Abstract
Mantle cell lymphoma (MCL) is an especially aggressive and highly heterogeneous mature B-cell lymphoma. Heat shock protein 90 (HSP90) is considered an attractive therapeutic target in a variety of cancers, including MCL, but no HSP90 inhibitors have succeeded in the clinical trials to date. Exploring fine mechanisms of HSP90 inhibition in cancer cells may shed light on novel therapeutic strategies. Here, we found that HSP90 knockdown and continuous inhibition with ganetespib inhibited growth of MCL cells in vitro and in vivo. To our surprise, transient exposure over 12 h was almost as efficient as continuous exposure, and treatment with ganetespib for 12 h efficiently inhibited growth and induced G1 cell cycle arrest and apoptosis of MCL cells. Transcriptome analysis complemented by functional studies was performed to define critical MCL signaling pathways that are exceptionally sensitive to HSP90 inhibition and vital to cell fate. Six genes (cell division cycle 6, cell division cycle 45, minichromosome maintenance 4, minichromosome maintenance 7, RecQ-mediated genome instability 2, and DNA primase polypeptide 1) involved in DNA replication and repair were identified as consistently downregulated in three MCL cell lines after transient ganetespib treatment. E2F1, an important transcription factor essential for cell cycle progression, was identified as a ganetespib target mediating transcriptional downregulation of these six genes, and its stability was also demonstrated to be maintained by HSP90. This study identifies E2F1 as a novel client protein of HSP90 that is very sensitive and worthy of targeting and also finds that HSP90 inhibitors may be useful in combination therapies for MCL.
Keywords: HSP90, ganetespib, mantle cell lymphoma, E2F1, DNA replication, DNA repair
Abbreviations: ALK, anaplastic lymphoma kinase; BTK, Bruton's tyrosine kinase; CDC, cell division cycle; CDK, cyclin-dependent kinase; CHX, cycloheximide; co-IP, coimmunoprecipitation; EdU, 5-ethynyl-2′-deoxyuridine; FBS, fetal bovine serum; HSP90, heat shock protein 90; IF, immunofluorescence; MCL, mantle cell lymphoma; MCM, minichromosome maintenance; mTOR, mechanistic target of rapamycin; ORC, origin recognition complex; PRIM1, DNA primase polypeptide 1; RMI2, RecQ-mediated genome instability 2; TF, transcription factor; qRT-PCR, quantitative RT-PCR
Mantle cell lymphoma (MCL) is a distinct and highly heterogeneous mature B-cell lymphoma characterized by aberrant expression of cyclin D1 (1), which represents about 3 to 10% of non-Hodgkin's lymphoma (2). Most patients with MCL are diagnosed at stage III to IV with symptoms, including lymphadenopathy, hepatosplenomegaly, bone marrow involvement, and leukemic spread. Despite impressive advances in the clinical outcomes of other types of lymphoma, MCL is still considered as an aggressive and incurable lymphoma with a median survival of 5 to 7 years (3). Therefore, there is an urgent need for discovery of novel effective treatments against MCL.
Heat shock protein 90 (HSP90) was an attractive therapeutic target in a variety of cancers including lymphomas (4, 5, 6, 7, 8). As an ATP-dependent molecular chaperone, HSP90 supports proper folding, stabilizes conformation, and aids in the degradation of myriad proteins (9, 10). Importantly, HSP90 was demonstrated to be moderately or strongly expressed in subsets of patients with non-Hodgkin's lymphoma, including MCL (11). Colomer et al. (12) demonstrated that the HSP90 inhibitor IPI-504 overcame bortezomib resistance in MCL in part by downregulating the prosurvival chaperone Grp78 (78 kDa glucose-regulated protein). More recently, Jacobson et al. (13) have reported that HSP90 inhibition with AUY922 overcame ibrutinib resistance in MCL cell lines by inducing the complete degradation of both Bruton's tyrosine kinase (BTK) and IκB kinase α. These studies nominated HSP90 inhibitors as promising drugs for bortezomib or ibrutinib-based combination therapy against MCL. However, in the context of the broad range of HSP90 client proteins, these studies did not thoroughly profile the roles of HSP90 in MCL.
Ganetespib (STA-9090) is a well-characterized second-generation HSP90 inhibitor that eliminates much toxicity of earlier agents and showed favorable potent anticancer activities against a variety of tumor types and tolerability (14, 15, 16). Ganetespib was successfully applied preclinically and in early clinical trials as the single agent in client protein-derived tumors, such as non–small-cell lung cancer with anaplastic lymphoma kinase (ALK) translocations, human epidermal growth factor receptor 2–overexpressing breast cancer, and other cancers (15). Ganetespib has also been demonstrated to increase the efficacy of standard-of-care chemotherapeutics and targeted agents in diverse cancers, providing compelling rationales for the exploration of ganetespib as part of novel combinatorial treatment strategies (15). One of the most intriguing characteristics of ganetespib was its capacity to overcome multiple forms of intrinsic and acquired resistance to other clinically relevant targeted agents, including tyrosine kinase inhibitors (15, 17, 18). But till now, ganetespib have not succeeded in phase 3 clinical trials because of poor selectivity, unsatisfied efficacy, or high toxicity. So, searching for the novel client protein of HSP90, which is the most sensitive and worthy for targeting and exploring the combination treatment to enhance the efficacy and reduce the toxicity, will help to take advantages of such HSP90 inhibitors. We have here evaluated the effects of ganetespib on MCL in vitro and in vivo, demonstrating strong growth inhibitory effects. Importantly, we have used transcriptome analysis complemented by functional studies to define critical MCL signaling pathways that are most sensitive to HSP90 inhibition and also vital to cell fate.
Results
HSP90 inhibition suppressed growth of MCL cells in vitro and in vivo
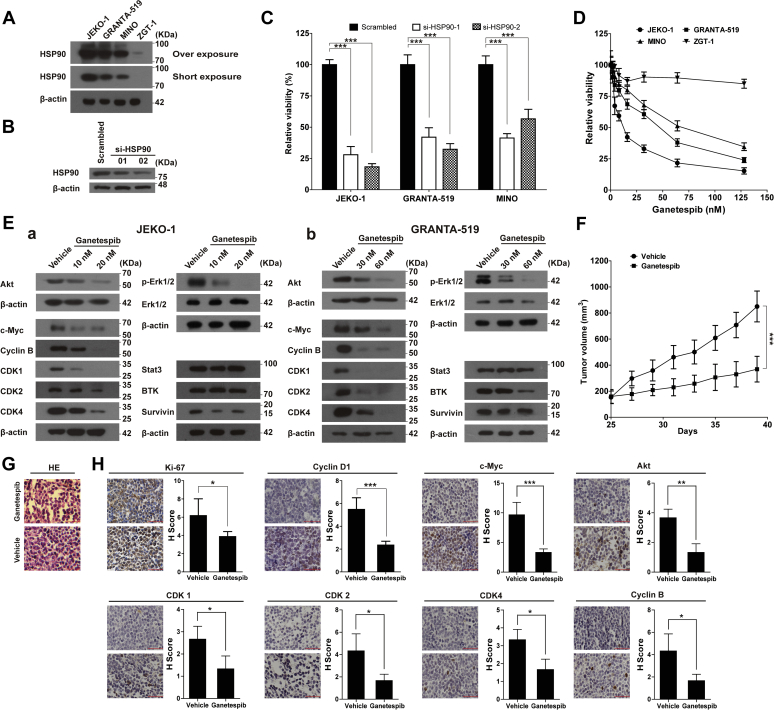
HSP90 protein was strongly expressed in the JEKO-1, GRANTA-519, and MINO cell lines when compared with the peripheral blood mononuclear cells from the healthy volunteer (Fig. 1A). siRNA inhibition of HSP90 in JEKO-1 (Fig. 1B), GRANTA-519, and MINO cells (Fig. S1) in each case dramatically reduced cell viability within 72 h, indicating an essential role of this protein in these MCL cell lines (Fig. 1C). We then assessed growth inhibitory activities of ganetespib in MCL cell lines by treating the cells with ganetespib for 72 h. The IC50 values of ganetespib were 12.7, 47.3, and 68.2 nM for JEKO-1, GRANTA-519, and MINO, respectively (Fig. 1D). In contrast, ganetespib did not inhibit the growth of mononuclear ZGT-1 cells derived from the peripheral blood of a healthy volunteer (Fig. 1D). The protein levels of key client proteins of HSP90, such as cyclin-dependent kinase 1 (CDK1), CDK2, CDK4, c-Myc, cyclin B, ALK, BTK, AKT, phosphorylated extracellular signal–regulated kinase 1/2, were reduced with ganetespib treatment in cell lines JEKO-1 (Fig. 1Ea) and GRANTA-519 (Fig. 1Eb).
Figure 1.
HSP90 inhibition inhibited growth of MCL cells in vitro and in vivo.A, the protein levels of HSP90 in JEKO-1, GRANTA-519, and MINO cells. ZGT-1, the mononuclear cells in peripheral blood from a healthy volunteer, was used as the control cells. B, the protein levels of HSP90 with or without HSP90 knockdown in JEKO-1 cells. C, the results of the cell viability assays on MCL cells with or without HSP90 knockdown (MTT assays were performed 72 h after electroporation). D, inhibiting HSP90 with ganetespib inhibits the viability of MCL cell lines. MCL cell lines JEKO-1, GRANTA-519, and MINO were treated with increasing concentrations of ganetespib for 72 h, and the cell viability was assessed by MTT assays. ZGT-1, the mononuclear cells in peripheral blood from a healthy volunteer, was used as the control cells. E, the protein levels of Akt, CDK1, CDK2, CDK4, c-Myc, cyclin B, p-Erk1/2, Erk1/2, BTK, STAT3, survivin in JEKO-1 (a) and GRANTA-519 cells (b) with or without 72-h ganetespib treatment at the indicated concentration. F, HSP90 inhibition with ganetespib significantly inhibited tumor growth in a xenograft mouse model of MCL. Over a period of 3 weeks, mice bearing JEKO-1 xenografts (n = 6/group) were i.v. dosed with 100 mg/kg ganetespib or vehicle starting at the 25th day once weekly. G and H, the representative pictures and statistics of H&E-stained tumor sections (G), and immunohistochemistry detections of Ki67, cyclin D1, c-Myc, Akt, CDK1, CDK2, CDK4, and cyclin B (H) of tumors in experiment F. The scale bar represents 50 μm. Data indicate the mean ± SD, calculated from triplicate samples from multiple independent experiments (n ≥ 3). Erk, extracellular signal–regulated kinase; HSP90, heat shock protein 90; MCL, mantle cell lymphoma; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide.
