Abstract
Objective
Multitask learning (MTL) using electronic health records allows concurrent prediction of multiple endpoints. MTL has shown promise in improving model performance and training efficiency; however, it often suffers from negative transfer – impaired learning if tasks are not appropriately selected. We introduce a sequential subnetwork routing (SeqSNR) architecture that uses soft parameter sharing to find related tasks and encourage cross-learning between them.
Materials and Methods
Using the MIMIC-III (Medical Information Mart for Intensive Care-III) dataset, we train deep neural network models to predict the onset of 6 endpoints including specific organ dysfunctions and general clinical outcomes: acute kidney injury, continuous renal replacement therapy, mechanical ventilation, vasoactive medications, mortality, and length of stay. We compare single-task (ST) models with naive multitask and SeqSNR in terms of discriminative performance and label efficiency.
Results
SeqSNR showed a modest yet statistically significant performance boost across 4 of 6 tasks compared with ST and naive multitasking. When the size of the training dataset was reduced for a given task (label efficiency), SeqSNR outperformed ST for all cases showing an average area under the precision-recall curve boost of 2.1%, 2.9%, and 2.1% for tasks using 1%, 5%, and 10% of labels, respectively.
Conclusions
The SeqSNR architecture shows superior label efficiency compared with ST and naive multitasking, suggesting utility in scenarios in which endpoint labels are difficult to ascertain.
Keywords: Machine Learning, Deep Learning, Multitask Learning, Electronic Health Records, Intensive Care
INTRODUCTION
The intensive care unit (ICU) manages a heterogeneous population of complex, medically vulnerable patients, requiring a range of organ support therapies. Predicting the clinical trajectories of ICU patients can inform conversations about limits of care and potentially guide preventative interventions. Risk predictions can also assist with resource allocation of staff and equipment across the department.
The traditional approach to risk stratification of ICU patients has been to use severity scores. First developed in the 1980s, these scores are typically designed to predict in-hospital mortality and have been refined through multiple editions. They include the APACHE (Acute Physiology, Age and Chronic Health Evaluation) score,1 the SAPS (Simplified Acute Physiology Score),2 and the SOFA (Sequential Organ Failure Assessment) score.3 These scoring tools are limited in that they use a small subset of the available patient data, often at a single time point during the ICU admission, and use fixed scoring thresholds that are not contextualized to the patient. Although they show strong discriminative performance for mortality at a population level, they are often poorly calibrated for patient-level outcome prediction.4,5
The widespread adoption of electronic health records (EHRs) creates an opportunity to use machine learning methods on routinely collected data for more accurate and personalized risk modeling. In recent years, there has been growing interest in the use of deep learning approaches to cater for the high-dimensional longitudinal data in the ICU, with numerous studies outperforming traditional risk scores at predicting mortality,6,7 specific organ dysfunctions or syndromes,8–10 and life-support interventions.11 One shortcoming is that the majority of models are examples of single-task (ST) learning—trained on a specific adverse event. By contrast, the mental model of a clinician is more holistic and typically involves concurrent prediction of multiple adverse events. This leverages the interdependencies between different organ systems and their corresponding pathophysiologies.12,13
Multitask learning (MTL) is a method for concurrent outcome prediction that has shown promising results across a range of domains including speech recognition, bioinformatics, computer vision, and natural language processing.14,15 By learning a shared representation across related tasks, MTL architectures have demonstrated several advantages over ST models including improved discriminative performance, computational efficiency,16 robustness,17 and a requirement for less labeled training data.18,19 MTL may also facilitate real-world deployment by having a single model serving multiple functions.20
There have been promising results in the EHR domain suggesting similar benefits from MTL.10,21–27 Harutyunyan et al22 applied a long short-term memory (LSTM)–based MTL architecture to benchmark tasks on MIMIC-III (Medical Information Mart for Intensive Care-III), including adverse event prediction and clinical phenotyping. They demonstrated that MTL provided consistent, though modest, improvements over ST discriminative performance for 3 of 4 tasks, concluding that it serves as an important regularizer. More recently, McDermott et al23 used the same ICU dataset to show that only highly related tasks result in effective cross-learning, with a high risk for negative transfer (reduced performance with MTL) when certain task combinations were used. Negative transfer happens when dissimilar tasks introduce conflicting inductive biases in the shared layers thereby hurting performance.28 Furthermore, McDermott et al23 found that MTL pretraining with finetuning on a new task significantly outperforms ST in few-shot learning (scarce training data) scenarios, especially on continuous (rolling) outcome prediction tasks.
In this work, we introduce a sequential deep MTL architecture, sequential subnetwork routing (SeqSNR), that automatically learns how to control parameter sharing across tasks and apply it to a diverse set of ICU endpoints. SeqSNR is a time series adaptation of the SNR architecture proposed by Ma et al28 as a method for flexible parameter sharing between tasks. We hypothesize that SeqSNR may show benefits over ST and shared-bottom (SB) (ie, traditional MTL with hard parameter-sharing) architectures. The main contributions of this article are the following:
We produce benchmark results on a diverse set of clinical endpoints using multiple feature sets extracted from MIMIC-III.
We demonstrate that SB MTL on clinical prediction tasks shows inferior performance to ST models, owing to negative transfer across tasks.
We propose a novel architecture to mitigate negative transfer by flexible parameter sharing.
We show that the proposed MTL architecture outperforms its ST counterparts in low-label scenarios.
MATERIALS AND METHODS
Data description
The EHR dataset used in this study is the open access, de-identified MIMIC-III dataset.29 The patient cohort consisted of 36 498 adult patients across 52 038 admissions to critical care units at the Beth Israel Deaconess Medical Center between 2001 and 2012. Patients were randomly split into training (80%), validation (10%), and test (10%) sets.
