Abstract
The skin typically tolerates exposure to various microbes and chemicals in the environment. Here we investigated how the epidermis maintains this innate immune tolerance to stimuli that are recognized by Toll-like receptors (TLRs). Loss of tolerance to TLR ligands occurred after silencing of the histone deacetylases (HDACs) HDAC8 and HDAC9 in keratinocytes. Transcriptional analysis identified MAP2K3 as suppressed by HDAC8/9 activity and a potential key intermediary for establishing this tolerance. HDAC8/9 influenced acetylation at H3K9 and H3K27 marks in the MAP2K3 promoter. Proteomic analysis further identified SSRP1 and SUPT16H as associated with HDAC8/9 and responsible for transcriptional elongation of MAP2K3. Silencing of MAP2K3 blocked the capacity of HDAC8/9 to influence cytokine responses. Relevance in vivo was supported by observations of increased MAP2K3 in human inflammatory skin conditions and the capacity of keratinocyte HDAC8/9 to influence dendritic cell maturation and T-cell proliferation. Keratinocyte-specific deletion of HDAC8/9 also increased inflammation in mice after exposure to ultraviolet radiation, imiquimod or S. aureus. These findings define a mechanism for the epidermis to regulate inflammation in the presence of ubiquitous TLR ligands.
One Sentence Summary:
Inhibition of HDAC8 and HDAC9 impairs keratinocyte tolerance of TLR ligand stimulation, leading to cutaneous inflammation.
Introduction
The human body is constitutively exposed to various environmental stimuli, and these stimuli are diverse and highly variable with time and location. As the most external organ, the skin is primarily responsible for protection and appropriate recognition of danger signals in this context (1, 2), but has also established tolerance mechanisms to adapt to these various challenges from the environment without unwanted inflammation that would promote disease (3, 4). For example, failure of immune tolerance mechanisms results in classic allergic responses to specific antigens and common human inflammatory skin disorders such as psoriasis, atopic dermatitis and acne reflect dysfunction of tolerance to innate immune stimuli. However, despite the importance of innate immune tolerance to overall immune homeostasis, and its potential relevance to multiple human diseases, the mechanisms which confer innate tolerance of the epidermis to diverse environmental stimuli remain incompletely understood.
Microbial products such as those that trigger activation of Toll-like receptors (TLRs) are a classic example of environmental stimuli that are a potent trigger of inflammation in immunocytes but are well tolerated by keratinocytes in the epidermis. Instead of triggering an immune defense response, the epidermis provides a large surface for positive interactions with microbes and has established symbiotic relationships with some members of the skin microbiome (5, 6). Subsequent to the communication between microbes and the epidermis, these epithelial cells then closely interact with classical immunocytes to shape the overall immune response (7–9). Therefore, understanding how keratinocytes interact with external environmental triggers is an important goal that can enable improved therapy of immune disorders.
A common mechanism for environmental control of cell function is through epigenetic control of gene expression. An important class of molecules involved in epigenetic control are short-chain fatty acids (SCFAs) that can increase acetylation of histones through inhibition of histone deacetylases (HDACs). SCFAs are generated from bacteria under anaerobic conditions (10) and can accumulate in hair follicles due to fermentation by C. acnes (11). Although SCFAs such as butyrate have been known to inhibit inflammatory responses by bone-marrow derived immunocytes (10, 12), an important clue to understand innate immune tolerance of the skin came with the observation that SCFAs increase inflammatory responses by keratinocytes to TLR ligands. Inhibition of HDAC8 and HDAC9 by SCFAs produced by C. acnes amplified inflammatory responses in keratinocytes (13). These observations suggested microbes could inhibit HDACs and that HDACs are a mechanism through which keratinocytes can tolerate exposure to the varied TLR ligands. This study confirms this hypothesis and provides a detailed understanding of mechanisms by which epigenetic events drive innate immune tolerance in the skin. Our findings suggest that the activity of HDAC8 and HDAC9 in keratinocytes is a central regulator of innate immune tolerance.
Results
HDAC8 and 9 inhibits the inflammatory response to TLR ligands and expression of MAP2K3 in keratinocytes
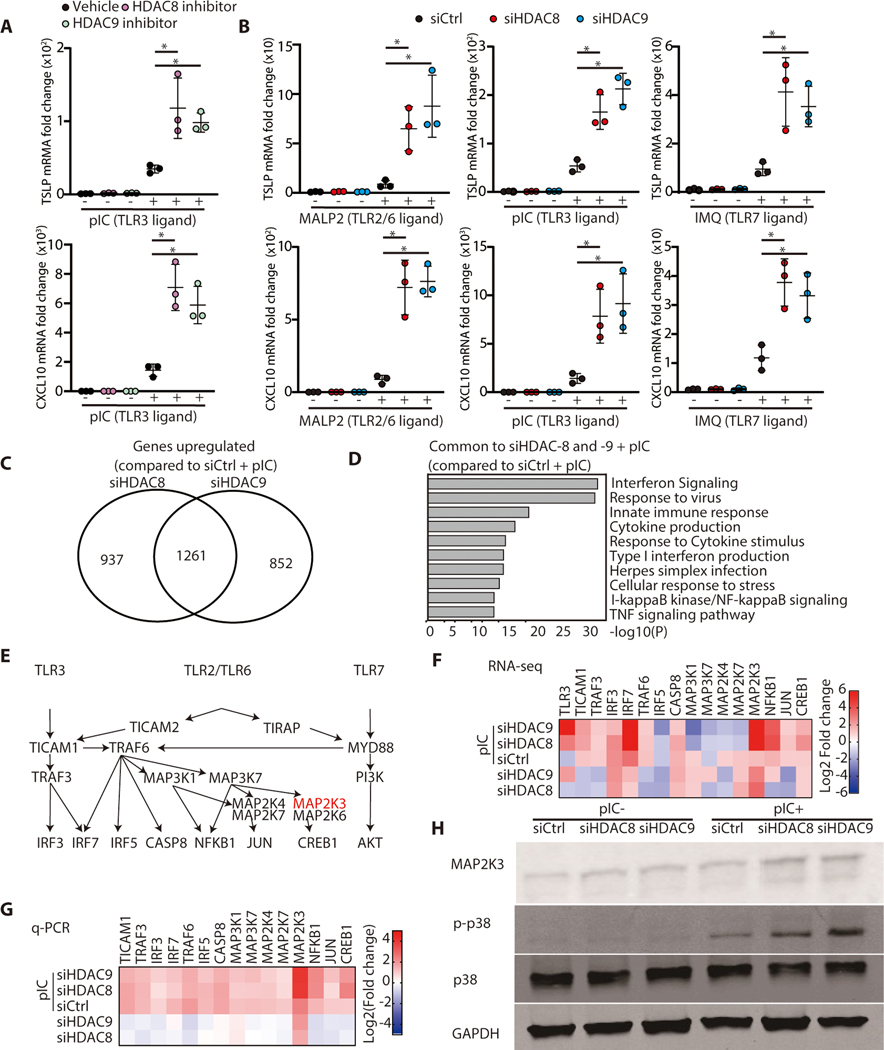
The capacity of microbes to promote inflammation by inhibition of histone deacetylation (13) has demonstrated the need to better understand how HDACs may maintain tolerance at the skin surface to innate stimuli of inflammation such as TLR ligands. Inhibition of HDAC8/9, but not HDAC1–7, was previously observed to result in greatly increased cytokine expression in the skin in response to microbial products (13). Addition of selective chemical inhibitors for HDAC8 and HDAC9 to primary cultures of normal human keratinocytes confirmed these prior observations and increased expression of TSLP and CXCL10 in the presence of a TLR3 ligand (Figure 1A). Targeting of HDAC8 or HDAC9 by RNA silencing of normal human keratinocytes confirmed this result and also increased expression of TSLP and CXCL10 when cells were exposed to ligands for TLR3, TLR2/6 and TLR7 (Figure 1B). Since no change in cytokine expression was observed following inhibition of HDACs without a TLR stimulus, we hypothesized that HDAC8 and 9 may act to suppress downstream signaling in keratinocytes that was triggered by TLRs.
Figure 1. HDAC8 and HDAC9 inhibit cytokine response to TLR 2, 3 and 7 ligands by suppression of MAP2K3 expression.
(A) Normal human keratinocytes (nHEKs) were pretreated for 1 hour with PCI34051 (10μM) or TMP269 (1μM) as HDAC8 or HDAC9 inhibitors respectively, and then cultured for 4 hours with poly I:C (1 μg/ml). mRNA expression of TSLP or CXCL10 was measured by qPCR. (N=3; one-way ANOVA). (B) nHEKs were treated for 24 hours with silencing RNAs to HDAC8 or HDAC9, or control scrambled siRNA, then cultured for 3 hours with MALP2 (200 ng/ml) or poly I:C (1 μg/ml) or 4 hours with IMQ (30 μg/ml). mRNA expression of TSLP or CXCL10 was measured by qPCR. (N=3; one-way ANOVA) (C) Total RNA sequencing after silencing of HDAC-8 or HDAC-9 and addition of poly I:C compared to control siRNA and poly I:C. (D) Gene ontology analysis of genes induced by poly I:C after HDAC 8,9 silencing with compared to genes induced by poly I:C after control siRNA. (E) Illustration of signaling pathways for TLR3, TLR2/6 and TLR7. (F) Heat map of TLR signaling-related genes that increased after HDAC8/9 silencing as determined by RNA sequencing and (G) validated by qPCR. (H) Immunoblotting for phospho-p38MAPK, p38MAPK, MAP2K3, or GAPDH from nHEKs treated with poly I:C following HDAC8/9 silencing. Results are expressed as the mean ± standard deviation (SD). *, P < 0.05. Data are representative of three independent experiments.