We also evaluated the effects of ganetespib on JEKO-1 xenograft tumors (Fig. 1F). Weekly administration of ganetespib (100 mg/kg) dramatically inhibited the growth of xenografts, reducing final volume by 56% (p < 0.001). H&E staining showed that ganetespib induced vacuoles in tumors in ganetespib-treated group (Fig. 1G). Immunohistochemical staining revealed that molecular markers of cell proliferation such as Ki-67 and cyclin D1 and typical client proteins of HSP90 c-Myc, AKT, CDK1, CDK2, CDK4, and cyclin B (Fig. 1H) were significantly lower in ganetespib-treated tumors. Together, these data revealed that HSP90 inhibition using either siRNA or ganetespib efficiently suppressed growth of MCL cells both in vitro and in vivo.
12-h treatment of ganetespib efficiently inhibited growth of MCL cells
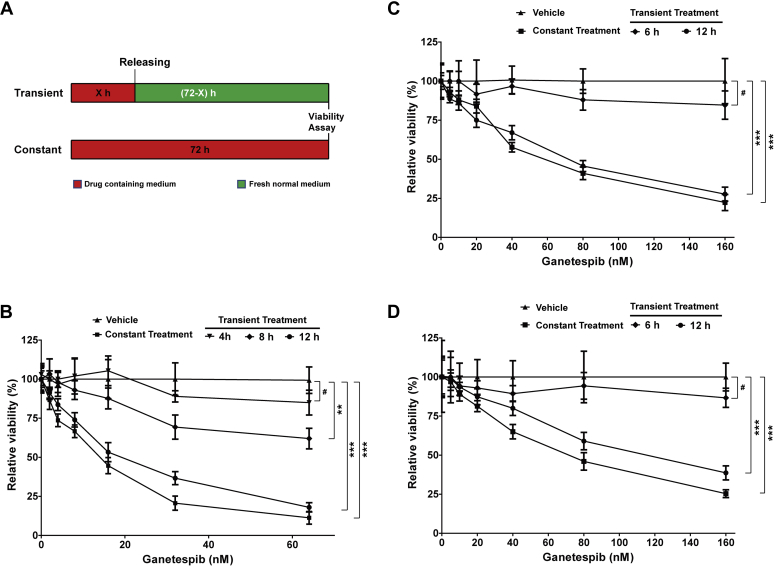
To determine whether short periods of exposure to ganetespib were sufficient to inhibit cell growth, we treated JEKO-1 cells with ganetespib for 4, 8, or 12 h followed by replacement with drug-free media, compared with continuous exposure for 72 h (Fig. 2A). We found that 4-h treatment had very limited effects on cell viability, whereas 8-h treatment modestly inhibited (−36%) cell growth (Fig. 2B). However, 12-h treatment dramatically inhibited growth of the cells and was almost as efficient as the constant treatment for 72 h (13.9 versus 18.8 nM, as compared by IC50, Table S1). Similar results were obtained in GRANTA-519 and MINO cells, with results indicating that 12-h treatment of ganetespib efficiently inhibited growth of MCL cells (Fig. 2, C and D and Table S1). These results are potentially significant for clinical use of HSP90 inhibitors. A short-term treatment may provide the opportunities to identify the signaling pathways that are the most sensitive to HSP90 inhibition and also vital to cell fate in MCL and avoid affecting a large number of client proteins of HSP90.
Figure 2.
Transient ganetespib treatment efficiently inhibited growth of MCL cells.A, the scheme of experiment settings. Cells were treated with either ganetespib or vehicle constantly for 72 h or treated with ganetespib or vehicle transiently for indicated hours and then washed and cultured in fresh normal media. B, the relative viability of JEKO-1 cells with ganetespib treatment at indicated concentration or with vehicle for 0, 4, 8, 12, or 72 h and viability was determined using MTT reagents. The relative viability was normalized to the respective vehicle-treated values for the same period. C and D, the relative viabilities of GRANTA-519 and MINO cells with transient or contentious treatment. Mean ± SD, n = 3. MCL, mantle cell lymphoma; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide.
Transient ganetespib treatment induced G1 cell cycle accumulation and apoptosis of MCL cells
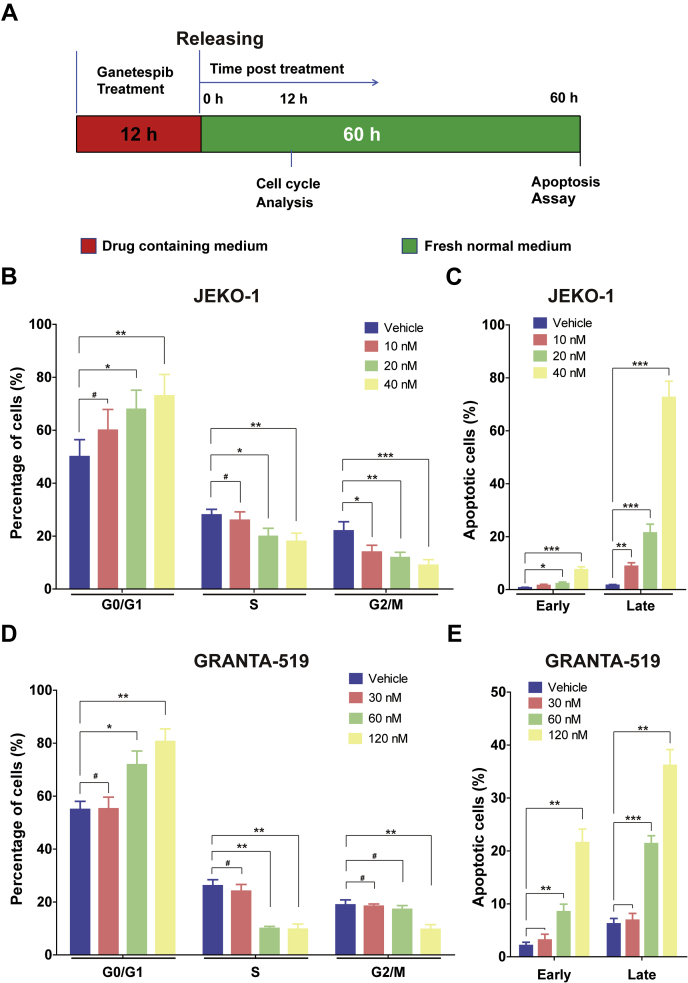
To understand the mechanism of ganetespib action, JEKO-1 cells were treated with ganetespib at different concentrations for 12 h and then released in fresh medium for 12 h (Fig. 3A). Ganetespib treatment increased the proportion of cells in G0/1 phase and decreased the proportion of cells in S and G2/M phases in a dose-dependent manner at 12 h following release (Fig. 3, B and C). This brief drug treatment also induced significant levels of apoptosis, in a dose-dependent manner, at 60 h following release (Fig. 3, D and E). Similar results were obtained when GRANTA-519 cells were treated (but with higher concentrations of drug) for 12 h (Fig. 3, F and G). Higher drug concentrations for GRANTA-519 cells were used because these cells are less responsive than JEKO-1 as seen from the data in Figure 1D.
Figure 3.
Transient ganetespib treatment induced cell cycle alteration and apoptosis in MCL cells.A, the scheme of the experiment settings. Cells were treated with either ganetespib at indicated concentration or vehicle for 12 h and then washed and cultured in fresh normal media for 12 h before cell cycle analysis. Cells were treated with either ganetespib at indicated concentration or vehicle for 12 h and then washed and cultured in fresh normal media for 60 h before apoptosis assays. B, statistics of cell cycle distribution of flow cytometry plots of JEKO-1 cells with or without 12-h ganetespib treatment. Cells were stained with propidium iodide. C, analyzed results of the presence of annexin V (+)/PI (−) (early apoptosis) and annexin V (+)/PI (+) (late apoptosis) of flow cytometry plots of JEKO-1 cells with or without 12-h ganetespib treatment. Cells were stained with annexin V–FITC/propidium iodide and analyzed for apoptosis distribution. D, the statistics of cell cycle distribution of GRANTA-519 cells with or without 12-h ganetespib treatment. E, the statistics of apoptosis assay of GRANTA-519 cells with or without 12-h ganetespib treatment. Mean ± SD, n = 3. MCL, mantle cell lymphoma.
Transient ganetespib treatment downregulated genes in DNA replication and cell cycle progression networks
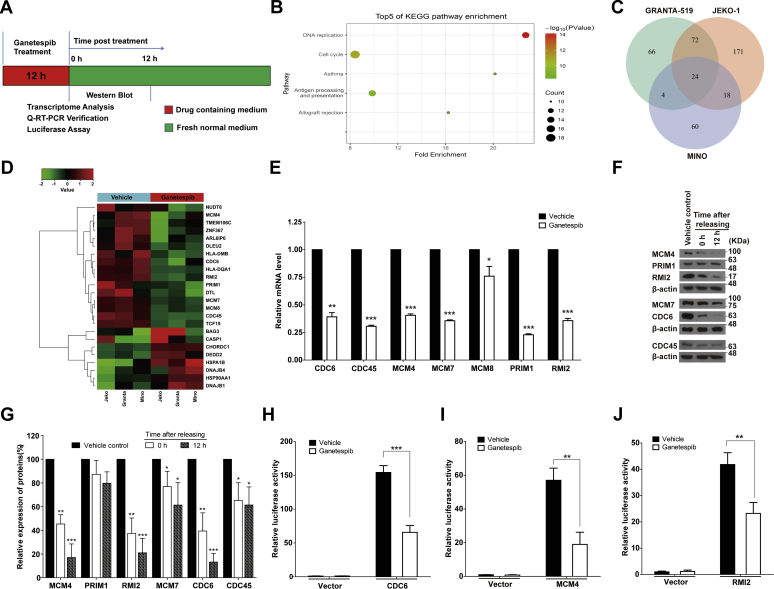
To identify the molecular pathways important for the cytostatic and cytotoxic activity of ganetespib in MCL, we examined the effect of 12 h of drug treatment on the expression of HSP90 client proteins that have been reported to be important in the development of lymphoma, including AKT, ALK, BCL2, BCL6, Janus kinase 1, signal transducer and activator of transcription 3, BTK, CDK1, CDK2, CDK4, CDK6, cyclin B, cyclin D, extracellular signal–regulated kinase 1/2, and c-Myc (19, 20, 21, 22, 23, 24). JEKO-1 cells were treated with 20 nM of ganetespib for 12 h, and protein levels were measured 0 or 12 h after medium change (Fig. 4A). For most of these lymphoma-related client proteins of HSP90, expression levels were unaffected at both time points, except for a minor decrease in CDK4 (Fig. S2). These results strongly suggested that an alternative mechanism mediated the observed cytostatic/cytotoxic activity of ganetespib. To gain insight into this mechanism, we performed transcriptome analysis.