We used a version of the MIMIC-III dataset mapped to the Fast Healthcare Interoperability Resource (FHIR) standard as described in Rajkomar et al30 and GitHub code.31 FHIR data is organized as a collection of timestamped “resources” (eg, Medication Administration or Observation), each of which has an associated clinical code (which we use as a feature ID) and, where applicable, a value. We used the following FHIR resources: Patient (demographic information: age and gender), Encounter (admission and ward location), Observation (labs and vitals), Medication Prescription, Medication Administration, Procedure, Condition (diagnosis).
Data preprocessing
FHIR resources were converted to sparse feature vectors via the following steps:
Clipping and standardization: The outlier values were clipped to the first and 99th percentile values and continuous features standardized based on the clipped data.
Time bucketing: Features were aggregated into hourly time buckets using the median for repeated values.
Addition of presence features: Similar to Tomašev et al,10 we added presence features for all continuous variables to explicitly encode missingness. No numerical feature imputation was used.
One-hot encoding: Categorical features were one-hot encoded.
Feature selection
The full feature set (n = 70 770) was designed to maximize the information available to the model, by including the majority of structured data elements with the following exclusion criteria: the features present only in nonadult cohorts (<18 years of age) were filtered; rare features recorded only once in the entire dataset were removed; unstructured data were excluded.
The reduced feature set (n = 123) consisted of a manually curated list of common laboratory tests, observations, and interventions (no medications). The rationale was to identify a subset of clinically relevant features that may be more generalizable across health systems. Expert-guided feature selection is widespread in the EHR literature22,32 and therefore useful as a benchmark comparator. The list used here is very similar to the features proposed in the MIMIC-Extract preprocessing pipeline,11 which harmonizes MIMIC data into 93 semantic features; however, our list is augmented with a number of additional common variables (see Supplementary Appendix).
Benchmark tasks
We defined a diverse suite of prediction endpoints, ranging from specific organ dysfunctions and critical care interventions to more general markers of deterioration. All tasks were formulated as continuous predictions, triggered every hour during eligible admissions as in previous benchmark studies.11 Inference was only triggered during ICU admission. There were fixed prediction horizons chosen for each task based on clinical judgment about the window of actionability (shown in Table 1). All tasks were set up as repeated classification tasks—predicting the onset of the label within the prediction window. Task definitions are the following (see the Supplementary Appendix for further details):
Table 1.
Patient characteristics for the full cohort and positively labeled cohorts for each end point
| All | AKI | CRRT Dialysis | Vasoactive Medications | |
|---|---|---|---|---|
| Organ system | — | Renal | Renal | Cardiovascular |
| Prediction horizon, h | — | 48 | 12 | 12 |
| Patients | 36 498 (100) | 17 381 (47.6) | 1165 (3.2) | 14 539 (39.8) |
| ICU admissions | 52 038 (100) | 14 918 (28.7) | 1308 (2.5) | 16 601 (31.9) |
| Time steps | 5 116 931 (100) | 71 306 (1.4) | 13 6423 (2.7) | 662 786 (13.0) |
| Age, y | 64 (52-76) | 69 (57-78) | 63 (51-73) | 67 (57-77) |
| Female | 15 414 (42.2) | 7344 (42.3) | 433 (37.2) | 5696 (39.2) |
| ICU LoS, d | 2.08 (1.17-4.08) | 2.58 (1.33-5.17) | 3.79 (1.75-9.67) | 2.88 (1.46-5.92) |
| In-ICU mortality | 4096 (7.9) | 3092 (10.4) | 560 (18.7) | 3000 (12.4) |
| Fraction of admission with positive label, % | — | 3.2 (1.6-5.7) | 28.1 (8.3-50.6) | 20.5 (8.4-45.5) |
| Contiguous label duration, d | — | 0.75 (0.33-1.46) | 1.92 (0.21-4.63) | 0.63 (0.21-1.63) |
| Time to first label, d | — | 0.54 (0.13-1.50) | 2.02 (0.83-4.46) | 0.13 (0.04-0.33) |
|
| ||||
| First MV | Mortality | Remaining LoS ≤ 2d | — | |
|
| ||||
| Organ system | Respiratory | — | — | — |
| Prediction horizon | 12 hours | 48 hours | 48 hours | — |
| Patients | 13933 (38.2) | 5129 (14.1) | — | — |
| ICU admissions | 18716 (36.0) | 4096 (7.9) | — | — |
| Time steps | 2793417 (54.6) | — | — | — |
| Age, y | 63 (49-76) | 71 (58-80) | — | — |
| Female | 6100 (43.8) | 2293 (44.7) | — | — |
| ICU LoS, d | 2.92 (1.54-6.29) | 2.88 (1.33-6.58) | — | — |
| In-ICU mortality | 3181 (14.3) | — | — | — |
| Fraction of admission labeled, % | 100 (97.2-100) | 3.4 (1.2-11.5) | 96.1 (49.5-100) | — |
| Time to first label, d | 0.00 (0.00-0.08) | 3.17 (1.08-8.04) | — | — |
Values are n (%) or median (interquartile range), unless otherwise indicated.
AKI: acute kidney injury; CRRT: continuous renal replacement therapy; ICU: intensive care unit; LoS: length of stay; MV: mechanical ventilation.
Acute kidney injury (AKI): AKI was defined using the Kidney Disease Improving Global Outcomes guidelines33 excluding the urine output criterion. AKI of stages 1 and above was included. Periods of dialysis (including continuous renal replacement therapy [CRRT], intermittent hemodialysis, and peritoneal dialysis) were censored from the AKI prediction because it is redundant to predict AKI during active dialysis.
CRRT: CRRT is a form of acute dialysis used in critically ill patients. All intervals of CRRT were separately labeled using the codes and logic summarized in the Supplementary Appendix. Where there was no explicit end timestamp for CRRT, the label was clipped 4 hours after the latest code suggestive of ongoing CRRT. Intervals within 24 hours of each other were concatenated.
