip60 is a global regulator of gene expression that contributes to the stress response, learning and memory, sleep, and resistance to arboviral infection (Kusch et al., 2014; Pirooznia et al., 2012; Xu et al., 2014; Yasunaga et al., 2014). It has been implicated in IMD signaling as well (Fukuyama et al., 2013). However, the latter study in S2 cells identified an interaction of the Tip60 complex with the IKK complex, while our results suggest that the Tip60 complex acts upstream of the most proximal point in the signaling cascade, the receptor PGRP-LC.
Our findings support a model in which EcR forms a complex with Tip60 and 20E at Ec-REs and activates PGRP-LC transcription via acetylation of H2Av. This is consistent with a previous report of a genetic interaction between the Tip60 complex and EcR and the observation that H2Av immunoprecipitates with Drosophila Tip60 complex (Kusch et al., 2004; Kwon et al., 2013). EcR is homologous to the mammalian family of nuclear steroid receptors. There is ample evidence that these receptors, which include the α and β1 estrogen receptors, the progesterone receptor, the androgen receptor, interact directly with the Tip60 complex to acetylate H2A.Z in nearby nucleosomes (Bakshi et al., 2017; Brady et al., 1999; Georgiakaki et al., 2006; Lee et al., 2013; van Beekum et al., 2008). The ultimate result is transcriptional activation (Jaiswal and Gupta, 2018).
Studies suggest that metabolic sensing and steroid hormones may also regulate the innate immune response of mammalian EECs. These cells express active TLRs 1, 2, and 4 and release TNF, MIP-2, and CCK in response to stimulation with specific microbe-associated ligands (Bogunovic et al., 2007). L cells, EECs that are found in regions of the intestine that house the microbiota, express estrogen receptors and respond to estrogen by releasing the EEP glucagon-like peptide 1 (Glp-1) (Handgraaf et al., 2018). This may contribute to the gender-specific differences in innate immune and metabolic responses to the commensal microbiota (Handgraaf and Philippe, 2019; Pace and Watnick, 2020). Therefore, while much remains to be elucidated, circumstantial evidence suggests that the signaling pathway that connects microbial and host metabolism in Drosophila EECs is also present in mammals.
Here we have identified an EEC-specific acetate transporter, the Tip60 complex, the steroid receptor EcR, and H2Av as components of a pathway that facilitates a dialog between the host and intestinal microbes. As might be predicted, this pathway involves innate immune signaling. The intestinal microbiota has been associated with myriad chronic host diseases including obesity, diabetes, rheumatologic illness, and even behavioral disturbance (Bundgaard-Nielsen et al., 2020; Pascale et al., 2019; Salem et al., 2019). Here we describe a mechanism in the model invertebrate host Drosophila that may accelerate our understanding of the interaction of mammalian hosts with their microbiota. With such an understanding, we may begin to design microbe-independent therapies that correct dysbiosis-induced chronic host afflictions.
Limitations of study: There are several components of our model that require further support. First of all, while we provide data implicating Targ in acetate import, we have not formally excluded the possibility that Targ functions as receptor to transduce an extracellular acetate signal. Second, we have not provided direct evidence for a physical interaction between Tip60 and EcR or acetate-induced acetylation of H2Av. Detecting these occurrences in the small numbers of Tk+ EECs of the AMG will require the development of new and more sensitive techniques and tools.
Star Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Dr. Paula I. Watnick (Paula.watnick@childrens.harvard.edu).
Material availability
This study did not generate new unique reagents.
