Significance
Prostate infections and inflammation are potential initiating factors in prostate cancer development. Here, we investigated whether bacterial infections are associated with the presence of the most common prostate cancer oncogenic gene fusion, TMPRSS2:ERG, in early precursor lesions. We provide evidence that TMPRSS2:ERG (ERG+) gene fusions can initiate in early prostate cancer risk factor lesions, including proliferative inflammatory atrophy in the setting of prostate infection. We further demonstrate that these infection-associated ERG+ precursor lesions are transitioning to early invasive cancer. Overall, we provide evidence that, in at least a subset of cases, infection-induced TMPRSS2:ERG gene fusions are an early alteration in the carcinogenic process.
Keywords: prostate cancer, infection, inflammation, TMPRSS2:ERG fusion, colibactin
Abstract
Prostate adenocarcinoma is the second most commonly diagnosed cancer in men worldwide, and the initiating factors are unknown. Oncogenic TMPRSS2:ERG (ERG+) gene fusions are facilitated by DNA breaks and occur in up to 50% of prostate cancers. Infection-driven inflammation is implicated in the formation of ERG+ fusions, and we hypothesized that these fusions initiate in early inflammation-associated prostate cancer precursor lesions, such as proliferative inflammatory atrophy (PIA), prior to cancer development. We investigated whether bacterial prostatitis is associated with ERG+ precancerous lesions in unique cases with active bacterial infections at the time of radical prostatectomy. We identified a high frequency of ERG+ non–neoplastic-appearing glands in these cases, including ERG+ PIA transitioning to early invasive cancer. These lesions were positive for ERG protein by immunohistochemistry and ERG messenger RNA by in situ hybridization. We additionally verified TMPRSS2:ERG genomic rearrangements in precursor lesions using tricolor fluorescence in situ hybridization. Identification of rearrangement patterns combined with whole-prostate mapping in three dimensions confirmed multiple (up to eight) distinct ERG+ precancerous lesions in infected cases. We further identified the pathogen-derived genotoxin colibactin as a potential source of DNA breaks in clinical cases as well as cultured prostate cells. Overall, we provide evidence that bacterial infections can initiate driver gene alterations in prostate cancer. In addition, our observations indicate that infection-induced ERG+ fusions are an early alteration in the carcinogenic process and that PIA may serve as a direct precursor to prostate cancer.
Prostate cancer is the most commonly diagnosed cancer (excluding skin cancer) in men in the United States, and currently afflicts ∼1.3 million men worldwide (1). The only known risk factors for prostate cancer are advanced age, family history, and African ancestry; the cause of the disease is, to date, unknown. Prostate infections and inflammation are potential initiating factors in prostate cancer development (2–5). Specifically, prostate infection or other inflammatory stimuli may drive the formation of proliferative inflammatory atrophy (PIA), a putative prostate cancer precursor lesion (6–8). The atrophic luminal epithelial cells in PIA are markedly proliferative compared to that of normal-appearing epithelium, are enriched with cells of an intermediate phenotype that have properties of both basal and luminal epithelial cells (9), and are sometimes observed in direct transition with prostatic intraepithelial neoplasia (PIN, the most accepted direct precursor to prostate cancer) and adenocarcinoma (10). The intermediate phenotype luminal epithelial cells in PIA, termed “intermediate cells,” are purported tumor-initiating cells in prostate cancer (9, 11).
Depending on the population, up to 50% of prostate cancers harbor gene fusions between the androgen-regulated gene TMPRSS2 and erythroblast transformation–specific (ETS) transcription factor ERG, an alteration that leads to overexpression of the oncogenic transcription factor ERG (12, 13). TMPRSS2:ERG gene fusions are an early clonal oncogenic event, as essentially all invasive cancer cells within an ERG-positive (ERG+) cancer share the same rearrangement (14). Likewise, in mouse models, transgenic expression of ERG in prostate epithelial cells induces PIN (15, 16) and carcinoma when combined with other genomic alterations (17–19). Androgen signaling induces three-dimensional (3D) spatial proximity between the TMPRSS2 and ERG gene loci, and DNA double-strand breaks (DSB) facilitate subsequent fusion events (20–22). The etiologic factors causing DSB that lead to TMPRSS2:ERG rearrangements are not completely defined. Importantly, inflammation induced by bacterial lipopolysaccharide (LPS) has been demonstrated in both in vitro and in vivo models to induce the formation of TMPRSS2:ERG gene fusions (23). In these models, inflammation-induced oxidative stress serves as a major source of DSB.
In addition to stimulating inflammation, bacteria can produce potent genotoxins that incite DNA damage, as has been described in the pathogenesis of colorectal cancer (24–30). Colibactin is a cryptic bacterial genotoxin produced by a nonribosomal peptide synthetase and polyketide synthase gene cluster (pks island) carried in some strains of Escherichia coli and closely related species (24). Infection of human cells with colibactin-producing E. coli strains induces DNA damage [i.e., DNA adducts, DSB, DNA cross-links, and genomic instability (24–28)]. Colibactin-producing bacteria are present in the human gastrointestinal microbiota and are associated with colorectal cancer (24, 25, 29–31). Colibactin-producing E. coli are also known uropathogens and are common among isolates from patients with urinary tract infections (32) and men with bacterial prostatitis (33).
Since prostate infections and infection-associated inflammation have been implicated in both the formation of PIA and the formation of TMPRSS2:ERG gene fusions, we questioned whether bacterial infections are associated with oncogenic TMPRSS2:ERG gene fusions in human prostate specimens. Furthermore, we hypothesized that infection-associated TMPRSS2:ERG fusions may initiate in early inflammation-associated prostate cancer risk factor lesions, such as PIA, prior to cancer development.
Results
Identification of Radical Prostatectomy Specimens Harboring Bacterial Infections.
We began by identifying a series of radical prostatectomy specimens that were suspicious for prostatic infections at the time of surgery. Our rationale was that even though these specimens had preexisting cancer, we could examine the nonneoplastic regions of these highly inflamed cases to investigate the effects of infection-induced inflammation on the human prostate. Florid acute or granulomatous chronic inflammation observed at radical prostatectomy is rare and likely indicative of an active prostate infection. In screening 1,341 cases (Materials and Methods), we identified 15 cases that were suspicious for the presence of an active prostate infection at the time when the radical prostatectomy was performed (SI Appendix, Fig. S1 and Table S1). For all of these cases, the entire prostate gland was processed for sampling and we mapped all invasive carcinoma foci in 3D.
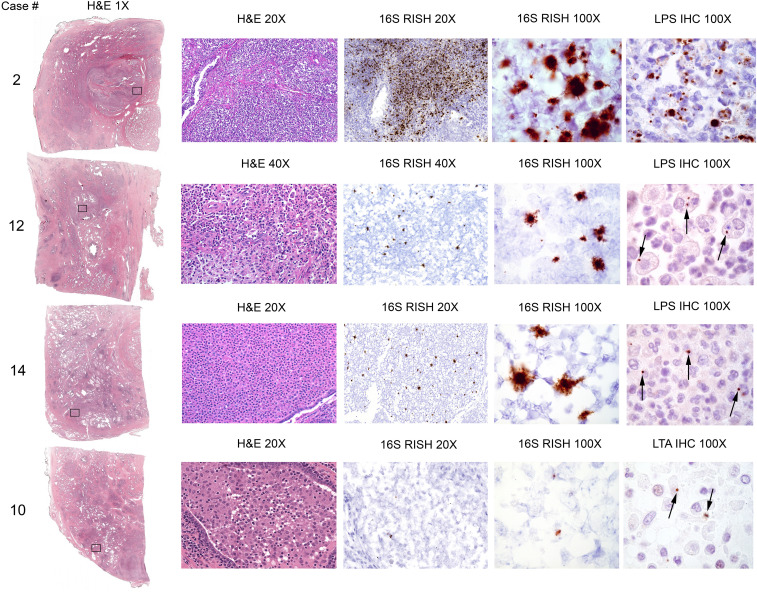
To further evaluate whether these cases harbored bacterial infections, we assessed all blocks that contained acute inflammation (defined as the presence of neutrophils, average nine blocks per case) using RNA in situ hybridization (RISH) with universal probes for bacterial 16S ribosomal RNA (rRNA) as well as immunohistochemistry (IHC) for LPS (Gram-negative bacteria) and lipoteichoic acid (LTA, Gram-positive bacteria). RISH and IHC assay validation data are in SI Appendix, Fig. S2. We detected bacteria in seven of the cases, six of which were LPS positive and one that was LTA positive (Fig. 1 and SI Appendix, Table S1 and Fig. S3). Importantly, even though bacteria were not identified in the remaining cases, the cases are still highly suspicious for infection due to the presence of florid inflammation. It is possible that the infectious agent cleared prior to the radical prostatectomy since in the positive cases, only a small number of glands were bacteria positive, despite florid inflammation across the bacteria-negative glands. Special stains for acid fast bacilli (auramine/rhodamine) and fungi (methenamine silver) were performed on a subset of the cases during the initial diagnostic workup and were negative (SI Appendix, Table S1). Noninflamed regions of normal-appearing prostate and prostate adenocarcinoma were all negative for bacteria (SI Appendix, Fig. S4). When present, bacteria were largely confined to acinar lumens where neutrophils and macrophages were present and were often observed to be internalized by these cells (Fig. 1 and SI Appendix, Fig. S3).
