Highlights
-
•
Global RNA editing levels or ADAR expression do not correlate with response to immunotherapy in melanoma patients.
-
•
RNA editing signatures within genes can segregate patients that will respond to immunotherapy across patient cohorts.
-
•
Recurrent RNA editing sites, those that are shared between melanoma patients, provide accurate prognostic predictions.
Abstract
Immunotherapy has improved the prognosis for many melanoma patients; however, our capacity to predict patient responses and to understand the biological differences between patients who will or will not respond is limited. Gene expression profiling of tumors from patients who respond to immunotherapy has focused on deriving primarily immune-related signatures; however, these have shown limited predictive power. Recent studies have highlighted the role of RNA editing in modulating resistance to immunotherapy. To evaluate the utility of RNA editing activity as a discriminative tool in predicting immunotherapy response, we conducted a retrospective analysis of RNA-sequencing data from melanoma patients treated with Pembrolizumab or Nivolumab. Here, we developed RNA editing signatures that can identify patients who will respond to immunotherapy with very high accuracy and confidence. Our analysis demonstrates that RNA editing is a strong discriminative tool for examining sensitivity of melanoma patients to immunotherapy.
Introduction
Melanoma is a highly aggressive and frequently lethal cancer that develops from the oncogenic transformation of melanocytes. Each year, over 300,000 people are diagnosed with melanoma [1], and this number continues to increase. The advent of immunotherapy as a frontline therapy has significantly improved the prognosis for a significant proportion of melanoma patients [2,3]. However, our understanding of which patients will or will not respond to these treatments is limited and represents a significant clinical hurdle [2–5]. A number of groups have profiled the tumors of melanoma patients on immunotherapy in clinical trials by utilizing RNA-sequencing (RNA-Seq) in an attempt to identify gene expression signatures associated with patient response [2,[3], [4],5]. Each of these studies have provided important biological insights into the genomic and transcriptional changes that drive melanoma; however, the discriminative power of these signatures is limited. These data are a rich resource for investigating potential biomarkers in patients.
Recent studies in a number of cancer types have identified RNA editing as an important regulator of the interferon response of cells, which is critical for effective immune killing. Much of this research has focused on the role of the adenosine deaminase acting on RNA (ADAR) family of proteins. These proteins have established functions in modulating RNA editing events that enable cells to distinguish between endogenous RNA structures and viral RNAs. In this role, the ADAR proteins are important for the immune response and T-cell activation [[6], [7], [8], [9]–10]. The ADAR protein family (ADAR1-3 (ADAR3: is enzymatically inactive)) catalyzes the deamination of Adenines (A) within double-stranded regions of RNA (dsRNA) into Inosines (I), in a process known as A to I editing [11]. The resulting I-U base-pair is significantly less stable than the replaced A-U interaction, resulting in destabilization of dsRNA regions [11]. These modifications have central roles in cellular homeostasis, as unedited dsRNA regions are recognized by human cells as viral contaminants and trigger strong immune responses [8,11]. Importantly, for our study, the Inosine (I) base is recognized as a Guanine (G) during RNA-Seq library construction. This enables the identification of A-I editing sites from RNA-Seq data by using specialized computational tools [[12], [13]–14].
Based on the real clinical need to identify patients who may respond to immunotherapy and to establish a strong link between RNA editing and the immune response, we tested the utility of RNA editing as a predictive biomarker of response by mining publicly available RNA-Seq datasets [2,3,12].
Methods
Datasets
The primarily Pembrolizumab-treated cohort (GSE78220) consisted of 15 responders and 13 nonresponders with response assignments based on Immune-related Response Evaluation Criteria In Solid Tumors (irRECIST) criteria[2]. The Nivolumab-treated cohort (GSE91061) consisted of 23 nonresponders, 10 responders, and 16 stable disease by RECIST criteria [3] with an additional 2 patients being of unknown response designation [3]. In addition to the immunotherapy-treated cohorts, we sampled 50 skin cutaneous melanoma from The Cancer Genome Atlas (TCGA-SKCM cohort) [15,16].
Bioinformatics and statistical analysis
RNA editing sites pipeline: RNA-Seq files for the Pembrolizumab and Nivolumab cohorts were imported from Gene Expression Omnibus (GSE78220 and GSE91061) by using SRAToolkit 2.9.0. TrimGalore 0.6.0 was used to perform adaptor and quality trimming of RNA-Seq reads with the additional specification of 6 bases removed from the 5’-end of each read [17]. Bowtie2 was used to remove contaminating rRNA and tRNA reads [18]. STAR 2.5.2a was used to align the reads to the GRCh38 p12 genome release 31 and for obtaining gene counts and BAM alignment files [19,20]. The gene counts were imported into the R environment using DESeq2, a Bioconductor package for differential expression analysis, and the resulting data were log-regularized [21]. Additionally, a differential expression analysis was conducted between responders and nonresponders.
SNP-free RNA editing IdeNtification Toolkit (Sprint) was used on the resulting BAM alignment files to identify RNA editing sites (RES) de novo [12]. The changesammapq.py script from Sprint was used to convert the BAM files to the correct format for Sprint [12]. The sprint_from_bam.py script was used to identify regular RES sites from the resulting BAM file using the GRCh8 p12 genome release 31 [20] and Sprint-provided hg38 repeat annotations. Annotating of the resulting RES positions for genes, genomic regions, and repeats was done using functions and hg38 annotations from annotatr, a Bioconductor package for investigating intersecting genomic annotations [22]. Sprint-provided hg38 repeat annotations were also used in annotating the RES sites.
