Abstract
DNA nanotechnology provides a versatile and powerful tool to dissect the structure-function relationship of biomolecular machines like the nuclear pore complex (NPC), an enormous protein assembly that controls molecular traffic between the nucleus and cytoplasm. To understand how the intrinsically disordered, Phe-Gly-rich nucleoporins (FG-nups) within the NPC establish a selective barrier to macromolecules, we built a DNA-origami NanoTrap. The NanoTrap comprises precisely arranged FG-nups in an NPC-like channel, which sits on a baseplate that captures macromolecules that pass through the FG network. Using this biomimetic construct, we determined that the FG-motif type, grafting density and spatial arrangement are critical determinants of an effective diffusion barrier. Further, we observed that diffusion barriers formed with cohesive FG-interactions dominate in mixed-FG-nup scenarios. Finally, we demonstrated that the nuclear transport receptor, Ntf2, can selectively transport model cargo through NanoTraps composed of FxFG but not GLFG Nups. Our NanoTrap thus recapitulates the NPC’s fundamental biological activities, providing a valuable tool for studying nuclear transport.
INTRODUCTION
Nuclear pore complexes (NPCs) reside in the nuclear envelope where they control the bidirectional exchange of molecules between the nucleus and cytoplasm.1–2 An individual NPC is composed of ~30 proteins (nucleoporins or nups) that build an 8-fold radially symmetric scaffold that houses a 40–50 nm wide central channel.2–3 The channel is filled with intrinsically disordered nups rich in phenylalanine (F) and glycine (G) amino acids in repetitive motifs.2–4 These “FG-nups” control the passage of molecules through both passive and active mechanisms that ultimately serve to ensure proper nuclear-cytoplasmic compartmentalization.5 First, they establish a diffusion barrier that restricts the passive diffusion of macromolecules larger than ~40 kD.6–8 Second, they provide binding sites for nuclear transport receptors (NTRs)9 that rapidly ferry cargo molecules through the NPC, with energy and directionality contributed by the Ran GTPase.10–11 A mechanistic understanding of the NPC’s gatekeeping function is not only essential to the study of cellular compartmentalization, but also provides design inspirations for synthetic macromolecule-sorting machines.
Work over the last few decades has sought to conceptualize the FG-nups as forming higher-order assemblies bearing a physicochemical resemblance to “hydrogels”,12–14 “polymer brushes”15–19 and most recently, liquids.20 Such models differ based largely on the relative importance attributed to cohesive interactions between FG-nups, which have been observed in vitro8, 12–13 but may be mitigated in vivo due to non-specific competition.19 Some in silico models suggest that cohesive interactions among the FG network exist in a regime that allows for reversible condensation, a putative key element of the gating mechanism.21 The lack of consensus here is also driven by the observations that cohesive and non-cohesive nups occupy different regions of the central channel, which may suggest discrete local environments that may or may not impact the entire collective.22–24 Such local FG-networks may also help to explain why mutations of select FG motifs can differentially impact specific NTR-transport pathways.8, 16 Exploring the potential function of FG-nups requires more elaborate NPC mimics, which so far have been limited to lining nanometer-sized artificial channels with a single type of FG-nup.25–27 As impressive as the current NPC mimics are, it remains challenging to delineate the contributions of precise numbers of different individual FG-nups, in particular their amino-acid compositions and positioning within the nuclear pore, to the overall permeability and selectivity of the FG-nup collective that makes the NPC function.
To reveal the structure-function relationship of the NPC’s central channel, namely to address how FG-nup configurations affect the formation of a size-selective diffusion barrier, we have built a DNA-origami-based biomimetic assembly that we term NanoTrap. NanoTrap allows the precise positioning of FG-nups with diverse FG-motifs (e.g., FxFG, GLFG, FG, SAFG, and PSFG)5, 28 within a DNA channel that gate the entry of proteins into a sealed chamber (Figure 1). This work thus takes advantage of the well-defined shape, programmable assembly and the chemically addressable surface of DNA-origami structures,29–32 giving a transformative upgrade to our recently reported NuPOD (NucleoPorins Organized on DNA) platform.33 The latter (and a similar platform)34 enabled the study of the morphology and dynamics of FG-nup collectives anchored on DNA-origami channels.33 Here we systematically vary the organization of two representative yeast FG-nups, Nsp1 (FxFG-rich) and Nup100 (GLFG-rich), to study how cohesiveness, density, and spatial organization of these FG-nups impact their selective sieving behaviors in a confined NPC-channel-like space. Unlike the first-generation NuPODs, the NanoTrap has a macromolecule-trapping baseplate that allows the direct assessment of the ability of the FG-nup collectives to both filter biomolecules of different molecular weights (i.e., size selectivity) and support NTR-mediated translocation of model cargo (i.e., biochemical selectivity). Thus, the NanoTraps provide an experimental platform to assay FG-nup function within a well-controlled nanopore-like environment that models the NPC. We expect this adaptable platform to help better define the properties of the NPC central channel, and ultimately contribute to elucidating the underlying mechanisms of nuclear transport.
Figure 1.
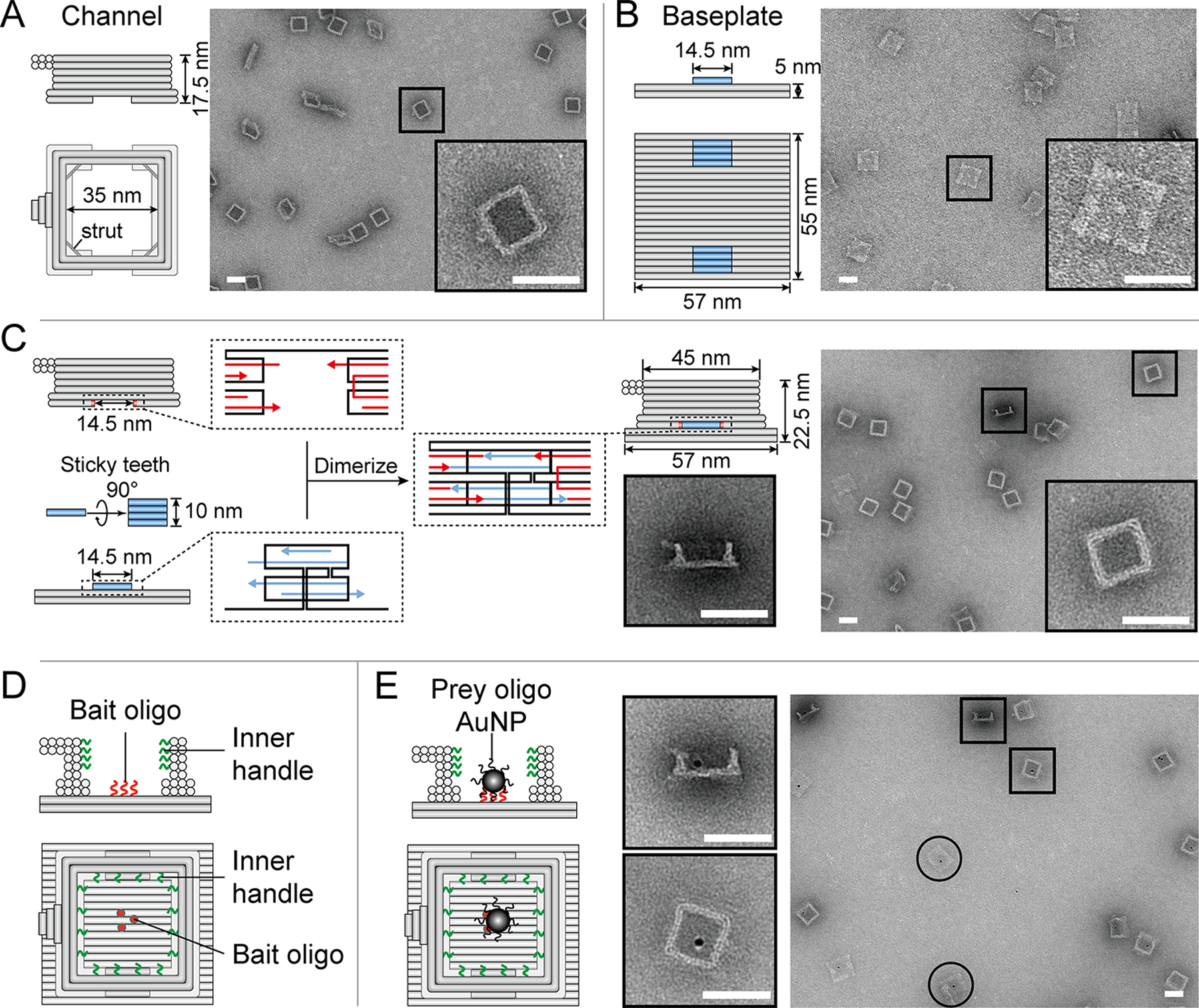
A DNA-origami NanoTrap built from two pre-assembled parts, a channel and a baseplate. (A) Cartoon models and negative-stain EM images of the channel. (B) Cartoon models and negative-stain EM images of the baseplate. (C) Two teeth (blue) with sticky ends (blue arrows) mediate the baseplate attachment onto the channel, which contains two cavities with complementary sticky ends (red arrows), resulting in the formation of a NanoTrap (cartoon model and negative-stain EM images shown on the right). (D) Up to four layers of DNA handles (12 handles per layer) protrude from the channel wall (green curls), serving as anchor points for anti-handle-conjugated proteins. The baseplate displays three “bait” oligonucleotides (red curls/dots) to capture nanoscale objects carrying “prey” oligonucleotides. (E) Cartoon models and EM images showing the immobilization of prey-oligo-modified AuNPs (dark spots) inside the NanoTrap. Circles indicate standalone baseplates. Scale bars = 50 nm.
