Abstract
Background
Hepatocellular carcinoma (HCC) is one of the most common malignancies in the world, and due to its complex pathogenic factors, its prognosis is poor. N6-methyladenosine (m6A) RNA methylation plays an important role in the tumorigenesis, progression, and prognosis of many tumors. The m6A RNA methylation regulator small nuclear ribonucleoprotein polypeptide C (SNRPC), which encodes one of the specific protein components of the U1 small nuclear ribonucleoprotein (snRNP) particle, has been proven to be related to the prognosis of patients with HCC. However, the effect of SNRPC on the tumor microenvironment and immunotherapy in HCC remains unclear.
Case presentation
The HCC RNA-seq profiles in The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) databases, including 421 LIHC and 440 LIRI-JP samples, respectively, were used in this study. Both the expression of SNRPC in HCC was upregulated in the TCGA and ICGC databases compared to normal tissues. Next, the expression of SNRPC was validated as a risk factor for prognosis by Kaplan-Meier analysis and employed to establish a nomogram with T pathologic stage. By gene set variation (GSVA) analysis and gene set enrichment (GSEA) analysis, we found that SNRPC was mainly related to protein metabolism and the immune process. Furthermore, the estimation of stromal and immune cells in malignant tumor tissues using expression (ESTIMATE), microenvironment cell population counter (MCP-counter), and single sample GSEA (ssGSEA) algorithms revealed that the high-SNRPC group had a lower stromal score, lower abundance of endothelial cells and fibroblasts, and lower immune infiltration. Ultimately, a tumor immune dysfunction and exclusion (TIDE) analysis revealed that patients in the low-SNRPC group may be more sensitive to immune checkpoint inhibitor therapy.
Conclusion
SNRPC could serve as a promising prognostic and immunotherapeutic marker in HCC and might contribute to new directions and strategies for HCC treatment.
Keywords: SNRPC, Hepatocellular carcinoma, Prognosis, Immune checkpoint, Immunotherapy
Background
Liver cancer was the third leading cause of cancer death worldwide in 2020 [1]. Hepatocellular carcinoma (HCC) is the predominant form of primary liver cancer and accounts for 75–85% of cases. Chronic infection with hepatitis B virus (HBV) or hepatitis C virus (HCV), heavy alcohol intake, type 2 diabetes, and smoking have been related to the occurrence of HCC [1]. More than 1 million liver cancer deaths are estimated by 2030 [2]. Although surgical resection of early HCC is still the preferred treatment, the recurrence rate at 5 years can be as high as 70% [3]. In addition, even if liver transplantation can reduce the recurrence rate, the number of available donor organs is always insufficient relative to the demand [4]. Moreover, the majority (> 70%) of patients with advanced disease do not benefit from surgery [5]. For these patients, immunotherapy is a potential option. In recent years, emerging immunotherapies, such as immune checkpoint blockade and chimeric antigen receptor t cell (CAR-T) immunotherapy, have shown better therapeutic outcomes for some tumors [6, 7]. Nevertheless, responses to immunotherapy typically occur in a minority of patients, ranging from 20 to 50% depending on the tumor type [8]. Thus, it is especially important to identify candidate biomarkers to target patients who have the greatest likelihood of benefiting from immunotherapy [8]. In 2017, the Food and Drug Administration (FDA) approved the PD-1 inhibitor nivolumab (OPDIVO®), which marked the beginning of the immunotherapy era for HCC [9]. Compared with certain tumors, immunotherapy for liver cancer is still in its infancy. Additionally, knowledge of the biomarkers for predicting the responses or primary resistance to immunotherapeutic agents is crucial for establishing immunotherapy criteria for HCC patients in the future.
Genome-wide analysis of mRNA expression profiles has been used to screen prognostic and treatment-related markers in many cancers in recent years [10–13]. The N6-methyladenosine (m6A) methylation modification occurs at the 6th N atom of adenine, and it is the most common posttranscriptional modification on RNA and mediates > 60% of RNA methylation [14, 15]. Studies have shown that m6A RNA methylation plays an important role in tumor occurrence, development, and prognosis [16]. As an important m6A RNA methylation regulator, small nuclear ribonucleoprotein polypeptide C (SNRPC) encodes one of the specific protein components of the U1 small nuclear ribonucleoprotein (snRNP) particle, which is required for the formation of the spliceosome [17]. A recent study showed that SNRPC has the potential to promote HCC cell motility by inducing epithelial-mesenchymal transition [18]. In addition, SNRPC also contributes to sex bias in systemic autoimmune diseases and plays a role in spinal muscular atrophy pathogenesis [19, 20]. However, to our knowledge, the interaction between SNRPC and immunotherapy in HCC has not been reported. Therefore, in this study, we aimed to analyze the potential of SNRPC as a prognostic biomarker and predictor of immunotherapy responses in HCC to provide reference data for the clinical treatment of HCC.
In this study, we found that SNRPC expression was upregulated in HCC samples compared to normal samples. Next, the expression of SNRPC was validated as a risk factor for prognosis by a Kaplan-Meier analysis and employed to establish a nomogram with T pathologic stage. Furthermore, we investigated the correlation between SNRPC and immunotherapeutic reactions through a tumor immune dysfunction and exclusion (TIDE) analysis. These findings may contribute to new directions and strategies for HCC treatment.
Materials and methods
Public transcriptome profiling
In this study, we used 861 HCC cases from two public databases. After excluding samples without clinical annotation and survival information, 421 LIHC samples (50 normal samples labeled with 11A and 364 HCC samples labeled with 01A) were obtained from The Cancer Genome Atlas (TCGA) database and used as a training set. Additionally, 440 LIRI-JP cases (197 normal and 243 HCC) were obtained from the International Cancer Genome Consortium (ICGC) database and used as a validation set. The detailed clinical data of these samples are listed in Table 1. Moreover, both the expression data from the TCGA and ICGC databases were standardized by the log2 function in R.
Table 1.