Vasoactive medications: Vasopressors and inotropes are medications used to manage circulatory function by modifying cardiac contractility and systemic vascular resistance, used in heart failure and certain shock syndromes. The label was based on the onset of any of the following 7 vasopressors and inotropes: norepinephrine, epinephrine, phenylephrine, vasopressin, dopamine, dobutamine, and milrinone.
Mechanical ventilation (MV): Labels were based on the SQL query provided on the MIMIC GitHub repository.34 We only label the onset of the first instance of MV during an ICU admission (first MV)—all timestamps after the first evidence of ventilation were labeled positive. We censored the event sequences of patients who were admitted to the Cardiac Surgery Recovery Unit because the overwhelming majority of these patients arrived in the unit already intubated.
Mortality: Mortality was timestamped using the “EXPIRE” flag included in the MIMIC-III dataset, which included both in- and out-of-hospital mortality.
Length of stay (LoS): The LoS task was defined as the remaining LoS from the time of inference. This was set up as a binary classification based on whether the remaining LoS was greater than 48 hours.
In addition, for the multitask models, we included a set of 13 common laboratory tests and vital signs: hemoglobin, platelets, white blood cells, sodium, potassium, creatinine, total bilirubin, arterial partial pressure of oxygen, arterial partial pressure of carbon dioxide, arterial pH, lactate, C-reactive protein, and serum glucose as secondary endpoints or auxiliary tasks. We computed the mean and SD of these labs and vitals over 24-, 48-, and 72-hour prediction horizons and added them as regression tasks at each time step (hourly). Where a particular lab value was not measured in the lookahead window, the model loss was set to zero.
Models
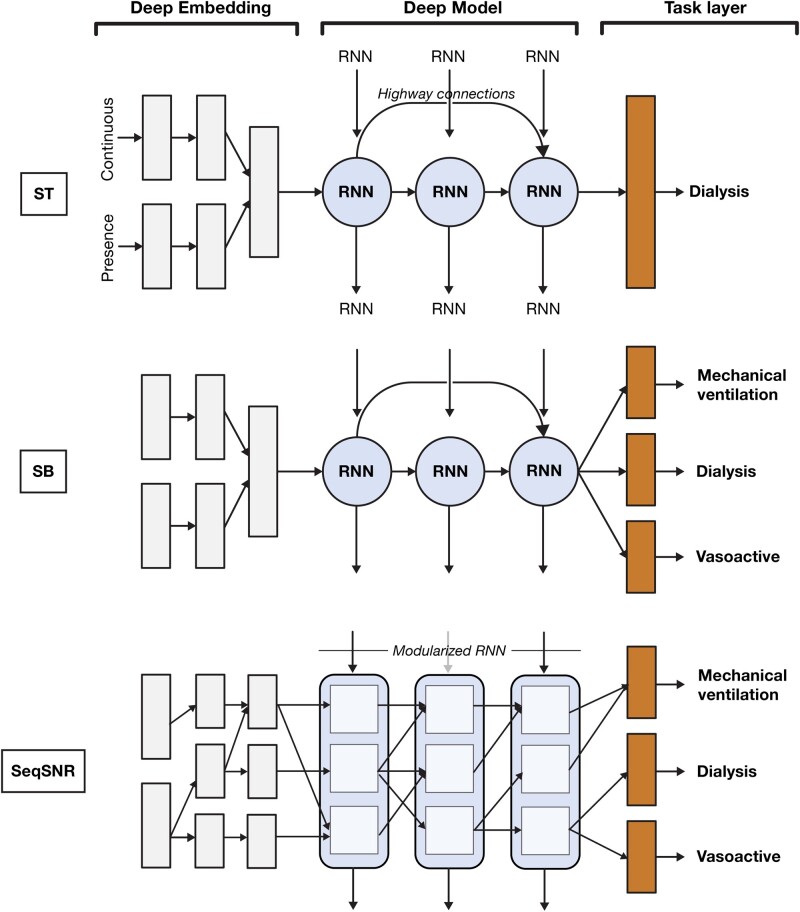
Our models extend on the recurrent neural network (RNN) architecture with highway connections introduced in Tomašev et al.10 For all architectures, the input tensor is fed through a sparse lookup embedding layer followed by a feed-forward neural network that forms the encoder, an RNN stack, and another task-specific feed-forward layer. Each feature type (continuous or categorical) has its own encoder and the representations obtained are concatenated before being fed into the model. We compared the following 3 configurations (illustrated in Figure 1):
Figure 1.
Comparison of single-task (ST), shared-bottom (SB) multitask, and sequential subnetwork routing (SeqSNR) architectures, showing simplified depictions of the embedding, recurrent neural network (RNN), and task modules.
ST: traditional approach in which a separate model is trained for each task.
SB multitask: all tasks trained concurrently, with a joint loss as described previously. SB is the most commonly used approach to MTL in neural networks and is achieved by sharing the hidden layers between all tasks (hard parameter sharing), while keeping several task-specific output layers.
SeqSNR: trains all tasks concurrently but also uses a layer-wise modularization of the encoder and RNN stack based on work described in Johnson et al.29
For both MTL architectures, 2 variants of each model are evaluated. In avg_best, all tasks, except labs and vitals, are considered as primary tasks and a single model is chosen based on the average area under the precision-recall curve (AUPRC) (%) across all tasks. In task_best, the model is optimized for a single endpoint (index task) and the other tasks act as auxiliaries. We show the results of task_best, as this tended to show superior performance on the validation set. The previous recurrent models are also compared against classical nonsequential benchmarks (logistic regression and XGBoost) in the Supplementary Appendix.
Sequential subnetwork routing
Subnetwork routing enables flexible parameter sharing through the use of learned Boolean connections that can “turn off” parts of the network for a given task. As shown in Figure 1, we split the encoder and deep model into multiple modules each of size de and ds, respectively, connected by learned routing variables. The routing connections are always created between blocks in one layer and the next, and are sampled from a hard concrete distribution35 with log α being a learned parameter and β, γ, and ζ being distribution hyperparameters. We experimented both with Boolean connections obtained via a hard sigmoid, and with scalar connections by using log α directly—the latter showed better performance.
