Abstract
Osteosarcoma, the most common primary malignant tumour of bone, affects both children and adults. No fundamental biological differences between paediatric and adult osteosarcoma are known. Here, we apply multi‐region whole‐genome sequencing to an index case of a 4‐year‐old child whose aggressive tumour harboured high‐level, focal amplifications of MYC and CCNE1 connected by translocations. We reanalysed copy number readouts of 258 cases of high‐grade osteosarcoma from three different cohorts and identified a significant enrichment of focal MYC, but not CCNE1, amplifications in children. Furthermore, we identified four additional cases of MYC and CCNE1 coamplification, highlighting a rare driver event which warrants further investigation. Our findings indicate that amplification of the MYC oncogene is a major driver of childhood osteosarcoma, while CCNE1 appears recurrently amplified independent of age.
Keywords: MYC, CCNE1, osteosarcoma, genomics, copy number variants
Introduction
Osteosarcoma is the most common primary cancer of bone, typically arising in long bones during periods of rapid bone growth in puberty, with a second peak in old age often associated with radiation and Paget's disease. The prognosis of osteosarcoma does not vary significantly across the age range, although some studies have demonstrated poorer outcomes in adolescence compared with younger patients [1, 2, 3]. These differences may be accounted for by the tolerability of systemic cytotoxic chemotherapy, which, for example, is thought to be reduced in adolescents compared to young children [1]. Over the past decade, studies have shown that there are no fundamental differences in the osteosarcoma genome of children (<18 years) and adults [4, 5].
The outlook of osteosarcoma has not substantially improved since the addition of multiagent cytotoxic chemotherapy to treatment protocols in the early 1980s [6]. With current approaches, the 5‐year survival stands at 60–70% in those presenting without metastatic disease [7]. Predictors of poor outcome include metastatic disease at presentation, incomplete resection, and poor histological response to neoadjuvant chemotherapy [7]. Apart from the amplification of specific oncogenes including FGFR1 [8] and MYC [9, 10, 11] that have been associated with poor clinical outcomes, there has been limited success in identifying potential prognostic biomarkers for osteosarcoma.
Based on an index case of a young child whose aggressive disease was driven by coamplification of CCNE1 and MYC, we investigated the prevalence of amplification and coamplification of these two genes in multiple osteosarcoma data sets, and whether these genes herald a particularly aggressive disease course.
Materials and methods
Genomic analysis of index case
We subjected five samples of the resection specimen (Figure 1A) and two blood samples from the index case to whole‐genome sequencing (coverage ranging 32–102×), as previously described [4] (see supplementary material, Supplementary materials and methods). Sequencing reads were analysed using the extensively validated analysis pipeline of the Wellcome Sanger Institute, to call substitutions (Caveman algorithm [12]), indels (Pindel [13]), and rearrangements (BRASS [14]). The clonal composition of tissues was determined using the Canopy algorithm [15] (see supplementary material, Table S1). The patient's legal guardian provided informed consent for participation in this study and ethical approval for this study was obtained from the Cambridge Central Research Ethics Committee (National NHS Research Ethics Service [reference 16/EE/0394]).
Figure 1.