To understand the mechanism by which the activity of HDAC8 and 9 suppress cytokine expression in response to TLR ligands, RNA-sequencing of keratinocytes was performed. We identified 1261 genes that were commonly upregulated when HDAC8 or HDAC9 were silenced and also exposed to poly I:C (Figure 1C). The gene sets which increased after silencing of HDAC8 or 9 in the presence of a TLR3 ligand were primarily associated with inflammatory responses. (Figure 1D). RNA sequencing of cultured normal human keratinocytes did not detect increased expression of inflammatory genes after silencing of HDAC8/9 without addition of poly I:C (fig S1A,B). These data supported the conclusion that HDAC8/9 does not act directly on the expression of inflammatory genes and provided candidate genes that may mediate the suppressive effect.
TLR2/6, TLR3 and TLR7 have both distinct and overlapping canonical signaling pathways that promote cytokine expression (Figure 1E). Interrogation of the RNA-sequencing results identified MAP2K3 and IRF3 from among this signaling cascade as highly induced by silencing of either HDAC8 or 9 (Figure 1F). Independent RT-qPCR validated that MAP2K3 was increased from among these genes (Figure 1G), and therefore we focused analysis on the activity of this important kinase. The function of MAP2K3 was confirmed to be increased since the substrate of this kinase (p38MAPK) showed increased phosphorylation after silencing of HDAC8 and HDAC9 and stimulation by poly I:C (Figure 1H, Fig S2). These observations suggested HDAC8 or HDAC9 inhibits MAP2K3 expression and function. The convergence of TLR signaling through this key kinase could explain the effects of HDAC inhibition on inflammatory cytokines.
HDAC8 and HDAC9 interact with SSRP1 and SUPT16H to influence keratinocyte cytokine and MAP2K3 expression
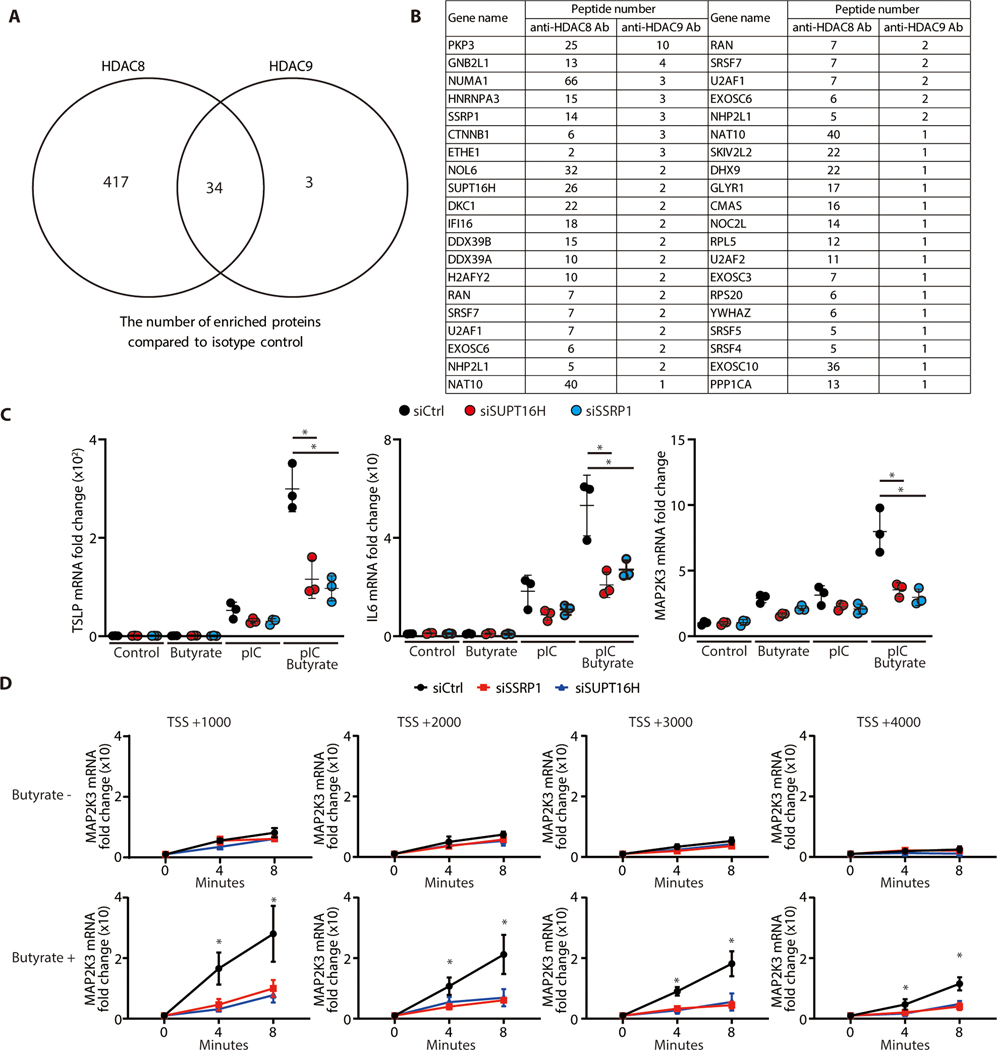
Through assembly into protein complexes and deacetylation at specific histone marks, HDACs are powerful regulators of expression for many target genes (13–21). Understanding the components of these protein complexes is important to elucidate the molecular mechanism for gene regulation by HDACs. To clarify this regulatory mechanism for HDAC8 and HDAC9 in human keratinocytes, immunoprecipitation and subsequent mass spectrometry analysis was performed to identify proteins that associate with HDAC8 and HDAC9. Specificity of anti-HDAC8 and anti-HDAC9 relative to the isotype control antibody was verified by Western blot (Fig S1C). Among known nucleosome proteins, total of 34 proteins were observed associated with both HDAC8 and HDAC9 (Figure 2A, B). In particular, HDAC8 and HDAC9 were both found to be associated with the SSRP1 and SUPT16H proteins, components of the facilitates chromatin transcription (FACT) complex that is known to act as a major regulator of transcription through effects on elongation (22, 23).
Figure 2. Transcription elongation complex is associated with HDAC 8/9 and is required for MAP2K3 gene expression following HDAC inhibition.
(A) Proteins identified by mass spectrometry of nHEK extracts enriched by immunoprecipitation with anti-HDAC8 or HDAC9 compared with control IgG. (B) The common enriched nucleosome proteins precipitated by anti-HDAC8 and HDAC9. (C) Transcriptional elongation FACT proteins SSRP1 and SUPT16H that were identified in B were silenced in nHEKs and then HDAC activity chemically inhibited by butyrate treatment (2 mM) for 1 hour. nHEKs were subsequently cultured with or without poly I:C (1 μg/ml) for 4 hours and gene expression measured by qPCR. (N=3; one-way ANOVA) (D) MAP2K3 transcriptional elongation assay. The transcriptional elongation rate of MAP2K3 was measured after SSRP1 or SUPT16H silencing with or without butyrate treatment (2mM) for 1 hour. Data show relative abundance of pulse-labeled RNA for indicated genes at the indicated times after removal of transcriptional inhibitor 6-Dichlorobenzimidazole 1-β-d-ribofuranoside (DRB). (N=3; one-way ANOVA). The results are expressed as the mean ± standard deviation (SD). *, P < 0.05. Data are representative of three independent experiments.
To our knowledge, a role for the FACT complex in regulation of inflammatory cytokine production in keratinocytes had not been previously reported. Therefore, to determine if the association we observed with HDAC8/9 could be relevant to the effects we had observed on innate immune tolerance, we examined keratinocyte inflammatory responses after silencing of SSRP1 or SUPT16H. Keratinocytes were stimulated with poly I:C and HDAC activity was inhibited with butyrate, a naturally occurring SCFA and pan-HDAC inhibitor in the skin that is produced by microorganisms through fermentation (13). Silencing of SSRP1 and SUPT16H blocked the capacity of butyrate to enhance TSLP, IL-6 and MAP2K3 expression in normal human keratinocytes (Figure 2C, fig S1D, E). To directly measure the impact of the FACT complex on MAP2K3 gene elongation, transcriptional elongation assays were performed. Butyrate treatment increased MAP2K3 gene expression and promoted transcriptional elongation, and these functions were inhibited by SSRP1 and SUPT16H silencing (Figure 2D). These results demonstrated how HDAC activity can act through the FACT complex to regulate transcription of MAP2K3.