Sex, tumor stage, and age of the samples from the ICGC database
| Number | Ratio | |
|---|---|---|
| Gender | ||
| Female | 68 | 0.261538462 |
| Male | 192 | 0.738461538 |
| Stage | ||
| Stage1 | 40 | 0.153846154 |
| Stage2 | 117 | 0.45 |
| Stage3 | 80 | 0.307692308 |
| Stage4 | 23 | 0.088461538 |
| Age | ||
| > 60 | 205 | 0.788461538 |
| < 60 | 55 | 0.211538462 |
Validation of the expression of SNRPC in HCC based on the TCGA and ICGC databases
To further investigate the expression of SNRPC in HCC, the Wilcoxon test was used to verify the significant difference in mRNA expression levels of SNRPC between the tumor and normal tissue samples based on the TCGA and ICGC databases. Moreover, the expression of SNRPC was further examined between 50 HCC tissue samples and 50 paired paracancerous tissue samples from the TCGA database (the ICGC database does not include paired cancer tissue and paracancerous tissue). Finally, the protein expression of SNRPC between HCC tissues and normal tissues was verified in the Human Protein Atlas (HPA) database (https://www.proteinatlas.org/).
Survival analysis of SNRPC
To evaluate whether SNRPC could impact the survival probability of HCC, all of the HCC samples in the TCGA and ICGC databases were divided into a high-SNRPC group and a low-SNRPC group based on the median expression value of SNRPC. Subsequently, survival probabilities were analyzed by a Kaplan-Meier analysis and log-rank test [21], with a p value < 0.05 considered statistically significant. In addition, time-dependent receiver operating characteristic (ROC) curves were plotted to assess the prognostic prediction performance of SNRPC, and the areas under the curve (AUCs) for overall survival (OS) were calculated using the survival ROC R package [22].
Construction of a nomogram for prognostic prediction
For better clinical application of SNRPC, we established a nomogram by univariate and multivariate Cox regression analysis to evaluate the probability of 1-, 3-, and 5-year OS for HCC patients via the rms R package [23] by integrating the clinical factors and SNRPC expression.
Functional annotation
To investigate the potential biological function of SNRPC in HCC, all of the differentially expressed genes between the high-SNRPC group and the low-SNRPC group in the TCGA database were ranked based on log2FC values. Then, a gene set enrichment analysis (GSEA) was implemented using the clusterProfiler package [24] to estimate the pathway activity changes in the above two groups. Similarly, the gene set variation (GSVA) package [25] was utilized to find the predominant pathways associated with SNRPC. Terms with values of FDR < 0.25 and p < 0.05 were retained.
ESTIMATE analysis
The Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression (ESTIMATE) algorithm can calculate immune scores and stromal scores via the estimate package in R [26], allowing us to quantify the immune and stromal components in a tumor. Therefore, the differences in immune scores, stromal scores, and tumor purity between the high-SNRPC group and the low-SNRPC group in the TCGA database were assessed by the Wilcoxon test.
Analysis of the tumor microenvironment
To gain further insights into the composition of the tumor microenvironment between the high-SNRPC group and low-SNRPC group in the TCGA database, a microenvironment cell population counter (MCP-counter) was used to quantify the numbers of immune cells, fibroblasts, and epithelial cells from each HCC sample based on marker genes [27]. Then, the single sample GSEA (ssGSEA) algorithm was used to calculate the immune infiltration of each sample, and significant differences in immune cell numbers were identified by analysis of variance (ANOVA).
Tumor immune dysfunction and exclusion analysis
Immune checkpoint pathways are associated with tumor immune escape. Therefore, immune checkpoint inhibitors can enhance anticancer immunity. Thus, we also employed the tumor immune dysfunction and exclusion (TIDE) algorithm [28] and subclass mapping [29] to predict the clinical response to immune checkpoint inhibitors between the high-SNRPC group and the low-SNRPC group. Moreover, we also calculated the correlation between SNRPC expression and TIDE scores.
Statistical analysis
All statistical tests were performed using R (version 3.5.2) software. Continuous data were analyzed using the Wilcoxon test or ANOVA. A p value of less than 0.05 was considered statistically significant in all analyses.
Results
SNRPC is upregulated in HCC
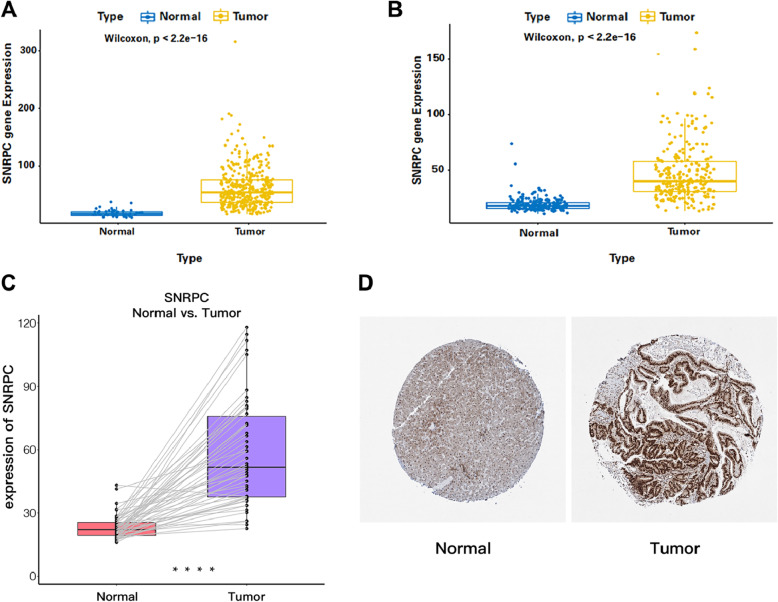
To further investigate the expression of SNRPC in HCC, SNRPC expression between the HCC and normal samples was analyzed using data from the TCGA and ICGC databases. Interestingly, the results of both the TCGA and ICGC databases suggested that SNRPC expression was upregulated in patients with HCC compared to normal samples (Fig. 1A, B). Moreover, the analysis of paired HCC and paracancerous tissues also revealed that SNRPC expression was upregulated in HCC tissues (Fig. 1C). Finally, a deeper staining level of SNRPC in HCC tissues compared to normal tissues revealed a higher protein expression of SNRPC in HCC (Fig. 1D).
Fig. 1.
