Abstract
Objective
Rheumatic heart disease (RHD) affects an estimated 39 million people worldwide and is the most common acquired heart disease in children and young adults. Echocardiograms are the gold standard for diagnosis of RHD, but there is a shortage of skilled experts to allow widespread screenings for early detection and prevention of the disease progress. We propose an automated RHD diagnosis system that can help bridge this gap.
Materials and Methods
Experiments were conducted on a dataset with 11 646 echocardiography videos from 912 exams, obtained during screenings in underdeveloped areas of Brazil and Uganda. We address the challenges of RHD identification with a 3D convolutional neural network (C3D), comparing its performance with a 2D convolutional neural network (VGG16) that is commonly used in the echocardiogram literature. We also propose a supervised aggregation technique to combine video predictions into a single exam diagnosis.
Results
The proposed approach obtained an accuracy of 72.77% for exam diagnosis. The results for the C3D were significantly better than the ones obtained by the VGG16 network for videos, showing the importance of considering the temporal information during the diagnostic. The proposed aggregation model showed significantly better accuracy than the majority voting strategy and also appears to be capable of capturing underlying biases in the neural network output distribution, balancing them for a more correct diagnosis.
Conclusion
Automatic diagnosis of echo-detected RHD is feasible and, with further research, has the potential to reduce the workload of experts, enabling the implementation of more widespread screening programs worldwide.
Keywords: echocardiography, rheumatic heart disease, deep learning, meta-learning, low-cost imaging, screening
INTRODUCTION
Cardiovascular disease (CVD) is the leading cause of mortality worldwide, with an estimated number of deaths of 17.8 million individuals per year and a 21.1% increase over the last 10 years.1 Even though CVDs are considered an expanding threat to global health, socioeconomic, racial, and ethnic differences still play a crucial role in access to cardiovascular care.2,3 Rheumatic heart disease (RHD)—damaged heart valves due to recurrent acute rheumatic fever (ARF)—affects an estimated 39 million people worldwide4 and is the most common acquired heart disease in children and young adults.5,6 Even though RHD ranks as a leading cause of death and disability in low-income and middle-income countries, it can be treated if detected in its early stages.7 Secondary prophylaxis in the form of regular penicillin injections can be initiated to prevent new episodes of ARF, avoiding further valve damage and progression of the disease. In 2013, the Brazilian Public Health System reported 5169 hospitalizations linked to ARF and 8841 linked to chronic RHD, at a cost of 33 million USD, mostly related to cardiovascular surgeries.8
Thanks to recent technological advances, echocardiography has become more cost-effective and widely available.9 Echocardiography is crucial for diagnosing a range of heart conditions9 and reducing CVD-related deaths.9–12 In particular, echocardiograms have emerged as an effective screening tool for early detection of latent RHD, identifying 10 times more subclinical disease cases when compared with auscultation.13,14 Following guidelines published by the World Heart Federation (WHF) in 2012,10 a skilled cardiologist can leverage findings related to valve regurgitation and stenosis on the mitral valve (MV) and aortic valve (AV) to issue a diagnosis for RHD.
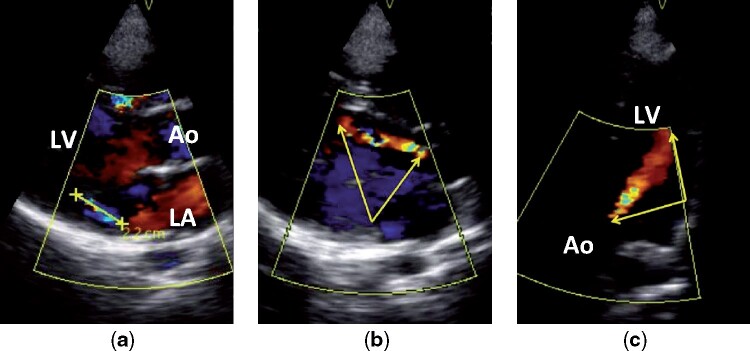
Three-quarters of children worldwide live in regions with a high prevalence of RHD.15 This age group is 1 of the most affected and, at the same time, the least likely to manifest sufficient echocardiographic features to meet a more certain diagnosis of the disease. Because of that, the WHF Guidelines—based on the severity and number of functional and morphological findings—delineate the Borderline RHD category for subclinical cases, with Definite RHD being the more severe form. The criteria for subclinical diagnosis applies only to patients aged ≤ 20 years, considering that this group benefits the most from early detection and secondary prevention of the disease. There are, however, individuals in that age range who suffer from significant progressions of the disease. Figure 1 depicts the process of diagnosing an 18-year old boy with Definite RHD. The different views observed are obtained by varying the position of the transducer in the patient’s chest.
Figure 1.
Echocardiographic images from an 18-year-old boy with definite RHD. A shows a > 2 cm jet of mitral regurgitation in parasternal long axis Doppler view; B and C show a > 2 cm jet of aortic insufficiency (yellow arrows) in parasternal long axis Doppler and apical 4 chamber Doppler views. Ao, aorta; LA, left atrium; LV, left ventricle.