Fig. 1.
Detection of bacteria in highly inflamed radical prostatectomy specimens. Examples of detection of bacteria by 16S RISH (universal detection of bacteria), LPS IHC (specific for Gram-negative bacteria, case 2, 12, and 14), and LTA IHC (specific for Gram-positive bacteria, case 10). Bacterial cells were often observed within infiltrating immune cells (arrows). Objective magnification denoted.
High Frequency of Isolated ERG+ Precursor Lesions in Association with Prostate Infection.
Exposing androgen-responsive prostate cancer cells to tumor necrosis factor alpha (TNF-α) as a means to recapitulate prostatic inflammation induces TMPRSS2:ERG gene fusion formation in vitro (23). We therefore hypothesized that in these highly inflamed cases, early molecular alterations indicative of cancer initiation (e.g., TMPRSS2:ERG fusion events) would be present in a subset of inflammation-associated precursor lesions, such as PIA. Isolated ERG+ high-grade PIN (HGPIN) that is not adjacent to ERG+ cancer occurs at a rare frequency between 2% in a series of radical prostatectomies (34) and 7% in a series of cystoprostatectomies (35). Other ERG+ lesions, such as low-grade PIN (LGPIN) and PIA, that are not directly adjacent to ERG+ cancers, are rare in radical prostatectomy specimens. In fact, TMPRSS2:ERG fusions were absent in 38 PIA samples examined on tissue microarrays (36). The analysis of over 200,000 benign glands in cross sections of 132 whole-mount radical prostatectomy specimens identified small single aggregates (3 to 5 glands) of ERG+ LGPIN in three (2.3%) of the cases (37). Likewise, a study using ERG IHC on 169 whole tissue sections from 41 prostatectomies identified a small focus of ERG+ benign acini in one case (2.4%) that was 0.4 cm away from an ERG+ cancer (34). We also performed ERG IHC (SI Appendix, Fig. S5) on 110 whole tissue sections from 64 unselected (not selected based on amount of inflammation) radical prostatectomy specimens and found isolated ERG+ LGPIN in one case, ERG+ PIA in one case, and both ERG+ LGPIN and PIA in one case (three cases total, 4.7%, SI Appendix, Fig. S6). We define “isolated” ERG+ PIA or LGPIN as glands found on blocks (as well as the adjacent blocks) with no ERG+ cancer or >9 mm away from ERG+ cancer. Additional studies have also reported a low frequency of ERG+ benign glands in biopsy or prostatectomy specimens but do not account for the proximity of these glands to ERG+ cancer (38, 39), an important consideration in establishing that the cells in the “benign” glands are not actually representative of retrograde invasion of cancer cells into benign acini (40). Our 3D mapping was used to avoid this possibility.
In stark contrast to the studies reported by Young et al. (34), Furusato et al. (37), and our assessment of whole tissue sections from unselected cases, we identified isolated ERG+ luminal epithelial cells in PIA and/or LGPIN in 7 (46.7%) of the 15 mapped cases in the highly inflamed/infection cohort (P < 0.0001 compared to unselected cases, χ2 test, Fig. 2A and SI Appendix, Fig. S7) when we examined ERG IHC on 126 whole tissue sections. We subsequently performed ERG IHC on all remaining blocks from the seven cases where we found ERG+ PIA and/or LGPIN to determine the spatial orientation of the ERG+ PIA or LGPIN foci to ERG+ or ERG− cancer in 3D (SI Appendix, Figs. S8–S14). We identified multiple (up to eight) spatially distant (>2 mm apart) foci of ERG+ PIA and/or LGPIN in five of the seven cases (Table 1). A total of 4 foci of ERG+ isolated HGPIN, 9 foci of isolated ERG+ LGPIN, and 15 foci of isolated ERG+ PIA were identified (Table 1). These data strongly suggest infection-induced TMPRSS2:ERG fusions in PIA and LGPIN foci due to the high frequency and multiplicity of ERG+ foci found in these infected cases.
Fig. 2.
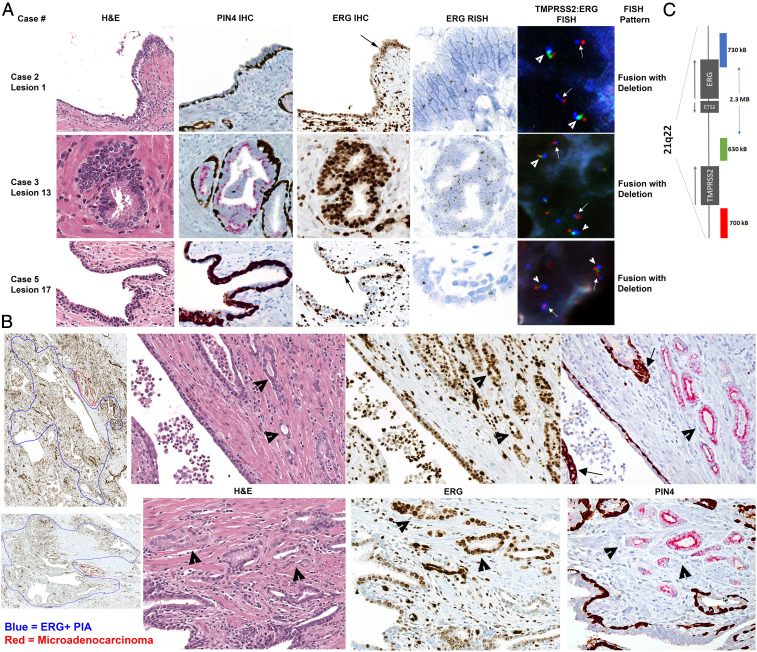
ERG expression from TMPRSS2:ERG gene fusions present in luminal epithelial cells in PIA (lesion 1 and 17) and LGPIN (lesion 13) in highly inflamed radical prostatectomy specimens. (A) Representative images from case 2, 3, and 5 showing intact basal cells indicative of a nonneoplastic gland (PIN4 IHC) and positive ERG IHC (200× magnification), RISH, and FISH in luminal epithelial cells. Black arrows point to magnified (400× magnification) regions in ERG RISH. White arrowheads point to normal allele, and white arrows point to TMPRSS2:ERG genomic rearrangements detected via FISH as denoted (1,000× magnification). Loss of green signal on one allele denotes the fusion with deletion rearrangement pattern. (B) Large regions of ERG+ PIA (circled in blue in first panel of ERG IHC, 40× magnification) with small regions of adjacent microadenocarcinoma (circled in red) from case 2 (Top Row) and case 5 (Bottom Row). Infiltrating immune cells and endothelial cells are also ERG+. Hematoxylin and eosin, ERG IHC, and PIN4 IHC (all 200× magnification) demonstrate ERG+ PIA next to ERG+ microadenocarcinoma (arrowheads). Arrows denote intermediate cells in PIN4 IHC. (C) Schematic of probe locations for TMPRSS2:ERG FISH probe set. Red probe is located in the distal TMPRSS2 gene region, green probe is located in the proximal TMPRSS2 gene region, and blue probe is located in the ERG (21q22) gene region.