MAP2K3 is directly influenced by HDAC8 and HDAC9 and is necessary for inhibition of cytokine expression by HDAC
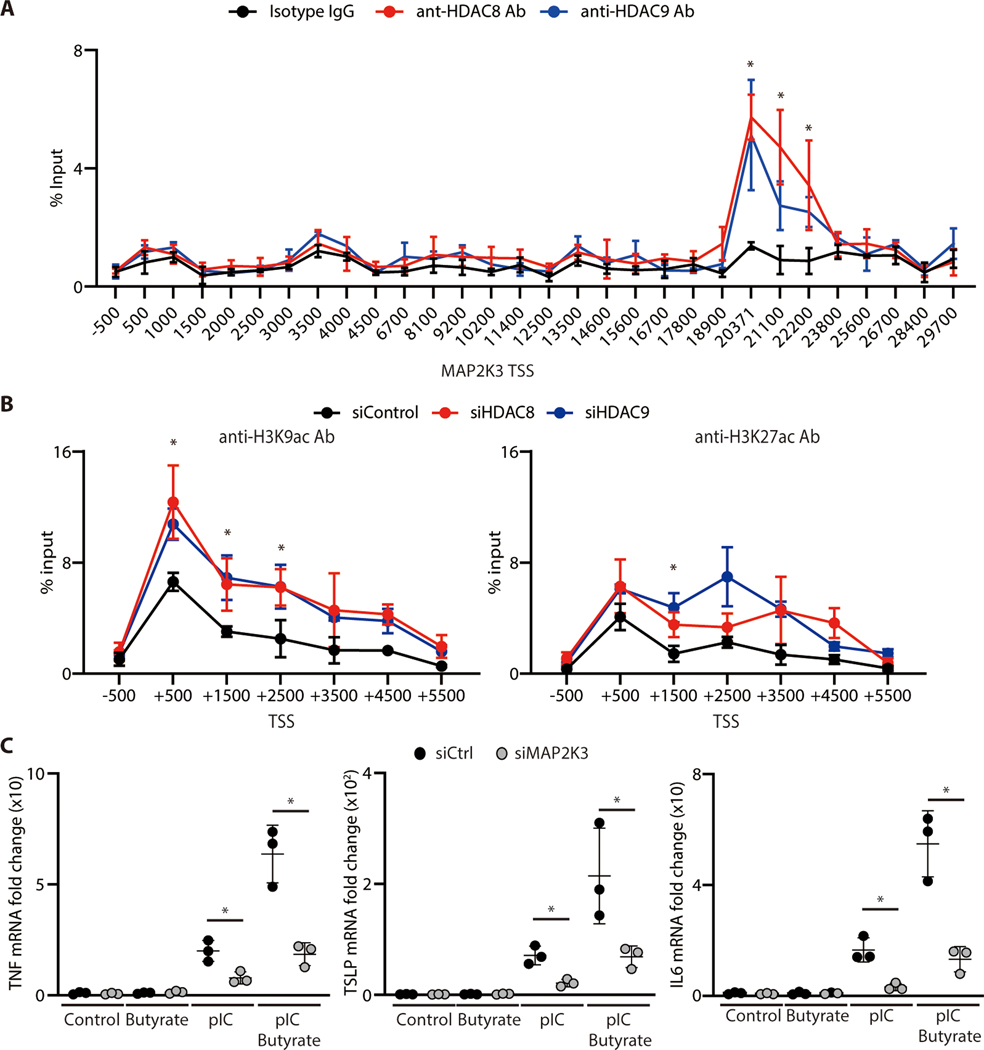
To test if HDAC8 or HDAC9 associate with MAP2K3, primary human keratinocytes were subjected to chromatin immunoprecipitation using anti-HDAC8 and HDAC9 antibodies and then evaluated by ChIP-qPCR. Compared to the isotype control, a significant peak was observed within the MAP2K3 gene following immunoprecipitation with either anti-HDAC8 or anti-HDAC9 (Figure 3 A). Furthermore, silencing of HDAC8 and HDAC9 in human keratinocytes resulted in an increase in histone acetylation in MAP2K3 as assessed by ChIP-qPCR using anti-H3K9ac and anti-H3K27ac (Figure 3B). These data show how both HDAC8 and HDAC9 can associate with the gene for MAP2K3, and acetylation of H3K9 and H3K27 marks within the MAP2K3 promoter will increase when the deacetylation activity from HDAC8 and HDAC9 is decreased (i.e. acetylation is increased) by inhibition of these HDACs.
Figure 3. HDAC8/HDAC9 associates with and acetylates MAP2K3 to influence cytokine expression by keratinocytes.
(A) ChIP-qPCR for sites within the MAP2K3 gene relative to the transcriptional start site (TSS) following precipitation with anti-HDAC8 or anti-HDAC9. (N=3; one-way ANOVA) (B) ChIP-qPCR for MAP2K3 gene relative to the transcriptional start site (TSS) following precipitation with anti-H3K9ac or anti-H3K27ac. Data are shown in nHEKs after silencing of HDAC8, HDAC9 or control siRNA. (N=3; one-way ANOVA) (C) TNF-α, TSLP or IL-6 mRNA expression in nHEKs measured by qPCR 4 hours after poly I:C stimulation and following silencing of MAP2K3 or control siRNA with or without pretreatment with butyrate (2 mM) for 1 hour. (N=3; Student’s t-test). The results are expressed as the mean ± standard deviation (SD). *, P < 0.05. Data are representative of three independent experiments.
To confirm the requirement for MAP2K3 to enable HDAC8 or 9 to influence cytokine responses, we next examined the effect of silencing of MAP2K3 in keratinocytes. Control keratinocytes showed increased inflammatory cytokine expression (TNF-α, TSLP, IL-6) when cells were activated with poly I:C and HDAC activity was inhibited by addition of butyrate, but the increase induced by HDAC inhibition was largely lost if MAP2K3 was silenced (Figure 3C, figure S1F). This observation supports the conclusion that MAP2K3 mediates the increase in inflammatory cytokine expression after inhibition of HDAC activity. Together with our findings that specific HDAC8 and HDAC9 silencing recapitulates the effects of butyrate, and that HDAC8 and HDAC9 associate with and alter histone acetylation of MAP2K3, our findings show that HDAC8 and HDAC9 confer tolerance to TLR stimuli by inhibition of MAP2K3 expression.
Expression of HDAC8/HDAC9 influences inflammatory responses of the skin
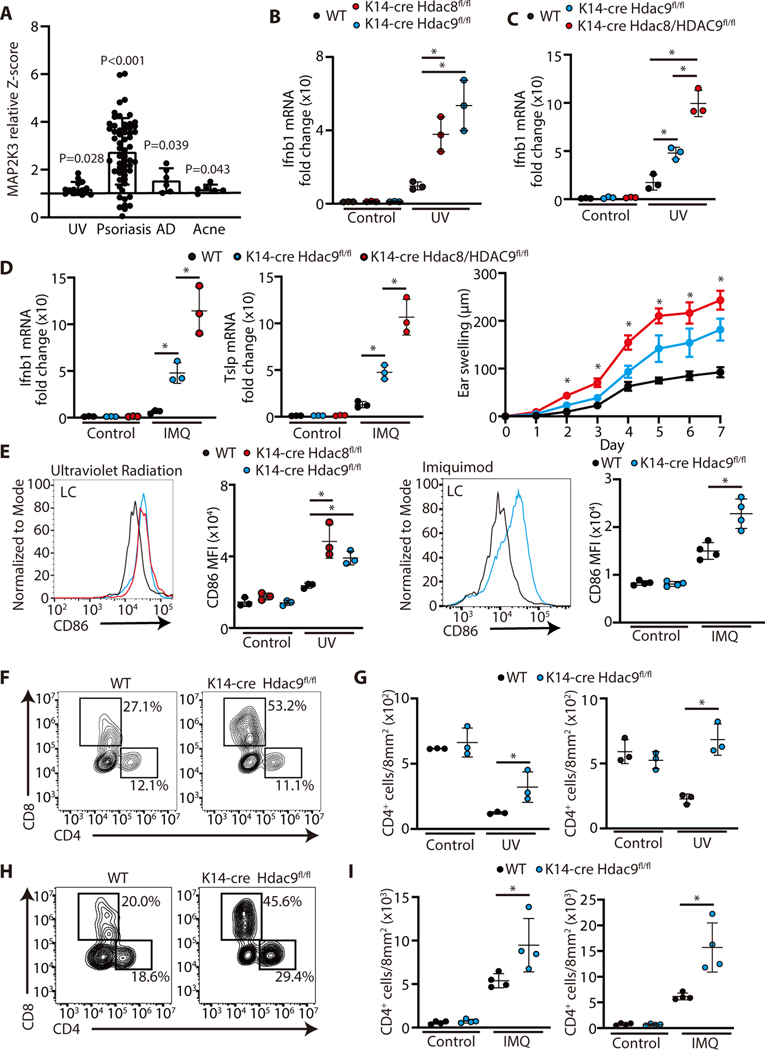
MAP2K3 has been proposed to be involved in inflammatory signaling in pulmonary epithelial cells through its action on p38MAPK (24–27). However, a role for regulation of MAP2K3 to control cytokine production by the skin was unknown. Analysis of expression of MAP2K3 in public microarray data sets from human inflammatory skin conditions (28–32) revealed that the expression of MAP2K3 was significantly upregulated in skin exposed to ultraviolet radiation and in lesional skin of patients with atopic dermatitis, psoriasis, and acne vulgaris if compared with healthy skin (Figure 4A). This observation in human clinical tissue supported the physiological relevance of MAP2K3 to skin inflammation and inspired us to further determine if HDAC8 or HDAC9 directly influence MAP2K3 expression in the skin.
Figure 4. Increased inflammatory response to UV light and imiquimod in mice after targeted deletion of HDAC 8 or HDAC9.
