Abstract
Background
Babesia species are intraerythrocytic Apicomplexan parasites that infect a wide range of vertebrate hosts. These pathogens are typically transmitted either by tick vectors or by direct blood-to-blood contact, and may cause life-threatening clinical disease, such as thrombocytopenia, hemolytic anemia and acute renal failure, in canine hosts. While Babesia vogeli and Babesia gibsoni infections have both been reported in Oklahoma, reports of Babesia conradae infections have been limited to California.
Methods
Four separate kennels of coyote-hunting dogs were identified in Oklahoma after the kennels had consulted with Oklahoma State University Boren Veterinary Medical Teaching Hospital (antemortem cases) or the Oklahoma Animal Disease Diagnostic Lab (postmortem cases). Upon owner consent, every accessible dog from each of the four kennels was briefly examined for ectoparasites, particularly ticks, and whole blood samples were collected in EDTA tubes. Clinically ill dogs were examined by a practicing veterinarian, and clinical signs included anorexia, vomiting, lethargy, fever and anemia. DNA was extracted from each blood sample, and a nested PCR was performed using general apicomplexan primers for the partial 18S rRNA gene. PCR products were electrophoresed in agarose matrix, and appropriately sized amplicons were sequenced. Sequences were compared to reference 18S rRNA gene sequences available in GenBank, and samples with > 98% homology to B. conradae (GenBank: AF158702) were considered positive. Babesia conradae-positive dogs were then treated with atovaquone (13.5 mg/kg three times per day) and azithromycin (10 mg/kg once daily) for 10 days and retested at 30 and 60 days post-treatment by PCR.
Results
Of 40 dogs tested, 15 (37.5%) were positive for B. conradae with 98–99% sequence homology to B. conradae from California. All positive cases were coyote-hunting Greyhounds. Ectoparasites were not identified on any of the dogs at the time of blood collection. Treatment of clinically ill dogs with atovaquone and azithromycin resulted in complete clinical recovery in all treated dogs with negative follow-up PCR at 30 and 60 days post-treatment.
Conclusions
Collectively, this study (i) documents the occurrence of B. conradae in Oklahoma, (ii) highlights this pathogen as a differential to be considered when clinical signs are present, (iii) supports the use of atovaquone and azithromycin as effective treatment in these cases and (iv) demonstrates chronic subclinical carrier dogs serving as potential reservoirs of B. conradae infection to naïve dogs.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-021-04897-x.
Keywords: Babesia conradae, Babesiosis, Canine, Dog, Oklahoma, Greyhound, Anemia, Thrombocytopenia, 18S rRNA
Background
Babesia species are intraerythrocytic protozoan parasites in the phylum Apicomplexa that are transmitted by the bite of an infected tick or by passage of contaminated blood to a susceptible, naïve host. There are over 100 species described, which are divided into two broad categories: small Babesia (measuring 1–3 µm) and large Babesia (measuring 3–7 µm). There are currently five named Babesia spp. enzootic to the USA known to infect dogs: Babesia vogeli (large), Babesia sp. (Coco isolate, large), B. gibsoni (small), B. vulpes (formerly Theileria annae, small) and B. conradae (small) [1–4].
Babesia conradae was first reported in California in 1991 as Babesia gibsoni, given that B. gibsoni was the only small Babesia sp. known to infect dogs at the time [5]. Further characterization of the piroplasm in 2006 revealed that the California organism was a distinct species, and the name was subsequently changed to B. conradae after the first reporting author, Dr. Patricia Conrad [6]. The transmission dynamics of B. conradae remain unknown as attempts at tick transmission of this piroplasm have not been successful [7].
Dogs infected with B. conradae exhibit typical clinical signs of babesiosis, including anorexia, hemolytic anemia, splenomegaly, thrombocytopenia and vomiting [5]. Similar to infections caused by other canine Babesia spp., clinical signs resulting from B. conradae infection can vary and range from mild to life-threatening. Severe complications, such as acute renal failure/renal disease, cardiac related alterations, acute respiratory distress syndrome and acute pancreatitis, may result, particularly in dogs with B. conradae infection [2, 5]. Membranoproliferative glomerulonephritis has been reported in one patient. Case fatality rate in B. conradae-infected patients can reach up to 40% without timely and appropriate therapeutic intervention [5]. While various medications have been attempted, combination therapy with atovaquone and azithromycin is the only treatment regimen shown to successfully clear B. conradae infection in dogs [8].
Babesia vogeli and B. gibsoni infections have been previously reported throughout the USA and in the state of Oklahoma; however, B. conradae infection has not yet been documented outside of the state of California. In the current study, we surveyed kenneled coyote-hunting dogs for B. conradae and other apicomplexan parasites in Oklahoma and evaluated the efficacy of atovaquone and azithromycin therapy [8, 9] in treating dogs with clinical disease.