Figure 4.
Transient ganetespib treatment induced downregulation of genes involved in DNA replication and cell cycle progression.A, the scheme of the experiment settings. Cells were treated with either ganetespib (20 nM for JEKO-1, 45 nM for GRANTA-519, and 60 nM for MINO) or vehicle for 12 h and then subjected to transcriptome analysis. B, KEGG pathway enrichment plots of transcriptome analysis of JEKO-1 cells with or without 12-h ganetespib treatment. C, the Venn diagram created based on the genes whose expression levels were changed in three different cell lines. D, clustering analysis on 24 genes that changed in the same direction in all three cell lines and identified in D. E, the mRNA levels of CDC6, CDC45, MCM4, MCM7, MCM8, PRIM1, and RMI2 in JEKO-1 cells with or without 12-h ganetespib treatment. JEKO-1 cells were treated with either ganetespib or vehicle for 12 h and then subjected to quantitative RT-PCR verification. F, the protein expression levels of CDC6, CDC45, MCM4, MCM7, PRIM1, and RMI2 in JEKO-1 cells with or without 12-h ganetespib treatment. JEKO-1 cells were treated with either ganetespib or vehicle for 12 h, washed, and immediately subjected to Western blot or washed and cultured in fresh normal media for 12 h before Western blot. The time points chosen here are 0 and 12 h after drug treatment. G, the quantified statistics of F. H–J, the luciferase assays on promoters of CDC6, MCM4, and RMI2 genes. JEKO-1 cells were treated with either ganetespib or vehicle for 12 h and then subjected to luciferase assays. Mean ± SD, n = 3. KEGG, Kyoto Encyclopedia of Genes and Genomes; MCM, minichromosome maintenance 4; PRIM1, DNA primase polypeptide 1; RMI2, RecQ-mediated genome instability 2.
JEKO-1 cells were treated with ganetespib (20 nM) for 12 h, and then total mRNA was collected and used for transcriptome analysis (Fig. 4A). The result of Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that the differentially expressed genes were significantly enriched in networks such as DNA replication, cell cycle, antigen processing and presentation, and allograft rejection (Fig. 4B). The results from reactome pathway analysis showed that the differentially expressed genes were enriched in networks including DNA replication, cell cycle, DNA repair, immune system, and cellular responses to external stimuli (Figs. S3 and S4). When GRANTA-519 (45 nM) and MINO cells (60 nM) were treated with ganetespib for 12 h, similar results were obtained (Fig. S3).
Overall comparison of gene expression changes in the three MCL cell lines showed that the expression of 24 genes changed in the same direction in all three cell lines (Fig. 4C), with eight genes upregulated and 16 genes downregulated (Fig. 4D). These genes were divided into four groups based on currently known gene functions (Table S2): HSP-related, DNA replication and cell cycle, B cell specific, and unknowns (e.g., with poor functional characterization in the literature). Five genes including HSP90AA1, HSPA1B, BAG3, DNAJB1, and DNAJB4 are relevant to heat shock and other stress responses and were upregulated when cells were treated with HSP90 inhibitor. HLA-DQA1 and HLA-DMB, the molecular markers of B cells, were downregulated by HSP90 inhibitor treatment. Importantly, cell division cycle 6 (CDC6), CDC45, minichromosome maintenance 4 (MCM4), MCM7, MCM8, RecQ-mediated genome instability 2 (RMI2), and DNA primase polypeptide 1 (PRIM1), which are involved in DNA replication and cell cycle progression through S phase, were consistently downregulated in all three cell lines. Consistently, based on reactome pathway analysis, the most prominent functions of affected genes are DNA replication and cell cycle (Figs. S3 and S4).
We used quantitative RT-PCR (qRT-PCR) to further confirm the results obtained from transcriptome analysis. As shown in Figure 4E, the mRNA levels of CDC6, CDC45, MCM4, MCM7, MCM8, PRIM1, and RMI2 decreased after cells were treated with ganetespib for 12 h. Similarly, when GRANTA-519 (45 nM) and MINO cells (60 nM) were treated with ganetespib for 12 h, the mRNA levels of CDC6, CDC45, MCM4, MCM7, PRIM1, and RMI2 significantly decreased except for MCM8 (Fig. S5). The results from qRT-PCR were highly consistent with those from transcriptome analysis.
We further measured the protein levels of the genes with the consistent changes in all three cell lines. As shown in Figure 4, F and G, the protein levels of PRIM1 did not significantly change. The protein levels of MCM7 and CDC45 only mildly decreased when the cells were transiently treated and released for 12 h. Importantly, the protein levels of CDC6, MCM4, and RMI2 significantly decreased just after 12-h treatment and almost exhausted after the cells were released for 12 h. Compared with the other proteins, CDC6, MCM4, and RMI2 decreased more rapidly and thoroughly. Since the mRNA levels can be affected by either transcription or mRNA stability, we used promoter-luciferase constructs for a subset of these genes to analyze consequences for transcription (Fig. 4, H–J). We determined that promoter activities of CDC6, MCM4, and RMI2 decreased dramatically after cells were treated with ganetespib for 12 h.
Transient ganetespib treatment resulted in decreased DNA synthesis and increased DNA damage
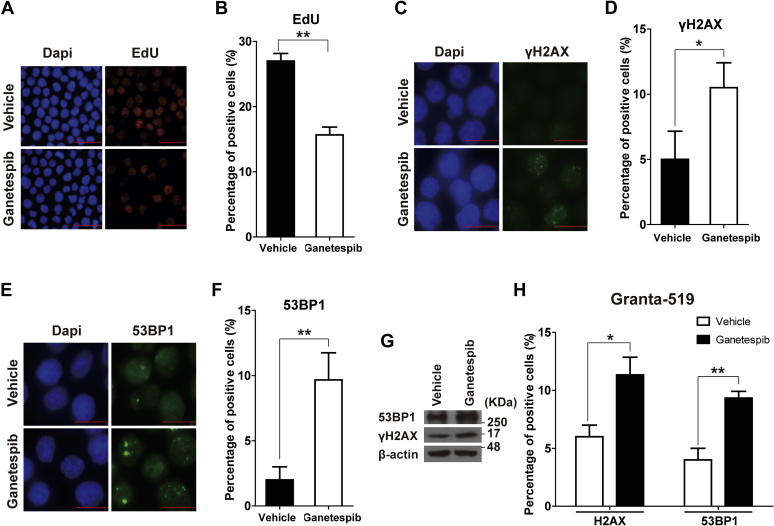
CDC6, MCM4, and RMI2 are important in DNA replication, with roles in unwinding DNA helices, maintaining DNA replication fork, and facilitating DNA damage repair (25, 26, 27). We analyzed 5-ethynyl-2′-deoxyuridine (EdU) incorporation to evaluate the DNA synthesis in JEKO-1 cells treated with ganetespib for 12 h and released for another 12 h. Ganetespib treatment significantly decreased DNA synthesis in JEKO-1 cells (Fig. 5, A and B). Furthermore, immunofluorescence (IF) analysis showed that ganetespib treatment caused significant accumulation of DNA damage, as manifested by increased γH2AX and 53BP1 foci (Fig. 5, C–F). Consistent with the IF results, Western analysis also showed the upregulation of γH2AX and 53BP1 levels in cells with ganetespib treatment (Fig. 5G). Similarly, increased numbers of γH2AX and 53BP1 foci were observed when they were treated with ganetespib (45 nM) for 12 h and released for another 12 h in GRANTA-519 cells (Fig. 5H).
Figure 5.
Transient ganetespib treatment resulted in decreased DNA synthesis and increased DNA damage.A and B, the representative pictures and the statistics of EdU staining of JEKO-1 cells with or without 12-h ganetespib treatment. JEKO-1 cells were first treated with either ganetespib (20 nM) or vehicle for 12 h and then cultured in fresh media for 24 h before EdU staining. The scale bar represents 25 μm. C–G, JEKO-1 cells were treated with either ganetespib or vehicle for 12 h before immunofluorescence (IF) staining or Western blot of γH2AX and 53BP1. The scale bar represents 10 μm. C and D, the representative IF pictures and the statistics of γH2AX. E and F, the representative IF pictures and the statistics of 53BP1. Mean ± SD, n = 3. G, representative Western blot results of γH2AX and 53BP1 in JEKO-1 cells with or without 12-h ganetespib treatment. H, the statistics of γH2AX and 53BP1 IF experiments on GRANTA-519 cells with or without 12-h ganetespib treatment. GRANTA-519 cells were treated with either ganetespib (45 nM) or vehicle for 12 h before IF staining of γH2AX and 53BP1. Mean ± SD, n = 3. EdU, 5-ethynyl-2′-deoxyuridine.
E2F1 mediated transcriptional downregulation of genes regulating DNA replication and repair after ganetespib treatment
To gain insight into the mechanism by which ganetespib downregulated transcription of cell cycle–related genes, we tested the hypothesis that transcription factors (TFs) dependent on HSP90 were downregulated by ganetespib treatment. We used the PROMO algorithm (28) to predict the TF candidates for the six genes confirmed in qRT-PCR. Weight matrices are constructed from known binding sites extracted from TRANSFAC and used for the identification of potential binding sites in sequences in PROMO with a number of unique features. Among the TFs identified by this approach, E2F1, AR, and c-Myc were selected for further analysis because they were the highest ranked from the PROMO analysis, and the existing literature shows that they are closely related to the transcriptional regulation of those six genes (Table S3). TP53 was excluded since JEKO-1 lacks p53 expression (29, 30).