RESULTS AND DISCUSSION
A DNA-origami NanoTrap mimics key features of the NPC central channel.
To engineer a molecular environment that recapitulates the essential structural and biochemical properties of the NPC central channel, we designed a NanoTrap consisting of two DNA-origami components: a channel and a baseplate (Figure 1 and S1). The cuboid-shaped channel has an inner width of 35 nm and a height of 17.5 nm (Figure 1A), forming the entryway of the NanoTrap. A double-stranded DNA strut is placed at each corner (near the bottom) to enhance rigidity (Figure S1). On the interior wall of the channel extend DNA oligonucleotides (handles) for hybridization with complementary DNA (anti-handles) that are conjugated to FG-nups. Each DNA channel is equipped with up to 48 handles, distributed in four layers (12 handles/layer) with radial symmetry (Figure 1D). The handle sequences are independently tunable, allowing us to decorate designated positions (layers) of the channel with selected FG-nups to build different NanoTraps, which we denote according to the protein type, stoichiometry, and anchor position (e.g., Nup10024Nsp124 for a channel with top two layers of handles occupied by a total of 24 copies of Nup100 and the bottom two layers by 24 copies of Nsp1).
To detect macromolecules passing through such an NPC mimic, we sealed the channel bottom by building a DNA-origami baseplate (57 nm×55 nm×5 nm, Figure 1B) with two raised stacking interfaces or ‘teeth’ (Figure 1B, blue bars). Each tooth comprises four parallel DNA helices, with a shape and sticky-end sequences complementary to a corresponding cavity on the bottom of the DNA channel that allows assembly of the complete NanoTrap (Figure 1C). The baseplate displays three single-stranded DNA extensions (termed ‘bait’ oligonucleotides) to immobilize entering molecules modified by a ‘prey’ oligonucleotide. As a result, macromolecules may only enter the NanoTrap through the entryway, subject to filtration by the FG-nups, and those that do cross the FG-nup barrier to reach the baseplate will be captured for easy detection by fluorescence-based assays.
We verified the proper formation of the DNA channel, baseplate, and the NanoTrap using well established DNA-origami assembly, purification, and characterization methods.33, 35 Gel electrophoresis (Figure S2) and negative-stain electron microscopy (EM) (Figure 1) clearly showed the correctly formed DNA-origami structures with expected shapes and dimensions. Importantly, stable NanoTraps formed with high efficiency (~80% dimerization yield) via the straight teeth and cavities, in contrast to an antecedent design of a cylindrical channel with curved stacking interfaces that led to inefficient dimerization (Figure S3). We further examined the chamber’s molecular trapping ability using gold nanoparticles (AuNPs) to mimic entering nanoscale entities. These 5 nm AuNPs were first conjugated with prey oligonucleotides (Figure S4) and then incubated with bait-displaying baseplates, either standalone (Figure S5) or pre-assembled into empty NanoTraps (without FG-nup grafting, Figure 1E & S6). Negative-stain EM confirmed that in both cases, nearly all baseplates (>97%) had AuNP attachment, demonstrating efficient capture of prey-modified molecules and good molecular accessibility to the NanoTrap baseplates.
The NPC’s central channel comprises a variety of nucleoporins with diverse FG-repeats that also differ in their propensity to interact with themselves and other FG-repeats.8, 36 Previous studies have shown that cohesive FG-nups (e.g., Nup98, the human orthologue of Nup100) are essential to the NPC’s function as a permeability barrier.13, 37 The DNA-origami-based NanoTrap enables a bottom-up approach to test the role of different FG-repeats in forming a size-selective barrier, owing to the complexity-reduced in vitro system and the exquisite control over FG-nup placement. To demonstrate the concept, we tested two common FG-motifs, GLFG, and FxFG. The more cohesive GLFG-rich domain38–39 of Nup100 (amino acids 2–610) and the less cohesive FxFG-rich domain39–40 of Nsp1 (amino acids 2–603) were produced in E.coli and affinity purified with an MBP-SUMO tag (to improve stability and solubility) and a SNAP tag (for conjugation with benzylguanine labeled DNA anti-handles). After conjugation with the DNA anti-handle, the MBP-FG-nup-domain-DNA conjugates (FG-nups hereafter for simplicity) were purified by size-exclusion chromatography: the conjugation yield was above 90% as verified by SDS-polyacrylamide gel electrophoresis (SDS-PAGE; Figure 2A and S7). The FG-nup-gated NanoTraps were assembled by incubating purified, undecorated NanoTraps (3 nM) with anti-handle-conjugated nucleoporins (~300 nM). With all 48 handles occupied, the NanoTrap reaches a grafting density of ~3 FG-nups per 100 nm2 and an FG-repeat concentration of up to 150 mM, similar to those in the NPC central channel.16, 41–42 The final assemblies were characterized by SDS-agarose electrophoresis and negative-stain EM (Figure 2B, S8 and S9). Electron micrographs that showed a top-view of Nup10048-NanoTraps were more likely to have a high-contrast protein mass occluding the channel compared to the Nsp148-NanoTraps, where the FG-nups consistently stained with less contrast and appeared as a light haze in the channel. These morphological differences were similar to those previously reported in the NuPODs.33
Figure 2.