The intuition of SeqSNR was to connect intermediate RNN states and tune connections for the endpoint of interest, thereby creating subnetworks. We achieve this by multiplying the cell activations with routing variables, passing the combined information to the cells in the next layer. Thus, for a given layer l and timestep t, with a number of subnetworks per layer defined as S, the input for a subnetwork c becomes:
| (1) |
Each subnetwork collects activations from the different tasks, which then need to be combined before passing through the next layer. We experimented with both concatenation and summation across tasks, but concatenation (yielding a vector of size ) consistently yielded better performance and is used throughout.
Training and hyperparameters
The validation split was used to tune a variety of hyperparameters including embedding size, regularization techniques, RNN stack size, and RNN cell type—LSTM,36 GRU,37 and UGRNN.38 We report here the optimal hyperparameter configuration. All models used an embedding layer of size 400 for the numerical and presence features. For the SeqSNR model, the embedding dimensionality was split among 2, 3, or 4 subnetworks per layer. All models were trained with a total of 3 layers. ST and SB have an LSTM cell size of 400, while SeqSNR used size 200 because it performed better than the larger size, and all have 3 layers. We used Xavier initialization39 and Adam40 optimization with a batch size of 128, and an initial learning rate of 0.0001 decaying every 12 000 steps by a factor of 0.85 replicating the setup in Tomašev et al.10 Additionally, we used state, input, and output variational dropout,41 with a probability of 0.4 for the RNN layers.
Loss
For multitask setups, we used 2 alternate approaches for weighting the losses across tasks and optimized on a per-task basis. One approach involved using predefined values for the task loss weights obtained through manual tuning; the second involved learning the weights during training using the uncertainty weighing technique described in Kendall et al.42 The loss can therefore be specified as , where ci can be either a predefined constant, or and Li represents cross-entropy for the binary tasks and L2 for the regression tasks.
Performance metrics and statistical significance
We report both AUPRC and area under the receiver-operating characteristic curve (AUROC) given the class imbalance for most tasks.43 For all reported results, we compute the 95% confidence intervals (CIs) using the pivot bootstrap estimator44 by sampling patients from the test dataset with replacement 200 times. A higher bootstrapping sample size (up to 500) was trialed for a subset of cases, and the conclusions were consistent (Supplementary Table 14). Two hundred was ultimately selected as a balance between precision and computational efficiency. Moreover, we performed the 2-sided Wilcoxon signed rank test45 to pairwise compare ST, SB, and SeqSNR on the bootstrapped samples. We chose the critical value α = 0.05 and used false discovery rate correction to adjust the P values for multiple hypotheses considering all the experiments performed in this study.
Label efficiency
We constructed prediction tasks in which only a fraction of the training labels were available for the primary prediction task, but the full dataset was available for the auxiliaries. We simulated this for AKI, MV, CRRT, and vasoactive medications as primary endpoints using 1%, 5%, and 10% of the training labels—with mortality, LoS, and labs and vitals as auxiliaries with 100% of labels. The 4 primary tasks are harder to timestamp, as they rely on multiple variables that are heterogeneously encoded in the EHR. The auxiliary tasks are straightforward to timestamp because they are reliably encoded in the EHR. Label efficiency experiments compared SeqSNRtask_best and ST on the full input feature set.
RESULTS
Patient characteristics
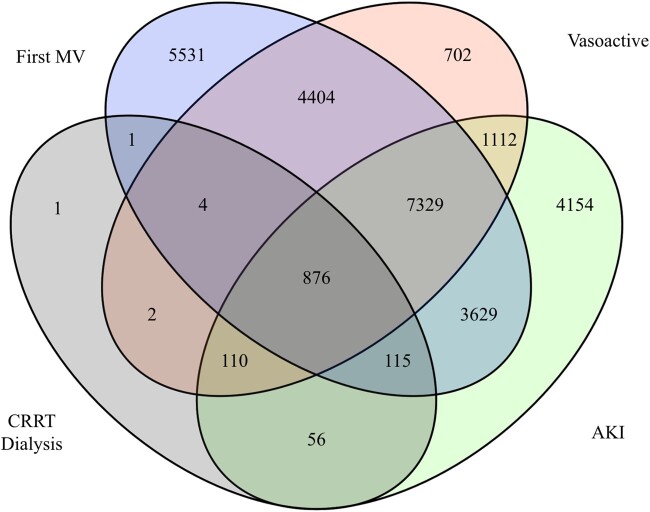
Table 1 shows descriptive statistics for the study population, and the subpopulations with positive labels for the 6 endpoints. Figure 2 shows a Venn diagram of patients with at least 1 positive label for AKI, first MV, CRRT, and vasoactive medications.
Figure 2.
Patient-level overlap of acute kidney injury (AKI), first mechanical ventilation (MV), continuous renal replacement therapy (CRRT), and vasoactive medication labels.
Model comparison
Table 2 summarizes the discriminative performance (AUPRC and AUROC) of each architecture for the full and reduced feature sets. In Table 3, we report the outcome of the Wilcoxon signed rank tests for pairwise comparison of results obtained by ST, SB, and SeqSNR. Outcome prevalence denotes the percentage of the positive class in the test set (timestep-level prevalence). PR and ROC curves are provided in the Supplementary Appendix.
Table 2.