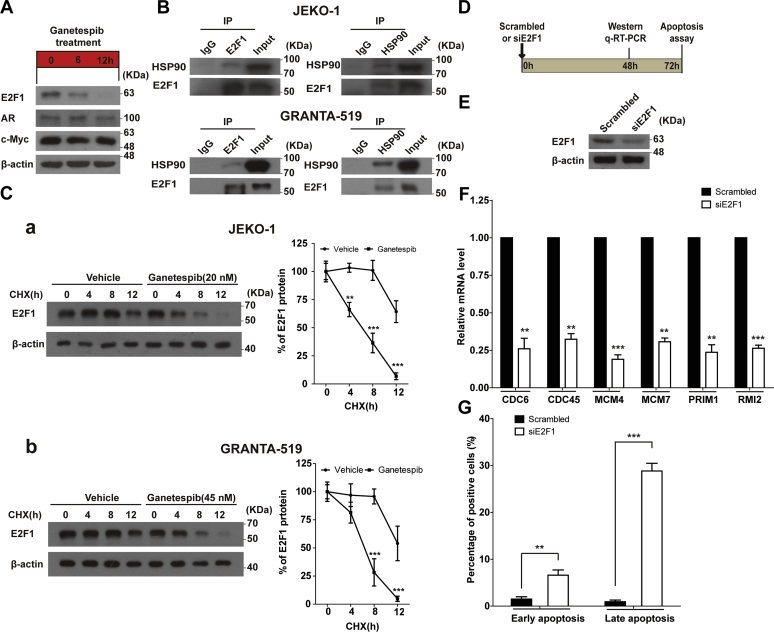
In JEKO-1 cells, expression of neither AR nor c-Myc was affected by ganetespib treatment (Fig. 6A). However, expression of E2F1 significantly decreased within 6 h and was almost depleted when cells were treated for 12 h (Fig. 6A). Based on the aforementioned information, we inferred that E2F1 may mediate the effect of HSP90 inhibition on transcription of genes regulating DNA replication and repair.
Figure 6.
E2F1 mediated transcriptional downregulation of genes involved in DNA replication and cell cycle progression under 12-h ganetespib treatment.A, protein expression levels of E2F1, AR, and c-Myc in JEKO-1 cells without ganetespib treatment or with ganetespib treatment at the indicated period of treatment. The cells were collected right after the drug treatment. B, E2F1 was coimmunoprecipitated with HSP90, and HSP90 was coimmunoprecipitated with E2F1 using the lysates from the nuclei. Indicated cells were subjected to coimmunoprecipitation experiments using the anti-HSP90 or anti-E2F1 antibody. An isotype-matched IgG was used as a negative control. Levels of immunoprecipitated HSP90 and E2F1 were determined by immunoblotting using the indicated antibodies. C, results of cycloheximide (CHX)-chase assay. Indicated cells were treated with either vehicle or ganetespib (20 nM of ganetespib for JEKO-1 (a) and 45 nM of ganetespib for GRANTA-519 (b)) for 0, 4, 8, and 12 h in the presence of CHX (5 mg/ml). Treated cells were used for immunoblot experiments, and protein levels of E2F1 were determined. Blots were quantified using the National Institutes of Health ImageJ software. The amounts of E2F1 were normalized to β-actin of the same sample. D, the scheme of experiments of E–G. JEKO-1 cells were knockdowned with scrambled siRNAs or siE2F1, and cells were harvested after 48 h for Western blot or quantitative RT-PCR experiments or harvested after 72 h for apoptosis assays. E, protein expression levels of E2F1 in JEKO-1 cells with or without E2F1 knockdown. F, mRNA levels of CDC6, CDC45, MCM4, MCM7, PRIM1, and RMI2 in JEKO-1 cells with or without E2F1 knockdown. G, the results of apoptosis assays of JEKO-1 cells with or without E2F1 knockdown. HSP90, heat shock protein 90; MCM, minichromosome maintenance 4; PRIM1, DNA primase polypeptide 1; RMI2, RecQ-mediated genome instability 2.
To explore whether there is interaction between HSP90 and E2F1, immunoprecipitation experiments were performed. It was found that E2F1 can be immunoprecipitated by HSP90 and vice versa (Fig. 6B). Whether HSP90 normally maintains E2F1 levels is also an interesting question. Cycloheximide (CHX), the inhibitor of protein biosynthesis, was used to solve this problem. We found that HSP90 does normally maintain E2F1 levels, and ganetespib treatment accelerates degradation of E2F1 (Fig. 6C). To further examine whether E2F1 can regulate transcription of these six genes, the expression of E2F1 was silenced using siRNAs (Fig. 6, D and E). E2F1 knockdown significantly downregulated the mRNA levels of those six genes (Fig. 6F). E2F1 depletion significantly induced early and late apoptosis (Fig. 6G), similar to ganetespib treatment (Fig. 3, D and E).
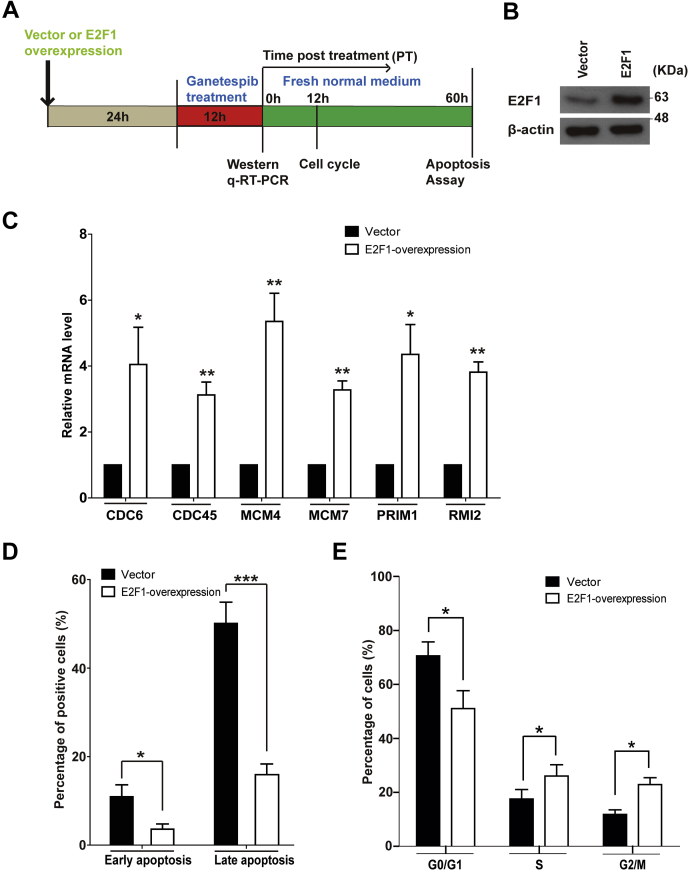
To further test our hypothesis, we then transiently overexpressed E2F1 in JEKO-1 cells (Fig. 7, A and B) and treated these cells with ganetespib for 12 h (Fig. 7A). As expected, E2F1 overexpression increased the mRNA levels of the six ganetespib-inhibited genes (Fig. 7C). In addition, E2F1 overexpression significantly rescued cells from apoptosis and cell cycle arrest induced by ganetespib treatment (Fig. 7, D and E). These results demonstrated that E2F1 overexpression attenuated the effects of 12 h of ganetespib treatment.
Figure 7.
The effects of E2F1 overexpression on 12-h ganetespib treatment.A, the scheme of experiments. JEKO-1 cells were transfected via electroporation with either empty vectors or E2F1 overexpression vectors for 24 h. Then cells were treated with ganetespib (20 nM) for 12 h and subjected to Western blot or quantitative RT-PCR experiments. After 12-h ganetespib treatment, cells were washed and cultured in fresh media for 60 h before apoptosis assays. B, protein expression levels of E2F1 in JEKO-1 cells with or without E2F1 overexpression. C, mRNA levels of CDC6, CDC45, MCM4, MCM7, PRIM1, and RMI2 in JEKO-1 cells with or without E2F1 overexpression under 12-h ganetespib treatment. D, the results of apoptosis assays of JEKO-1 cells with or without E2F1 overexpression under 12-h ganetespib treatment. E, the results of cell cycle assays of JEKO-1 cells with or without E2F1 overexpression under 12-h ganetespib treatment. Mean ± SD, n = 3. MCM, minichromosome maintenance 4; PRIM1, DNA primase polypeptide 1; RMI2, RecQ-mediated genome instability 2.
Pretreatment of ganetespib sensitize MCL cells to mechanistic target of rapamycin inhibitors
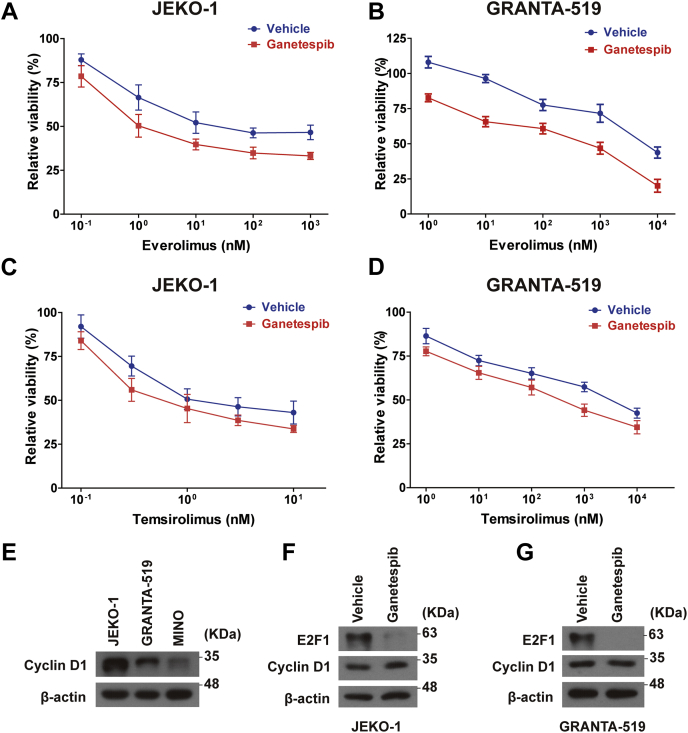
As a class of powerful drugs for targeted therapy, mechanistic target of rapamycin (mTOR) inhibitors have been widely used in treating multiple solid tumors and hematologic malignancies (31, 32). Recent studies have revealed great potential of mTOR inhibitors in treating MCL (33, 34, 35). More importantly, recent studies have showed synergistic effects between mTOR inhibitors and CDK inhibitors in many cancers including MCL (36, 37). Since transient treatment of ganetespib can significantly reduce E2F1 protein in MCL cells, the major downstream effector of CDKs, we hypothesized that transient treatment of ganetespib can increase sensitivities of MCL cells to mTOR inhibitors. In our experiments, JEKO-1 and GRANTA-519 cells were first treated with ganetespib (or vehicle) at 20 and 45 nM, respectively, for 12 h, and then ganetespib was removed, and they were treated with everolimus or temsirolimus for another 60 h. As shown in Figure 8A and Table S4, IC50 of everolimus reached 8.147 nM with ganetespib pretreatment compared with IC50 of 108.2 nM without pretreatment in JEKO-1 cells (p = 0.042). Similarly, IC50 of everolimus was 243.0 nM with ganetespib pretreatment compared with IC50 of 5026 nM without pretreatment in GRANTA-519 cells (Fig. 8B; p = 0.015). In addition, IC50 of temsirolimus was 1.286 nM with ganetespib pretreatment, whereas it was 2.755 nM without ganetespib pretreatment in JEKO-1 cells (Fig. 8C; p = 0.094). IC50 of temsirolimus was 374.4 nM with ganetespib pretreatment, whereas it was 2409 nM without ganetespib pretreatment in GRANTA-519 cells (Fig. 8D; p = 0.009).