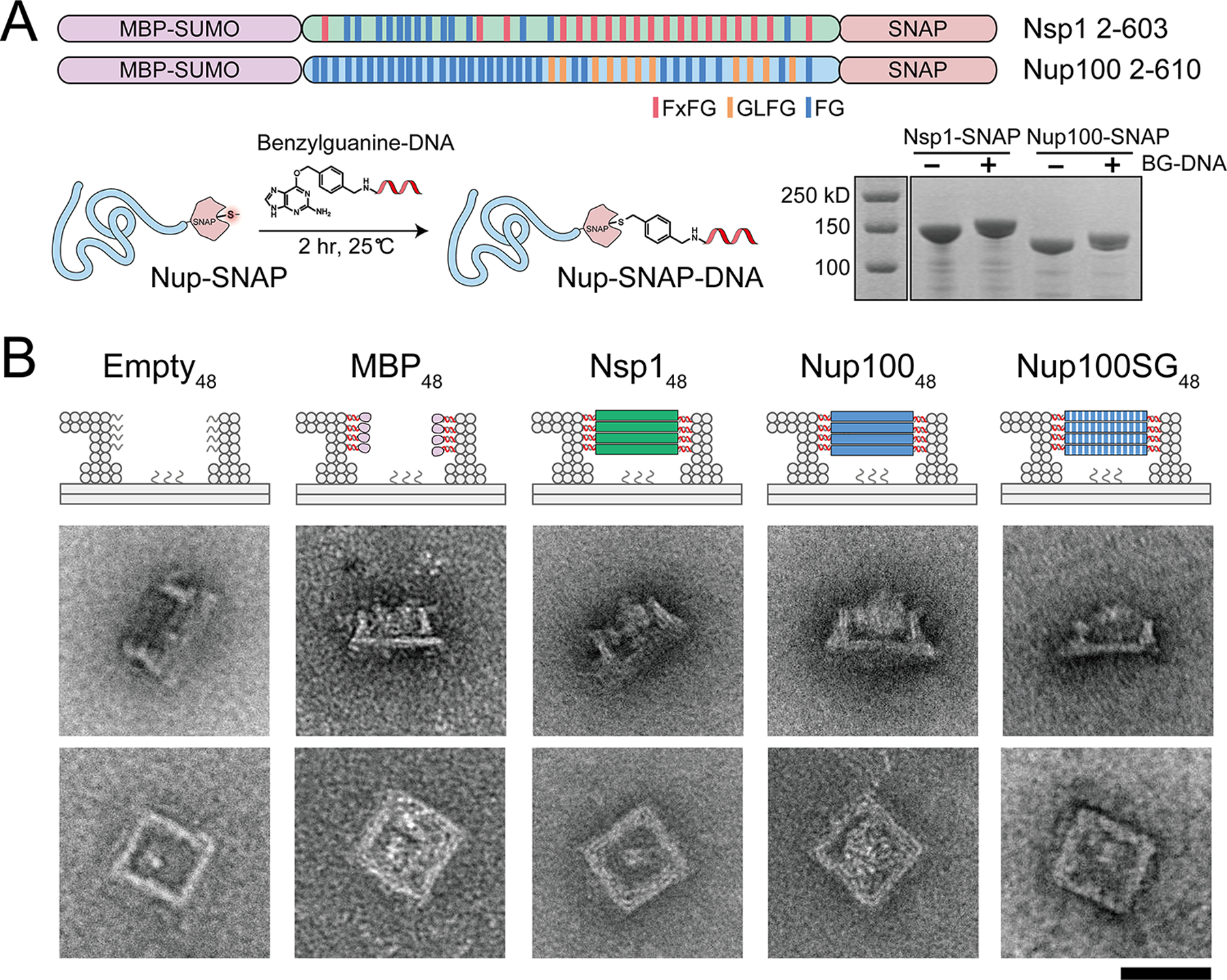
Assembly and characterization of nucleoporin-gated NanoTraps. (A) Two central channel FG-nup domains of yeast origin, Nsp1 (FxFG-rich) and Nup100 (GLFG-rich), were cloned and expressed in E.coli as MBP-SUMO-nup-SNAP fusions. Such SNAP-tag bearing nucleoporins were conjugated with benzylguanine modified DNA oligonucleotides (BG-DNA) and verified by SDS-PAGE. (B) Cartoon models (top) and negative-stain EM images (bottom) of an ungated (empty) and various protein-gated NanoTraps. Scale bar = 50 nm.
Type of FG-repeat affects the strength of diffusion barrier.
The NanoTrap design facilitates convenient ensemble measurement of the amount of macromolecules that cross the FG-nup barrier, thereby enabling systematic testing and unambiguous ranking of barrier strengths. We started by testing the diffusion barriers formed by the different FG-nups by incubating the NanoTraps (3 nM) with a GFP-tagged prey-oligo-conjugated protein (GFP-SNAP-prey, 53 kD, 1 μM), which would be immobilized on the baseplate upon entering the trap (Figure 3A). A given barrier’s permeability was determined by separating the reaction mixture by gel electrophoresis and normalizing the NanoTrap’s GFP fluorescence (from the trapped molecules) against its ethidium bromide fluorescence. We first ensured that the GFP-SNAP-prey conjugate had unfettered access to the baseplate when no FG-nups decorated the channel. Indeed, after incubation with the GFP-SNAP-prey conjugate (Figure S10), the undecorated trap exhibited clear GFP fluorescence (Figure 3B), which we normalized to the trap’s ethidium bromide fluorescence to account for loading errors and set as the maximal penetration (100%) for subsequent quantification of barrier permeability. For example, we found that 48 copies of the maltose-binding proteins (MBP, ~40 kD) did not hinder access of the 53 kD protein, as the MBP48 channel allowed nearly as much (~95%) GFP-binding on the baseplate as an empty channel did (Figure 3B). In contrast, the NanoTrap with 48 copies of Nsp1 gating its entryway (Nsp148) halved the penetration of the 53 kD protein, suggesting that it imposed a barrier to this molecule. Interestingly, the Nup10048-gated trap was virtually impermeable with only ~7% penetration of the GFP-SNAP-prey reporter. These data suggest that compared with the well-folded MBPs, the intrinsically disordered FG-nups can impose diffusion barriers to at least a 53 kD protein, albeit with different characteristics. We note that such a difference in permeability is due to the intrinsic properties of the FG-nups rather than Nsp1 degradation during NanoTrap preparation and the permeability assay, as has been verified by SDS-PAGE (Figure S11). Indeed, the relative permeability of the NanoTraps could be predicted from our studies of the first-gen NuPODs, which showed that while MBP tended to occupy space near the channel wall, Nsp1 could sample volume both inside and outside of the channel and Nup100 formed a plug-like condensate.33 Thus, these morphological differences correlate with the permeability of the NanoTraps and suggest that the propensity to form a cohesive network contributes to barrier strength.
Figure 3.
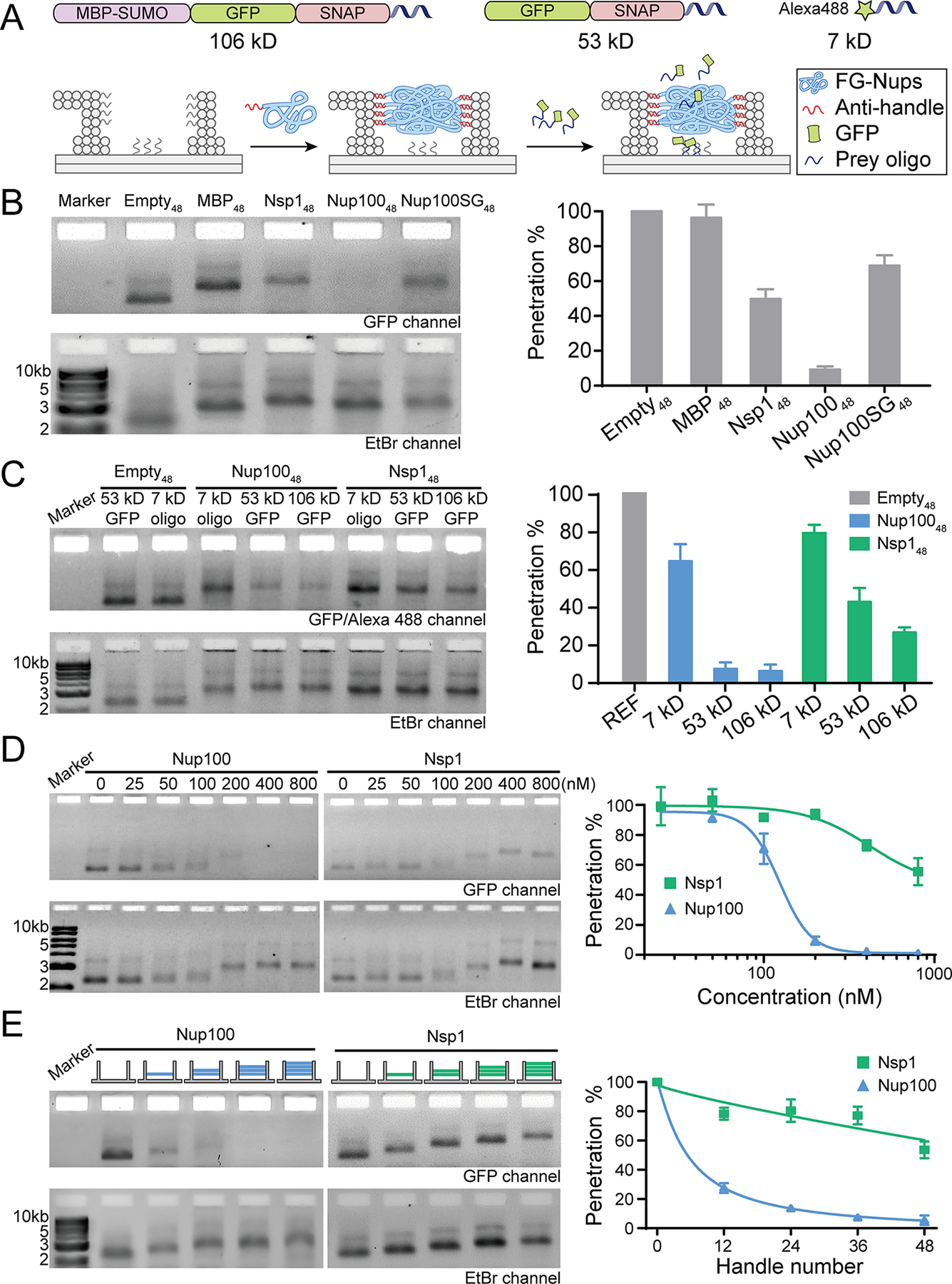
The influence of FG-nup type, density and geometric distribution on the barrier permeability. (A) Schematic diagrams showing the permeability assay using an FG-nup-gated NanoTrap and fluorescently tagged macromolecules of different sizes (106 kD MBP-GFP-SNAP-prey, 53 kD GFP-SNAP-prey, and 7 kD Alexa488-prey). (B) The barrier strengths of different gating proteins (48 copies per NanoTrap) against the 53 kD GFP-SNAP-prey. (C) Size-selective diffusion barriers formed by 48 copies of nucleoporins (Nup100 or Nsp1) within a NanoTrap. The penetration levels of the GFP-SNAP-prey (53 kD GFP) and Alexa488-prey (7 kD oligo) into empty NanoTraps were both set as 100%, serving as references (REF) for quantifying penetration levels of the GFP (53 and 106 kD) and Alexa488 labeled (7 kD) molecules into nup-gated NanoTraps, respectively. (D) Permeability of NanoTraps (4 nM, 48×handles per trap) formed at different nucleoporin (Nup100 or Nsp1) concentrations (0–800 nM), tested against the 53 kD GFP-SNAP-prey. Fitted curves are guides to the eye. (E) Permeability of NanoTraps containing 12–48 copies of Nup100 or Nsp1 tested against the 53 kD GFP-SNAP-prey. The exact nup arrangement is shown by the schematic drawing at the top of each lane (blue: Nup100, green: Nsp1, 12 nups/layer). Fitted curves are guides to the eye. Statistical data are plotted to show mean ± standard error of the mean (SEM) from three trials.