For the TCGA-SKCM dataset, BAM files were downloaded through the GDC Data Transfer Tool Client. Samtools bam2fq was used to convert the BAM files to FASTQ files [23]. The FASTQ files were processed through the main Sprint pipeline to identify RNA editing sites [12]. The resulting RNA editing sites were annotated and processed in a similar manner to the Pembrolizumab and Nivolumab cohorts.
Identifying differential and response specific RNA editing: For each gene, we developed an RNA editing score defined as the number of RNA editing sites per gene [24]. For our RES scores, we only focused on A to G or T to C transitions for a gene. The RES scores were log2-transformed with a pseudo-count of 1 to normalize the data. A two-tailed t-test was performed on genes that contained non-zero RES scores for >20% samples to determine differential RNA editing scores for each gene between responding and nonresponding patients with significance criteria being two-tailed p-value < 0.05 and log2-fold change > 0.3785. Additionally, we overlapped the significant genes with DESeq2 results and removed genes that were differentially expressed at the nominal level of significance (Wald's Test two-tailed p-values < 0.05).
In addition to differential editing, we also identified responder-specific and nonresponder-specific genes for each cohort that had RNA editing within one response group and not in the other. We overlapped results from both cohorts to identify common responder and non-responder specific genes. Additionally, we overlapped differential editing and response specific results to develop a common RNA editing signature. A functional enrichment analysis using ToppFun from the ToppGene suite [25] was conducted on genes from the common editing gene signature with significant annotations having FDR-adjusted p-value < 0.20.
RNA editing signatures: The mean RES score of the signature upregulated and downregulated genes was used as a measure of the RNA editing signatures for each patient sample. Two-tailed pairwise t-tests were conducted between responders and nonresponders to determine how significantly each signature discriminated between response groups. Additionally, a t-test was also conducted on the corresponding log-regularized transcript levels to determine how the transcript levels of the signature genes significantly discriminated between patient response groups. The mean RES and transcript signature was calculated for the upregulated and downregulated genes signature for each cohort as well as the common RNA editing signature.
Logistic regression modeling: The means of the significant log2-tranformed RNA editing scores from cohort-specific upregulated and downregulated genes as well as the common editing signature were used as input to a set of logistic regression models using the glm functions in R where response ∼ mean RNA editing score. Two-tailed p-values of the mean RNA editing score coefficient were used to assess significance and model accuracy was calculated for correctly classified patient samples. The pROC R package was used for receiver operating characteristic (ROC) analysis [26].
Recurrent RNA editing sites: AG/TC RNA editing sites from all samples in each cohort were stratified into sites identified in only responders, sites identified in only nonresponders, and those identified in both responders and nonresponders. A two-tailed Fisher's Exact test was performed to determine the contingency of RNA editing sites being enriched in responders or nonresponders. Significant recurrent RNA editing sites had a two-tailed p-value < 0.05 and were only annotated to genes with DESeq2 nominal Wald's Test p-values > 0.05. Significant sites whose contingency odds ratio favored responders were responder-enriched and significant sites whose odds ratios favored non-responders were non-responder enriched. For the responder and nonresponder enriched RES, we determined the annotated genes for these sites using functions derived from annotatr [22]. The responder and nonresponder enriched RES and their annotated genes were compared across cohorts.
Survival analysis: Survival analyses were performed to determine the effects of RNA editing signatures and recurrent RNA editing sites on patient survival. Analyses were performed on all patient samples within a cohort. The survival and survminer R packages were used [27]. A survival object or response variable was created from survival data using the Surv function. Kaplan-Meier curves were created for visualization using the survfit function, and Cox proportional hazards regression modeling was performed via the coxph function [28]. The ggsurvplot function was used for visualizing survival curves.
Results
RNA editing or ADAR levels do not predict patient sensitivity to immunotherapy
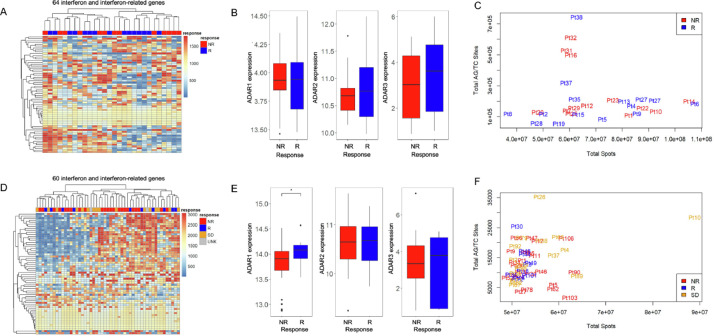
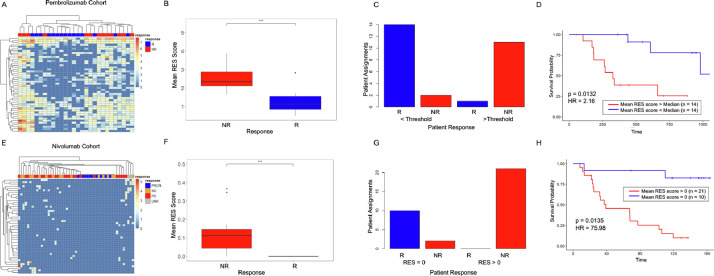
By using published data from patients treated with Pembrolizumab and Nivolumab [2], (PEM cohort), we tested whether the levels of interferon response or ADAR genes correlated with patient response to immunotherapy. These results showed no association between interferon (Fig. 1A) and ADAR gene expression (Fig. 1B) and patient outcome (Non-Responder (NR) vs. Responder (R)). We next investigated ADAR-mediated RNA editing levels in each tumor using A-I calling software, Sprint [12]. This analysis revealed that the overall levels of RNA A-I editing events, as measured by AG/TC transitions in RNA-Seq data, do not correlate with patient response (Fig. 1C, Supp Table 1). To evaluate whether these results were also present in additional cohorts, we conducted the same analysis on independent data from Nivolumab (NIV cohort) treated melanoma patients. In agreement with our initial findings, an interferon response signature (Fig. 1D), ADAR levels (Fig. 1E), and gross A-I editing sites (Fig. 1F, Supp Table 2) were not discriminative.