Comparison of ST, SB, and SeqSNR performance on the full and reduced feature sets
| Task | Prediction Horizon | Outcome Prevalence (%) | Feature Set |
Model | AUPRC (%) | AUROC (%) |
|---|---|---|---|---|---|---|
| AKI | 48 h | 12.6 | Full |
|
|
|
| Reduced |
|
|
|
|||
| CRRT dialysis | 12 h | 0.4 | Full |
|
|
|
| Reduced |
|
|
|
|||
| Vasoactive medications | 12 h | 1.8 | Full |
|
|
|
| Reduced |
|
|
|
|||
| First MV | 12 h | 3.4 | Full |
|
|
|
| Reduced |
|
|
|
|||
| Mortality | 2 d | 3.3 | Full |
|
|
|
| Reduced |
|
|
|
|||
| Remaining LoS | ≤48 h | 40.0 | Full |
|
|
|
| Reduced |
|
|
|
AKI: acute kidney injury; AUPRC: area under the precision-recall curve; AUROC: area under the receiver-operating characteristic curve; CRRT: continuous renal replacement therapy; ICU: intensive care unit; LoS: length of stay; MV: mechanical ventilation; SB: shared bottom; SeqSNR: sequential subnetwork routing; ST: single task.
Table 3.
Wilcoxon signed rank test for pairwise comparison of performance obtained by ST, SB, and SeqSNR on the full and reduced feature sets.
| Task | Feature Set | Pairwise Comparison | P Value for AUPRC | P Value for AUROC |
|---|---|---|---|---|
| AKI | Full | ST vs SB | <.001 | <.001 |
| SeqSNR vs SB | <.001 | <.001 | ||
| SeqSNR vs ST | .002 | <.001 | ||
| Reduced | ST vs SB | <.001 | <.001 | |
| SeqSNR vs SB | <.001 | <.001 | ||
| SeqSNR vs ST | .635 | .060 | ||
| CRRT dialysis | Full | ST vs SB | .003 | <.001 |
| SeqSNR vs SB | .035 | .739 | ||
| SeqSNR vs ST | <.001 | <.001 | ||
| Reduced | ST vs SB | <.001 | .019 | |
| SeqSNR vs SB | .002 | .679 | ||
| SeqSNR vs ST | <.001 | .063 | ||
| Vasoactive medications | Full | ST vs SB | <.001 | <.001 |
| SeqSNR vs SB | <.001 | .888 | ||
| SeqSNR vs ST | <.001 | <.001 | ||
| Reduced | ST vs SB | .020 | <.001 | |
| SeqSNR vs SB | <.001 | <.001 | ||
| SeqSNR vs ST | <.001 | <.001 | ||
| First MV | Full | ST vs SB | .330 | <.001 |
| SeqSNR vs SB | <.001 | .006 | ||
| SeqSNR vs ST | <.001 | <.001 | ||
| Reduced | ST vs SB | .149 | .005 | |
| SeqSNR vs SB | <.001 | .011 | ||
| SeqSNR vs ST | <.001 | <.001 | ||
| Mortality | Full | ST vs SB | .081 | <.001 |
| SeqSNR vs SB | .019 | <.001 | ||
| SeqSNR vs ST | <.001 | <.001 | ||
| Reduced | ST vs SB | <.001 | <.001 | |
| SeqSNR vs SB | .203 | <.001 | ||
| SeqSNR vs ST | <.001 | <.001 | ||
| Remaining LoS | Full | ST vs SB | .021 | <.001 |
| SeqSNR vs SB | .271 | .021 | ||
| SeqSNR vs ST | .025 | <.001 | ||
| Reduced | ST vs SB | <.001 | .186 | |
| SeqSNR vs SB | <.001 | <.001 | ||
| SeqSNR vs ST | <.001 | <.001 |
To adjust for multiple hypothesis testing, we perform false discovery rate correction considering all experiments performed in this study and report the false discovery rate–adjusted P values. The P values marked in bold are statistically significant on the 95% confidence limit (α = 0.05).
AKI: acute kidney injury; AUPRC: area under the precision-recall curve; AUROC: area under the receiver-operating characteristic curve; CRRT: continuous renal replacement therapy; LoS: length of stay; MV: mechanical ventilation; SB: shared bottom; SeqSNR: sequential subnetwork routing; ST: single task.
For the full feature set, when compared with ST, SB shows equivalent performance for MV and mortality (2 of 6 tasks), positive transfer for CRRT Dialysis (1 of 6 tasks), and negative transfer for AKI, vasoactive medications, and LoS (3 of 6 tasks) (Tables 2 and 3). SeqSNR outperforms SB on AKI, CRRT, vasoactives, and mortality (4 of 6 tasks). Both show equivalent performance on LoS (1 of 6 tasks), while SeqSNR underperforms on first MV (1 of 6 tasks). Compared with ST, SeqSNR demonstrates positive transfer on AKI, CRRT, mortality, and LoS (4 of 6 tasks) and negative transfer on MV and vasoactives (2 of 6 tasks). In summary, SeqSNR shows a modest performance boost relative to SB and ST for the majority of tasks.
For the reduced feature set, compared with ST, SB shows positive transfer for CRRT and mortality (2 of 6 tasks), equivalent performance on MV and vasoactive medications (2 of 6 tasks), and negative transfer for AKI and LoS (2 of 6 tasks). Compared with SB, SeqSNR demonstrates better performance on AKI, MV, vasoactives, and LoS (4 of 6 tasks); equivalent performance on mortality (1 of 6 tasks); and worse performance on CRRT (1 of 6 tasks). Comparing SeqSNR with ST, we find that SeqSNR outperforms ST on CRRT dialysis, vasoactives, MV, mortality, and LoS (5 of 6 tasks), and both architectures show equivalent performance on AKI (1 of 6 tasks). The results demonstrate trends similar to the experiments on the full feature set (ie, while SB shows similar performance to ST, overall SeqSNR outperforms both SB and ST).
There were significant advantages from using the full vs the reduced feature set for MV, CRRT, vasoactive medications, mortality, and LoS (absolute AUPRC uplifts of 41.8%, 44.9%, 25.6%, 16.9%, and 5.8%, respectively, in the ST formulation).
Label efficiency
Performance of both architectures decreased as the percentage of labels for the index task was reduced. SeqSNRtask_best outperformed ST across all tasks at the 10%, 5%, and 1% training data reductions, and the absolute boost of performance was statistically significant for all cases (Table 4). There were large improvements for specific tasks, eg, first MV at 1% (AUPRC and AUROC boosts of 4.9%). We excluded SB from these experiments because, as shown in previously, SeqSNR outperforms it. CRRT dialysis was excluded for the 1% label scenario because the models do not converge during training, likely due to the low label prevalence of CRRT Dialysis.
