Abstract
eIF5-mimic protein (5MP) is a translational regulatory protein that binds the small ribosomal subunit and modulates its activity. 5MP is proposed to reprogram non-AUG translation rates for oncogenes in cancer, but its role in controlling non-AUG initiated synthesis of deleterious repeat-peptide products, such as FMRpolyG observed in Fragile-X associated tremor ataxia syndrome (FXTAS), is unknown. Here we show that 5MP can suppress both general and repeat-associated non-AUG (RAN) translation by a common mechanism in a manner dependent on its interaction with eIF3. Essentially, 5MP displaces eIF5 through the eIF3c subunit within the PIC, thereby increasing the accuracy of initiation. In Drosophila, 5MP/Kra represses neuronal toxicity and enhances lifespan in a FXTAS disease model. These results implicate 5MP in protecting cells from unwanted byproducts of non-AUG translation in neurodegeneration.
Introduction
During eukaryotic translation initiation, Met-tRNAiMet is recruited by eIF2, a heterotrimeric factor that binds the tRNA in a GTP-dependent manner (Hinnebusch et al., 2007; Sonenberg and Hinnebusch, 2009). The resulting ternary complex (TC) and three other eIFs eIF1, eIF3 and eIF5 form the multifactor complex (MFC) and bind the 40S ribosomal subunit, thereby generating the 43S preinitiation complex (PIC) (Asano et al., 2000). The 43S is then recruited to mRNA by eIF4F that binds its 5’ cap (Singh et al., 2012) and allowed to migrate along the mRNA in a 5’-to-3’ direction until it encounters an AUG start codon (mRNA scanning). Prior to this event, eIF5 catalyzes GTP hydrolysis for eIF2, while the products, GDP and Pi, stay bound to eIF2 during scanning. When the AUG start codon base-pairs with the Met-tRNAiMet anticodon in the P-site, the 40S subunit stalls, eIFs are released in conjunction with Pi release from eIF2, and the 60S subunit joins the 40S subunit (Algire et al., 2005; Singh et al., 2006). The resulting 80S initiation complex accepts an amino-acyl tRNA in the A-site to begin the translation elongation cycle.
The major regulator of the start codon fidelity is eIF1 which binds the 40S subunit P-site and stabilizes the open, scanning-competent conformation of the PIC by physically impeding the mismatched codon-anticodon base-pairs to bind the ribosomal P-site (Asano and Sachs, 2007; Cheung et al., 2007). Upon start codon selection by the PIC, eIF1 is released to stabilize the closed conformation, which promotes 60S subunit joining (Llacer et al., 2015). Importantly, the N-terminal domain (NTD) of eIF5, originally shown to be responsible for eIF2 GTPase activating protein (GAP) function (Algire et al., 2005), has a structure homologous to that of eIF1 (Asano, 2013; Conte et al., 2006) (Fig. 1A) and thereby interacts with the ribosomal P-site (Llacer et al., 2018). In this way, eIF5-NTD impedes eIF1 binding to the PIC, ensuring a unidirectional transition to the closed state (Llacer et al., 2018). A number of the PIC-bound eIFs, including the c-subunit of eIF3 (Obayashi et al., 2017) and the C-terminal domain (CTD) of eIF5 (Luna et al., 2012), directly interact with eIF1 to control the balance of its binding and release, thereby regulating the level of initiation accuracy, and hence modulating the frequency of near-cognate (non-AUG) translation initiation, in a manner appropriate for eukaryotic cell function (Luna et al., 2013; Singh et al., 2012); for review, see (Asano, 2014).
Fig. 1. 5MP1 carries basic and acidic surfaces responsible for binding eIF3 and eIF2.
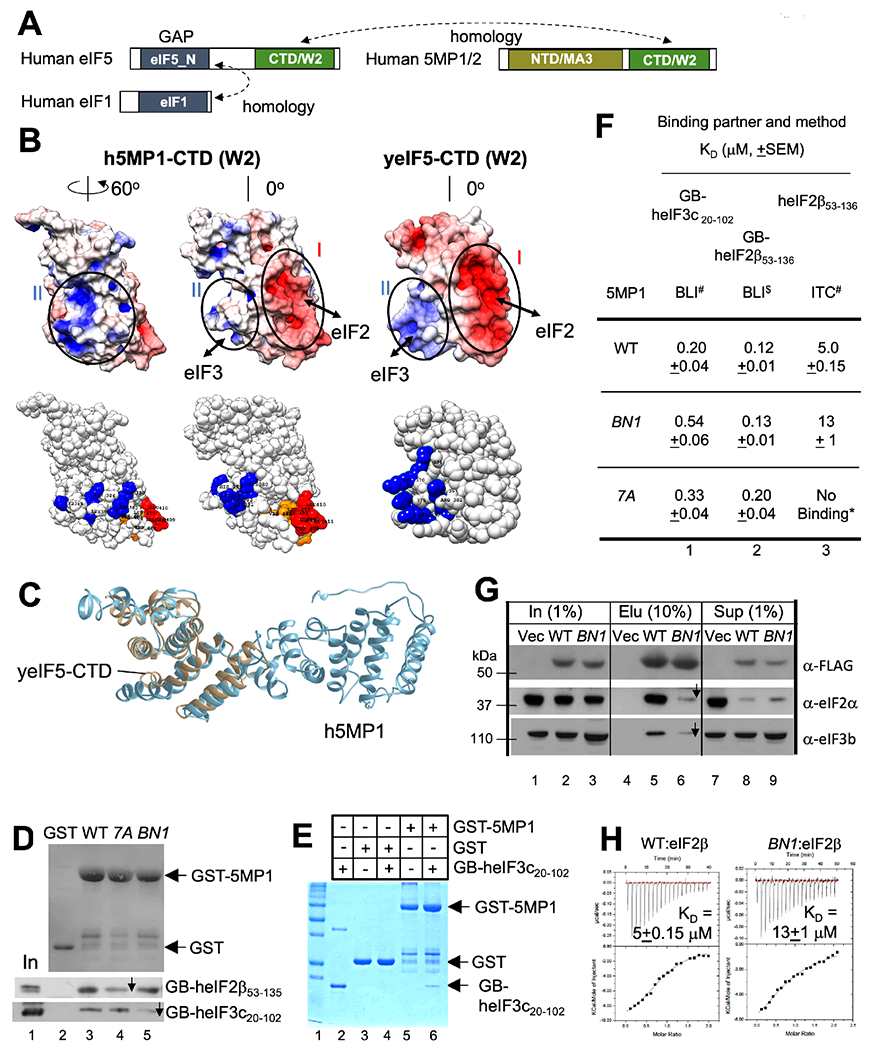
(A) Homology between eIF1, eIF5 and 5MP. Boxes indicate primary structure with domains defined by Pfam in distinct colors. (B) Yeast (y) eIF5 and human (h) 5MP1 mutations used in this study. Top, surface charge presentation of h5MP1-CTD (homology model, left and middle) and yeIF5-CTD (crystal structure, right) (Wei et al., 2006). h5MP1, middle, is viewed from the same angle as yeIF5. h5MP1, right, is viewed after 60° rotation. Red and blue indicate the areas of negative and positive charges, respectively. Bottom, spherical presentations indicating the location of amino acids altered by BN1 (K380Q K383Q H386Q K389Q K391Q for h5MP1-BN1) in blue and by 7A (F401A V402A W404A L405A EEE409-411AAA for h5MP1-7A) in red (glutamate) or orange (hydrophobic residues). The 5MP1 homology model was generated with I-TASSER. (C) Homology model of 5MP1 (blue) superimposed with crystal structure of yeIF5-CTD (gray) (PBD ID: 2FUL) (Wei et al., 2006). (D) and (E) GST pull down assays. 10 μg GST-h5MP1, GST-h5MP1-BN1, or ~5 μg GST as control, was incubated with (D) GB-heIF2β53-136 or GB-heIF3c20-102 expressed in E. coli or (E) purified GB-heIF3c20-102 (20 μg), precipitated with glutathione resin and analyzed by SDS-PAGE along with input (2.5% in lane 1, panel D; 10% in lane 2, panel E). (D) top, Ponceau staining; bottom two gels, immunoblot with anti-His antibodies. (E), Coomassie Blue staining. (F) Effect of 5MP1 surface mutations on interactions in vitro. KD values obtained with BLI or ITC is tabulated for 5MP1 (WT or its BN1 or 7A mutant) interaction with binding partners listed across the top. *No binding detected (Fig. S1F). See also Fig. S1D–E and S2A. # n=4; $ n=3. (G) Immunoblot analyses of FLAG-h5MP1 complexes affinity-purified from HEK293T transfected with expression plasmids (Key Resource Table). 10% of WT (lane 5) and BN1 mutant (lane 6) 5MP1 complexes or mock purified fraction from the empty vector transfectant (lane 4) were analyzed along with 1% input amount (lanes 1-3) and 1 % supernatant fractions (lanes 7-9). Antibodies used for detection of the human antigen are indicated to the right (Santa Cruz Biotechnology) (Kozel et al., 2016). (H) ITC experiments. The top panels represent the raw data from ITC titrations. Bottom panels show the non-linear least square fit of the integrated heats as function of molar ratio of titration of heIF2β53-136 (200 μM) with (top) h5MP1 (WT) or (bottom) 5MP1-BN1 at 20 μM with the estimated KD shown inside (n=4). *p=0.008, compared to WT.
Accumulating evidence indicates that carcinogenesis is tightly associated with altered rates of translation from near-cognate start codons (Asano, 2014; Ingolia et al., 2011; Sendoel et al., 2017). Recently, both the two genes in human encoding the translational regulatory protein, eIF5-mimic protein (5MP1/BZW2 and 5MP2/BZW1), were characterized as oncogenes whose increased gene dosage promotes various types of cancer (Cheng et al., 2017; Huang et al., 2020; Kozel et al., 2016; Li et al., 2009). 5MP is named due to its homology with the CTD of eIF5, which contains an α-helical HEAT domain termed W2, and its conserved eIF2 and eIF3 binding sites (Singh et al., 2011) (Fig. 1A–C). However, the NTD of 5MP bears no homology with that of eIF5 and folds into a HEAT domain of MA3-type (Fig. 1A–C), implicating 5MP as a competitive inhibitor (Hiraishi et al., 2014). We recently showed that 5MP can suppress non-AUG translation by competing with eIF5 (Tang et al., 2017), whose overexpression otherwise decreases the stringency of start codon selection (Nanda et al., 2013). In colorectal cancer, 5MP1/BZW2 reprograms translation of the transcription factor c-Myc by repressing its longer, CUG-initiated isoform relative to its more oncogenic AUG-initiated isoform (Sato et al., 2019). Along with its paralog 5MP2/BZW1, the role of 5MP in regulating non-AUG-initiated genes in cancer is currently under investigation.
Non-AUG initiation has also been implicated in a number of neurodegenerative disorders caused by GC-rich repeat expansions. These repeats are translated from non-AUG start codons located upstream of, or sometimes within the repeat, resulting in an aberrant process termed repeat associated non-AUG (RAN) translation (Ash et al., 2013; Mori et al., 2013; Zu et al., 2011). In the neurodegenerative disorder Fragile-X associated tremor ataxia syndrome (FXTAS), RAN translation through a CGG repeat expansion within the 5’ UTR of FMR1 (CGG RAN) leads to the production of three toxic, aggregate-prone homopolypetides, which accumulate into ubiquitinated neuronal inclusions in patient brains (Todd et al., 2013). RAN translation of CGG repeats contributes to their toxicity in model systems (Green et al., 2017; Kearse et al., 2016; Sellier et al., 2017 ; Todd et al., 2013). The most abundant CGG RAN product is a polyglycine protein, FMRpolyG, whose translation is initiated at three near cognate codons, two GUGs and an ACG, upstream of the repeat (Kearse et al., 2016; Zhang et al., 2020). Similar to translation of other non-AUG initiated genes, CGG RAN initiation is influenced by availability of specific canonical initiation factors. Activation of the integrated stress response, which phosphorylates eIF2α and prevents TC recycling, leads to an increase in CGG RAN (Green et al., 2017). Overexpression of eIF1, which promotes AUG start codon fidelity, causes a decrease in CGG RAN, while overexpression of eIF5, which relaxes the fidelity, increases CGG RAN (Linsalata et al., 2019).
The present work aims to investigate the molecular basis of 5MP regulation of canonical non-AUG translation and RAN translation. Here, we show that eIF3 is 5MP’s crucial partner in suppressing both the modes of non-AUG translation. Based on the recently solved cryo-EM PIC structures, we propose a common mechanism by which 5MP binding to eIF3 modulates general and repeat-associated non-AUG translation.
Results
5MP-CTD is functionally homologous to eIF5-CTD
5MP was previously shown to bind eIF2 and eIF3, but the exact binding location on its structure to eIF3 has not been determined. To determine the residues required for this interaction, we generated a 3D homology model of human 5MP1 (h5MP1) (Fig. 1B–C, see STAR Methods for detail). As shown in Fig. 1B, h5MP1-CTD has basic and acidic surfaces in opposite sides (left two columns), whose homologous locations in eIF5-CTD are responsible for eIF3(c) and eIF2(β) binding, respectively (right column). 5MP1’s acidic surface was previously shown to bind eIF2β (Kozel et al., 2016; Singh et al., 2011), as the 5MP1-7A mutation altering this site reduced binding to eIF2 (made of α, β, and γ subunits) and to a lysine-rich segment (K-boxes) of eIF2β (also see Fig. 1D and S1D) without altering eIF3 binding (Singh et al., 2011). Using the homology model as a guide, we generated a basic-to-neutral (BN1) mutation altering five of its basic residues (Fig. 1B). To study eIF3c binding, we generated a GB1 (solubility enhancer tag)-fusion protein, GB-heIF3c20-102, to the minimal 5MP1-binding segment in human eIF3c (Fig. 1E and S1A–B). Circular dichroism (CD) analysis of recombinant h5MP1 proteins show that both the BN1 and 7A mutations do not change overall 5MP1 structure or folding (Fig. S1C).
As expected, BN1 reduced GST-h5MP1 binding to GB-heIF3c20-102 in both the pull-down assay (Fig. 1D, 3rd gel and S1D) and bio-layer interferometry (BLI) (Fig. 1F and S1E). Similarly, BN1 strongly reduced the amount of eIF3 co-immunoprecipitated with FLAG-h5MP1 in cultured human cells (Fig. 1G, gel labeled α-eIF3b). As a control for specificity, we determined whether the BN1 mutation affected binding of eIF2. We did not observe reduced GST-h5MP1 binding to GB-heIF2β53-136 (Fig. 1D, 3rd gel, 1G and S1D), although isothermal titration calorimetry (ITC) indicated a minor but significant increase in Kd between h5MP1 and untagged heIF2β53-136 (Fig. 1H) (as 7A almost eliminated this interaction, as shown in Fig. S1F, more drastic mutational effects by ITC may be due to removing the solubility enhancer tag and the high concentrations required in this technique, causing the 7A to precipitate. See Fig. S2A and Limitation of the Study).