Applications of artificial neural networks to conventional 2-dimensional echocardiographic data date back to 1990.16 In recent years the number of publications in the field has risen considerably due to the popularity of deep learning (DL).17 Many medical fields, such as oncology and pneumology, have seen successful applications of DL methods for disease detection.18–21 Concerning conventional echocardiograms, the DL literature mainly comprises studies on echocardiogram viewpoint (view) identification,22–25 heart chamber segmentation25,26 and classification of heart disease24,25,27,28 and primarily applied for structural rather than functional abnormalities. None of the disease-related works directly address valve abnormalities, let alone RHD, and virtually all of the research works use a frame by frame (2D) approach to process images, discarding the temporal relation encoded in video clips. Echocardiography identification of RHD, especially the subtle findings of subclinical disease, is highly dependent on the behavior of cardiac structures across sequences of frames in a video, and, therefore, it is unlikely that such an atemporal approach would achieve the best performance.
In this article, we address the challenges of RHD identification with a 3D convolutional neural network. Our model receives echocardiogram videos as input data and takes into account spatio-temporal information to improve the accuracy of predictions. In echocardiogram exams, multiple videos with different viewpoints of the heart are captured. To take advantage of these multiple videos for an improved exam classification, we also propose a supervised aggregation technique built on top of a decision tree-based meta-classifier. Our aggregation component tries to capture underlying biases in the neural network output distribution and balance them for a more correct diagnosis.
Although the proposed methodology may be applied to echocardiographic exams obtained by a range of devices with different features, resolutions, and file formats, the videos we work with come from handheld devices, applied in screening programs focused on the early detection of subclinical disease and prevention of RHD progression in underdeveloped areas of Brazil and Uganda. While the functionality of handheld devices is limited by resolution, poorer signal-to-noise ratio, and absence of spectral Doppler, their affordable price, practicality, ease of use, and small file size led to their common adoption in RHD screening programs.29–31 Moreover, due to a shortage of experienced echocardiographers, most of the exams were collected by nonphysicians, and telemedicine was used for remote diagnosis. In this context, the quality of the images and the fact that they were acquired by a nonexpert make the problem much more challenging for computational methods.
The experiments show that our video-based RHD classifier is significantly better than a frame-based one and that the complete proposed architecture, which includes the meta-classifier, significantly increases classification accuracy for exams when compared to a simple majority voting strategy. This automated diagnosis system has the potential to address even further the prohibitive financial and workforce barriers to widespread RHD screening by reducing the workload dependence on experts. Moreover, if embedded in screening devices or made available as a cloud-based application, it could inform prioritization of follow-up in near real time, therefore increasing the chance of patients seeking specialized care.
MATERIALS AND METHODS
Dataset description
Our dataset is composed of 11 646 echocardiographic videos in MP4 format (resolution 320 × 240 pixels), taken with VSCAN devices (GE Healthcare, Milwaukee, WI, USA) that sum up to 912 complete exams of unique patients. The data were acquired as part of screening programs in Uganda29,30 (359 exams) and the PROVAR screening program in Brazil31 (553 exams). The programs were conducted between 2012 and 2016 and screened children attending public schools. It focused on the early detection and prevention of disease progression, and screenings were performed mostly by trained nonexperts (584 exams). Table 1 presents the demographic profile of our dataset. Note that only a subset of 528 exams, all of which came from the PROVAR study, have complete demographic data annotated. The observed discrepancy in the prevalence of rheumatic valve disease by gender, with a remarkably higher prevalence in females, is also noted in other studies.31–33 The studies were approved by the institutional review boards of both the Children’s National Health System and Universidade Federal de Minas Gerais. In both studies informed consent was collected during visits to schools. After the intervention, a letter explaining the study procedures was sent to families with the consent and assent terms. Patients were only included after returning the signed consents, and their echocardiograms were deidentified.
Table 1.
Demographic data of subjects present in the dataset
| Overall | Negative | Positive | Definite | |
|---|---|---|---|---|
| (N = 912) | (n = 456, 50%) | (n = 349, 38.3%) | (n = 107, 11.7%) | |
| Patient demographics | 528 (100%) | 265 (50.2%) | 231 (43.8%) | 32 (6%) |
| Age, years (SD) | 13.1 (3.1) | 12.6 (3.1) | 13.6 (3.0) | 13.1 (3.4) |
| Gender, n female (%) | 316 (59.9%) | 145 (54.7%) | 150 (64.9%) | 21 (65.6%) |
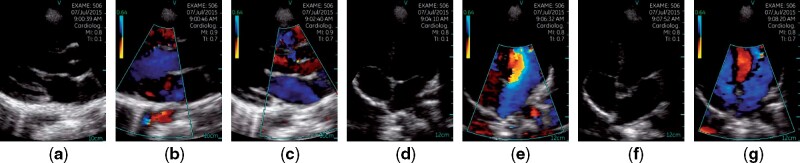
The estimated RHD prevalence in the examined regions and age group is ≤ 4.2%.29,31 However, due to the sensibility of the evaluated learning methods to extreme imbalances in the distribution of labels,34 the dataset comprises 456 (50%) RHD negative and 456 (50%) RHD positive exams, which are composed of Borderline RHD and Definite RHD diagnosis. Each exam contains, on average, 12.77 (3.59) videos, each possibly representing 1 of 7 different views of the patient’s heart. The viewpoints include Parasternal long axis, Parasternal long axis with Doppler on the MV level, Parasternal long axis with Doppler on the AV level, Apical 4 chambers, Apical 4 chambers with Doppler, Apical 5 chambers, and Apical 5 chambers with Doppler, as depicted in Figure 2. Apart from the RHD diagnosis, 314 videos (2.7%) have viewpoint labels, but no other metadata have been provided.