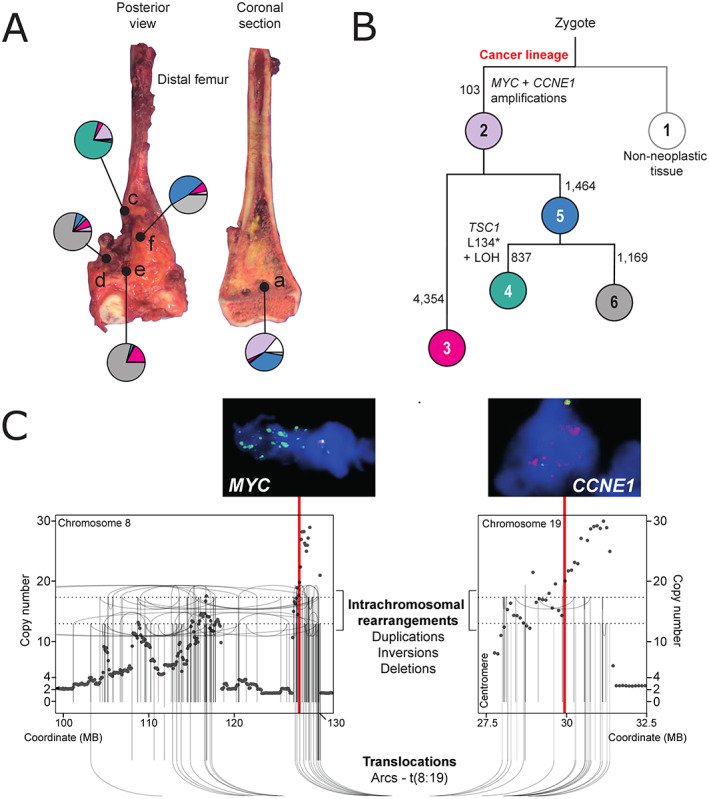
Multi‐sample whole‐genome analysis of the index case. (A) Image of resected tumour highlighting sampled tumour regions with pie charts (colouring as per B) demonstrating the relative proportion of clones comprising each region, as identified through phylogenetic analysis (Canopy algorithm). Letters refer to sample IDs. (B) Tumour phylogeny. Each circle represents a clone (number in circle = clone ID). The number next to each branch represents branch defining substitutions. The lengths of branches represent these only qualitatively. Driver events are highlighted next to each branch. (C) Consensus rearrangements and copy number across tumour samples with corresponding FISH validation. Vertical red lines highlight the position of MYC and CCNE1. Each dot represents the copy number (y‐axis) of a specific genomic coordinate (x‐axis). Lines and arc show the position and relationship of breakpoints.
Analysis of copy number data
We reanalysed array‐based copy number readouts of high‐grade osteosarcomas from three cohorts: International Cancer Genome Consortium (ICGC) data set (n = 54; segment files provided in publication [4]; Affymetrix Genome‐Wide Human SNP Array 6.0 (Applied Biosystems, Foster City, CA, USA) and in silico arrays derived from whole‐genome sequences), German‐Swiss published data set [16] (n = 124; Affymetrix Cytoscan HD array (Applied Biosystems, Foster City, CA, USA) raw data files [ArrayExpress accession E‐MTAB‐4815]; segment files generated with ASCAT algorithm [17]), and Therapeutically Applicable Research to Generate Effective Treatments (TARGET) Osteosarcoma data set (n = 80; publicly available segment files [dbGaP Sub‐study ID phs000468] generated from Affymetrix Human SNP Array 6.0 data). We applied a widely used definition [4] of focal amplifications, i.e. a total copy number to tumour ploidy ratio of over 2, and an amplicon that was smaller than 2 Mb.
Analysis of MYC and CCNE1 copy number by FISH
Twenty‐three cases of paediatric osteosarcoma diagnosed at the Royal National Orthopaedic Hospital (RNOH) between 2011 and 2018 were subjected to fluorescence in situ hybridization (FISH) analysis of MYC and CCNE1 as described previously (see supplementary material, Supplementary materials and methods) [18]. Probes used for MYC and CCNE1 were MYC/CEN8 dual colour probe (Zytovision, Bremerhaven, Germany) and CCNE1/CEN19p dual colour probe (Abnova, Taipei, Taiwan), respectively.
Statistical analyses
Binomial regression analysis was performed to measure the association between patient age and our determined MYC and CCNE1 copy states using the ‘glm’ function of the R stats package. Survival analysis was performed using the Kaplan–Meier method and survival curves were plotted using the ‘survival’ and ‘survminer’ R packages.