Data and code availability
The published article includes all datasets generated or analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila Husbandry and Strains
All experimental flies were reared on standard fly food (16.5 g L−1 yeast, 9.5 g L−1 soy flour, 71 g L−1 cornmeal, 5.5 g L−1 agar, 5.5 g L−1 malt, 7.5% corn syrup and 0.4% propionic acid) and kept in incubators at 25°C, 70% humidity in a 12 hr light/dark cycle. The following control and mutant fly lines were obtained from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu/): yw (BL 1495), Targ8536 (BL 26121), Targ9036/+ (BL 51248), Targ deficiency (BL 7749 and BL 27928), RelE20 (BL 9457), PGRP-LCΔ5 (BL 36323), PGRP-LE112 (BL 33055). The following RNAi and overexpression strains were also obtained from the Bloomington Stock Center: TargRNAi (BL 57427), ACSRNAi (BL 41917), ACCRNAi (BL 34885), CSRNAi (BL 36740), ACLYRNAi (BL 65175), AconRNAi (BL 34028), HDAC1RNAi (BL 31616), HDAC3RNAi (BL 31633), HDAC3 overexpression (BL 55078), HDAC4RNAi (BL 28549), HDAC6RNAi (BL 31053), Tip60RNAi (BL 35243), PontRNAi (BL 50972), Ada3RNAi (BL 32451), Gcn5RNAi (BL 33981), NejRNAi (BL 31728), HAT (CG1894)RNAi (BL 34925), ShadeRNAi (BL 67356), EcRRNAi (BL 58286), PGRP-LERNAi (BL 60038), PGRP-LCRNAi (BL 33383), and PGRP-LCx overexpression (BL 30919), PGRP-LCa overexpression (BL 30917), TkRNAi (BL 25800), DomARNAi (BL 65873), DomBRNAi (BL 55917) and H2AvRNAi (BL 44056). The RelRNAi line (VDRC49413) was obtained from Vienna Drosophila Resource Center (http://stockcenter.vdrc.at). The Actin-Gal4, TK-Gal4 and NP1-Gal4 (Myo1A-Gal4) driver flies were kind gifts from Norbert Perrimon. The PGRP-LC mutant parental strain PGRP-LCE12 (A113) and rescue lines in this background: PGRP-LCaxy (K83), PGRP-LCa (K85), and PGRP-LCy (K86) were kind gifts from Bruno Lemaitre. RNAi of the gene targeted by each RNAi was quantified by RT-qPCR and confirmed in most cases (Table S1). Microbe-depleted flies were generated by supplementing fresh LB medium with an antibiotic cocktail of 500 μg mL−1 Ampicillin (Thermo Fisher Scientific 69-52-3), 50 μg mL−1 Tetracycline (IBI Scientific 64-75-5), 200 μg mL−1 Rifampicin (Calbiochem 557303), and 100 μg mL−1 Streptomycin (Sigma-Aldrich 3810-74-0). Where noted, 50 mM sodium acetate (Thermo Fisher Scientific 127-09-3), 10 μM trichostatin (TSA; CAYMAN, 58880-19-6) or 10 μM 20-hydroxyecdysone (20E; Sigma, H5142-5MG) were used.
METHOD DETAILS
Bioinformatics
Proposed membrane topology of Targ protein was generated with Protter software (http://wlab.ethz.ch/protter/start/).
Immunofluorescence
The midguts and fat bodies of 5- to 7-day-old female flies were dissected and fixed in PBS containing 4% formaldehyde (Thermo Fisher Scientific NC9658705) at room temperature for at least 30 min. After fixation, tissues were washed with fresh PBST (PBS supplemented with 0.1% Triton X-100 (Sigma-Aldrich 9002-93-1)) three times for 10 min, followed by incubation for 1 hr at room temperature in blocking buffer (PBS with 2% BSA and 0.1% Triton X-100). The incubation of the gut tissues with primary antibodies in PBST containing 2% BSA was performed overnight at 4°C. Samples were then washed in PBST three times for 10 min and incubated in the blocking buffer containing a fluorophore-conjugated secondary antibody, DAPI (1:1000, Invitrogen D1306) and BODIPY 493/503 (1 mg mL−1, Invitrogen D3922) for 2 hr at room temperature, and washed again in PBST three times for 10 min. The tissues were then mounted in Vectashield mounting medium (Vector Laboratories H-1000). Confocal images were acquired using a Zeiss LSM 780 confocal microscope. Primary and secondary antibodies used in this study were rabbit anti-Tk (1:500, kind gift from Jan Adrianus Veenstra) and Alexa 594 conjugated anti-rabbit secondary antibody (1:500, Thermo Fisher Scientific A-11012). Tachykinin-expressing cells in the anterior and posterior midgut were counted manually. For assessment of lipid accumulation in fat bodies or intestines, ImageJ (FIJI) was used to quantify total fluorescence. This was normalized by dividing by the total area included in the measurement. Background fluorescence was subtracted from this measurement. A minimum of 5 midguts or fat bodies were quantified for each condition and genotype.
Generation of Microbe-depleted Animals
For gut microbiota depletion, female adult flies aged 5- to 7-days-old were distributed into fly vials containing a cellulose acetate plug infiltrated with 3 mL of LB broth containing an antibiotic cocktail, consisting of 500 μg mL−1 Ampicillin (Thermo Fisher Scientific 69-52-3), 50 μg mL−1 Tetracycline (IBI Scientific 64-75-5), 200 μg mL−1 Rifampicin (Calbiochem 557303), and 100 μg mL−1 Streptomycin (Sigma-Aldrich 3810-74-0), for 3 days.
Acetate, 20-hydroxyecdysone (20E), and Trichostatin Supplementation
For oral acetate, 20E and trichostatin treatment, flies were placed in vials containing a cellulose acetate plugs infused with 3 mL of LB broth alone or supplemented with 50 mM sodium acetate (Thermo Fisher Scientific 127-09-3), 10 μM trichostatin (TSA; CAYMAN, 58880-19-6) or 10 μM 20E (Sigma, H5142-5MG) with or without antibiotics. 30 female flies were used per condition and divided evenly between three vials. Flies were exposed to plugs with fresh solutions every day for 48 hr (TSA or 20E) or 72 hr (acetate). The fly midguts were then harvested for microscopic or RT-qPCR analysis.