Figure 2.
Examples of echocardiogram viewpoints present in our dataset. Images were sampled from different videos of a single exam. (a) Parasternal Long Axis; (b) Parasternal Long Axis with Doppler on the Mitral Valve Level; (c) Parasternal Long Axis with Doppler on the Aortic Valve Level; (d) Apical 4 Chambers; (e) Apical 4 Chambers with Doppler; (f) Apical 5 Chambers; (g) Apical 5 Chambers with Doppler.
For the PROVAR exams, 5 cardiologists with expertise in RHD examined all morphological and functional changes in MVs and AVs according to the WHF criteria. All abnormal echocardiograms were independently reviewed by 2 readers, and discrepancies were resolved by consensus between 3 readers. The inter-reviewer agreement was 0.89 (95% CI 0.86–0.92), and the between-reviewer agreement 92%.31 A similar reviewing process was executed for exams performed in Uganda. The self-agreement ranged between 71.4% and 94.1% (κ 0.47–0.84), and the between-reviewer agreement ranged from 66.7% to 82.8% (κ 0.34–0.46).29,30
When considering the usefulness of the collected data for computer-aided diagnosis, however, the measures taken to make screenings more widespread pose some challenges. As previously mentioned, handheld devices present a poor signal-to-noise ratio that scales up, as the environments where screening takes place are many times improvised and have substandard infrastructure. Pediatric imaging has the potential to aggravate these errors even further, due to the smaller size of hearts, higher heart rates, and a limited ability to have the patients voluntarily hold their breath.
Another point that should also be taken into account is that the average number of videos per exam is well above the count of unique viewpoint classes, and this happens due to 2 behaviors reproduced by the professionals that performed the exams: videos with recording problems (eg, bad positioning of the transducer) were not deleted and, without any type of tag to differentiate and remove them afterwards, noisy instances populate our final dataset; also, some videos did not correspond directly to any of the specified viewpoints. This happens when a point of interest that would later help identify the presence of RHD is perceived, another video, zooming into the area, is recorded, changing the scale of cardiac structures to an unknown pattern.
Proposed architecture
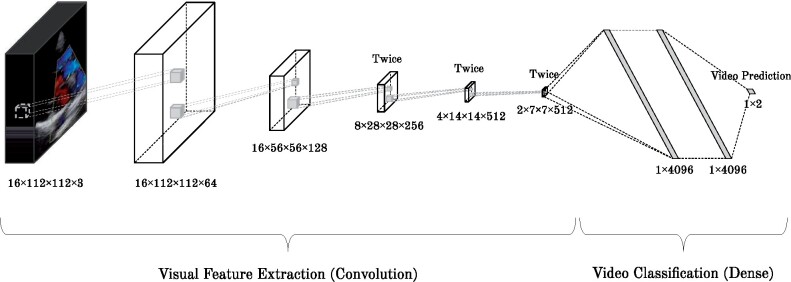
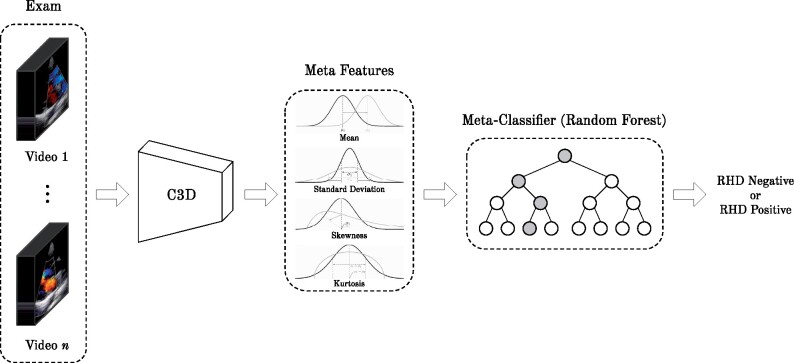
This section introduces the 2 main components of our proposed methodology: i) a deep convolutional neural network (CNN) to classify the videos as RHD-positive or -negative and ii) an aggregation method, which accounts for the results of all videos of a single patient, as shown in Figures 3 and 4, respectively. The methodology starts by feeding a 3D CNN (C3D)35 with videos from all viewpoints of the patient exam. Then the outputs of the networks for all videos of a single patient are combined using a meta-classifier, as detailed next. It is noteworthy that information regarding the patient is not provided during the training phase (ie, the CNN does not know which views correspond to the same exam).
Figure 3.
C3D network architecture for video classification.
Figure 4.
Proposed supervised meta-classifier for result aggregation toward exam classification.
Convolutional 3D network
We use the C3D as the backbone network of our method, as illustrated in Figure 3. The C3D network is a deep CNN that can learn from the temporal information by applying 3-dimensional convolution operations. The network receives a tensor of 112 × 112 pixels × 3 color channels × 16 frames. The initial 16 frames of each video are used to train the network. Since some videos contain less than 16 frames, we add padding frames that are a balanced number of duplicates of the first and last frames, until the required length is achieved. In a transfer learning fashion, we used the model pretrained on the Sports1M dataset.36
Initially, visual features are extracted by convolution layers with small 3 × 3 × 3 kernels combined with the max pooling operation. These features are then fed to a fully connected set of layers, with the last layer composed of only 2 neurons and the softmax function as activation, outputting the probability of the video belonging to 1 out of 2 classes: RHD Negative or RHD Positive. In order to simplify the problem, the Borderline RHD and Definite RHD diagnosis were grouped into a single class, named RHD Positive. All other layers use ReLU as the activation function. To prevent overfitting and improve generalization, dropout37 with a probability of 0.5 is implemented within the first 2 dense layers.