Consistent with cohesive interactions contributing to barrier strength, our results showed that the impermeability of the Nup100-NanoTraps could be overcome by disrupting cohesive interactions mediated by the GLFG repeats. We generated a NanoTrap gated by 48 copies of a Nup100 variant where every phenylalanine (F) in the 44 FG repeats was mutated to serine (S) that is known to reduce the cohesiveness of FG-repeats.13 Interestingly, while these mutations led to an increase in permeability of the Nup100SG48-NanoTraps with ~70% penetration, they were able to form a diffusion barrier with similar permeability as Nsp148 (Figure 3B). In fact, Nup100SG48-NanoTraps bear more resemblance to Nsp148 than to Nup10048 under negative-stain EM (Figure 2B and S9), confirming the cohesiveness-dependent morphology and barrier strength of the GLFG networks. Further, while GLFG-mediated cohesive interactions impact the relative strength of the diffusion barrier, FG-repeats per se are not required to impede the passage of the GFP-SNAP-prey.
GLFG and FxFG nups have unique size-selective filtering properties.
To gain more insight into the diffusion barriers established by Nsp1 and Nup100, we next tested a series of reporters that ranged from 7 kD to 106 kD (Figure 3A and 3C). Both Nsp148 and Nup10048 imposed only slight impedance to the passage of the 7 kD DNA molecule (~70–80% penetration); importantly, these data also established that the FG-nups minimally interfered with binding of the prey oligonucleotide to the baseplate. Interestingly, while Nup100-NanoTraps were essentially impermeable to both the 53 and 106 kD reporters, Nsp1-NanoTraps allowed passage of both in a manner that correlated with their molecular weights (the 53 kD and 106 kD reporters penetrated to ~50% and ~30%, respectively). Thus, while Nup100 appears to establish a diffusion barrier with a more stringent molecular weight cutoff (<53 kD), suggesting the formation of a sieve-like meshwork, Nsp1’s permeability to macromolecules is more consistent with an entropy-driven barrier, in that molecules with increasing molecular weight are met with more resistance. These results suggest the NanoTrap’s size selectivity, either “soft” or “hard”, is heavily influenced by the properties of the constituent FG-nup, making the programmable FG-nup-gated NanoTraps an ideal in vitro platform for studying the molecular underpinnings of the NPC’s size selectivity.
Densely grafted nucleoporins under spatial confinement lead to strong barriers.
In the experiments described above, we crowded 48 copies of FG-nups into the NanoTrap to match the FG-repeat concentration in a natural NPC. To understand how FG-repeat density could influence barrier properties, we performed two sets of experiments. First, we titrated 4 nM of NanoTraps bearing 48-handles with increasing concentrations (0–800 nM) of Nsp1 or Nup100 as a means to gradually increase the average FG-nup density within the NanoTraps until saturation. The resulting FG-nup-gated NanoTraps were subjected to the permeability assay (Figure 3D). Increasing FG-nup concentration led to slower mobility of the NanoTraps in the gels as well as decreased penetration of the 53 kD GFP-SNAP-prey. Notably, both the NanoTrap mobility and permeability of the Nup100-gated NanoTraps remained mostly unchanged when the Nup100 concentration was below 50 nM (i.e., on average ≤12.5 Nup100 per trap), but dropped dramatically when the Nup100 concentration was raised from 100 nM to 200 nM (i.e., on average ≥25 Nup100 per trap). Further increasing nucleoporin concentration produced diminishing improvements to the barrier strength, reducing penetration from ~10% to below the detection limit. This sharp transition suggested a possible concentration-dependent, cooperative association among the FG-nups within a nanopore — only when FG-repeats reach a minimal density does a functional barrier form. At every FG-nup concentration tested, Nsp1 formed a weaker barrier than Nup100, further supporting a cohesiveness-dependent permeability barrier.
Second, we varied the number of handles in the NanoTrap to deterministically control the FG-nup organization (Figure 3E). We thus derived eight versions of NanoTraps, each with 12, 24, 36, or 48 copies (i.e., 1–4 layers) of Nup100 or Nsp1 gating the entryway of a NanoTrap. Consistent with the data presented above, Nsp1 alone could not form a sufficient barrier to block the 53 kD GFP-SNAP-prey from entering the NanoTrap, except at the highest FG-nup densities tested where a modest impact was observed (Figure 3B–D). In contrast, merely one layer (12 copies) of Nup100 placed near the bottom of the DNA channel (Empty36Nup10012) reduced GFP-SNAP-prey penetration to ~30%. Additional Nup100 produced an enhancement of the barrier strength, with a near-complete rejection of the GFP-SNAP-prey with 24 or 36 copies of Nup100. This result is in qualitative agreement with the sharp transition of permeability observed when titrating the Nup100-to-NanoTrap ratio (Figure 3D).