Triacylglyceride quantification
Three cohorts of five female flies per genotype (5-7 days old) representing independent experimental replicates were collected, removed from their food source for 1 hr, and homogenized in 100 μL Tris-EDTA buffer supplemented with 0.1% Triton X-100. Homogenates were then heat-treated at 95°C for 20 min to inactivate enzymatic activity before proceeding with triglyceride (TAG) quantification. 20 μL of homogenate was used in the following TAG assay. Following a 1 hr incubation at 37°C with either 20 μL buffer or triglyceride reagent (Sigma-Aldrich T2449) and centrifugation for 3 min at maximum speed, 30 μL of each sample was mixed with 100 μL free glycerol reagent (Sigma-Aldrich F6428). The mixtures were incubated at 37°C for 5 min, and absorbance was measured at 540 nm. For quantification, a standard curve was generated from glycerol standards (Sigma-Aldrich G7793) (0.125-0.5 mg/mL).
Vibrio cholerae infections
Oral Vibrio cholerae infections were carried out in an arthropod containment level 2 facility. Three cohorts of ten female flies per genotype (5-7 days old) representing independent experimental replicates were infected with quorum sensing-competent V. cholerae strain C6706 as follows. Fly cohorts were placed in fly vials containing cellulose acetate plugs infused with a 3 mL suspension of an overnight V. cholerae culture diluted 1:10 in LB broth. For RT-qPCR experiments, midguts were harvested after exposure to V. cholerae for 3 days. For survival assays, the number of viable animals was recorded twice daily.
For V. cholerae burden assays, randomly chosen six flies were removed from each vial following 72 hours of pathogen exposure. These were placed singly in microfuge tubes and maintained on ice until plating. After addition of 200 μl of sterile PBS, flies were homogenized, and the supernatant was serially diluted in PBS. Dilutions were plated on LB agar supplemented with 100 μg/ml streptomycin, and colony forming units per fly were assessed after overnight incubation at 27 °C.
Quantification of commensal Acetobacter and Lactobacillus sp. in the Drosophila intestine
Three cohorts of ten female flies per genotype (5-7 days old) were washed with 70% ethanol and PBST to lyse bacteria attached to the external surface of the fly, and then flies were homogenized. Serial dilutions of the homogenates were plated on deMan, Rogosa and Sharpe (MRS) plates (Sigma-Aldrich 69966). Colonies of Acetobacter and Lactobacillus sp. were identified by morphology, and the number of colony forming units (CFU) per fly was calculated.
RNA extraction and RT-qPCR
Total RNA was extracted from a minimum of 10 midguts or whole flies using TRIzol reagent (Thermo Fisher Scientific 15596026), followed by further purification using the Direct-zol RNA MiniPrep Plus kit (Zymo Research R2070), according to the supplier’s instructions. RNA was quantified using a NanoDrop 1000 Spectrophotometer. cDNA was synthesized from 500 ng of total RNA using the iScript cDNA Synthesis Kit (Bio-Rad 1708891) as described by the manufacturer. RT-qPCR was performed using iTaq SYBR Green (Bio-Rad 1725121) on a StepOnePlus real-time PCR system (Applied Biosystems). Three biological replicates were performed for each experiment, and duplicate technical measurements were included for each of the three experimental replicates. Relative expression of target genes was calculated using the comparative CT method normalized to rp49. Primers used for RT-qPCR analyses are listed in Supplementary Table S2.
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism 9.0 software was used for statistical analyses and graph generation. Measurements represent the mean of at least three biological replicates in all graphs. Error bars represent mean ± the standard deviation. As appropriate, a two-tailed students t-test, ordinary one-way ANOVA or Brown-Forsythe ANOVA with post-hoc Dunnett’s test was used to calculate significance. The statistical test applied is noted in the Figure Legend. For all tests, statistical significance was calculated as a p value. Asterisks indicate the following p value ranges: * for p <0.05, ** for p < 0.01, *** for p <0.001, **** for p <0.0001, and ns (non-significant) for p>0.05.