(A) MAP2K3 expression in human inflammatory skin disorders. Bar plots of Z scores (Y-axis) for skin taken from human healthy controls and lesional skin taken from patients with UV radiation, psoriasis, AD, and Acne Vulgaris were analyzed. Data were obtained from a public data set (GEO accession no. GDS2381, GSE13355, GDS968, and GDS2478). (Student’s t-test, two-sided). (B) Ifnb1 and Tslp gene expression in the skin of K14-Cre Hdac8 fl/fl and K14-Cre Hdac9 fl/fl mice or controls as measured by qPCR 24 hours after UVB exposure (200 mJ/cm2). (N=3; one-way ANOVA). (C) Ifnb1 gene expression in the skin of controls, K14-Cre Hdac9 fl/fl and double knockout K14-Cre Hdac8/9 fl/fl 24 hours after UV radiation. (N=3; one-way ANOVA). (D) Ifnb1 gene expression (N=3; one-way ANOVA) and ear swelling response after 7 days of topical imiquimod application to controls, K14-Cre Hdac9 fl/fl and double knockout K14-Cre Hdac8/9 fl/fl mice. (controls N=3, K14-Cre Hdac9 fl/fl mice N=3 and double knockout K14-Cre Hdac8/9 fl/fl mice N=4; one-way ANOVA). (E) Flow cytometry analysis of CD86 expression in LCs isolated from the skin 24 hours after UVB exposure or after 7 days of IMQ application. Representative flow cytometry plots are shown to the left and the mean fluorescence intensity of CD86 is shown to the right. (UV radiation experiment (N=3); one-way ANOVA, IMQ application experiment (N=4); Student’s t-test) (F,G) Flow cytometry analysis of cells gated from CD45+TCRβ1+ cells in the skin 24 hours after UVB exposure. Representative flow cytometry plots are shown as the number of CD4+ or CD8+ T-cells in the skin is shown to the right (N=3; Student’s t-test). (H,I) Flow cytometry analysis of CD45+TCRβ1+ cells in the skin 24 hours after the final IMQ application on day 7. Representative flow cytometry plots are shown in (H), and the number of CD4+ or CD8+ cells in the skin is shown in (I) (N=4; Student’s t-test).The results are expressed as the mean ± standard deviation (SD). Each dot represents an individual mouse. *, P < 0.05. Data are representative of three independent experiments.
To test the in vivo consequences of loss of HDAC8 and 9 on skin inflammation, we generated mice with skin-specific loss HDAC8 or HDAC9, or both, by breeding Hdac8fl/fl and Hdac9fl/fl with mice with a skin-specific keratin 14 Cre driver (33, 34). To test responsiveness to activation of TLR3, mice were exposed to a 200 mJ/cm2 of ultraviolet B radiation (UV). UV radiation was chosen as the initial model since this form of nonionizing radiation results in release of non-coding double stranded RNA that is recognized by TLR3 (35), and the initial inflammatory signal is focused on surface epithelial keratinocytes. To test responsiveness to TLR7, imiquimod (IMQ) was applied to the skin of the ear for 7 days. Under conventional specific pathogen-free housing conditions we did not observe a significant difference in IFN-β, TSLP, TNF-α or CCL20 expression between control mice and mice lacking HDAC8 and 9 in the skin (Figure 4B–D and fig S3A,B). However, after UV radiation or IMQ application, inflammatory cytokine gene expression was significantly increased in mice without HDAC8 and 9 in keratinocytes (Figure 4B–D and fig S3A,B). Mice lacking both HDAC8 and HDAC9 had a greater increase than loss of HDAC9 alone (Figure 4C,D). These data suggested that tolerance to inflammation by UV damage or IMQ was lost in the absence of HDAC8 and 9.
To further define the cell inflammatory responses in vivo to loss of HDAC8 or 9, dendritic cell (DC) activation in the skin was assessed by FACS analysis of CD86. No change in Langerhans cells, Langerin+ dermal DCs, and Langerin- dermal DCs was observed in the absence of an inflammatory challenge (Figure 4E, fig S3C,D). However, consistent with prior observations of inflammatory cytokine responses and tissue inflammation, UV or IMQ exposure of K14-Cre Hdac9fl/fl and K14-Cre Hdac8fl/fl mice showed an increase in each DC subset compared with WT mice (Figure 4E, fig S3C–F and fig S4).
We next examined the T cell response in mouse skin lacking HDAC9. In normal skin, UV radiation results in an expected decrease in T cells (36, 37), however the number of CD4+ and CD8+ cells increased in K14-Cre Hdac9fl/fl after UV radiation (Figure 4F,G). Loss of HDAC9 in the skin also resulted in an increase in CD4+ and CD8+ cells in mice exposed to topical IMQ (Figure 4H,I). Together, these observations show that the expression of HDAC8/9 in keratinocytes decreased the inflammatory response of the skin to UV radiation (triggered in part by TLR3), and IMQ, a TLR7-specific chemical ligand.
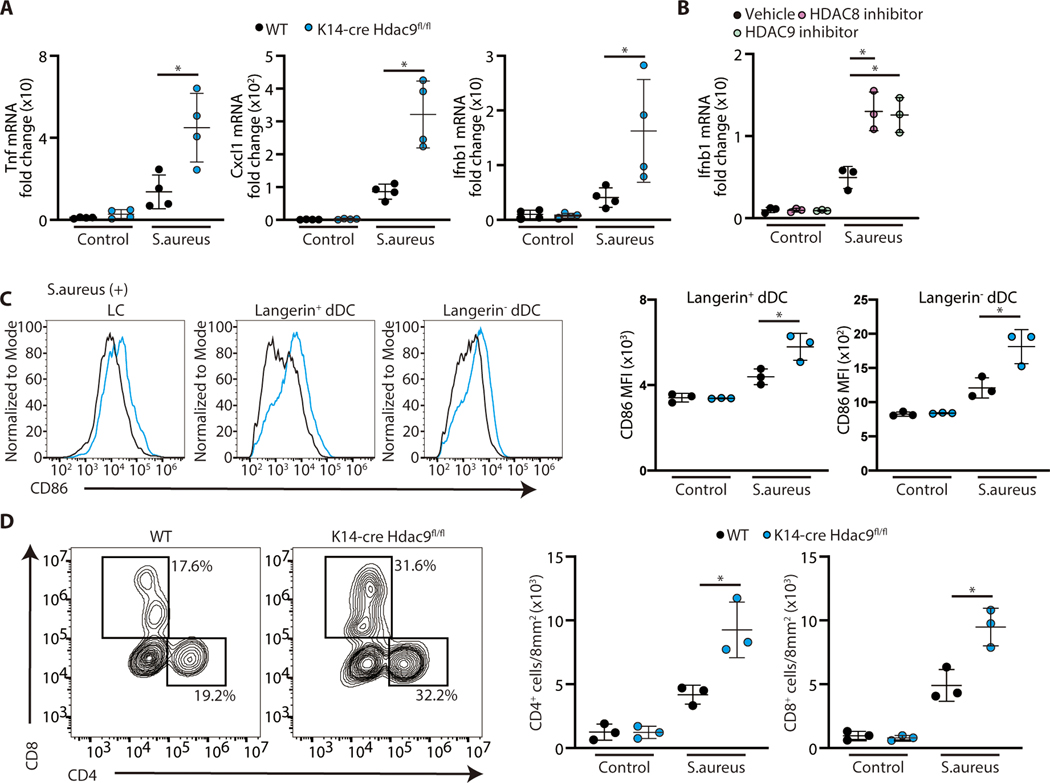
We next sought to determine if the specific activity of HDAC8/9 influences responses to challenge by an infectious pathogen that can be detected by TLR2. To do this we examined inflammation in response to S. aureus (Figure 5). Similar to the responses to UV and IMQ, topical application of S. aureus to K14-Cre Hdac9fl/fl mice resulted in increased cytokine expression (Figure 5A,). Topical application of HDAC8 or HDAC9 selective inhibitors also increased IFN-β expression by the skin (Figure 5B). In addition K14-Cre Hdac9fl/fl mice showed evidence of increased DC activation (Figure 5C) and accumulation of CD4 and CD8+ T cells (Figure 5D) compared to wild type mice. Taken together, observations from 3 distinct mouse models of inflammation confirmed a role for HDAC8 and HDAC9 to suppress cutaneous inflammation and promote tolerance to a variety of inflammatory stimuli.
Figure 5. Increased inflammatory response to S. aureus after targeted deletion of HDAC 8 or HDAC9.
(A) Inflammatory gene expression in skin measured by qPCR 24 hours after topical S. aureus application (N=3; Student’s t-test). (B) IFN-β gene expression in PCI34051 or TMP269 -treated skin by qPCR 24 hours after topical S. aureus application (N=3; one-way ANOVA). (C) Flow cytometry analysis of CD86 expression in skin 24 hours after topical S. aureus application. Representative flow cytometry plots are shown in (C, left), and the mean fluorescence intensity of CD86 is shown in (C, right) (N=3; Student’s t-test). (D) Flow cytometry analysis of CD45+TCRβ1+ cells in the skin 24 hours after topical S. aureus application. Representative flow cytometry plots are shown in (D, left), and the number of CD4+ or CD8+ cells in the skin is shown in (D, right) (N=3; Student’s t-test). The results are expressed as the mean ± standard deviation (SD). Each dot represents an individual mouse. *, P < 0.05. Data are representative of three independent experiments.