In contrast, BN1 strongly reduced eIF2 binding following FLAG-h5MP1 immunoprecipitation (IP) (Fig. 1G, lane 6, gel labeled α-eIF2α). The strong depletion of eIF2 in its supernatant fraction (lane 9) indicates that eIF2 is bound to FLAG-h5MP1 but dissociated while washing the affinity resin bound by these proteins prior to its immune-detection. Thus, the eIF2/h5MP1 complex appears to be unstable in the presence of BN1, or when h5MP1 cannot bind eIF3c. This observation motivated us to study h5MP1 interactions in vivo more carefully (see below). However, based on the in vitro binding studies, we conclude that h5MP1-CTD bind eIF2 and eIF3 through binding sites homologous to those of eIF5-CTD.
5MP requires its binding partner, eIF3, to function in stringent start codon selection
S. cerevisiae does not encode 5MP (Hiraishi et al., 2014), thus it provides a unique setting to test the effect of heterologous expression in the absence of endogenous WT competitors. Previously, we showed that h5MP1 overexpression (OE) in yeast inhibited non-AUG initiation imposed by yeast eIF5/Tif5p OE but did not suppress basal non-AUG initiation. Yet, in human cells, h5MP1 overexpression alone can suppress basal non-AUG initiation (Loughran et al., 2018; Tang et al., 2017). We reasoned that there are two levels of competition by which 5MP counteracts with eIF5 and thereby modulates non-AUG translation, one outside of the ribosome for eIF2 and the other for eIF2 within the PIC involving a distinct factor. The first level of competition explains why h5MP1 OE in yeast can reverse the effect of Tif5p OE on non-AUG initiation. The second level of competition allows overexpressed h5MP1 to displace eIF5 from the PIC by mass action, making initiation accurate in human cells. In agreement, such displacement was observed in rabbit reticulocyte lysate by sucrose gradient analysis of the PIC (Singh et al., 2011). Moreover, mutual exclusivity was observed between eIF5 and h5MP1 in the their IP complex with eIF2 and eIF3 in human cells (Kozel et al., 2016) (also see below). By contrast, due to its inability to bind the presumed critical PIC component in yeast, h5MP1 OE alone might not allow the displacement of eIF5 from the PIC.
As h5MP1 binds human eIF3 tightly, but only weakly associates with yeast eIF3 in vivo (Kozel et al., 2016; Singh et al., 2011), we hypothesized that eIF3, the large 40S binding factor, is the key binding partner within the PIC. To assess the model that h5MP1 displacement of eIF5 requires a h5MP1 interaction within the PIC, we examined the effect of h5MP1 OE on yeast PIC formation in vivo. As shown in Fig. 2A, the WT h5MP1, but not the h5MP1-7A mutant defective in eIF2 binding, associated with yeast PIC (Fig. 2A, panels 2 and 3), demonstrating eIF2-dependent h5MP1 recruitment to the PIC. However, we observed a striking accumulation of Tif5p and h5MP1 within the 48S PIC as well as ribosome-free MFC fractions in yeast with WT h5MP1 OE (Fig. 2B). Interestingly, our quantification shows that h5MP1 found in the 48S PIC is roughly equivalent in abundance to eIF5 found in the same fraction (Fig. S2B–C). Thus, the h5MP1 defective in binding yeast eIF3 is unable to displace eIF5, in contrast to the h5MP1 displacement of rabbit eIF5 from the PIC observed in reticulocyte lysates (Singh et al., 2011).
Fig. 2. In vivo PIC assembly analysis in yeast suggests that eIF3 is the partner of 5MP1 for its function in accurate initiation.
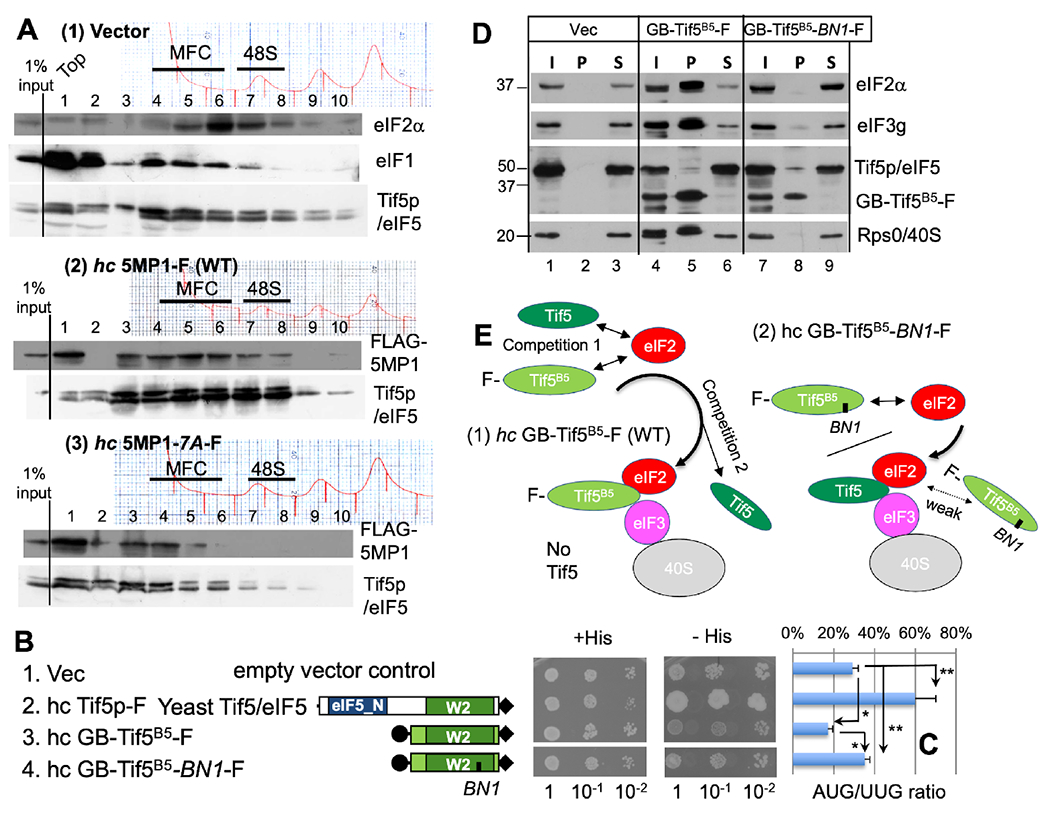
(A) Cell extracts of S. cerevisiae strain KAY1027 (sui1-G107R, Sui−) bearing high-copy (hc) plasmids expressing human 5MP1 or its 7A mutant and grown in a synthetic complete (SC) medium were analyzed by sucrose gradient fractionation and immunoblotting with antibodies against proteins listed to the right (see Fig. S2D legend). Top graphs, A260 absorbance trace. Lane 7-8, 48S complex (PIC); lanes 4-6, MFC fractions free of 40S. (B) Construction and examination of GB-TIf5B6-F, an “yeast version” of 5MP made from Tif5p. Schematics in the middle describe the domain structure of WT Tif5p/eIF5 and its Tif5B5 (aa. 201-405) derivatives employed here. Filled circle, GB1-tag. Filled diamonds, FLAG (F)-tag. Fixed amounts of KA1121 (sui1-K60E, Sui−) transformants bearing indicated constructs were spotted on minimal media with full (+ His) or trace amounts (10 μM) of histidine (- His) and their growth was monitored after 2 and 13 days, respectively. (C) UUG/AUG initiation ratio from the transformants carrying UUGhis4-lacZ and AUGHIS4-lacZ plasmids was measured in the strain used in (B), as described previously (Lee et al., 2007). *, p<0.002; **, p=0.02. (D) Co-IP with FLAG-tagged GB-Tif5B5. Strain KAY1027 (sui1-G107R, Sui−) bearing hc plasmids encoding WT GB-Tif5B5-F or its BN1 mutant or vector control (Vec) were grown in synthetic complete media lacking leucine at 30° C and used to prepare WCEs. Aliquots of WCEs were incubated with anti-FLAG affinity resin and after washing, the bound proteins were analyzed by SDS-PAGE and immunoblotting with antibodies listed to the right. I, 10% input amounts of WCE. P, 100% pellet fraction. S, 10% supernatant fraction. (E) Model of GB-Tif5B5-F interactions deduced from panel D. Top, GB-Tif5B5-F (light green oval) competes with eIF5 (dark green oval) for eIF2 (red oval) (1st level of competition). Under hc GB-Tif5B5-F, GB-Tif5B5-F is predominantly incorporated into PIC by mass action, displacing eIF5 through its ability to bind eIF3c (panel 1) (2nd level of competition). Under hc GB-Tif5B5-F BN1 mutant, the mutant form is recruited to the PIC through eIF2, but cannot be stably incorporated into the PIC, due to defective binding to eIF3 (panel 2). The mutant GB-Tif5B5-F is tethered loosely to the PIC and hence detached from the PIC during the FLAG-IP reaction.
An alternative explanation for why human 5MP1 cannot regulate start codon stringency in yeast is that the h5MP1 is not able to interact with yeast proteins via its NTD. To determine whether 5MP interaction with eIF3 is ultimately responsible for its function in accurate initiation and to gain insights into its role in 5MP recruitment to the PIC, we generated the yeast version of “5MP”, using a yeast eIF5 deletion mutant. This construct, GB-Tif5B5-F, carries an N-terminal deletion construct termed Tif5-B5 (aa. 201-405) that retains the Tif5p-CTD (Asano et al., 1999): Stable expression of Tif5B5 required GB1-tagging (Fig. S2B). GB-Tif5B5-F is able to bind yeast eIF2 and eIF3 (Asano et al., 1999), and thus is homologous to 5MP (see Fig. 2B, row 3 for its primary structure). We also generated GB-Tif5B5-BN1-F carrying a basic-to-neutral mutation in its eIF3c binding domain (Yamamoto et al., 2005) (Fig. 2B, row 4; see Fig. 1B, bottom right, for mutation sites). Both constructs were expressed at similar levels to full length Tif5p expressed from the same vector – ~20 fold more than endogenous Tif5p (Fig. S2B) (Singh et al., 2007). Importantly, the tif5-BN1 mutation alone did not disrupt binding to eIF2β (Yamamoto et al., 2005), but decreased eIF5 loading to the 43S/48S PIC without decreasing loading of eIF3, eIF2 or eIF1 (Fig. S2D), demonstrating that eIF3 contributes to eIF5 recruitment to the PIC.
To determine whether GB-Tif5B5-F repressed non-AUG initiated translation, we implemented a his4 reporter assay in which the his4 gene is initiated with a UUG codon. We used a yeast eIF1 mutant that increases UUG initiation (suppressor of initiation codon mutation, or Sui-) (Asano and Sachs, 2007), in order to examine the ability of GB-Tif5B5-F to suppress UUG initiation. Increased frequencies of initiation at this UUG will confer survival to yeast grown on His– media (Fig. 2B, row 1). Yeast survival decreased upon expression of GB-Tif5B5-F (row 3), compared to a vector only control, and was unaffected in the GB-Tif5B5-BN1-F mutant (row 4). Similarly, expression of GB-Tif5B5-F caused a decrease in a UUG-initiated lacZ reporter assay, compared to vector alone, while GB-Tif5B5-BN1-F had no effect (Fig. 2C). Together, this indicates that expression of the Tif5p-CTD capable of binding eIF2 and eIF3 was sufficient to repress UUG initiation, and this repression was dependent on an intact eIF3-binding site. In contrast, full-length eIF5/Tif5p overexpression increased UUG initiation (Fig. 2B–C, row 2), indicative of relaxed stringency of initiation as reported previously (Nanda et al., 2013; Tang et al., 2017). Thus, the Tif5p-NTD has an opposite role in accurate initiation compared to that of its CTD (see Discussion).
To directly determine whether GB-Tif5B5-F prevents UUG initiation through eIF3c, we performed co-IP with anti-FLAG antibodies. We found that GB-Tif5B5-F associates with eIF2, eIF3, and the 40S ribosome, but not with eIF5 (Fig. 2D, panel 1, lanes 5). In contrast, GB-Tif5B5-BN1-F failed to associate with eIF2, eIF3, eIF5/Tif5p, or the 40S ribosome (Fig. 2D). Together these data support the model that GB-Tif5B5-F competes with eIF5 for eIF2 at the level of the MFC and the 43S PIC formation (Fig. 2E). The lack of eIF2 in the GB-Tif5B5-BN1-F complex is similar to what we observed in human cells expressing h5MP1-BN1 (Fig. 1F) and is most likely an indirect effect due to loss of Tif5B5:eIF3c binding (Fig. 2E). The eIF2-5MP/eIF5 complex is therefore likely to be stabilized through eIF3c, which would position eIF2 and 5MP/eIF5 into proximity (see below Fig. 4D and Discussion). Thus, combined with the previous works in mammalian cells, we propose that 5MP1 and GB-Tif5B5-F efficiently compete with eIF5/Tif5p for binding eIF2 within the PIC through their interaction with eIF3.
Fig. 4. MS analysis of 5MP-eIF complexes shows that 5MP is recruited to the PIC through eIF3c in humans and flies.