ADDITIONAL RESOURCES
None
Supplementary Material
Key Resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor® 594 conjugate | Thermo Fisher Scientific | Cat# A-11012 |
| Rabbit anti-Tachykinin (DTK) | Kind Gift from Jan Adrianus Veenstra | https://www.ncbi.nlm.nih.gov/pubmed/18972134 |
| Chemicals | ||
| Bacto-Agar | VWR | Cat# 214010 |
| DeMan, Rogosa and Sharpe (MRS) | Sigma-Aldrich | Cat# 69966 |
| Formaldehyde | Thermo Fisher Scientific | Cat# NC9658705 |
| Phosphate buffered saline (PBS) | TEKnova | Cat# P0191 |
| Bovine Serum Albumin | Sigma-Aldrich | Cat# A8022 |
| Triton™ X-100 | Sigma-Aldrich | Cat# 9002-93-1 |
| DAPI (4′,6-diamidino-2-phenylindole,Dihydrochloride) | Invitrogen | Cat# D1306 |
| BODIPY® 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl- 4-Bora-3a,4a-Diaza-s-Indacene) | Invitrogen | Cat# D3922 |
| Vectashield Antifade Mounting Medium | Vector Laboratories | Cat# H-1000 |
| Molecular Biology Grade Ethanol | Thermo Fisher Scientific | Cat# BP28184 |
| Tween20 | American Bioanalytical | Cat# 9005-64-5 |
| TRIzol Reagent | Thermo Fisher Scientific | Cat# 15596026 |
| Paraformaldehyde | EMS | Cat# 19208 |
| Sodium Acetate | Thermo Fisher Scientific | Cat# 127-09-3 |
| Trichostatin | CAYMAN | Cat# 58880-19-6 |
| 20-hydroxyecdysone | Sigma | Cat# H5142-5MG |
| Antibiotics | ||
| Ampicillin, Sodium Salt | Thermo Fisher Scientific | Cat# 69-52-3 |
| Tetracycline Hydrochloride | IBI Scientific | Cat# 64-75-5 |
| Streptomycin, Sulfate | Sigma-Aldrich | Cat# 3810-74-0 |
| Rifampicin | Calbiochem | Cat# 557303 |
| Critical Commercial Assays | ||
| Direct-zol RNA MiniPrep Plus | Zymo Research | Cat# R2070 |
| iScript cDNA Synthesis Kit | Bio-Rad | Cat# 1708891 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat# 1725121 |
| Triglyceride Reagent | Sigma-Aldrich | Cat# T2449 |
| Glycerol Reagent | Sigma-Aldrich | Cat# F6428 |
| Glycerol Standard | Sigma-Aldrich | Cat# G7793 |
| Experimental Models: Drosophila Strains | RNAi Stock Validation & Phenotypes (RSVP) https://www.flyrnai.org/cgi-bin/RSVP_search.pl |
||
|---|---|---|---|
| y1w1 | Bloomington Drosophila Stock Center (BDSC) | BL 1495 | |
| TargRNAi line | BDSC | BL 57427 | HMC04734 |
| Targ 8536 | BDSC | BL 26121 | |
| Targ 9036/+ | BDSC | BL 51248 | |
| Targ deficiency line | BDSC | BL 7749 and BL 27928 | |
| ACSRNAi line | BDSC | BL 41917 | HMS02314 |
| ACCRNAi line | BDSC | BL 34885 | HMS01230 |
| CSRNAi line | BDSC | BL 36740 | HMS01631 |
| ACLYRNAi line | BDSC | BL 65175 | HMC06049 |
| HDAC1RNAi line | BDSC | BL 31616 | JF01401 |
| HDAC3RNAi line | BDSC | BL 31633 | JF01420 |
| UAS-HDAC3 | BDSC | BL 55078 | |
| HDAC4RNAi line | BDSC | BL 28549 | HM05035 |
| HDAC6RNAi line | BDSC | BL 31053 | JF01501 |
| Tip60RNAi line | BDSC | BL 35243 | GL00130 |
| PontRNAi line | BDSC | BL 50972 | HMJ21078 |
| Ada3RNAi line | BDSC | BL 32451 | HMS00450 |
| Gcn5RNAi line | BDSC | BL 33981 | HMS00941 |
| NejRNAi line | BDSC | BL 31728 | HM04037 |
| HAT (CG1894) RNAi line | BDSC | BL 34925 | HMS01274 |
| ShadeRNAi line | BDSC | BL 67356 | HMC06461 |
| EcrRNAi line | BDSC | BL 58286 | HMJ22371 |
| rel E20 | BDSC | BL 9457 | |
| PGRP-LC Δ5 | BDSC | BL 36323 | |
| PGRPLE 112 | BDSC | BL 33055 | |
| PGRP-LERNAi line | BDSC | BL 60038 | HMC05031 |
| PGRP-LCRNAi line | BDSC | BL 33383 | HMS00259 |
| UAS-PGRP-LCx | BDSC | BL 30919 | |
| UAS-PGRP-LCa | BDSC | BL 30917 | |
| DomARNAi line | BDSC | BL 65873 | GLC01875 |
| DomBRNAi line | BDSC | BL 55917 | HMC04203 |
| H2AvRNAi line | BDSC | BL 44056 | HMS02773 |
| RelRNAi line | VDRC | VDRC49413 | |
| TachykininRNAi line | BDSC | BL25800 | JF01818 |
| PGRP-LC rescue lines (PGRP-LCaxy, PGRP-LCa, and PGRP-LCy in PGRP-LC E12 background) | Kind gifts from Bruno Lemaitre | N/A | |
| Actin-Gal4 | Kind gift from Norbert Perrimon | N/A | |
| Tk-Gal4 | Kind gift from Norbert Perrimon | N/A | |
| Myo1A-Gal4 | Kind gift from Norbert Perrimon | N/A | |
| Software and Algorithms | ||
|---|---|---|
| BRB-Array tools v4.5.1 | Harvard Medical School | N/A |
| Graphpad Prism 7.00 | Boston Children’s Hospital | N/A |
Highlights.