Results and discussion
The impetus for this study was the case of a 4‐year‐old child treated for osteosarcoma of the distal femur with an unusually aggressive disease course. At the time of diagnosis, she had a rapidly growing primary tumour with a skip lesion in the affected bone and lung metastases. She underwent neoadjuvant MAP chemotherapy (cisplatin, methotrexate, and doxorubicin) followed by surgical resection and two cycles of post‐operative MAP chemotherapy, as per the European and American Osteosarcoma study group (EURAMOS) protocol [19]. She developed bulky metastatic lung disease within 8 weeks of cessation of treatment, to which she succumbed. Histological examination of the resection specimen revealed extensive viable high‐grade osteosarcoma, from which we subjected five samples to whole‐genome sequencing (Figure 1A). Analysis of all classes of somatic variants revealed typical somatic features of osteosarcoma, including complex structural variants [4, 20] (Figure 1B,C and supplementary material, Table S2). The clonal composition of different tumour regions showed spatial diversity (Figure 1A,B and supplementary material, Table S1). Two somatic driver events were common to all tumour regions – high‐level gains of both MYC and CCNE1 (Figure 1C). We determined that these amplifications were generated by a single chromothripsis‐driven amplification event on chromosomes 8q (MYC) and 19q (CCNE1), interconnected by multiple translocations (Figure 1C and supplementary material, Figure S1). Chromothripsis‐driven amplification is a recognised mechanism of oncogene amplification in a subset of tumours across several cancer types [21], and has been previously described as a recurrent event in osteosarcoma [4].
MYC amplifications are enriched in childhood osteosarcoma
We next reanalysed copy segment data from our previously published data set of 54 high‐grade osteosarcoma exomes and genomes (ICGC cohort [4]). Focal amplifications of MYC and CCNE1 were detected in 15 and 13% of the ICGC cohort, respectively. These events were particularly common in children, with 40 and 20% of patients under the age of 13 (n = 15) harbouring MYC and CCNE1 amplifications, respectively, compared with 5% (p = 0.002, chi‐square test) and 10% (p = 0.6, chi‐square test) in the older population (n = 39). Looking at driver alterations across the cohort, only TERT mutations showed a notable difference in frequency between MYC‐amplified and non‐amplified cases; however, this was not statistically significant after multiple hypothesis testing (false discovery rate (FDR) adjusted P value: 0.54).
We then analysed copy number data of osteosarcomas from two publicly available osteosarcoma data sets: the US TARGET Osteosarcoma cohort (TARGET OS, n = 80) and a German‐Swiss cohort [16] (n = 124). In these, MYC was amplified in 1 and 10%, and CCNE1 was amplified in 5 and 13%, respectively. Statistical analysis performed on all three data sets combined confirmed a significant association between young age and the presence of MYC amplifications (p < 0.02, binomial model, see Materials and methods), but not CCNE1 gains (p > 0.05). Next, we examined the age distribution, copy number, and size of genomic segments harbouring MYC or CCNE1 across the three cohorts and observed that they shared a consistent pattern of high‐level, focal copy number gains of MYC and CCNE1 in patients younger than 18 years (Figure 2A,B). Finally, we employed FISH in a fourth tumour collection from 23 anonymised paediatric osteosarcomas assembled from the archives of the Pathology Department of the RNOH (Stanmore, UK). Informative readouts were obtained from 22 out of 23 cases (see supplementary material, Table S3). Of these, one tumour exhibited coamplification of MYC and CCNE1, five cases amplification of CCNE1 alone, and another five cases MYC amplification, corroborating the genomic finding of these alterations in paediatric osteosarcoma.
Figure 2.