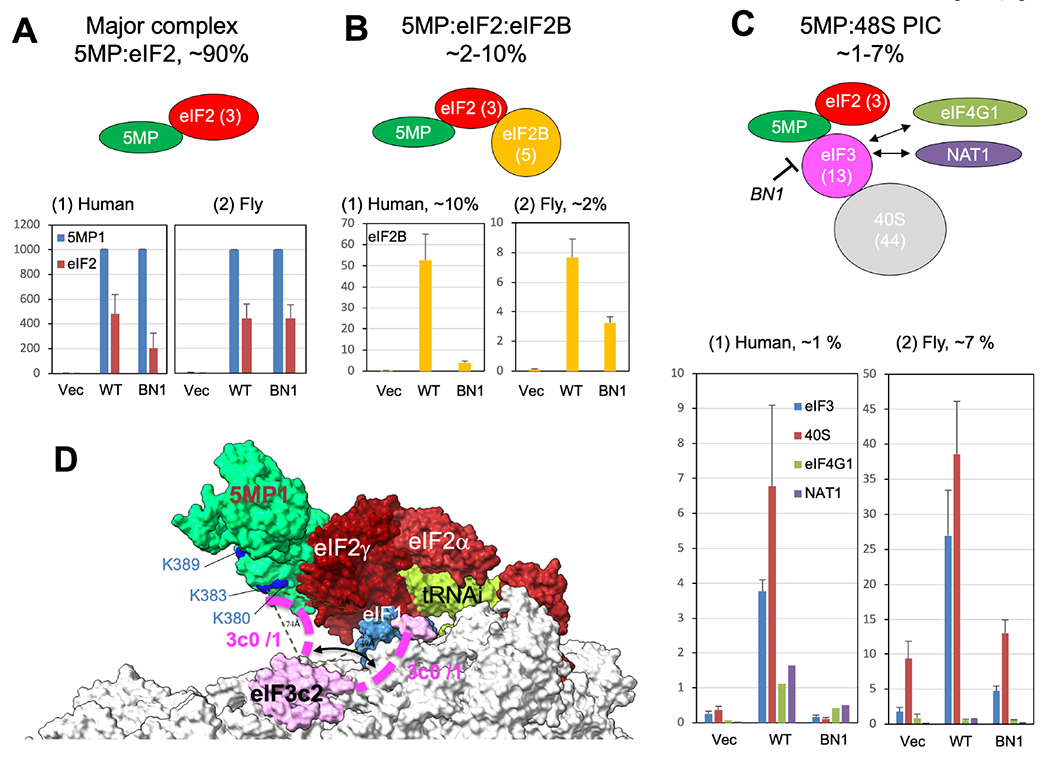
(A)-(C) 5MP-eIF complexes were purified from HEK293T (panel 1) or fly S2 cells (panel 2) and subjected for whole-lane MS analysis, as described previously (Kozel et al., 2016) (also see Supplementary Text and Fig. S3A). Graphs show emPAI values in y axis, a proxy for relative molecular amounts of proteins detected (Ishihama et al., 2005), obtained from the whole lane of the mock-treated sample (Vec), WT FL-5MP1 complex (WT) and FL-5MP1-BN1 complex (BN1), averaged for all the experiments and subunits (except human experiment 2 for C, panel 1). Data from co-purified components were presented based on their membership in the following complexes, 5MP1-eIF2 (panel 1), 5MP1-eIF2-eIF2B (panel 2) and 5MP1-PIC (panel 3). Their relative ratios in the panel title were measured by % compared to average emPAI values of eIF2 in WT. Bars indicate SEM. (D) A 5MP1-PIC docking model. A view from the mRNA-exit side is presented to describe the location of 5MP1 (green) relative to the locations of 3c2 segment of eIF3c (pink) and eIF1 (blue). The lysine residues mutated by BN1 are highlighted in blue and labeled if visible. Purple dotted line, possible alternative locations of 3c0/1 segment of eIF3c. See supplemental text for generation of the docking model.
5MP1-eIF3c binding ensures accurate initiation from AUG codons in human cells
We next sought to determine whether binding to eIF3 is critical for 5MP-mediated repression of non-AUG translation in humans. Since the exclusion of eIF5 appears to be critical for 5MP’s role in accurate initiation, the h5MP1-BN1 mutation disrupting eIF3 binding is expected to eliminate its ability to govern accurate initiation. WT h5MP1 OE suppressed CUG and GUG initiated translation in HEK293T, while h5MP1-7A disrupting eIF2 binding partially alleviated its effect on translation from these codons (Fig. 3A) (Tang et al., 2017). In contrast, h5MP1-BN1 had no effect compared to vector (Fig. 3A), even though its expression is equivalent to WT (Fig. 1F, lanes 1-3). However, when both h5MP and eIF5 were co-expressed, only WT h5MP1 was able to suppress non-AUG translation (Fig. 3B). Thus, while binding of h5MP1 to eIF2 helps promote repression of non-AUG initiation, binding of h5MP1 to eIF3c through its basic surface is more crucial for stringent AUG selection in human cells, in agreement with the idea of the two levels of competition, one outside of the ribosome for eIF2 and the other within the PIC involving eIF3c. Conversely, the knock down of h5MP1 or h5MP2 (Kozel et al., 2016) increased non-AUG translation from the ACG-initiated luciferase reporter (Fig. 3C). This indicates that basal levels of non-AUG translation are kept low by both the copies of human 5MP through active competition with eIF5 (Tang et al., 2017).
Fig. 3. Effect of 5MP1, its BN1 mutant or knockdown on general non-AUG translation in human cells.
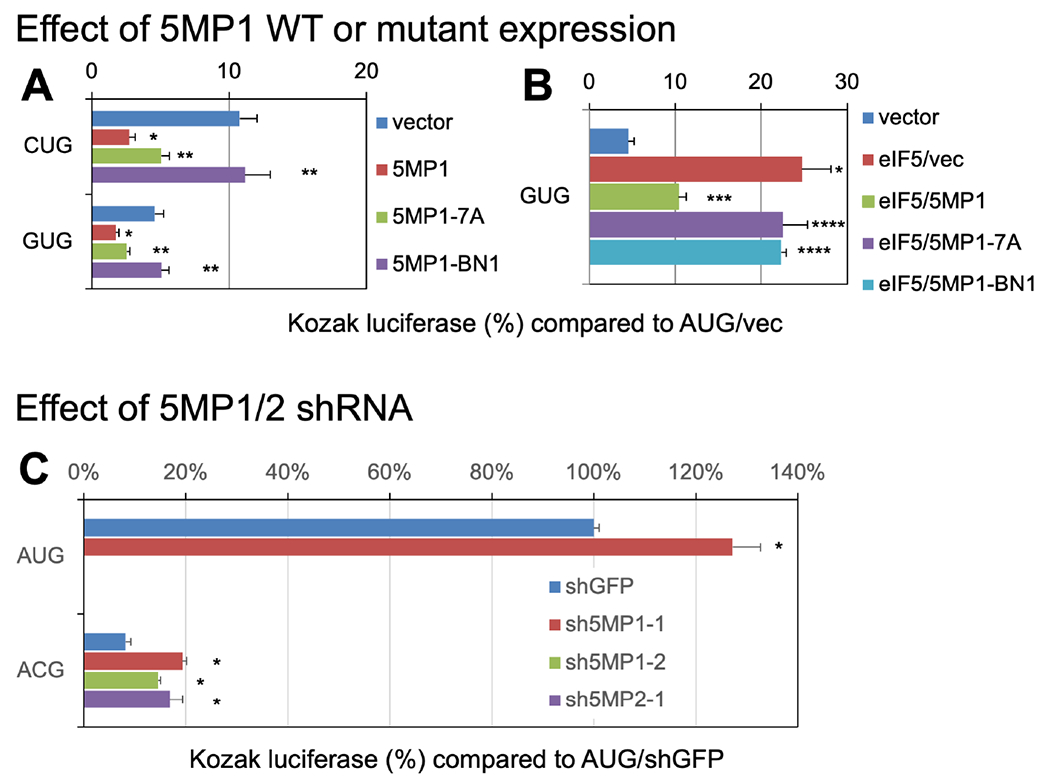
HEK293T was transfected with the indicated non-AUG firefly luciferase (Fluc) reporter plasmid, AUG Renilla luciferase (Rluc) control plasmid, and a pEF1A derivative expressing indicated proteins under the EF1A promoter (Key Resource Table) or an sh5MP plasmid (5:1 compared to firefly plasmid: sh5MP1-1, p1462; sh5MP1-2, p1465; sh5MP2-1, p1469 [Key Resource Table]), and assayed for firefly and Renilla luciferase activities with DualGlo™ dual luficerase assay system (Promega). All the experiments were conducted in parallel with cells transfected with the combination of AUG-Fluc, AUG-Rluc and an appropriate expression control plasmid (an empty vector in A and B, or shGFP plasmid in C). The firefly/Renilla expression ratios were then presented after normalizing to the ratio obtained with this AUG-Fluc control experiment. In (A and B), except for results with BN1, data were adapted from (Tang et al., 2017). Asterisks denote statistical significance (P < 0.05) compared to relevant controls; (A and B) *, p<0.05 compared to vector control (n=4); **, p<0.05 compared to 5MP1 (n=4); ***, p<0.05 compared to eIF5/vec (n=6); ****, p<0.05 compared to eIF5/5MP1 (n=6 with 7A, n=4 with BN1); (C) *, p<0.05 compared to vector control (n=4). The transient transfection of the sh5MP plasmids used in this study reduces expression of corresponding targets, h5MP1 and h5MP2, to ~10-20% and ~20-50%, respectively, as measured with immunoblotting (Kozel et al., 2016).
5MP is recruited to the PIC through eIF3 in humans and flies
Next, we attempted to examine the effect of the BN1 mutation on 5MP recruitment to the PIC by combining FLAG-affinity purification and semi-quantitative mass spectrometry (MS), as we previously described (Kozel et al., 2016). The molecular amount of each associated protein was deduced from the total whole-lane emPAI value as proxy for molarity (Ishihama et al., 2005; Kozel et al., 2016) (Tables S1 and S2). The sizes of intact associated proteins were verified by a peak of peptide counts in the predicted locations within the gel (Fig. S3). Fig. 4A–C summarizes the emPAI values for FLAG-tagged 5MP1 (human, panel 1) or 5MP/Kra (fly, panel 2) and their major associated proteins, averaged from all the experiments (however, Fig. 4C, panel 1, shows the average from Experiment 2 only, as described below). As shown in Fig. 4A, panel 1, and Table S3, rows 9-11, ~1000 emPAI units of WT h5MP1 were found associated with nearly half the amount (480 units) of eIF2 (column O, row 9), while ~1000 emPAI units of h5MP1-BN1 associated with reduced amount (~200 units) of eIF2 (p=0.05, n=6 with 3 subunits in 2 experiments). Thus, BN1 reduced h5MP1-eIF2 binding, in agreement with the FLAG-IP experiment (Fig. 1G). A smaller amount (52 units) of eIF2B was associated with WT h5MP1 (Fig. 4B, panel 2; Table S3, column O, row 31), again in a manner reduced by BN1 (p=0.02, n=5 for subunit averages or p=0.004, n= 10, from 5 subunits in 2 expts). Most likely, h5MP1:eIF2B interaction is bridged by eIF2, similar to eIF5:eIF2:eIF2B complex previously observed (Kozel et al., 2016; Singh et al., 2006). In contrast, little or no eIF5 or h5MP2 was associated with WT h5MP1 (Table S3, rows 4 and 29), indicating the specificity of these experiments and in agreement with the idea that h5MP1 competes with eIF5 for eIF2.
As shown in Fig. 4C, panel 1, WT h5MP1 bound roughly equivalent amounts of eIF3 and the 40S subunit and smaller amounts of eIF4G1 and NAT1/eIF4G2 in one (Exp. 2) of the two sets of IP experiments. The amount of h5MP1:eIF3:40S(:eIF2) complex is therefore ~1 % of total h5MP1:eIF2 complex abundance. BN1 almost eliminated these interactions, indicating that the basic surface altered by BN1 is the primary binding site for eIF3(c). In the other biological replicate (Exp. 1), a larger (statistically significant) amount of eIF3 bound to WT h5MP1 (23 unit in Exp 1 vs 4 units in Exp 2; Table S3, column O, rows 17 and 19) at the expense of non-specific binding (6 units) observed in the vector control experiment. This trend was even stronger with 40S binding (Table S3, column O, rows 41 and 43), with 93 vs 133 emPAI units on average bound to mock-treated vs h5MP1 samples, respectively, in Exp 1. Nevertheless, on average, ~13 emPAI units of eIF3 was associated with WT h5MP1, ~10 units above the background level (~3 units) of eIF3 binding (Table S3, row 13), equivalent to ~2% of the amount of h5MP1:eIF2 complex. Again, BN1 strongly reduced eIF3 binding to h5MP1, with only 0.5 units bound to h5MP1-BN1.
In summary, as shown in Fig. 4A–C, panels 1, eIF2, eIF2B, eIF3 and 40S were associated with ~50%, ~5%, ~1%, and ~1% of purified h5MP1, respectively (also see Fig. S3, Tables S1 and S3). Thus, the majority (~90%) of h5MP1:eIF2 complex are free of other eIFs. Equivalent amounts of eIF3 and 40S co-precipitated with 5MP1 and were primarily absent in the BN1 mutant pulldowns, supporting a model in which 5MP1 binding to the 40S subunit is mediated by mutual interaction between eIF3 and the basic surface of 5MP1 (Fig. 4C). Low levels of eIF4G1 and NAT1 (aka eIF4G2/p97/DAP5) co-precipitated with WT 5MP1 but not with 5MP1-BN1 in human cells, again supporting a role for eIF3 in bridging these eIF4 factors (schematics in Fig. 4C) (Brito Querido et al., 2020).
To determine whether the 5MP interaction observed in human is conserved in flies, we expressed FLAG-tagged Drosophila melanogaster (Dme) 5MP/Kra and its BN1 mutant in S2 cells and analyzed their affinity-purified products by MS (Fig. S3B–D and Tables S2 and S4). We likewise found that the majority of 5MP:eIF2 complex are free of eIFs, and their minor proportions are partitioned into 5MP:eIF2:eIF2B and 5MP:43S/48S PIC complexes, although in the latter the binding of eIF4G1 was obscured by the fact that it was also found in the mock-treated fraction (Fig. 4A–C, panels 2). Importantly, equivalent amounts of the 40S and eIF3 (~20-30 emPAI units) were associated with 5MP above the background (vector control) level, dependent on the 5MP basic surface altered by BN1 (p=0.0008, n= 39 from 13 subunits in 3 experiments) (Table S4 and Fig. 4C, panel 2).
Encouraged by these findings, we performed docking simulations to deduce the 5MP binding site within the PIC (see Supplementary text for details). Our results using a homology model of human 5MP1 (Fig. 1B–C) and yeast 48S PIC open structure (Llacer et al., 2015) showed that 5MP is recruited to eIF2 through a basic surface of eIF2γ (Fig. 4D, S4A–B), even though the PIC structure lacked eIF2β-NTD crucial for 5MP binding to eIF2 (Hiraishi et al., 2014; Singh et al., 2011). This is remarkable as the predicted location of 5MP is homologous to the recently discovered location of eIF5 within Trypanosoma PIC with eIF2β-NTD bound near the acidic face of its CTD (Fig. S4B) (Bochler et al., 2020). The docking study places the 5MP basic surface (blue highlights in green) and eIF1 (blue) at 74 and 49 Å, respectively, from the eIF3c2 segment (pink), allowing the more N-terminally located eIF3c0/1 segment (purple dotted line) to reach these alternative targets (Fig. 4D). Thus, this model depicts how 5MP is recruited to the PIC through the NTDs of eIF2β and eIF3c and, once it is recruited, how it prevents the eIF3c0/1 segment from touching eIF1 and thereby destabilizing the open PIC structure (Obayashi et al., 2017). This promotes stringent initiation at AUGs. 5MP and eIF5 bind to the same sites of eIF2 and eIF3, thus eIF5 is excluded from the PIC in the presence of 5MP (Fig. 2E and 4C). This favors accurate initiation, as eIF5-NTD would otherwise destabilize the open PIC by excluding eIF1 from the P-site (Fig. 1A and 4C) (Llacer et al., 2018).