Table 1.
Pathologic assessment of all ERG+ PIA, LGPIN, and HGPIN foci
| Case no. | Block designation | Erg+ lesion no. | Notes |
| 2 | RAP | 1 | PIA, some reactive nuclei with nucleolar enlargement not diagnostic of LGPIN or HGPIN |
| LAP | 2 | PIA, atypia*, some early invasive carcinoma (microadenocarcinoma†) | |
| RBP | 3 | PIA with nuclear reactive changes and microadenocarcinoma | |
| 4 | PIA with nuclear reactive changes | ||
| RCP | 5 | PIA | |
| RDP | 6 | PIA and microadenocarcinoma | |
| 7 | PIA | ||
| LDUMB | 8 | PIA with nuclear reactive changes and microadenocarcinoma | |
| 3 | RAP | 9 | PIA |
| RBP | 10 | LGPIN | |
| RCP | 11 | LGPIN | |
| 12 | LGPIN | ||
| 13 | LGPIN | ||
| 14 | LGPIN, atypia, some glands microadenocarcinoma | ||
| LCP | 15 | HGPIN | |
| 4 | LDA | 16 | Merging PIA, LGPIN, and HGPIN. Atypia and microadenocarcinoma. |
| 5 | RDA | 17 | PIA |
| RE | 18 | PIA and microadenocarcinoma | |
| 7 | LCP | 19 | LGPIN |
| 10 | LA | 20 | LGPIN and HGPIN |
| 21 | LGPIN and HGPIN | ||
| 22 | PIA merging with atypia | ||
| 13 | RDA | 23 | PIA with nuclear atypia not diagnostic of LGPIN or HGPIN |
| REA | 24 | PIA |
Atypia defined as small foci of atypical glands suspicious for but not diagnostic of carcinoma.
Microadenocarcinoma as defined by McNeal (41).
A dual IHC stain for Alpha-Methylacyl-CoA Racemase (AMACR) combined with high–molecular weight cytokeratin (PIN4 IHC) confirmed the presence of basal cells in the ERG+ PIA or LGPIN foci (Fig. 2A), further indicating that these ERG+ foci are not cancer. In three of the cases, ERG+ PIA was identified in the absence of any ERG+ cancer in the blocks from the prostatectomy (SI Appendix, Figs. S10, S12, and S14). ERG messenger RNA (mRNA) expression was also observed using RISH (SI Appendix, Fig. S5) in the ERG+ PIA and LGPIN foci (Fig. 2A and SI Appendix, Fig. S15). We also confirmed ERG staining on a subset of the glands using IHC with a separate ERG antibody that targets a different epitope (C versus N terminus of ERG, SI Appendix, Fig. S16).
Detection of Early Invasive Carcinoma Budding from ERG+ Precursors.
We noted with interest seven foci in cases 2, 3, 4, and 5 where ERG+ PIA or LGPIN was directly adjacent to small clusters of glands that lacked basal cells and were apparently in the process of initial invasion (Fig. 2B and SI Appendix, Fig. S17) consistent with early invasive carcinoma [also called “microadenocarcinoma” (41)]. In all but one instance, there was no other ERG+ cancer on the blocks with these lesions or on either adjacent block in 3D. Furthermore, PIN4 IHC indicated that the ERG+ cells in the PIA lesions were AMACR negative, but the ERG+ cells in the budding adenocarcinoma were AMACR positive (Fig. 2B). These results suggest that the fusion event occurred in the PIA lesion and that TMPRSS2:ERG fusion events represent a very early event in prostate carcinogenesis, although requiring the acquisition of further oncogenic events to progress to adenocarcinoma. Intriguingly, we observed the presence of intermediate cells (luminal cells expressing the typical basal cell cytokeratins) in the PIA lesions that were adjacent to early invasive carcinoma (Fig. 2B).
Verification of ERG Rearrangement by Fluorescence In Situ Hybridization Reveals Multiple Independent ERG+ PIA and LGPIN Foci.
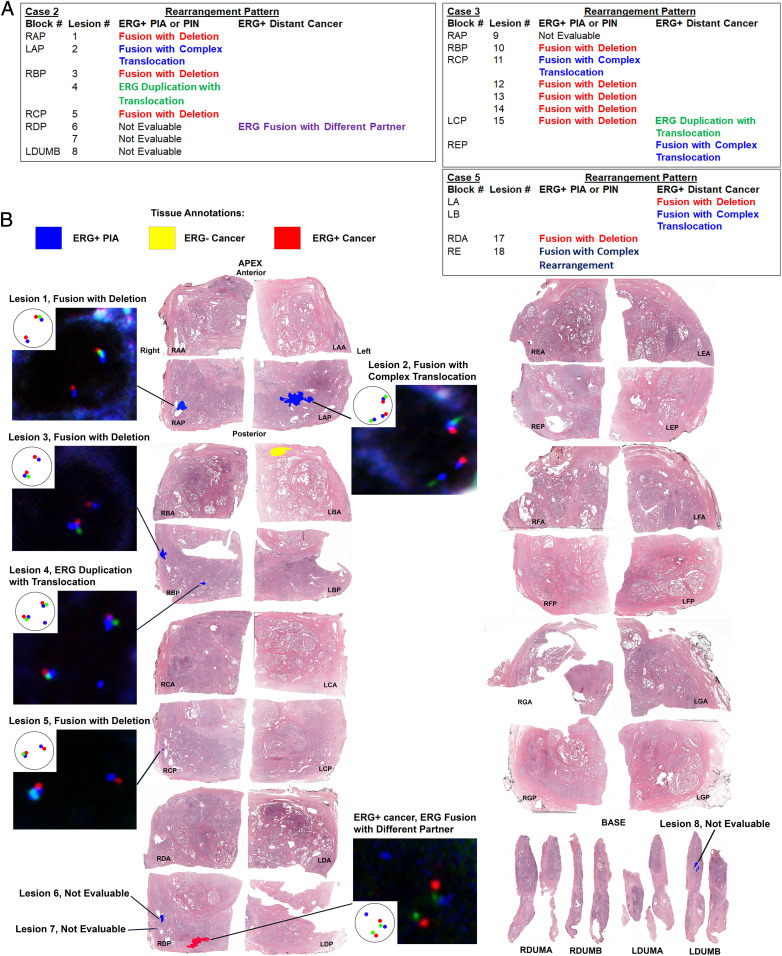
Since it is possible that ERG mRNA and protein expression could be up-regulated by mechanisms that do not involve structural genomic alterations, we further verified that TMPRSS2:ERG genomic rearrangements were present in the ERG+ PIA and LGPIN glands. We used a triple color fluorescence in situ hybridization (FISH) assay designed to detect multiple scenarios of genomic alteration between the TMPRSS2 and ERG loci on 21q22 (Fig. 2C and SI Appendix, Figs. S18 and S19) in three of the cases that had multiple ERG+ PIA and/or LGPIN foci (cases 2, 3, and 5). Non–neoplastic-appearing luminal epithelial cells within the PIA or LGPIN foci that were positive by ERG IHC and ERG RISH were likewise positive for genomic alterations in the TMPRSS2 and ERG loci (Figs. 2A and 3 and SI Appendix, Figs. S20–S23 and Table S2). Each of these cases had small ERG+ cancer that was distant to any of the ERG+ PIA or LGPIN foci (SI Appendix, Figs. S8, S9, and S11). In all three cases, the ERG+ cancer, in addition to being spatially distant to ERG+ PIA and LGPIN, contained a different rearrangement pattern than most or all of the ERG+ PIA and LGPIN foci in that same case (Fig. 3 A and B and SI Appendix, Fig. S21 and Table S2). This finding further verifies that all or at least a subset of the ERG+ PIA and LGPIN foci were distinct from any preexisting ERG+ cancer present in a given case.