Figure 8.
Transient ganetespib treatment sensitizes MCL cells to mTOR inhibitors. Cells were pretreated with or without ganetespib for 12 h, before they were treated with everolimus or temsirolimus for 60 h. A and B, relative viabilities of JEKO-1 and GRANTA-519 cells with everolimus with or without transient ganetespib pretreatment. C and D, relative viabilities of JEKO-1 and GRANTA-519 cells with temsirolimus with or without transient ganetespib pretreatment. For transient ganetespib treatment, ganetespib (DMSO was used in the vehicle group) was applied on cells (20 nm for JEKO-1 and 45 nM for GRANTA-519) for 12 h and then washed. After that, cells were cultured in media with everolimus or temsirolimus at different concentrations for another 60 h. Mean ± SD, n = 3. E, protein levels of cyclin D1 in JEKO-1, GRANTA-519, and MINO cells. F, protein levels of E2F1 and cyclin D1 in JEKO-1 cells with or without transient ganetespib treatment. G, protein levels of E2F1 and cyclin D1 in GRANTA-519 cells with or without transient ganetespib treatment. DMSO, dimethyl sulfoxide; MCL, mantle cell lymphoma; mTOR, mechanistic target of rapamycin.
Often overexpressed in MCL cell lines (Fig. 8E), cyclin D1 is a known client of HSP90. Our further experiments showed that transient ganetespib treatment had very limited effects on cyclin D1 protein levels but almost eliminated E2F1 in JEKO-1 (Fig. 8F) and GRANTA-519 (Fig. 8G) cells. Considering that transient ganetespib treatment had little impact on expressions of other cyclins and CDKs (Fig. S2), these results further supported that transient ganetespib treatment sensitizes MCL cells to mTOR inhibitors through E2F1 depletion, which will in turn deactivate the relevant CDK-involving prosurvival pathways.
Discussion
Our data revealed that ganetespib efficiently inhibited the growth of MCL cells in vitro and in vivo, with IC50 values lower than HSP90 inhibitors AT13387, PU-H71, HSP990, 17-AAG, and comparable to AUY922 in these MCL cell lines and similar to the IC50 values of ganetespib on other B-cell lymphoma cell lines (13, 17, 38). More importantly, we found that transient inhibition of HSP90 using ganetespib shows potent growth inhibitory effects on cells. Considering the clinical limitation of HSP90 inhibitors caused by their toxic side effects, a strategy using short-term treatment might light new ideas to decrease the toxicity of the drugs. Since HSP90 has hundreds of protein clients, continuous HSP90 inhibition is expected to broadly disrupt cellular signaling. A transient inhibition of HSP90 may help to identify the most sensitive and quick-acting pathways. In our study, early transcriptome responses to ganetespib treatment indicate that inhibition of HSP90 limits E2F1-dependent transcription to induce cell cycle arrest and apoptosis through downregulation of proteins important for DNA replication and repair, namely CDC6, MCM4, and RMI2 (Fig. S6).
The replication of genomic DNA starts with the formation of the origin recognition complex (ORC) and the binding of CDC6 and Cdt1 to ORC. After that, MCM complex proteins, a family of ATP-dependent helicases, are recruited to the ORC to form prereplication complex. The joining of CDC45, GINS complex, and other additional factors finally complete the formation of CDC45–MCM–GINS helicases, which “license” DNA synthesis (39). In our study, transient ganetespib treatment depressed DNA synthesis, whereas decreased the protein levels of CDC6 and MCM4. More and more studies revealed that the components of prereplication complex are essential for DNA repair and genome stability (40, 41). In addition, RMI2, an essential component of the Bloom helicase, was proven necessary for DNA homologous recombination repair (42, 43). Consistently, we found that transient ganetespib treatment significantly increased DNA damage accumulation, whereas decreased the protein levels of CDC6, MCM4, and RMI2 in cells. HSP90 inhibitors (especially ganetespib) have shown significant effects on cell cycle regulatory genes and signaling pathways and DNA repair genes. We found that ganetespib treatment decreased expression of cyclin B1 and CHK1 earlier in gastric cancer cell lines (44). Guan et al. (45) found that ganetespib treatment caused an increase in cell cycle–dependent kinase inhibitors p15 and p21 in c-Myc–dependent esophageal squamous cell carcinoma. Ganetespib significantly represses CDK1 expression and caused G2 block and mitotic arrest in thyroid cancer cells (46). Proia et al. (47) found that ganetespib treatment selectively altered the expression of a large set of genes involved in cell cycle–related activities, including DNA replication and repair (BRCA1/2), cell cycle regulation (CDC2 and CDC25), centrosome/spindle activities (BUB1/3, CENPE/M, KIF14, and FAM33A), chromosome condensation (TOP2A and NCAPG), and replication (RFC3/4, MCM family). Connolly et al. (48) also found that ganetespib treatment destabilized HSP90 client proteins involved in DNA damage response, cell cycle checkpoint, and DNA repair and exhibited prominent dose-dependent decreases in BRCA1, BRCA2, CDK1, CHK1, ATM serine/threonine kinase, homologous recombination protein RAD51, MRE11, and c-Myc in ovarian cancer cells. In addition, levels of p21, p27, CHK1, RAD51, and phosphorylated form of histone H2AX changed, and activation of CHK2 was observed following ganetespib exposure in colorectal cancer (49).
MCM genes are well-known E2F-inducible genes (50, 51, 52). An early study by Yan et al. (53) showed that transcription of CDC6 is positively regulated by E2F1. Several previous studies have demonstrated the high expression of E2F1 in MCL (54, 55). Our data support that E2F1 positively regulates the transcription of six genes, including CDC6, CDC45, MCM4, MCM7, PRIM1, and RMI2 in MCL cells. Amere Subbarao et al. (56) reported that HSP90 interacts with E2F1 and E2F2 in breast cancer cells with coimmunoprecipitation (co-IP) experiments earlier this year. Interestingly, they observed that E2F1 and E2F2 stability at the protein level was affected by the 17-AAG treatment in MCF-7 cells, whereas only E2F2 stability was affected in human embryonic kidney 293T cells. They found a significant decrease of the nuclear accumulation of E2F1 and E2F2 in MCF-7 cells, but they were not affected by 17-AAG treatment in human embryonic kidney 293T cells, which indicated that HSP90-dependent E2F regulation appeared to be specific to cancer cells. We think E2F1 can work as an amplifier for transient ganetespib treatment to induce cell cycle arrest and apoptosis. Our data strongly propose E2F1 to be a promising therapeutic target in MCL.
Cyclin D1 is overexpressed in virtually all patients with MCL and was proved to be highly sensitive to CDK inhibitors (36, 57). Since ganetespib can greatly downregulate E2F1, which is the main downstream effector of CDKs in MCL cells, transient treatment of ganetespib can be considered as an effective strategy for CDK inhibition. There were around 40 clinical trials in phase I/II/III involving ganetespib as a single agent or in combination therapies on solid tumors, leukemia, myeloproliferative disorders, bone marrow diseases, and other diseases. Although randomized phase III study of ganetespib with docetaxel versus docetaxel in advanced non–small-cell lung cancer (GALAXY-2) trial did not result in improved survival for salvage therapy of patients with advanced-stage lung adenocarcinoma (58), there has been clear evidence of inhibition of vascular endothelial growth factor, signal transducer and activator of transcription 3, and hypoxia-inducible factor 1α with ganetespib in patient tumors (58, 59). Given that a combination of CDK and mTOR inhibitors may exert synergic effects on MCL cells, it is not unanticipated that in our study transient treatment of ganetespib significantly sensitized MCL cells to mTOR inhibitors.
So far, there is no standard frontline therapy for MCL. Today, many patients with MCL are commonly treated with combination of chemotherapy and immunotherapy. Recent studies have proposed new molecular targets in MCL, such as proteins contributing to protein homeostasis, DNA damage processes, cell cycle regulation, cell proliferation, and apoptosis (33). Encouraged by the success of consecutive usage of transient treatment with ganetespib and mTOR inhibitors, we believed that further understanding of the molecular mechanisms will greatly improve the development of combination therapies with HSP90 inhibitors and pave the ways for proposing novel treatment strategies for MCL.
Experimental procedures
Cell lines and reagents
The MCL cell lines JEKO-1 and MINO (American Type Culture Collection) were maintained in RPMI1640 supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (100 units/ml and 100 g/ml, respectively) (all from Thermo Fisher Scientific). The cell line GRANTA-519 was grown in medium containing 90% minimum essential media and 10% FBS, supplemented with penicillin/streptomycin (cell line and media from Thermo Fisher Scientific). All cell lines were obtained from the American Type Culture Collection and cultured at 37 °C in a humidified, 5% carbon dioxide atmosphere, and tested for mycoplasma contamination before experiments.
Cell treatment
Cells were treated with either ganetespib or vehicle constantly for 72 h or treated with ganetespib or vehicle transiently for indicated hours and then washed and cultured in fresh normal media. IC50 values were calculated using CompuSyn (ComboSyn).