Methods
Study population
Four separate kennels (groups) of coyote-hunting dogs were identified following presentation of severe acute illness in at least one dog in the kennel or in a closely associated kennel. Groups 1, 3 and 4 sought consultation at either the Oklahoma State University Boren Veterinary Medical Teaching Hospital/College of Veterinary Medicine (antemortem cases) or the Oklahoma Animal Disease Diagnostic Laboratory (postmortem cases). Group 2 volunteered for testing given their working association with Group 1. All kenneled dogs in each group were included in the study. All kennels were located in Oklahoma.
Group 1 consisted of six Greyhounds and four Treeing Walker Coonhounds from a kennel in Crescent, OK, sampled in March 2014; Group 2 consisted of 16 Greyhounds from a kennel in Kingfisher, OK, also sampled in March of 2014; Group 3 consisted of ten Greyhounds from a kennel in Vinita, OK, sampled in February 2019; and Group 4 consisted of four Greyhounds from a kennel in Hobart, OK, sampled in May 2020 (Fig. 1). One dog from Group 1 displayed clinical signs of lethargy, fever and anemia (Dog 1) while the remaining dogs were subclinical (bright, alert, responsive, good body condition, good appetite, hunted effectively and had no apparent clinical signs, such as fever/lethargy). Clinical signs were not apparent in any of the dogs in Group 2. In Group 3, two dogs (Dogs 27 and 28) showed clinical signs, including vomiting, lethargy, anorexia and anemia, while the remaining dogs were subclinical. Group 4 had one dog showing clinical signs, including profound lethargy, anorexia, icterus and splenomegaly, which died prior to definitive diagnosis and treatment (Dog 37) and one subclinical dog.
Fig. 1.
Geographical distribution of Babesia conradae cases in Oklahoma. A total of 40 dogs were included in this study from four separate kennels in Oklahoma: 6 Greyhounds and 4 Treeing Walker Coonhounds from Crescent, OK (Group 1), 16 Greyhounds from Kingfisher, OK (Group 2), 10 Greyhounds from Vinita, OK (Group 3) and 4 Greyhounds from Hobart, OK (Group 4)
Housing varied from kennel to kennel. Group 1 dogs were housed in individual horse stalls, dogs in Groups 2 and 4 were kept in outdoor chain link runs with dirt bedding and dogs in Group 3 were in outdoor wood panel runs on a combination of dirt and gravel. At initial presentation, each dog was briefly examined for ectoparasites, particularly ticks. Patient temperament, however, impeded an extensive dermatological evaluation. For tick prevention, Group 1 received Certifect (Merial, Duluth, GA, USA) monthly, Groups 2 and 3 were not on tick prevention and Group 4 received Simparica (Zoetis, Kalamazoo, MI, USA), treatment duration unknown. Whole blood (1–5 ml) was collected in EDTA tubes from all animals (n = 40). Blood was transported on ice to the Oklahoma State University College of Veterinary Medicine (OSU-CVM) and stored at 4 °C prior to processing.
Serologic testing and blood smear evaluation
All Group 3 samples were tested using a standard SNAP® 4Dx® Plus test according to manufacturer’s instructions (IDEXX, Westbrook, Maine, USA). Blood smears were prepared for Group 1 samples upon arrival to Oklahoma State University using a previously published method and evaluated microscopically [10]. Blood samples for blood smear evaluation were not available for Groups 2–4 and were therefore not evaluated.
DNA extraction and PCR
For Groups 1 and 2, DNA was extracted from whole blood using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA). For Groups 3 and 4, DNA was extracted using the Illustra™ blood genomic Prep Mini Spin Kit (GE Healthcare, Piscataway, New Jersey, USA). All extractions were performed according to the manufacturers’ instructions. Dedicated laboratory areas were utilized for DNA extractions, primary and secondary PCR amplifications and PCR product purifications to prevent contamination events. Separate ultra-purified water samples (no template control [NTC]) were included as negative controls in DNA extractions and PCR amplifications. DNA extracts from Groups 1–4 were analyzed by previously described nested PCR methods which amplify a 460- to 520-bp hypervariable region of the 18S rRNA gene of Babesia spp. and other apicomplexans (Hepatozoon spp., Sarcocystis spp. and Toxoplasma spp.) (Table 1) [11, 12], except for Dog 27 for which the PCR and sequencing was carried out by the North Carolina State Vector Borne Disease Diagnostics Lab (NCSVBDDL) per their standard operating procedures. NCSVBDDL utilizes primers that also amplify the 18S rRNA gene. DNA extracts from known piroplasm-positive blood samples by microscopy were included as positive controls for each sample set. Dogs in Group 3 with antibodies to tick-borne bacteria (namely Ehrlichia spp.) as detected by the SNAP 4Dx assay were tested by PCR to potentially amplify and identify circulating organism as previously described [13].
Table 1.