IFN-β is responsible for DC activation by keratinocytes after HDAC8 and HDAC9 silencing
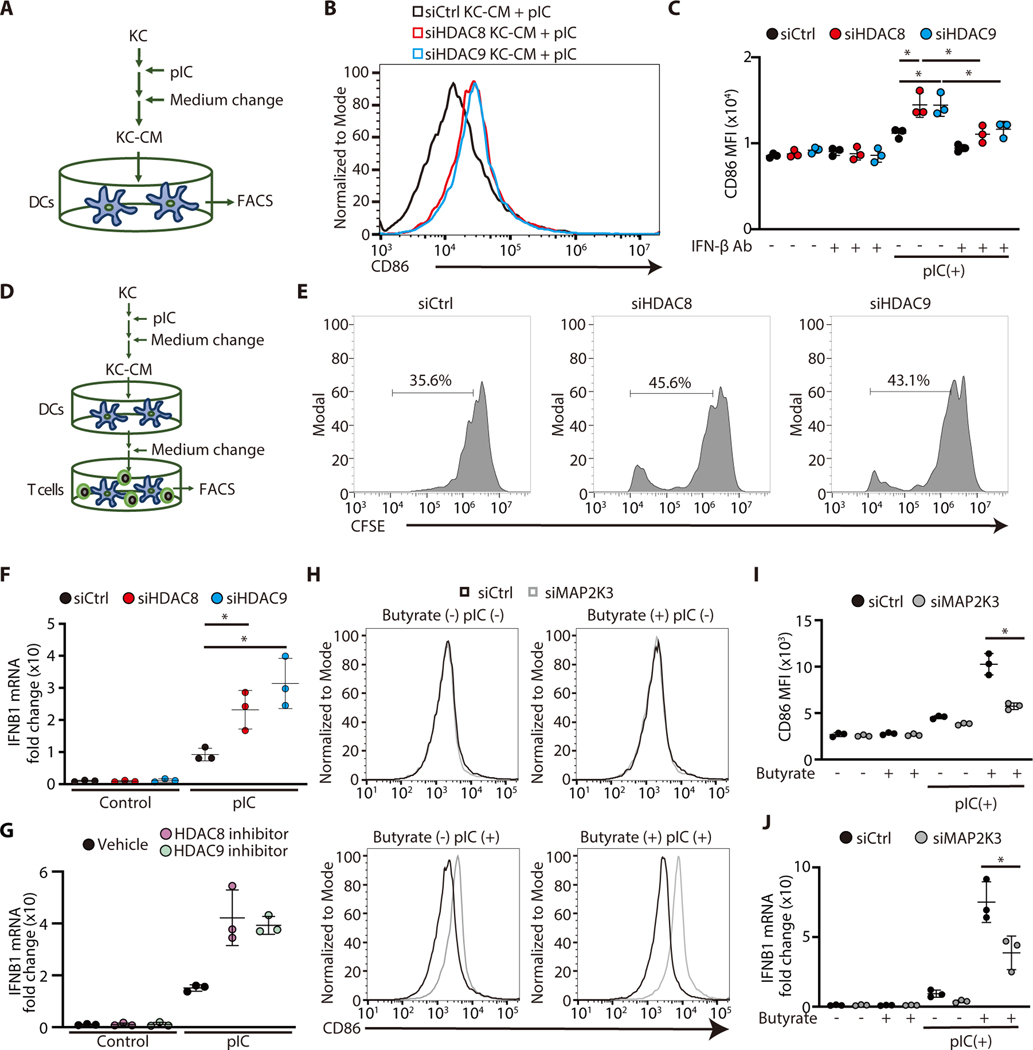
To further clarify how the activity of HDAC8 or 9 in keratinocytes can exert an effect on immune cells, human monocyte-derived DCs (MoDCs) were generated from peripheral blood then cultured with the condition media (CM) obtained from keratinocytes after silencing of HDAC8 or HDAC9 (Figure 6A). CD86 expression was increased on MoDCs after exposure to CM from keratinocytes after silencing of HDAC8 or 9 (Figure 6B, C). Addition of a neutralizing antibody to IFN-β abrogated this effect of keratinocyte CM on MoDCs (Figure 6C). This finding is consistent with observations that IFN-β is one of the factors derived from keratinocytes responsible for activation of DCs (8). Furthermore, we tested if these activated DC could influence T cell proliferation (Figure 6D). An increase in T-cell proliferation was observed in cells exposed to MoDC CM after activation by the CM of keratinocytes lacking HDAC8 or 9 (Figure 6E). Consistent with the earlier finding that neutralization of IFN-β could decrease DC activation, gene expression of IFN-β was significantly increased after silencing or application of inhibitors of HDAC8 and HDAC9 (Figure 6F, G). However, while CD86 expression on DC was significantly increased by CM derived from keratinocytes after inhibition of HDAC activity, this effect was canceled after silencing of MAP2K3 in keratinocytes (Figure 6H, I). Furthermore, IFNB1 gene expression was significantly suppressed by MAP2K3 silencing (Figure 6J). These findings provide a new model to explain how keratinocytes regulate immune responses by the action of HDAC8 and HDAC9 on MAP2K3 activity and subsequent production of IFN-β (Figure 7).
Figure 6. HDAC8/HDAC9 inhibits DCs activation by keratinocyte IFN-β.
(A) Schematic showing experimental approach; nHEKs (KC) were activated by poly I:C, then KC-conditioned media (KC-CM) was transferred to human monocyte-derived DCs for analysis by FACS. (B) Representative FACS histogram of CD86 expression in human monocyte-derived DCs 24 hours after stimulation with KC-CM from HDAC8 or HDAC9 silenced nHEKs. (C) Mean fluorescence intensity from experiments shown in B (N=3; one-way ANOVA). (D) Schematic showing experimental approach: as in A with transfer of DC to CD3+ cells for T cell proliferation assay. (E) The proliferation of CFSE-labeled CD3+ cells as analyzed by flow cytometry. (F, G) IFNB1 gene expression in nHEKs measured by qPCR after silencing of HDAC8/9 in (F) or treatment with PCI34051 (10μM) or TMP269 (1μM) in (G), followed by culture with or without poly I:C (1 μg/ml) (N=3; one-way ANOVA). (H, I) Flow cytometry analysis of CD86+ human monocyte-derived DCs stimulated with CM from nHEK after silencing of MAP2K3. nHEK CM was cocultured with peripheral blood CD3+ cells for 5 days. Representative flow cytometry histogram is shown in (H), and the mean fluorescence intensity is shown in (I) (N=3; Student’s t-test). (J) IFNB1 mRNA expression in nHEK measured by qPCR following MAP2K3 silencing and treatment with Poly I:C for 4 hours and after butyrate treatment (2mM) (N=3; Student’s t-test). The results are expressed as the mean ± standard deviation (SD). *, P < 0.05. Data are representative of three independent experiments.
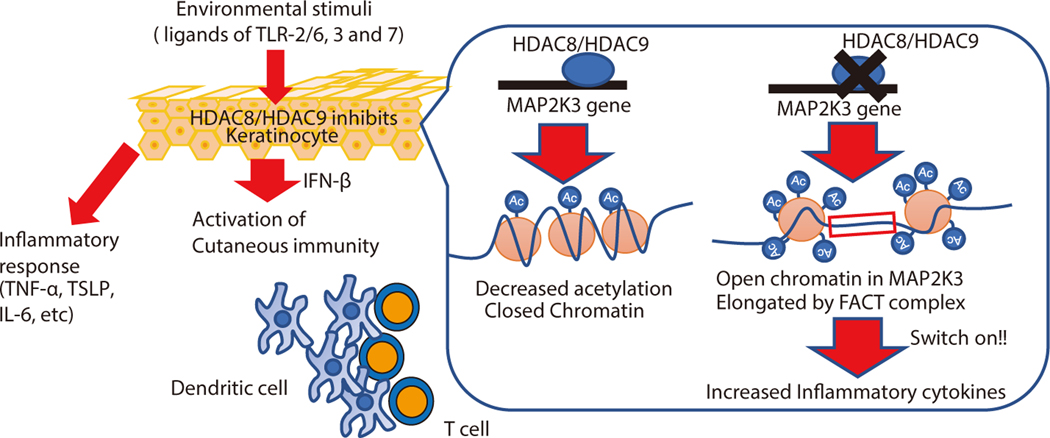
Figure 7. Model for how HDAC8 and 9 maintain immune tolerance in the skin.
HDAC8 and HDAC9 are expressed in keratinocytes and decrease cytokine expression in response to commonly encountered inflammatory stimuli. When these HDACs are inhibited, keratinocytes mount a greater inflammatory response to stimulation by TLR ligands. This response includes release of IFN-β that enables a DC maturation and subsequent T cell proliferation. HDAC8 and HDAC9 act to decrease acetylation of the MAP2K3 gene. Inhibition of HDAC8 and HDAC9 leads to increased acetylation, activation of the FACT complex and subsequent enhanced transcription of MAP2K3. This primes keratinocytes to increase cytokine expression in response to TLR stimuli.
Discussion
Our study provides new insight into how the skin surface tolerates constant exposure to environmental and microbial compounds that would otherwise be expected to initiate a brisk inflammatory response. Inappropriate inflammatory responses of the epidermis are a hallmark of several human diseases, yet the factors that distinguish keratinocyte innate immune responses from classical immunocytes such as resident macrophages and dendritic cells are poorly understood. Here, we show that transcriptional expression of MAP2K3 is a target for suppression by the action of HDAC8 and HDAC9. Our observations also illustrate how the FACT proteins, SSRP1 and SUPT16H, are critical for the expression of MAP2K3 in keratinocytes, thus defining additional checkpoints in this system. Furthermore, we show that HDAC8- and HDAC9-deficient keratinocytes produce increased IFN-β that then activates DC function to subsequently lead to T cell proliferation. These observations provide new insight into maintenance of innate immune tolerance.