5MP selectively suppresses CGG RAN translation
RAN translation at CGG repeats is regulated in an initiation codon-dependent manner and enhanced by eIF5 OE, suggesting that 5MP could regulate this aberrant process (Green, 2017; Linsalata, 2019) (also see Fig. 5B, panels 2 and 4, columns labeled eIF5). To determine if 5MP has an impact on RAN translation, we co-transfected HEK293T cells with empty, h5MP1 or h5MP2 OE vectors and our CGG-Nluc RAN reporters in multiple reading frames (Fig. 5A). 5MP1/2 OE reduced RAN translation 2 fold in both the +1 (polyglycine) and +2 (polyalanine) reading frames (Fig. 5A–B and S5A). Consistent with these observations, knockdown of either 5MP1 or 5MP2 significantly enhanced CGG RAN translation in the +1 and +2 reading frames while having no effect on a canonically AUG-initiated Nluc control (Fig. 5C). Thus, both the two copies of human 5MP, 5MP1 and 5MP2, are involved in regulating RAN translation. This 5MP-mediated inhibition of RAN translation in both the +1 and +2 reading frames was dependent on interactions with eIF3c, as the BN1 mutant was unable to repress RAN translation (Fig. 5D and S5B). The 7A mutant less strongly inhibited 5MP repression of RAN translation, again supporting the idea that interaction with eIF3c, and not eIF2, is key for inhibition of non-AUG initiation (Fig 5D). The similar effects of BN1 and 7A mutations to those for regular non-AUG translation (Fig. 3A) support the notion that 5MP regulates RAN and canonical non-AUG initiated translation through eIF3 by the same mechanism.
Fig. 5. 5MP represses RAN translation through eIF3c.
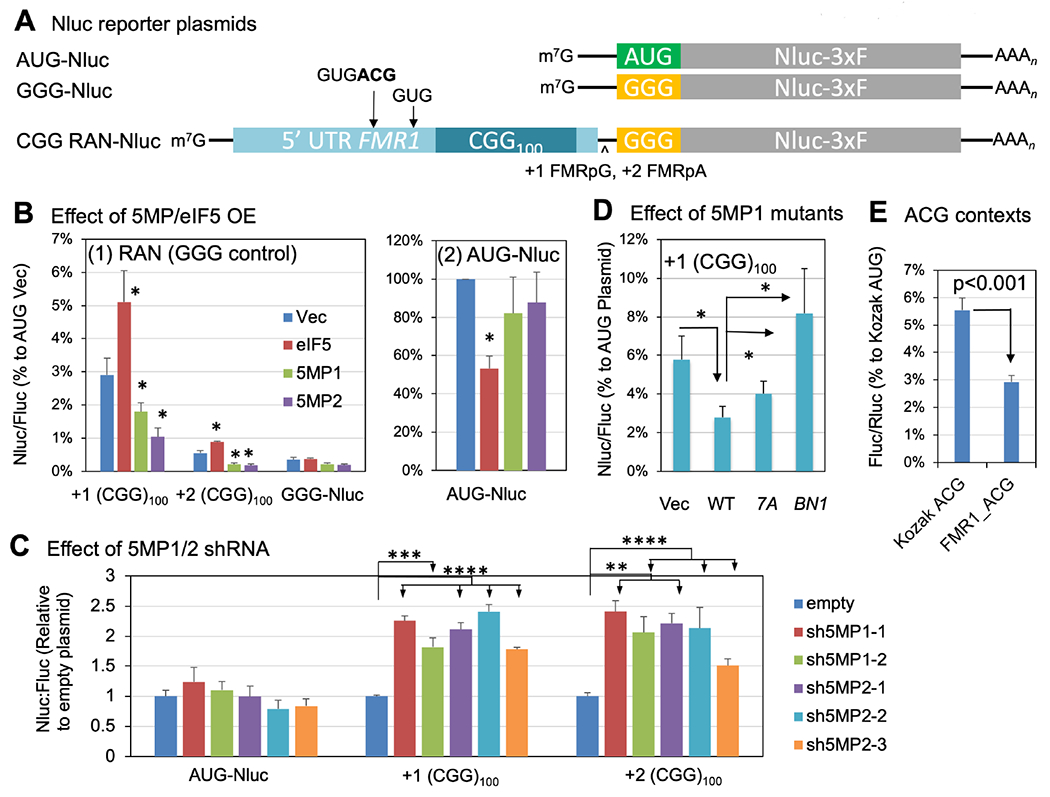
(A) NanoLuciferase (Nluc)-3xF plasmids used in this study. Arrows indicate the location of non-AUG start codon initiating translation of the +1 CGG reading frame (Kearse et al., 2016); the ACG codon studied in panel (E) was boldfaced. ^, 1 or 2 base-insertions to make FMR1-polyG or -polyA in frame to Nluc. (B) HEK293T transfected with indicated NanoLuciferase-3xF plasmid, a eIF5 or 5MP1/2-expressing plasmid and a firefly luciferase plasmid was assayed for nano- and firefly luciferase activities. Nluc/Fluc ratio was normalized to the values with AUG vector control transfectant. Bars indicate standard errors. *, p<0.05 compared to vector control in 4 or more independent experiments. (C) Expression of Nluc RAN translation and AUG Fluc control reporters coexpressed with empty plasmid or plasmid expressing sh5MPs (sh5MP1-1, p1462; sh5MP1-2, p1465; sh5MP2-1, p1467; sh5MP2-2, p1469; sh5MP2-3, p1470, see Key Resource Table) in HEK293Ts. Bars represent mean +/− SEM, N=6. 2 way ANOVA:p<0.0001. Two tailed Student’s T-test with Bonferroni correction, **p<0.01, ***p<0.001, ****p<0.0001. (D) Effect of 5MP1 mutations. Assays were done similar to (B) with 5MP1 plasmids with indicated mutations. Nluc/Fluc ratio was normalized to the value from AUG-Nluc transfectant with the same expression plasmid [e. g. the value from +1(CGG)100/5MP1 compared to that from AUG/5MP1] to correct for changes in AUG expression by each plasmid. *p<0.05 in 7 or more experiments. (E) ACG initiation frequency from its isolated FMR1 context located outside of the repeats. Firefly luciferase activity from the reporter bearing the 24-nt-long region upstream of the ACG codon was assessed along with a control Rluc reporter and compared to the values from Kozak AUG and ACG contexts (n=6), as in Fig. 3, after normalizing to one from Kozak AUG.
To test if the FMR1 CGG RAN translation is explained by non-AUG initiation independent of the repeats, we used an ACG-Fluc reporter with the 24-nt-long sequence context (GUG ACG G) that exists before the ACG codon within the FMR1 5’UTR but absent in the repeat (Kearse et al., 2016). Initiation at this ACG start codon was 2.9+0.2 % (n=6) compared to AUG under normal Kozak context (ACC AUG G) and smaller than the ACG initiation from the same Kozak context (Fig. 5E). Since this level of ACG initiation is equivalent to the frequency of +1 CGG translation as compared to the AUG/Vec control (Fig. 5B), the majority of +1 CGG translation appears to be explained by translation of the ACG start codon upstream of the repeat reading frame, consistent with previous works (Kearse et al., 2016).
5MP/Kra expression suppresses FMRpolyG-mediated toxicity in flies
FMRpolyG accumulates in ubiquinated inclusions in FXTAS patient neurons and FMRpolyG is sufficient to induce inclusion formation in transfected cells. RAN translation of FMRpolyG from CGG repeats is required for their toxicity in multiple model systems, including Drosophila (Bonapace et al., 2019; Hoem et al., 2019; Krans et al., 2019; Oh et al., 2015; Rodriguez et al., 2020; Sellier et al., 2017; Todd et al., 2013). To determine if 5MP/Kra (encoded by krasavietz in Drosophila) expression reduces RAN translation in Drosophila, we crossed two UAS-5MP/Kra OE lines to a GMR-Gal4, UAS-(CGG)103-EGFP line (Todd et al., 2013). Importantly, Kra OE resulted in an almost complete loss of EGFP positive aggregates in the fly eye of both the lines (Fig. S6A). Kra OE significantly reduced FMRpolyG-EGFP levels in the Kra-5 line, and the levels of EGFP that initiates from a downstream ATG codon (Fig. S6B).
To determine if Kra OE alters CGG repeat-induced rough eye phenotypes in a Drosophila eye model, we crossed a GMR-Gal4 driven UAS-(CGG)90-EGFP line, previously shown to have a robust eye phenotype (Jin et al., 2003; Todd et al., 2013), to three UAS-5MP/Kra OE lines, a Kra heterozygous loss of function mutant (P[lacW] element insertion: Kraj9B6), and 2 siKra lines. All three 5MP/Kra OE lines had no overt rough eye phenotype on their own, but rescued the overall rough eye phenotype in males compared to a GFP control (Fig. 6A–C). No rescue was observed in females, however, this is likely due to the fact that females have a milder phenotype to begin with (Fig. S6D–E).
Fig. 6. Drosophilla 5MP/Kra rescues FMRpolyG-induced neuronal toxicity.
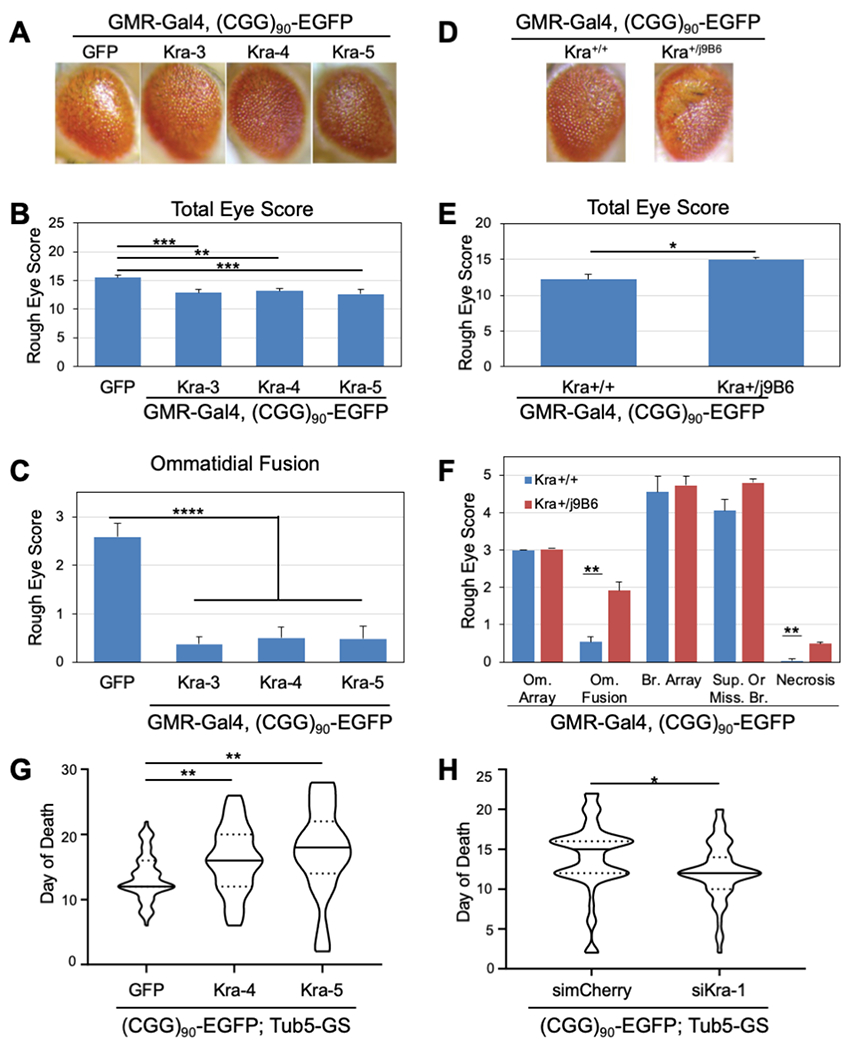
(A) Representative images of male fly eyes expressing (CGG)90-EGFP under GMR-GAL4 with Kra OE. B-C) Quantification of male B) total eye phenotype and C) ommatidial fusion phenotype of GMR-GAL4, (CGG)90-EGFP crossed to Kra OE lines. GFP serves as a control. Bars represent mean +/− stdev for 3 individual experiments (N≥25/genotype/experiment with the exception of 1 experiment for Kra-5 (N=10). One-way ANOVA B)***, C)**** with Dunnett’s multiple-comparison test. D) Representative images of male fly eyes expressing (CGG)90-EGFP under GMR-GAL4 with WT or heterozygous Kraj9B6 mutant. E-F) Quantification of male E) total eye phenotype and F) Individual eye category phenotype of GMR-GAL4, (CGG)90-EGFP crossed to WT or heterozygous Kraj9B6 mutant lines. Bars represent mean +/− stdev for 3 individual experiments (N≥20/genotype/experiment with the exception of 1 experiment (N≥11). Two-tailed Welch’s t-test with Bonferonni correction. G-H) Quantification of survival of flies expressing (CGG)90-EGFP under GMR-GAL4 with G) Kra OE or H) siKra. Solid lines represent median day of death, dotted lines represent 25% and 75% quartiles, for N≥35/genotype (from 2-3 experiments/genotype). Two-tailed Welch’s T-test with Bonferonni correction. *p<0.05, **p<0.01., ***p<0.001, ****p<0.0001.GFP and simCherry are controls.
GMR-driven knockdown of Kra alone had no rough eye phenotype, but in combination with the (CGG)90-EGFP was synthetic lethal, suggesting a direct and important contribution of this factor to basal suppression of CGG repeat associated toxicity. However, a hypomorphic Kraj9B6 mutant was viable in context with (CGG)90-EGFP. The (CGG)90-EGFP rough eye phenotype was significantly enhanced by Kraj9B6 for both male and females (Figure 6D–F and S6F–G).
Given that Kra could impact on developmental phenotypes that might elicit rough eye effects observed with CGG repeats, we evaluated a second disease-relevant phenotype. Activation of ubiquitous (CGG)90-EGFP expression after eclosion significantly reduces adult lifespan in Drosophila (Linsalata, 2019; Jin et al, 2003). To determine whether Kra expression influences adult lifespan in (CGG)90-EGFP expressing flies, we co-expressed (CGG)90-EGFP post-eclosion with either Kra OE or siKra with transgene induction activated, using a Tub5 Geneswitch inducible driver. Kra OE significantly increased lifespan in our (CGG)90-EGFP line compared to a GFP control in both male and females (Fig. 6G and S6F). In contrast, Kra knockdown significantly reduced lifespan of our (CGG)90-EGFP expressing flies compared to a simCherry control in males (Fig. 6H and S6I). These results indicate that 5MP/Kra negatively regulates RAN translation in vivo.