PCR primers used to amplify partial 18S rRNA gene fragments of Babesia spp. and other apicomplexans
For Groups 1 and 2, primary PCR reactions were performed in 25-µl volumes containing 0.25 U Taq polymerase (Promega, Madison, WI, USA), 10× Taq buffer (Promega), 1.5 mM MgCl2, 0.8 mM dNTP mixture (Promega), 0.8 µM each primer BABA-F and BABA-R and 5 µl template DNA. Primary reaction conditions were as follows: 94 °C, 5 min; then 94 °C/1 min, 56.6 °C/1 min, 72 °C/2 min for 35 cycles; and a final extension step of 72 °C for 5 min. Nested PCR was carried out using 1 µl of the primary product and primers RLB-F and RLB-R. Nested reaction conditions were as follows: 94 °C, 5 min; then 94 °C/1 min, 50 °C/1 min, 72 °C/2 min for 35 cycles; and a final extension step of 72 °C for 5 min.
For Groups 3 and 4, primary PCR reactions were prepared in 25-µl volumes containing 0.075 U Accuprime™ Taq HIFI (Thermo Fisher Scientific, Waltham, MA, USA), 1× AccuPrime™ PCR Buffer II (Thermo Fisher Scientific), 1.5 mM MgSO4 (ThermoFisher), 0.2 µM each primer 3.1 and 5.1, and 5 µl of DNA extract. Primary reaction conditions were as follows: 94 °C for 2 min followed by 30 cycles of 94 °C for 1 min, 55 °C for 1 min, 68 °C for 1.5 min, and a final extension step of 72 °C for 10 min. Nested PCR was again carried out using 1 µl of primary product and primers RLB-F and RLB-R, but cycling conditions were different than above. For Group 3 and 4 dogs, nested PCR reaction conditions were as follows: 94 °C, 2 min; then 94 °C/1 min, 50 °C/1 min, 68 °C/1.5 min for 40 cycles; and a final extension step of 72 °C for 10 min.
PCR product purification and sequencing
PCR products were electrophoresed in a 2% agarose matrix containing either GelRed® QIAquick® Gel Extraction Kit (Qiagen) or the Wizard® SV Gel and PCR Clean-Up System (Promega) according to manufacturers’ instructions. DNA sequencing was performed at the Oklahoma State University Molecular Core Facility (Stillwater, OK, USA) with an ABI 3730 DNA Analyzer (Applied Biosystems, Thermo Fisher Scientific), except for Dog 27. Forward and reverse sequences were aligned with ClustalW (Bioinformatics Center, Kyoto, Japan) and compared with sequence data available in the National Center for Biotechnology Information database (GenBank) for B. conradae (AF158702), B. gibsoni (AF205636) and B. vogeli (AY371198) to determine percent homology. Samples were considered positive if they were ≥ 98% homologous to B. conradae (GenBank: AF158702). The sequences from Dogs 1, 28, 29, 33, 34, 35, 37 and 38 have been deposited in GenBank under the accession numbers MW147022, MT430944, MW145168, MW145196, MW145199, MW145504, MW145505 and MW145506, respectively.
Phylogenetic tree and percent identity matrix construction
All partial 18S rRNA gene sequences obtained from canines in the current study were entered into MacVector, then aligned and trimmed along with sequences from various piroplasm reference sequences available in GenBank which were used in previous analyses [1, 6, 14]. A maximum likelihood phylogenetic tree was constructed using unweighted pair group method with arithmetic mean (UPGMA) analysis. Bootstrap values are based on 1000 replicates and only bootstraps > 50% are indicated. The percent homology matrix was also produced in MacVector using the same 18S rRNA gene full and partial sequences.
Treatment protocol
Upon owner consent, surviving B. conradae PCR-positive dogs from each cohort were treated with a previously described treatment regimen shown to eliminate infection in vivo [8], except for Dog 4 which was sold and lost to follow-up. Atovaquone (GlaxoSmithKline, Research Triangle Park, NC, USA) and azithromycin (Pfizer, New York, NY, USA) were compounded in an oral suspension and administered to all surviving PCR-positive dogs at a dose of 13.5 mg/kg TID (atovaquone) and 10 mg/kg SID (azithromycin) for 10 days. Whole blood (1–5 ml) was collected in EDTA tubes prior to treatment (day 0) and at 30 and 60 days post-treatment. DNA was extracted from each blood sample and tested by PCR to detect Babesia sp. infection as previously described.
Statistics
The prevalence of B. conradae infection was calculated according to Bush et al. [15] and 95% confidence intervals (CIs) were calculated using QuickCalcs GraphPad (https://www.graphpad.com/quickcalcs/). The prevalence of B. conradae in hunting dogs among kennel age groups were compared using chi-square (X2) tests [16].
Results
PCR analysis of dogs screened for B. conradae infection
From the 40 coyote-hunting dogs screened for B. conradae infection, 15 (37.5%; 95% CI 24.1–53.0%) were positive by PCR and sequencing for B. conradae infection (Additional file 1: Table S1). This included three of ten (30.0%; 95% CI 10.3‒60.8%) dogs from Group 1 (Crescent, OK; all Greyhounds), four of 16 (25%; 95% CI 9.7‒50.0%) dogs from Group 2 (Kingfisher, OK; all Greyhounds), six of ten (60.0%; 95% CI 31.2‒83.3%) dogs from Group 3 (Vinita, OK; all Greyhounds) and two of four (50.0%; 95% CI 15.0–85.0%) dogs from Group 4 (Hobart, OK; all Greyhounds). A significant difference in the prevalence of B. conradae among the kennels was not detected (X2 = 3.733, df = 2, P = 0.292).