A Expression of SNRPC in the TCGA-LIHC dataset. B Expression of SNRPC in the ICGC-LIRI dataset. C Expression of SNRPC between paired HCC and paracancerous tissues in the TCGA database. D Protein expression of SNRPC between HCC tissues and normal tissues in the HPA database
SNRPC is related to the survival of HCC
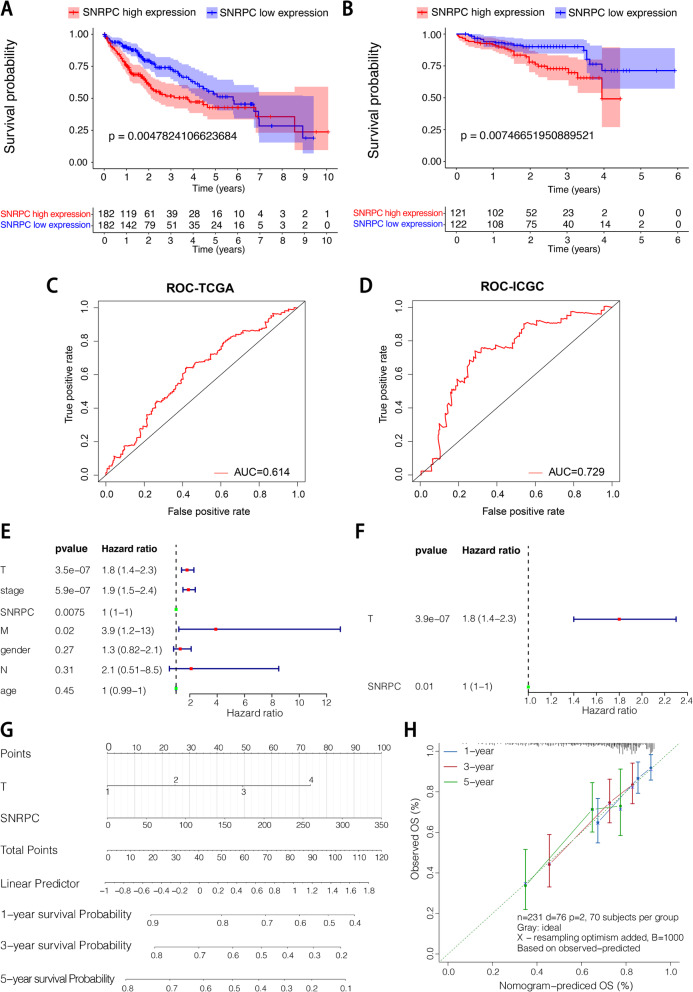
A Kaplan-Meier survival curve was drawn, and the results showed that in both the TCGA and ICGC databases, the low-SNRPC groups had a high survival probability relative to the high-SNRPC groups (p < 0.05) (Fig. 2A, B). In addition, as shown in the ROC curves, the AUCs of SNRPC were 0.614 and 0.729 in the TCGA and ICGC databases, respectively (Fig. 2C, D).
Fig. 2.
A KM survival curve of total survival time based on the SNRPC in the TCGA-LIHC dataset. B KM survival curve of total survival time based on the SNRPC in the ICGC-LIRI dataset. C ROC curve using samples from the TCGA database. D ROC curve using samples from the ICGC database. E Univariate Cox regression analysis. F Multivariate Cox regression analysis. G Nomogram for HCC 1-, 3- and 5-year prognosis. H Calibration curve
Construction of a nomogram for predicting HCC prognosis
The univariate and multivariate Cox regression analysis results suggested that the pathological T stage and SNRPC were significantly correlated with the prognosis of HCC (Fig. 2E, F). Therefore, a nomogram including T clinical factors and SNRPC expression values was constructed for predicting the 1-, 3-, and 5-year survival probabilities of HCC using samples from the TCGA database (Fig. 2G). A calibration curve demonstrated that our nomogram could accurately estimate the mortality of HCC (Fig. 2H).
Identification of SNRPC-related functional annotation
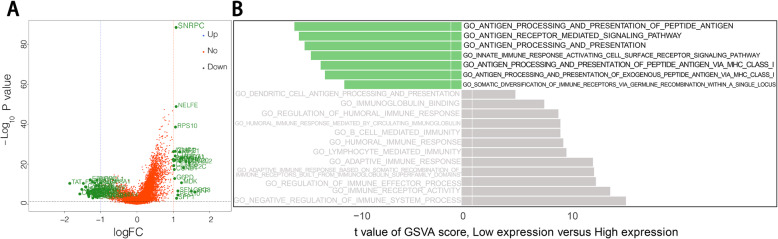
To gain insights into the potential functions related to SNRPC, we first identified 64 differentially expressed genes (DEGs) between the HCC samples of the high-SNRPC group and the low-SNRPC group (Fig. 3A). We found that catalytic activity-related biological processes, including catalytic_activity_acting_on_DNA, catalytic_step_2_spliceosome, and catalytic_activity_acting_on_RNA, were mainly activated in the high-SNRPC group (Table 2). Conversely, metabolism-related biological processes, including alcohol metabolism, amino acid metabolism, lipid metabolism, etc., were mainly activated in the low-SNRPC group (Table 2). Among the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, the cell cycle-, spliceosome-, ribosome-, and complement-related pathways were mainly activated in the high-SNRPC group while the metabolism-related pathways were mainly activated in the high-SNRPC group (Table 3). In addition, the GSVA results suggested that a large number of immune-related processes were disordered. For example, antigen processing-related biological processes were mainly upregulated in the high-SNRPC group while immune response-related biological processes were mainly upregulated in the low-SNRPC group (Fig. 3B, Table 4).
Fig. 3.
A Volcano map showing the low- and high-SNRPC groups. B Nineteen immune-related items enriched in the high- and low-SNRPC groups by GSVA
Table 2.
