Abstract
Objective
PubMed has suffered from the author ambiguity problem for many years. Existing studies on author name disambiguation (AND) for PubMed only used internal metadata for development. However, some of them are incomplete (eg, a large number of names are only abbreviated and their full names are not available) or less discriminative. To this end, we present a new disambiguation method, namely AggAND, by aggregating information from external databases.
Materials and Methods
We address this issue by exploring Microsoft Academic Graph, Semantic Scholar, and PubMed Knowledge Graph to enhance the built-in name metadata, and extend the internal metadata with some external and more discriminative metadata.
Results
Experimental results on enhanced name metadata demonstrate comparable performance to 3 author identifier systems, as well as show superiority over the original name metadata. More importantly, our method, AggAND, incorporating both enhanced name and extended metadata, yields F1 scores of 95.80% and 93.71% on 2 datasets and outperforms the state-of-the-art method by a large margin (3.61% and 6.55%, respectively).
Conclusions
The feasibility and good performance of our methods not only help better understand the importance of external databases for disambiguation, but also point to a promising direction for future AND studies in which information aggregated from multiple bibliographic databases can be effective in improving disambiguation performance. The methodology shown here can be generalized to broader bibliographic databases beyond PubMed. Our code and data are available online (https://github.com/carmanzhang/PubMed-AND-method).
Keywords: PubMed, author name disambiguation, bibliographic database, digital library
INTRODUCTION
Background and significance
As a special case of the more general problem of entity resolution,1–4 the author ambiguity problem is ubiquitous in large-scale bibliographic databases. It is usually not easy to determine whether 2 citations with similar names are authored by the same individual, and this uncertainty is even higher for abbreviated names. Nowadays, scholarly databases/libraries, eg, PubMed (https://pubmed.ncbi.nlm.nih.gov), provide easy-to-use interfaces for academic search, allowing researchers to quickly identify the latest literature despite the fast proliferation of scientific research. However, most of them do not disambiguate authors.5 This problem not only hinders the communication of valuable discoveries produced by others, but also restricts many downstream pieces of research or applications, such as funding fairness or gender inequality studies for scientists. Zhang et al6 claimed that the author identifier (ID) constructed by a disambiguation algorithm is a core component of AMiner (https://www.aminer.cn), a large-scale bibliographic database covering many journals and conference papers. A study of the query log analysis in PubMed suggested that nearly a quarter of queries used author names exclusively.7
This article focuses on the name ambiguity problem in PubMed. To alleviate this problem, the research team at the National Library of Medicine has provided a useful functionality: ranking citations that contain the query name in the PubMed interface.8 In addition, many academic services have been developed for researchers, such as ORCID (https://orcid.org), ResearcherID (https://www.researcherid.com), and ResearchGate (https://www.researchgate.net),9,10 but none of them are widely used in today’s author community. For example, only 15.4% of 214 million publications have an ORCID iD. Recent computational author name disambiguation (AND) methods for PubMed fall into 3 groups: rule based, graph based, and machine learning (ML) based. The principle of the rule-based approaches is to define a set of rules in advance and to use them for heuristically determining authors.11–13 The major drawback is the lack of supportive evidence; a preset threshold is manually chosen without clarification. In recent years, some graph-based disambiguation approaches14,15 have been proposed. Although not specifically designed for PubMed, most of them can be extended to AND in PubMed. However, high computational cost with graph representation learning techniques makes such methods inefficient to disambiguate on a PubMed-scale database. Others approach AND using ML-based methods.16–18 Models are trained through a learning process based on prior observations (ie, features extracted from metadata) and can therefore be used to infer the class of unseen data.19 However, the metadata sparsity problem has not been adequately considered. Existing ML-based approaches only used metadata within PubMed, and for some of those frequently used by priorworks,16 they may be not always available for all the PubMed authors. For example, only 58.03% of all PubMed authors have full names, and the low percentage has been reported as a limitation of PubMed data by many studies.5,20,21 Such a problem has raised even more ambiguities, as authors who were originally distinguishable by name variance become indistinguishable.
Objective
To resolve name ambiguities in PubMed, this study tries to handle the metadata-related issue by leveraging multiple databases. Specifically, 3 well-known databases, Microsoft Academic Graph (MAG),22 Semantic Scholar (S2),23 and PubMed Knowledge Graph (PKG)24 are exploited to aggregate additional information to enhance the internal metadata and extend with new, discriminative metadata. This work extends a previous study: Vishnyakova et al17 However, different from them, our work tries to boost AND methods using the external databases beyond PubMed and explore their impact on PubMed author disambiguation.