Fig. 1.
Interferon-related genes and global RNA editing are unable to segregate patients by response. (A) Heat map of interferon and interferon-related gene expression changes in nonresponder (NR) and responder (R) patients treated with Pembrolizumab. (B) ADAR1, ADAR2, and ADAR3 gene expression levels in nonresponder and responder patients from patients treated with Pembrolizumab. (C) Total AG/TC RNA editing sites vs. total spots in nonresponder (red) and responder (blue) patients. (D) Heat map of interferon and interferon-related gene expression changes in nonresponder (NR), responder (R), stable disease (SB), and unknown (UNK) patients treated with Nivolumab. (E) ADAR1, ADAR2 and ADAR3 gene expression levels in nonresponder and responder patients treated with Nivolumab. (F) Total AG/TC RNA editing sites vs. total spots in nonresponder (red), responder (blue), and stable disease (orange) patients (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Table 1.
Common RES sites.
| Chromosome | Start | End | Strand | Transition | Genes | Regions |
|---|---|---|---|---|---|---|
| chr1 | 155733924 | 155733925 | + | AG | DAP3 | Intron |
| chr2 | 151477764 | 151477765 | + | AG | RIF1 | 3UTR |
| chr2 | 24002029 | 24002030 | + | AG | UBXN2A | 3UTR |
| chr22 | 42383507 | 42383508 | - | TC | NFAM1 | 3UTR |
| chr7 | 92200451 | 92200452 | - | TC | KRIT1 | 3UTR |
RNA editing signatures can retrospectively identify patients who respond to immunotherapy
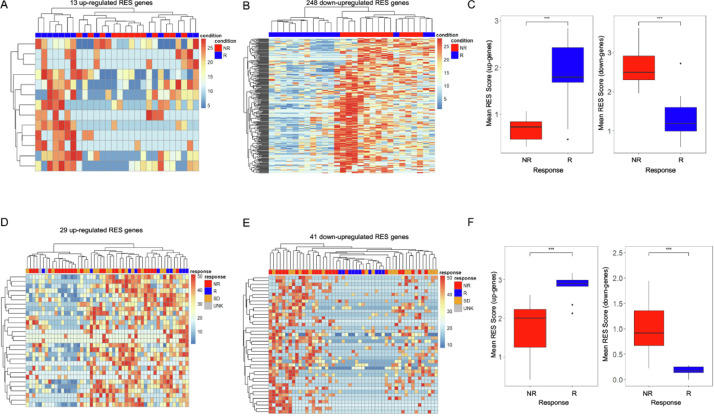
We next investigated whether RNA editing sites (RES) in independent genes, rather than global levels, were associated with patient response. Our analysis of RES levels identified significant variations in the number of RES sites per tumor and cohort (Supp. Fig. 1) primarily due to differences in sequencing coverage, with only a fraction of A-I editing sites being identified in most patients (Supp. Figs. 2, 3). As a result, we focused on editing events within each gene rather than those at a particular site. For this, we tabulated the number of RES identified within each gene to generate a gene-specific RES score [24]. By using this approach, we identified genes with RES scores that were significantly associated with response (t-test p < 0.05) and were not differentially expressed (Wald p > 0.05) [21,29]. By excluding genes with altered gene expression, we focused on RES changes that were due specifically to alterations in A-I editing levels. From this analysis, we identified 13 upregulated (Figs. 2A, Supp Fig. 4A) and 248 downregulated RES scores (Figs. 2B, Supp 4B) that correlated with patient response to Pembrolizumab (Supp Tables 3, 4). This approach enabled immunotherapy response to be identified independent of driver mutation, as equal levels of BRAF and RAS mutations were found in both responders and nonresponders (Supp Fig. 4C, D). Downregulated RES events provided the cleanest clustering of patients based on outcome (Fig. 2B). To evaluate the heterogeneity between patients, we compared the means of the signature RES scores and found a striking and statistically significant difference between nonresponding vs responding patients (Fig. 2C). These findings suggest that RES scores may provide the basis for more accurately discriminating patients by their response to immunotherapy.
Fig. 2.