Top 10 GO terms annotated in the high- and low-SNRPC expression groups
| Terms | NES | p value | p-adjust |
|---|---|---|---|
| GO_ACUTE_INFLAMMATORY_RESPONSE | − 2.320055852 | 1.00E-10 | 5.02E-09 |
| GO_ALCOHOL_METABOLIC_PROCESS | − 1.785043835 | 1.00E-10 | 5.02E-09 |
| GO_ALPHA_AMINO_ACID_CATABOLIC_PROCESS | − 2.682453887 | 1.00E-10 | 5.02E-09 |
| GO_ALPHA_AMINO_ACID_METABOLIC_PROCESS | − 2.355743228 | 1.00E-10 | 5.02E-09 |
| GO_BLOOD_MICROPARTICLE | − 2.56496589 | 1.00E-10 | 5.02E-09 |
| GO_CATALYTIC_ACTIVITY_ACTING_ON_DNA | 1.82665719 | 1.00E-10 | 5.02E-09 |
| GO_CATALYTIC_ACTIVITY_ACTING_ON_RNA | 1.681035365 | 1.00E-10 | 5.02E-09 |
| GO_CATALYTIC_STEP_2_SPLICEOSOME | 1.970594215 | 1.00E-10 | 5.02E-09 |
| GO_CELLULAR_AMINO_ACID_CATABOLIC_PROCESS | − 2.691409978 | 1.00E-10 | 5.02E-09 |
| GO_CELLULAR_LIPID_CATABOLIC_PROCESS | − 2.125968674 | 1.00E-10 | 5.02E-09 |
Table 3.
Top 10 KEGG terms enriched in the high- and low-SNRPC expression groups
| Terms | NES | p value | p-adjust |
|---|---|---|---|
| KEGG_CELL_CYCLE | 1.858825048 | 6.01E-10 | 1.38E-08 |
| KEGG_COMPLEMENT_AND_COAGULATION_CASCADES | − 2.985586938 | 1.00E-10 | 3.07E-09 |
| KEGG_DNA_REPLICATION | 1.97287282 | 2.06E-08 | 3.78E-07 |
| KEGG_DRUG_METABOLISM_CYTOCHROME_P450 | − 2.545914368 | 1.00E-10 | 3.07E-09 |
| KEGG_FATTY_ACID_METABOLISM | − 2.639833598 | 1.00E-10 | 3.07E-09 |
| KEGG_PPAR_SIGNALING_PATHWAY | − 2.409646016 | 5.09E-10 | 1.34E-08 |
| KEGG_RETINOL_METABOLISM | − 2.808138608 | 1.00E-10 | 3.07E-09 |
| KEGG_RIBOSOME | 2.545828247 | 1.00E-10 | 3.07E-09 |
| KEGG_SPLICEOSOME | 2.04347142 | 1.00E-10 | 3.07E-09 |
| KEGG_VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION | − 2.486033153 | 4.87E-09 | 9.96E-08 |
Table 4.
Nineteen immune-related items enriched in the high- and low-SNRPC expression groups by GSVA
| Type | Average GSVA score | p | |
|---|---|---|---|
| Low_SNRPC | High_SNRPC | ||
| GO_HUMORAL_IMMUNE_RESPONSE | 0.243174558 | − 0.171333299 | < 2.2e−16 |
| GO_REGULATION_OF_HUMORAL_IMMUNE_RESPONSE | 0.217516904 | − 0.152275441 | < 2.2e−16 |
| GO_HUMORAL_IMMUNE_RESPONSE_MEDIATED_BY_CIRCULATING_IMMUNOGLOBULIN | 0.240056081 | − 0.194481039 | < 2.2e−16 |
| GO_REGULATION_OF_IMMUNE_EFFECTOR_PROCESS | 0.231855592 | − 0.168855904 | < 2.2e−16 |
| GO_ADAPTIVE_IMMUNE_RESPONSE_BASED_ON_SOMATIC_RECOMBINATION_OF_IMMUNE_RECEPTORS_BUILT_FROM_IMMUNOGLOBULIN_SUPERFAMILY_DOMAINS | 0.225524044 | − 0.17393338 | < 2.2e−16 |
| GO_B_CELL_MEDIATED_IMMUNITY | 0.240056081 | − 0.194481039 | < 2.2e−16 |
| GO_INNATE_IMMUNE_RESPONSE_ACTIVATING_CELL_SURFACE_RECEPTOR_SIGNALING_PATHWAY | − 0.261145258 | 0.190182919 | < 2.2e−16 |
| GO_ADAPTIVE_IMMUNE_RESPONSE | 0.227846975 | − 0.175474417 | < 2.2e−16 |
| GO_LYMPHOCYTE_MEDIATED_IMMUNITY | 0.234004969 | − 0.178173068 | < 2.2e−16 |