Several independent lines of evidence support the conclusion that MAP2K3 is directly affected by HDAC8 and HDAC9, thus providing epigenetic control of keratinocyte immune function. There are few reported studies focused on MAP2K3-related skin diseases (38). Our analysis of previously published transcriptional data from human skin diseases showed expression of MAP2K3 increases in major inflammatory skin diseases such as atopic dermatitis, psoriasis, and acne vulgaris. This association is consistent with our conclusions that regulation of MAP2K3 function may be a key checkpoint for maintaining homeostasis. Furthermore, additional studies have shown that inhibition of the MAP2K3 downstream target, p38MAPK, impairs inflammatory responses in IMQ-induced psoriasis inflammation (39), C. acnes-induced skin inflammation (40) and UV radiation-induced skin inflammation (41). Thus, both MAP2K3 and p38MAPK inhibition are potential therapeutic strategies for control of unwanted skin inflammation.
A putative binding site for HDAC8 and HDAC9 was identified in a location distant from the promoter region of MAP2K3 gene and may represent an enhancer location. However, both H3K9ac and H3K27ac marks in the promoter region of MAP2K3 were increased after silencing of HDAC8 and HDAC9. Therefore, we believe both HDAC8 and HDAC9 can influence acetylation in promoter region of MAP2K3. We speculate that a DNA loop structure may be involved that would lead to contact between the HDAC8/9 binding region and the MAP2K3 promoter region (42). Although we could not identify its mechanism in this study, further analysis of the structure of the MAP2K3 gene is warranted.
It remains unclear what the natural impact is of HDAC inhibition by SCFAs derived from skin commensal bacteria. Under normal physiological conditions, SCFAs produced in the skin should easily penetrate into the dermis due to their low molecular weight less of than 500 Da (43). Indeed, the topical application of butyrate impairs acquired immune responses in the skin (44), This observation is consistent with the opposing effects that inhibition of HDACs have on bone marrow-derived immunocytes such as macrophages (13). Unlike keratinocytes that become less tolerant of TLR stimuli upon inhibition of HDAC, macrophage responses are inhibited due to increased expression of repressor genes. A complex contextual immune regulatory system within the 3D structure of the skin must tolerate danger signals at the surface and appropriately respond to danger. In the example of the skin disease acne, it has been assumed that occluded hair follicles under fermentation conditions metabolize lipid-enriched materials secreted from sebaceous glands into SCFAs. Because the lipid-enriched molecules are able to retain epidermis or stratum corneum (45), SCFAs in hair follicles at sufficient concentrations can then exert cell-dominant effects on the keratinocytes rather than the dermal cells. From the perspective of microbial ecology, production of SCFAs might be have some advantages for commensal bacteria to survive at the expense of other bacteria if an increased host inflammatory response selectively targets the competition.
The different inflammatory responses between epithelial cells and bone marrow cells illustrate how epigenetic modifications are cell type-dependent and can have opposite effects. In macrophages, the presence of the corepressor and the nucleosome remodeling and deacetylase complex (NuRD) suppresses expression of inflammatory cytokines (46) (47). Inhibition of HDAC activity will increase NuRD expression and therefore suppress expression of inflammatory cytokines. In contrast, this complex is not active in keratinocytes (13), and inhibition of HDAC8 and HDAC9 positively drives inflammation, a response that is opposite from macrophages. Therefore, similar epigenetic changes can have either a pro-inflammatory or anti-inflammatory response depending on the cell type that is targeted.
The FACT complex is involved in transcription-related chromatin dynamics (48), promotes transcriptional elongation, and contributes to the development of liver cancer (49). Our observations in keratinocytes suggest HDAC8/9 and the FACT complex are powerful genetic regulators that have opposite effects. This may enable rapid acceleration of inflammatory responses upon exposure to SCFAs. Prior reports have suggested inhibition of the FACT complex is beneficial in an atopic dermatitis model (50), an IMQ-induced psoriasis model (51) and following UV radiation (52). A clinical trial in psoriasis patients of oral administration of a FACT complex inhibitor was not therapeutically successful (53). Clearly, more work is needed to understand the role of components of the FACT complex, but our findings suggest that local topical application of agents targeting the pathways defined in this study could be useful for the treatment of inflammatory skin diseases.
Methods and Methods
Study design
The aim of this study was to investigate the role of HDAC8 and HDAC9 in skin inflammation. We performed inhibited expression of HDAC8 or HDAC9 in keratinocytes and evaluated inflammatory responses after exposure to TLR2/6, TLR3, or TLR7 ligands. ChIP-seq and signal pathway analysis by RNA-seq, as well as antibody pull-down and mass spectrometry were used to identify the targets of HDAC8 and HDAC9. Mice deficient in HDAC8 and/or HDAC9 were used to assess the in vivo contribution of HDAC8/9 to inflammatory triggers including UV radiation, IMQ and topical S. aureus. Keratinocytes were assessed in vitro to determine how loss of HDAC8/9 activity can activate dendritic cells and subsequent T-cell proliferation.
Animals and animal care
All experiments involving mice were done with the approval of the Institutional Animal Care and Use Guidelines of the University of California, San Diego (Protocol number: S09074). Eight- to 12-week-old mice were used in this study. All animals were in the C57BL/6 genetic background. K14-Cre mice were purchased from Jackson Laboratory (Bar Harbor, ME). Hdac8fl/fl mice were purchased from Taconic Biosciences (Hudson, NY) under MTA. Hdac9fl/fl mice were provided from Augusta University and originally generated by Prof. Neal L. Weintraub. To prepare keratinocyte-specific Hdac8- and Hdac9-deficient mice, mice bearing the Cre recombinase transgene, under the control of the keratin 14 promoter (K14-Cre), were mated with Hdac8fl/fl or Hdac9fl/fl mice.
Mouse inflammation induced by UV radiation, IMQ application or S. aureus
UV radiation was performed as previously described with some modification (54). Briefly, 92 hours after the treatment of hair shave (Wahl, Sterling, IL)) and chemical hair remover (Church & Dwight Co., Inc., Princeton, NJ), 200 mJ/cm2 of UV was radiated on back skin by a UV radiator (Spectroline Model EB-280C UV lamp; Spectronics Corp., Westbury, NY). The total amount of UV was measured by a solar meter (Solartech Inc., Harrison Township, MI). The skin sample was collected for further experiments. IMQ (10 mg) was daily applied on the ear skin for 6 consecutive days and the ear thickness was measured by a thickness gauge (Teclock, Japan). The skin sample was collected on Day 7 for further experiments.
After shaving of back skin and application of a chemical depilatory agent, S. aureus USA300 (106 CFU) was placed on 8mm2 TSB agar, which was applied on the back skin by the fixation of Tegaderm (3M Healthcare, Germany). The skin sample was collected 24 h after the treatment for further experiments.
Topical HDAC inhibitor treatment was conducted for 24 hours before S. aureus application. Agar plates (2%) were made by using UltraPure Agarose (Invitrogen) containing PCI34051 (1 mM) and TMP269 (100 μM). 8 mm agar discs were made to include 10 μl of distilled H2O (dH2O) or PCI34051 (1 mM) or TMP269 (100 μM). These discs were then topically applied to the dorsal skin and fixed in the place using Tegaderm (3M Company).
Keratinocyte culture
Normal human epidermal keratinocytes (nHEKs) (Thermo Fisher Scientific, Waltham, MA) were cultured with EpiLife medium supplemented with 60 mM CaCl2 (Thermo Fisher Scientific), 1×EpiLife Defined Growth Supplement (Thermo Fisher Scientific), and 1× antibiotic/antimycotic (Thermo Fisher Scientific).
Silencing HDAC8, HDAC9, MAP2K3, SSRP1, and SUPT16H by siRNA
HDAC8, HDAC9, and MAP2K3 in nHEKs were knocked down using Silencer Select siRNA (Thermo Fisher Scientific) according to the manufacturer’s protocol with some modifications. Briefly, 100 nM siRNA (Thermo Fisher Scientific) in Opti-MEM (Thermo Fisher Scientific) with 2.5% Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) were incubated at room temperature for 5 min and then, diluted in 1:10 ratio in EpiLife culture medium supplemented with 1×EpiLife Defined Growth Supplement, and 1× antibiotic/antimycotic to set a final concentration of siRNA and Lipofectamine as 10 nM and 0.25%, respectively. After siRNA treatment for 16 hours, siRNA-treated was recovered for 48 to 72 hours before further experiments.
Quantitative real-time PCR
Total RNA was extracted from the skin using a Trizol RNA extraction reagent (Invitrogen, Carlsbad, CA). Cultured cells were lysed, and RNA was isolated using the PureLink RNA Isolation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Complementary DNA (cDNA) was reverse transcribed from the total RNA samples using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Berkeley, CA). cDNA was used in quantitative real-time PCR using TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) and TaqMan Gene Expression Assays (Thermo Fisher Scientific) or iTaq Universal SYBR Green Supermix (Bio-Rad). The gene expression was measured by a CFX96 Real-Time System (Bio-Rad) and was determined by ΔΔCt. The results were normalized to those of the housekeeping glyceraldehyde-3-phosphate dehydrogenase mRNA.