The C3D model minimizes the binary cross-entropy loss function as follows:
where is the label of the -th sample, the predicted probability of the positive class, and the number of samples. In summary, the network minimizes the distance between the confidence in its predictions and the true diagnosis for each echocardiogram.
Aggregation with a supervised meta-classifier
In the first step of our methodology, images from an exam are given to C3D independently. Next, the output of the CNN can be used in different ways to provide a diagnosis to a single patient. A standard approach to aggregate the results of all frames is to use a majority vote strategy, where each predicted video class counts as a single vote. However, a binary view of each prediction (positive RHD or negative RHD) disregards a great deal of information that could be useful for counterbalancing biases that evolved during the training of our model and help to improve the accuracy of our prediction. Thus, we propose a new aggregation strategy that uses a supervised classifier to predict the diagnosis (see Figure 4). This aggregation strategy is based on a set of meta-features extracted from the probability distribution output by CNN over videos per exam, namely, the mean, standard deviation, skewness, and kurtosis.
Stacked generalization,38 now commonly referred to as stacking, is 1 of the most used ensemble learning techniques in machine learning. It consists of combining multiple classification or regression models via a meta-classifier or a meta-regressor that leverages the output of the base models to give a final prediction. In the context of our study, there is only 1 base predictor, but multiple instances that should be aggregated into a single output. As the number of videos per exam is not fixed, we use the statistical moments of our classifier’s confidence distribution as inputs for the meta-model.
The proposed aggregation strategy is agnostic to both the base classifier and the meta-classifier, as long as the first can output its prediction as a probability. The meta-classifier of choice for this article was the decision tree-based random forest,39 due to its notorious efficacy when little is known about the domain being evaluated.40
Baseline, hyperparameters, and implementation
We consider that 1 of the novelties of the methodology proposed here is the use of a C3D that received videos as inputs instead of a 2D CNN, which works with images. First, in order to verify that the video-based approach contributes to the success of our methodology, we compare it against a frame-based method that uses VGG-16 as the backbone neural network. The VGG-1641 architecture is similar to the ones used in 2 related works24,25 and it is well-established in the computer vision community. VGG-16 is a 2D CNN which receives as input a still frame of 224 × 224 pixels × 3 color channels. Following a similar methodology as the one in,25 we have sampled 10 random frames from each video to create instances for the network, and the network predictions are then aggregated using a majority vote strategy per video and then per exam, giving preference to the positive class in case of a draw. The model used was pretrained on the ImageNet dataset. The loss function used was also the binary cross-entropy.
In the next step, to measure the contribution of the proposed meta-classifier, we have also run experiments where the C3D results were aggregated using a majority voting to give the patient’s final diagnosis.
We have trained in our dataset the VGG-16 network (pretrained in the ImageNet dataset) with the Adam optimizer, a learning rate of 1e-5, batch size of 32 and 25 epochs, using early stopping with a patience value of 10. For C3D we used an SGD optimizer, a learning rate of 1e-3, batch size of 16 and 25 epochs also, but with 5 as the patience for early stopping. The random forest model was trained with 200 estimators in the forest and a max depth of 75. Unlisted hyperparameters for all models were left to their default values. The set of hyperparameters for each method were chosen through a random search setup with 30 iterations for the neural networks and 500 for the random forest.
Our code was written in Python 3.6, and executed in a machine with Intel Core i7-9700K CPU and an NVIDIA GeForce RTX 2080 Ti GPU. All the neural networks were implemented using Keras with TensorFlow 1.12 as the back end. The used random forest classifier is packed within version 0.20 of scikit-learn. The code is freely available for download (https://github.com/joaofbsm/rhd-classification).
Experimental setup
We have performed a binary classification with the Borderline RHD and Definite RHD diagnosis grouped into a single class, named RHD Positive. All echocardiograms were deidentified by applying a mask of black pixels to the area outside of the ultrasound beam during preprocessing, therefore omitting the metadata present in the images. As all echocardiograms in the dataset were collected using the same equipment and software, the size of this area was fixed. For the C3D inputs, videos were first rotated 90 degrees and then resized to 128 × 171 × 3 × 16. This was done to obtain a better aspect ratio when removing the mean cube of the original training data, a preprocessing step called whitening.42 A centered cropping was then applied to generate the final data. As for the VGG-16, videos were directly resized to the expected input dimensions.
In order to train the model, tune hyperparameters and then diagnose new exams, we randomly split the dataset into training, validation, and test in an approximate 80:10:10 ratio. The splits were stratified, and videos from the same exam were always in the same data partition. Each patient has only 1 exam. Hyperparameter tuning for both neural networks and the random forest meta-classifier used for aggregation was done using only the train and validation sets to prevent information leakage from the test partitions. To assess accuracy, we have used a 10-fold cross-validation procedure, making each video go through the validation and test partitions only once. Folds are the same for all evaluated methods.