Interestingly, 12× Nup100 formed a better diffusion barrier when grafted in one layer as opposed to when it was randomly distributed across all 4 layers as would be predicted to be the case in the Nup-to-NanoTrap titration experiments. These data suggest that, additional confinement endowed by spatial positioning may also play a role in the formation of a diffusion barrier. To test this hypothesis, we attached the same copy numbers of nucleoporins at different axial positions (i.e., layers) along the entryway of the NanoTrap. As shown in Figure 4A, providing there were at least 24 copies of Nup100, changing their axial positioning did not lead to an observable impact on reporter penetration. However, we observed a significant permeability difference when only 12 copies of Nup100 were grafted at either the top or bottom of the chamber, with Nup10012Empty36 showing ~70% penetration compared to Empty36Nup10012 with only ~30% penetration. We interpret these data in a model in which Nup100 near the channel opening is less likely to form a tightly-knit network because of its ability to sample larger solvent volume. We also considered the possibility that the FG-nups at the bottom layer could sterically hinder prey-bait hybridization and thus artificially inflate the barrier strength measurement of Empty36Nup10012. However, the data support that such a steric effect is negligible as the 7 kD prey oligonucleotide efficiently bound the baseplate of Nup10048 or Nsp148 (Figure 3C). Thus, despite their proximity, FG-nups near the baseplate do not substantially interfere with the prey-capturing function of the bait. Together, our data highlight the importance of geometric constraints in forming an FG-nup-based diffusion barrier, at least in one that relies on cohesive interactions, which are likely favored under confinement.
Figure 4.
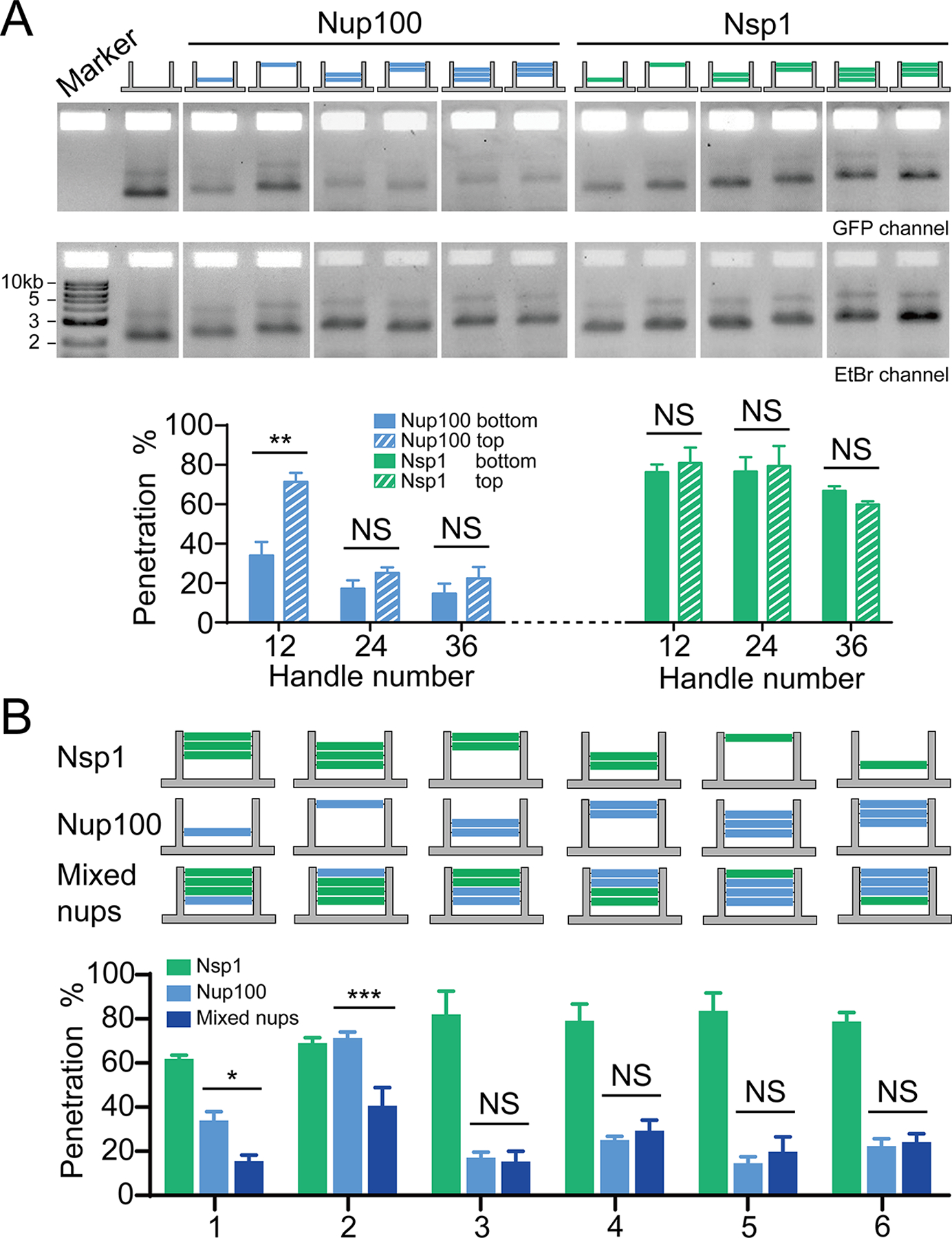
Different nucleoporin arrangements affect barrier permeability. (A) Permeability of NanoTraps with the FG-nups located near the entrance (top) or the baseplate (bottom) of the NanoTrap, tested against the 53kDa GFP-SNAP-prey. The exact nup arrangement is shown by the schematic drawing at the top of each lane (blue: Nup100, green: Nsp1, 12 nups/layer). Statistical data are plotted to show mean ± SEM. Statistical significance was determined by a two-tailed Student’s t-test; n=3; NS: not significant (P≥0.05); **: P<0.01. (B) Permeability of NanoTraps containing single or mixed types of FG-nups, tested against the 53kDa GFP-SNAP-prey. The exact nup arrangements are shown by the schematic drawings at the top of each group of bars (blue: Nup100, green: Nsp1, 12 nups/layer). Data are plotted to show mean ± SEM. Difference between mixed-nup and Nup100-NanoTraps was analyzed by two-way ANOVA and Tukey’s multiple comparison; n=3; NS: not significant (P≥0.05); *: P<0.05; ***:P<0.001.
The more cohesive Nup100 dictates the barrier strengths of mixed FG-nups.
The formation of distinct diffusion barriers by the two different FG-nups raises the question of how, or whether, they impact each other within the NPC-like nanopore confinement. Addressing this question will shed light on how the physiological in vivo NPC diffusion barrier is ultimately formed from the collective of a dozen FG-nups with unique biophysical characteristics. To begin to explore how combinations of distinct FG-nups might alter the properties of a diffusion barrier, we anchored both the GLFG-dominated Nup100 and FxFG-dominated Nsp1 into a single NanoTrap. In total, we designed 6 different NanoTrap handle arrangements to facilitate the attachment of mixed FG-nups, as illustrated in Figure 4B. Each handle arrangement has three subtypes: one containing Nup100 and Nsp1, one with Nup100 only, and one with Nsp1 only.
Using the SDS-agarose gel assay to detect the penetration efficiency of the 53 kD GFP-SNAP-prey, we observed that in scenarios in which there were 24 copies of Nup100, the resulting cohesive network dominated and was not impacted by the introduction of even 24 copies of Nsp1. This phenomenon is in qualitative agreement with the prediction of a computational simulation that models FG-nups as polymers with varying cohesion strengths.21 Interestingly, however, in cases with only 12 copies of Nup100, the introduction of Nsp1 strengthened the barrier (Figure 4B). This effect was most significant when Nup100 was grafted at the entrance to the channel, where Nup10012Empty36 itself did not form an efficient barrier (Figure 4A). These results suggest that Nsp1 might promote cohesive interactions of relatively low copy-number networks of Nup100 or that Nup100 promotes the confinement of Nsp1 in a way that establishes a stronger barrier, both of which can be attributed to volume exclusion. For example, the likely dynamic Nsp1 chains could limit the space in the channel that Nup100 can occupy, perhaps pushing Nup100 chains together, which would be predicted to increase their cohesive interactions.21
Ntf2 mediates cargo transport through FG-nup-gated NanoTraps.