Table 4.
Label efficiency results showing discriminative performance when the training dataset for the index task is reduced to 1%, 5%, and 10% while the auxiliary tasks have access to all training labels.
| Label, Patients | Task | Predicted Horizon (h) | Model | AUPRC (%) | AUROC (%) |
|---|---|---|---|---|---|
| 1%, 365 | AKI | 48 |
|
|
|
| Vasoactive medications | 12 |
|
|
|
|
| First MV | 12 |
|
|
|
|
| 5%, 1825 | AKI | 48 |
|
|
|
| CRRT dialysis | 12 |
|
|
|
|
| Vasoactive medications | 12 |
|
|
|
|
| First MV | 12 |
|
|
|
|
| 10%, 3650 | AKI | 48 |
|
|
|
| CRRT dialysis | 12 |
|
|
|
|
| Vasoactive medications | 12 |
|
|
|
|
| First MV | 12 |
|
|
|
We perform the Wilcoxon signed rank test for pairwise comparison of SeqSNR and ST for each case and report the false discovery rate–corrected P values for both AUPRC (%) and AUROC (%).
AKI: acute kidney injury; AUPRC: area under the precision-recall curve; AUROC: area under the receiver-operating characteristic curve; CRRT: continuous renal replacement therapy; LoS: length of stay; MV: mechanical ventilation; SeqSNR: sequential subnetwork routing; ST: single task.
DISCUSSION
This study is a proof of concept for SeqSNR with EHRs, demonstrating that this flexible framework for multitask prediction has benefits over traditional multitask and ST learning. While there were modest boosts in discriminative performance relative to naive multitasking on certain tasks, the main advantage of SeqSNR is its performance in low-training-label scenarios (label efficiency).
Label efficiency is a useful property given the challenges of assigning endpoint labels in EHR datasets, often requiring manual review by clinicians. The ability to exploit multiple endpoints, some of which may be more straightforward to label (eg, LoS or mortality), could reduce the requirements for manual curation on more challenging endpoints that are encoded heterogeneously (eg, MV). Notably, this approach is different from the classical transfer learning paradigm of pretraining and fine tuning. Instead of a 2-step process, we use a single-step process in which all the tasks are jointly trained under a multitask framework. The improved label efficiency of SeqSNR corresponds with the few-shot learning experiments conducted by McDermott et al,23 which found that MTL pretraining preserved performance at subsampling rates as low as 0.1% of training data.
Besides the low-label scenario, there is also the issue of negative transfer across EHR prediction tasks, which was reported by McDermott et al23 for most common MIMIC-III endpoints. Our results corroborate these findings, demonstrating that SB MTL tends to show a performance drop relative to ST learning. We find that the degree of negative transfer varies depending on the index task and is more common when using the full feature set. McDermott et al23 propose a solution involving multistage training (MTL pretraining followed by ST fine tuning); however, this carries the risk of catastrophic forgetting.46 We propose SeqSNR as an alternative approach for mitigating negative transfer via soft parameter sharing, which allows the network to optimize for cross-learning between related tasks. Although the performance boost from SeqSNR relative to SB was modest, these results suggest that flexible parameter sharing may be a promising mitigation strategy for negative transfer and should be further investigated for multitask modeling with EHR data.
Because most of the EHR literature uses a manually curated set of clinically relevant features, rather than the entire EHR, we demonstrate results on both a full and a reduced feature set. Across all tasks and architectures, there was a significant performance drop when using the reduced feature set. The dimensionality of this feature set is several orders of magnitude lower than the complete raw EHR (123 features vs over 70 000 including all medications and interventions). This reinforces the findings from Tomašev et al10 and Rajkomar et al30 that a complete embedding of the EHR can yield significant performance improvements. However, there is likely a trade-off between performance and generalizability because the full EHR contains many operational factors that are site-specific.
As a benchmarking exercise, this paper presents state-of-the-art or near-state-of-the-art performance across the 6 ICU endpoints when the full feature set is used. Although static predictions (triggered at a single time point during an admission [eg, 24 hours]) are more commonplace in the literature, comparable continuous prediction results on MIMIC-III are presented in other studies,11,22,23,47 although these all use more limited, manually curated input features (ranging from 17 to 136 features). Our results for mortality in 48 hours on both feature sets exceed the mortality in 24 hours results presented in Harutyunyan et al,22 independent of the ST/SB/SeqSNR architecture used. Wang et al11 predicted vasopressor and ventilator onset with a different formulation, framing it as a 4-class multilabel classification over a 4-hour prediction window offset by 6 hours from the time of inference. Our results on the full feature set exceeded the performance on the onset prediction task for both vasoactive medications and MV.
This study has a number of important limitations. First, we demonstrate these results on a single EHR dataset with ICU-related endpoints. While this is a valuable proof of concept, further investigation is warranted on other datasets and task combinations to evaluate the generalizability of SeqSNR. Second, several of the tasks (eg, MV) typically have very early onset in the ICU admission (because respiratory support is often the reason for ICU transfer), meaning that the prediction window was extremely short. Future work could evaluate SeqSNR on endpoints with longer prediction horizons and more straightforward interdependencies. Third, there is a lack of consensus on how best to report confidence bounds in EHR studies. We have used the conservative approach of patient-level bootstrapping22; however, this leads to wide confidence intervals due to the heterogeneity in the patient population. To combat this issue, we performed the Wilcoxon signed rank test to pairwise compare ST, SB, and SeqSNR on the bootstrapped samples and have drawn conclusions based on the outcome of these tests. Finally, we emphasize that these are prototype models to demonstrate methods. In order to translate these models into deployment, more rigorous evaluation would be needed including prospective validation and detailed case review.