MATERIALS AND METHODS
Problem definition and method overview
In this section, we describe the materials and methods used in our method. We first give a formal definition of the disambiguation problem and then describe our method in a detailed manner. Our work follows the ML-based approach because it is more effective than rule-based approaches and is more efficient than graph-based approaches. For scholarly name disambiguation, the task is to characterize a function that can map 2 citations and with a similar name to a binary value , designating whether and are authored by the same person in the real world.
| (1) |
In doing so, a classification model with parameters and extracted features is used to contextualize the function , where is derived from author profiles. Here, author profiles are used to characterize authors in author level (ie, author name, email, organization, and location) and paper level (ie, paper title and abstract). Author profiles in citation can be represented by a sequence of metadata. For instance, , where is the number of PubMed internal metadata in use. Then, the disambiguation result can be formalized as:
| (2) |
, where represents the similarity measurement of the metadata.
In this study, we address the metadata-related problem by considering external databases; the associated metadata are utilized to enhance or extend the internal author profiles. As shown in equation 3, and are joined with the internal feature vector to form a new representation of ambiguous authors, where and denote the feature vector of enhanced metadata and extended metadata, respectively. The range of enhanced metadata is from to , and the range of extended metadata is from to .
| (3) |
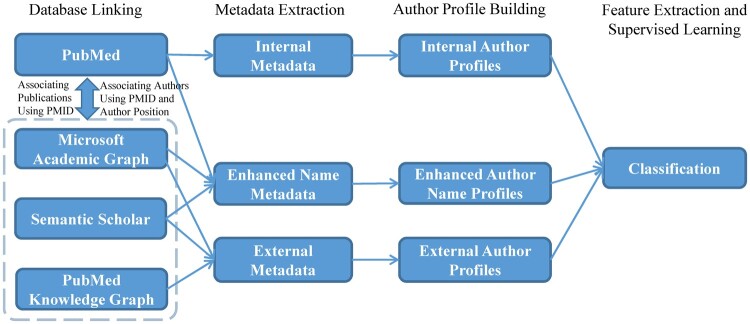
In this way, more discriminative information can be obtained and thus helps the AND task. Our research framework is illustrated in Figure 1. To obtain such metadata, multiple comprehensive databases are jointly used through linking databases; the associated metadata are incorporated to build more accurate author profiles, and . Then, by modeling the profiles with a supervised learning model on the training set, model predictions on the test set for each pair of ambiguous authors can be determined, and performance can be measured with respect to ground truth labels.
Figure 1.
Overview of our method for PubMed author name disambiguation; external databases are jointly used to boost author profiles for better disambiguation. PMID: PubMed publication identifier.
Database linking
In order to enrich author profiles, we made considerable efforts to link PubMed to MAG, S2, and PKG. These databases have opened their corpus either in full or in part for availability. We downloaded, parsed, and stored them in a database management system (note that the versions of the 4 databases are all 2019 versions). All of the external databases contain comprehensive publication records and have their own author ID system, of which MAG and S2 include over 170 million articles and have high coverage of PubMed papers (MAG-PubMed: 80.76%; S2-PubMed: approximately 100%) and authors (MAG-PubMed: 77.28%; S2-PubMed: 96.66%). As demonstrated in Figure 1, the linking approach includes 2 steps: publication linking and author linking. The publication linkages are established on the official PubMed publication identifier (PMID), which can be extracted from the “outbound links” of the databases. For example, “Paper Urls” (https://docs.microsoft.com/en-us/academic-services/graph/reference-data-schema#paper-urls) in MAG schema refers to the original links to the publications that MAG has obtained, and these links usually contain the identifiable metadata, PMID. The author linkages are based on the publication linkages. Because the author sequence or position is explicitly noted within each citation, we can connect the authorships between PubMed and these databases when there are the same PMID and same author position. Details of the linking approach are provided in our prior study.25 Note that the amount of data involved in the linking process is the entire PubMed, rather than the evaluation datasets, because the densities of the metadata obtained in this way are computed globally, so it is more appropriate to demonstrate the feasibility of our method when generalizing to the entire PubMed.
Metadata extraction
Table 1 summarizes the metadata harvested from the respective databases. For PubMed, we extract a variety of metadata. For the external metadata, we extract the corresponding author name and identifiers. The author names collected here are used to enhance the built-in name metadata, and the identifiers are used as an extension to PubMed metadata. We introduce such metadata extensions for several reasons. First, they are discriminating by nature. By borrowing the ideology of ensemble meta-algorithms in machine learning,26 the homogeneous IDs allow us to develop more powerful disambiguation methods. This ideology has been successfully applied in many machine learning models, like random forest (RF), which consists of multiple, less impressive trees. Second, they can be easily retrieved from the aforementioned databases; also, it is very convenient to transform them into features by determining whether the author ID is exactly the same or the intersections of coauthor IDs.