Fig. 3.
Evaluation and spatial mapping of TMPRSS2:ERG genomic rearrangement patterns in separate PIA and PIN foci versus distant ERG+ cancer. (A) Results summary of TMPRSS2:ERG FISH analysis on each ERG+ lesion found in case 2, 3, and 5. Note that ERG+ PIA and LGPIN/HGPIN foci are often a different fusion pattern than the distant ERG+ cancer in the case, even if on the same block. Adjacent ERG− benign glands to ERG+ glands were assessed for TMPRSS2:ERG fusions in an identical manner to the ERG+ glands. All ERG− benign glands were negative for TMPRSS2:ERG rearrangements. (B) Spatial mapping of TMPRSS2:ERG rearrangement patterns in case 2. Pattern shown in example nuclei is depicted in the Inset. Red probe is located in the distal TMPRSS2 gene region, green probe is located in the proximal TMPRSS2 gene region, and blue probe is located in the ERG (21q22) gene region.
Colibactin Detection in Clinical Cases and as a Cause of DSB in Prostate Cells.
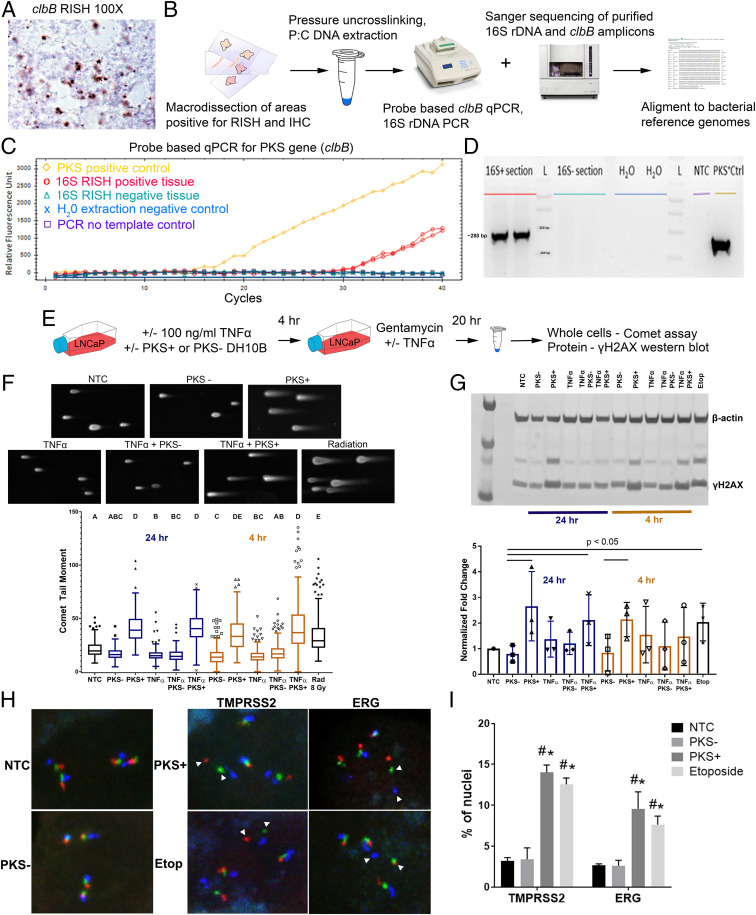
Two of the cases (case 2 and case 12) had bacteria present in large enough regions that we could macrodissect the tissue, extract DNA, and further identify the infecting species. Sequencing of a partial region of the 16S rRNA gene from these cases identified a bacterium in the family Enterobacteriaceae, which would also be consistent with the LPS IHC results (Fig. 1). RISH for the colibactin clbB gene indicated that the infecting bacteria in both case 2 and case 12 were colibactin producers (Fig. 4A). The presence of the pks island in bacteria from these cases was further confirmed by probe-based qPCR and sequencing (Fig. 4 B–D and SI Appendix, Fig. S24).
Fig. 4.
Exposure of prostate cancer cells to colibactin (PKS) induces DNA DSB as assessed by comet assay, Western blot for γ-H2AX, and TMPRSS2:ERG FISH. (A) Example of PKS expression in the bacteria in case 2 (1,000× magnification) as visualized by cblB RISH. (B) Strategy for bacterial DNA isolation from FFPE tissues, 16S rDNA sequencing, and PKS qPCR. (C) Detection of colibactin clbB gene by qPCR in DNA extracted from a macrodissected 16S RISH–positive tissue area from case 2. (D) Agarose gel image of PCR products from PKS qPCR. (E) Experimental outline for in vitro colibactin exposure experiments. (F, Top) Representative images of comet moments for each experimental condition. Comet moments are visibly induced in LNCaP cells by 8-Gy ionizing radiation exposure or 4-h infection with PKS+ E. coli. NTC = no-treatment control. (Bottom) Quantification of comet tail moments after exposure of LNCaP cells to PKS+ E. coli induces DNA DSB at or greater than exposure to 8-Gy radiation 4 and 24 h postexposure. The addition of TNF-α does not increase DSB in this model system. Representative results of the experiment are shown. One-way nonparametric ANOVA with Kruskal–Wallis multiple comparison test was performed. Differing letters denote statistical significance (P < 0.05). The statistical significance of PKS+ E. coli–treated groups at 4 h postinfection was variable across three biological replicates. The trend at 24 h was consistent across four biological replicates. (G, Top) Western blot for γ-H2AX likewise demonstrates a >twofold increase in DSB repair at 4 and 24 h after PKS+ E. coli exposure. Etop = etoposide (positive control). (Bottom) Densitometry quantification of Western blot images. The results are normalized to β-actin levels and no-treatment control (NTC) and are representative of three biological replicates. Statistically significant differences noted (P < 0.05, one-tailed Student’s t test). (H) Representative images of TMPRSS2:ERG FISH performed on LNCaP cells after NTC or 4-h treatment with PKS− and PKS+ E.coli and etoposide (positive control). LNCaP cells are hypotetraploid with four sets of normal signals. White arrowheads denote DSB as visualized by split apart of the red and green signals at TMPRSS2 (labeled as TMPRSS2) or as split apart of the green and blue signals between TMPRSS2 and ERG (labeled as ERG) in the PKS+ and Etop–treated groups. (I) Percentage of cells (n ≥ 200) showing split apart of FISH probes at TMPRSS2 and ERG. Results presented as mean ± SEM of three biological replicates. #P < 0.05 versus NTC and *P < 0.05 versus PKS−, two-way ANOVA with multiple comparisons.
We noted with interest that one of the cases found to harbor colibactin-producing bacteria was also the case identified with eight distinct ERG+ PIA lesions (case 2, Fig. 3B). We therefore questioned whether bacterial genotoxins such as colibactin, along with inflammation, can contribute to genomic damage that promotes the formation of TMPRSS2:ERG gene fusions. We exposed LNCaP cells to the E. coli strain DH10B hosting a bacterial artificial chromosome (BAC) bearing the pks island that produces colibactin (PKS+) or hosting the empty pBeloBAC11 vector (PKS−) as previously described (25) with or without the addition of TNF-α to simulate inflammation (Fig. 4E) and then assessed DSB by comet assay. Transient treatment of LNCaP cells with the PKS+ colibactin-producing bacteria, with or without the addition of TNF-α, induced DSB breaks at or greater than that of 8 Gy of ionizing radiation at 4 and 24 h after exposure (Fig. 4F). A similar trend was observed for induction of phosphorylated H2A histone family member X (γ-H2AX), as an indicator of DSB break repair (Fig. 4G). Cleaved caspase-3 was increased in the TNF-α–treated groups but not comparably in the PKS+ colibactin-alone–treated groups, indicating that the DNA damage observed was not due to apoptotic cells (SI Appendix, Fig. S25). Finally, we observed an induction of apparent chromosomal breaks at the TMPRSS2 and ERG loci as evidenced by split apart of FISH signals when LNCaP cells were treated for 4 h with PKS+ colibactin–producing bacteria or etoposide (positive control) compared with untreated or PKS− bacteria–treated cells (Fig. 4 H and I).