| GO_NEGATIVE_REGULATION_OF_IMMUNE_SYSTEM_PROCESS | 0.229845123 | − 0.166309991 | < 2.2e−16 |
| GO_IMMUNE_RECEPTOR_ACTIVITY | 0.196420292 | − 0.141311054 | < 2.2e−16 |
| GO_SOMATIC_DIVERSIFICATION_OF_IMMUNE_RECEPTORS_VIA_GERMLINE_RECOMBINATION_WITHIN_A_SINGLE_LOCUS | − 0.222668922 | 0.155772825 | 1e−13 |
| GO_IMMUNOGLOBULIN_BINDING | 0.186800511 | − 0.125978607 | < 2.2e−16 |
| GO_ANTIGEN_PROCESSING_AND_PRESENTATION_OF_EXOGENOUS_PEPTIDE_ANTIGEN_VIA_MHC_CLASS_I | − 0.237508396 | 0.180189519 | < 2.2e−16 |
| GO_ANTIGEN_PROCESSING_AND_PRESENTATION_OF_PEPTIDE_ANTIGEN | − 0.259010315 | 0.210540976 | < 2.2e−16 |
| GO_ANTIGEN_PROCESSING_AND_PRESENTATION_OF_PEPTIDE_ANTIGEN_VIA_MHC_CLASS_I | − 0.240473386 | 0.183322272 | < 2.2e−16 |
| GO_ANTIGEN_PROCESSING_AND_PRESENTATION | − 0.230240714 | 0.187319947 | < 2.2e−16 |
| GO_ANTIGEN_RECEPTOR_MEDIATED_SIGNALING_PATHWAY | − 0.231518181 | 0.178373361 | < 2.2e−16 |
| GO_DENDRITIC_CELL_ANTIGEN_PROCESSING_AND_PRESENTATION | 0.180625588 | 0.005442914 | 6.8e−07 |
ESTIMATE analysis
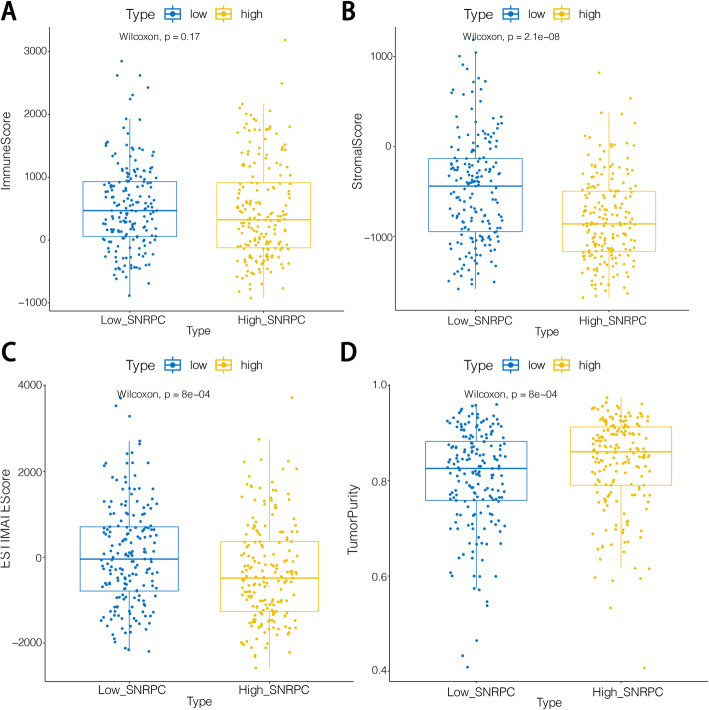
In our study, we found that except for the immune scores, the stromal scores, ESTIMATE scores, and tumor purity were significantly different between the high- and low-SNRPC groups (Fig. 4). We also found that a lower stromal score and higher tumor purity were associated with poor survival in HCC patients.
Fig. 4.
A Box diagram showing the immune scores in the high- and low-SNRPC groups. B Box diagram showing the stromal scores in the high- and low-SNRPC groups. C Box diagram demonstrating the ESTIMATE scores in the high- and low-SNRPC groups. D Box diagram demonstrating the tumor purity in the high- and low-SNRPC groups
Correlation between SNRPC expression and tumor microenvironment
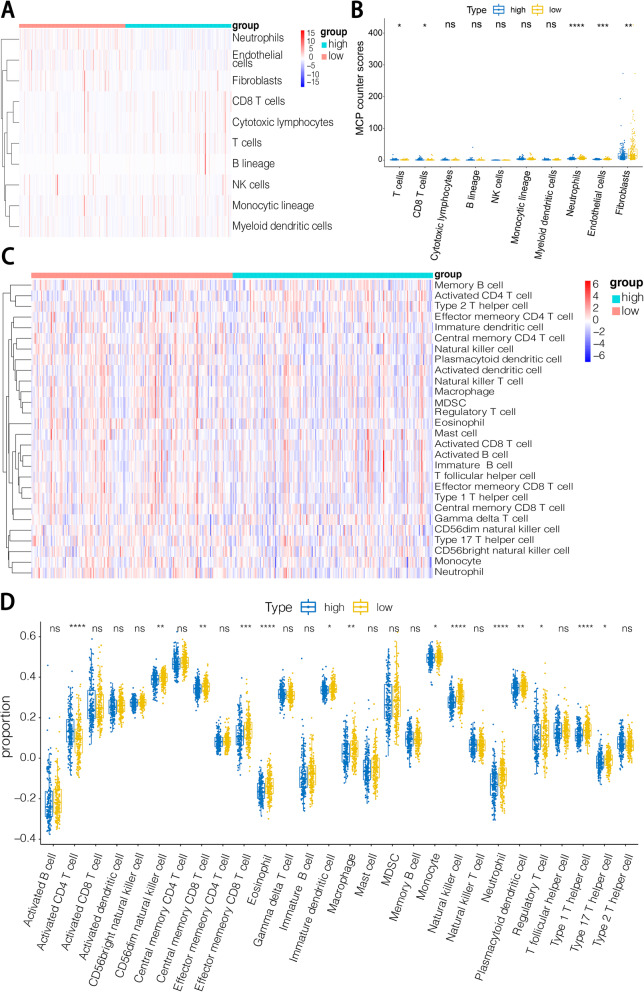
MCP-counter and ssGSEA analyses were conducted to clarify the abundance of various cells in the tumor microenvironment (TME). Immune cells, such as T cells, CD8+T cells, neutrophils, and stromal cells, such as endothelial cells and fibroblasts, were significantly different between the high- and low-SNRPC groups. Among these cell types, neutrophils, endothelial cells, and fibroblasts were increased in the low-SNRPC group while other types were decreased (Fig. 5A, B). Through the ssGSEA, additional subtypes of immune cells were counted. In the low-SNRPC group, only activated CD4+T cells were significantly decreased while CD56dim natural killer cells, central memory CD8+T cells, effector memory CD8+T cells, eosinophils, immature dendritic cells, macrophages, memory B cells, monocytes, natural killer T cells, natural killer T cells, plasmacytoid dendritic cells, regulatory T cells, type 1 T helper cells, and type 17 T helper cells were all increased (Fig. 5C, D).
Fig. 5.