Table 1.
Internal and external metadata
| Source | Metadata |
|---|---|
| PubMed (Internal) | Author Name, Affiliation, Coauthor, Paper Title, Abstract, Keyword, MeSH Heading, Publication Date, Journal Title, Language, Reference |
| S2/MAG/PKG (External) | Author Name, Author ID, Coauthor IDs |
MAG: Microsoft Academic Graph; PKG: PubMed Knowledge Graph; S2: Semantic Scholar.
Author profile building
Most author profiles can be represented directly from metadata, whereas some profiles, such as enhanced name, affiliation-related profiles (Organization, Location, Country, City), and content-related profiles (Journal Descriptors, Semantic Types), require further refinement or the assistance of pretrained models.
In terms of the enhanced name profile, we consider the longest first name among the collected names as the well-formed first name to improve its density. For the affiliation-related profiles, we follow the same settings as that are described in the competing methods—Song et al16 and Vishnyakova et al17—to build these profiles. Specifically, named entity recognition provided in Stanford CoreNLP27 and Natural Language Toolkit (http://www.nltk.org/) are utilized to detect geographic information because various variants of valuable information are mixed together. The disambiguation features, Journal Descriptors (JDs), and Semantic Types (STs), proposed by the studies17,28 are used to capture the content information of the works. We utilize the Journal Descriptor Indexing tool29 to generate a ranked list of JDs or STs as an output to a given textual content (ie, title, abstract, and other textual matter of a citation where available).
Features and supervised learning
Based on the author profiles, we calculate the similarity of pairwise author profiles (hereafter referred to as features) and divide them into 4 groups according to the type of metadata or data source: Inner Group (metadata from PubMed only), InnerName Group (name metadata from PubMed only), EnhancedName Group (name metadata enhanced by external databases), and ExtendedIDs Group (ID-related metadata from external databases only). Table 2 describes the feature groups and the corresponding author profiles, metadata, and data sources. Note that we no longer itemize the Inner Group used by the competing methods,16,17 as it has been sufficiently described in their studies.
Table 2.
Feature group settings
| Feature Group | Feature Name | Similarity Measurement | Author Profile | Metadata | Source |
|---|---|---|---|---|---|
| InnerName/EnhancedName | Full Name Similarity | Full Name () | Author Name | PubMed/(PubMed, MAG, S2) | |
| Last Name Length | Last Name () | Author Name | PubMed/(PubMed, MAG, S2) | ||
| Full Name Difference | Full Name | Author Name | PubMed/(PubMed, MAG, S2) | ||
| Last Name Ambiguity Score | Last Name | Author Name | PubMed/(PubMed, MAG, S2) | ||
| ExtendedIDs | MAG Author ID Consistency | MAG Author ID () | Author ID | MAG | |
| S2 Author ID Consistency | S2 Author ID () | Author ID | S2 | ||
| PKG Author ID Consistency | PKG Author ID () | Author ID | PKG | ||
| MAG Coauthor IDs Overlap | MAG Coauthor IDs () | Coauthor IDs | MAG | ||
| S2 Coauthor IDs Overlap | S2 Coauthor IDs () | Coauthor IDs | S2 | ||
| PKG Coauthor IDs Overlap | PKG Coauthor IDs () | Coauthor IDs | PKG |
To demonstrate feature group contributions in the Results, the features are organized into several groups.
MAG: Microsoft Academic Graph; PKG: PubMed Knowledge Graph; S2: Semantic Scholar.
We derive these features from author profiles via the similarity measurements in Table 2. By definition, and mean extracting characters or word sequences from the input string. denotes the number of occurrences of a particular last name in the entire PubMed authors, and in this formulation is the number of unique last names.
Among all classification algorithms, RF is the most popular algorithm for AND tasks.5,16 Extensive evaluations indicated that the intrinsic complexity of AND is relatively high, and some simple models (eg, logistic regression, bayesian classifier) have a limited ability to handle this. Hence, in this article, we employ an RF classifier as a way of learning to disambiguate.