RESULTS
Table 2 reports the mean specificity, sensitivity, precision, and accuracy for the test partitions in the 10-fold cross-validation procedure. We performed a Wilcoxon signed-rank test with 95% of confidence to compare the results of the 3 different methods, and the best method for each metric is highlighted in bold in the table. In cases where there is no evidence of statistical difference, both results are highlighted. The specificity and sensitivity obtained by the best model were 70.59 (95% CI, 66.53–74.65) and 74.94 (95% CI, 70.10–79.77), respectively. Its accuracy, averaged over the test partitions for each of the 10 folds, was 72.77 (95% CI, 69.28–76.26).
Table 2.
Mean specificity, sensitivity, precision, and accuracy (with 95% confidence intervals) for RHD classification on the test set over a 10-fold cross-validation procedure for different levels of result aggregation
| Aggregation | Metric | VGG-16 | C3D + Majority Vote | C3D + Meta-Classifier |
|---|---|---|---|---|
| Frame | Specificity | 54.37 (51.78, 56.97) | — | — |
| Sensitivity | 56.90 (53.98, 59.82) | |||
| Precision | 57.65 (56.27, 59.04) | |||
| Accuracy | 55.70 (54.58, 56.81) | |||
| Video | Specificity | 59.59 (56.29, 62.90) | 52.67 (47.00,58.34) | — |
| Sensitivity | 55.17 (51.19, 59.15) | 67.71 (62.60,72.83) | ||
| Precision | 59.86 (58.12, 61.61) | 60.69 (58.59,62.78) | ||
| Accuracy | 57.29 (55.83, 58.75) | 60.42 (58.76,62.07) | ||
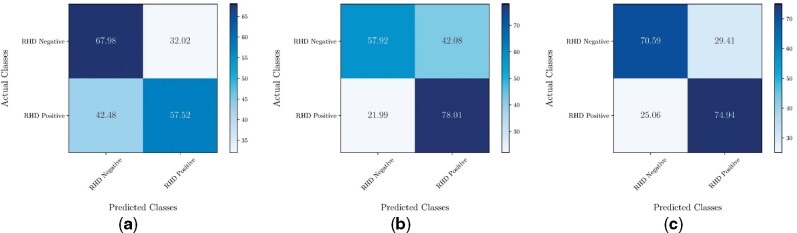
| Exam | Specificity | 67.98 (62.68, 73.28) | 57.92 (47.99,67.85) | 70.59 (66.53,74.65) |
| Sensitivity | 57.52 (52.40, 62.63) | 78.01 (71.53,84.49) | 74.94 (70.10,79.77) | |
| Precision | 64.65 (61.76, 67.53) | 66.10 (61.80,70.39) | 71.88 (68.41,75.35) | |
| Accuracy | 62.80 (60.69, 64.91) | 67.95 (64.92,70.98) | 72.77 (69.28,76.26) |
Note: Results in bold are the best for that metric according to a 95% confidence Wilcoxon signed-rank test. In cases where there was no evidence of difference, both results are highlighted.
In Table 3 we break the results for the RHD Positive class considering its original subclasses: Definite RHD and Borderline RHD.
Table 3.
Sensitivity values (with 95% confidence intervals) for classification of the 2 subclasses aggregated as RHD Positive in our dataset
| Diagnosis | Subclass Exam Sensitivity |
|---|---|
| Borderline RHD | 71.90 (66.71, 77.09) |
| Definite RHD | 85.78 (79.37, 92.18) |
Table 4 presents the average feature importance detected by the meta-classifier across folds, indicating that the distribution moments used as features were indeed relevant for a better prediction.
Table 4.
Average meta-feature importance percentage observed across folds using the C3D network as the base classifier
| Meta-feature | Importance |
|---|---|
| Confidence Mean | 76.4 |
| Confidence Std | 6.3 |
| Confidence Skewness | 12.6 |
| Confidence Kurtosis | 4.7 |
Figure 6 shows examples of 4 frames extracted from 4 videos where the proposed model classified an exam as RHD positive or negative with a high confidence (these 4 videos and 57 others that are part of the exams they belong to are available as Supplementary Material). They can help understand the model's decisions.
Figure 6.
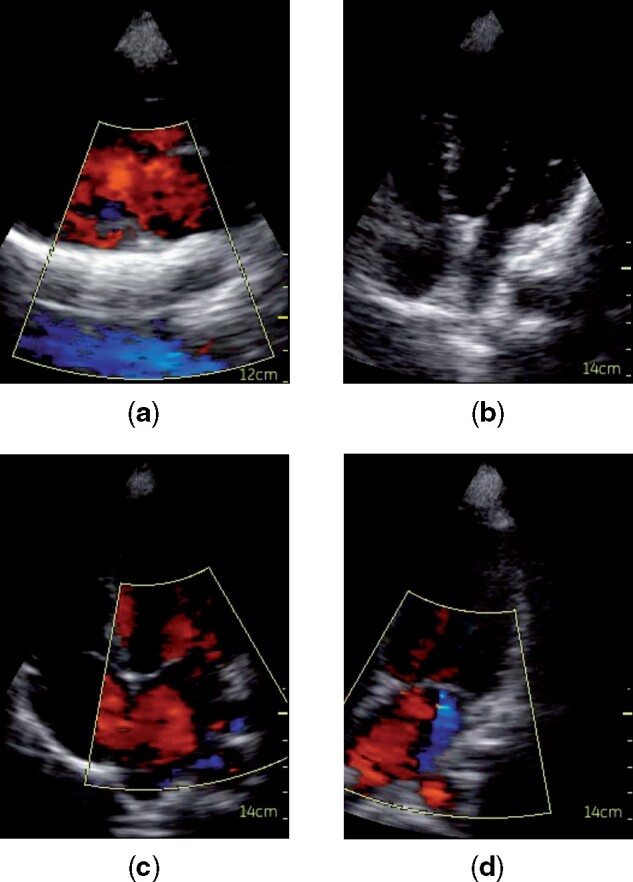
Examples of frames extracted from 4 videos where the model made the predictions with high confidence. Videos are from different exams, and we consider their predictions when in the test set. (a) RHD negative misclassified as RHD positive; (b) RHD positive misclassified as RHD negative; (c) RHD negative correctly classified; (d) RHD positive correctly classified.