Discussion
We herein report a high frequency of isolated foci of PIA and LGPIN with TMPRSS2:ERG genomic rearrangements in prostate specimens harboring evidence of active bacterial infections. Mapping these ERG+ foci in relation to any ERG+ cancer (or lack of ERG+ cancer in three cases) along with FISH analysis of the TMPRSS2:ERG rearrangement patterns suggest that the TMPRSS2:ERG fusion event can be initiated within a PIA or LGPIN lesion. Furthermore, in several instances, we observed ERG+ PIA and/or LGPIN in direct transition with microadenocarcinoma. Since TMPRSS2:ERG fusion is currently regarded as an early somatic genetic oncogenic event driving prostate carcinogenesis, we submit that our findings represent evidence in human tissues that bacterial infections can promote early prostate cancer development. We speculate that the inflammation induced by infection, in combination with DNA damage induced by bacterial genotoxins, contributes to the development of precancerous lesions and oncogenic gene fusions and promotes early prostate carcinogenesis (Movie S1). Importantly, our study does not address the frequency by which infections contribute to prostate cancer initiation, and the findings of this study may be representative of only a subset of cases. Our study does not rule out the possibility that androgen receptor–mediated or alternate mechanisms contribute to TMPRSS2:ERG gene fusions in the setting of infection or in other settings. Likewise, we did not find evidence of persistent bacterial presence within prostate cancer, as bacteria were never observed within cancerous regions in the cases in our 16S RISH and LPS and LTA IHC assays (SI Appendix, Fig. S4). This represents an important epidemiologic challenge in linking prostate infections to prostate cancer risk, as the initiating infection likely often occurs and is then cleared many years prior to the cancer diagnosis.
Prostate infections and inflammation may contribute to other oncogenic events in addition to TMRPSS2:ERG gene fusions and may play a role in the pathogenesis of ERG− cancers as well. Our study was limited to examining the contribution of bacterial infections to ERG+ cancer, as TMPRSS2:ERG fusion events are one of the few known early oncogenic events in prostate cancer. It is of keen interest that we found ERG+ PIA and/or ERG+ LGPIN in apparent direct transition with ERG+ early invasive adenocarcinoma in the absence of any ERG+ HGPIN. Currently, the most accepted direct precursor to prostate cancer is HGPIN (42), and it is hypothesized that PIA serves as a risk factor lesion that can directly transition to HGPIN (10). It has also been previously hypothesized that prostate atrophy may give rise to carcinoma directly (7, 43–46), and our current study would support this hypothesis in at least a subset of cases. LGPIN is not currently regarded as a risk factor lesion for prostate cancer based on studies showing that when present on a prostate biopsy, LGPIN is not associated with a higher risk of cancer on rebiopsy than the risk after a benign diagnosis on initial biopsy (47), although results are varied (48, 49). Herein, we provide evidence of ERG+ LGPIN merging with early invasive adenocarcinoma, prompting further scrutiny of at least ERG+ LGPIN as a risk factor lesion. Finally, whereas not all ERG+ precursor lesions may progress to invasive cancer, our finding of multiple foci of ERG+ precursors in infected cases would be in line with the multifocal nature of prostate cancer (50).
A colibactin DNA damage signature has been identified that is detected in a subset of colorectal cancers as well as other cancer types including bladder cancer, stomach cancer, and cancer of the uterine corpus (29, 30, 51). Interestingly, this mutational signature has also been previously detected in prostate cancer samples, albeit at a low frequency (29, 51). These studies provide further evidence of a potential causal role of colibactin-producing bacteria in prostate carcinogenesis in at least a subset of cases. Our in vitro assay demonstrated that colibactin can likewise induce chromosomal breaks particularly at TMPRSS2 and to a lesser extent between the TMPRSS2 and ERG loci in the LNCaP prostate cancer cell line (Fig. 4 H and I). These results are in accordance with previous in vitro studies of DSB induced by dihydrotestosterone (22) and TNF-α (23) in prostate cancer cells. Our study implicates colibactin-producing bacteria in the induction of gene rearrangements leading to gene fusion events as a mechanism of promoting carcinogenesis in addition to the previously described role in induction of DNA mutations. We propose that colibactin may not be the only bacterial genotoxin that can cause DNA damage that promotes prostate carcinogenesis, and indeed there may be many types of infectious agents that contribute to the same end.
Overall, our study suggests that bacterial prostatitis should be considered as a legitimate risk factor for prostate carcinogenesis in at least a subset of cases and prompts the development of methodologies to detect undiagnosed prostate infections, as well as to mitigate infections and inflammation in the prostate as a prostate cancer prevention strategy.
Materials and Methods
Clinical Specimens.
Deidentified specimens were obtained and studied under a Johns Hopkins Medicine Institutional Review Board–approved protocol. We searched the Johns Hopkins Hospital radical prostatectomy pathology reports from January 2016 through January 2018 for the terms prostatitis/cystitis, moderate or extensive acute inflammation, and granuloma. Hematoxylin and eosin slides were reviewed to confirm that the degree/extent of inflammation contained in the radical prostatectomy specimens was highly atypical (reference SI Appendix, Fig. S1). As these potential infections were undiagnosed, we cannot comment on when or how the infection occurred, and it is possible that the infection was from the diagnostic biopsy.
16S rRNA, clbB, and ERG RISH.
RISH was performed on formalin-fixed paraffin-embedded (FFPE) tissues using the Advanced Cell Diagnostics (ACD) RNAscope 2.5 High Definition Assay (ACD, Cat. No. 322370). The probes used for the RISH assays were as follows: 1) 16S rRNA (Cat. No. 427731); 2) colibactin (clbB, Cat. No. 561691); 3) ERG (Cat. No. 604028); 4) peptidyl prolyl isomerase B (positive control, Cat. No. 313901); and 5) Zea mays superal1 (negative control, Cat. No. 316381). RISH was performed according to a modified manufacturer’s suggested protocol. Briefly, the slides were deparaffinized on a 60 °C heat block followed by xylene treatments. Slides were then dehydrated in absolute alcohol and air dried followed by hydrogen peroxidase blocking treatment at room temperature (RT) for 10 min. The slides were treated with the antigen retrieval buffer for 15 min using the steaming method (temperature ≥ 99 °C) followed by a brief rinse with deionized water (dH2O). Two enzyme treatment steps with lysozyme and achromopeptidase were performed exclusively in RISH targeting 16S rRNA to digest the cell wall of bacteria. Slides are first treated with lysozyme at 10 mg/mL (Sigma Aldrich) followed by treatment with achromopeptidase at 30 U/mL (Sigma Aldrich) for 10 min at 37 °C. Slides were then digested using protease plus pretreatment for 30 min at 40 °C. Finally, prewarmed probes (40 °C) were hybridized onto the slides with 2-h incubation at 40 °C. The slides were washed using wash buffer and then the amplification steps were performed in the following order: amplification buffer 1, amplification buffer 2, amplification buffer 3, and amplification buffer 4 for alternating 30 and 15 min at 40 °C with wash buffer between each amplification step. Then, amplification 5 was performed for 60 min at RT followed by a brief wash and amplification 6 for 30 min at RT. After a brief wash, a 1:1 solution of 3,3'-Diaminobenzidine A and B was applied to the tissue and incubated for 10 min at RT. This was then rinsed with dH2O and counterstained with hematoxylin for 2 min. The slides were rinsed, dehydrated, and coverslipped.
LPS and LTA IHC.