Antibodies and chemical reagents
The commercial sources of antibodies used are listed in Table S5. Ganetespib was purchased from Selleck Chemicals, Inc. siRNAs targeting HSP90 and E2F1 (siHSP90s and siE2F1) and scrambled siRNAs were synthesized by Shanghai GenePharma Co (Table S6). All chemicals were purchased from Sigma–Aldrich unless otherwise specified.
Isolation and culture of primary cells
Approval for the study was obtained from the Ethics Committee at Jiangsu University. The informed consent form was signed by the health volunteer. Mononuclear cells from peripheral blood of this healthy donor (ZGT-1) were isolated using Ficoll gradient centrifugation (Sigma–Aldrich). The obtained cells were cultured in RPMI1640 medium, supplemented with 10% FBS at 37 °C, in a humidified atmosphere containing 5% carbon dioxide.
Electroporation
Cells (1.5 × 105 cells/well) were transfected via electroporation with the Etta X-Porator H1 (Etta Biotech) at a final concentration of 1000 nM siRNA. Manufacturer's protocol was followed, and the electroporation program was optimized in our laboratory as voltage of 150 V, pulse duration 100 μs, pulse number 6, and interval of 1000 ms.
Cell viability
Cells were plated in 96-well plates (12,000 cells/well), and ganetespib was added into the wells to meet the desirable concentrations. Cell viability was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide reagents (KeyGen Biotech). In drug combination experiments, cell lines were pretreated with or without ganetespib for 12 h, before they were treated with everolimus or temsirolimus for 60 h. The cell viabilities were measured at least in three independent experiments.
In vivo analysis
All procedures involving mice were approved by the Institutional Animal Care and Use Committee of Jiangsu University. All Nu/Nu nude mice were provided with sterilized food and water and housed in a barrier facility under a 12-h light/dark cycle. Six-week-old Nu/Nu nude mice were injected subcutaneously in the left flank with 2 × 106 JEKO-1 cells. Tumors were measured every 2 days with vernier calipers, and tumor volumes were calculated using the formula: volume = 0.52 × W2 × L, where L and W represent the length and width of tumors, respectively. When tumor size reached approximately 150 mm3 (25 days after initial transplantation), the mice were randomized into two groups (six mice/group, ganetespib and vehicle groups), and drug treatment was started. Ganetespib (100 mg/kg formulated in 10/18 DRD [10% dimethyl sulfoxide, 18% Cremophor RH 40, 3.6% dextrose, and 68.4% water] or 10/18 DRD [vehicle]) was administered once a week by tail vein injection for 3 weeks. At the end of the observation, mice were sacrificed and autopsied. Tumors were removed and measured. Differences between two groups were compared by the Mann–Whitney test, with p < 0.05 considered significant.
Tissue preparation, histology, and immunohistochemistry
Tumors were paraffin embedded, sectioned (5 μm thick), and deparaffinized as previously described (44). Sections were either H&E stained or subjected to antigen retrieval for immunohistochemistry. Antigen retrieval was conducted by pepsin digestion at 37 °C for 15 to 30 min or by heating slides in EDTA buffer (pH 9.0) for 30 min. Endogenous peroxidase was blocked by preincubation with hydrogen peroxide, and 10% goat serum was used to block the nonspecific antigens. The following primary antibodies at the indicated dilutions was used: anti-Ki-67 antibody (1:100) and anti-cyclin D1 antibody (1:50). Sections were exposed to appropriate species-specific secondary antibodies and exposed to 3,3′-diaminobenzidine (KeyGen Biotech). Then sections were lightly counterstained with hematoxylin (KeyGen Biotech) and coverslipped.
Immunoreactivity was evaluated by two independent researchers blinded to tumor origins. The evaluation was based on the extent of staining and the staining intensity. The staining extent was dependent on the percentage of positive-stained cells and scored as 4 (76–100%), 3 (51–75%), 2 (26–50%), 1 (1–25%), and 0 (0%). The staining intensity was scored as 4 (extremely strong), 3 (strong), 2 (moderate), 1 (weak), and 0 (negative). The sum of the staining extent and intensity was used as the final immunoreactivity score. In the case of a scoring discrepancy, the slides were re-examined by both researchers.
Apoptosis and cell cycle assays
Annexin V-FITC/propidium iodide staining reagent (Yeasen Biotech) was used to evaluate apoptosis. For cell cycle assays, cells were harvested and stained with propidium iodide (Sigma–Aldrich) as previously reported (44). Apoptosis and cell cycle samples were analyzed using a Gallios Flow Cytometer (Beckman Coulter) available in the Analytical and Testing Center at Jiangsu University. Data were analyzed using FlowJo software (FlowJo).
Transcriptome analysis
Total RNA was extracted from cultured cells by using Trizol reagent (Thermo Fisher Scientific) according to manufacturer's protocol. RNA amplification, labeling, hybridization, and microarray imaging were performed by CapitalBio Corporation. A gene was defined as differentially expressed when the difference of mRNA expression was more than 2-fold and the p value was less than 0.05. Differentially expressed genes were analyzed using the gplots package in R language (60, 61). Venn and heat map diagrams were plotted using the Venn and heatmap.2 functions in the gplots package, respectively. For Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis, the differentially expressed genes were analyzed by DAVID Bioinformatics Resources 6.8 (62, 63) and the online Reactome platform (64).
TF prediction
The potential TFs were predicted using an online software, PROMO (65), with the promoter sequences of six candidate genes (−2000 to +500 bp). The species was selected as human, and the maximum matrix dissimilarity rate was set to 5%.
Luciferase reporter assays
The proximal promoter regions (−2000 to +500) of CDC6, MCM4, and RMI2 genes were amplified from the genomic DNA of JEKO-1 cells and inserted into pGL3 Luciferase Reporter Vector (Promega Corporation) as previously described (66). The promoter-driven luciferase reporter (3 μg) was electroporated into JEKO-1 cells, whereas the original vector was used as the control. A β-galactosidase plasmid (0.5 μg) was electroporated into cells with luciferase reporter plasmids. Cell debris was removed using low-speed centrifugation, and surviving cells were treated with either vehicle or 20 nM of ganetespib for 12 h. Luciferase assays were performed following the protocol from the manufacturer, whereas β-galactosidase activities of the cells were used as a control of electroporation efficiency and cell numbers (67).
EdU staining
Cells were incubated with 12.5 μM of EdU for 2 h, transferred to polylysine-treated adhesive slides (Liusheng), and then incubated at 37 °C for 30 min. Thereafter, the EdU staining was performed using the Cell-Light EdU Apollo488 In Vitro Imaging Kit (Ribobio).
CHX-chase assay
Cells were treated with either vehicle or ganetespib for 0, 4, 8, and 12 h in the presence of CHX (5 mg/ml). Treated cells were collected for immunoblot experiments.
Overexpression of E2F1
JEKO-1 cells (5 × 106 cells/well) were transfected via electroporation with 10 μg of pcDNA3.1(+)-E2F1 (synthesized by Genescript) or pcDNA3.1(+) as the control. The electroporation program was optimized in our laboratory for MCL cell lines.
Immunoblot, qRT-PCR, and IF
These experiments were performed as previously described (67, 68, 69). Primes for qRT-PCR are listed in Table S7.
Co-IP experiments
The Co-IP experiments were carried out as described previously (68). Briefly, cells were lysed in the buffer (10 mM Hepes–KOH [pH = 8.0], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol [pH = 7.5], and 0.1% Triton X-100) for 5 min, and centrifuged at 1300g for 5 min at 4 °C. The supernatant was removed, and the nuclei pellet was washed once with PBS, and then lysed using a buffer (50 mM Tris–HCl [pH 8.0], 1 mM EDTA, 0.5% NP-40, and 150 mM NaCl with the proteinase inhibitors) for 30 min at 4 °C. The supernatant was collected by centrifuging the cell lysates at 12,000g for 10 min at 4 °C, and protein concentrations were determined using the Bicinchoninic Acid Protein Assay Kit (Beyotime Biotech). Then, 1 mg of total protein was subjected to IP with 2 μg antibody or an isotype-matched IgG control. The protein was incubated with antibodies for 3 h at 4 °C, and the protein A/G agarose resin was added, then incubated for 1 h at 4 °C. The IP products were collected after being washed for four times with lysis buffer and used in immunoblot.
Statistical analysis
One-way ANOVA followed by Bonferroni post-testing was used to determine statistically significant differences between different groups (∗p < 0.05; ∗∗p < 0.01; and ∗∗∗p < 0.001).
Data availability
All data are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
H. L., E. A. G., and Z. T. conceptualization; Z. L. and X. S. methodology; L. L. and P. Z. validation; H. L. and Z. L. formal analysis; H. L., X. S., and Z. T. investigation; X. S. resources; H. L., Z. L., and P. Z. data curation; H. L., Z. T., and E. A. G. writing-original draft; H. L., Z. L., X. S., L. L., P. Z., E. A. G., and Z. T. writing-review and editing; H. L., E. A. G., and Z. T. supervision; H. L. and E. A. G. project administration; H. L., E. A. G., and Z. T. funding acquisition.
Funding and additional information
The authors and this work were supported by the National Natural Science Foundation of China 81672582 (to H. L.) and 31771521 (to Z. T.); Natural Science Foundation of Jiangsu Province for Distinguished Young Scholars BK20160013 (to H. L.); Natural Science Foundation of Jiangsu Province BE2016718 (to Z. T.); Top Talent of Innovative Research Team of Jiangsu Province (to H. L. and Z. T.); Six Talent Peak Project from Government of Jiangsu Province 2015-SWYY-019 (to H. L.) and 2016-SWYY-011 (to Z. T.); Start-up Scientific Research Fund for the Returned Oversea Scholars from Chinese Ministry of Education (to Z. T.); and P30 CA006927 (to Fox Chase Cancer Center).
Edited by Patrick Sung
Contributor Information
Erica A. Golemis, Email: Erica.Golemis@fccc.edu.
Zhigang Tu, Email: zhigangtu@ujs.edu.cn.