Discussion
Despite the progress in understanding detailed molecular functions in translation initiation, the role of 5MP is among the least understood. Here, by investigating similarities between 5MP and eIF5, we have uncovered the molecular mechanism by which 5MP regulates start codon stringency. The amino terminal domain of eIF5 was originally identified as the GAP for eIF2 (Algire et al., 2005), functioning early in the 43S/48S PIC formation. In contrast, the carboxyl terminal domain of eIF5 is integral to MFC assembly and function, playing a key role in initiating scanning (Fig. 7A) and maintaining the scanning-competent conformation of the PIC directly and indirectly (Fig. 7B) (Asano et al., 2000). More recently, eIF5 was shown to destabilize eIF1 through directly binding its NTD to the P-site, placing its function late initiation pathway (Fig. 7C) (Llacer et al., 2018). These activities are mediated by eIF2β-NTD tethering eIF5-CTD through its acidic surface, and placing eIF5-CTD in proximity to eIF2γ within the PIC (Bochler et al., 2020). Being corroborated with the role of its CTD as a core of MFC assembly (Yamamoto et al., 2005), this tethering in turn allows eIF5-CTD to bind other MFC partners located away from eIF2γ, letting it regulate the binding and release of eIF1 (Luna et al., 2012; Singh et al., 2012).
Fig. 7. Model of translation initiation and its control by 5MP.
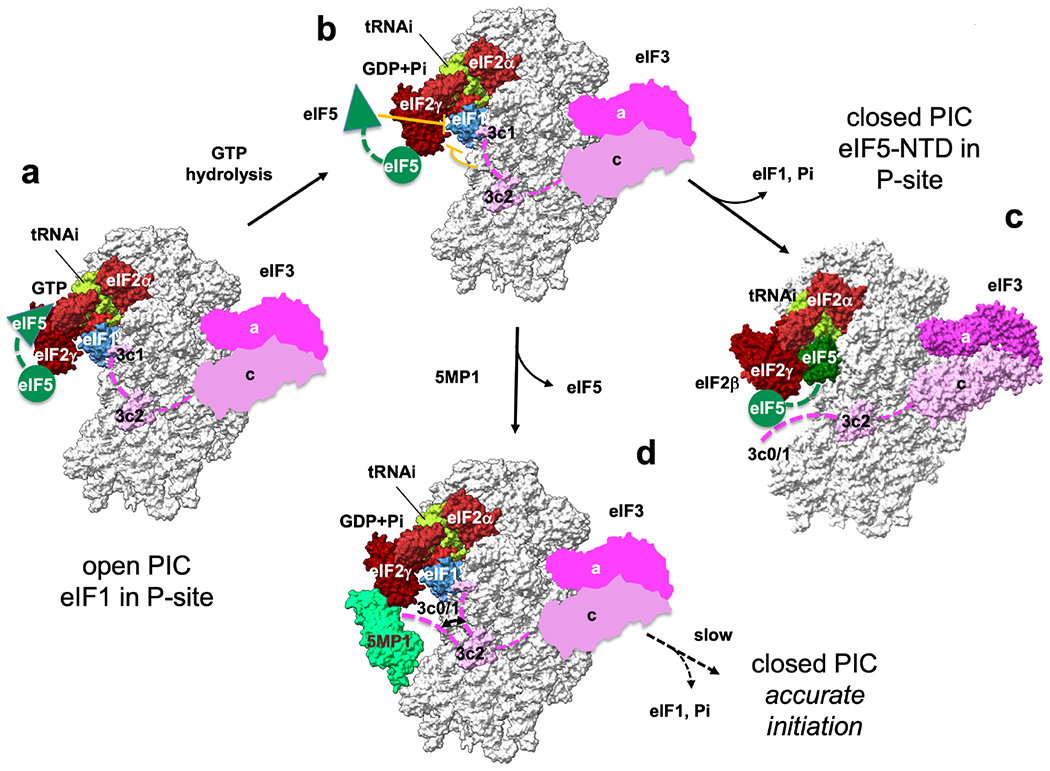
(a) MFC components stimulate a change to the scanning-competent open PIC conformation, while eIF5-NTD catalyzes GTP hydrolysis for eIF2. (b) During scanning, eIF1 plays the central role in keeping the open conformation necessary for accurate initiation at AUG codon. However, eIF5-NTD (Llacer et al., 2018) and eIF3c N-terminal tail termed eIF3c0 (Obayashi et al., 2017) intrinsically destabilize this conformation (stop bars in orange). For this reason, eukaryotes allow a certain level of general and repeat-associated non-AUG translation. (c) On AUG recognition, eIF1 and Pi are released to trigger the conformational change to the closed state, leading to translation initiation. eIF5-NTD plays a pivotal role in this process by physically excluding eIF1 from the P-site and binding to the P-site (Llacer et al., 2018). (d) 5MP can increase the accuracy of initiation by antagonizing the destabilizing effects caused by both eIF5-NTD and eIF3c0. Open and closed PIC structures are based on 3JAQ and 6FYX, respectively. In open PICs, the location of eIF3 is based on 6FYX. The location of eIF5-CTD (green circle) is based on (Bochler et al., 2020). Green triangle denotes eIF5-NTD responsible for GTP hydrolysis for eIF2 and docking into P-site on AUG recognition. Green dotted line, the linker between NTD and CTD of eIF5. Pink dotted line, eIF3c-N-terminal tail.
Within the context of this previous work and our own findings, we propose that 5MP displaces eIF5 through direct competition with eIF2, mediated by eIF3c and potentially other parts of eIF3 (Fig. 7D). eIF2β-NTD (Lysine-boxes) (Asano et al., 1999) and the whole external surface of eIF2γ are basic (Fig. S4A), and hence ideal for interacting with the acidic face of eIF5 or 5MP (Fig. 1B–C). The other side of eIF5 or 5MP is basic and binds eIF3c. Similar to what has been observed for eIF2:eIF5 interactions within the PIC (Bochler et al., 2020), our data supports a model in which a 5MP-eIF2β-NTD interaction allows 5MP to face eIF2γ and form a stable 5MP:eIF2 complex. Our data also support an essential role for a 5MP:eIF3c interaction in mediating 5MP:eIF2 binding within the PIC and recruiting 5MP to the PIC, particularly when eIF5 is present. The eIF3c N-terminal tail normally interacts with the ribosome-binding face of eIF1 to impede scanning (Obayashi et al., 2017), but 5MP binding to eIF3c prevents this interaction. Furthermore, 5MP displacement of eIF5 would in principle inhibit eIF5-NTD from interfering with eIF1 binding to the P-site (Llacer et al., 2018). As eIF5 OE suppresses non-AUG translation through its NTD (Fig. 2B), eIF5 in this context would otherwise decrease the accuracy of initiation. Thus, the presence of 5MP within the PIC promotes accurate start site selection by two mechanisms (Fig. 7D). Further works are warranted to define the precise molecular roles of eIF3c, eIF1 and potentially other parts of the MFC in the proposed conformational change model of 5MP-mediated accuracy of translation initiation.
We previously showed that 5MP could suppress non-AUG initiation when eIF2 binding was compromised (Tang et al., 2017) (Fig. 3A). Here we show that 5MP inhibition of non-AUG initiation requires binding to eIF3c. 5MP was also able to suppress RAN translation at CGG repeats within the 5’ UTR of FMR1, in an eIF3c-binding dependent manner in multiple reading frames (Fig. 5). While the +1 reading frame harbors a number of non-AUG start codons, the +2 reading frame does not. The exact mechanism by which fly 5MP inhibits translation in this frame is not clear; however, numerous examples of N-termini beginning with a GCG codon, particularly those that code for polyalanine stretches, have recently been identified, suggesting that initiation could occur at this codon, despite having little in common with the canonical AUG start codon (Na et al., 2018). This suggests that 5MP could be inhibiting RAN translation in both the +1 and +2 reading frames via similar mechanisms as for regular non-AUG translation and thus might have relevance at other disease-causing repeats. Intriguingly, 5MP OE suppressed neuronal toxicity and improved lifespan in Drosophila expressing the expanded CGG repeat (Fig. 6 and S6). Taken together, these data suggest that modulation of 5MP levels could be of viable therapeutic target by which to selectively reduce RAN translation in repeat expansion disorders.
In conclusion, we propose that 5MP suppresses general and repeat-associated non-AUG translation by effectively outcompeting eIF5 within the PIC after eIF5 activates GTP hydrolysis for eIF2 and with a direct recruitment role for eIF3c. Moreover, the demonstration that 5MP enhances the lifespan of flies expressing the toxic CGG repeats capable of supporting RAN translation suggests a potential therapeutic avenue for selective enhancement of start-codon stringency in conditions where RAN translation is thought to contribute to disease pathogenesis. This detailed mechanistic understanding of the role of 5MP in non-AUG initiation should provide a path towards such developments in both neurodegenerative disorders and in other conditions where non-canonical initiation contributes to disease.
Limitation of the Study
While we used 5MP/eIF5-BN1, a five-amino acid substitution mutation, to eliminate the interaction with eIF3 throughout this paper, a minor concern remains whether the effect of this mutation in 5MP is limited to eIF3 binding in human or fly cells. It is also unclear if eIF3c-NTD is the sole binding partner of the basic surface of 5MP within eIF3. Given that numerous other factors (eIF3g, eIF3i, eIF3b and ABCE1) interact with the interface side of the 40S (Llacer et al., 2015; Simonetti et al., 2020) besides eIF1, eIF1A and Met-tRNAiMet-bound eIF2 shown in Fig. 7, it is very likely that there is a mutual cooperativity and induced-fit mechanism(s) to stabilize 5MP recruitment to the PIC. More work is warranted to elucidate the mechanism of initiation accuracy involving 5MP.
The mechanism of 5MP regulation of RAN translation requires more work as well, related to the control of RAN translation from a triplet codon within the repeat. Our recent works show rather very low initiation frequencies from such repeat codons, typically less than 1% compared to normal AUG initiation (e. g. Fig. 5). With expression so low, it may be even difficult to dissect defined mechanisms leading to that event. However, the common 5MP involvement in RAN translation of both the +1 and +2 reading frames and similar mutational effects (by 7A and BN1) strongly suggest that RAN translation is mediated by a faulty initiation that escapes from the rigorous cap-dependent mechanism. It should be emphasized, however, that the subtle expression of the repeat peptides does impact human life through their accumulation over time in long-lived cells such as neurons.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Katsura Asano (kasano@ksu.edu).
Materials Availability Statements
All the materials generated in this study, including plasmids, yeast strains, and fly and cell lines, are available upon request (Materials Transfer Agreement may be necessary).
Data and Code Availability Statement
The raw data of MS analyses were deposited on jPOST under accession JPST001188 (ProteomeXchange under accession PXD026284).
The 5MP homology model structure and its model docked with eIF2γ or PIC are available at https://github.com/10ackert/5MP-PIC.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Human cell lines HEK293T was maintained under sterile culture conditions in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum and recommended amounts of penicillin and streptomycin at 37°C/5% CO2 in air. Drosophila S2 cells were maintained in Schneider’s medium supplemented with 10% fetal bovine serum and recommended amounts of penicillin and streptomycin at 27° C.
Fly lines
The two FMRpolyG-EGFP fly lines, GMR-Gal4, UAS (CGG)90-EGFP, and UAS (CGG)103-EGFP (#168), have been previously published (Jin et al., 2003; Todd et al., 2013). Kra OE lines were made by inserting d5MP-Flag into pUAST between BglII and KpnI sites (see below for sequence), followed by PhiC31 integrase-mediated insertion on the second chromosome (BestGene Inc.). Kraj9B6 (10216) and Kra KD lines (27248, 38918) were all purchased from Bloomington.
Yeast strains
The yeast Saccharomyces cerevisiae Sui− mutant strains KAY1027 [his4-301(ACG) trp1 leu1 ura3 sui1Δ p(sui1-mof2-1 TRP1)] and KAY1027 [his4-301(ACG) trp1 leu1 ura3 sui1Δ p(sui1-K60E TRP1)] were transformed to Leu+ with appropriate expression plasmids and grown in synthetic complete (SC) medium lacking leucine (Lee et al., 2007). S. cerevsiae KAY1121 was constructed by transforming JCY03 (Cheung et al., 2007) to Trp+ with YCpW-SUI1-K60E (p1782, TRP1 SUI1-K60E), followed by the eviction of the resident SUI1+ URA3 plasmid by selecting against URA3 using the drug 5-flouroorotic acid. Permanent stocks were generated in 30% glycerol and stored at −80° C.
METHODS DETAILS
5MP1 homology modeling and docking studies
The homology-model structure of h5MP1 was obtained using I-TASSER (Iterative Threading ASSEmbly Refinement) server (Roy et al., 2010; Yang et al., 2015; Yang and Zhang, 2015). The human 5MP1-CTD and yeast eIF5-CTD (PBD ID: 2FUL) were prepared for electrostatics calculations by PDB2PQR (Dolinsky et al., 2007; Dolinsky et al., 2004). Electrostatic potential distribution on solvent-accessible surfaces of 5MP1 and yeast eIF5 CTD were then calculated by Adaptive Poisson-Boltzmann Solver (APBS) (Baker et al., 2001). The electrostatic potentials and solvent-accessible surfaces were visualized by Chimera (Pettersen et al., 2004).
The 5MP1 homology model was used to dock initially with yeast 48S preinitiation open complex (PDB ID: 3JAQ). Since 5MP1 tended to bind eIF2γ within the PIC in multiple attempts albeit at its different but mostly basic sites, yeast eIF2γ structure taken from the PIC structure was used to dock 5MP1, resulting in a refined model shown in Fig. 4D, 7, S4 and S7. Both 5MP1 and eIF2γ were prepared by using the Schrödinger Protein Preparation Wizard (Sastry et al., 2013) and subjected for the protein-protein docking process implemented in BioLuminate (Chuang et al., 2008; Kozakov et al., 2006) (Schrödinger, 2020-3). All of the figures were made by Chimera (Pettersen et al., 2004) and ChimeraX (Goddard et al., 2018). The closed state of preinitiation complex structure is based on the yeast 48S preinitiation complex with eIF5 N-terminal domain (PDB ID: 6FYX).