DISCUSSION
As expected, C3D with the majority vote is significantly better than VGG-16 for all metrics except specificity and precision (where the results of both methods present no statistically significant difference) at the video level, showing the already stated importance of spatio-temporal information to the task at hand. Considering the exam level, which provides the final diagnosis, the proposed methodology significantly outperforms the other 2 methods with regards to accuracy, which is our final classification objective. The meta-classifier significantly outperforms the majority voting in terms of specificity, but for the other metrics there is no statistically significant difference. During a screening, it is preferable that a healthy patient is wrongly referred to a better equipped health facility to perform follow-up exams than that an unhealthy one receives a normal diagnosis and progresses to more severe forms of the disease; hence, better sensitivity is desirable.
An analysis to assess if Definite RHD cases are easier to identify than Borderline cases—which is expected—was also performed, and the reported sensitivities corroborate the expected results. The sensitivity obtained for the Definite RHD class is comparable to the 83 (95% CI, 76–89) overall sensitivity achieved by nonexpert users in RHD identification after following a computer-based 3-week training curriculum, as reported by Beaton et al.43
Regarding the proposed aggregation method, it achieved significantly better results than the baselines. We took advantage of the interpretability of the decision tree method it is built upon to explore even further the functionality of the meta-classifier and also compare its effects against the solo C3D model. An analysis of feature importance showed all distribution moments were indeed relevant for a better prediction. For a simple comparison, training the same model only with the Confidence Mean feature, responsible for most of the feature importance, the cross-validation accuracy of the C3D + Meta-Classifier would be 70.47, which is not statistically better than the C3D with the majority vote strategy (confidence of 95%).
We assumed that the meta-classifier counterbalances biases acquired during the training of the base model. If this holds true, results from a majority vote ensembling strategy should be more unbalanced in nature. Figure 5 shows the confusion matrices obtained for both aggregation methods along with the C3D network. By comparing Figure 5b with Figure 5c, one can observe a loss of sensitivity around 0.03 with a compensatory increase in specificity of almost 0.13. This same pattern appears in executions with different hyperparameters.
Figure 5.
Resulting confusion matrices for each method on RHD classification of echocardiographic exams. (a) VGG16 with Majority Vote; (b) C3D with Majority Vote ;(c) C3D with Meta-Classifier.
This indicates that the proposed aggregation strategy possibly identifies when there are noisy videos in an exam, through unexpected disruptions in the confidence distribution, and filters the noise out, obtaining more accurate predictions overall.
Analyzing the images classified as RHD-positive or -negative in Figure 6, we notice that images (a) and (b) have quality problems. In Figure 6a the blood flow from the abdomen (in blue) was captured by the Doppler, which probably confused the network due to a pattern similar to a valve regurgitation, and led to the classification of a negative example as positive. Figure 6b shows a video of low quality where heart structures are poorly visible—which can be caused, for instance, by adipose tissue thicker than normal between the transducer and the patient’s heart. Without any clearly detectable anomalies, the network classified an RHD positive as negative. In Figures 6c and 6d, the images are clear. Figure 6c shows the absence of mitral regurgitation during systole to represent the lack of abnormalities in the video, which led the model to correctly predict the exam as RHD negative. In Figure 6d we can observe, also during systole, the presence of mitral regurgitation as the blue Doppler jet, which is 1 of the main factors for the detection of RHD, and therefore probably led the model to classify the video as RHD-positive.
These examples show that the quality of images directly affects the performance of the model. However, as the main motivation for this work is to process the images as they come, a preprocessing step to remove this type of noise from the dataset can greatly improve the performance of the model.
CONCLUSION
This article lays the foundations for automatically detecting RHD in echocardiographic exams through machine learning algorithms. We have used 2 different deep neural network models and proposed a supervised meta-classifier for the aggregation of video predictions into a single patient diagnosis, with the later significantly outperforming the baselines with an accuracy of 72.77 over a 10-fold cross-validation procedure. RHD diagnosis is very difficult due to different types of data that are mixed together with noise. Moreover, existing literature is very limited, and no previous related works used methods suitable for the task approached in this study. Nonetheless, automatic diagnosis of echo-detected RHD seems feasible and, with further research, has the potential to reduce the workload on cardiologists and experts, enabling the implementation of more widespread screening programs that can reduce the disease burden in the underdeveloped world. More than the simple point-of-care diagnosis of subclinical RHD, the proposed system, embedded in screening devices or made available as a cloud-based application, also has the potential for allowing low-cost identification of patients at higher risk for other valvulopathies and cardiovascular diseases.
We plan to explore preprocessing methods to identify and remove noise instances, which are probably making the training process more difficult and reducing the method’s performance. Also, recent developments in neural networks introduced attention mechanisms, components which can detect tiny variations in the data—such as the format of a valve in a specific frame—and give more importance to it during classification. This seems to fit very well with our task, as punctual variations hold great value to diagnose the disease, given WHF’s criteria. Furthermore, these mechanisms greatly improve the explainability of a machine learning model, a very relevant characteristic for widespread acceptance of the application by the physicians and the population.