Supporting information
References
- 1.Sharma S., Kelly T.K., Jones P.A. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jares P., Campo E. Advances in the understanding of mantle cell lymphoma. Br. J. Haematol. 2008;142:149–165. doi: 10.1111/j.1365-2141.2008.07124.x. [DOI] [PubMed] [Google Scholar]
- 3.Perez-Galan P., Dreyling M., Wiestner A. Mantle cell lymphoma: Biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood. 2011;117:26–38. doi: 10.1182/blood-2010-04-189977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solier S., Kohn K.W., Scroggins B., Xu W., Trepel J., Neckers L., Pommier Y. Heat shock protein 90alpha (HSP90alpha), a substrate and chaperone of DNA-PK necessary for the apoptotic response. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12866–12872. doi: 10.1073/pnas.1203617109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrarini M., Heltai S., Zocchi M.R., Rugarli C. Unusual expression and localization of heat-shock proteins in human tumor cells. Int. J. Cancer. 1992;51:613–619. doi: 10.1002/ijc.2910510418. [DOI] [PubMed] [Google Scholar]
- 6.Tsutsumi S., Neckers L. Extracellular heat shock protein 90: A role for a molecular chaperone in cell motility and cancer metastasis. Cancer Sci. 2007;98:1536–1539. doi: 10.1111/j.1349-7006.2007.00561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamal A., Thao L., Sensintaffar J., Zhang L., Boehm M.F., Fritz L.C., Burrows F.J. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 8.Cerchietti L.C., Lopes E.C., Yang S.N., Hatzi K., Bunting K.L., Tsikitas L.A., Mallik A., Robles A.I., Walling J., Varticovski L., Shaknovich R., Bhalla K.N., Chiosis G., Melnick A. A purine scaffold Hsp90 inhibitor destabilizes BCL-6 and has specific antitumor activity in BCL-6-dependent B cell lymphomas. Nat. Med. 2009;15:1369–1376. doi: 10.1038/nm.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neckers L., Workman P. Hsp90 molecular chaperone inhibitors: Are we there yet? Clin. Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trepel J., Mollapour M., Giaccone G., Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valbuena J.R., Rassidakis G.Z., Lin P., Atwell C., Georgakis G.V., Younes A., Jones D., Medeiros L.J. Expression of heat-shock protein-90 in non-Hodgkin's lymphomas. Mod. Pathol. 2005;18:1343–1349. doi: 10.1038/modpathol.3800459. [DOI] [PubMed] [Google Scholar]
- 12.Roue G., Perez-Galan P., Mozos A., Lopez-Guerra M., Xargay-Torrent S., Rosich L., Saborit-Villarroya I., Normant E., Campo E., Colomer D. The Hsp90 inhibitor IPI-504 overcomes bortezomib resistance in mantle cell lymphoma in vitro and in vivo by down-regulation of the prosurvival ER chaperone BiP/Grp78. Blood. 2011;117:1270–1279. doi: 10.1182/blood-2010-04-278853. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson C., Kopp N., Layer J.V., Redd R.A., Tschuri S., Haebe S., van Bodegom D., Bird L., Christie A.L., Christodoulou A., Saur A., Tivey T., Zapf S., Bararia D., Zimber-Strobl U. HSP90 inhibition overcomes ibrutinib resistance in mantle cell lymphoma. Blood. 2016;128:2517–2526. doi: 10.1182/blood-2016-04-711176. [DOI] [PubMed] [Google Scholar]
- 14.Liu H., Xiao F., Serebriiskii I.G., O'Brien S.W., Maglaty M.A., Astsaturov I., Litwin S., Martin L.P., Proia D.A., Golemis E.A., Connolly D.C. Network analysis identifies an HSP90-central hub susceptible in ovarian cancer. Clin. Cancer Res. 2013;19:5053–5067. doi: 10.1158/1078-0432.CCR-13-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proia D.A., Bates R.C. Ganetespib and HSP90: Translating preclinical hypotheses into clinical promise. Cancer Res. 2014;74:1294–1300. doi: 10.1158/0008-5472.CAN-13-3263. [DOI] [PubMed] [Google Scholar]
- 16.Jhaveri K., Modi S. Ganetespib: Research and clinical development. Onco Targets Ther. 2015;8:1849–1858. doi: 10.2147/OTT.S65804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ying W., Du Z., Sun L., Foley K.P., Proia D.A., Blackman R.K., Zhou D., Inoue T., Tatsuta N., Sang J., Ye S., Acquaviva J., Ogawa L.S., Wada Y., Barsoum J. Ganetespib, a unique triazolone-containing Hsp90 inhibitor, exhibits potent antitumor activity and a superior safety profile for cancer therapy. Mol. Cancer Ther. 2012;11:475–484. doi: 10.1158/1535-7163.MCT-11-0755. [DOI] [PubMed] [Google Scholar]
- 18.Shimamura T., Perera S.A., Foley K.P., Sang J., Rodig S.J., Inoue T., Chen L., Li D., Carretero J., Li Y.C., Sinha P., Carey C.D., Borgman C.L., Jimenez J.P., Meyerson M. Ganetespib (STA-9090), a nongeldanamycin HSP90 inhibitor, has potent antitumor activity in in vitro and in vivo models of non-small cell lung cancer. Clin. Cancer Res. 2012;18:4973–4985. doi: 10.1158/1078-0432.CCR-11-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hertlein E., Wagner A.J., Jones J., Lin T.S., Maddocks K.J., Towns W.H., 3rd, Goettl V.M., Zhang X., Jarjoura D., Raymond C.A., West D.A., Croce C.M., Byrd J.C., Johnson A.J. 17-DMAG targets the nuclear factor-kappaB family of proteins to induce apoptosis in chronic lymphocytic leukemia: Clinical implications of HSP90 inhibition. Blood. 2010;116:45–53. doi: 10.1182/blood-2010-01-263756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopalakrishnan R., Matta H., Chaudhary P.M. A purine scaffold HSP90 inhibitor BIIB021 has selective activity against KSHV-associated primary effusion lymphoma and blocks vFLIP K13-induced NF-kappaB. Clin. Cancer Res. 2013;19:5016–5026. doi: 10.1158/1078-0432.CCR-12-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki M., Takeda T., Nakagawa H., Iwata S., Watanabe T., Siddiquey M.N., Goshima F., Murata T., Kawada J., Ito Y., Kojima S., Kimura H. The heat shock protein 90 inhibitor BIIB021 suppresses the growth of T and natural killer cell lymphomas. Front. Microbiol. 2015;6:280. doi: 10.3389/fmicb.2015.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin L., Tabe Y., Kimura S., Zhou Y., Kuroda J., Asou H., Inaba T., Konopleva M., Andreeff M., Miida T. Antiproliferative and proapoptotic activity of GUT-70 mediated through potent inhibition of Hsp90 in mantle cell lymphoma. Br. J. Cancer. 2011;104:91–100. doi: 10.1038/sj.bjc.6606007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding H., Peterson K.L., Correia C., Koh B., Schneider P.A., Nowakowski G.S., Kaufmann S.H. Histone deacetylase inhibitors interrupt HSP90∗RASGRP1 and HSP90∗CRAF interactions to upregulate BIM and circumvent drug resistance in lymphoma cells. Leukemia. 2017;31:1593–1602. doi: 10.1038/leu.2016.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z., Graf N., Herrmann K., Junger A., Aichler M., Feuchtinger A., Baumgart A., Walch A., Peschel C., Schwaiger M., Buck A., Keller U., Dechow T. FLT-PET is superior to FDG-PET for very early response prediction in NPM-ALK-positive lymphoma treated with targeted therapy. Cancer Res. 2012;72:5014–5024. doi: 10.1158/0008-5472.CAN-12-0635. [DOI] [PubMed] [Google Scholar]
- 25.Sun M., Feng X., Liu Z., Han W., Liang Y.X., She Q. An Orc1/Cdc6 ortholog functions as a key regulator in the DNA damage response in Archaea. Nucleic Acids Res. 2018;46:6697–6711. doi: 10.1093/nar/gky487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheu Y.J., Kinney J.B., Stillman B. Concerted activities of Mcm4, Sld3, and Dbf4 in control of origin activation and DNA replication fork progression. Genome Res. 2016;26:315–330. doi: 10.1101/gr.195248.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hudson D.F., Amor D.J., Boys A., Butler K., Williams L., Zhang T., Kalitsis P. Loss of RMI2 increases genome instability and causes a Bloom-Like syndrome. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farre D., Roset R., Huerta M., Adsuara J.E., Rosello L., Alba M.M., Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amin H.M., McDonnell T.J., Medeiros L.J., Rassidakis G.Z., Leventaki V., O'Connor S.L., Keating M.J., Lai R. Characterization of 4 mantle cell lymphoma cell lines. Arch. Pathol. Lab. Med. 2003;127:424–431. doi: 10.5858/2003-127-0424-COMCLC. [DOI] [PubMed] [Google Scholar]
- 30.Drakos E., Atsaves V., Li J., Leventaki V., Andreeff M., Medeiros L.J., Rassidakis G.Z. Stabilization and activation of p53 downregulates mTOR signaling through AMPK in mantle cell lymphoma. Leukemia. 2009;23:784–790. doi: 10.1038/leu.2008.348. [DOI] [PubMed] [Google Scholar]
- 31.Dancey J. mTOR signaling and drug development in cancer. Nat. Rev. Clin. Oncol. 2010;7:209–219. doi: 10.1038/nrclinonc.2010.21. [DOI] [PubMed] [Google Scholar]
- 32.Li J., Kim S.G., Blenis J. Rapamycin: One drug, many effects. Cell Metab. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parekh S., Weniger M.A., Wiestner A. New molecular targets in mantle cell lymphoma. Semin. Cancer Biol. 2011;21:335–346. doi: 10.1016/j.semcancer.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang M., Popplewell L.L., Collins R.H., Jr., Winter J.N., Goy A., Kaminski M.S., Bartlett N.L., Johnston P.B., Lister J., Fanning S.R., Tuscano J.M., Beck J.T., Kaya H., Robeva A., Fan J. Everolimus for patients with mantle cell lymphoma refractory to or intolerant of bortezomib: Multicentre, single-arm, phase 2 study. Br. J. Haematol. 2014;165:510–518. doi: 10.1111/bjh.12780. [DOI] [PubMed] [Google Scholar]