IHC was performed manually with the following antibodies: 1) LTA (Thermo Fisher Scientific, Cat. No. MA1-7402, Clone G43J) and 2) LPS (Abcam, Cat. No. 35654, Clone 2D7/1). Slides were heated at 60 °C for 10 min and deparaffinized by xylene treatment followed by rehydration in ethanol gradient and a rinse in dH2O. After quick rinses in 0.1% Tween-20 in dH2O solution and citrate buffer (Vector Laboratories, Cat. No. H-3300), antigen retrieval was conducted using steaming method in ethylene-diamine-tetraacetic acid (EDTA) buffer (Thermo Fisher Scientific, Cat. No. BP2473500) for 45 min for LPS antibody and Target Retrieval Solution (Agilent Technologies, Cat. No. S169984-2) for LTA antibody. The slides were rinsed with tris-buffered saline with Tween 20 (TBST), and blocking step was conducted using Dako REAL peroxidase-blocking solution (Agilent Technologies, Cat. No. S202386-2) for 5 min at RT. The primary antibody was applied at 1:100 for LPS and 1:25 for LTA antibodies for 45 min at RT or overnight at 4 °C, respectively. After a rinse, anti-mouse secondary antibodies were applied (Leica Biosystems, Cat. No. PV6110) for 40 min at RT. The slides were rinsed, and DAB (Sigma Aldrich, Cat. No. D4293) was applied for 20 min at RT. Counterstaining was done with Mayer’s hematoxylin (Agilent Technologies, Cat. No. S330930-2) for 2 min, and the slides were mounted with Cytoseal-60 (Thermo Fisher Scientific, Cat. No. 8310-16) after dehydration through an alcohol gradient and xylene.
FFPE DNA Extraction.
DNA was extracted from macrodissected FFPE tissues from areas that were positive for both 16S rRNA RISH and LPS IHC in cases 2 and 12. DNA extraction was conducted using a phenol:chloroform–based method. Briefly, 5-µm tissue sections from areas with positive bacterial signature on adjacent cuts were macrodissected using a sterile scalpel and placed in a 2-mL Eppendorf tube. The tissue was autoclaved to 120 °C for 25 min in an alkali digestion buffer (0.1 M NaOH in 1% sodium dodecyl sulfate (SDS) solution) for reversal of formalin-induced crosslinking. After a brief cooling of the tissue, 500 µL 25:24:1 phenol:chloroform:isoamyl alcohol mixture was added. The mixture was agitated for 5 min at RT and centrifuged at 10,000 × g for 5 min at RT. The upper aqueous layer was transferred to a new tube. The agitation and centrifugation was repeated twice. Then, 1 volume of isopropanol and 0.1 volume of 3 M sodium acetate was added to the aqueous layer and mixed. The mixture was then centrifuged at 10,000 × g for 30 min at RT. The supernatant was discarded, and the pellet was rinsed gently with 1 mL 85% ethanol. The resultant washed pellet was air dried and resuspended in molecular-grade dH2O.
Colibactin clbB qPCR.
qPCR was conducted using a primer set (31) and probe targeting the clbB gene in the colibactin PKS island as follows: pks-F 5′- GCG-CATCCTCAAGAGTAAATA-3′, pks-R 5′- GCGCTCTATGCTCATCAACC-3′, pks probe 5′-FAM-TATTCGACACAGAACAACGCCGGT-BHQ1-3′ (probe designed by Julia Drewes in the laboratory of Cynthia Sears at Johns Hopkins). qPCR was performed using Bio-Rad iTaq Universal Probe Supermix (Bio-Rad) system following the manufacturer suggested protocol with 7 to 20 ng input DNA. The cycling conditions for the assay were 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s, 60 °C for 1 min, and a final extension at 72 °C. qPCR was conducted on the Bio-Rad CFX Connect Real Timer PCR Detection System, and the data were analyzed using Bio-Rad CFX Manager 3.1 software. The amplified product from the qPCR (∼280 base pairs) was visualized via gel electrophoresis, gel extracted, and purified using the QiaQuick Gel Extraction kit (Qiagen). The eluted products were then Sanger sequenced at the Johns Hopkins Genetic Resources Core Facility and aligned with bacterial genomes using National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST).
16S rRNA PCR.
DNA extracted from case 2 and case 12 was amplified via PCR with the following primer sets designed as universal primers against the 16S rRNA gene: 27F 5′-AGAGTTTGATCMTGGCTCAG-3′ + 519R 5′- GWATT-ACCGCGGCKGCTG-3′, 533F 5′- GTGCCAGCAGCCGCGGTAA-3′ + 907R 5′- CC-GTCAATTCMTTTRAGTTT-3′, and V6-F 5′- CAACGCGWRGAACCTTACC-3′ and V6-R 5′-CRRCACGAGCTGACGAC-3′. The amplified product from the PCR was visualized via gel electrophoresis and purified using the QiaQuick PCR Purification kit (Qiagen). The eluted products were then Sanger sequenced at the Johns Hopkins Genetic Resources Core Facility and aligned with bacterial genomes using NCBI BLAST.
ERG and PIN4 IHC.
IHC was performed on the Ventana Discovery Ultra Immunohistochemistry/In Situ Hybridization system (Roche Diagnostics) with the following antibodies 1) ERG (Roche Diagnostics, Cat. No. 790-4576, Clone EPR3864) and 2) PIN4–Cytokeratin1/5/10/14 (Enzo Life Sciences, Cat. No. ENZ-C34903), p63 (Biocare Medical, Cat. No. SKU: 163, Clone 4A4), and AMACR (Zeta Corporation, Cat. No. Z2001, Clone 13H4). IHC was performed per the manufacturer’s protocol. The slides were steamed for 32 min (for ERG) and 48 min (for PIN4) in Cell Conditioning 1 solution (Roche Diagnostics, Cat. No. 950-124) for antigen retrieval. Then, the corresponding primary antibodies were applied with the following conditions: ERG (prediluted) for 32 min at RT and PIN 4—a combination of 1:50 dilution of Cytokeratin 1/5/10/14 and P63 for 40 min at RT followed by 1:50 dilution of AMACR for 32 min at RT. The Discover 3-hydroxy-2-quinoxaline (HQ) horseradish peroxidase (HRP) hapten-linked multimer detection kit (Roche Diagnostics, Cat. No. 760-4602) and the Discovery Amp HQ kit (Roche Diagnostics, Cat. No. 760-052) were used to develop ERG staining, while the PIN4 staining was developed in the Discover HQ HRP hapten-linked multimer detection kit.
As a confirmation of the ERG staining, we also performed ERG IHC with a separate antibody (Biocare Medical, SKU: 421, Clone 9FY) and manual staining with the Power Vision+ Poly-HRP IHC kit (Leica Biosystems, Cat. No PV6109).
TMPRSS2:ERG FISH and Imaging.
FISH was conducted on FFPE radical prostatectomy tissues and LNCaP cells prepared as cytospins using the TMPRSS2-ERG (21q22) Deletion, Break Triple color FISH assay (Leica Biosystems, Cat. No. KI-10726). The assay was conducted using a modified manufacturer’s method. FFPE tissues were deparaffinized at 60 °C for 10 min followed by 2× xylene treatment. The tissue was rehydrated through an alcohol gradient and dH2O. The slides were then treated with 0.2N HCl for 15 min at RT followed by antigen retrieval using a 10 mM sodium citrate solution (Vector Laboratories, Cat. No. H-3300) at 80 °C for 40 min. The slides were then treated with 2× SSC, dH2O, and 0.2N HCl at RT for 2 min each. Protease digestion was conducted using in situ hybridization protease 3 (Roche Diagnostics, Cat. No. 780-4149) on the Benchmark Ultra IHC/ISH system (Roche Diagnostics) for 40 min for tissues and 15 min for cells at 37 °C. The slides were washed 2× in dH2O and fixed in 10% neutral-buffered formalin. After 2× brief dH2O wash, the slides were dehydrated and air dried. The slides were stored at −20 °C until the probe hybridization.