RES scores can be used across datasets to sub-classify immunotherapy response. (A) Heat map of 13 upregulated RES scores in genes in nonresponder (NR) and responder (R) Pembrolizumab-treated patients. (B) Heat map of 248 downregulated RES scores in genes in non-responder (NR) and responder (R) Pembrolizumab-treated patients. (C) Means of RES scores of up-regulated and downregulated genes from nonresponder (NR) and responder (R) Pembrolizumab-treated patients. (D) Heat map of 29 upregulated RES scores in genes in nonresponder, responder, stable disease, and unknown Nivolumab-treated patients. (E) Heat map of 41 downregulated RES scores in genes in nonresponder (NR), responder (R), stable disease, (SB) and unknown (UNK) Nivolumab-treated patients. (F) Means of RES scores of upregulated and downregulated genes from nonresponder (NR) and responder (R) Nivolumab-treated patients. ***p < 0.001.
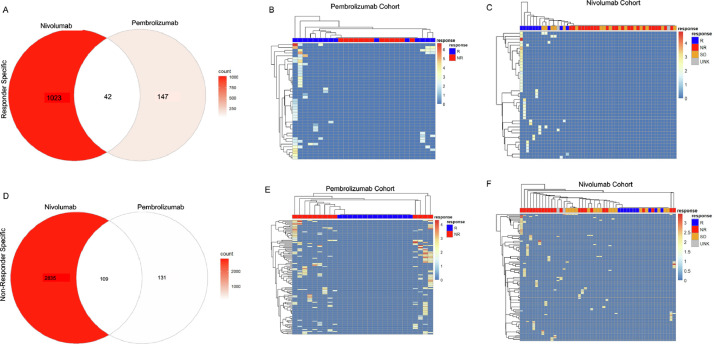
Fig. 4.
Genes can be specific for responding or nonresponding patients. (A) Venn diagram of Responder-specific genes from Pembrolizumab- and Nivolumab-treated patients. (B) Heatmap of common Responder-specific genes from Pembrolizumab-treated patients. (C) Heatmap of common Responder-specific genes from Nivolumab-treated patients. (D) Venn diagram of Nonresponder-specific genes from Pembrolizumab- and Nivolumab-treated patients. (E) Heatmap of common Nonresponder-specific genes from Pembrolizumab-treated patients. (F) Heatmap of common Nonresponder-specific genes from Nivolumab-treated patients.
The utility of the RES score to segregate patients by response in our PEM dataset compelled us to investigate whether RNA editing signatures could stratify patients from an additional patient cohort. For this, we used a dataset that included RNA-Seq for melanoma patients treated with Nivolumab from the following groups: responder (R), nonresponder (NR), and stable disease (SD) patients [3]. By using our RES criteria, we identified 29 upregulated and 41 downregulated gene RES scores that correlated with patient sensitivity to immunotherapy in this independent NIV melanoma cohort and is not driver mutation dependent (Figs. 2D, E, Supp 4E–H, Supp Table 5). We again found that downregulated RES sites enable the best clustering of patients who respond to NIV therapy (Fig. 2E). The mean RES scores across these patient groups also showed strong separation in responding patients (Fig. 2F, Supp Table 6). Importantly, patients with stable disease primarily clustered with RES scores of nonresponders (Fig. 2D, E) and show similar RES means.
Logistic regression modeling of RES score correlates with response and patient outcome
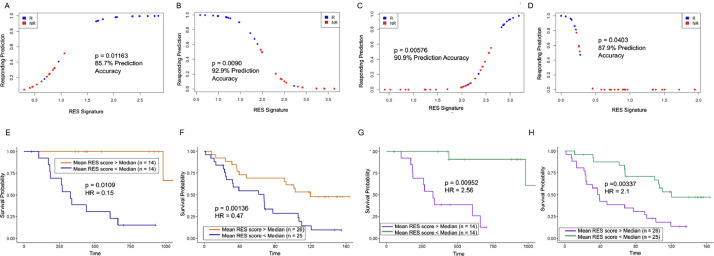
Because our RES scores correlate well with patient response to immunotherapy, we next investigated the discriminative potential of this pipeline for patients. For this, we used logistic regression models from upregulated and downregulated mean RES scores from each cohort. In the PEM-treated patients, we found that upregulated RES scores had a ∼85% capacity to retrospectively predict patient response to immunotherapy (Figs. 3A, S5A). In accordance, the downregulated RES scores have a ∼93% predictive capacity and clearly segregate responding patients (Figs. 3B, S5B). The sole responding patient that our model does not segregate (blue dot below 0.2 in Fig. 3B) is patient 38 (Pt38), who had very high AG/TC levels and is an outlier in Fig. 1C. In the independent NIV-treated cohort, our logistic regression model of RES scores accurately segregates patients based on response. Both the upregulated (91%) (Figs. 3C, Supp 5C) and downregulated (88%) RES genes have a very strong predictive capacity (Figs. 3D, Supp 5D). Based on these analyses, we find that the downregulated RES scores represent the most accurate model for determining patient sensitivity to either treatment. These results highlight the discriminative power of this approach for identifying patients who are likely to respond to immunotherapy.
Fig. 3.