PCR-positive dogs ranged in age from 1.5–6 years ( = 3.7 years; 95% CI 2.6–4.8), with age unknown in one positive dog. PCR-negative dogs ranged in age from 5 months to 10 years ( = 2.8 years; 95% CI 1.68–3.9). There was no significant difference in age groups of PCR-positive and -negative dogs (X2 = 3.911, df = 3, P = 0.271). Of the PCR-positive dogs in Group 3, four were females and two were males. Of the PCR-positive dogs in Group 4, both were males. Hepatozoon canis and Ehrlichia canis were confirmed based on sequence in dogs 11 and 31, respectively.
SNAP® 4Dx® Plus results
Of the dogs tested by SNAP® 4Dx® Plus test (Group 3, n = 10), only Dog 31 had antibodies to Ehrlichia spp. All other Group 3 dogs were negative for Anaplasma spp., Dirofilaria immitis, Ehrlichia spp. and Borrelia burgdorferi antibodies by the SNAP® 4Dx® Plus test.
Clinicopathologic findings
Clinicopathologic data (serum biochemistry, complete blood counts) were available for two clinically ill dogs who survived infection (Dogs 1 and 27), with the clinical signs of regenerative anemia, thrombocytopenia, hyperglycemia and hypocalcemia in both animals. One dog from the same kennel as Group 3 dogs exhibited the aforementioned changes as well as severe azotemia. This dog passed away prior to diagnosis and was the only dog submitted for postmortem examination. We were unable to obtain a sequence on this dog, and it is therefore not included in the total sample set; however, postmortem microscopic lesions included membranoproliferative glomerulonephritis, tubular proteinosis, myocardial necrosis and necrosuppurative opportunistic bronchopneumonia that cultured Escherichia coli, Staphylococcus pseudintermedius and Streptococcus minor. The renal and myocardial lesions have both been reported in canine Babesiosis [2]. Evaluation of blood smears of infected dogs from Group 1 revealed mild polychromasia (indicative of regenerative anemia) and numerous basophilic, intraerythrocytic piroplasms consistent with Babesia sp. (Fig. 2). Blood smears were not evaluated for dogs from Groups 2–4.
Fig. 2.
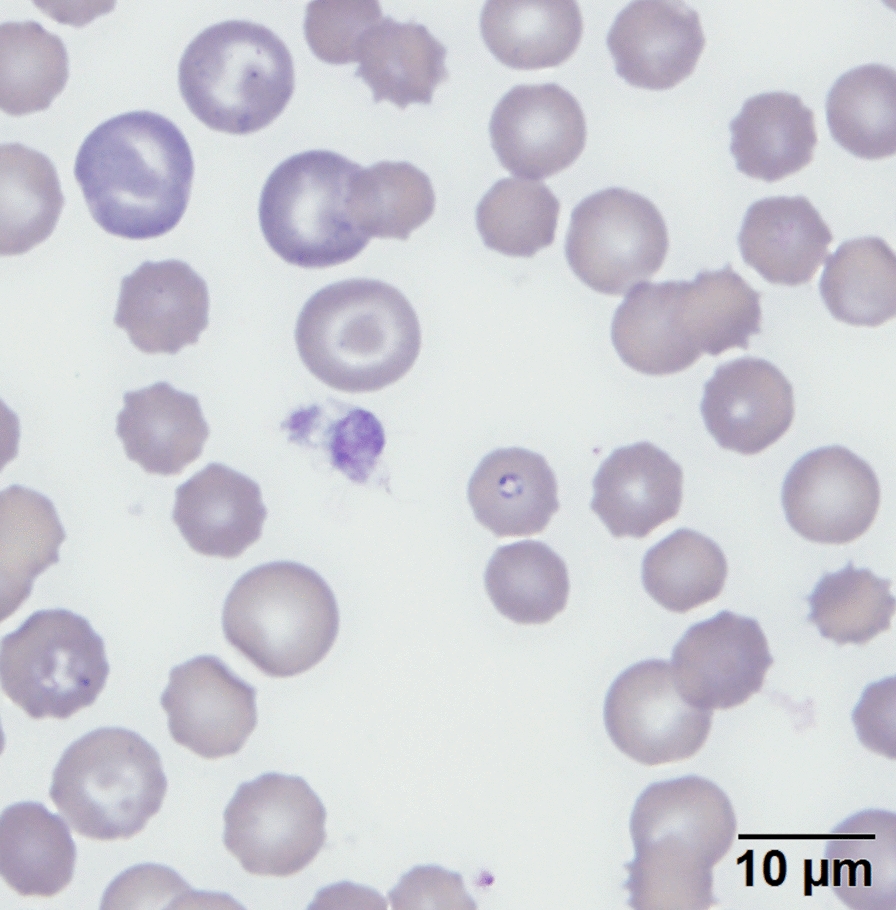
Blood smear from a Babesia conradae-positive Greyhound in Group 1. Approximately 10% of the erythrocytes contain pale basophilic intracellular protozoa, consistent with intraerythrocytic B. conradae piroplasms. A mild degree of polychromasia is also present within the specimen, indicating regenerative anemia. Scale bar: 10 µm
Sequence analysis
The 18S rRNA gene sequences exhibited 98–99% homology to B. conradae (GenBank: AF158702) and 97–100% homology with each other, and shared 78% homology with B. gibsoni and B. vogeli (Table 2). Phylogenetic analysis demonstrated a distinct relationship between the Oklahoma Babesia sp. and the California B. conradae strains; sequences from Oklahoma dogs clustered with B. conradae sequences documented from California dogs (GenBank: MK256976 and AF158702) and were more distant to other Babesia spp. sequences used in the comparison (Fig. 3).