The transporter Tarag is required for the enteroendocrine cell response to acetate.
Acetate increases acetyl-CoA pools in tachykinin-expressing enteroendocrine cells.
Acetyl-CoA pools modulate the activity of Tip60 histone acetylase complex.
The Tip60 complex augments ecdysone-dependent transcription of PGRP-LC.
Acknowledgements:
This work was supported by NIH R21 AI109436 and R01 AI162701 (P.I.W.). Stocks as noted were obtained from the Bloomington Drosophila stock center (NIH P40OD018537) or provided by Dr. Bruno Lemaitre. The Tk antibody was a gift of Dr. Jan-Adrianus Veenstra. Microscopy was performed at the Microscopy Resources on the North Quad (MicRoN) core facility at Harvard Medical School. The graphical abstract was created with BioRender.com. We thank Daniela Barraza for careful reading of the manuscript and helpful suggestions.
Footnotes
Declaration of interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, and Wu C (2005). The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev 19, 2540–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi K, Ranjitha B, Dubey S, Jagannadham J, Jaiswal B, and Gupta A (2017). Novel complex of HAT protein TIP60 and nuclear receptor PXR promotes cell migration and adhesion. Sci Rep 7, 3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benguettat O, Jneid R, Soltys J, Loudhaief R, Brun-Barale A, Osman D, and Gallet A (2018). The DH31/CGRP enteroendocrine peptide triggers intestinal contractions favoring the elimination of opportunistic bacteria. PLoS Pathog 14, e1007279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkey CD, Blow N, and Watnick PI (2009). Genetic analysis of Drosophila melanogaster susceptibility to intestinal Vibrio cholerae infection. Cell Microbiol 11, 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodai L, Zsindely N, Gaspar R, Kristo I, Komonyi O, and Boros IM (2012). Ecdysone induced gene expression is associated with acetylation of histone H3 lysine 23 in Drosophila melanogaster. PLoS One 7, e40565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic M, Dave SH, Tilstra JS, Chang DT, Harpaz N, Xiong H, Mayer LF, and Plevy SE (2007). Enteroendocrine cells express functional Toll-like receptors. Am J Physiol Gastrointest Liver Physiol 292, G1770–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsos BN, Pankotai T, Kovacs D, Popescu C, Pahi Z, and Boros IM (2015). Acetylations of Ftz-F1 and histone H4K5 are required for the fine-tuning of ecdysone biosynthesis during Drosophila metamorphosis. Dev Biol 404, 80–87. [DOI] [PubMed] [Google Scholar]
- Brady ME, Ozanne DM, Gaughan L, Waite I, Cook S, Neal DE, and Robson CN (1999). Tip60 is a nuclear hormone receptor coactivator. J Biol Chem 274, 17599–17604. [DOI] [PubMed] [Google Scholar]
- Bundgaard-Nielsen C, Knudsen J, Leutscher PDC, Lauritsen MB, Nyegaard M, Hagstrom S, and Sorensen S (2020). Gut microbiota profiles of autism spectrum disorder and attention deficit/hyperactivity disorder: A systematic literature review. Gut Microbes 11, 1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, and Deisenhofer J (2006). Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science 311, 1761–1764. [DOI] [PubMed] [Google Scholar]
- Chang CI, Ihara K, Chelliah Y, Mengin-Lecreulx D, Wakatsuki S, and Deisenhofer J (2005). Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition. Proc Natl Acad Sci U S A 102, 10279–10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KM, Lee H, and Anderson KV (2005). Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci U S A 102, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KM, Werner T, Stoven S, Hultmark D, and Anderson KV (2002). Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296, 359–362. [DOI] [PubMed] [Google Scholar]
- Drucker DJ (2016). Evolving Concepts and Translational Relevance of Enteroendocrine Cell Biology. J Clin Endocrinol Metab 101, 778–786. [DOI] [PubMed] [Google Scholar]
- Dutta D, Dobson AJ, Houtz PL, Glasser C, Revah J, Korzelius J, Patel PH, Edgar BA, and Buchon N (2015). Regional Cell-Specific Transcriptome Mapping Reveals Regulatory Complexity in the Adult Drosophila Midgut. Cell Rep 12, 346–358. [DOI] [PubMed] [Google Scholar]