A Abundance of TME cells between the high- and low-SNRPC groups calculated by MCP-counter. B Box plot showing the proportion of TME cells between the high- and low-SNRPC groups analyzed by MCP-counter. C, D Heat map and box plot showing the infiltration of immune cells between the high- and low-SNRPC groups calculated by ssGSEA
SNRPC expression could predict the immunotherapeutic response of HCC
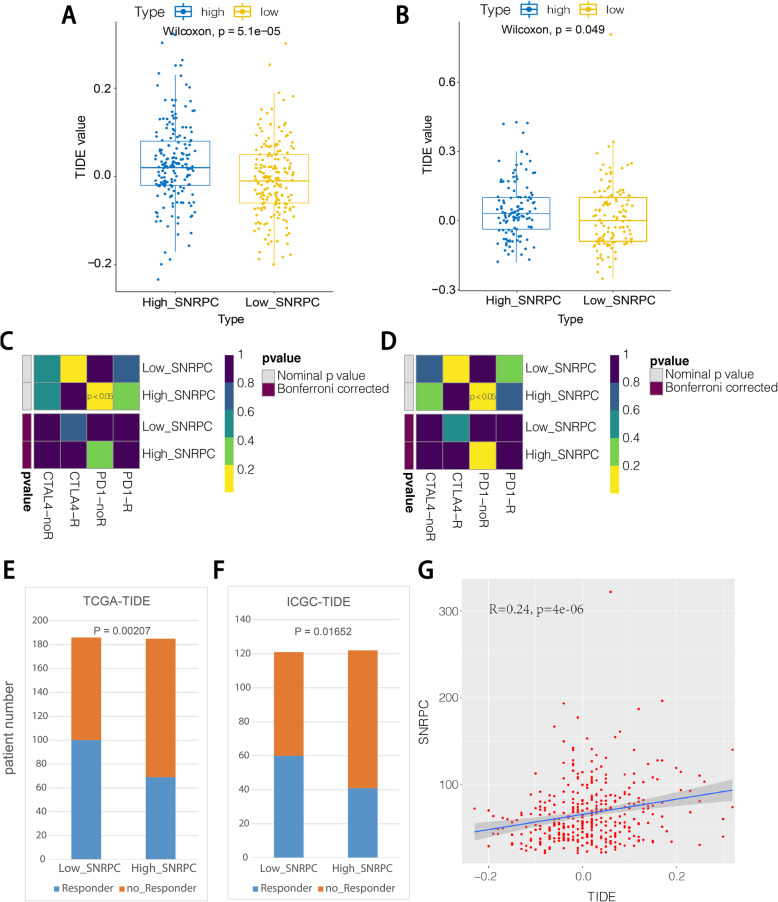
The TIDE score is used to predict the efficacy of anti-PD1 and anti-CTLA4 treatments [27]. As shown in Fig. 6A, B, both the high-SNRPC groups showed higher TIDE scores in the TCGA and ICGC databases. Consistently, both the high-SNRPC groups showed no response to anti-PD1 therapy in the TCGA and ICGC databases by subclass mapping analysis (Fig. 6C, D). Furthermore, both the low-SNRPC groups presented more patients with the response to immune checkpoint inhibitor therapy in the TCGA and ICGC databases (Fig. 6E, F). More importantly, the correlation analysis suggested that the expression of SNRPC was positively correlated with the TIDE scores (Fig. 6G). Thus, these findings revealed that patients in the low-SNRPC group may be more sensitive to immune checkpoint inhibitor therapy.
Fig. 6.
A TIDE scores of the high- and low-SNRPC groups using samples from the TCGA database. B TIDE scores of the high- and low-SNRPC groups using data from the ICGC database. C Immunotherapy responses between the high- and low-SNRPC groups in the TCGA database. D Immunotherapy responses between the high- and low-SNRPC groups in the ICGC database. E Proportion of patients who responded to immunotherapy in the high-SNRPC and low-SNRPC groups in the TCGA. F Proportion of patients who respond to immunotherapy in the high-SNRPC and low-SNRPC groups in the ICGC. G Correlation analysis between SNRPC expression and TIDE score
Discussion
HCC accounts for 75–85% of liver cancers and is also the third leading cause of cancer-related death globally, which has a poor prognosis [1]. Studies have shown that multiple genes play an important role in HCC progression [30–34]. Currently, immunotherapy has already become a viable option for advanced patients. However, not all patients show complete responses to such immunotherapies [35]. As a result, a reliable biomarker for predicting HCC prognosis and immunotherapy response is urgently needed.
In the present study, we further analyzed the role of SNRPC in HCC. SNRPC encodes one of the specific protein components of U1snRNP, which plays an essential role in the formation of the spliceosome. U1snRNP is reported to suppress the migration and invasion of multiple types of tumor cells [36]. Additionally, genes of the snRNP polypeptide family also exert an important role in the tumorigenesis and prognosis of cancers [37, 38]. For example, the high expression of snRNP polypeptide G, which is another indispensable component in the formation of snRNPs, is positively associated with the occurrence, progression and severity of breast, lung, and colon cancers [39–41]. Moreover, the snRNP polypeptide N is highly expressed in colorectal cancer tissues and involved in the progression of colorectal cancer [42]. A recent study demonstrated that SNRPC has the potential to enhance HCC cell motility by promoting epithelial-mesenchymal transition, which might contribute to a poor prognosis [18]. However, few studies have focused on the role of SNRPC in the immunotherapy of tumors. Therefore, this study aimed to reveal the effect of SNRPC on the prognosis and immunotherapy of HCC. In our study, we first found that SNRPC was overexpressed in HCC and that higher expression of SNRPC was associated with a poorer prognosis. Then, we constructed a nomogram containing T clinical factors and SNRPC expression values, which can precisely predict 1-, 3-, and 5-year survival probabilities. To gain detailed insights into the mechanisms of SNRPC, we performed functional analyses and found that SNRPC was mainly related to protein metabolism and the immune process. Interestingly, a TME analysis indicated that natural killer cells, CD8+T cells, macrophages, neutrophils, endothelial cells, and fibroblasts were less infiltrated in tissues with high SNRPC expression. These results implied that both stromal cells and immune cells were abundant, which was beneficial for HCC prognosis. Ultimately, we employed the TIDE analysis method to predict the efficacy of anti-PD1 and anti-CTLA4 treatments. The results showed that the high-SNRPC group failed to respond to immune checkpoint inhibitor (ICI) therapy, especially to a PD-1 inhibitor.