Table 2.
Percent identity matrix of Oklahoma Babesia conradae 18S rRNA gene sequences compared to reference sequences in GenBank
| Dog ID | Dog 1 | Dog 27 | Dog 28 | Dog 29 | Dog 33 | Dog 34 | Dog 35 | Dog 37 | Dog 38 | Babesia conradae (AF158702) | Babesia gibsoni (AF205636) | Babesia vogeli (AY371198) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dog 1 | 100 | 98 | 99 | 99 | 98 | 98 | 98 | 98 | 98 | 98 | 78 | 78 |
| Dog 27 | 98 | 100 | 98 | 98 | 98 | 97 | 98 | 98 | 98 | 98 | 78 | 78 |
| Dog 28 | 99 | 98 | 100 | 100 | 98 | 99 | 99 | 98 | 99 | 99 | 78 | 78 |
| Dog 29 | 99 | 98 | 100 | 100 | 98 | 99 | 99 | 98 | 97 | 99 | 78 | 78 |
| Dog 33 | 98 | 98 | 98 | 98 | 100 | 98 | 98 | 99 | 98 | 98 | 78 | 78 |
| Dog 34 | 98 | 97 | 99 | 99 | 98 | 100 | 99 | 98 | 98 | 98 | 78 | 78 |
| Dog 35 | 98 | 98 | 99 | 99 | 98 | 99 | 100 | 98 | 98 | 99 | 78 | 78 |
| Dog 37 | 98 | 98 | 98 | 98 | 99 | 98 | 98 | 100 | 98 | 98 | 78 | 78 |
| Dog 38 | 98 | 98 | 99 | 99 | 97 | 98 | 98 | 98 | 100 | 99 | 78 | 78 |
| Babesia conradae (AF158702) | 98 | 98 | 99 | 99 | 98 | 98 | 99 | 98 | 99 | 100 | 79 | 78 |
| Babesia gibsoni (AF205636) | 78 | 78 | 78 | 78 | 78 | 78 | 78 | 78 | 78 | 79 | 100 | 89 |
| Babesia vogeli (AY371198) | 78 | 78 | 78 | 78 | 78 | 78 | 78 | 78 | 78 | 78 | 89 | 100 |
GenBank accession numbers are given in parentheses
Fig. 3.
Phylogenetic comparison of Babesia conradae isolates from Oklahoma to other piroplasms. Maximum likelihood phylogenetic tree constructed using unweighted pair group method with arithmetic mean analysis of 18S rRNA gene sequences extracted from GenBank. Clustering of study samples within a distinct subclade (red lines) illustrates the homology of these Oklahoma strains to the original (California) B. conradae strains (GenBank: MK256976 and AF158702) and highlights the distinct separation of study samples from other Babesia spp. Bootstrap values are based on 1000 replicates and only bootstraps < 50% are indicated
Response to therapy
Of the PCR-positive dogs in Groups 1–4, 13 dogs (Additional file 1: Table S1) were administered a combination therapy of atovaquone and azithromycin for 10 days as previously described and then re-tested by PCR at 30 and 60 days post-treatment. In one case (Dog 27) three blood transfusions were required to stabilize the patient during treatment. Dog 4 was sold and lost to follow-up. Dog 37 passed away shortly after blood was collected for testing and was therefore not in the treatment group. Complete resolution of clinical signs was observed in all sick dogs by day 4 of treatment, and B. conradae DNA was not detected in the blood of any dog at 30 or 60 days post-treatment.
Case follow-up was conducted via telephone conversation with the owners approximately 1 year post-treatment. Previously subclinical dogs reportedly continued to do well with no change noted by the owners. Clinically ill dogs recovered well, gained weight back that had been lost and returned to hunting the following season. Dog 38 (Group 4, subclinical) notably improved in body condition, energy level and coat quality as noted by the first and second authors at the 60 day blood collection.