Immunoblotting
nHEKs were lysed by RIPA buffer (Thermo Fisher Scientifics) with protease inhibitor (Sigma-Aldrich). After passing the lysate 20 times through a 21-gauge needle attached to an RNase-free syringe (BD, Franklin Lakes, NJ), lysates were sonicated in ice water for 20 min. After centrifuging for 20 min 16000 rpm, the supernatant was collected. Protein concentration was calculated by a BCA assay (Pierce Biotechnology, Rockford, IL). Samples were mixed with 4x Laemmli sample buffer (Bio-Rad) and were incubated 95°C for 10 min. Proteins from the total cell lysate were separated by Mini-Protein TGX precast gel (Bio-Rad), transferred to the Trans-blot Turbo mini PVDF membrane (Bio-Rad) using Trans-Blot Turbo Instrument (Bio-Rad). The membrane was washed by Odyssey blocking solution (LI-COR Biosciences, Lincoln, NE) for 1 hour at room temperature, and then was stained with anti-MAP2K3 (Abcam, Cambridge, MA), anti-phospho-p38MAPK, anti-p38MAPK (Cell Signaling, Danvers, MA), anti-HDAC8 or anti-HDAC9 antibodies (Abcam) and subsequently treated with secondary antibody (LI-COR Biosciences). GAPDH was used as internal control and was stained by anti-GAPDH Ab (Fitzgerald Industries, Concord, MA). The membrane was washed and the immunoblot signals were detected by the Odyssey infrared imaging system (LI-COR Biosciences).
ChIP-PCR
ChIP-qPCR was performed as previously described (13). Samples were cross-linked in 1% formaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature, and the reaction was quenched by 125 mM glycine for 5 min at room temperature. After washing with PBS twice, adhesive cells were scraped by cell scraper (Thermo Fisher Scientific) and the sample pellet was collected into 10 ml PBS in a conical tube by centrifuge 600g for 5 min. The cell pellet was lysed by lysis buffer (5 mM PIPES (pH 8.0), 85 mM KCl, 0.5% NP-40) with 1x protease inhibitor (Sigma-Aldrich) on ice for 5 min and the pellet was collected by centrifuge 400g for 5 min. After resuspended by RIPA buffer with 1x protease inhibitor (Sigma-Aldrich), the sample was sonicated by a Diagenode Bioruptor sonicator (4 × 7.5-min cycles of 30 s on, 60 s off) (Diagenode Inc., Sparta, NJ), and centrifuge at 16,000g for 15 min at 4°C. The supernatant was collected and then was used for immunoprecipitation treatment. The sonicated sample was incubated with beads conjugated anti-HDAC8, anti-HDAC9, anti-H3K9ac, anti-H3K27ac Abs (Abcam), or isotype control IgG at 4°C overnight. After washed with wash buffer (100 mM tris (pH 7.5), 500 mM LiCl, 1% NP-40, 1% sodium deoxycholate) and once with Tris-EDTA, beads were suspended with 200 μL immunoprecipitation elution buffer (1% sodium deoxycholate, 0.1 M NaHCO3). The cross-linked chromatin was eluted from the beads by incubation at 65oC for 1 hour and centrifugation at 16000g for 3 min. the supernatant was collected and cross-link reversal treatment was performed by incubation at 65oC overnight. Immunoprecipitated DNA was purified using a Qiagen cleanup kit (Qiagen, Hilden, Germany) and samples were used for ChIP-seq or qPCR along with input DNA.
FACS analysis
To prepare single-cell suspensions, skin sample was taken from back skin by 8 mm2 disposable biopsy punches (Integra-Miltex, York, PA) and was treated by 2.5 mg/ml collagenase D (Sigma-Aldrich) and 30 ng/mL DNAse1 (MP Biomedicals, Solon, OH) for 2 hours at 37°C. After filtration by 40-μm of cell strainer (Thermo Fisher Scientific), skin suspension cells were stained by various combinations of fluorescence-conjugated monoclonal antibodies and were analyzed by ZE5 Cell Analyzer (Bio-Rad) and FlowJo software (TreeStar, San Carlos, CA). For cell analysis, the following antibodies were used: APC-conjugated anti-CD11c and Langerin mAbs (Biolegend), APC-eFluor 780-conjugated anti-CD4 mAb (Invitrogen), BV-conjugated anti-CD45 mAb (Biolegend), BV680-conjugated anti-CD8 mAb (Biolegend), FITC-conjugated anti-CD86 (Biolegend), HLA-DR (Biolegend), and MHC class II (eBioscience, San Diego, CA) mAbs, PE-conjugated anti-EpCAM and CD86 mAbs (eBioscience). Per-CP5.5-conjugated anti-CD11c (eBioscience), and TCRβ (Invitrogen) mAbs. The cell number was counted using CountBright Absolute Counting Beads (Thermo Fisher Scientific).
RNA-seq analysis
RNA extracts taken from nHEKs were submitted to the University of California, San Diego (UCSD), Institute for Genomic Medicine core facility for quality analysis, library preparation, and high-throughput sequencing. The RNA quality was assessed using the Agilent TapeStation instrument (Agilent Technologies, Santa Clara, CA). Libraries were constructed using TruSeq Stranded mRNA Library Prep Kits (Illumina, San Diego, CA) and were analyzed on a HiSeq 4000 instrument (Illumina). The raw data obtained from UCSD, Institute for Genomic Medicine core facility, were analyzed using Partek Flow software (Partek Inc., St Louis, MO) to determine transcript abundance and identify differentially expressed genes.
MoDC preparation
Monocytes were isolated from peripheral blood mononuclear cells (PBMCs) obtained from healthy blood donors by the Ficoll-Paque method (Pharmacia, Uppsala, Sweden). CD14+ cells purified with MACS bead (Miltenyi Biotec, Auburn, CA) were cultured in complete RPMI (RPMI 1640; Sigma-Aldrich, St. Louis, MO) containing 10% heat-inactivated fetal calf serum (Gemini Bioproducts, Calabasas, CA), 50 μM 2-mercaptoethanol (Sigma-Aldrich), 2 mM l-glutamine (Corning), 25 mM N-2-hydroxyethylpiperazine-N′−2-ethanesulfonic acid, 1 mM nonessential amino acids (Life Technologies), 1 mM sodium pyruvate (GE Healthcare), 100 U/ml penicillin, and 100 μg/ml streptomycin (Life Technologies), supplemented with 100 ng/ml recombinant human GM-CSF (R&D Systems, Minneapolis, MN) and 50 ng/ml recombinant human IL-4 (R&D Systems) for 7 days.
DC activation and T cell proliferation
MoDC was cocultured with the CM from nHEKs with poly I:C (Invitrogen) and/or a neutralizing anti-IFN-β antibody (Biolegend, San Diego, CA) for 24 hours and then the degree of CD86 expression was counted by flow cytometry as mean fluorescence intensity (MFI). In addition, to measure the degree of T cell proliferation, autologous naïve CD3+ cells were sorted from human peripheral blood using MACS bead (Miltenyi Biotec) were stained by CellTrace CFSE (Thermo Fisher Scientific). After stimulation with KC-CM and/or IFN-β neutralizing antibody for 24 hours, MoDCs were then washed to remove KC-CM before adding to CD3+ cells in a ratio of 1:5 (DCs: T cells) under KC-CM stimulation. Five days after the culture, the intensity of CFSE fluorescence was determined by flow cytometry.
Immunoprecipitation for mass spectrometry
nHEKs were washed with cold PBS and scraped from the plates using a cell scraper. After transferring the cells from the plates to a 15 ml conical tube, samples were centrifuged for 5 mins 500g at 4°C. After the aspiration of supernatant, cell pellets were frozen on dry ice and were transferred to −80°C for storage until ready for cell lysis and affinity purification. Pellets were treated with lysis buffer (50 mM Hepes-NaOH pH 8.0, 100 mM KCl, 2 mM EDTA, 0.1% NP-40 and 10% glycerol) with protease inhibitor (Sigma-Aldrich). After passing the lysate 20 times through a 21-gauge needle attached to an RNase-free syringe (BD, Franklin Lakes, NJ), lysates were performed two freeze-thaw cycles treatment by incubating the tube on dry ice for 10 min, then transfer it to a 37°C water bath. After centrifuging for 20 min 16000 rpm at 4°C, the supernatant was collected and used for immunoprecipitation treatment by incubation with bead-conjugated anti-HDAC8, anti-HDAC9 (Abcam), or isotype control IgG at 4°C overnight. After three washes with lysis buffer containing 50 mM ammonium bicarbonate, bead-conjugated proteins were used for mass spectrometry analysis.
4sUDRB elongation assay
First, the gene transcription was inhibited by 5,6-dichlorobenzimidazole 1-β-D-ribofuranoside (DRB) (Sigma-Aldrich) at a final concentration of 100 μM for 3 hours. After 4-thiouridine (Sigma-Aldrich) was pulsed at a final concentration of 1 mM to label newly transcript RNA. Total RNA was extracted with the miRNeasy kit (Qiagen) 0 min, 4 min, and 8 min after DRB removal. 4sU-labeled RNA was biotinylated by EZ-Link Biotin-HPDP (Thermo Fisher Scientific) dissolved in dimethylformamide (Thermo Fisher Scientific) and purified by Dynabeads MyOne Streptavidin T1 beads (Life Technologies). 4sU-labeled RNA was eluted with 100 mM dithiothreitol (Thermo Fisher Scientific). After cleaning up by RNeasy MinElute Kit (Qiagen), eluted RNA was used for qPCR amplifications by using specific primers.