At present, many clinical prognostic biomarkers are available for HCC, such as alpha-fetoprotein (AFP), alpha-fetoprotein lens culinaris agglutin-3 (AFP-L3), and des-γ-carboxy prothrombin (DCP). The relationship between AFP and prognosis has been extensively studied [43]. Increasing AFP values are associated with lower survival and a higher tumor recurrence rate in patients at very early or early stages as well as poor prognosis in patients undergoing advanced HCC [43]. By comparison, overexpressed SNRPC is correlated with low infiltration of immune cells and stromal cells, poor prognosis, and poor immunotherapy response. However, SNRPC expression remains relatively complex to detect compared with AFP, which can be detected in serum. We hope that in the future, SNRPC could be used in combination with other biomarkers for the clinical staging system and prognosis prediction of HCC.
At present, numerous biomarkers have been employed to select candidates for immunotherapy. PD-L1 expression is the most widely adopted predictor of the immunotherapeutic response rate. High PD-L1 expression is associated with an increased response rate and clinical benefit in anti-PD-1/anti-PD-L1 therapy [44]. However, PD-L1 expression is not associated with treatment response in some patients. Additionally, the TIDE score can also directly guide the use of ICIs. Patients with higher TIDE scores have higher antitumor immune escape opportunities, resulting in lower ICI treatment response rates [28]. Moreover, the TIDE score has been shown to have higher accuracy than the PD-L1 expression level in predicting the survival outcome of cancer patients treated with ICI agents [45–47]. In the current study, the TIDE analysis revealed that patients in the low-SNRPC group may be more sensitive to immune checkpoint inhibitor therapy. Therefore, we speculated that SNRPC might be associated with the immunotherapeutic response of HCC.
Recently, novel strategies have been identified to enhance the effect of anti-PD-1/PD-L1 by promoting T cell infiltration, such as an anti-TGF-β/PD-L1 bispecific antibody and anti-PD-1 combining anti-CTLA-4 [48, 49]. Combining immunotherapy with anti-VEGF agents also displayed a good therapeutic effect in some HCC cases. Consequently, SNRPC might contribute to improving the effectiveness of anti-PD-1/PD-L1 by regulating the expression of SNRPC.
Conclusion
In this study, we showed that the HCC biomarker SNRPC was overexpressed in tumor tissues and linked to a worse prognosis in patients with HCC. Functional analysis showed that SNRPC was mainly related to protein metabolism and the immune process. In addition, we found that upregulated SNRPC may be associated with a less sensitive response to immune checkpoint inhibitor therapy. Therefore, SNRPC may be a promising prognostic and immunotherapeutic marker for HCC. Nevertheless, we only included anti-PD1 and anti-CTLA4 treatments in our TIDE analysis rather than considering more promising immunotherapy methods such as combination therapies. In addition, we did not choose a proposed cutoff for predicting poor prognosis and immunotherapeutic response. Hence, follow-up experiments and clinical validation are extremely necessary.
Acknowledgements
We thank LetPub (www.letpub.com) and American Journal Experts (http://bit.ly/AJE-HS) for their linguistic assistance during the preparation of this manuscript.
Abbreviations
- HCC
Hepatocellular carcinoma
- HBV
Hepatitis B virus
- HCV
Hepatitis C virus
- CAR-T
Chimeric antigen receptor T cell
- FDA
Food and Drug Administration
- m6A
N6-methyladenosine
- SNRPC
Small nuclear ribonucleoprotein polypeptide C
- snRNP
Small nuclear ribonucleoprotein
- TIDE
Tumor immune dysfunction and exclusion
- TCGA
The Cancer Genome Atlas
- ICGC
International Cancer Genome Consortium
- HPA
Human Protein Atlas
- ROC
Receiver operating characteristic
- AUCs
Areas under the curve
- OS
Overall survival
- GSEA
Gene set enrichment analysis
- GSVA
Gene set variation
- ESTIMATE
Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression
- MCP-counter
Microenvironment Cell Population counter
- ssGSEA
Single sample GSEA
- ANOVA
Analysis of variance
- DEGs
Differential expression genes
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- TME
Tumor microenvironment
- AFP
Alpha-fetoprotein
- AFP-L3
Alpha-fetoprotein lens culinaris agglutin-3
- DCP
Des-γ-carboxy prothrombin
- ICIs
Immune checkpoint inhibitors
Authors’ contributions
All authors were responsible for the analysis, interpretation of data, and graphing. JH.C drafted the manuscript. JH.C and ML.Z revised the manuscript. All authors read and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
The datasets analyzed during the current study are available in the TGCA [https://portal.gdc.cancer.gov], ICGC [https://dcc.icgc.org/] public database, and HPA [https://www.proteinatlas.org/].
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. 10.3322/caac.21660 [DOI] [PubMed]
- 2.Karb, D.B. and S.N. Sclair, Hepatocellular carcinoma, in liver disease. 2019. 141-154. Springer International Publishing, Cham.