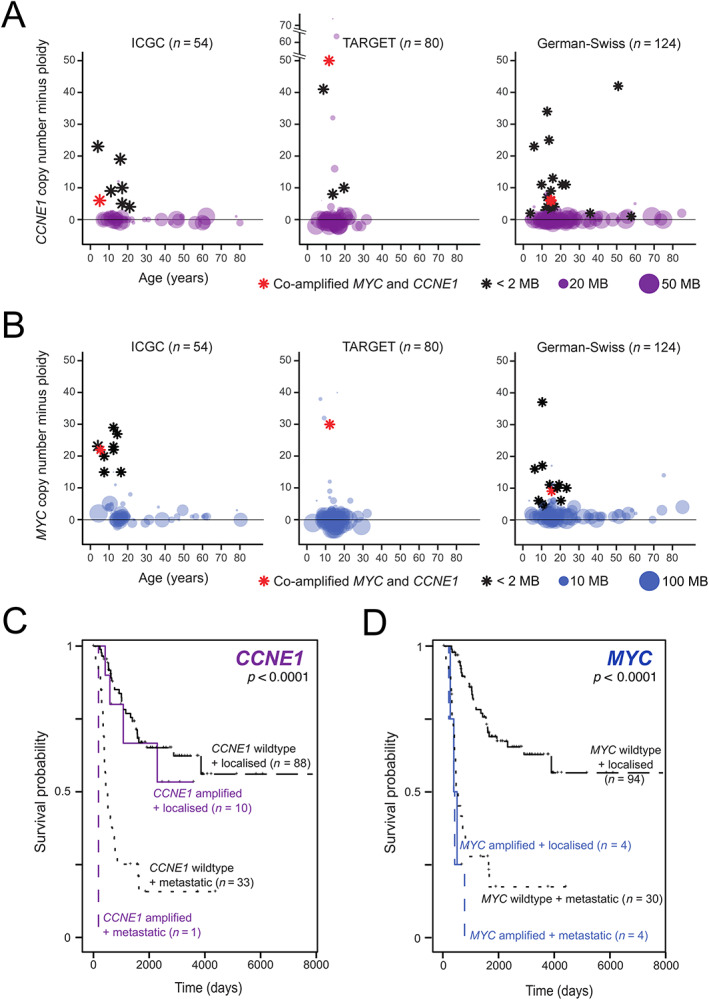
MYC and CCNE1 amplifications in osteosarcoma. Genomic segments harbouring (A) CCNE1 or (B) MYC are shown across three independent cohorts. In each plot, circles or stars represent data from an individual tumour. The position on the x‐axis shows the age. The y‐axis position represents absolute gene copy number minus tumour ploidy. The sizes of circles represent the segment size with focal amplifications (total copy number to ploidy ratio > 2 and amplicon size ≤2 Mb) represented by black stars. Red stars identify tumours with coamplification of MYC and CCNE1. Effects of (C) CCNE1 or (D) MYC amplification on survival (Kaplan–Meier analysis) by metastatic status. Survival analysis performed on ICGC and TARGET‐OS cohorts only.
Survival analysis jointly performed on the ICGC and TARGET OS data sets demonstrated an association between MYC amplification and inferior outcomes (Figure 2C) confirming previous reports. In contrast, CCNE1 amplification did not correlate with adverse survival (Figure 2D). No significant association was found between survival and amplification of either gene in our FISH cohort (see supplementary material, Figure S2), possibly as these results were not subject to the focal amplification criteria applied to our copy number data.
Coamplification of MYC and CCNE1 is a rare event in osteosarcoma
Four further tumours demonstrating gains in both MYC and CCNE1 were identified in addition to the index case in patients aged 5–15 years. Each of the three copy number data sets included a single case with this coamplification (Figure 2A,B and supplementary material, Tables S4 and S5). Clinical outcome data were available for two of these cases (PD9980a and PAMLKS) and both demonstrated aggressive behaviour with early metastatic disease and subsequent death. The low frequency of these events suggests that these are rare events possibly occurring as part of the complex structural alterations which characterise osteosarcoma. A case harbouring both MYC and CCNE1 amplifications was also identified in our FISH cohort (R010), which arose in a 8‐year‐old with no adverse events on follow‐up (see supplementary material, Table S3). Greater numbers and detailed analysis of genomic data are therefore required to evaluate this finding fully.