- Fabbiano S, Suarez-Zamorano N, and Trajkovski M (2017). Host-Microbiota Mutualism in Metabolic Diseases. Front Endocrinol (Lausanne) 8, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast D, Kostiuk B, Foley E, and Pukatzki S (2018). Commensal pathogen competition impacts host viability. Proc Natl Acad Sci U S A 115, 7099–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast D, Petkau K, Ferguson M, Shin M, Galenza A, Kostiuk B, Pukatzki S, and Foley E (2020). Vibrio cholerae-Symbiont Interactions Inhibit Intestinal Repair in Drosophila. Cell Rep 30, 1088–1100 e1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T, Heyland A, Rus F, Porpiglia E, Sherlock C, Yamamoto R, Garbuzov A, Palli SR, Tatar M, and Silverman N (2008). Hormonal regulation of the humoral innate immune response in Drosophila melanogaster. J Exp Biol 211, 2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama H, Verdier Y, Guan Y, Makino-Okamura C, Shilova V, Liu X, Maksoud E, Matsubayashi J, Haddad I, Spirohn K, et al. (2013). Landscape of protein-protein interactions in Drosophila immune deficiency signaling during bacterial challenge. Proc Natl Acad Sci U S A 110, 10717–10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiakaki M, Chabbert-Buffet N, Dasen B, Meduri G, Wenk S, Rajhi L, Amazit L, Chauchereau A, Burger CW, Blok LJ, et al. (2006). Ligand-controlled interaction of histone acetyltransferase binding to ORC-1 (HBO1) with the N-terminal transactivating domain of progesterone receptor induces steroid receptor coactivator 1-dependent coactivation of transcription. Mol Endocrinol 20, 2122–2140. [DOI] [PubMed] [Google Scholar]
- Giaimo BD, Ferrante F, Herchenrother A, Hake SB, and Borggrefe T (2019). The histone variant H2A.Z in gene regulation. Epigenetics Chromatin 12, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LI (2004). Halloween genes encode P450 enzymes that mediate steroid hormone biosynthesis in Drosophila melanogaster. Mol Cell Endocrinol 215, 1–10. [DOI] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, and Royet J (2002). The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416, 640–644. [DOI] [PubMed] [Google Scholar]
- Handgraaf S, Dusaulcy R, Visentin F, Philippe J, and Gosmain Y (2018). 17-beta Estradiol regulates proglucagon-derived peptide secretion in mouse and human alpha- and L cells. JCI Insight 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handgraaf S, and Philippe J (2019). The Role of Sexual Hormones on the Enteroinsular Axis. Endocr Rev 40, 1152–1162. [DOI] [PubMed] [Google Scholar]
- Hang S, Purdy AE, Robins WP, Wang Z, Mandal M, Chang S, Mekalanos JJ, and Watnick PI (2014). The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe 16, 592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iatsenko I, Kondo S, Mengin-Lecreulx D, and Lemaitre B (2016). PGRP-SD, an Extracellular Pattern-Recognition Receptor, Enhances Peptidoglycan-Mediated Activation of the Drosophila Imd Pathway. Immunity 45, 1013–1023. [DOI] [PubMed] [Google Scholar]
- Jaiswal B, and Gupta A (2018). Modulation of Nuclear Receptor Function by Chromatin Modifying Factor TIP60. Endocrinology 159, 2199–2215. [DOI] [PubMed] [Google Scholar]
- Kamareddine L, Robins WP, Berkey CD, Mekalanos JJ, and Watnick PI (2018a). The Drosophila Immune Deficiency Pathway Modulates Enteroendocrine Function and Host Metabolism. Cell Metab 28, 449–462 e445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamareddine L, Wong ACN, Vanhove AS, Hang S, Purdy AE, Kierek-Pearson K, Asara JM, Ali A, Morris JG Jr., and Watnick PI (2018b). Activation of Vibrio cholerae quorum sensing promotes survival of an arthropod host. Nat Microbiol 3, 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, and Silverman N (2004). Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20, 637–649. [DOI] [PubMed] [Google Scholar]
- Kleino A, Ramia NF, Bozkurt G, Shen Y, Nailwal H, Huang J, Napetschnig J, Gangloff M, Chan FK, Wu H, et al. (2017). Peptidoglycan-Sensing Receptors Trigger the Formation of Functional Amyloids of the Adaptor Protein Imd to Initiate Drosophila NF-kappaB Signaling. Immunity 47, 635–647 e636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleino A, and Silverman N (2014). The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol 42, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleino A, and Silverman N (2019). Regulation of the Drosophila Imd pathway by signaling amyloids. Insect Biochem Mol Biol 108, 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle MR, Talbot WS, Segraves WA, Bender MT, Cherbas P, and Hogness DS (1991). The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell 67, 59–77. [DOI] [PubMed] [Google Scholar]
- Koh A, De Vadder F, Kovatcheva-Datchary P, and Backhed F (2016). From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165, 1332–1345. [DOI] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR 3rd, Abmayr SM, Washburn MP, and Workman JL (2004). Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306, 2084–2087. [DOI] [PubMed] [Google Scholar]