Discussion
Multiple outbreaks of B. conradae have been previously reported in dogs from southern California since 1991 [5, 8, 17, 18], but infection has not yet been reported outside this core, initial area. A single report of B. gibsoni-like parasite genetically similar to B. conradae was documented in Oklahoma in 2001 (GenBank: AF205636) [19], but further BLAST analysis by the authors showed 100% alignment with numerous B. gibsoni sequences and significant genetic divergence from B. conradae (76.8% homologous to B. conradae, GenBank: AF158702). Full travel and family histories are unavailable for all dogs in the current study; however, none of the dogs originated from or had been transported to California. To the authors’ knowledge, this is the first published report of B. conradae infection outside of California and represents the emergence of an important pathogen in Oklahoma that is capable of causing significant disease in kenneled dogs.
Diagnosis of babesiosis is typically based on clinical signs and/or observation of intraerythrocytic piroplasms on blood smear (Fig. 2). Ancillary testing (PCR and DNA sequencing) is required for accurate identification given that small piroplasms are microscopically indistinguishable from each other, both those within the Babesia genus and those from Theileria spp., and there is cross reactivity with immunofluorescence antibody (IFA) testing [20]. Additionally, B. conradae parasitemia can be low (< 1% of erythrocytes with piroplasms), making detection of the organism on blood smear difficult. PCR has become widely available and is both sensitive and specific for detection of B. conradae DNA using the 18S rRNA gene [21]. Differentiation between the small Babesia spp. is important, given that B. conradae can be more pathogenic than B. gibsoni [5, 8, 22].
Testing for disease and identification of the disease have previously and primarily been conducted in clinically ill dogs. However, in the current study, we observed four B. conradae-positive dogs with clinical babesiosis and ten with subclinical infections. Our observation of ten subclinical dogs suggests that some dogs may be chronic carriers and possibly serve as a reservoir of infection for naïve animals. Further research should determine if B. conradae is more prevalent in high-risk groups than previously thought. It remains to be determined if there are any negative long-term health problems associated with a chronic carrier state and whether atovaquone and azithromycin treatment would be beneficial in these animals.
Babesia spp. are primarily transmitted by ticks worldwide; however, transmission mechanisms of Babesia spp. in the USA differ. For example, B. vogeli is transmitted by Rhipicephalus sanguineus (s.l.) (brown dog tick), while B. gibsoni is largely considered to be transmitted by passage of contaminated blood during dog fights [5]. Comparatively, the source of B. conradae infection in domestic dogs is still unknown, and it is unclear whether transmission occurs via a tick vector, by transfer of contaminated blood between dogs, trans-placentally from an infected dam or by a previously undocumented route [7].
There is some evidence that other Babesia spp. can be transmitted trans-placentally, but for this study a detailed family history was not available to assess the possibility for vertical B. conradae infection in these Greyhounds [23]. Kennel owners in the present study suspected that their dogs were becoming infected with B. conradae by fighting coyotes while hunting. A previous report of B. conradae infection in coyote-hunting dogs in southern California also documented that infection was associated with a history of aggressive interactions with coyotes [17]. A serosurvey performed in California in 1994 showed that three of nine coyotes were seropositive for Babesia gibsoni, suggesting these wild canids may be a reservoir host for Babesia sp. [18]. At that time B. conradae had not yet been recognized, highlighting the potential for these seropositive coyotes to have been actually infected with multiple Babesia spp., including B. conradae. Moreover, these studies used IFA for organism detection, which is also subject to cross-reactivity [20]. It is also possible to have a subclinical carrier dog transmit the disease to a naïve dog during hunting, as the dogs may bite each other during the interaction with the coyote. Although speculative at this time, all of these scenarios represent possible reservoirs for B. conradae infection in these study animals, and studies are currently underway to investigate the presence of B. conradae in wild tick populations, free-ranging coyotes and other domestic canids via contact tracing to determine the source of this pathogen in the Oklahoma region.
Some Babesia spp. can infect multiple vertebrate species, such as B. microti, which has been identified in both rodents and humans, and B. divergens, identified in both cattle and gerbils [21]. Phylogenetic analyses showed B. conradae to be closely related to piroplasms from humans, bighorn sheep and mule deer [1]. Determining the suitability of various hosts for B. conradae, particularly humans, is an important area needing further research, especially when the route of transmission remains undetermined. A maximum likelihood phylogenetic tree (Fig. 3) constructed from samples with sequence data and other sequences available in GenBank demonstrates a close relationship of the B. conradae strains from Oklahoma with those from California. It also demonstrates a clear distinction from B. gibsoni [1, 14].
Babesia conradae infection in these animals resulted in a variety of clinical signs, including fever, vomiting, anorexia, regenerative anemia, and thrombocytopenia—all of which were mitigated by treatment with atovaquone and azithromycin. The source of B. conradae infection in Oklahoma remains unknown. At this time, B. conradae infection in Oklahoma has been documented in only Greyhounds used for hunting coyotes.