Microarray data analysis
Expression data for patients with atopic dermatitis, psoriasis, allergic contact dermatitis, and UV light radiation were obtained from a public data set deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (GEO accession no. GDS2381, GSE13355, GDS968, and GDS2478), and Patient and sample information was previously described (29, 31, 32, 55).
Statistical analysis
All statistical analyses were calculated using GraphPad Prism 6.05 (GraphPad Prism Software Inc., La Jolla, CA). The Student’s t-test was used to calculate significant differences between the two groups. Statistical significance between multiple groups was analyzed using the ANOVA comparisons test. All P-values less than 0.05 were considered statistically significant.
Supplementary Material
Table S1. Raw data file (Excel spreadsheet)
Acknowledgments:
We appreciate Prof. Neal L. Weintraub in Augusta University for providing Hdac9fl/fl mice.
Funding: RLG is funded by National Institutes of Health grants R01AR069653, R01AR074302, R01AR076082, R37AI052453 and U01AI52038. YS is supported by JSPS overseas fellowship.
Footnotes
Competing Interests: RLG is a co-founder, scientific advisor, consultant and has equity in MatriSys Bioscience and is a consultant, receives income and has equity in Sente Inc. The other authors declare no competing interests.
Data and Materials Availability: RNA-seq data were deposited in the NCBI GEO database (GEO accession no. GSE154163). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Dainichi T. et al. , The epithelial immune microenvironment (EIME) in atopic dermatitis and psoriasis. Nat Immunol 19, 1286–1298 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Genuis SJ, Sensitivity-related illness: the escalating pandemic of allergy, food intolerance and chemical sensitivity. Sci Total Environ 408, 6047–6061 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Alferink J. et al. , Control of neonatal tolerance to tissue antigens by peripheral T cell trafficking. Science 282, 1338–1341 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Zhivaki D, Lo-Man R, Unique aspects of the perinatal immune system. Nat Rev Immunol 17, 495–507 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Chen YE, Fischbach MA, Belkaid Y, Skin microbiota-host interactions. Nature 553, 427–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scharschmidt TC, Establishing Tolerance to Commensal Skin Bacteria: Timing Is Everything. Dermatol Clin 35, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kashiwagi M. et al. , Direct control of regulatory T cells by keratinocytes. Nat Immunol 18, 334–343 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang LJ et al. , Antimicrobial Peptide LL37 and MAVS Signaling Drive Interferon-beta Production by Epidermal Keratinocytes during Skin Injury. Immunity 45, 119–130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volpe E. et al. , Thymic stromal lymphopoietin links keratinocytes and dendritic cell-derived IL-23 in patients with psoriasis. J Allergy Clin Immunol 134, 373–381 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Arpaia N. et al. , Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu M. et al. , Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PloS one 8, e55380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furusawa Y. et al. , Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Sanford JA et al. , Inhibition of HDAC8 and HDAC9 by microbial short-chain fatty acids breaks immune tolerance of the epidermis to TLR ligands. Sci Immunol 1, (2016). [DOI] [PubMed] [Google Scholar]
- 14.Boucheron N. et al. , CD4(+) T cell lineage integrity is controlled by the histone deacetylases HDAC1 and HDAC2. Nat Immunol 15, 439–448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L. et al. , FOXP3+ regulatory T cell development and function require histone/protein deacetylase 3. J Clin Invest 125, 1111–1123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Zoeten EF et al. , Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3(+) T-regulatory cells. Mol Cell Biol 31, 2066–2078 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarro MN, Goebel J, Feijoo-Carnero C, Morrice N, Cantrell DA, Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat Immunol 12, 352–361 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanford JA, O’Neill AM, Zouboulis CC, Gallo RL, Short-Chain Fatty Acids from Cutibacterium acnes Activate Both a Canonical and Epigenetic Inflammatory Response in Human Sebocytes. J Immunol 202, 1767–1776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahiya S. et al. , HDAC10 deletion promotes Foxp3(+) T-regulatory cell function. Sci Rep 10, 424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du T, Zhou G, Khan S, Gu H, Roizman B, Disruption of HDAC/CoREST/REST repressor by dnREST reduces genome silencing and increases virulence of herpes simplex virus. Proc Natl Acad Sci U S A 107, 15904–15909 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laherty CD et al. , Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89, 349–356 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D, The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400, 284–288 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Pathak R. et al. , Acetylation-Dependent Recruitment of the FACT Complex and Its Role in Regulating Pol II Occupancy Genome-Wide in Saccharomyces cerevisiae. Genetics 209, 743–756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derijard B. et al. , Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science 267, 682–685 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ, MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol 16, 1247–1255 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mannam P. et al. , Endothelial MKK3 is a critical mediator of lethal murine endotoxemia and acute lung injury. J Immunol 190, 1264–1275 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otterbein LE et al. , MKK3 mitogen-activated protein kinase pathway mediates carbon monoxide-induced protection against oxidant-induced lung injury. Am J Pathol 163, 2555–2563 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esaki H. et al. , Identification of novel immune and barrier genes in atopic dermatitis by means of laser capture microdissection. J Allergy Clin Immunol 135, 153–163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nair RP et al. , Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet 41, 199–204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen MB, Skov L, Menne T, Johansen JD, Olsen J, Gene expression time course in the human skin during elicitation of allergic contact dermatitis. J Invest Dermatol 127, 2585–2595 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Rieger KE et al. , Toxicity from radiation therapy associated with abnormal transcriptional responses to DNA damage. Proc Natl Acad Sci U S A 101, 6635–6640 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trivedi NR, Gilliland KL, Zhao W, Liu W, Thiboutot DM, Gene array expression profiling in acne lesions reveals marked upregulation of genes involved in inflammation and matrix remodeling. J Invest Dermatol 126, 1071–1079 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Kuraguchi M. et al. , Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS genetics 2, e146 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vassar R, Rosenberg M, Ross S, Tyner A, Fuchs E, Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc Natl Acad Sci U S A 86, 1563–1567 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernard JJ et al. , Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat Med 18, 1286–1290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caricchio R, Reap EA, Cohen PL, Fas/Fas ligand interactions are involved in ultraviolet-B-induced human lymphocyte apoptosis. J Immunol 161, 241–251 (1998). [PubMed] [Google Scholar]
- 37.Ozawa M. et al. , 312-nanometer ultraviolet B light (narrow-band UVB) induces apoptosis of T cells within psoriatic lesions. J Exp Med 189, 711–718 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W. et al. , The p38-MAPK/SAPK pathway is required for human keratinocyte migration on dermal collagen. J Invest Dermatol 117, 1601–1611 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Sakurai K. et al. , Cutaneous p38 mitogen-activated protein kinase activation triggers psoriatic dermatitis. J Allergy Clin Immunol 144, 1036–1049 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Weber JS et al. , Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol 16, 375–384 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Hildesheim J, Awwad RT, Fornace AJ Jr., p38 Mitogen-activated protein kinase inhibitor protects the epidermis against the acute damaging effects of ultraviolet irradiation by blocking apoptosis and inflammatory responses. J Invest Dermatol 122, 497–502 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Brown JM et al. , A tissue-specific self-interacting chromatin domain forms independently of enhancer-promoter interactions. Nat Commun 9, 3849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bos JD, Meinardi MM, The 500 Dalton rule for the skin penetration of chemical compounds and drugs. Exp Dermatol 9, 165–169 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Schwarz A, Bruhs A, Schwarz T, The Short-Chain Fatty Acid Sodium Butyrate Functions as a Regulator of the Skin Immune System. J Invest Dermatol 137, 855–864 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Savic V. et al. , Tacrolimus-loaded lecithin-based nanostructured lipid carrier and nanoemulsion with propylene glycol monocaprylate as a liquid lipid: Formulation characterization and assessment of dermal delivery compared to referent ointment. Int J Pharm 569, 118624 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Ramirez-Carrozzi VR et al. , Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev 20, 282–296 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Musselman CA et al. , Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem J 423, 179–187 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D, FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92, 105–116 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Shen J. et al. , Histone chaperone FACT complex mediates oxidative stress response to promote liver cancer progression. Gut 69, 329–342 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Sharma S, Sethi GS, Naura AS, Curcumin Ameliorates Ovalbumin-Induced Atopic Dermatitis and Blocks the Progression of Atopic March in Mice. Inflammation 43, 358–369 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Sun J, Zhao Y, Hu J, Curcumin inhibits imiquimod-induced psoriasis-like inflammation by inhibiting IL-1beta and IL-6 production in mice. PloS one 8, e67078 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H. et al. , Protective Effect of Curcumin Against Acute Ultraviolet B Irradiation-induced Photo-damage. Photochem Photobiol 92, 808–815 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Kurd SK et al. , Oral curcumin in the treatment of moderate to severe psoriasis vulgaris: A prospective clinical trial. J Am Acad Dermatol 58, 625–631 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kulkarni NN et al. , IL-1 Receptor-Knockout Mice Develop Epidermal Cysts and Show an Altered Innate Immune Response after Exposure to UVB Radiation. J Invest Dermatol 137, 2417–2426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plager DA et al. , Early cutaneous gene transcription changes in adult atopic dermatitis and potential clinical implications. Exp Dermatol 16, 28–36 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Raw data file (Excel spreadsheet)