RES scores accurately predict response and survival of melanoma patients to immunotherapy. (A) Logistic regression models from Pembrolizumab-treated patients for 13 upregulated RES score means and responding prediction for responder (R, blue) and non-responder (NR, red) patients. (B) Logistic regression models from Pembrolizumab-treated patients for 248 downregulated RES score means and responding prediction for responder (R, blue) and nonresponder (NR, red) patients. (C) Logistic regression models from Nivolumab-treated patients for 29 upregulated RES score means and responding prediction for responder (R, blue) and non-responder (NR, red) patients. (D) Logistic regression models from Nivolumab-treated patients for 41 downregulated RES score means and responding prediction for responder (R, blue) and non-responder (NR, red) patients. (E) Survival analysis of patients stratified by upper 50% (red) and lower 50% (blue) means of upregulated RES scores for genes in the Pembrolizumab cohort. (F) Survival analysis of patients stratified by upper 50% (red) and lower 50% (blue) means of upregulated RES scores for genes in the Nivolumab cohort. (G) Survival analysis of patients stratified by upper 50% (purple) and lower 50% (green) means of downregulated RES scores for genes in the Pembrolizumab cohort. (H) Survival analysis of patients stratified by upper 50% (purple) and lower 50% (green) means of downregulated RES scores for genes in the Nivolumab cohort (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Fig. 5.
Common 46 downregulated signature genes segregate patients by response and survival across 2 cohorts. (A) Heat map of signature genes from Pembrolizumab-treated patients. (B) Means of RES scores from nonresponder (NR) and responder (R) Pembrolizumab-treated patients. (C) Assignments of Pembrolizumab-treated patients as nonresponder (NR) and responder (R) by predictions from signature genes. (D) Survival analysis of Pembrolizumab-treated patients with mean signature RES scores above and below the median. (E) Heat map of signature genes from Nivolumab-treated patients. (F) Means of RES scores from nonresponder (NR) and responder (R) Nivolumab-treated patients. (G) Assignments of patients from Nivolumab-treated patients as nonresponder (NR) and responder (R) by predictions from signature genes. (H) Survival analysis of Nivolumab-treated patients with mean signature RES scores above and below the median. ***p < 0.001.
We next evaluated whether these RES scores correlated with patient survival. Cox proportional hazards modeling was used for this analysis as it enables assessment of how continuous mean RES scores influenced patient survival. Kaplan-Meier curves, which dichotomized mean RES scores by their medians, were used for visualization. For patients with elevated RES scores in the upregulated group, we found improved survival periods for patients from the PEM cohort (Fig. 3E). In agreement, patients with elevated RES score means within the NIV dataset also showed significant survival differences to those with lower means (Fig. 3F). Although each of these models strongly support the discriminative power of our assay, the downregulated RES score means show the most significant differences in patient survival (Fig. 3G–H). To ensure that there were no cohort-specific differences in survival, we performed a survival analysis between all patients in PEM and NIV cohorts and found that they had comparable survival (Supp Fig. 5E). To determine whether RES scores could also identify patient sensitivity to other treatments, we analyzed melanoma TCGA data and found that RES score does not correlate with patient survival (Supp Fig. 5F). These data show that RES scores can be used to identify patients who may have improved survival and benefit from immunotherapy treatments.
In summary, based on the logistic regression model and survival analysis of these patients, the downregulated RES score enables the most accurate sub-classification of melanoma patients across cohorts. These data highlight the discriminative nature of RES scores for understanding the clinical benefit for patients on immunotherapy.
RNA editing can be used to segregate patients
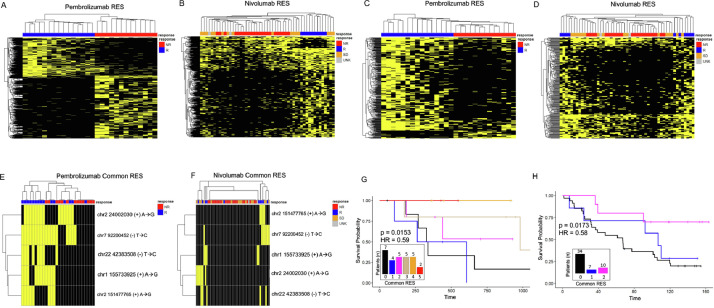
Based on our results, the downregulated RNA editing levels show promise as a resource for determining sensitivity to immunotherapy. While the downregulated RES genes are highly predictive in each cohort, the overlap between cohorts was limited. Many of these effects are due to differences in RNA sequencing coverage between tumors and cohorts. To determine whether RNA A-I editing events could cluster in patients based on their sensitivity to immunotherapy, we identified all RES gene sites that were only present in responders or nonresponders. From this approach, we identified 1023 and 147 responder specific commonly annotated RES modified genes. Of these shared gene sites, 42 were found in both cohorts (Fig. 4A). We then evaluated whether these genes could subclassify patients, based on their sensitivity to immunotherapy. As shown in Fig. 4B, and C, responder specific signatures accurately divide patients. We next conducted this analysis on nonresponder specific genes and identified 109 shared genes (Fig. 4D). Utilizing these sites, the RES genes identified in non-responders can also segregate patients (Fig. 4E, F). This unfiltered approach highlights the utility and accuracy of this analysis for separating patients according to their sensitivity to immunotherapy.