EXPERIMENT SETUP
Evaluation datasets
In recent years, 2 manually annotated disambiguation datasets of PubMed have been available for performance evaluation, they were created by Song et al16 and Vishnyakova et al,17 respectively; hereafter referred to as SONG and GS. In this article, we use them to examine the effectiveness of AND models. Both datasets were carefully curated. In SONG, multiple annotation iterations were conducted to ensure quality. It is worth mentioning that SONG was specifically designed for the highly productive first author (7.5 average citations per author), as the authors argued that first author–based disambiguation is more important than other positions, and it is also easier to disambiguate as more information about the first author can be obtained. GS is the first unbiased gold standard based on 1900 publication pairs. Though small in size, the gold standard shows close similarities to PubMed in terms of chronological distribution and information completeness (eg, coverage of East Asian last names). Note that, in SONG, citations are grouped into “blocks” by names. For evaluation, we transformed the blocks into pairwise form as Vishnyakova et al17 did. After transformation, 74.6% of samples in SONG are negatives, identical to the figure reported by them.17
Metrics
In this article, we use accuracy, precision, recall, and F1 metrics for evaluation, as they have been widely used in classification-based AND research.16,17,30 Each instance (paired citations) in the 2 datasets is predicted to be a binary value (0/1), designated as the same person or not. The ground truth is also a binary value (0/1), indicating whether 2 authors are the same person in the real world. Then, a confusion matrix can be constructed by considering predicted labels and ground truth labels, and the performance can be thus calculated.
Comparable methods
We introduce 7 baselines for comparison and further categorize them into 2 groups: ID Group and Internal Group. The ID Group consists of 3 types of author IDs disambiguated by respective databases and the Internal Group comprises 4 disambiguation methods based on PubMed metadata only.
ID Group:
MAG Author ID:22 Author identifier of Microsoft Academic Graph.
S2 Author ID:23 Author identifier of Semantic Scholar.
PKG Author ID:24 Author identifier of PubMed Knowledge Graph.
Internal Group:
MinSong16: A disambiguation method designed for the first author.
Vishnyakova17: Current state-of-the-art (sota) method for all author positions.
Vishnyakova&MinSong: A combination of the MinSong and Vishnyakova methods. Features of MinSong and Vishnyakova are jointly used to develop this method.
InnerName: Corresponding to the InnerName group in Table 2. Features are restricted to those derived from internal name metadata.
For comparison, our methods are the following.
EnhancedName: Corresponding to the feature group of EnhancedName in Table 2. Features are extracted from the enhanced name metadata for the purpose of examining the effect of the enhanced name metadata exclusively.
ExtendedIDs: Corresponding to the ExtendedIDs group in Table 2. Features are extracted from the extended metadata for the purpose of examining the effect of the external ID metadata exclusively.
AggAND: Assumed to be the most effective disambiguation method, corresponding to the combination of the Vishnyakova, EnhancedName, and ExtendedIDs methods. Features are largely derived from external metadata.
RESULTS
PubMed author name disambiguation benchmark
We conducted experiments on 2 gold standard datasets. Results are shown in Table 3. Based on it, several findings can be outlined.
Table 3.
Evaluation results on 2 PubMed-related datasets
| Method Group | Methods | GS |
SONG (only for the first author) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Accuracy (%) | Precision (%) | Recall (%) | F1 (%) | Accuracy (%) | Precision (%) | Recall (%) | F1 (%) | ||
| ID group | MAG Author ID | 71.77 | 99.07a | 56.32 | 71.81 | 94.22 | 96.8a | 78.45 | 86.66 |
| S2 Author ID | 85.16 | 94.01 | 81.77 | 87.46 | 90.55 | 96.06 | 65.9 | 78.17 | |
| PKG Author ID | 89.76 | 92.39 | 91.28 | 91.83 | 93.31 | 95.6 | 77.2 | 85.42 | |
| Internal group | MinSong | 80.28 | 82.38 | 86.9 | 84.52 | 93.5 | 80.3 | 86.99 | 83.18 |
| Vishnyakova (sota) | 90.02 | 91.8 | 92.6 | 92.19 | 95.04 | 86.72 | 88.05 | 87.16 | |
| Vishnyakova&MinSong | 89.95 | 91.17 | 93.02 | 92.06 | 94.94 | 85.37 | 87.95 | 86.17 | |
| InnerName | 79.11 | 83.35 | 84.65 | 83.98 | 86.95 | 65.9 | 70.32 | 67.36 | |
| Our | EnhancedName | 85.22 | 88.36 | 87.99 | 88.14 | 89.53 | 68.56 | 88.16 | 76.18 |
| ExtendedIDs | 92.07 | 92.56 | 95.67 | 94.07 | 96.93 | 95.93 | 90.28 | 92.83 | |
| AggAND | 94.66a | 94.61 | 97.04a | 95.8a | 97.1a | 94.21 | 93.43a | 93.71a | |
Note that all results generated by a learning algorithm in Table 3 were calculated by 10-fold cross-validation. In other words, those metrics were averaged over 10 consecutive runs.
MAG: Microsoft Academic Graph; PKG: PubMed Knowledge Graph; S2: Semantic Scholar.