CCNE1 is a cell cycle regulator controlling G1/S transition, and plays a role in tumourigenesis by contributing to cell proliferation and chromosomal instability [22]. CCNE1 is a recognised driver in many malignancies including osteosarcoma [16, 23], and has proven to be an independent prognostic factor in both triple‐negative breast cancer and ovarian cancer [24, 25, 26]. As part of the many MYC‐regulated pathways which contribute to tumourigenesis, MYC has been shown to directly regulate CCNE1 gene expression as well as CCNE1‐CDK2 complex activity [27, 28], and mouse models have demonstrated the co‐operative action of MYC and CCNE1 in contributing to tumour formation in hepatocellular cancer [29]. However, while recurrent coamplification of MYC with other driver genes including ERRB2 and EGFR has previously been reported [30, 31], and in breast cancer is associated with a poorer prognosis [30], coamplification with CCNE1 has not been previously described.
In this analysis of oncogenic MYC and CCNE1 amplifications in osteosarcoma, we identified a somatic genomic feature that appears to be enriched in paediatric cases. Previous investigations of the osteosarcoma genome have not revealed differences between age groups, bar the expected excess of age‐related base substitutions associated with age, and the greater contribution of pathogenic germline variants to childhood tumours. Furthermore, we report the rare occurrence of coamplification of the known oncogenic drivers MYC and CCNE1, a finding which warrants further studies. We also provide additional evidence for the utility of MYC alterations as a prognostic factor in osteosarcoma.
Author contributions statement
JI performed sequence analysis of the index case. SDN performed copy number profiling of tumour data sets and clinical data collection. THHC and IL contributed to bioinformatic analyses. HY, AS and FA performed FISH analysis. SDN and JI wrote the manuscript, with contributions from AMF and SB. AMF and SB were responsible for study design and direction of research.
Supporting information
Supplementary materials and methods
Figure S1. Circos plots showing structural rearrangements detected in sequenced distinct regions from the index tumour
Figure S2. Survival analysis showing the impact of (A) MYC and (B) CCNE1 amplifications as detected by FISH in the RNOH cohort (n = 22)
Table S1. Somatic alterations for phylogenetic analysis (Canopy)
Table S2. Index case BRASS detected structural rearrangements
Table S3.MYC and CCNE1 FISH results (RNOH cohort)
Table S4.MYC and CCNE1 copy number profiles for published data sets
Table S5. Survival data, ICGC cohort
Acknowledgements
We thank the clinical teams of the London Sarcoma Service and Cambridge University Hospitals NHS Foundation Trust. The results published here are in part based on data generated by the TARGET (https://ocg.cancer.gov/programs/target) initiative, phs000218. We are grateful to our little and older patients and their families for participating in our research. This study was funded by the Wellcome Trust (fellowship to SB), The Tom Prince Cancer Trust (AMF), and the Bone Cancer Research Trust. AMF is supported by the National Institute for Health Research, UCLH Biomedical Research Centre, and the UCL Experimental Cancer Centre. SDN holds a Jean Shanks Foundation – Pathological Society Clinical Fellowship. IL is supported by the Lundbeck Foundation (grant: R303‐2018‐3018).
Conflict of interest statement: AMF is currently President of The Pathological Society, which is owner of the Journal of Pathology and Journal of Pathology: Clinical Research. No other potential conflicts of interest were declared.
Contributor Information
Adrienne M Flanagan, Email: a.flanagan@ucl.ac.uk.
Sam Behjati, Email: sb31@sanger.ac.uk.