Conclusions
In conclusion, the current study documents the emergence of B. conradae infection in 15 coyote-hunting Greyhounds in Oklahoma. Treatment with atovaquone and azithromycin promoted the mitigation of clinical signs and also reduced B. conradae DNA to undetectable levels. Further research is needed to determine the regional prevalence, reservoir host(s), mode(s) of transmission and diversity of vertebrate host species. Similarly, surveillance should be expanded to include other areas of the Midwest and Southern geographical regions of the USA.
Supplementary Information
Additional file 1: Table S1. PCR results of coyote hunting dogs in Oklahoma tested for infection with Babesia conradae.
Acknowledgements
We would like to thank the NC State Vector Borne Disease Diagnostics Laboratory for their cooperation and sequence contribution, Dr. Jim Meinkoth, DVM, PhD, DACVP for Fig. 2, Dr. Kathryn Duncan, DVM, for help constructing the Phylogenetic tree in MacVector, as well as Dr. Brent Hancock, DVM, of Southwest Veterinary Clinic for cooperation, coordination and consultation with the Group 4 kennel and more kennels to come. Thanks also to Victoria Saccomagno for preparing the graphical abstract.
Abbreviations
- EDTA
Ethylenediaminetetraacetic acid
- rRNA
Ribosomal ribonucleic acid
- SID
Once daily
- TID
Three times daily
Authors' contributions
MR and JT performed the sample collection, PCR and sequence analysis for Groups 1 and 2. GY was the primary clinician involved in sample collection and treatment administration for Group 1 while TB was the clinician for Group 3. ML performed DNA extraction, PCR and sequence analysis for Groups 3 and 4. RC, KA, CM and MR provided consultation and sample coordination while ES drafted the manuscript. All authors collaborated on manuscript assembly revision and submission. All authors read and approved the final manuscript.
Funding
Funding to cover diagnostics and treatment costs in these cases were provided by IDEXX, Pfizer and internal grants through the College of Veterinary Medicine, Oklahoma State University.
Availability of data and materials
The sequences from dogs 1, 28, 29, 33, 34, 35, 37 and 38 have been deposited in GenBank under the accession numbers MW147022, MT430944, MW145168, MW145196, MW145199, MW145504, MW145505 and MW145506, respectively.
Declarations
Ethics approval and consent to participate
All samples were ethically collected from client-owned animals with owner-informed verbal consent that the samples will be used in research. No animals experienced undue pain or stress during routine sample collection.
Consent for publication
All samples and results were obtained, tested and prepared for publication with owner consent.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mason Reichard and Craig Miller contributed equally to this article
Contributor Information
Erin Stayton, Email: erin.m.roberts@okstate.edu.
Megan Lineberry, Email: megan.wohltjen@okstate.edu.
Jennifer Thomas, Email: jennifer.e.thomas@okstate.edu.
Tina Bass, Email: countrysideanimaldoctor@gmail.com.
Kelly Allen, Email: kelly.allen10@okstate.edu.
Ramaswamy Chandrashekar, Email: Ramaswamy-Chandrashekar@idexx.com.
Gene Yost, Email: Gene.yost@gmail.com.
Mason Reichard, Email: mason.reichard@okstate.edu.
Craig Miller, Email: craig.miller@okstate.edu.
References
- 1.Kjemtrup A, Kocan A, Whitworth L, Meinkoth J, Birkenheuer A, Cummings J, et al. There are at least three genetically distinct small piroplasms from dogs. Int J Parasitol. 2000;30:1501–1505. doi: 10.1016/S0020-7519(00)00120-X. [DOI] [PubMed] [Google Scholar]
- 2.Greene C. Infectious diseases of the dog and cat. 4. St. Louis: Elsevier; 2012. [Google Scholar]
- 3.Baneth G, Florin-Christensen M, Cardoso L, Schnittger L. Reclassification of Theileria annae as Babesia vulpes sp. nov. Parasites Vectors. 2015;8:207. doi: 10.1186/s13071-015-0830-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sikorski L, Birkenheuer A, Holowaychuk M, McCleary-Wheeler A, Davis J, Littman M. Babesiosis caused by a large Babesia species in 7 immunocompromised dogs. J Vet Int Med. 2010;24:127–131. doi: 10.1111/j.1939-1676.2009.0440.x. [DOI] [PubMed] [Google Scholar]
- 5.Conrad P, Thomford J, Yamane I, Whiting J, Bosma L, Uno T, et al. Hemolytic anemia caused by Babesia gibsoni infection in dogs. J Am Vet Med Assoc. 1991;199:601–605. [PubMed] [Google Scholar]