The best value of each metric (column).
First, among the 3 ID systems, PKG author ID presents the best performance overall. Though MAG is slightly better than PKG on SONG, it performs less well on the full author positions-based dataset (GS). Note that MAG shows the highest precision overall and a relatively lower recall, suggesting that MAG is able to precisely attribute a proportion of citations to the correct authors.
Second, regarding the combination method: Vishnyakova&MinSong, we did not observe performance gains when MinSong was incorporated with Vishnyakova, suggesting that this combination strategy does not contribute more to this task. Thus, in the following evaluation phrase, the Vishnyakova method is joined as a part of AggAND instead of MinSong and Vishnyakova&MinSong, because not only is it more effective, but also it contains necessary internal information that is not present in the external databases.
Third, 2 name-based approaches, InnerName and EnhancedName, show significant performance differentiation. The EnhancedName approach gains additional F1 scores of 4.16% and 8.82% on the 2 datasets, respectively. Also, we found that the enhanced name–based approach could achieve comparable performance with state-of-the-art author ID systems. Interestingly, the ExtendedIDs method alone outperforms all baseline methods and was found to be the second-best performer. We also observed that, on top of Vishnyakova method, our method achieved an even better performance when the feature group of EnhancedName and ExtendedIDs joined. Our method AggAND, based on metadata enhancement and extension, produced an F1 score of 95.8%, an absolute 3.61% improvement over the current state-of-the-art method (Vishnyakova) on GS. A similar trend can be observed on SONG: a 6.55% improvement on F1 than the state-of-the-art method.
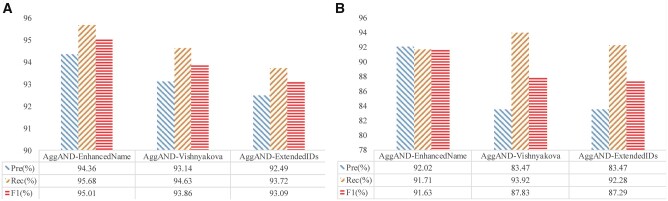
Feature group importance
To investigate the importance of the 3 parts of AggAND, we conducted a leave-one-out feature experiment (see Figure 2); this step helps to better understand the importance of external databases to disambiguation works as 2 parts of AggAND are established on external databases. From Figure 2, we can identify that ExtendedIDs and Vishnyakova are the 2 most important feature groups on both GS and SONG. This is especially the case for ExtendedIDs, which has the highest impact on performance.
Figure 2.
Leave-one-out feature experiments of AggAND; the evaluation results on (A) GS and (B) SONG gold standards. “X-Y” represents removing Y from X. The degree of performance reduction caused by removing an individual feature group indicates the importance of that feature group.
DISCUSSION
Enhanced Name vs Original Name
The Enhanced Name disambiguation method (EnhancedName) led the Original Name method (InnerName) by a large margin, which can be explained by the strong evidence in Table 4, in which ∼32% additional abbreviated names were restored to their full names. This exercise makes disambiguation algorithms work easily because more authors with restored full names can be distinguished directly, and may not need further judgment by their affiliation, and research topics. Therefore, it can be understood that increasing the density of full name could substantially improve the ML-based disambiguation method.
Table 4.
Metadata density at author level or paper level
| Metadata Group | Metadata | Density on the Whole PubMed (%) | Density on GS (%) | Density on SONG (%) | Level |
|---|---|---|---|---|---|
| Enhanced metadata | Enhanced full name | 90.85 | 89.01 | 98.78 | Author level |
| Extended metadata | MAG Author ID | 78.78 | 92.86 | 93.67 | Author level |
| S2 Author ID | 98.53 | 99.13 | 98.96 | Author level | |
| PKG Author ID | 99.13 | 98.95 | 99.86 | Author level | |
| Internal metadata | Paper title | 99.94 | 99.97 | 100 | Paper level |
| Vernacular title | 12 | 8.73 | 0.14 | Paper level | |
| Original full name | 58.03 | 59.05 | 77.6 | Author level | |
| 7.28 | 7.14 | 57.15 | Author level | ||
| Affiliation | 38.5 | 35.85 | 94.92 | Author level | |
| Coauthor | 77.42 | 95.73 | 86.53 | Paper level | |
| Keyword | 20.56 | 22.56 | 4.25 | Paper level | |
| MeSH Heading | 86.67 | 88.03 | 93.56 | Paper level | |
| Abstract | 65.54 | 82.82 | 89.38 | Paper level | |
| Reference | 18.12 | 26.91 | 26.21 | Paper level | |
| Journal title | 100 | 100 | 100 | Paper level | |
| Publication date | 100 | 100 | 100 | Paper level | |
| Databank | 1.18 | 2.11 | 1.64 | Paper level | |
| Grant support | 10.56 | 12.73 | 21.58 | Paper level | |
| Publication type | 100 | 100 | 100 | Paper level |
MAG: Microsoft Academic Graph; PKG: PubMed Knowledge Graph; S2: Semantic Scholar.