Plasmid construction
Human eIF3c cDNA segments with defined ends were amplified by PCR using the oligos listed in Key Resource Table and cloned into pGBfusion1 (Reibarkh et al., 2008) after BamHI digestion to generate pGB-heIF3c plasmids (Key Resource Table). These plasmids were then used to produce in E. coli the heIF3c fragments employed in Fig. 1 and S1. The BN1 mutation (K380Q K383Q H386Q K389Q K391Q in Homo sapiens and R380Q K383Q H386Q K389Q K391Q in Drosophila melanogaster) was introduced by QuickChange™ (Promega) with oligos listed in Key Resource Table to the relevant h5MP1- or Dme5MP-expression plasmids (Key Resource Table). YEpL-GB-TIF5B5-F and its BN1 mutant derivative were generated by subcloning into YEpL-TIF5-F (Key Resource Table) a 0.8-kb NdeI-SalI fragment encoding N-terminal GB1-fusion to Tif5p (aa. 201-405) WT or BN1 (Yamamoto et al., 2005), respectively, FLAG-tagged at its C-terminus. To produce the ACG firefly luciferase reporter plasmid p1785 (Key Resource Table), the complimentary oligos listed in Key Resource Table were annealed and inserted between the PstI and BamHI sites of pSV40 firefly Kozak ACG (p1525).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| h5MP1 and its 7A and BN1 mutant forms | This paper | N/A |
| GST-h5MP1 and its 7A and BN1 mutant forms | This paper | N/A |
| GB-heIF3c20-102 | This paper | N/A |
| GB- heIF2β53-136 | This paper | N/A |
| Critical commercial assays | ||
| Dual-Glo® Luciferase Assay System | Promega | E2940 |
| Nano-Glo® Luciferase Assay System | Promega | N1110 |
| Deposited data | ||
| Raw MS data | This paper (Fig. 4A–C) | JPST001188 |
| Homology model structure of h5MP1 and its docking simulations | This paper (Fig. 4D&S7) | https://github.com/10ackert/5MP-PIC |
| Experimental models: Cell lines | ||
| Human embryonic kidney (HEK) 293T | John A. Chiorini | N/A |
| D. melanogaster cell line S2 | Erika Geisbrecht | N/A |
| Experimental models: Organisms/strains | ||
| S. cerevisiae S288C: his4-301(ACG) trp1 leu1 ura3 sui1Δ p(sui1-mof2-1 TRP1) | (Tang et al., 2017) | Asano lab KAY1027 |
| S. cerevisiae S288C: his4-301(ACG) trp1 leu1 ura3 sui1Δ p(sui1- K60E TRP1) | This paper | Asano lab KAY1121 |
| D. melanogaster: GMR-Gal4, (CGG)90-eGFP BD/Cyo | (Jin et al., 2003; Todd et al., 2010) | Todd Lab #34 |
| D. melanogaster: UAS-CGG90-eGFP-BD/CyO; Tub5/TM3-Ser line B | (Todd et al., 2010) | Todd Lab #503 |
| D. melanogaster: y[1] w[1118]; P{w[+mC]=lacW}kra[j9B6]/TM3, Sb[1] | Bloomington Drosophila Stock Center |
10216 |
| D. melanogaster: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02556}attP2 | Bloomington Drosophila Stock Center |
27248 |
| D. melanogaster: y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8]=TRiP.GL00690}attP2 | Bloomington Drosophila Stock Center |
38918 |
| D. melanogaster: UAS-d5MP/Cyo | BestGene, this paper | 28098-1-M3-M-Ch2, Todd Lab #1218 |
| D. melanogaster: UAS-d5MP/Cyo | BestGene, this paper | 28098-1-M4-M-Ch2, Todd Lab #1219 |
| D. melanogaster: UAS-d5MP/Cyo | BestGene, this paper | 28098-1-M5-M-Ch2, Todd Lab # 1220 |
| Oligonucleotides | ||
| gcGGATCCGAATTCatgtcgcggtttttcaccacc | This paper (p1834 & p1835) | Asano lab 3c0_N |
| gcGGATCCGAATTCgaggagctcgtcaccaaac | This paper (p1836 & p1837) | Asano lab 3c1_N |
| ccGGATCCctcgctcagcaacaatgg | This paper (p1834 & p1836) | Asano lab 3c1_C |
| ggGGATCCgacaccttctttgtccacaatg | This paper (p1835 & p1837) | Asano lab 3c2_C |
| gcaatactgcaatggtatcaggaagcacaggttgctcaaggccaa agtgtttttcttgac |
This paper (p1659 & p1818) | Asano lab h5MP1-BN1/S |
| aaaaacactttggccttgagcaacctgtgcttcctgataccattgcagtattgcttcttc | This paper (p1659 & p1818) | Asano lab h5MP1-BN1/AS |
| ggagatcatcttgcagtggtaccaggagggccagtcaaaccagggccaaatgcatttcc | This paper (p1964) | Asano lab Dme5MP-BN1/S |
| ggaaatgcatttggccctggtttgactggccctcctggtaccactgcaagatgatctcc | This paper (p1964) | Asano lab Dme5MP-BN1/AS |
| GtagcggcgagcgcgggcggcggcggtgacggagG | This paper (p1785) | Asano lab hFMR1_1/S |
| GATCCctccgtcaccgccgccgcccgcgctcgccgctaCTG CA |
This paper (p1785) | Asano lab hFMR1_1/AS |
| Recombinant DNA | ||
| pGEX-h5MP1; GST-fusion to h5MP1 | (Singh et al., 2011), (Fig. 1 & S1) | Asano lab p1477 |
| pGEX-h5MP1-7A; pGEX-h5MP1 carrying 7A | (Singh et al., 2011), (Fig. 1 & S1) | Asano lab p1116 |
| pGEX-h5MP1-BN1; pGEX-h5MP1 carrying BN1 | This paper (Fig. 1&S1) | Asano lab p1818 |
| pGB-heIF2β-K2K3; GB1-fusion to heIF2β53-136 | (Luna et al., 2012) (Fig. 1&S1) | Asano lab p1336 |
| pGB-heIF3c12-His; GB1-fusion to heIF3c20-102 | This paper (Fig. 1&S1) | Asano lab p1837 |
| pGB-heIF3c01-His; GB1-fusion to heIF3c1-40 | This paper (Fig. S1) | Asano lab p1834 |
| pGB-heIF3c02-His; GB1-fusion to heIF3c1-102 | This paper (Fig. S1) | Asano lab p1835 |
| pGB-heIF3c1-His; GB1-fusion to heIF3c20-40 | This paper (Fig. S1) | Asano lab p1836 |
| pEF1A-h5MP1; 3xF-h5MP1 under the eEF1A promoter | (Kozel et al., 2016) (Fig. 1, 3–5) | Asano lab p1556 |
| pEF1A-h5MP1-BN1; pEF1A-h5MP1carrying BN1 |
This paper (Fig. 1, 3–5) | Asano lab p1659 |
| pEF1A-h5MP1-7A; pEF1A-h5MP1carrying 7A | (Kozel et al., 2016) (Fig. 3–5) | Asano lab p1667 |
| pEF1A-h5MP2; 3xF-h5MP2 under the eEF1A promoter | (Kozel et al., 2016) (Fig. 3–5) | Asano lab p1660 |
| pEF1A-heIF5; 3xF-heIF5 under the eEF1A promoter | (Kozel et al., 2016) (Fig. 3–5) | Asano lab p1558 |
| YEpL-TIF5-F; hc TIF5-F LEU2 plasmid | (Asano et al., 1999) (Fig. S2) | Asano lab p313 |
| YEpL-GB-TIF5B5-F; hc LEU2 plasmid encoding GB1-fusion to yeIF5201-405-F under TIF5 promoter | This paper (Fig. 2 & S2) | Asano lab p1206 |
| YEpL-GB-TIF5B5 -BN1-F; YEpL-GB-TIF5B5-F carrying BN1 | This paper (Fig. 2 & S2) | Asano lab p1263 |
| YEpL-h5MP1-F; h5MP1-F LEU2 plasmid | (Tang et al., 2017) (Fig. 2 & S2) | Asano lab p1587 |
| YEpL-h5MP1-7A-F; hc h5MP1-7A-F LEU2 plasmid | Tang et al., 2017) (Fig. 2 & S2) | Asano lab p1621 |
| pCDNA-h5MP1; h5MP1 under the CMV promoter | (Singh et al., 2011) (Fig. S5) | Asano lab p1910 |
| pAC-Dme5MP; Drosophila Kra under the fly actin promoter | (Kozel et al., 2016) (Fig. 4) | Asano lab p1708 |
| pAC-Dme5MP-BN1; pAc-Dme5MP carrying BN1 |
This paper (Fig. 4) | Asano lab p1964 |
| pLKO-sh5MP1 0140053 (# 2) | (Kozel et al., 2016) (Fig. 3 & 5) | Asano lab p1462 |
| pLKO-sh5MP1 0139465 (# 5) | (Kozel et al., 2016) (Fig. 3 & 5) | Asano lab p1465 |
| pLKO-sh5MP2 0147346 (# 7) | (Kozel et al., 2016) (Fig. 3 & 5) | Asano lab p1467 |
| pLKO-sh5MP2 0148768 (# 9) | (Kozel et al., 2016) (Fig. 3 & 5) | Asano lab p1469 |
| pLKO-sh5MP2 0147812 (# 10) | (Kozel et al., 2016) (Fig. 3 & 5) | Asano lab p1470 |
| pLKO-shGFP | (Kozel et al., 2016) (Fig. 3 & 5) | Asano lab p1471 |
| pSV40-firefly Kozak AUG; AUG under a typical Kozak context | (Ivanov et al., 2010) (Fig. 3) | Asano lab p1523 |
| pSV40-firefly Kozak CUG; CUG under a typical Kozak context | (Ivanov et al., 2010) (Fig. 3) | Asano lab p1521 |
| pSV40-firefly Kozak GUG; GUG under a typical Kozak context | (Ivanov et al., 2010) (Fig. 3) | Asano lab p1520 |
| pSV40-firefly Kozak ACG; ACG under a typical Kozak context | (Ivanov et al., 2010) (Fig. 3) | Asano lab p1525 |
| pSV40-Renilla Kozak AUG; Renilla control plasmid: AUG under a typical Kozak context | (Ivanov et al., 2010) (Fig. 3) | Asano lab p1526 |
| pSV40 firefly FMR1_24; ACG under FMR1_24 context | This paper (Fig. 5E) | Asano lab p1785 |
| AUG-nLuc -3xF; AUG-Nluc control plasmid | (Kearse et al., 2016) (Fig. 5 & S5) | Asano lab p1810 |
| GGG-nLuc -3xF; GGG-Nluc control plasmid | (Kearse et al., 2016) (Fig. 5 & S5) | Asano lab p1811 |
| +1 (CGG)100-nLuc -3xF; +1 (CGG)100-nLuc -3xF | (Kearse et al., 2016) (Fig. 5 & S5) | Asano lab p1812 |
| +2 (CGG)100-nLuc -3xF; +2 (CGG)100-nLuc -3xF | (Kearse et al., 2016) (Fig. 5 & S5) | Asano lab p1813 |
| pGW 5mp-2A-mapple #10 | This paper (Fig. S6B) | Todd Lab #BG109 |
| pGW 5mp-7a-2A-mapple #10 | This paper (Fig. S6B) | Todd Lab #BG111 |
| pGW 5MP-BN1-2a-mApple | This paper (Fig. S6B) | Todd Lab #BG284 |
Recombinant protein expression, purification and circular dichroism (CD)
All the constructs expressing h5MP1 wild-type, BN1 and 7A mutants were transformed into Escherichia coli strain BL21(DE3) cells and grown in Luria–Bertani medium containing 50 ug/ml kanamycin and incubated at 37°C until OD600=0.6 was reached. At this point IPTG was added to a final concentration of 0.5 mM and incubated further for 3-4 hours. Cells were harvested by centrifugation at 7500 rpm min−1 for 15 min and stored at −80°C. For purification, cell pellets were thawed and resuspended in 20ml binding buffer [20 mM Tris pH 8, 500 mM NaCl, 5 mM Immidazole 1 mM TCEP] per gram (wet weight) and lysed by sonication. The resulting cell lysate was centrifuged at 20 000 rev min−1 for 40 min at 4°C. The supernatant was filtered using a 0.22 μm filter prior to loading onto a 5 ml Ni-NTA affinity column equilibrated with binding buffer. The column was washed with binding buffer followed by 5% elution buffer [20 mM Tris pH 8, 500 mM NaCl, 1 M Immidazole, 1 mM TCEP] and fusion protein was eluted with 50% elution buffer. The protein fractions were pooled and subjected for overnight thrombin digestion (20mM Tris pH8.0; 150 mM NaCl; 1mM TCEP) to remove poly-histidine tail. The His6-tag cleavage efficiency was checked on a 12% SDS–PAGE gel and protein was loaded onto a 5 ml Ni–NTA column pre-equilibrated with binding buffer to remove undigested protein. The His6 tag-free protein was concentrated to a final volume of ~3 ml using an Amicon Ultra concentrator with a 10 kDa molecular-weight cutoff (Millipore) and loaded onto a HiLoad 16/60 Superdex 75 prep-grade (GE Healthcare) column pre-equilibrated with size-exclusion buffer (10 mM HEPES pH 7.5; 125 mM NaCl; 1 mM TCEP). The fractions with target protein were pooled, concentrated to ~2 mg/ml and stored at −80°C.
The plasmid encoding GB-heIF2β53-136 (previously called GB-eIF2β-K2K3) was transformed into E. coli BL21 (DE3) and cells were grown at 37°C in Luria–Bertani medium containing 100 mg/ml ampicillin and incubated at 37°C until OD600=0.6 was reached. Cells were induced for T7 RNA polymerase expression with 0.5 mM IPTG and incubated further for 3-4 hours, cells were harvested by centrifugation and stored at −80°C. Purification was carried out as mentioned above for h5MP1, followed by overnight TEV digestion (20mM Tris pH8.0; 100 mM NaCl; 1mM TCEP). Post digestion efficiency check, protein was further polished as described above, concentrated to 2 mg/ml and stored at −80°C. CD spectra are measured with Jasco J-1500 CD spectropolarimeter.
Isothermal Titration Calorimetry (ITC)
20 μM h5MP1 (WT), h5MP1-7A, and h5MP1-BN1 in 10 mM HEPES pH7.5; 125 mM NaCl; 1mM TCEP buffer (in cell; 200 μl) were titrated with 200 μM heIF2β53-136 (in syringe) at an equilibrium temperature of 25°C in a Microcal ITC200 instrument (20 injections with first injection 0.5μL and remaining 19 injections of 2μL each with 150 seconds spacing). h5MP1-7A was titrated again in the same way in a VP-ITC Microcalorimeter instrument at 25°C (in cell; 1.4 ml) (30 injections with first injection 4μL and rest 29 injections 10 μl each with 200 seconds spacing time). All the data were processed and plotted using Origin software.