The slides were warmed to 45 °C, and 5 µL Leica TMPRSS2:ERG break apart probe was added to the area of interest. A coverslip was placed over the probe and sealed. The probes were hybridized in a Thermobrite slide denaturation and hybridization system (Leica Microsystems, Cat. No. 23-021-580) at 80 °C for 5 min followed by 37 °C for 16 to 24 h. Posthybridization, the coverslip was removed, and the slides were washed with prewarmed 2XSSC/0.3%IgePal CA-60 wash (Thermo Fisher Scientific, Cat. No. 15557044 and Sigma Aldrich, Cat. No. I8896, respectively) for 2 min at 72 °C. This was followed by a second wash of 2× SSC/0.1% IgePal CA-60 for 1 min at RT. After a brief rinse in dH2O, the slides were dehydrated and air dried. The tissue was counterstained with DAPI (Leica Biosystems, Cat. No. LK-095A) at a dilution of 1:5 in counterstain diluent (Leica Biosystems, LK-097A) and coverslipped. The slides were stored in 4 °C in the dark until they were imaged.
The slides were initially visualized using a Nikon Eclipse 2000 fluorescence microscope under 40× objective magnification with channels for fluorescein isothiocyanate (FITC, green), Cy3 (red), and cyan fluorescent protein (CFP, aqua) markers, respectively. For individual cell analysis, the slides were imaged using the TissueFAXS Plus (Tissue Gnostics) automated microscopy workstation equipped with a Zeiss Z2 Axio Imager microscope. First, each slide was imaged with a 10× objective with the DAPI channel to generate a preview image of the slide. Following the preview, specific regions of interest were identified and imaged using the 63× oil objective. The ranges of exposure time for each filter was determined for each individual sample: DAPI (50 ms, 0 to 2,500), FITC (250 ms, 0 to 4,095), Cy3 (175 ms, 0 to 4,095), and ET-A (175 ms, 0 to 4095). TissueFAXS Viewer (TissueGnostics) software was used to visualize the composite images, and Photoshop (Adobe) was used to further reduce background levels. Assignment of TMPRSS2:ERG gene status in individual cells in areas of interest was conducted by two reviewers. Each region was assessed for a minimum of 100 nonoverlapping distinct cells. Each cell was scored if both sets of TMPRSS2 (red) and ERG (blue) signals were present. If either of those signals were missing or a clear score was unable to be assigned, an unquantifiable score (*) was assigned to the cell. A region was defined as fusion positive if >20% of the cells showed the same fusion pattern. For analysis of FISH performed on the LNCaP cell line, a minimum of 200 cells were assessed per treatment group where all four sets of TMPRSS2 and ERG signals could be visualized. The average pixel distance between red, green, and blue signals in 20 randomly selected cells in the untreated group was determined. Split-apart signals were assigned if the distance between signals was greater than 2× the average distance + 1 pixel. LNCaP cells and select tissues were imaged for figure purposes using a Zeiss LSM 880 confocal microscope and 63× objective.
In Vitro Cell Culture Modeling.
LNCaP cells (obtained from American Type Culture Collection, Cat. No. CRL‐1740) were cultured in RPMI medium with 10% fetal bovine serum in 6-well plates or T25 flask to 80% confluence. PKS-producing bacteria were obtained courtesy of Jean-Phillipe Nougayrède, Université de Toulouse, Toulouse, France. Briefly, for the in vitro assay E. coli strain DH10B hosting a bacterial artificial chromosome bearing the pks island (PKS+) was used as a colibactin-producing strain, while DH10B hosting the empty pBeloBAC11 vector (PKS−) was used as the negative bacterial control. PKS+ and PKS− bacteria were grown shaking overnight at 37 °C in Luria-Bertani (LB) broth. Prior to the start of the experiment, the number of LNCaP cells per well/flask was measured in a proxy flask using an automated cell counter (Invitrogen Countess II). Fresh media was changed and PKS+/PKS− bacteria (multiplicity of infection 100:1), and TNF-α (100 ng/mL) was added to the cells. The treatment groups were no-treatment control, TNF-α only, PKS+ only, PKS− only, TNF-α & PKS+, and TNF-α & PKS−. At 4 h postinfection, cells were harvested for a 4-h time point or the cells were washed with Dulbecco's phosphate-buffered saline (DPBS), and media with gentamycin (20 µg/mL) was added to the cells. TNF-α (100 ng/mL) was added to the applicable groups. At 24 h, the cells were washed and resuspended in DPBS and used for comet assay directly and stored at −20 °C for Western blot. Etoposide- (10 µM) and ionizing radiation (8 Gy)–treated cells were used as positive controls for DNA damage.
Comet Assay.
Neutral comet assay was performed using the Trevigen CometAssay kit (Trevigen, Cat. No. 4250-050-K). Cells were washed with 1× DPBS, trypsinized for 5 min at 37 °C, washed, and resuspended in 1× DPBS to a final concentration of 1 × 105 cells per mL. The comet assay was performed according to the manufacturer’s protocol. Briefly, 50 µL washed 1 × 105 cells/mL were mixed with 500 µL low melting agarose at 37 °C. Next, 30 µL of the cell agarose mixture was put onto warmed comet slides to form a uniform layer which was set on a cool flat metal surface at 4 °C. Cell membrane lysis was conducted using the precooled Trevigen lysis solution overnight at 4 °C. The slides were washed in 1× Tris/Borate/EDTA buffer followed by gel electrophoresis at a fixed voltage of 21 V for 45 min at 4 °C. Slides were rinsed in dH2O and 70% ethanol and dried in a 37 °C incubator until the agarose dehydrated forming a flat surface (∼2 h). Then, the slides were stained with 1× SYBR green solution (Invitrogen, Cat. No. S7585) for 30 min at RT in the dark and visualized under a Nikon fluorescence microscope in the FITC channel. Images were taken using Roper scientific image software at 4× for analysis. CometScore 2.0 was used to analyze the images from the Comet assay and the tail moment was used as a measure of DNA damage. The analyzer was blinded to the treatment groups. The software was used according to the developer’s recommendations. A minimum of 50 individual comets were analyzed for each group. Statistical analysis was conducted individually for each biological replicate. A nonparametric one way ANOVA followed by post hoc analysis (Dunn’s test) was used to compare the means of the different groups.
Western Blot.
Cells from the in vitro assay were used for Western blot. γ-H2AX protein was detected from whole-protein isolate. Briefly, cells were harvested and lysed in using lysing buffer supplemented with phosphatase, benzoase, and proteinase inhibitors as per the manufacturer’s protocol. The protein concentration was measured using a Pierce bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, Cat. No. 23225) and run on an 8% Bis-Tris SDS-polyacrylamide gel electrophoresis (PAGE) gel (Thermo Fisher Scientific, Cat. No. NW00105BOX). The gel was transferred onto a nitrocellulose membrane and blocked with a blocking buffer (LI-COR, Cat. No. 927-50000). The membrane was stained with 1:1,000 dilution of γ-H2AX (MilliporeSigma, Cat. No. 05-636, Clone JBW301) and 1:5,000 dilution of β-Actin (Cell Signaling Technologies, Cat. No. 3700S, Clone 8H10D10) primary antibodies shaking overnight at 4 °C. The membranes were washed and incubated in the anti-mouse secondary (LI-COR, Cat. No. 925-68070) shaking for 45 min at RT. The membranes were washed and visualized using an Odyssey scanner at 562-nm absorbance. Densitometry quantification was conducted using the ImageJ Gel analysis algorithm. For one of the biological replicates, the membrane was also stained with 1:1,000 dilution of cleaved caspase-3 (Cell Signaling Technologies, Cat. No. 9664S, Clone 5A1E) primary antibody followed by incubation with anti-rabbit secondary (LI-COR, Cat. No. 925-32211).