A shared gene editing signature can sub-classify patients
By cross-referencing gene lists, we found that 51 nonresponder specific genes in the NIV dataset were also found in the 248 down-regulated genes in PEM patients. We then filtered out differentially expressed genes, which resulted in a signature of 46 shared genes (Supp Table S7). These genes were then used as a gene editing signature for evaluating the responders and non-responders across the independent datasets. By measuring the RES levels in these shared 46 genes, we could separate patients who will respond to PEM (Fig. 5A, Supp Table 8). When we examine the mean RES scores (Fig. 5B, Supp Table 8) and logistic regression predictions of these genes (Figs. 5A–C, Supp 6A, B), responders have a reduced mean signature score and are clearly separated from nonresponders. In addition, patients with low RES gene signatures show extended overall survival (Fig. 5D), and the RES gene signature is associated with an elevated hazard ratio. Importantly, the overall transcript levels of the genes within this signature cannot separate patients by response (Supp Fig. 6C). We next investigated how this RES gene signature worked in patients treated with NIV. Again, this approach can accurately separate patients (Figs. 5E–G, Supp Table 8, Supp 6D–F) with responders having no detectable RNA editing events within these genes. In contrast, non-responding patients show varying levels of RNA editing. In agreement with our data from the PEM cohort, we found improved survival for patients who have no RNA editing (Fig. 5H), and RNA editing is associated with a higher hazard ratio in nonresponders. These data show that this approach may provide a platform for understanding the drivers of immune response and in predicting patient response to immunotherapy.
Fig. 6.
Recurrent and common RES sites occur and predict outcomes in melanoma tumors. (A) Heat map of 1308 responder enriched and 2148 nonresponder enriched RES sites in nonresponder (NR) and responder (R) patients from Pembrolizumab-treated patients. (B) Heat map of 370 responder enriched and 42 nonresponder enriched RES sites in non-responder (NR), responder (R), stable disease (SD) and unknown (UNK) patients from Nivolumab-treated patients. (C) Heat map of 302 responder enriched and 26 non-responder enriched RES from shared genes in non-responder (NR) and responder (R) patients from Pembrolizumab-treated patients. (D) Heat map of 207 responder enriched and 19 nonresponder enriched RES shared RES genes in non-responder (NR), responder (R), stable disease (SD) and unknown (UNK) patients from Nivolumab treated patients. (E) Heat map of significant 5 shared RES sites in nonresponder (NR) and responder (R) patients from Pembrolizumab-treated patients. (F) Heat map of significant 5 shared RES sites in nonresponder (NR), responder (R), stable disease (SD), and unknown (UNK) patients from Nivolumab-treated patients. (G) Survival analysis of patients stratified by common RES number 0–5 from Pembrolizumab-treated patients (0 common RES - black, 1 common RES – blue, 2 common RES – magenta, 3 common RES – tan, 4 common RES – orange, 5 common RES - red). (H) Survival analysis of patients stratified by common RES number 0–2 from Nivolumab-treated patients (0 common RES - black, 1 common RES – blue, 2 common RES - magenta) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
While processing these data, we found significant differences in sequencing coverage between tumors and cohorts that limits our capacity to conduct multiple comparison analysis. To evaluate the accuracy of our approach, we tested the capacity of random sets of RES genes to separate patient response in each cohort. For this, we selected 1000 groups of 46 random genes (equal size of RNA editing signature) and examined their ability to segregate patient sensitivity to immunotherapy. From this analysis, none of the datasets (0/1000) were able to significantly separate patients based on their response to immunotherapy in either cohort. These controls support our approach and strongly suggest that A-I editing events can be used to segregate patients based on their response to immunotherapy. Gene Ontology profiling of these common signature genes identified genes with “nucleotide binding” function as enriched [25]. The 46 genes within our profile were found to have a number of interactions within key DNA damage and oncogenic proteins including p53, EGFR, and FANCD2 (Supp Table 9).
Logistic regression modeling predicts stable disease patients to be primarily nonresponders
We then tested how this approach would cluster patients with stable disease following NIV treatment. This analysis found that stable patients clustered primarily with nonresponders. (Fig. 5E, Supp Table 10). Logistic regression of NIV upregulated genes and the shared gene signature predicted that 11 of the 16 stable patients would cluster with nonresponders. Logistic regression of NIV downregulated genes accurately predicted all 16 to be nonresponders. While stable disease is an intermediate state, the models primarily cluster it with nonresponders.
Recurrent RNA editing sites are present in patient tumors and cluster with response
The above analyses focused on shared RES scores across genes; however, we next wanted to determine the discriminative power of recurrent RES sites. For this, we sub-classified RES sites enriched in responder or nonresponder patients. By using recurrent RES sites, we could completely separate patients within the PEM cohort based on clinical response (Fig. 6A). This is also true for patients within the NIV group (Fig. 6B), suggesting that recurrent RNA editing within genes is contributing to immunotherapy response. To focus on this, we compared the recurrent RES from common genes from each dataset. RES sites from common genes were also strongly discriminative of patient response and could segregate responders from nonresponders or stable disease (Fig. 6C, D). We then examined whether recurrent RES sites existed between patient cohorts. For this analysis, we compared RES sites from both groups and identified 5 RES sites within DAP3, RIF1, UBXN2A, NFAM1, and KRITI that were present and significant in each (Table 1). We next tested whether these recurrent RES events could separate patients by response and found that although the number of RES sites is small, these sites correlated with patient response (Fig. 6E, F). By using the cumulative levels of RES sites within these genes, we found that the elevated numbers of these RES sites correlate with improved patient survival in both patient cohorts (Fig. 6G, H). We then used Cox Proportional Hazards modeling [28] to determine hazard potential of these events and found a significant association between patient response and the number of common RES sites. Our findings strongly implicate recurrent RES sites within melanoma tumors as predictive of patient outcome and survival in a retrospective study.