Bio-layer interferometry (BLI)
BLI experiments were performed with the single channel BLItz system (Forte Bio) equipped with anti-GST biosensors and analyzed with the associated software BLItz Pro 1.3.0.5. This method detects in real-time the dynamic changes in reflectance interference wave pattern due to the change in protein mass on the biosensor tip. The binding steps were performed in the standard PBS supplemented with 0.1% bovine serum albumin and 0.1% tween-20 as binding buffer. Biosensors were hydrated using the binding buffer for 10 min following the manufacturing protocol. GST tagged 5MP1 or its mutants was immobilized to anti-GST coated biosensor. The attachment of GST-5MP1 to the biosensor surface is intended to avoid the avidity effect potentially caused by GST-fusion protein dimerization (Ladbury et al., 1995). The immobilized 5MP1 was incubated at increasing concentration of its binding partners using a stirring speed of 2000 rpm to give optimal signal responses.
Other protein protein interaction studies
GST pulldown assays were performed by attaching GST-fused h5MP1 and its derivatives to Glutathione Sepharose 4B resin (Millipore Sigma GE17–0756–01) and incubating with the partner proteins in GST-pulldown Binding Buffer (Singh and Asano, 2007). After washing the resin, the bound proteins were analyzed by SDS-PAGE followed by western blotting, along with a portion of in-put proteins. Co-IP in yeast was performed by preparing whole-cell extracts (WCE) from yeast expressing FLAG-tagged proteins in IP Buffer A and by immunoprecipitating proteins in the WCE through anti-FLAG M2 affinity resin (Millipore Sigma A2220) (Asano et al., 2000). The in-put and supernatant fractions were kept for the western blot analysis with immunoprecipitated fractions. In density gradient-velocity sedimentation analysis of eIF complexes in yeast, we cross-reacted the complexes by HCHO in culture, prepared and resolved the WCE fraction by fractionating on a 15-40% sucrose gradient and centrifugation at 39000 rpm for 4.5 hr (Singh et al., 2004). Gradient fractions were ethanol-precipitated and analyzed by western blotting with appropriate antibodies.
Luciferase assays
Firefly luciferase (Fluc) assays on general non-AUG translation and nanoLuciferease (Nluc) assays on RAN translation were performed as described previously (Tang et al., 2017; Todd et al., 2013). Specifically, ~80-90% confluent HEK293T cells in 75 μl medium loaded on a 96-well assay plate were transfected with 250 ng of plasmid DNA mixture using 0.25 μg polyethylenimine (PEI). We transfected cells in duplicate with the 1:5 mixture of the firefly vs. Renilla reporter plasmid mixture (5:1) and a pEF1A-derivative plasmid DNA. On the next day (Day 2), firefly and Renilla luciferase activities were measured with the Dual Glo reagents (Promega) using Victor 3 Plate Reader (Perkin Elmer). The ratio of average firefly to Renilla activities was used as a specific firefly luciferase activity. Nluc activities from the RAN reporter plasmids were likewise determined, except that the Nluc versus Fluc reporter DNA mixture (5:1) was used, and that the transfectants were lysed in the RIPA buffer. Portions of the lysates were then used for Dual-Glo firefly and Nano-Glo luciferase assay systems.
Anti-FLAG affinity purification and mass spectrometry
FLAG-tagged 5MP (h5MP1/BZW2 from human and 5MP/Kra from fly) was ectopically expressed in relevant host cells (HEK293T for human and S2 for fly) by transient transfection with PEI and HilyMax (Dojindo), respectively, and one-step affinity-purified by anti-FLAG M2 affinity resin (Millipore Sigma), as described previously (Kozel et al., 2016). We minimized the number of washes during purification to retain intact 5MP:eIF2 complex and specific association of other proteins. Purification of WT 5MP (WT) was done with a mock vector transfection control (Vec) and an experiment with the BN1 mutant (BN1). Two and three biological replicates of this set were performed for human and fly complexes, respectively (although the third replicate of fly Vec experiment was omitted). The one-step affinity purification products were resolved by SDS-PAGE, silver-stained and subjected for MS after dividing the whole lane into 12 to 16 zones (Fig. S3A) at a facility in Institute of Molecular Embryology and Genetics, Kumamoto University. The molecular amount of each associated protein was computed from the values obtained from all the slices per lane and presented as the total whole-lane emPAI value as proxy for molarity (Ishihama et al., 2005; Kozel et al., 2016). The list of all the detected proteins in human 5MP1 and fly 5MP/Kra experiments, along with their total emPAI values, was provided in Tables S1 and S2, respectively. Compilation of emPAI data from each gel slice verified sizes of intact subunits of associated proteins (see Fig. S3B–D for FLAG-tagged 5MP, eIF2 and eIF3 as examples).
In-gel analysis of affinity-purified proteins was performed essentially as described (Kozel et al., 2016) with the following modifications. The one-step affinity purification products were resolved by SDS-PAGE, silver-stained and subjected for MS after dividing the whole lane into 12 to 15 zones (12-15 gel slices). Further, the gel slices were cut approximately 1mm sized pieces (for efficient in-gel trypsin digestion). Proteins in the gel pieces were reduced with 10m M DTT (Thermo Fisher Scientific) in 25 mM ammonium bicarbonate, alkylated with 55mM iodoacetamide (Thermo Fisher Scientific) in 25 mM ammonium bicarbonate, and digested with trypsin and lysyl endopeptidase (Promega) in a buffer containing 40 mM ammonium bicarbonate, pH 8.0, overnight at 37°C. The digested peptides from gel pieces were extracted with 50% acetonitrile (FUJIFILM Wako), 0.1% formic acid (FUJIFILM Wako), and 70% acetonitrile, 0.1% formic acid. Supernatants were combined in a fresh vial, and were concentrated to 15 μL in a centrifugal evaporator. The concentrated samples were 2-fold diluted with 2% acetonitrile, 0.1% trifluoroacetic acid (FUJIFILM Wako), applied to LC-MS/MS analysis (10 μl/injection). Specifically, the resultant peptides were analyzed on an Advance UHPLC system (Microm BioResources) coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific) processing the raw mass spectrum using Xcalibur (Thermo Fisher Scientific). The raw LC-MS/MS data was analyzed against the SwissProt or Uniprot protein database restricted to Homo sapiens or Drosophila melanogaster using Proteome Discoverer version 1.4 (Thermo Fisher Scientific) with the Mascot search engine version 2.5-2.7 (Matrix Science). The resulting datasets was further analyzed using Scaffold 4.8.4 (Proteome Software Inc.) for calculating emPAI values. The obtained MS data was deposited to jPOST.
Fly genetics on RAN translation: Drosophila toxicity assays
Survival assays under FMRpolyG expression were performed according to previously published methods with minor modifications (Linsalata et al., 2019). In brief, following crosses, flies were placed on SY10 supplemented with 200uM RU486 and flipped to fresh tubes containing 200uM RU486 every 48 hrs. Dead flies were counted and removed every 48 hrs.
Rough eye phenotype assays were performed according to previously published methods (Linsalata et al., 2019) with minor modifications to scoring. For each aberrant morphological category: disorganized ommatidial array, ommatidial fusion, abnormal bristle orientation, supernumerary inter-ommatidial bristles, and necrosis and/or collapse, one point was given for less than 5% of the eye affected, 3 points were given if 5-50% of the eye was affected, and 5 points were given if greater than 50% of the eye was affected. Scores for individual eyes ranged from 0-25, ranging from no phenotype to severe phenotype. Eye images were captured using a Leica M125 stereomicroscope and a Leica DFC425 digital camera.
GFP aggregate immunohistochemistry (IHC) was performed according to previously published methods (Todd et al., 2013). In brief, 10-12 flies/genotype were positioned in a fly collar and fixed in OTC over dry ice. 12 um sections were transferred to slides, dried at RT for 10 minutes, fixed in 4% PFA for 10 minutes, and mounted in Prolong Gold with DAPI. Sections were imaged on an Olympus FV1000 confocal microscope equipped with a 40 × dry objective.
Drosophila western blots were performed according to previously published methods (Linsalata et al., 2019). In brief, 20 fly heads/sample from 1-3 day eclosed flies were hand homogenized in RIPA buffer and passed through a 28.5G insulin syringe. 20ug of lysate were loaded/lane. Membranes were blotted with mouse α GFP (Roche 11814460001) 1:1000, and mouse α β-tubulin (DSHB E7) 1:1000, and visualized using HRP-conjugated secondary antibody Goat α Mouse at 1:5000.
Drosophila Kra overexpression line sequence:
AGATCTCCCAAAATGAGTCAAAAAACTGAAAGACCAGTGCTATCGGGTCAACGCATCAAGACCAGAAAAAGAGATGAAAGAGAAAAATATGACCCAACGGGATTCCGTGATGCGGTCATTGCTGGTCTCGAAAAAACTGAAGGCGACCTGGAACAAATCTCTAAATATCTAGACAGCGCGGGCAACAAGCTCGACTATCGTCGCTACGGTGAAGTTCTCTTTGACATATTGATTGCAGGAGGTCTACTAGTGCCTGGTGGTTCTATATCTCAGGATGGCGAAAAGCCGCGCACAAGCTACTGCATCTTCGATGCGCCCGAGAGCATGGAGAGCATGCGCAACCATGAGCAGGTATTTGTCAAGCTCATCAGACGGTACAAGTACCTTGAGAAAATGTTCGAAGAGGAGATGGGAAAGGTTCTATTATTCGTAAAGGGCTTTACTCCTAGTGAACGCATCAAGCTTGCGCGTATGACAGCGCTCTGGTTAGTCAACGGTTCTGTACCGCCAAATGTTTTACTGGTACTAAACAACGAGCACCTAATCAAGGATGGCATTGCATTGGAGTTTCTACTAGAGCTGTTCCAGACGTTCAAGCAAGAGAAGGGCATTGCATATCTGATTCAGGCGCTTAAGAAGGGTGGACTGGAAAGCAAACTTATGGACTTTTTCCCACCGAACAAACGTACTGAGGAATATTTTAAACAAGTTTTTCTTGACAAAGAACTTAACGAGATCGTCAAGCTACATAAGGCTCAGGCTAGCCAGGAGGCGAAGCGTGAGTTGCAGCAGGCGCTCATCGATGATATCAACGATGAGAAGCCATATAATGAGATAACCTCAGACATTAAGGACTTCTCGCAGCGCACCAACATTCCCGATCATGAAATAATTGTTATTATTTGGTCCACCATCATGTCATTGGGCGAATGGAACAAGAAGGAGGAGCTGGTTACCGATCAGGCTGTTCGTCATCTGAAGAACTACTGCCCACTGTTGCAGGCTTTCGCCTCGACCGACCGTTCGGAATTGGCCCTAATCTTGAAGGTGCAGGAATTCTGCTACGAGAACATGAACTTCATGAAGGCCTTCCAGAAAATCATTTTGTTGTTCTACAAGACCGAGGTCCTGTCCGAGGAGATCATCTTGCGCTGGTACAAGGAGGGCCACTCAAACAAGGGCAAAATGCATTTCCTTGAACAAATGCGAAAGTTTGTCGAATGGCTTCAGTCAGCCGAAGAAGAATCCGAATCTGAGGACGAACAAAAGAATGGCGAGGACTACAAAGATGATGACGACAAGAACTAGGATCCCTCGAGGGTACC
QUANTIFICATION AND STATISTICAL ANALYSIS
Image quantification was performed and analyzed using ImageJ software. Statistical analysis of reporter studies used students’ T-test. Fly statistics data was analyzed using Graphpad Prism 7.0 with analysis of variance (ANOVA) models. Error bars represent S.E.M. (or S.D. where indicated). We did not use methods to determine whether the data meet assumptions of the statistical approach.
Supplementary Material
Table S3, list of selected proteins associated with human 5MP1 involved in translation initiation.Table S3, related toFigure 4.
Average emPAI values for vector control, WT h5MP1 or h5MP1-BN1 experiments are provided for selected initiation factors, 5MP, and 40S and 60S ribosomal subunit proteins. Averages for the holo complex (eIF2, eIF3, eIF2B, 40S and 60S) were also provided, with p values for difference between WT and BN1 experiments.
Table S4, list of selected proteins associated with fly 5MP/Kra involved in translation initiation.Table S4, related toFigure 4.
Average emPAI values for vector control, WT 5MP/Kra or 5MP/Kra-BN1 experiments are provided for selected initiation factors, 5MP, and 40S and 60S ribosomal subunit proteins. Averages for the holo complex (eIF2, eIF3, eIF2B, 40S and 60S) were also provided, with p values for difference between WT and BN1 experiments.
Table S1, list of proteins associated with human 5MP1 anti-FLAG affinity purification experiments.Table S1, related toFigure 4.
Average emPAI values for vector control, WT h5MP1 or h5MP1-BN1 experiments are provided for all the proteins detected.
Table S2, list of proteins associated with fly 5MP/Kra anti-FLAG affinity purification experiments.Table S2, related toFigure 4.
Average emPAI values for vector control, WT fly 5MP/Kra or 5MP/Kra-BN1 experiments are provided for all the proteins detected.
Acknowledgement
We thank Hitomi Yagi (Kindai University, Japan), Takuya Ohshima (Kindai University, Japan) and Rho Murakami (Niigata University, Japan) for technical help on yeast assays and Indranil Malik (University of Michigan) for assistance with Drosophila lifespan assays. This work was supported by a pilot grant from the K-INBRE program P20 GM103418, National Institutes of Health; Innovative Research Awards from the Kansas State University (KSU) Terry Johnson Cancer Center; National Science Foundation Research Grant (No. 1412250), National Institutes of Health grant GM125671, the Joint Usage/Research Center for Developmental Medicine, IMEG, Kumamoto University, and JSPS International Collaboration Enhancement Grant 18K19963 (to K.A.); KSU Arts and Science Research Scholarship (to M. H., A. C., A. T., and E. A.); K-INBRE Semester Scholarship P20 GM103418 (to S. G., E. A., M. H. and M. T); K-INBRE Star Trainee program P20 GM103418 (to C. M. and M. T.) and Goldwater Scholarship (to M. T.); National Institute of Health grants NS099280, NS086810, and P50HD104463 and VA BLRD BX004842 (to P. K. T.) and T32NS00722237 to M. R. G.. M. R. G. was supported by the A2A3 Susan Ross Award.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects.