Additionally, validating the methodology on new data coming from the same screening program or other similar programs is necessary to assess the robustness and generalization of the methodology. There are many other interesting directions to follow providing we obtain more data. One is to address the problem of RHD diagnosis considering the 3 classes of RHD, namely positive, borderline, and definite. We currently do not work with all classes due to data scarcity for the definite class. Another interesting point is to evaluate the impact of demographic patient data, such as race, producing a model agnostic to these factors.
FUNDING
This work was supported by FAPEMIG (through the grant no. CEX-PPM-00098-17), MPMG (through the project Analytical Capabilities), CNPq (through the grant no. 310833/2019-1), CAPES, MCTIC/RNP (through the grant no. 51119) and H2020 (through the grant no. 777154). Dr Nascimento was supported in part by CNPq (312382/2019-7), by the Edwards Lifesciences Foundation (Every Heartbeat Matters Program 2020) and by FAPEMIG (APQ-000627-20). Dr Ribeiro was supported in part by CNPq (310679/2016-8 and 465518/2014-1) and by FAPEMIG (PPM-00428-17 and RED-00081-16).
AUTHOR CONTRIBUTIONS
BRN, CAS, AZB, and ALR were responsible for the conception of the screening projects and for the labeling of the data. JFBSM, ERN, and GLP designed the methods and the experimental setup. JFBSM wrote the source code and ran all the experiments under the supervision of ERN, GLP, and WMJ. ERN and GLP were also responsible for the conceptualization, methodology, and validation. All authors were engaged in the writing of the article.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
DATA SHARING STATEMENT
The data underlying this article will be shared on reasonable request to the corresponding author. A sample of exams was provided as Supplementary Material to help readers assess the quality of the videos.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
REFERENCES
- 1.Roth GA, Abate D, Abate KH, et al. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the global burden of disease study 2017. Lancet 2018; 392 (10159): 1736–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis AM, Vinci LM, Okwuosa TM, et al. Cardiovascular health disparities. Med Care Res Rev 2007; 64 (5 Suppl): 29S–100S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen MG, Fonarow GC, Peterson ED, et al. Racial and ethnic differences in the treatment of acute myocardial infarction. Circulation 2010; 121 (21): 2294–301. [DOI] [PubMed] [Google Scholar]
- 4.James SL, Abate D, Abate KH, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the global burden of disease study 2017. Lancet 2018; 392 (10159): 1789–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mondo C, Musoke C, Kayima J, et al. Presenting features of newly diagnosed rheumatic heart disease patients in Mulago Hospital: a pilot study. Cardiovasc J Afr 2013; 24 (2): 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okello E, Wanzhu Z, Musoke C, et al. Cardiovascular complications in newly diagnosed rheumatic heart disease patients at Mulago hospital, Uganda. Cardiovasc J Afr 2013; 24 (3): 76–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steer AC, Danchin MH, Carapetis JR.. Group A streptococcal infections in children. J Paediatr Child Health 2007; 43 (4): 203–13. [DOI] [PubMed] [Google Scholar]
- 8.Ribeiro ALP, Duncan BB, Brant LCC, et al. Cardiovascular health in Brazil: trends and perspectives. Circulation 2016; 133 (4): 422–33. [DOI] [PubMed] [Google Scholar]
- 9.Douglas PS, Garcia MJ, Haines DE, et al. ; Society for Cardiovascular Magnetic Resonance. ACCF/ASE/AHA/ASNC/HFSA/HRS/SCAI/SCCM/SCCT/SCMR 2011 appropriate use criteria for echocardiography: a report of the American College of Cardiology Foundation appropriate use criteria task force, American Society of Echocardiography, American Heart Association, American Society of Nuclear Cardiology, Heart Failure Society of America, Heart Rhythm Society, Society for Cardiovascular Angiography and Interventions, Society of Critical Care Medicine, Society of Cardiovascular Computed Tomography, and Society For Cardiovascular Magnetic Resonance Endorsed by the American College of Chest Physicians. J Am Coll Cardiol 2011; 57 (9): 1126–66. [DOI] [PubMed] [Google Scholar]
- 10.Remenyi B, Wilson N, Steer A, et al. World Heart Federation criteria for echocardiographic diagnosis of rheumatic heart disease—an evidence-based guideline. Nat Rev Cardiol 2012; 9 (5): 297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papolos A, Narula J, Bavishi C, et al. US hospital use of echocardiography: insights from the nationwide inpatient sample. J Am Coll Cardiol 2016; 67 (5): 502–11. [DOI] [PubMed] [Google Scholar]
- 12.Ziaeian B, Fonarow GC.. Epidemiology and aetiology of heart failure. Nat Rev Cardiol 2016; 13 (6): 368–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marijon E, Ou P, Celermajer DS, et al. Prevalence of rheumatic heart disease detected by echocardiographic screening. N Engl J Med 2007; 357 (5): 470–6. [DOI] [PubMed] [Google Scholar]
- 14.Roberts KV, Brown ADH, Maguire GP, et al. Utility of auscultatory screening for detecting rheumatic heart disease in high-risk children in Australia’s northern territory. Med J Aust 2013; 199 (3): 196–9. [DOI] [PubMed] [Google Scholar]
- 15.Vos T, Barber RM, Bell B, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the global burden of disease study 2013. Lancet 2015; 386 (9995): 743–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cios KJ, Chen K, Langenderfer RA.. Use of neural networks in detecting cardiac diseases from echocardiographic images. IEEE Eng Med Biol Mag 1990; 9 (3): 58–60. [DOI] [PubMed] [Google Scholar]
- 17.Martin-Isla C, Campello VM, Izquierdo C, et al. Image-based cardiac diagnosis with machine learning: A review. Front Cardiovasc Med 2020; 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua KL, Hsu CH, Hidayati S, et al. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. Onco Targets Ther 2015; 8: 2015–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, Khosla A, Gargeya R, et al. Deep learning for identifying metastatic breast cancer. arXiv preprint2016; arXiv:1606.05718.