Discussion
This analysis highlights the utility of RNA editing sites in determining the response of melanoma patients to immunotherapy. Previous studies have focused on understanding the transcriptional changes in patients to identify immune-related signatures in pathways including interferon signaling [30], MHC antigen presentation [3,31,32], and innate anti-PD1 resistance [2]. Although innovative, the discriminative power of these signatures to identify patients that will respond to immunotherapy has been limited. In this study, we investigated the capacity of RNA A-I editing events to accurately segregate patients by outcomes and response to either Pembrolizumab [2] and Nivolumab [3] immunotherapy. Our approach has incorporated new knowledge generated by a number of groups that have identified the role of ADAR-mediated RNA editing as important in the development of cancer [6–8,11,33,34]. These studies have linked ADAR activity or ADAR-loss to immune response levels and immune checkpoint blockade [6–8]. In agreement with a number of other studies, we found no correlation between interferon signatures, ADAR levels, or total A-I editing events to immunotherapy response [30,35,36]. However, by systematically identifying A-I RNA editing changes within genes, those solely within genes of responders and nonresponders, shared gene editing events, and recurrent RNA editing sites between tumor cohorts, we found that RNA editing may be important for improving immunotherapy responses and for identifying sensitive patients. In addition, by using TCGA-SKCM, we highlight that these RNA editing signatures are specific for anti-PD1 immunotherapy treatments. Despite the success of our analysis, these datasets have significant variations in their RNA-seq coverage between tumors and cohorts that limit multiple comparison analysis. These differences are common in RNA-Seq data generated by different groups using alternate protocols and with diverse patients. Despite this, it is clear that this approach does have utility in segregating melanoma patients and demonstrates that expanding this analysis into other tumor types may be fruitful.
One of the interesting observations from this research is the under-representation in immune-related RNA editing events. Our initial hypothesis centered on ADAR-mediated RNA editing of immune sensing or immune response genes as key for patient sensitivity. However, our analysis only identified one immune-related gene as having high RES levels, but the predictive capacity of this gene was low. In contrast, many of the most prognostic RES events were found on genes that interact with key oncogenes and DNA damage repair proteins including FANCD2 [37], EGFR [38], NTRK1, and p53. These results are supported by clinical data from lung cancer studies that show that mutations within EGFR and NTRK1 are common in patients resistant to immunotherapy [39,40]. In contrast, p53 disruptions have been linked to improved sensitivity to these therapies [41]. These common RNA editing events suggest that aberrant growth signaling and genome stability may be key contributors to patient response in the melanoma patients we analyzed.
Additionally, we found that ADAR-mediated RNA editing of the DAP3 gene (chr1 155733925) is recurrent in the responder-enriched RES group. DAP3 has previous been shown to repress RNA-editing, while fueling tumor progression [35]. Interestingly, many nucleotide binding proteins show strong levels of downregulated RES sites in both cohorts. In PEM-treated patients, ZNF124, 490 and 827 show statistically lower RES levels in responders and are predictive of immunotherapy sensitivity. In the NIV cohort, ZNF 226, 329, 426 and 836 act in a similar manner. Although the functions of these proteins are not well understood, there is a clear thread from this analysis that suggests that reductions in the levels of RNA-editing within nucleotide-binding genes is recurrent in tumors of patients sensitive to immunotherapy.
Collectively, this analysis highlights the utility of understanding RNA editing changes in melanoma patients. It suggests that the in-depth profiling and sub-categorizing of events, rather than bulk A-I levels, can shed new light on the biology and responsiveness of these tumors to immunotherapy. These results identify pathways previously linked to immunotherapy and new alterations that have under-explored roles in tumor biology and immune sensitivity. Further work utilizing specialized RNA-seq platforms that maximize gene coverage and minimize inter-tumoral and cohort variation are required before these ideas are ready for clinical predictions of treatment. However, these results suggest that this an area of real opportunity for improving immunotherapy outcomes for patients.
Data availability
Data presented in this manuscript are available from the corresponding author upon request.
CRediT authorship contribution statement
Jalal Siddiqui: Methodology, Software, Writing – original draft. Wayne O. Miles: Conceptualization, Resources, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to acknowledge the support of the Damon Runyon Cancer Research Foundation award (51-18) (W.O.M), an American Cancer Society Research Scholar award (RSG-20-060-01-RMC) (W.O.M), National Cancer Institute R01 (R01CA251753) (W.O.M) and the support of a P30 award (CA016058) to the OSUCCC from the National Cancer Institute, Bethesda, MD. The results here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101197.