References
- Algire MA, Maag D, and Lorsch JR (2005). Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 20, 251–262. [DOI] [PubMed] [Google Scholar]
- Asano K (2013). Evolution of eukaryotic translation initiation factors. In Encyclopedia of Systems Biology, Dubitzky W, Wolkenhauser O, Cho K-H, and Yokota H, eds. (New York: Springer; ), pp. 682–687. [Google Scholar]
- Asano K (2014). Why is start codon selection so precise in eukaryotes? Translation 2, e28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, and Hinnebusch AG (2000). A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev 14, 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Krishnamoorthy T, Phan L, Pavitt GD, and Hinnebusch AG (1999). Conserved bipartite motifs in yeast eIF5 and eIF2Be, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J 18, 1673–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, and Sachs MS (2007). Translation factor control of ribosome conformation during start codon selection. Genes Dev 21, 1280–1287. [DOI] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R, et al. (2013). Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, and McCammon JA (2001). Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochler A, Querido JB, Prilepskaja T, Soufari H, Simonetti A, Del Cistia ML, Kuhn L, Ribeiro AR, Valášek LS, and Hashem Y (2020). Structural Differences in Translation Initiation between Pathogenic Trypanosomatids and Their Mammalian Hosts. Cell Reports 33, 108534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonapace G, Gullace R, Concolino D, Iannello G, Procopio R, Gagliardi M, Arabia G, Barbagallo G, Lupo A, Manfredini LI, et al. (2019). Intracellular FMRpolyG-Hsp70 complex in fibroblast cells from a patient affected by fragile X tremor ataxia syndrome. Heliyon 5, e01954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito Querido J, Sokabe M, Kraatz S, Gordiyenko Y, Skehel JM, Fraser CS, and Ramakrishnan V (2020). Structure of a human 48S translational initiation complex. Science 369, 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D.-d., Li S.-j., Zhu B, Yuan T, Yang Q.-c., and Fan C.-y. (2017). Downregulation of BZW2 inhibits osteosarcoma cell growth by inactivating the Akt/mTOR signaling pathway. Oncology Rep, 10.3892/or.2017.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung Y-N, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, and Hinnebusch A (2007). Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev 21, 1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang G-Y, Kozakov D, Brenke R, Comeau SR, and Vajda S (2008). DARS (Decoys As the Reference State) potentials for protein-protein docking. Biophysical journal 95, 4217–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte MR, Kelly G, Babon J, Sanfelice D, youell J, Smerdon SJ, and Proud CG (2006). Structure of the eukaryotic initiation factor 5 reveals a fold common to several translation factors. Biochemistry 45, 4550–4558. [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, and Baker NA (2007). PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic acids research 35, W522–W525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, and Baker NA (2004). PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic acids research 32, W665–W667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Science 27, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KM, Glineburg MR, Kearse MG, Flores BN, Linsalata AE, Fedak SJ, Goldstrohm AC, Barmada SJ, and Todd PK (2017). RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat Commun 8, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Dever TE, and Asano K (2007). Mechanism of translation initiation in the yeast Saccharomyces cerevisiae. In Translational Control in Biology and Medicine, Mathews MB, Sonenberg N, and Hershey JWB, eds. (Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; ), pp. 225–268. [Google Scholar]
- Hiraishi H, Oatman J, Haller S, Blunk L, McGivern B, Morris J, Papadopoulos E, Guttierrez W, Gordon M, Bokhari W, et al. (2014). Essential role of eIF5-mimic protein in animal development is linked to control of ATF4 expression Nucl Acids Res 42, 10321–10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoem G, Bowitz Larsen K, Overvatn A, Brech A, Lamark T, Sjottem E, and Johansen T (2019). The FMRpolyGlycine Protein Mediates Aggregate Formation and Toxicity Independent of the CGG mRNA Hairpin in a Cellular Model for FXTAS. Front Genet 10, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Chen S, Fan H, Ai F, and Sheng W (2020). BZW2 promotes the malignant progression of colorectal cancer via activating the ERK/MAPK pathway. J Cell Physiol 235, 4834–4842. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, and Weissman JS (2011). Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity of Mammalian Proteomes. Cell 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, and Mann M (2005). Exponentially Modified Protein Abundance Index (emPAI) for Estimation of Absolute Protein Amount in Proteomics by the Number of Sequenced Peptides per Protein. Mol Cell Proteomics 4, 1265–1272. [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Sachs MS, and Atkins JF (2010). Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc Natl Acad Sci USA 107, 18056–18060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, and Warren ST (2003). RNA-Mediated Neurodegeneration Caused by the Fragile X Premutation rCGG Repeats in Drosophila. Neuron 39, 739–747. [DOI] [PubMed] [Google Scholar]
- Kearse MG, Green KM, Krans A, Rodriguez Caitlin M., Linsalata AE, Goldstrohm AC, and Todd PK (2016). CGG Repeat-Associated Non-AUG Translation Utilizes a Cap-Dependent Scanning Mechanism of Initiation to Produce Toxic Proteins. Mol Cell 62, 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D, Brenke R, Comeau SR, and Vajda S (2006). PIPER: an FFT - based protein docking program with pairwise potentials. Proteins: Structure, Function, and Bioinformatics 65, 392–406. [DOI] [PubMed] [Google Scholar]
- Kozel C, Thompson B, Hustak S, Moore C, Nakashima A, Singh CR, Reid M, Cox C, Papadopoulos E, Luna RE, et al. (2016). Overexpression of eIF5 or its protein mimic 5MP perturbs eIF2 function and induces ATF4 translation through delayed re-initiation. Nucl Acids Res 44, 8704–8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krans A, Skariah G, Zhang Y, Bayly B, and Todd PK (2019). Neuropathology of RAN translation proteins in fragile X-associated tremor/ataxia syndrome. Acta Neuropathol Commun 7, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladbury JE, Lemmon MA, Zhou M, Green J, Botfield MC, and Schlessinger J (1995). Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proceedings of the National Academy of Sciences 92, 3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Udagawa T, Singh CS, and Asano K (2007). Yeast phenotypic assays on translational control. Methods Enzymol 429, 139–161. [DOI] [PubMed] [Google Scholar]
- Li S, Chai Z, Li Y, Liu D, Bai Z, Li Y, Li Y, and Situ Z (2009). BZW1, a novel proliferation regulator that promotes growth of salivary mucoepidermoid carcinoma. Cancer Lett 284, 86–94. [DOI] [PubMed] [Google Scholar]
- Linsalata AE, He F, Malik AM, M.R.G., Green KM, Natla S, Flores BN, Krans A, Archbold HC, Fedak SJ, Barmada SJ, et al. (2019). DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation. EMBO Rep 20, e47498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llacer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG, and Ramakrishnan V (2015). Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex. Mol Cell 59, 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llacer JL, Hussain T, Saini AK, Nanda JS, Kaur S, Gordiyenko Y, Kumar R, Hinnebusch AG, Lorsch JR, and Ramakrishnan V (2018). Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. eLife 7, e39273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G, Firth AE, Atkins JF, and Ivanov IP (2018). Translational autoregulation of BZW1 and BZW2 expression by modulating the stringency of start codon selection. PLOS ONE 13, e0192648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RE, Arthanari H, Hiraishi H, Akabayov B, Tang L, Cox C, Markus MA, Luna LE, Ikeda Y, Watanabe R, et al. (2013). The interaction between eukaryotic initiation factor 1A and eIF5 retains eIF1 within scanning preinitiation complexes. Biochemitry 52, 9510–9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RE, Arthanari H, Hiraishi H, Nanda J, Martin-Marcos P, Markus M, Arabayov B, Milbradt A, Luna LE, Seo H-C, et al. (2012). The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2β Cell Reports 1, 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. (2013). The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338. [DOI] [PubMed] [Google Scholar]
- Na CH, Barbhuiya MA, Kim M-S, Verbruggen S, Eacker SM, Pletnikova O, Troncoso JC, Halushka MK, Menschaert G, Overall CM, et al. (2018). Discovery of noncanonical translation initiation sites through mass spectrometric analysis of protein N termini. Genome research 28, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda J, Saini AK, Munoz AM, Hinnebusch AG, and Lorsch JR (2013). Coordinated Movements of Eukaryotic Translation Initiation Factors eIF1, eIF1A and eIF5 Trigger Phosphate Release from eIF2 in response to Start Codon Recognition by the Ribosomal Pre-initiation Complex. J Biol Chem 288, 5316–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi E, Luna RE, Nagata T, Martin-Marcos P, Hiraishi H, Singh CR, Erzberger JP, Zhang F, Arthanari H, Morris J, et al. (2017). Molecular landscape of the ribosome pre-initiation complex during mRNA scanning: structural role for eIF3c and its control by eIF5. Cell Reports 18, 2651–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SY, He F, Krans A, Frazer M, Taylor JP, Paulson HL, and Todd PK (2015). RAN translation at CGG repeats induces ubiquitin proteasome system impairment in models of fragile X-associated tremor ataxia syndrome. Hum Mol Genet 24, 4317–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Reibarkh M, Yamamoto Y, Singh CR, Rio F.d., Fahmy A, Lee B, Luna RE, Ii M, Wagner G, and Asano K (2008). Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection J Biol Chem 283, 1094–1103. [DOI] [PubMed] [Google Scholar]
- Rodriguez CM, Wright SE, Kearse MG, Haenfler JM, Flores BN, Liu Y, Ifrim MF, Glineburg MR, Krans A, Jafar-Nejad P, et al. (2020). A native function for RAN translation and CGG repeats in regulating fragile X protein synthesis. Nat Neurosci 23, 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, and Zhang Y (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry GM, Adzhigirey M, Day T, Annabhimoju R, and Sherman W (2013). Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. Journal of computer-aided molecular design 27, 221–234. [DOI] [PubMed] [Google Scholar]
- Sato K, Masuda T, Hu Q, Tobo T, Gillaspie S, Niida A, Thornton M, Kuroda Y, Eguchi H, Nakagawa T, et al. (2019). Novel oncogene 5MP1 reprograms c-Myc translation initiation to drive malignant phenotypes in colorectal cancer. EBioMed 44, 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger (2020-3). BioLuminate version 4.0.139 (New York: Schrodinger, LLC; ). [Google Scholar]
- Sellier C, Buijsen RAM, He F, Natla S, Jung L, Tropel P, Gaucherot A, Jacobs H, Meziane H, Vincent A, et al. (2017). Translation of Expanded CGG Repeats into FMRpolyG Is Pathogenic and May Contribute to Fragile X Tremor Ataxia Syndrome. Neuron 93, 331–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, et al. (2017). Translation from unconventional 5′ start sites drives tumour initiation. Nature 541, 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A, Guca E, Bochler A, Kuhn L, and Hashem Y (2020). Structural Insights into the Mammalian Late-Stage Initiation Complexes. Cell Reports 31, 107497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, and Asano K (2007). Localization and characterization of protein-protein interaction sites. Methods Enzymol 429, 139–161. [DOI] [PubMed] [Google Scholar]
- Singh CR, Lee B, Udagawa T, Mohammad-Qureshi SS, Yamamoto Y, Pavitt GD, and Asano K (2006). An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J 25, 4537–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Udagawa T, Lee B, Wassink S, He H, Yamamoto Y, Anderson JT, Pavitt GD, and Asano K (2007). Change in nutritional status modulates the abundance of critical pre-initiation intermediate complexes during translation initiation in vivo. J Mol Biol 370, 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Watanabe R, Chowdhury D, Hiraishi H, Murai MJ, Yamamoto Y, Miles D, Ikeda Y, Asano M, and Asano K (2012). Sequential eIF5 binding to the charged disordered segments of eIF4G and eIF2β stabilizes the 48S pre-initiation complex and promotes its shift to the initiation mode. Mol Cell Biol 32, 3978–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Watanabe R, Zhou D, Jennings MD, Fukao A, Lee B-J, Ikeda Y, Chiorini JA, Fujiwara T, Pavitt GD, et al. (2011). Mechanisms of translational regulation by a human eIF5-mimic protein Nucl Acids Res 39, 8314–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Yamamoto Y, and Asano K (2004). Physical association of eukaryotic initiation factor 5 (eIF5) carboxyl terminal domain with the lysine-rich eIF2b segment strongly enhances its binding to eIF3. Journal of Biological Chemistry 279, 49644–49655. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, and Hinnebusch AG (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Morris J, Wan J, Moore C, Gillaspie S, Aube E, Fujita Y, Nanda J, Anderson A, Cox C, et al. (2017). Competition between translation initiation factor eIF5 and its mimic protein 5MP determines non-AUG initiation rate genome-wide. Nucl Acids Res 45, 11941–11953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, Renoux AJ, Chen K.-c., Scaglione KM, Basrur V, et al. (2013). CGG Repeat-Associated Translation Mediates Neurodegeneration in Fragile X Tremor Ataxia Syndrome. Neuron 78, 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Oh SY, Krans A, Pandey UB, Di Prospero NA, Min KT, Taylor JP, and Paulson HL (2010). Histone deacetylases suppress CGG repeat-induced neurodegeneration via transcriptional silencing in models of fragile X tremor ataxia syndrome. PLoS Genet 6, e1001240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Xue Y, Xu H, and Gong W (2006). Crystal structure of the C-terminal domain of S. cerevisiae eIF5. J Mol Biol 359, 1–9. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Singh CR, Marintchev A, Hall NS, Hannig EM, Wagner G, and Asano K (2005). The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3 and eIF4G. Proc Natl Acad Sci USA 102, 16164–16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, and Zhang Y (2015). The I-TASSER Suite: protein structure and function prediction. Nature methods 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, and Zhang Y (2015). I-TASSER server: new development for protein structure and function predictions. Nucleic acids research 43, W174–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Glineburg MR, Basrur V, Conlon K, Hall DA, and Todd PK (2020). Near-cognate initiation generates FMRpolyG from CGG repeats in Fragile X associated Tremor Ataxia Syndrome. bioRxiv, 2020.2010.2019.345736. [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. (2011). Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A 108, 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3, list of selected proteins associated with human 5MP1 involved in translation initiation.Table S3, related toFigure 4.
Average emPAI values for vector control, WT h5MP1 or h5MP1-BN1 experiments are provided for selected initiation factors, 5MP, and 40S and 60S ribosomal subunit proteins. Averages for the holo complex (eIF2, eIF3, eIF2B, 40S and 60S) were also provided, with p values for difference between WT and BN1 experiments.
Table S4, list of selected proteins associated with fly 5MP/Kra involved in translation initiation.Table S4, related toFigure 4.
Average emPAI values for vector control, WT 5MP/Kra or 5MP/Kra-BN1 experiments are provided for selected initiation factors, 5MP, and 40S and 60S ribosomal subunit proteins. Averages for the holo complex (eIF2, eIF3, eIF2B, 40S and 60S) were also provided, with p values for difference between WT and BN1 experiments.
Table S1, list of proteins associated with human 5MP1 anti-FLAG affinity purification experiments.Table S1, related toFigure 4.
Average emPAI values for vector control, WT h5MP1 or h5MP1-BN1 experiments are provided for all the proteins detected.
Table S2, list of proteins associated with fly 5MP/Kra anti-FLAG affinity purification experiments.Table S2, related toFigure 4.
Average emPAI values for vector control, WT fly 5MP/Kra or 5MP/Kra-BN1 experiments are provided for all the proteins detected.
Data Availability Statement
The raw data of MS analyses were deposited on jPOST under accession JPST001188 (ProteomeXchange under accession PXD026284).
The 5MP homology model structure and its model docked with eIF2γ or PIC are available at https://github.com/10ackert/5MP-PIC.