We next tested if the NanoTraps could recapitulate NTR-mediated transport of macromolecules that would otherwise be excluded by the diffusion barrier. We used Ntf2 as a model NTR, which imports its cargo (Ran-GDP) as a dimer. We therefore generated a prey-conjugated Ntf2-GFP-SNAP fusion protein and purified the dimer form for the permeation assay (Figure S12). As the dimerized GFP-SNAP “cargo” is ~106 kD, we used the MBP-GFP-SNAP-prey (106 kD) as a control (Figure 5A). Consistent with the idea that both Nsp1 and Nup100 NanoTraps formed effective diffusion barriers, the MBP-GFP-SNAP-prey was not detected passing through Nup100 NanoTraps and only marginally permeated those made with Nsp1 (Figure 5B and S12). In striking contrast, the prey-labeled Ntf2-GFP-SNAP freely passed through the Nsp1-NanoTrap barriers with little impedance, apparently reaching ~95% permeation. Interestingly, and consistent with prior work indicating that Ntf2 cannot interact with GLFG-repeats,26, 43–44 Ntf2-GFP-SNAP-prey was largely unable to cross the Nup100 GLFG-network. These data further reinforce that direct interactions between Ntf2 and the FxFG repeats mediate passage through the Nsp1 barrier.26, 44–46 Although Ntf2 has been shown to permeate phase separated Nup100 particles in vitro,47 in this work the FG-nups are confined in a nanometer-scale channel rather than forming micrometer-sized condensates in solution, thus residing in an environment that more closely resembles the native NPC. Additionally, here the Ntf2 is fused to a sizable cargo, which likely makes it more challenging to nonspecifically penetrate the FG-nup barriers. Overall, the data suggest that FG-nups within the NanoTraps recapitulate their established functional interactions that underlie barrier formation and selective NTR-mediated transport, further establishing the NanoTrap as an in vitro model system for studying the structure-function relationship of the NPC central channel.
Figure 5.
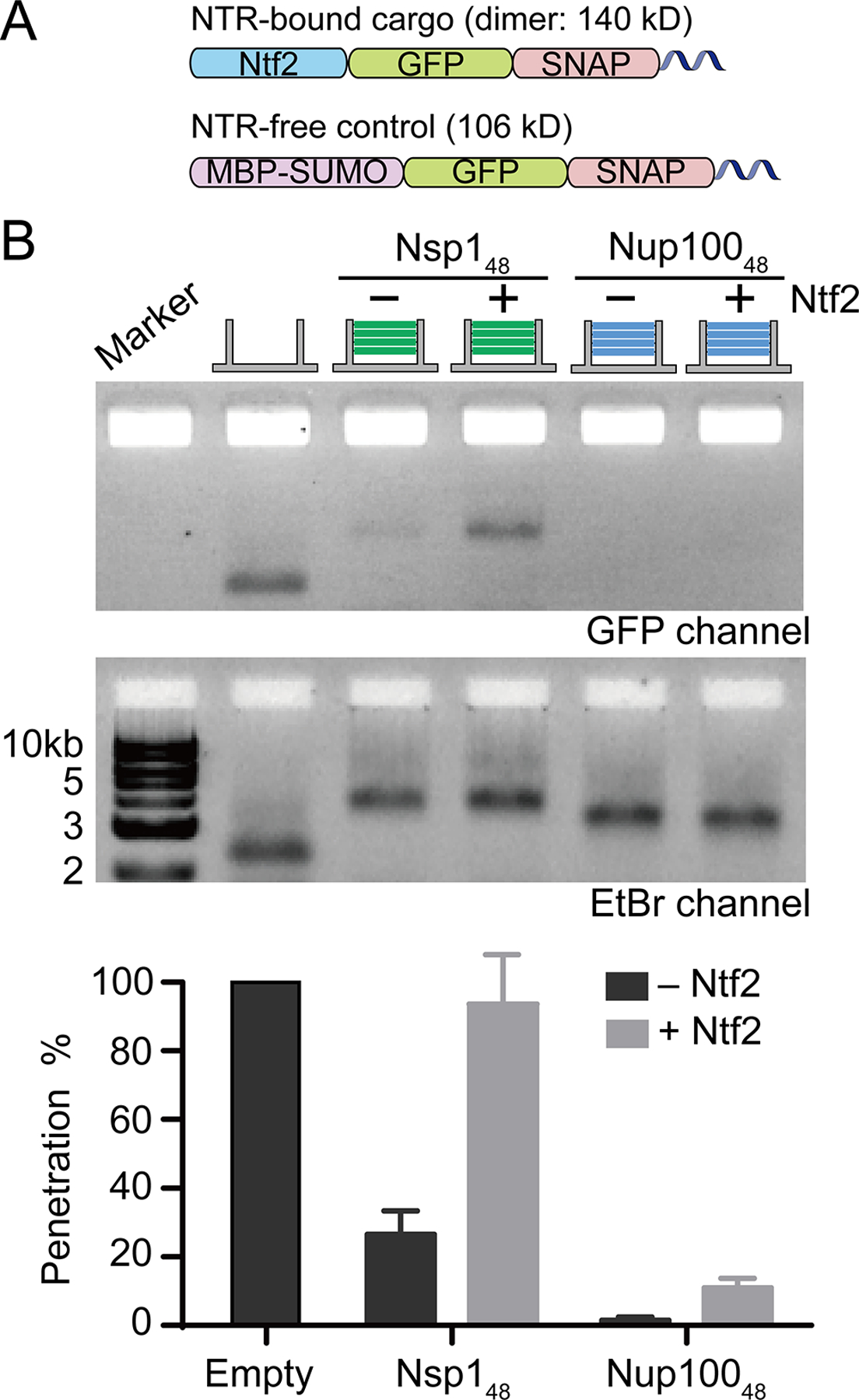
Ntf2-mediated cargo transport through FG-nup gated NanoTraps. (A) Schematics of an NTR-bound cargo (Ntf2-GFP-SNAP-prey, only a monomeric chain of the homodimer is shown) and an NTR-free control molecule (MBP-GFP-SNAP-prey). (B) Permeability of Nsp148 and Nup10048-NanoTraps to the Ntf2-fused molecule (140 kD), compared with the 106 kD NTR-free control. Data are plotted to show mean ± SEM from three trials.
CONCLUSION
The generation of an in vitro mimic capable of recapitulating the NPC’s complexity remains a significant challenge for synthetic biology. The NanoTrap system presented here has advanced our NuPOD platform to extend its utility beyond a morphological characterization of FG-nups confined to a nanochannel, to one that is now capable of assessing a key in vivo function of the FG-nups: the generation of a selective diffusion barrier. We draw the following conclusions from this work: First, cohesive FG-nups can form a much stronger barrier to molecular passage than their less-cohesive siblings. Indeed, as few as 12 copies of Nup100, provided they are confined within the channel walls, can form a diffusion barrier to macromolecules as small as 50 kD. Consistent with the notion that the cohesive properties of the GLFG repeats are responsible for this function, the cohesivity-ablating Nup100SG mutant fails to establish such a barrier.7, 13 These data are thus congruent with many other studies that have also compellingly established that the cohesivity of nups is a central feature of an effective diffusion barrier, at least in vitro.8, 13, 37 However, it remains to be established whether the abrupt molecular weight cutoff that we (Figure 3C) and others13 have observed with Nup100 ultimately reflects the permeability properties of NPCs in vivo. Indeed, recent work suggests that the diffusion barrier may be “soft” and permeable to even very large macromolecules.6, 48 Our recent work also showed that the tip of the megadalton-sized HIV capsid can at least insert into the NPC-mimicking NuPOD.49
The permeability properties of Nsp1 that we measured in the NanoTraps seem to be more in line with a soft-diffusion barrier type mechanism — one with a continuum of penetration rates that negatively correlates with increasing molecular weights of entering molecules. These observations are consistent with entropic barrier models where the dynamic nature of unstructured and flexible FG-nup filaments effectively occlude the channel.6, 50 However, a central challenge in the field is to reconcile the abundance of data surrounding the importance of cohesive interactions in controlling the function of the NPC with such an entropic barrier model of the FG-nups. For example, it is potentially interesting that the cohesive and non-cohesive nups seem to populate distinct parts of the central channel,8, 22–24, 39 but whether this relative positioning functionally matters remains elusive. Similarly, it remains uncertain how cohesive and non-cohesive nups interact to mitigate or enhance their individual cohesive properties. Our system provides an ideal platform for assessing these questions within the highly controlled environment afforded by the DNA NanoTraps. While we are just beginning these investigations, some interesting themes are emerging. First, a cohesive network of GLFG-nups will dominate over its non-cohesive siblings when at sufficiently high concentrations. These data suggest that non-cohesive interactions are inefficient to weaken a cohesive network. Consistent with this, and secondly, when cohesive nups are at low concentrations, the addition of a non-cohesive network can actually increase the strength of the barrier. Thus, collectively, these data support a model in which cohesive networks would dominate within the native NPC at least with respect to the formation of a diffusion barrier.