- Kusch T, Mei A, and Nguyen C (2014). Histone H3 lysine 4 trimethylation regulates cotranscriptional H2A variant exchange by Tip60 complexes to maximize gene expression. Proc Natl Acad Sci U S A 111, 4850–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon MH, Callaway H, Zhong J, and Yedvobnick B (2013). A targeted genetic modifier screen links the SWI2/SNF2 protein domino to growth and autophagy genes in Drosophila melanogaster. G3 (Bethesda) 3, 815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Leung YK, Chung I, Tarapore P, and Ho SM (2013). Estrogen receptor beta (ERbeta1) transactivation is differentially modulated by the transcriptional coregulator Tip60 in a cis-acting element-dependent manner. J Biol Chem 288, 25038–25052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone P, Bischoff V, Kellenberger C, Hetru C, Royet J, and Roussel A (2008). Crystal structure of Drosophila PGRP-SD suggests binding to DAP-type but not lysine-type peptidoglycan. Mol Immunol 45, 2521–2530. [DOI] [PubMed] [Google Scholar]
- Li X, Watanabe K, and Kimura I (2017). Gut Microbiota Dysbiosis Drives and Implies Novel Therapeutic Strategies for Diabetes Mellitus and Related Metabolic Diseases. Front Immunol 8, 1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk E, Ranjan A, Fitzgerald PC, Mizuguchi G, Huang Y, Wei D, and Wu C (2010). Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell 143, 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellroth P, Karlsson J, Hakansson J, Schultz N, Goldman WE, and Steiner H (2005). Ligand-induced dimerization of Drosophila peptidoglycan recognition proteins in vitro. Proc Natl Acad Sci U S A 102, 6455–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel-Aliaga I, Jasper H, and Lemaitre B (2018). Anatomy and Physiology of the Digestive Tract of Drosophila melanogaster. Genetics 210, 357–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllymaki H, Valanne S, and Ramet M (2014). The Drosophila imd signaling pathway. J Immunol 192, 3455–3462. [DOI] [PubMed] [Google Scholar]
- Neyen C, Poidevin M, Roussel A, and Lemaitre B (2012). Tissue- and ligand-specific sensing of gram-negative infection in Drosophila by PGRP-LC isoforms and PGRP-LE. J Immunol 189, 1886–1897. [DOI] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Muller S, and Wollscheid B (2014). Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886. [DOI] [PubMed] [Google Scholar]
- Overend G, Luo Y, Henderson L, Douglas AE, Davies SA, and Dow JA (2016). Molecular mechanism and functional significance of acid generation in the Drosophila midgut. Sci Rep 6, 27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace F, and Watnick PI (2020). The Interplay of Sex Steroids, the Immune Response, and the Intestinal Microbiota. Trends Microbiol. doi: 10.1016/j.tim.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahi Z, Kiss Z, Komonyi O, Borsos BN, Tora L, Boros IM, and Pankotai T (2015). dTAF10- and dTAF10b-Containing Complexes Are Required for Ecdysone-Driven Larval-Pupal Morphogenesis in Drosophila melanogaster. PLoS One 10, e0142226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T, Popescu C, Martin D, Grau B, Zsindely N, Bodai L, Tora L, Ferrus A, and Boros I (2010). Genes of the ecdysone biosynthesis pathway are regulated by the dATAC histone acetyltransferase complex in Drosophila. Mol Cell Biol 30, 4254–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale A, Marchesi N, Govoni S, Coppola A, and Gazzaruso C (2019). The role of gut microbiota in obesity, diabetes mellitus, and effect of metformin: new insights into old diseases. Curr Opin Pharmacol 49, 1–5. [DOI] [PubMed] [Google Scholar]
- Pirooznia SK, Chiu K, Chan MT, Zimmerman JE, and Elefant F (2012). Epigenetic regulation of axonal growth of Drosophila pacemaker cells by histone acetyltransferase tip60 controls sleep. Genetics 192, 1327–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhf ML, Braun A, Papoulas O, Tamkun JW, Randsholt N, and Meister M (2001). The domino gene of Drosophila encodes novel members of the SWI2/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development 128, 1429–1441. [DOI] [PubMed] [Google Scholar]