Data availability statement
The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files or from the corresponding author on reasonable request. Sequencing data generated by this study are available from the EGA (EGAD00001006859). Previously published sequencing data that support the findings of this study are available from the EGA: EGAD00001000107, EGAS00001000196, and EGAD00001000147 (DOI: 10.1038/ncomms15936), and ArrayExpress: E‐MTAB‐4815 (DOI: 10.1002/ijc.30778). The data used for the TARGET OS data set are available at https://portal.gdc.cancer.gov/projects.
References
- 1.Haddox CL, Han G, Anijar L, et al. Osteosarcoma in pediatric patients and young adults: a single institution retrospective review of presentation, therapy, and outcome. Sarcoma 2014; 2014: 402509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins M, Wilhelm M, Conyers R, et al. Benefits and adverse events in younger versus older patients receiving neoadjuvant chemotherapy for osteosarcoma: findings from a meta‐analysis. J Clin Oncol 2013; 31: 2303–2312. [DOI] [PubMed] [Google Scholar]
- 3.Janeway KA, Barkauskas DA, Krailo MD, et al. Outcome for adolescent and young adult patients with osteosarcoma: a report from the Children's Oncology Group. Cancer 2012; 118: 4597–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behjati S, Tarpey PS, Haase K, et al. Recurrent mutation of IGF signalling genes and distinct patterns of genomic rearrangement in osteosarcoma. Nat Commun 2017; 8: 15936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suehara Y, Alex D, Bowman A, et al. Clinical genomic sequencing of pediatric and adult osteosarcoma reveals distinct molecular subsets with potentially targetable alterations. Clin Cancer Res 2019; 25: 6346–6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer 2009; 115: 1531–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smeland S, Bielack SS, Whelan J, et al. Survival and prognosis with osteosarcoma: outcomes in more than 2000 patients in the EURAMOS‐1 (European and American Osteosarcoma Study) cohort. Eur J Cancer 2019; 109: 36–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernanda Amary M, Ye H, Berisha F, et al. Fibroblastic growth factor receptor 1 amplification in osteosarcoma is associated with poor response to neo‐adjuvant chemotherapy. Cancer Med 2014; 3: 980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen D, Zhao Z, Huang Z, et al. Super enhancer inhibitors suppress MYC driven transcriptional amplification and tumor progression in osteosarcoma. Bone Res 2018; 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng W, Dean DC, Hornicek FJ, et al. Myc is a prognostic biomarker and potential therapeutic target in osteosarcoma. Ther Adv Med Oncol 2020; 12: 1758835920922055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Scionti I, Michelacci F, Pasello M, et al. Clinical impact of the methotrexate resistance‐associated genes C‐MYC and dihydrofolate reductase (DHFR) in high‐grade osteosarcoma. Ann Oncol 2008; 19: 1500–1508. [DOI] [PubMed] [Google Scholar]
- 12.Jones D, Raine KM, Davies H, et al. cgpCaVEManWrapper: simple execution of CaVEMan in order to detect somatic single nucleotide variants in NGS data. Curr Protoc Bioinformatics 2016; 56: 15.10.1–15.10.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye K, Schulz MH, Long Q, et al. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired‐end short reads. Bioinformatics 2009; 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nik‐Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole‐genome sequences. Nature 2016; 534: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y, Qiu Y, Minn AJ, et al. Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next‐generation sequencing. Proc Natl Acad Sci U S A 2016; 113: E5528–E5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smida J, Xu H, Zhang Y, et al. Genome‐wide analysis of somatic copy number alterations and chromosomal breakages in osteosarcoma. Int J Cancer 2017; 141: 816–828. [DOI] [PubMed] [Google Scholar]