- 6.Kjemtrup AM, Wainwright K, Miller M, Penzhorn BL, Carreno RA. Babesia conradae, sp. nov., a small canine Babesia identified in California. Vet Parasitol. 2006;138:103–11. doi: 10.1016/j.vetpar.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 7.Yamane I, Gardner I, Telford S, Elward T, Hair J, Conrad PA. Vector competence of Rhipicephalus sanguineus and Dermacentor variabilis for American isolates of Babesia gibsoni. Exp Appl Acarol. 1993;17:913–919. doi: 10.1007/BF02328068. [DOI] [Google Scholar]
- 8.Di Cicco MF, Downey ME, Beeler E, Marr H, Cyrog P, Kidd L, et al. Re-emergence of Babesia conradae and effective treatment of infected dogs with atovaquone and azithromycin. Vet Parasitol. 2012;187:23–27. doi: 10.1016/j.vetpar.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Birkenheuer AJ, Levy MG, Breitschwerdt EB. Efficacy of combined atovaquone and azithromycin for therapy of chronic Babesia gibsoni (Asian genotype) infections in dogs. J Vet Int Med. 2004;18:494–498. doi: 10.1111/j.1939-1676.2004.tb02573.x. [DOI] [PubMed] [Google Scholar]
- 10.Allison RW, Meinkoth JH. Hematology without the numbers: in-clinic blood film evaluation. Vet Clin North Am Small Anim Pract. 2007;37:245–266. doi: 10.1016/j.cvsm.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Medlin L, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 12.Yabsley MJ, McKibben J, Macpherson CN, Cattan PF, Cherry NA, Hegarty BC, et al. Prevalence of Ehrlichia canis, Anaplasma platys, Babesia canis vogeli, Hepatozoon canis, Bartonella vinsonii berkhoffii, and Rickettsia spp. in dogs from Grenada. Vet Parasitol. 2008;151:279–85. doi: 10.1016/j.vetpar.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Little SE, O’Connor TP, Hempstead J, Saucier J, Reichard MV, Meinkoth K, et al. Ehrlichia ewingii infection and exposure rates in dogs from the southcentral United States. Vet Parasitol. 2010;172:355–360. doi: 10.1016/j.vetpar.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Lack JB, Reichard MV, Van Den Bussche RA. Phylogeny and evolution of the Piroplasmida as inferred from 18S rRNA sequences. Int J Parasitol. 2012;42:353–363. doi: 10.1016/j.ijpara.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Bush AO, Lafferty KD, Lotz JM, Shostak AW. Parasitology meets ecology on its own terms: Margolis et al. revisited. J Parasitol. 1997;83:575–83. doi: 10.2307/3284227. [DOI] [PubMed] [Google Scholar]
- 16.Sokal R, Rohlf F. The chi-square distribution. New York: WH Freeman and Co.; 1997. pp. 152–154. [Google Scholar]
- 17.Dear JD, Owens SD, Lindsay LL, Biondo AW, Chomel BB, Marcondes M, et al. Babesia conradae infection in coyote hunting dogs infected with multiple blood-borne pathogens. J Vet Int Med. 2018;32:1609–1617. doi: 10.1111/jvim.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamane I, Gardner I, Ryan C, Levy M, Urrico J, Conrad PA. Serosurvey of Babesia canis, Babesia gibsoni and Ehrlichia canis in pound dogs in California, USA. Prev Vet Med. 1994;18:293–304. doi: 10.1016/0167-5877(94)90054-X. [DOI] [Google Scholar]
- 19.Kocan AA, Kjemtrup A, Meinkoth J, Whitworth LC, Murphy G, Decker L, et al. A genotypically unique Babesia gibsoni-like parasite recovered from a dog in Oklahoma. J Parasitol. 2001;87:437–438. doi: 10.1645/0022-3395(2001)087[0437:AGUBGL]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 20.Yamane I, Thomford J, Gardner I, Dubey J, Levy M, Conrad PA. Evaluation of the indirect fluorescent antibody test for diagnosis of Babesia gibsoni infections in dogs. Am J Vet Res. 1993;54:1579–1584. [PubMed] [Google Scholar]
- 21.Zahler M, Rinder H, Schein E, Gothe R. Detection of a new pathogenic Babesia microti-like species in dogs. Vet Parasitol. 2000;89:241–248. doi: 10.1016/S0304-4017(00)00202-8. [DOI] [PubMed] [Google Scholar]
- 22.Meinkoth JH, Kocan AA, Loud SD, Lorenz MD. Clinical and hematologic effects of experimental infection of dogs with recently identified Babesia gibsoni-like isolates from Oklahoma. J Am Vet Med Assoc. 2002;220:185–189. doi: 10.2460/javma.2002.220.185. [DOI] [PubMed] [Google Scholar]
- 23.Abu M. Babesia infections in puppies: probably due to transplacental transmission. J Vet Med. 1973;609:203–206. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. PCR results of coyote hunting dogs in Oklahoma tested for infection with Babesia conradae.
Data Availability Statement
The sequences from dogs 1, 28, 29, 33, 34, 35, 37 and 38 have been deposited in GenBank under the accession numbers MW147022, MT430944, MW145168, MW145196, MW145199, MW145504, MW145505 and MW145506, respectively.