- 20.Rajpurkar P, Irvin J, Zhu K, et al. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint2017; arXiv:1711.05225.
- 21.Shen D, Wu G, Suk HI.. Deep learning in medical image analysis. Annu Rev Biomed Eng 2017; 19: 221–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao X, Li W, Loomes M, et al. A fused deep learning architecture for viewpoint classification of echocardiography. Information Fusion 2017; 36: 103–13. [Google Scholar]
- 23.Madani A, Arnaout R, Mofrad M, et al. Fast and accurate view classification of echocardiograms using deep learning. Npj Digital Med 2018; 1 (1): 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madani A, Ong JR, Tibrewal A, et al. Deep echocardiography: data efficient supervised and semi-supervised deep learning towards automated diagnosis of cardiac disease. Npj Digital Med 2018; 1 (1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Gajjala S, Agrawal P, et al. Fully automated echocardiogram interpretation in clinical practice: feasibility and diagnostic accuracy. Circulation 2018; 138 (16): 1623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carneiro G, Nascimento JC, Freitas A.. The segmentation of the left ventricle of the heart from ultrasound data using deep learning architectures and derivative-based search methods. IEEE Trans Image Process 2012; 21 (3): 968–82. [DOI] [PubMed] [Google Scholar]
- 27.Lu A, Dehghan E, Veni G, et al. Detecting anomalies from echocardiography using multi-view regression of clinical measurements. In proceedings of the IEEE 15th International Symposium on Biomedical Imaging; 2018: 1504–8; Washington, DC, USA.
- 28.Ghorbani A, Ouyang D, Abid A, et al. Deep learning interpretation of echocardiograms. NPJ Digit Med 2020; 3 (1): 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beaton A, Lu JC, Aliku T, et al. The utility of handheld echocardiography for early rheumatic heart disease diagnosis: a field study. Eur Heart J Cardiovasc Imaging 2015; 16 (5): 475–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ploutz M, Lu JC, Scheel J, et al. Handheld echocardiographic screening for rheumatic heart disease by non-experts. Heart 2016; 102 (1): 35–9. [DOI] [PubMed] [Google Scholar]
- 31.Nascimento BR, Beaton AZ, Nunes MCP, et al. Echocardiographic prevalence of rheumatic heart disease in Brazilian schoolchildren: data from the PROVAR study. Int J Cardiol 2016; 219: 439–45. [DOI] [PubMed] [Google Scholar]
- 32.Sudeep DD, Sredhar K.. The descriptive epidemiology of acute rheumatic fever and rheumatic heart disease in low and middle-income countries. Am J Epidemiol Infect Dis 2013; 1 (4): 34–40. [Google Scholar]
- 33.Vakamudi S, Wu Y, Jellis C, et al. Gender differences in the etiology of mitral valve disease. J Am Coll Cardiol 2017; 69 (11 Supplement): 1972. [Google Scholar]
- 34.Wang S, Liu W, Wu J, et al. Training deep neural networks on imbalanced data sets. In proceedings of the International Joint Conference on Neural Networks (IJCNN); 2016: 4368–74; Vancouver, BC, Canada.
- 35.Tran D, Bourdev L, Fergus R, et al. Learning spatiotemporal features with 3d convolutional networks. In Proceedings of the IEEE International Conference on Computer Vision; 2015: 4489–97; Santiago, Chile.
- 36.Karpathy A, Toderici G, Shetty S, et al. Large-scale video classification with convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2014: 1725–32; Columbus, OH, USA.
- 37.Srivastava N, Hinton G, Krizhevsky A, et al. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 2014; 15 (1): 1929–58. [Google Scholar]
- 38.Wolpert DH.Stacked generalization. Neural Netw 1992; 5 (2): 241–59. [Google Scholar]
- 39.Breiman L.Random forests. Mach Learn 2001; 45 (1): 5–32. [Google Scholar]
- 40.Fernandez-Delgado M, Cernadas E, Barro S, et al. Do we need hundreds of classifiers to solve real world classification problems? J Mach Learn Res 2014; 15 (1): 3133–81. [Google Scholar]
- 41.Simonyan K, Zisserman A.. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv preprint2014; arXiv:1409.1556.
- 42.Bishop CM.Neural Networks for Pattern Recognition. USA: Oxford University Press, 1995. [Google Scholar]
- 43.Beaton A, Nascimento BR, Diamantino AC, et al. Efficacy of a standardized computer-based training curriculum to teach echocardiographic identification of rheumatic heart disease to nonexpert users. Am J Cardiol 2016; 117 (11): 1783–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.