- 17.Van Loo P, Nordgard SH, Lingjærde OC, et al. Allele‐specific copy number analysis of tumors. Proc Natl Acad Sci U S A 2010; 107: 16910–16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amary MF, Berisha F, Bernardi Fdel C, et al. Detection of SS18‐SSX fusion transcripts in formalin‐fixed paraffin‐embedded neoplasms: analysis of conventional RT‐PCR, qRT‐PCR and dual color FISH as diagnostic tools for synovial sarcoma. Mod Pathol 2007; 20: 482–496. [DOI] [PubMed] [Google Scholar]
- 19.Meyers PA, Schwartz CL, Krailo M, et al. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high‐dose methotrexate. J Clin Oncol 2005; 23: 2004–2011. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Bahrami A, Pappo A, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep 2014; 7: 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortés‐Ciriano I, Lee JJ‐K, Xi R, et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole‐genome sequencing. Nat Genet 2020; 52: 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spruck CH, Won K‐A, Reed SI. Deregulated cyclin E induces chromosome instability. Nature 1999; 401: 297–300. [DOI] [PubMed] [Google Scholar]
- 23.Sayles LC, Breese MR, Koehne AL, et al. Genome‐informed targeted therapy for osteosarcoma. Cancer Discov 2019; 9: 46–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X, Wu HM, Zhu C, et al. Next generation sequencing reveals CCNE1 amplification as an independent prognostic factor for triple negative breast cancer (TNBC) patients. J Clin Oncol 2020; 38: 558–558.31821071 [Google Scholar]
- 25.Rosen DG, Yang G, Deavers MT, et al. Cyclin E expression is correlated with tumor progression and predicts a poor prognosis in patients with ovarian carcinoma. Cancer 2006; 106: 1925–1932. [DOI] [PubMed] [Google Scholar]
- 26.Chan AM, Enwere E, McIntyre JB, et al. Combined CCNE1 high‐level amplification and overexpression is associated with unfavourable outcome in tubo‐ovarian high‐grade serous carcinoma. J Pathol Clin Res 2020; 6: 252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beier R, Bürgin A, Kiermaier A, et al. Induction of cyclin E‐cdk2 kinase activity, E2F‐dependent transcription and cell growth by Myc are genetically separable events. EMBO J 2000; 19: 5813–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez‐Roger I, Solomon DL, Sewing A, et al. Myc activation of cyclin E/Cdk2 kinase involves induction of cyclin E gene transcription and inhibition of p27(Kip1) binding to newly formed complexes. Oncogene 1997; 14: 2373–2381. [DOI] [PubMed] [Google Scholar]
- 29.Sonntag R, Giebeler N, Nevzorova YA, et al. Cyclin E1 and cyclin‐dependent kinase 2 are critical for initiation, but not for progression of hepatocellular carcinoma. Proc Natl Acad Sci U S A 2018; 115: 9282–9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al‐Kuraya K, Schraml P, Torhorst J, et al. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res 2004; 64: 8534–8540. [DOI] [PubMed] [Google Scholar]
- 31.Mitsui F, Dobashi Y, Imoto I, et al. Non‐incidental coamplification of Myc and ERBB2, and Myc and EGFR, in gastric adenocarcinomas. Mod Pathol 2007; 20: 622–631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods
Figure S1. Circos plots showing structural rearrangements detected in sequenced distinct regions from the index tumour
Figure S2. Survival analysis showing the impact of (A) MYC and (B) CCNE1 amplifications as detected by FISH in the RNOH cohort (n = 22)
Table S1. Somatic alterations for phylogenetic analysis (Canopy)
Table S2. Index case BRASS detected structural rearrangements
Table S3.MYC and CCNE1 FISH results (RNOH cohort)
Table S4.MYC and CCNE1 copy number profiles for published data sets
Table S5. Survival data, ICGC cohort
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files or from the corresponding author on reasonable request. Sequencing data generated by this study are available from the EGA (EGAD00001006859). Previously published sequencing data that support the findings of this study are available from the EGA: EGAD00001000107, EGAS00001000196, and EGAD00001000147 (DOI: 10.1038/ncomms15936), and ArrayExpress: E‐MTAB‐4815 (DOI: 10.1002/ijc.30778). The data used for the TARGET OS data set are available at https://portal.gdc.cancer.gov/projects.