Supplementary Material
Acknowledgments
We thank Jessica Hicks for assistance with IHC, Tracy Jones for assistance with slide scanning and block requests, and Barbara Smith for assistance with confocal imaging. We thank and acknowledge Dr. William Sukov for review of FISH images and interpretation. We also thank Dr. Srinivasan Yegnasubramanian, Dr. William Nelson, Ajay Vaghasia, Dr. Cynthia Sears, and Dr. James Eshleman for helpful advice and discussion. We acknowledge Dr. Tim Phelps and Dr. David Rini for important input and guidance on Movie S1. We acknowledge and thank Dr. Jean-Philippe Nougayrède for providing the PKS+ and PKS− E. coli strains. Funding was provided by the Prevent Cancer Foundation, Department of Defense Prostate Cancer Research Program awards W81XWH-11-1-0521 and W81XWH-14-1-0364, the Patrick C. Walsh Prostate Cancer Research Fund, National Cancer Institute (NCI) Specialized Programs of Research Excellence (SPORE) in Prostate Cancer award P50CA058236, the Prostate Cancer Foundation Chris and Felicia Evensen Young Investigator Award, and the Prostate Cancer Foundation Neil DeFeo 16CHAL13 Challenge Award.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018976118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Bray F., et al., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Sfanos K. S., Isaacs W. B., De Marzo A. M., Infections and inflammation in prostate cancer. Am. J. Clin. Exp. Urol. 1, 3–11 (2013). [PMC free article] [PubMed] [Google Scholar]
- 3.Sfanos K. S., Yegnasubramanian S., Nelson W. G., De Marzo A. M., The inflammatory microenvironment and microbiome in prostate cancer development. Nat. Rev. Urol. 15, 11–24 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Doat S., et al., Prostatitis, other genitourinary infections and prostate cancer risk: Influence of non-steroidal anti-inflammatory drugs? Results from the EPICAP study. Int. J. Cancer 143, 1644–1651 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Sutcliffe S., Pontari M. A., “Inflammation and infection in the etiology of prostate cancer” in Prostate Cancer, Mydlo J. H., Godec C. J., Eds. (Academic Press, San Diego, ed. 2, 2016), pp. 13–20. [Google Scholar]
- 6.De Marzo A. M., et al., Inflammation in prostate carcinogenesis. Nat. Rev. Cancer 7, 256–269 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Marzo A. M., Marchi V. L., Epstein J. I., Nelson W. G., Proliferative inflammatory atrophy of the prostate: Implications for prostatic carcinogenesis. Am. J. Pathol. 155, 1985–1992 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peiffer L. B., et al., Inflammation-associated pathologies in a case of prostate schistosomiasis: Implications for a causal role in prostate carcinogenesis. Prostate 79, 1316–1325 (2019). [DOI] [PubMed] [Google Scholar]
- 9.van Leenders G. J., et al., Intermediate cells in human prostate epithelium are enriched in proliferative inflammatory atrophy. Am. J. Pathol. 162, 1529–1537 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Putzi M. J., De Marzo A. M., Morphologic transitions between proliferative inflammatory atrophy and high-grade prostatic intraepithelial neoplasia. Urology 56, 828–832 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Liu X., et al., Low CD38 identifies progenitor-like inflammation-associated luminal cells that can initiate human prostate cancer and predict poor outcome. Cell Rep. 17, 2596–2606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomlins S. A., et al., Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Mosquera J.-M., et al., Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin. Cancer Res. 15, 4706–4711 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerveira N., et al., TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia 8, 826–832 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klezovitch O., et al., A causal role for ERG in neoplastic transformation of prostate epithelium. Proc. Natl. Acad. Sci. U.S.A. 105, 2105–2110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomlins S. A., et al., Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 10, 177–188 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zong Y., et al., ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc. Natl. Acad. Sci. U.S.A. 106, 12465–12470 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King J. C., et al., Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat. Genet. 41, 524–526 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carver B. S., et al., Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat. Genet. 41, 619–624 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin C., et al., Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 139, 1069–1083 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mani R. S., et al., Induced chromosomal proximity and gene fusions in prostate cancer. Science 326, 1230 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haffner M. C., et al., Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat. Genet. 42, 668–675 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mani R. S., et al., Inflammation-induced oxidative stress mediates gene fusion formation in prostate cancer. Cell Rep. 17, 2620–2631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nougayrède J.-P., et al., Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Cuevas-Ramos G., et al., Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 107, 11537–11542 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson M. R., et al., The human gut bacterial genotoxin colibactin alkylates DNA. Science 363, eaar7785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue M., et al., Structure elucidation of colibactin and its DNA cross-links. Science 365, eaax2685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z.-R., et al., Macrocyclic colibactin induces DNA double-strand breaks via copper-mediated oxidative cleavage. Nat. Chem. 11, 880–889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dziubańska-Kusibab P. J., et al., Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 26, 1063–1069 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Pleguezuelos-Manzano C., et al., Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580, 269–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arthur J. C., et al., Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chagneau C. V., et al., Uropathogenic E. coli induces DNA damage in the bladder. PLoS Pathog. 17, e1009310 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krieger J. N., Dobrindt U., Riley D. E., Oswald E., Acute Escherichia coli prostatitis in previously health young men: Bacterial virulence factors, antimicrobial resistance, and clinical outcomes. Urology 77, 1420–1425 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Young A., et al., Correlation of urine TMPRSS2:ERG and PCA3 to ERG+ and total prostate cancer burden. Am. J. Clin. Pathol. 138, 685–696 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morais C. L., et al., ERG and PTEN status of isolated high-grade PIN occurring in cystoprostatectomy specimens without invasive prostatic adenocarcinoma. Hum. Pathol. 55, 117–125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perner S., et al., TMPRSS2-ERG fusion prostate cancer: An early molecular event associated with invasion. Am. J. Surg. Pathol. 31, 882–888 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Furusato B., et al., ERG oncoprotein expression in prostate cancer: Clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 13, 228–237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He H., et al., The diagnostic utility of novel immunohistochemical marker ERG in the workup of prostate biopsies with “atypical glands suspicious for cancer”. Am. J. Surg. Pathol. 35, 608–614 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Tsourlakis M. C., et al., Heterogeneity of ERG expression in prostate cancer: A large section mapping study of entire prostatectomy specimens from 125 patients. BMC Cancer 16, 641 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haffner M. C., et al., Molecular evidence that invasive adenocarcinoma can mimic prostatic intraepithelial neoplasia (PIN) and intraductal carcinoma through retrograde glandular colonization. J. Pathol. 238, 31–41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNeal J. E., Prostatic microcarcinomas in relation to cancer origin and the evolution to clinical cancer. Cancer 71(suppl.3), 984–991 (1993). [DOI] [PubMed] [Google Scholar]
- 42.De Marzo A. M., Haffner M. C., Lotan T. L., Yegnasubramanian S., Nelson W. G., Premalignancy in prostate cancer: Rethinking what we know. Cancer Prev. Res. (Phila.) 9, 648–656 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Franks L. M., Atrophy and hyperplasia in the prostate proper. J. Pathol. Bacteriol. 68, 617–621 (1954). [DOI] [PubMed] [Google Scholar]
- 44.Liavåg I., Atrophy and regeneration in the pathogenesis of prostatic carcinoma. Acta Pathol. Microbiol. Scand. 73, 338–350 (1968). [DOI] [PubMed] [Google Scholar]
- 45.Rich A. R., On the frequency of occurrence of occult carcinoma of the prostrate. 1934. Int. J. Epidemiol. 36, 274–277 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Nakayama M., et al., Hypermethylation of the human glutathione S-transferase-π gene (GSTP1) CpG island is present in a subset of proliferative inflammatory atrophy lesions but not in normal or hyperplastic epithelium of the prostate: A detailed study using laser-capture microdissection. Am. J. Pathol. 163, 923–933 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Epstein J. I., Herawi M., Prostate needle biopsies containing prostatic intraepithelial neoplasia or atypical foci suspicious for carcinoma: Implications for patient care. J. Urol. 175, 820–834 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Langer J. E., et al., Strategy for repeat biopsy of patients with prostatic intraepithelial neoplasia detected by prostate needle biopsy. J. Urol. 155, 228–231 (1996). [PubMed] [Google Scholar]
- 49.Goeman L., et al., Is low-grade prostatic intraepithelial neoplasia a risk factor for cancer? Prostate Cancer Prostatic Dis. 6, 305–310 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Andreoiu M., Cheng L., Multifocal prostate cancer: Biologic, prognostic, and therapeutic implications. Hum. Pathol. 41, 781–793 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Boot A., et al., Characterization of colibactin-associated mutational signature in an Asian oral squamous cell carcinoma and in other mucosal tumor types. Genome Res. 30, 803–813 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.