- 35.Witzig T.E., Reeder C.B., LaPlant B.R., Gupta M., Johnston P.B., Micallef I.N., Porrata L.F., Ansell S.M., Colgan J.P., Jacobsen E.D., Ghobrial I.M., Habermann T.M. A phase II trial of the oral mTOR inhibitor everolimus in relapsed aggressive lymphoma. Leukemia. 2011;25:341–347. doi: 10.1038/leu.2010.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Leary B., Finn R.S., Turner N.C. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 2016;13:417–430. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 37.Divakar S.K., Ramana Reddy M.V., Cosenza S.C., Baker S.J., Perumal D., Antonelli A.C., Brody J., Akula B., Parekh S., Reddy E.P. Dual inhibition of CDK4/Rb and PI3K/AKT/mTOR pathways by ON123300 induces synthetic lethality in mantle cell lymphomas. Leukemia. 2016;30:86–93. doi: 10.1038/leu.2015.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giulino-Roth L., van Besien H.J., Dalton T., Totonchy J.E., Rodina A., Taldone T., Bolaender A., Erdjument-Bromage H., Sadek J., Chadburn A., Barth M.J., Dela Cruz F.S., Rainey A., Kung A.L., Chiosis G. Inhibition of Hsp90 suppresses PI3K/AKT/mTOR signaling and has antitumor activity in Burkitt lymphoma. Mol. Cancer Ther. 2017;16:1779–1790. doi: 10.1158/1535-7163.MCT-16-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braun K.A., Breeden L.L. Nascent transcription of MCM2-7 is important for nuclear localization of the minichromosome maintenance complex in G1. Mol. Biol. Cel. 2007;18:1447–1456. doi: 10.1091/mbc.E06-09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shima N., Alcaraz A., Liachko I., Buske T.R., Andrews C.A., Munroe R.J., Hartford S.A., Tye B.K., Schimenti J.C. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- 41.Alvarez S., Diaz M., Flach J., Rodriguez-Acebes S., Lopez-Contreras A.J., Martinez D., Canamero M., Fernandez-Capetillo O., Isern J., Passegue E., Mendez J. Replication stress caused by low MCM expression limits fetal erythropoiesis and hematopoietic stem cell functionality. Nat. Commun. 2015;6:8548. doi: 10.1038/ncomms9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh T.R., Ali A.M., Busygina V., Raynard S., Fan Q., Du C.H., Andreassen P.R., Sung P., Meetei A.R. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoadley K.A., Xue Y., Ling C., Takata M., Wang W., Keck J.L. Defining the molecular interface that connects the Fanconi anemia protein FANCM to the Bloom syndrome dissolvasome. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4437–4442. doi: 10.1073/pnas.1117279109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H., Lu J., Hua Y., Zhang P., Liang Z., Ruan L., Lian C., Shi H., Chen K., Tu Z. Targeting heat-shock protein 90 with ganetespib for molecularly targeted therapy of gastric cancer. Cell Death Dis. 2015;6:e1595. doi: 10.1038/cddis.2014.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guan L., Zou Q., Liu Q., Lin Y., Chen S. HSP90 inhibitor ganetespib (STA-9090) inhibits tumor growth in c-Myc-dependent esophageal squamous cell carcinoma. Onco Targets Ther. 2020;13:2997–3011. doi: 10.2147/OTT.S245813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin S.F., Lin J.D., Hsueh C., Chou T.C., Yeh C.N., Chen M.H., Wong R.J. Efficacy of an HSP90 inhibitor, ganetespib, in preclinical thyroid cancer models. Oncotarget. 2017;8:41294–41304. doi: 10.18632/oncotarget.17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Proia D.A., Foley K.P., Korbut T., Sang J., Smith D., Bates R.C., Liu Y., Rosenberg A.F., Zhou D., Koya K., Barsoum J., Blackman R.K. Multifaceted intervention by the Hsp90 inhibitor ganetespib (STA-9090) in cancer cells with activated JAK/STAT signaling. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gabbasov R., Benrubi I.D., O'Brien S.W., Krais J.J., Johnson N., Litwin S., Connolly D.C. Targeted blockade of HSP90 impairs DNA-damage response proteins and increases the sensitivity of ovarian carcinoma cells to PARP inhibition. Cancer Biol. Ther. 2019;20:1035–1045. doi: 10.1080/15384047.2019.1595279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He S., Smith D.L., Sequeira M., Sang J., Bates R.C., Proia D.A. The HSP90 inhibitor ganetespib has chemosensitizer and radiosensitizer activity in colorectal cancer. Invest. New Drugs. 2014;32:577–586. doi: 10.1007/s10637-014-0095-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohtani K., Iwanaga R., Nakamura M., Ikeda M., Yabuta N., Tsuruga H., Nojima H. Cell growth-regulated expression of mammalian MCM5 and MCM6 genes mediated by the transcription factor E2F. Oncogene. 1999;18:2299–2309. doi: 10.1038/sj.onc.1202544. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida K., Inoue I. Regulation of Geminin and Cdt1 expression by E2F transcription factors. Oncogene. 2004;23:3802–3812. doi: 10.1038/sj.onc.1207488. [DOI] [PubMed] [Google Scholar]
- 52.Hayashi R., Goto Y., Haga A., Kobayashi D., Ikeda R., Yoshida K. Comparative genomics on MCM8 orthologous genes reveals the transcriptional regulation by transcription factor E2F. Gene. 2006;367:126–134. doi: 10.1016/j.gene.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Yan Z., DeGregori J., Shohet R., Leone G., Stillman B., Nevins J.R., Williams R.S. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiarle R., Budel L.M., Skolnik J., Frizzera G., Chilosi M., Corato A., Pizzolo G., Magidson J., Montagnoli A., Pagano M., Maes B., De Wolf-Peeters C., Inghirami G. Increased proteasome degradation of cyclin-dependent kinase inhibitor p27 is associated with a decreased overall survival in mantle cell lymphoma. Blood. 2000;95:619–626. [PubMed] [Google Scholar]
- 55.Korz C., Pscherer A., Benner A., Mertens D., Schaffner C., Leupolt E., Dohner H., Stilgenbauer S., Lichter P. Evidence for distinct pathomechanisms in B-cell chronic lymphocytic leukemia and mantle cell lymphoma by quantitative expression analysis of cell cycle and apoptosis-associated genes. Blood. 2002;99:4554–4561. doi: 10.1182/blood.v99.12.4554. [DOI] [PubMed] [Google Scholar]
- 56.Kotwal A., Suran S., Amere Subbarao S. Hsp90 chaperone facilitates E2F1/2-dependent gene transcription in human breast cancer cells. Eur. J. Cell Biol. 2021;100:151148. doi: 10.1016/j.ejcb.2020.151148. [DOI] [PubMed] [Google Scholar]
- 57.Marzec M., Kasprzycka M., Lai R., Gladden A.B., Wlodarski P., Tomczak E., Nowell P., Deprimo S.E., Sadis S., Eck S., Schuster S.J., Diehl J.A., Wasik M.A. Mantle cell lymphoma cells express predominantly cyclin D1a isoform and are highly sensitive to selective inhibition of CDK4 kinase activity. Blood. 2006;108:1744–1750. doi: 10.1182/blood-2006-04-016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pillai R.N., Fennell D.A., Kovcin V., Ciuleanu T.E., Ramlau R., Kowalski D., Schenker M., Yalcin I., Teofilovici F., Vukovic V.M., Ramalingam S.S. Randomized phase III study of ganetespib, a heat shock protein 90 inhibitor, with docetaxel versus docetaxel in advanced non-small-cell lung cancer (GALAXY-2) J. Clin. Oncol. 2020;38:613–622. doi: 10.1200/JCO.19.00816. [DOI] [PubMed] [Google Scholar]
- 59.Nagaraju G.P., Park W., Wen J., Mahaseth H., Landry J., Farris A.B., Willingham F., Sullivan P.S., Proia D.A., El-Hariry I., Taliaferro-Smith L., Diaz R., El-Rayes B.F. Antiangiogenic effects of ganetespib in colorectal cancer mediated through inhibition of HIF-1alpha and STAT-3. Angiogenesis. 2013;16:903–917. doi: 10.1007/s10456-013-9364-7. [DOI] [PubMed] [Google Scholar]
- 60.Smid M., Rodriguez-Gonzalez F.G., Sieuwerts A.M., Salgado R., Prager-Van der Smissen W.J., Vlugt-Daane M.V., van Galen A., Nik-Zainal S., Staaf J., Brinkman A.B., van de Vijver M.J., Richardson A.L., Fatima A., Berentsen K., Butler A. Breast cancer genome and transcriptome integration implicates specific mutational signatures with immune cell infiltration. Nat. Commun. 2016;7:12910. doi: 10.1038/ncomms12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chung W., Eum H.H., Lee H.O., Lee K.M., Lee H.B., Kim K.T., Ryu H.S., Kim S., Lee J.E., Park Y.H., Kan Z., Han W., Park W.Y. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 63.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., Milacic M., Roca C.D., Rothfels K., Sevilla C., Shamovsky V. The reactome pathway Knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei C.L., Lin Y.C., Chen T.A., Lin R.Y., Liu T.H. Respiration detection chip with integrated temperature-insensitive MEMS sensors and CMOS signal processing circuits. IEEE Trans. Biomed. Circuits Syst. 2015;9:105–112. doi: 10.1109/TBCAS.2014.2315532. [DOI] [PubMed] [Google Scholar]
- 66.Dougherty D.C., Sanders M.M. Comparison of the responsiveness of the pGL3 and pGL4 luciferase reporter vectors to steroid hormones. BioTechniques. 2005;39:203–207. doi: 10.2144/05392ST02. [DOI] [PubMed] [Google Scholar]
- 67.Tu Z., Aird K.M., Bitler B.G., Nicodemus J.P., Beeharry N., Xia B., Yen T.J., Zhang R. Oncogenic RAS regulates BRIP1 expression to induce dissociation of BRCA1 from chromatin, inhibit DNA repair, and promote senescence. Dev. Cell. 2011;21:1077–1091. doi: 10.1016/j.devcel.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tu Z., Zhuang X., Yao Y.G., Zhang R. BRG1 is required for formation of senescence-associated heterochromatin foci induced by oncogenic RAS or BRCA1 loss. Mol. Cell Biol. 2013;33:1819–1829. doi: 10.1128/MCB.01744-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shang D., Wu Y., Ding Y., Lu Z., Shen Y., Zhu F., Liu H., Zhu C., Tu Z. Identification of a pyridine derivative inducing senescence in ovarian cancer cell lines via P21 activation. Clin. Exp. Pharmacol. Physiol. 2018;45:452–460. doi: 10.1111/1440-1681.12891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.