- Rus F, Flatt T, Tong M, Aggarwal K, Okuda K, Kleino A, Yates E, Tatar M, and Silverman N (2013). Ecdysone triggered PGRP-LC expression controls Drosophila innate immunity. EMBO J 32, 1626–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem F, Kindt N, Marchesi JR, Netter P, Lopez A, Kokten T, Danese S, Jouzeau JY, Peyrin-Biroulet L, and Moulin D (2019). Gut microbiome in chronic rheumatic and inflammatory bowel diseases: Similarities and differences. United European Gastroenterol J 7, 1008–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacchetti A, Schauer T, Reim A, Apostolou Z, Campos Sparr A, Krause S, Heun P, Wierer M, and Becker PB (2020). Drosophila SWR1 and NuA4 complexes are defined by DOMINO isoforms. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RL, Trejo TR, Plummer TB, Platt JL, and Tang AH (2008). Infection-induced proteolysis of PGRP-LC controls the IMD activation and melanization cascades in Drosophila. FASEB J 22, 918–929. [DOI] [PubMed] [Google Scholar]
- Sedkov Y, Cho E, Petruk S, Cherbas L, Smith ST, Jones RS, Cherbas P, Canaani E, Jaynes JB, and Mazo A (2003). Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature 426, 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, and Lee WJ (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674. [DOI] [PubMed] [Google Scholar]
- Song W, Veenstra JA, and Perrimon N (2014). Control of lipid metabolism by tachykinin in Drosophila. Cell Rep 9, 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, and Boeke JD (2006). Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell 23, 207–217. [DOI] [PubMed] [Google Scholar]
- Tao D, Lu J, Sun H, Zhao YM, Yuan ZG, Li XX, and Huang BQ (2004). Trichostatin A extends the lifespan of Drosophila melanogaster by elevating hsp22 expression. Acta Biochim Biophys Sin (Shanghai) 36, 618–622. [DOI] [PubMed] [Google Scholar]
- Thomas HE, Stunnenberg HG, and Stewart AF (1993). Heterodimerization of the Drosophila ecdysone receptor with retinoid X receptor and ultraspiracle. Nature 362, 471–475. [DOI] [PubMed] [Google Scholar]
- van Beekum O, Brenkman AB, Grontved L, Hamers N, van den Broek NJ, Berger R, Mandrup S, and Kalkhoven E (2008). The adipogenic acetyltransferase Tip60 targets activation function 1 of peroxisome proliferator-activated receptor gamma. Endocrinology 149, 1840–1849. [DOI] [PubMed] [Google Scholar]
- Veenstra JA (2009). Peptidergic paracrine and endocrine cells in the midgut of the fruit fly maggot. Cell Tissue Res 336, 309–323. [DOI] [PubMed] [Google Scholar]
- Veenstra JA, Agricola HJ, and Sellami A (2008). Regulatory peptides in fruit fly midgut. Cell Tissue Res 334, 499–516. [DOI] [PubMed] [Google Scholar]
- Wang Z, Hang S, Purdy AE, and Watnick PI (2013). Mutations in the IMD pathway and mustard counter Vibrio cholerae suppression of intestinal stem cell division in Drosophila. MBio 4, e00337–00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick PI, and Jugder BE (2019). Microbial Control of Intestinal Homeostasis via Enteroendocrine Cell Innate Immune Signaling. Trends Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, and Thompson CB (2009). ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Borge-Renberg K, Mellroth P, Steiner H, and Hultmark D (2003). Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J Biol Chem 278, 26319–26322. [DOI] [PubMed] [Google Scholar]
- Worthington JJ (2015). The intestinal immunoendocrine axis: novel cross-talk between enteroendocrine cells and the immune system during infection and inflammatory disease. Biochem Soc Trans 43, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Wilf R, Menon T, Panikker P, Sarthi J, and Elefant F (2014). Epigenetic control of learning and memory in Drosophila by Tip60 HAT action. Genetics 198, 1571–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Jiang X, Denton D, and Kumar S (2020). Ecdysone controlled cell and tissue deletion. Cell Death Differ 27, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N, Rewitz KF, and O'Connor MB (2013). Ecdysone control of developmental transitions: lessons from Drosophila research. Annu Rev Entomol 58, 497–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga A, Hanna SL, Li J, Cho H, Rose PP, Spiridigliozzi A, Gold B, Diamond MS, and Cherry S (2014). Genome-wide RNAi screen identifies broadly-acting host factors that inhibit arbovirus infection. PLoS Pathog 10, e1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Rus F, Hernandez A, Kang P, Goldman W, Silverman N, and Tatar M (2018). Dehydration triggers ecdysone-mediated recognition-protein priming and elevated anti-bacterial immune responses in Drosophila Malpighian tubule renal cells. BMC Biol 16, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietek T, and Daniel H (2015). Intestinal nutrient sensing and blood glucose control. Curr Opin Clin Nutr Metab Care 18, 381–388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.