Advantages of our metadata extension method
The impressive result achieved by ExtendedIDs alone outperforms all baseline methods, suggesting that integrating less impressive author IDs can produce an even better ID, which confirms our previous hypothesis about the bagging ideology on external author identifiers. Apart from its effectiveness, we note that our ExtendedIDs method has other advantages. It is easy to obtain the dependent metadata (Extended Metadata in Table 4) from external databases and transform them into features. In addition, such metadata covers almost all PubMed authors, with densities ranging from 78% to 99%, and most of them over 90%, as Table 4 demonstrated. High density is critically important as it can not only show good performance on the evaluation dataset but, more importantly, provide a feasible and practical way to scale to the entire PubMed. Interested scholars can try to build a more accurate author ID system for PubMed using ExtendedIDs.
Disambiguation method based on metadata enhancement and extension
Experimental results demonstrate the superiority of AggAND over existing methods or systems. We attribute this to the rich metadata of PubMed. National Library of Medicine has made considerable efforts to strengthen PubMed for decades; many rich, discriminative metadata can be extracted for building accurate author profiles so that many excellent methods like Vishnyakova can be developed. In this work, a widely used metadata identifier, PMID, which is generally available for over 30 million PubMed citations, enables us to enhance and extend the internal metadata by linking to other large-scale databases.
Importance of external databases for disambiguation
The leave-one-out experiment shows that extended metadata (ExtendedIDs) has the most impact on performance. For the enhanced metadata (EnhancedName), although, the performance reduction when excluding the EnhancedName feature group is not as large as excluding other feature groups, we can still observe 0.79% and 2.08% reductions of F1 on the respective datasets. We believe it is a nontrivial performance margin as it is generally more difficult to improve the performance on top of an existing F1 score of more than 90%. Based on the analysis, it is clear that external databases are critically important for disambiguation works, and the methods incorporated with external metadata can be even more effective than the “internal” methods constructed by a variety of PubMed metadata.
Error analysis
Figure 3 shows the cases that AggAND fails to predict. As shown, the importance distribution clearly demonstrates “PKG Author ID Consistency” contributes the most, indicating that the dominant feature may be the most important factor in leading to the errors. For example, in cases 1 and 3, although other features have positive effects on correct predicting, eg, very close publication year gap and similar JDs in case 1, and different names in case 3, the dominant feature values of them are opposite to the ground truth.
Figure 3.
Randomly selected 2 false negatives and 2 false positives. Note that they were all selected from GS, because this dataset does not restrict to the first author and more importantly, is very proximate to PubMed. In each case, a prediction possibility denotes the confidence of it to be a positive sample (0.5 is the decision value). For simplicity, we only report the top 6 most important features (all feature importance is shown in Supplementary Table 1), the vertical bar above each feature represents the corresponding contribution calculated by random forest using the mean squared error criterion, which is a widely used tool for explainable analysis. The cells below each feature contain 3 lines; the first 2 lines are author profiles and the third line represents a feature value. MAG: Microsoft Academic Graph; PKG: PubMed Knowledge Graph; PMID: PubMed publication identifier.
For cases 2 and 4, values of the dominant feature are in line with the ground truth. The errors are caused by different reasons. In case 2, the paired authors share the same name; however, the ambiguity score is much higher than other cases, suggesting that “Huang” is a very popular last name, which further implies that they are more likely to be different authors, besides, the larger gap of publication years and the diverse JDs also exacerbate the matter. Similarly, in case 4, identical author names and close publication years tend to imply that the 2 authorships are the same author.
Suggestions for further improvement
Although our models achieved such success, over 5% of authors in the GS remain ambiguous. We note that the performance can be further improved by considering 1 of the following 2 aspects. First, future studies can explore other metadata like we did, for example, the affiliation metadata, which was proved to be one of the most effective metadata for AND when it is available for most authors.16 However, our evaluation shows that the density is only ∼38% (see Table 4). Second, future studies can consider author similarity from a semantic perspective. To measure the similarity, we simply used word-level measurements; it seems to be very straightforward, and the semantic information in this way was not exploited adequately. Therefore, some cutting-edge semantic-matching algorithms such as DSSM31 are also worth exploring.