Appendix. Supplementary materials
References
- 1.Samuel E., Moore M., Voskoboynik M., Shackleton M., Haydon A. An update on adjuvant systemic therapies in melanoma. Melanoma Manag. 2019;6(3):MMT28. doi: 10.2217/mmt-2019-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riaz N., Havel J.J., Makarov V., Desrichard A., Urba W.J., Sims J.S. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171(4):934–949. doi: 10.1016/j.cell.2017.09.028. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei S.C., Duffy C.R., Allison J.P. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8(9):1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 5.Van Allen E.M., Miao D., Schilling B., Shukla S.A., Blank C., Zimmer L. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhate A., Sun T., Li J.B. ADAR1: a new target for immuno-oncology therapy. Mol. Cell. 2019;73(5):866–868. doi: 10.1016/j.molcel.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 7.Ishizuka J.J., Manguso R.T., Cheruiyot C.K., Bi K., Panda A., Iracheta-Vellve A. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 2019;565(7737):43. doi: 10.1038/s41586-018-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mannion N.M., Greenwood S.M., Young R., Cox S., Brindle J., Read D. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9(4):1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu D., Schilling B., Liu D., Sucker A., Livingstone E., Jerby-Amon L. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2019;25(12):1916–1927. doi: 10.1038/s41591-019-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auslander N., Zhang G., Lee J.S., Frederick D.T., Miao B., Moll T. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med. 2018;24(10):1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Ann. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang F., Lu Y., Yan S., Xing Q., Tian W. SPRINT: an SNP-free toolkit for identifying RNA editing sites. Bioinformatics. 2017;33(22):3538–3548. doi: 10.1093/bioinformatics/btx473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q. Transcriptome Data Analysis. Springer; 2018. Analysis of RNA editing sites from RNA-Seq data using GIREMI; pp. 101–108. [DOI] [PubMed] [Google Scholar]
- 14.Picardi E., Pesole G. REDItools: high-throughput RNA editing detection made easy. Bioinformatics. 2013;29(14):1813–1814. doi: 10.1093/bioinformatics/btt287. [DOI] [PubMed] [Google Scholar]
- 15.Guan J., Gupta R., Filipp F.V. Cancer systems biology of TCGA SKCM: efficient detection of genomic drivers in melanoma. Sci. Rep. 2015;5(1):1–10. doi: 10.1038/srep07857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomczak K., Czerwińska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 2015;19(1A):A68. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krueger F. Trim galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. 2019;516:517. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ [Google Scholar]
- 18.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–DD73. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavalcante R.G., Sartor MA. Annotatr: genomic regions in context. Bioinformatics. 2017;33(15):2381–2383. doi: 10.1093/bioinformatics/btx183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10(2):giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chigaev M., Yu H., Samuels D.C., Sheng Q., Oyebamiji O., Ness S. Genomic positional dissection of RNA Editomes in tumor and normal samples. Front. Genet. 2019;10:211. doi: 10.3389/fgene.2019.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J., Bardes E.E., Aronow B.J., Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(suppl_2):W305–WW11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12(1):1–8. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore D.F. Springer; 2016. Applied survival analysis using R. [Google Scholar]
- 28.Lin H., Zelterman D. Taylor & Francis; 2002. Modeling survival data: extending the Cox model. [Google Scholar]
- 29.Anders S., Huber W. Differential expression analysis for sequence count data. Nat. Preced. 2010:1. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alavi S., Stewart A.J., Kefford R.F., Lim S.Y., Shklovskaya E., Rizos H. Interferon signaling is frequently downregulated in melanoma. Front. Immunol. 2018;9:1414. doi: 10.3389/fimmu.2018.01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M., Fritsche J., Roszik J., Williams L.J., Peng X., Chiu Y. RNA editing derived epitopes function as cancer antigens to elicit immune responses. Nat. Commun. 2018;9(1):1–10. doi: 10.1038/s41467-018-06405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S., He Z., Wang X., Li H., Liu X.S. Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction. Elife. 2019;8:e49020. doi: 10.7554/eLife.49020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H., Golji J., Brodeur L.K., Chung F.S., Chen J.T., deBeaumont R.S. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat. Med. 2019;25(1):95–102. doi: 10.1038/s41591-018-0302-5. [DOI] [PubMed] [Google Scholar]
- 34.Lehmann K.A., Bass B.L. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39(42):12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 35.Han J., An O., Hong H., Chan T.H.M., Song Y., Shen H. Suppression of adenosine-to-inosine (A-to-I) RNA editome by death associated protein 3 (DAP3) promotes cancer progression. Sci. Adv. 2020;6(25):eaba5136. doi: 10.1126/sciadv.aba5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deffit S.N., Hundley H.A. To edit or not to edit: regulation of ADAR editing specificity and efficiency. Wiley Interdiscip. Rev. RNA. 2016;7(1):113–127. doi: 10.1002/wrna.1319. [DOI] [PubMed] [Google Scholar]
- 37.Nepal M., Che R., Ma C., Zhang J., Fie P. FANCD2 and DNA damage. Int. J. Mol. Sci. 2017;18(8):1804. doi: 10.3390/ijms18081804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai J., Guo X.G., Bai X.P. Epidermal growth factor receptor-related DNA repair and radiation-resistance regulatory mechanisms: a mini-review. Asian Pac. J. Cancer Prev. 2012;13(10):4879–4881. doi: 10.7314/apjcp.2012.13.10.4879. [DOI] [PubMed] [Google Scholar]
- 39.Yu S., Liu D., Shen B., Shi M., Feng J. Immunotherapy strategy of EGFR mutant lung cancer. Am. J. Cancer Res. 2018;8(10):2106. [PMC free article] [PubMed] [Google Scholar]
- 40.Konen J.M., Rodriguez B.L., Fradette J.J., Gibson L., Davis D., Minelli R. Ntrk1 promotes resistance to PD-1 checkpoint blockade in mesenchymal Kras/p53 mutant lung cancer. Cancers. 2019;11(4):462. doi: 10.3390/cancers11040462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Assoun S., Theou-Anton N., Nguenang M., Cazes A., Danel C., Abbar B. Association of TP53 mutations with response and longer survival under immune checkpoint inhibitors in advanced non-small-cell lung cancer. Lung Cancer. 2019;132:65–71. doi: 10.1016/j.lungcan.2019.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data presented in this manuscript are available from the corresponding author upon request.