Furthermore, our work supports the concept that confinement of FG-nups within a channel impacts their collective properties and favors their condensation.33 The observation that best exemplifies this is that 12 copies of Nup100 can only form a barrier when grafted deep within the NanoTrap and not near its entrance, where the nucleoporins are exposed to the large surrounding volume. To reconcile these observations of an in vitro cohesive “hard” barrier with the observations of an in vivo “soft” barrier, future experiments will need to explore how other nucleoporins as well as factors like NTRs or non-specific competitors ultimately serve to weaken cohesive interactions, as has been suggested.51 Such considerations must also extend to explaining how certain NTRs exhibit preferences for only some FG-repeat types8, 52 yet are still able to cross the NPC. For example, our data, in the context of prior work, support that Ntf2 is unable to bind to GLFG-nups like Nup100 and in fact does not penetrate a GLFG-rich network. As Ntf2 can rapidly transit the NPC,53–55 a probable explanation is that it does not encounter such a GLFG-rich barrier in the native channel. Thus, much remains to be understood regarding how various types of FG-networks, NTRs and other factors work in concert to ultimately form a functional NPC.
The NanoTrap system provides an ideal platform to explore the complex interactions that establish NPC function. Our observations that “simple” NanoTraps can recapitulate the fundamentals of selective barriers coupled with the ability to systematically add-in components with control over copy number and anchor position will allow the NanoTrap system to explore future questions unanswerable with more traditional in vitro platforms. Beyond providing an adaptable framework for studying nuclear transport mechanisms, the NuPOD/NanoTrap systems can be viewed as prototypes of macromolecule sorting devices with tunable size-filtration behaviors. The impact of the gating biopolymers’ cohesivity and positioning on the NanoTrap’s permeability provides valuable design references for future engineering of DNA nanopores with increasing structural and functional diversity. Future development in this direction, in conjunction with the fast-evolving technologies that shape and perforate membranes with DNA nanostructures,56–58 may usher in a range of applications in biotechnology such as sensing viral pathogens and synthetic biology such as building artificial nuclei.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Rout, David Cowburn and Luke Davis and members of the Lin Lab for discussion; This work was supported by National Institutes of Health grants R01 GM132114 and P50 AI150481 (via a collaboration development program of Pittsburgh Center for HIV Protein Interactions) to C.L., R01 GM105672 to C.P.L., and R21 GM109466 to C.L., C.P.L. and T.J.M., as well as a Singapore Agency for Science, Technology and Research Graduate Scholarship to Q.X.
Footnotes
Supporting Information
Additional experimental details, materials and methods, including DNA-origami design, NanoTrap and protein characterizations, and data of statistical analysis.
The authors declare no competing financial interest.
REFERENCES
- 1.Strambio-De-Castillia C; Niepel M; Rout MP, The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Bio 2010, 11 (7), 490–501. [DOI] [PubMed] [Google Scholar]
- 2.Beck M; Hurt E, The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Bio 2017, 18 (2), 73–89. [DOI] [PubMed] [Google Scholar]
- 3.Lin DH; Hoelz A, The Structure of the Nuclear Pore Complex (An Update). Annu Rev Biochem 2019, 88, 725–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brohawn SG; Partridge JR; Whittle JRR; Schwartz TU, The Nuclear Pore Complex Has Entered the Atomic Age. Structure 2009, 17 (9), 1156–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heinss N; Sushkin M; Yu M; Lemke EA, Multifunctionality of F-rich nucleoporins. Biochem Soc Trans 2020, 48 (6), 2603–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timney BL; Raveh B; Mironska R; Trivedi JM; Kim SJ; Russel D; Wente SR; Sali A; Rout MP, Simple rules for passive diffusion through the nuclear pore complex. J Cell Biol 2016, 215 (1), 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labokha AA; Gradmann S; Frey S; Hulsmann BB; Urlaub H; Baldus M; Gorlich D, Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. Embo J 2013, 32 (2), 204–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel SS; Belmont BJ; Sante JM; Rexach MF, Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 2007, 129 (1), 83–96. [DOI] [PubMed] [Google Scholar]
- 9.Gorlich D; Kutay U, Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Bi 1999, 15, 607–660. [DOI] [PubMed] [Google Scholar]
- 10.Rout MP; Aitchison JD, The nuclear pore complex as a transport machine. J Biol Chem 2001, 276 (20), 16593–16596. [DOI] [PubMed] [Google Scholar]
- 11.Kalita J; Kapinos LE; Lim RYH, On the asymmetric partitioning of nucleocytoplasmic transport - recent insights and open questions. J Cell Sci 2021, 134 (7), jcs240382. [DOI] [PubMed] [Google Scholar]
- 12.Frey S; Richter RP; Goerlich D, FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 2006, 314 (5800), 815–817. [DOI] [PubMed] [Google Scholar]
- 13.Hulsmann BB; Labokha AA; Gorlich D, The Permeability of Reconstituted Nuclear Pores Provides Direct Evidence for the Selective Phase Model. Cell 2012, 150 (4), 738–751. [DOI] [PubMed] [Google Scholar]
- 14.Frey S; Gorlich D, A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 2007, 130 (3), 512–523. [DOI] [PubMed] [Google Scholar]
- 15.Rout MP; Aitchison JD; Magnasco MO; Chait BT, Virtual gating and nuclear transport: the hole picture. Trends Cell Biol 2003, 13 (12), 622–628. [DOI] [PubMed] [Google Scholar]
- 16.Strawn LA; Shen TX; Shulga N; Goldfarb DS; Wente SR, Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol 2004, 6 (3), 197–206. [DOI] [PubMed] [Google Scholar]
- 17.Lim RYH; Fahrenkrog B; Koser J; Schwarz-Herion K; Deng J; Aebi U, Nanomechanical basis of selective gating by the nuclear pore complex. Science 2007, 318 (5850), 640–643. [DOI] [PubMed] [Google Scholar]
- 18.Lim RYH; Deng J, Interaction Forces and Reversible Collapse of a Polymer Brush-Gated Nanopore. Acs Nano 2009, 3 (10), 2911–2918. [DOI] [PubMed] [Google Scholar]
- 19.Tetenbaum-Novatt J; Rout MP, The Mechanism of Nucleocytoplasmic Transport through the Nuclear Pore Complex. Cold Sh Q B 2010, 75, 567–584. [DOI] [PubMed] [Google Scholar]
- 20.Celetti G; Paci G; Caria J; VanDelinder V; Bachand G; Lemke EA, The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. J Cell Biol 2020, 219 (1), e201907157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis LK; Ford IJ; Saric A; Hoogenboom BW, Intrinsically disordered nuclear pore proteins show ideal-polymer morphologies and dynamics. Phys Rev E 2020, 101 (2–1), 022420. [DOI] [PubMed] [Google Scholar]
- 22.Ando D; Zandi R; Kim YW; Colvin M; Rexach M; Gopinathan A, Nuclear Pore Complex Protein Sequences Determine Overall Copolymer Brush Structure and Function. Biophys J 2014, 106 (9), 1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SJ; Fernandez-Martinez J; Nudelman I; Shi Y; Zhang W; Raveh B; Herricks T; Slaughter BD; Hogan JA; Upla P; Chemmama IE; Pellarin R; Echeverria I; Shivaraju M; Chaudhury AS; Wang J; Williams R; Unruh JR; Greenberg CH; Jacobs EY; Yu Z; de la Cruz MJ; Mironska R; Stokes DL; Aitchison JD; Jarrold MF; Gerton JL; Ludtke SJ; Akey CW; Chait BT; Sali A; Rout MP, Integrative structure and functional anatomy of a nuclear pore complex. Nature 2018, 555 (7697), 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang K; Tagliazucchi M; Park SH; Rabin Y; Szleifer I, Nanocompartmentalization of the Nuclear Pore Lumen. Biophys J 2020, 118 (1), 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kowalczyk SW; Kapinos L; Blosser TR; Magalhaes T; van Nies P; Lim RYH; Dekker C, Single-molecule transport across an individual biomimetic nuclear pore complex. Nat Nanotechnol 2011, 6 (7), 433–438. [DOI] [PubMed] [Google Scholar]