Limitations
Our first limitation is that efficiency is not fully considered. Parsing geographically relevant author profiles (used by Vishnyakova et al and our method) like organization or location from affiliation metadata using the named entity recognition model is performed inefficiently according to our empirical observation. In the widely accepted namespace-based disambiguation framework, in which ambiguous authors are aggregatedinto namespaces according to the same last name and first initial, there are nearly 10 000 namespaces with a size of over 1000 in PubMed. Thus, more computational resources will be required to build this kind of author profile. Another limitation we are aware of is that none of the available datasets addresses the name synonymy problem (ie, name variants of the same person) adequately. This is especially the case for SONG, in which all last names of the same author are the same. However, it is often the case that an author's names in bibliographic databases differ from their actual names due to many reasons, such as typographical errors, errors caused by the publishing process, etc. This problem limits our methods in examining the performance in a more realistic scenario.
CONCLUSION
Despite such limitations, our work still makes valuable contributions. We for the first time explored large-scale external databases to help disambiguate authors in PubMed. The new disambiguation method, AggAND, based on metadata enhancement and extension, outperformed the current state-of-the-art method by a large margin on the 2 gold standards. Moreover, the feature analysis revealed that external databases can be crucially important for PubMed AND. Though our research focuses only on PubMed, we believe that future disambiguation works on other large-scale bibliographic databases, such as Web of Science or DBLP, will benefit from our findings.
FUNDING
This work was supported by the Major Project of the National Social Science Foundation of China grant number 17ZDA292.
AUTHOR CONTRIBUTIONS
WL designed the study. LZ conducted this work and analyzed the data under WL’s supervision. LZ, YH, and JY wrote the manuscript. All authors revised and approved the manuscript for publishing.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge the National Center for Biotechnology Information team, the Microsoft Academic team, and the team of Allen Institute for AI for their data collections.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The data collections underlying this article were provided by the National Library of Medicine, Microsoft Research, the Allen Institute for AI, and the Texas Advanced Computing Center at the University of Texas at Austin under licenses, and they are publicly available at ftp://ftp.ncbi.nlm.nih.gov/pubmed/baseline, https://zenodo.org/record/2628216, http://s2-public-api-prod.us-west-2.elasticbeanstalk.com/corpus/, and http://er.tacc.utexas.edu/datasets/ped. The code for this article is available in GitHub at https://github.com/carmanzhang/PubMed-AND-method.
REFERENCE
- 1.Getoor L, Machanavajjhala A.. Entity resolution: theory, practice & open challenges. Proc VLDB Endow 2012; 5 (12): 2018–9. [Google Scholar]
- 2.Elmagarmid AK, Ipeirotis PG, Verykios VS.. Duplicate record detection: a survey. IEEE Trans Knowl Data Eng 2007; 19 (1): 1–16. [Google Scholar]
- 3.Christen P.A survey of indexing techniques for scalable record linkage and deduplication. IEEE Trans Knowl Data Eng 2012; 24 (9): 1537–55. [Google Scholar]
- 4.Shen W, Wang J, Han J.. Entity linking with a knowledge base: Issues, techniques, and solutions. IEEE Trans Knowl Data Eng 2015; 27 (2): 443–60. [Google Scholar]
- 5.Sanyal DK, Bhowmick PK, Das PP.. A review of author name disambiguation techniques for the PubMed bibliographic database. J Inf Sci 2021; 47 (2): 227–54. [Google Scholar]
- 6.Zhang Y, Zhang F, Yao P, et al. Name disambiguation in a miner: clustering, maintenance, and human in the loop. In: Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining 2018: 1002–11. [Google Scholar]
- 7.Herskovic JR, Tanaka LY, Hersh W, et al. A day in the life of PubMed: analysis of a typical day’s query log. J Am Med Inform Assoc 2007; 14 (2): 212–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, Islamaj Doğan R, Kim S, et al. Author name disambiguation for PubMed. J Assoc Inf Sci Technol 2014; 65 (4): 765–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lerchenmueller MJ, Sorenson O.. Author disambiguation in PubMed: evidence on the precision and recall of author-ity among NIH-funded scientists. PLoS One 2016; 11 (7): e0158731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison AM, Harrison AM.. Necessary but not sufficient: unique author identifiers. BMJ Innov 2016; 2 (4): 141–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varadharajalu A, Liu W, Wong W.. Author name disambiguation for ranking and clustering PubMed data using NetClus. In: Australasian Joint Conference on Artificial Intelligence; 2011: 152–61. [Google Scholar]