- 3.Llovet JM, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 4.Vivarelli M, et al. Liver transplantation for hepatocellular carcinoma under calcineurin inhibitors: reassessment of risk factors for tumor recurrence. Ann Surg. 2008;248(5):857–862. doi: 10.1097/SLA.0b013e3181896278. [DOI] [PubMed] [Google Scholar]
- 5.Thomas MB, et al. Hepatocellular carcinoma: consensus recommendations of the National Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol. 2010;28(25):3994–4005. doi: 10.1200/JCO.2010.28.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fournier C, et al. Trial Watch: adoptively transferred cells for anticancer immunotherapy. Oncoimmunology. 2017;6(11):e1363139. doi: 10.1080/2162402X.2017.1363139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smyth MJ, et al. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol. 2016;13(3):143–158. doi: 10.1038/nrclinonc.2015.209. [DOI] [PubMed] [Google Scholar]
- 8.Llovet JM, et al. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15(10):599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston MP, Khakoo SI. Immunotherapy for hepatocellular carcinoma: current and future. World J Gastroenterol. 2019;25(24):2977–2989. doi: 10.3748/wjg.v25.i24.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyault S, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45(1):42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 11.Chiang DY, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68(16):6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoshida Y, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69(18):7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JS, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 14.Du K, et al. m(6)A RNA methylation controls neural development and is involved in human diseases. Mol Neurobiol. 2019;56(3):1596–1606. doi: 10.1007/s12035-018-1138-1. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, et al. Roles of RNA methylation by means of N(6)-methyladenosine (m(6)A) in human cancers. Cancer Lett. 2017;408:112–120. doi: 10.1016/j.canlet.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, et al. Identification of a m(6)A RNA methylation regulators-based signature for predicting the prognosis of clear cell renal carcinoma. Cancer Cell Int. 2020;20:157. doi: 10.1186/s12935-020-01238-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Libri D, et al. A role for the Psi-U mismatch in the recognition of the 5' splice site of yeast introns by the U1 small nuclear ribonucleoprotein particle. J Biol Chem. 2002;277(20):18173–18181. doi: 10.1074/jbc.M112460200. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. SNRPC promotes hepatocellular carcinoma cell motility by inducing epithelial-mesenchymal transition. FEBS Open Bio. 2021;11(6):1757–70. 10.1002/2211-5463.13175 [DOI] [PMC free article] [PubMed]
- 19.Linden M, et al. Sex influences eQTL effects of SLE and Sjogren's syndrome-associated genetic polymorphisms. Biol Sex Differ. 2017;8(1):34. doi: 10.1186/s13293-017-0153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng D, et al. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25(1):71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Heagerty PJ, Saha-Chaudhuri P, Saha-Chaudhuri MP. Package ‘survivalROC’ [J] 2013. [Google Scholar]
- 22.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61(1):92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 23.Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26(8):1364–70. 10.1200/JCO.2007.12.9791 [DOI] [PubMed]
- 24.Yu G, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hänzelmann SH, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. Published 2013 Jan 16. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed]
- 26.Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. 10.1038/ncomms3612 [DOI] [PMC free article] [PubMed]
- 27.Becht E, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang P, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoshida Y, et al. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS One. 2007;2(11):e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao S, et al. Reduced expression of DACT2 promotes hepatocellular carcinoma progression: involvement of methylation-mediated gene silencing. World J Surg Oncol. 2013;11:57. doi: 10.1186/1477-7819-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, et al. Clinical significance of mitofusin-2 and its signaling pathways in hepatocellular carcinoma. World J Surg Oncol. 2016;14(1):179. doi: 10.1186/s12957-016-0922-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu M, et al. Identification and validation of three core genes in p53 signaling pathway in hepatitis B virus-related hepatocellular carcinoma. World J Surg Oncol. 2021;19(1):66. doi: 10.1186/s12957-021-02174-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo W, et al. The role of kinesin KIF18A in the invasion and metastasis of hepatocellular carcinoma. World J Surg Oncol. 2018;16(1):36. doi: 10.1186/s12957-018-1342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, et al. Low expression of organic anion-transporting polypeptide 1B3 predicts a poor prognosis in hepatocellular carcinoma. World J Surg Oncol. 2020;18(1):127. doi: 10.1186/s12957-020-01891-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber JS, et al. Toxicities of Immunotherapy for the Practitioner. J Clin Oncol. 2015;33(18):2092–2099. doi: 10.1200/JCO.2014.60.0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh JM, et al. U1 snRNP regulates cancer cell migration and invasion in vitro. Nat Commun. 2020;11(1):1. doi: 10.1038/s41467-019-13993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mabonga L, Kappo AP. The oncogenic potential of small nuclear ribonucleoprotein polypeptide G: a comprehensive and perspective view. Am J Transl Res. 2019;11(11):6702–16. Published 2019 Nov 15. [PMC free article] [PubMed]
- 38.Tu J, et al. Bioinformatics analysis of molecular genetic targets and key pathways for hepatocellular carcinoma. Onco Targets Ther. 2019;12:5153–5162. doi: 10.2147/OTT.S198802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, et al. PACT is a negative regulator of p53 and essential for cell growth and embryonic development. Proc Natl Acad Sci U S A. 2007;104(19):7951–7956. doi: 10.1073/pnas.0701916104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hull R, et al. The Drosophila retinoblastoma binding protein 6 family member has two isoforms and is potentially involved in embryonic patterning. Int J Mol Sci. 2015;16(5):10242–10266. doi: 10.3390/ijms160510242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ezkurdia I, et al. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum Mol Genet. 2014;23(22):5866–5878. doi: 10.1093/hmg/ddu309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji M, et al. Small nuclear ribonucleoprotein polypeptide N accelerates malignant progression and poor prognosis in colorectal cancer transcriptionally regulated by E2F8. Front Oncol. 2020;10:561287. Published 2020 Nov 2. 10.3389/fonc.2020.561287 [DOI] [PMC free article] [PubMed]
- 43.Pinero F, Dirchwolf M, Pessoa MG. Biomarkers in hepatocellular carcinoma: diagnosis, prognosis and treatment response assessment. Cells. 2020;9(6):1370. Published 2020 Jun 1. 10.3390/cells9061370 [DOI] [PMC free article] [PubMed]
- 44.Yi M, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer. 2018;17(1):129. doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaderbhai C, Tharin Z, Ghiringhelli F. The role of molecular profiling to predict the response to immune checkpoint inhibitors in lung cancer. Cancers (Basel). 2019;11(2):201. Published 2019 Feb 10. 10.3390/cancers11020201 [DOI] [PMC free article] [PubMed]
- 46.Keenan TE, Burke KP, Van Allen EM. Genomic correlates of response to immune checkpoint blockade. Nat Med. 2019;25(3):389–402. doi: 10.1038/s41591-019-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S, et al. Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction. Elife. 2019;8:e49020. Published 2019 Nov 26. 10.7554/eLife.49020 [DOI] [PMC free article] [PubMed]
- 48.Yi M, et al. The construction, expression, and enhanced anti-tumor activity of YM101: a bispecific antibody simultaneously targeting TGF-beta and PD-L1. J Hematol Oncol. 2021;14(1):27. doi: 10.1186/s13045-021-01045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu K, et al. The efficacy and safety of combination of PD-1 and CTLA-4 inhibitors: a meta-analysis. Exp Hematol Oncol. 2019;8:26. doi: 10.1186/s40164-019-0150-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available in the TGCA [https://portal.gdc.cancer.gov], ICGC [https://dcc.icgc.org/] public database, and HPA [https://www.proteinatlas.org/].