CONCLUSION
MTL is a promising approach for clinical predictions because it learns generalizable representations across tasks and mirrors the interdependencies of physiological systems. We show that naive multitasking has variable performance compared with ST learning, with the possibility for negative transfer. We introduce a time series adaptation of a recent subnetwork routing architecture that outperforms naive multitasking and ST learning in terms of label efficiency. SeqSNR should be considered for multitask predictive modeling using EHR data, especially in situations in which training data are limited or endpoint labels difficult to ascertain.
FUNDING
Outside of the author affiliations listed previously, this research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
AUTHOR CONTRIBUTIONS
SR, DM, AK, and MS conceived the study. SR and DM designed and implemented models. SR, DM, EL, AM, and IP conducted the data analysis and ran the experiments. NH, YX, JS, and NT provided technical guidance and support. MS, AC, HM, AK, EL, SR, AM, and IP validated the feature sets and endpoint labels. SR, DM, and MS led the writing of the manuscript. All authors contributed to result interpretation and manuscript editing, and approve of its final version.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Zhe Zhao, Kathryn Rough, Cian Hughes, Sebastien Baur, Megumi Morigami, and Doris Wong from Google Health for their input and review. They also thank the MIMIC team for curating this open access dataset for the research community.
DATA AVAILABILITY STATEMENT
MIMIC-III is a freely accessible dataset to which interested researchers can gain direct access, upon completing human subjects training and signing a data use agreement (https://mimic.physionet.org/gettingstarted/access/forinstructions). More information about MIMIC-III can be found on their website (https://mimic.mit.edu/about/mimic/). Modelling source code is available online (https://github.com/google/ehr-predictions).
CONFLICT OF INTEREST STATEMENT
HM was a paid consultant to Google Health. The authors have no other competing interests to disclose.
REFERENCES
- 1.Knaus WA, Zimmerman JE, Wagner DP, Draper EA, Lawrence DE.. APACHE-acute physiology and chronic health evaluation: a physiologically based classification system. Crit Care Med 1981; 9 (8): 591–7. [DOI] [PubMed] [Google Scholar]
- 2.Le Gall JR, Lemeshow S, Saulnier F.. A new Simplified Acute Physiology Score (SAPS II) based on a European/North American multicenter study. JAMA 1993; 270 (24): 2957–63. [DOI] [PubMed] [Google Scholar]
- 3.Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med 1996; 22 (7): 707–10. [DOI] [PubMed] [Google Scholar]
- 4.Poole D, Rossi C, Latronico N, Rossi G, Finazzi S, Bertolini G.. Comparison between SAPS II and SAPS 3 in predicting hospital mortality in a cohort of 103 Italian ICUs. Is new always better? Intensive Care Med 2012; 38 (8): 1280–8. [DOI] [PubMed] [Google Scholar]
- 5.Nassar AP, Mocelin AO, Baptiston Nunes AL, et al. Caution when using prognostic models: A prospective comparison of 3 recent prognostic models. J Crit Care 2012; 27 (4): 423.e1–7. [DOI] [PubMed] [Google Scholar]
- 6.Shickel B, Loftus TJ, Adhikari L, Ozrazgat-Baslanti T, Bihorac A, Rashidi P.. DeepSOFA: a continuous acuity score for critically ill patients using clinically interpretable deep learning. Sci Rep 2019; 9 (1): 1936–1946.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson AEW, Mark RG. Real-time mortality prediction in the Intensive Care Unit. AMIA Annu Symp Proc2017; 2017: 994–1003. [PMC free article] [PubMed]
- 8.Lauritsen SM, Kalør ME, Kongsgaard EL, et al. Early detection of sepsis utilizing deep learning on electronic health record event sequences. Artif Intell Med 2020; 104:101820. [DOI] [PubMed] [Google Scholar]
- 9.Liu R, Greenstein JL, Granite SJ, et al. Data-driven discovery of a novel sepsis pre-shock state predicts impending septic shock in the ICU. Sci Rep 2019; 9 (1): 6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomašev N, Glorot X, Rae JW, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 2019; 572 (7767): 116–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, McDermott MBA, Chauhan G, Hughes MC, Naumann T, Ghassemi M. MIMIC-Extract: A Data Extraction, Preprocessing, and Representation Pipeline for MIMIC-III. arXiv, http://arxiv.org/abs/1907.08322, 19 Aug 2020, preprint: not peer reviewed.
- 12.Zador Z, Landry A, Cusimano MD, Geifman N.. Multi-morbidity states associated with higher mortality rates in organ dysfunction and sepsis: A data-driven analysis in critical care. Crit Care 2019; 23 (1): 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue Y, Zhou D, Du N, et al. Deep state-space generative model for correlated time-to-event predictions. In: Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2020: 1552–62.
- 14.Sebastian R. An overview of multi-task learning in deep neural networks. arXiv, http://arxiv.org/abs/1706.05098, 15 Jun 2017, preprint: not peer reviewed.
- 15.Yu Z, Qiang Y. A Survey on Multi-Task Learning. arXiv, http://arxiv.org/abs/1707.08114, 29 Mar 2021, preprint: not peer reviewed.
- 16.Vandenhende S, Georgoulis S, Proesmans M, Dai D, Van Gool L. Revisiting Multi-Task Learning in the Deep Learning Era. arXiv, http://arxiv.org/abs/2004.13379, 24 Jan 2021, preprint: not peer reviewed.
- 17.Mao C, Gupta A, Nitin V, et al. Multitask Learning Strengthens Adversarial Robustness. arXiv, http://arxiv.org/abs/2007.07236, 11 Sep 2020, preprint: not peer reviewed.
- 18.Crichton G, Pyysalo S, Chiu B, Korhonen A.. A neural network multi-task learning approach to biomedical named entity recognition. BMC Bioinform 2017; 18 (1): 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobrescu A, Giuffrida MV, Tsaftaris SA.. Doing more with less: a multitask deep learning approach in plant phenotyping. Front Plant Sci 2020; 11: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miquel M, Atsuto M. A multitask deep learning model for real-time deployment in embedded systems. arXiv, http://arxiv.org/abs/1711.00146, 31 Oct 2017, preprint: not peer reviewed.