- 12.Strotmann A, Zhao D, Bubela T.. Author name disambiguation for collaboration network analysis and visualization. Proc Am Soc Info Sci Tech 2009; 46 (1): 1–20. [Google Scholar]
- 13.Johnson SB, Bales ME, Dine D, et al. Automatic generation of investigator bibliographies for institutional research networking systems. J Biomed Inform 2014; 51: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Wan R, Wen C, et al. Author name disambiguation on heterogeneous information network with adversarial representation learning. AAAI Proc 2020; 34 (1): 238–45. [Google Scholar]
- 15.Qiao Z, Du Y, Fu Y, et al. Unsupervised Author Disambiguation using Heterogeneous Graph Convolutional Network Embedding. In: 2019 IEEE International Conference on Big Data (Big Data); 2019. [Google Scholar]
- 16.Song M, Kim EH-J, Kim HJ.. Exploring author name disambiguation on PubMed-scale. J Informetr 2015; 9 (4): 924–41. [Google Scholar]
- 17.Vishnyakova D, Rodriguez-Esteban R, Rinaldi F.. A new approach and gold standard toward author disambiguation in MEDLINE. J Am Med Inform Assoc 2019; 26 (10): 1037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim K, Sefid A, Weinberg BA, et al. A web service for author name disambiguation in scholarly databases. In: 2018 IEEE International Conference on Web Services (ICWS); 2018. [Google Scholar]
- 19.Hussain I, Asghar S.. A survey of author name disambiguation techniques: 2010-2016. Knowl Eng Rev 2017; 32: e22. [Google Scholar]
- 20.Torvik VI, Smalheiser NR.. Author name disambiguation in MEDLINE. ACM Trans Knowl Discov Data TKDD 2009; 3 (3): 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, Kim J.. Effect of forename string on author name disambiguation. J Assoc Inf Sci Technol 2020; 71 (7): 839–55. [Google Scholar]
- 22.Sinha A, Shen Z, Song Y, et al. An overview of Microsoft Academic Service (mas) and applications. In: WWW ’15 Companion: Proceedings of the 24th International Conference on World Wide Web; 2015: 243–6. [Google Scholar]
- 23.Ammar W, Groeneveld D, Bhagavatula C, et al. Construction of the Literature Graph in Semantic Scholar, AWS Simple Cloud Storage [dataset]. http://s2-public-api-prod.us-west-2.elasticbeanstalk.com/corpus. Accessed December 1, 2019.
- 24.Xu J, Kim S, Song M, et al. Building a PubMed knowledge graph, Texas Advanced Computing Center Storage [dataset]. http://er.tacc.utexas.edu/datasets/ped. Accessed December 20, 2019.
- 25.Zhang L, Huang Y, Cheng Q, et al. Mining Author Identifiers for PubMed by Linking to Open Bibliographic Databases. In: 2020 IEEE 20th International Conference on Software Quality, Reliability and Security Companion (QRS-C); 2020. [Google Scholar]
- 26.Breiman L.Bagging predictors. Mach Learn 1996; 24 (2): 123–40. [Google Scholar]
- 27.Manning CD, Surdeanu M, Bauer J, et al. The Stanford CoreNLP natural language processing toolkit. In: Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics; 2014: 55–60. [Google Scholar]
- 28.Vishnyakova D, Rodriguez-Esteban R, Ozol K, et al. Author name disambiguation in MEDLINE based on journal descriptors and semantic types. In: Proceedings of the Fifth Workshop on Building and Evaluating Resources for Biomedical Text Mining; 2016: 134–42. [Google Scholar]
- 29.Humphrey SM, Lu CJ, Rogers WJ, et al. Journal descriptor indexing tool for categorizing text according to discipline or semantic type. AMIA Annu Symp Proc 2006; 2006: 960. [PMC free article] [PubMed] [Google Scholar]
- 30.Treeratpituk P, Giles CL.. Disambiguating authors in academic publications using random forests. In: Proceedings of the 9th ACM/IEEE-CS Joint Conference on Digital Libraries; 2009: 39–48. [Google Scholar]
- 31.Huang P-S, He X, Gao J, et al. Learning deep structured semantic models for web search using clickthrough data. In: Proceedings of the 22nd ACM International Conference on Information & Knowledge Management; 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data collections underlying this article were provided by the National Library of Medicine, Microsoft Research, the Allen Institute for AI, and the Texas Advanced Computing Center at the University of Texas at Austin under licenses, and they are publicly available at ftp://ftp.ncbi.nlm.nih.gov/pubmed/baseline, https://zenodo.org/record/2628216, http://s2-public-api-prod.us-west-2.elasticbeanstalk.com/corpus/, and http://er.tacc.utexas.edu/datasets/ped. The code for this article is available in GitHub at https://github.com/carmanzhang/PubMed-AND-method.