- 26.Jovanovic-Talisman T; Tetenbaum-Novatt J; McKenney AS; Zilman A; Peters R; Rout MP; Chait BT, Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature 2009, 457 (7232), 1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fragasso A; de Vries HW; Andersson J; van der Sluis EO; van der Giessen E; Dahlin A; Onck PR; Dekker C, A designer FG-Nup that reconstitutes the selective transport barrier of the nuclear pore complex. Nat Commun 2021, 12 (1), 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denning DP; Patel SS; Uversky V; Fink AL; Rexach M, Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci U S A 2003, 100 (5), 2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothemund PW, Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440 (7082), 297–302. [DOI] [PubMed] [Google Scholar]
- 30.Gerling T; Wagenbauer KF; Neuner AM; Dietz H, Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 2015, 347 (6229), 1446–1452. [DOI] [PubMed] [Google Scholar]
- 31.Hong F; Zhang F; Liu Y; Yan H, DNA Origami: Scaffolds for Creating Higher Order Structures. Chem Rev 2017, 117 (20), 12584–12640. [DOI] [PubMed] [Google Scholar]
- 32.Dey S; Fan C; Gothelf KV; Li J; Lin C; Liu L; Liu N; Nijenhuis MAD; Saccà B; Simmel FC; Yan H; Zhan P, DNA origami. Nature Reviews Methods Primers 2021, 1 (1), 13. [Google Scholar]
- 33.Fisher PDE; Shen Q; Akpinar B; Davis LK; Chun KKH; Baddeley D; Saric A; Melia TJ; Hoogenboom BW; Lin CX; Lusk CP, A Programmable DNA Origami Platform for Organizing Intrinsically Disordered Nucleoporins within Nanopore Confinement. Acs Nano 2018, 12 (2), 1508–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ketterer P; Ananth AN; Laman Trip DS; Mishra A; Bertosin E; Ganji M; van der Torre J; Onck P; Dietz H; Dekker C, DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex. Nat Commun 2018, 9 (1), 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin C; Perrault SD; Kwak M; Graf F; Shih WM, Purification of DNA-origami nanostructures by rate-zonal centrifugation. Nucleic Acids Res 2013, 41 (2), e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zahn R; Osmanovic D; Ehret S; Araya Callis C; Frey S; Stewart M; You C; Gorlich D; Hoogenboom BW; Richter RP, A physical model describing the interaction of nuclear transport receptors with FG nucleoporin domain assemblies. Elife 2016, 5, e14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labokha AA; Gradmann S; Frey S; Hulsmann BB; Urlaub H; Baldus M; Gorlich D, Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. Embo J 2013, 32 (2), 204–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halfmann R; Wright JR; Alberti S; Lindquist S; Rexach M, Prion formation by a yeast GLFG nucleoporin. Prion 2012, 6 (4), 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada J; Phillips JL; Patel S; Goldfien G; Calestagne-Morelli A; Huang H; Reza R; Acheson J; Krishnan VV; Newsam S; Gopinathan A; Lau EY; Colvin ME; Uversky VN; Rexach MF, A Bimodal Distribution of Two Distinct Categories of Intrinsically Disordered Structures with Separate Functions in FG Nucleoporins. Mol Cell Proteomics 2010, 9 (10), 2205–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ader C; Frey S; Maas W; Schmidt HB; Gorlich D; Baldus M, Amyloid-like interactions within nucleoporin FG hydrogels. P Natl Acad Sci USA 2010, 107 (14), 6281–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisele NB; Labokha AA; Frey S; Gorlich D; Richter RP, Cohesiveness tunes assembly and morphology of FG nucleoporin domain meshworks-Implications for nuclear pore permeability. Biophys J 2013, 105 (8), 1860–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapinos LE; Schoch RL; Wagner RS; Schleicher KD; Lim RYH, Karyopherin-Centric Control of Nuclear Pores Based on Molecular Occupancy and Kinetic Analysis of Multivalent Binding with FG Nucleoporins. Biophys J 2014, 106 (8), 1751–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarkson WD; Corbett AH; Paschal BM; Kent HM; McCoy AJ; Gerace L; Silver PA; Stewart M, Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran. J Mol Biol 1997, 272 (5), 716–730. [DOI] [PubMed] [Google Scholar]
- 44.Bayliss R; Leung SW; Baker RP; Quimby BB; Corbett AH; Stewart M, Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. Embo J 2002, 21 (12), 2843–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayliss R; Ribbeck K; Akin D; Kent HM; Feldherr CM; Gorlich D; Stewart M, Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J Mol Biol 1999, 293 (3), 579–593. [DOI] [PubMed] [Google Scholar]
- 46.Paschal BM; Gerace L, Identification of Ntf2, a Cytosolic Factor for Nuclear Import That Interacts with Nuclear-Pore Complex Protein P62. J Cell Biol 1995, 129 (4), 925–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt HB; Gorlich D, Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. Elife 2015, 4, e04251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Popken P; Ghavami A; Onck PR; Poolman B; Veenhoff LM, Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol Biol Cell 2015, 26 (7), 1386–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen Q; Xu C; Jang S; Xiong Q; Devarkar SC; Tian T; Bedwell GJ; Tripler TN; Hu Y; Yuan S; Temple J; Shi J; Aiken C; Engelman AN; Perilla JR; Lusk CP; Lin C; Xiong Y, A DNA-origami nuclear pore mimic reveals nuclear entry mechanisms of HIV-1 capsid. bioRxiv 2020, 2020.08.10.245522. [Google Scholar]
- 50.Aramburu IV; Lemke EA, Floppy but not sloppy: Interaction mechanism of FG-nucleoporins and nuclear transport receptors. Semin Cell Dev Biol 2017, 68, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tetenbaum-Novatt J; Hough LE; Mironska R; McKenney AS; Rout MP, Nucleocytoplasmic Transport: A Role for Nonspecific Competition in Karyopherin-Nucleoporin Interactions. Mol Cell Proteomics 2012, 11 (5), 31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allen NPC; Huang L; Burlingame A; Rexach M, Proteomic analysis of nucleoporin interacting proteins. J Biol Chem 2001, 276 (31), 29268–29274. [DOI] [PubMed] [Google Scholar]
- 53.Ribbeck K; Gorlich D, Kinetic analysis of translocation through nuclear pore complexes. Embo J 2001, 20 (6), 1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siebrasse JP; Peters R, Rapid translocation of NTF2 through the nuclear pore of isolated nuclei and nuclear envelopes. Embo Rep 2002, 3 (9), 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kubitscheck U; Grunwald D; Hoekstra A; Rohleder D; Kues T; Siebrasse JP; Peters R, Nuclear transport of single molecules: dwell times at the nuclear pore complex. J Cell Biol 2005, 168 (2), 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen Q; Grome MW; Yang Y; Lin C, Engineering Lipid Membranes with Programmable DNA Nanostructures. Adv Biosyst 2020, 4 (1), 1900215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Howorka S, Building membrane nanopores. Nat Nanotechnol 2017, 12 (7), 619–630. [DOI] [PubMed] [Google Scholar]
- 58.Darley E; Singh JKD; Surace NA; Wickham SFJ; Baker MAB, The Fusion of Lipid and DNA Nanotechnology. Genes (Basel) 2019, 10 (12), 1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