- 21.Ngufor C, Upadhyaya S, Murphree D, Kor DJ, Pathak J. Multi-task learning with selective cross-task transfer for predicting bleeding and other important patient outcomes. In: Proceedings of the 2015 IEEE International Conference on Data Science and Advanced Analytics, DSAA 2015; 2015. doi:10.1109/DSAA.2015.7344836.
- 22.Harutyunyan H, Khachatrian H, Kale DC, Ver Steeg G, Galstyan A.. Multitask learning and benchmarking with clinical time series data. Sci Data 2019; 6 (1): 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDermott MBA, Nestor B, Kim E, et al. A Comprehensive Evaluation of Multitask Learning and Multi-task Pre-training on EHR Time-series Data. arXiv, http://arxiv.org/abs/2007.10185, 20 Jul 2020, preprint: not peer reviewed.
- 24.Suresh H, Gong JJ, Guttag JV. Learning tasks for multitask learning: heterogenous patient populations in the ICU. In: Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2018: 802–10. doi:10.1145/3219819.3219930.
- 25.Razavian N, Marcus J, Sontag D. Multi-task Prediction of Disease Onsets from Longitudinal Lab Tests. arXiv, http://arxiv.org/abs/1608.00647, 20 Sep 2016, preprint: not peer reviewed.
- 26.Wang X, Wang F, Hu J, Sorrentino R. Exploring joint disease risk prediction. AMIA Annu Symp Proc, 2014; 2014: 1180–87. [PMC free article] [PubMed] [Google Scholar]
- 27.Choi E, Taha Bahadori M, Schuetz A, Stewart WF, Sun J. Doctor AI: predicting clinical events via recurrent neural networks. JMLR Workshop Conf Proc2016; 56: 301–18. [PMC free article] [PubMed]
- 28.Ma J, Zhao Z, Chen J, Li A, Hong L, Chi EH.. SNR: sub-network routing for flexible parameter sharing in multi-task learning. Proc AAAI Conf Artif Intell 2019; 33 (1): 216–23. [Google Scholar]
- 29.Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med 2018; 1 (1): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Google. google/fhir. https://github.com/google/fhir. Accessed March 9, 2020.
- 32.Cao X, Edward C, Jimeng S.. Opportunities and challenges in developing deep learning models using electronic health records data: a systematic review. J Am Med Inform Assoc 2018; 25 (10): 1419–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khwaja A.KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin Pract 2012; 120 (4): c179–84. [DOI] [PubMed] [Google Scholar]
- 34.Mit-Lcp. Mit-lcp/mimic-code. https://github.com/MIT-LCP/mimic-code. Accessed February 17, 2020.
- 35.Louizos C, Welling M, Kingma DP. Learning sparse neural networks through L0 regularization. In: 6th International Conference on Learning Representations, ICLR 2018 - Conference Track Proceedings; 2018: 1–13.
- 36.Hochreiter S, Schmidhuber J.. Long short-term memory. Neural Comput 1997; 9 (8): 1735–80. [DOI] [PubMed] [Google Scholar]
- 37.Cho K, Merriënboer BV, Gulcehre C, et al. Learning phrase representations using RNN encoder-decoder for statistical machine translation. In: EMNLP 2014 - 2014 Conference on Empirical Methods in Natural Language Processing, Proceedings of the Conference; 2014: 1724–34. doi:10.3115/v1/d14-1179.
- 38.Jasmine C, Jascha S-D, David S. Capacity and trainability in recurrent neural networks. In: 5th International Conference on Learning Representations, ICLR 2017 - Conference Track Proceedings; 2017: 1–17.
- 39.Glorot X, Bengio Y.. Understanding the difficulty of training deep feedforward neural networks. J Mach Learn Res 2010; 9: 249–56. [Google Scholar]
- 40.Kingma DP, Lei Ba J. Adam: A method for stochastic optimization. In: 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings; 2015: 1–15.
- 41.Kingma DP, Salimans T, Welling M.. Variational dropout and the local reparameterization trick. In: Cortes C, Lawrence N, Lee D, Sugiyama M, Garnett R, eds. Advances in Neural Information Processing Systems 28. San Diego, CA: Neural Information Processing Systems; 2015: 2575–83. [Google Scholar]
- 42.Kendall A, Gal Y, Cipolla R. Multi-task learning using uncertainty to weigh losses for scene geometry and semantics. arXiv, http://arxiv.org/abs/1705.07115, 24 Apr 2018, preprint: not peer reviewed.
- 43.Davis J, Goadrich M. The relationship between precision-recall and roc curves. In: Proceedings of the 23rd International Conference on Machine Learning; 2006: 233–240.
- 44.Bradley E, Tibshirani RJ.. An Introduction to the Bootstrap. Number 57 in Monographs on Statistics and Applied Probability. Boca Raton, FL: Chapman & Hall/CRC; 1993. [Google Scholar]
- 45.Demšar J.Statistical comparisons of classifiers over multiple data sets. J Mach Learn Res 2001; 7: 1–30. [Google Scholar]
- 46.Kirkpatrick J, Pascanu R, Rabinowitz N, et al. Overcoming catastrophic forgetting in neural networks. Proc Natl Acad Sci U S A 2017; 114 (13): 3521–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purushotham S, Meng C, Che Z, Liu Y.. Benchmarking deep learning models on large healthcare datasets. J Biomed Inform 2017; 83: 112–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MIMIC-III is a freely accessible dataset to which interested researchers can gain direct access, upon completing human subjects training and signing a data use agreement (https://mimic.physionet.org/gettingstarted/access/forinstructions). More information about MIMIC-III can be found on their website (https://mimic.mit.edu/about/mimic/). Modelling source code is available online (https://github.com/google/ehr-predictions).