Abstract
Turnover numbers (kcat values) quantitatively represent the activity of enzymes, which are mostly measured in vitro. While a few studies have reported in vivo catalytic rates (kapp values) in bacteria, a large-scale estimation of kapp in eukaryotes is lacking. Here, we estimated kapp of the yeast Saccharomyces cerevisiae under diverse conditions. By comparing the maximum kapp across conditions with in vitro kcat we found a weak correlation in log scale of R2 = 0.28, which is lower than for Escherichia coli (R2 = 0.62). The weak correlation is caused by the fact that many in vitro kcat values were measured for enzymes obtained through heterologous expression. Removal of these enzymes improved the correlation to R2 = 0.41 but still not as good as for E. coli, suggesting considerable deviations between in vitro and in vivo enzyme activities in yeast. By parameterizing an enzyme-constrained metabolic model with our kapp dataset we observed better performance than the default model with in vitro kcat in predicting proteomics data, demonstrating the strength of using the dataset generated here.
Keywords: Saccharomyces cerevisiae, turnover number, k cat , proteomics, metabolism
Enzyme turnover numbers, also termed kcat values, are fundamental parameters that specify the maximum rates of enzymatic reactions and hence determine the rates of biological processes such as metabolism. Determining kcat is therefore essential for quantitatively understanding, modeling, and engineering cells. Traditionally, kcat values are measured in vitro, which might differ from the in vivo situation. In addition, the coverage of measured kcat is poor even for well-studied organisms (1). To address these issues, an approach for estimating in vivo enzyme catalytic rates, also termed kapp values, is to use the equation
| [1] |
where is the metabolic flux through the enzyme and the enzyme abundance (2). This approach was used to estimate kapp for Escherichia coli using absolute proteomics and flux data from various sources (2, 3), and it was found that the maximum kapp values across conditions, defined as kmax values, correlate well with in vitro kcat values in log scale (2).
Here, we generate a kapp dataset for Saccharomyces cerevisiae under diverse conditions. We analyze our dataset and correlate in vivo with in vitro enzyme activities in yeast. Finally, we compare the predictive power of an enzyme-constrained metabolic model using in vivo and in vitro kinetic data.
Results and Discussion
To generate the yeast kapp dataset we collected absolute proteomics data of S. cerevisiae under diverse conditions (4–7) (Dataset S1). The absolute protein abundance can be directly adopted as enzyme abundance in Eq. 1. Note that we did not consider enzyme complexes composed of multiple distinct subunits due to the difficulty in calculating the abundance of catalytic sites (2). To determine the metabolic flux in Eq. 1, we performed flux balance analysis (FBA), as done previously (2), using the latest genome-scale metabolic model (GEM) of S. cerevisiae Yeast8 (8) (SI Appendix). Using the absolute proteomics and flux data, we calculated kapp for 358 metabolic reactions under 26 conditions (Dataset S2). By correlating the estimated kapp in log10 scale, we found that yeast kapp varied between conditions with the lowest R2 being around 0.4 (Dataset S3). This is different from the findings for E. coli, where log-transformed kapp values correlate strongly across conditions with the lowest R2 being above 0.9 (9).
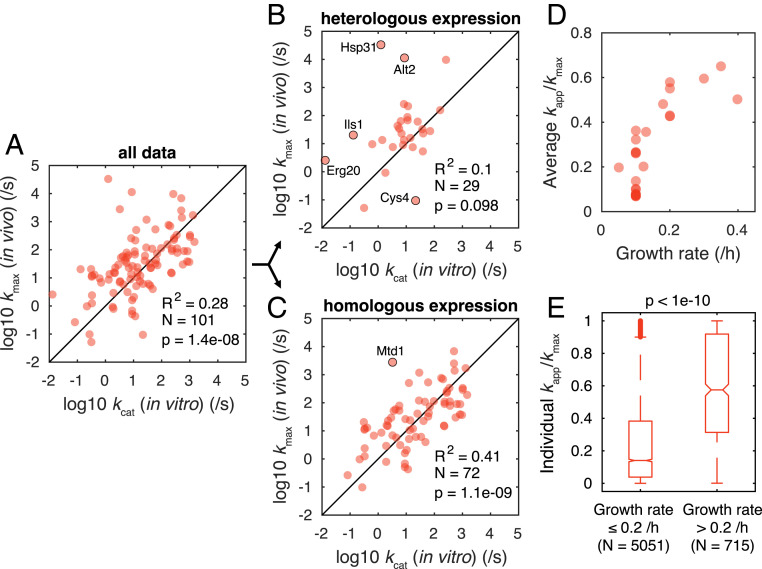
By comparing kmax (Dataset S4), i.e., maximum kapp across all the studied conditions, with the corresponding in vitro kcat (Dataset S5) we obtained a fairly weak correlation in log scale with R2 = 0.28 for S. cerevisiae (Fig. 1A), which is much lower than that of E. coli (R2 = 0.62) (2). A weak correlation was also reported for the plant Arabidopsis thaliana (10). By examining in vitro kcat, we found that some were estimated using purified enzymes obtained through heterologous expression in E. coli, with the others being estimated from yeast extracts. We therefore divided the in vitro kcat dataset into two groups, i.e., heterologous and homologous expression. We found that there was no correlation for the heterologous expression group (Fig. 1B), suggesting that in vitro kcat values obtained through heterologous expression poorly represent in vivo catalytic rates of yeast enzymes. This might be due to the lack of natural posttranslational modifications (PTMs) in the expression organism, which could regulate enzyme activity. Indeed, we found that 27 out of the 29 reactions have reported PTMs on the enzymes (Dataset S6), indicating that these PTMs could functionally affect enzyme activity (11). In the homologous expression group we observed an improved correlation of R2 = 0.41 in log scale (Fig. 1C) and thus identified the data obtained through heterologous expression as the main source of deviations.
Fig. 1.
Analysis of kapp and kmax of S. cerevisiae. Correlation in log scale between kmax and in vitro kcat for all data points (A) and for the data points in which in vitro kcat were measured using enzymes obtained through heterologous expression (B) and through homologous expression (C). The data points with deviations more than two orders of magnitude are labeled by the enzyme names. Student’s t test was used to calculate P value for Pearson’s correlation. (D) Change in average kapp/kmax of each condition with growth rate. (E) Comparison between kapp/kmax of individual reactions in two groups divided by a growth rate of 0.2/h. A two-sided Wilcoxon rank sum test was used to calculate P value.
To evaluate how uncertainties in the FBA-based flux may impact our dataset we first investigated the effect of flux variability on the estimated kmax (SI Appendix). We found that less than 8% of kmax values could differ, due to flux variability, by more than one order of magnitude (Dataset S4), and after removing these data we found the correlation between in vivo and in vitro values in log scale to be almost unchanged, i.e., R2 (all data) = 0.27, R2 (heterologous data) = 0.12, and R2 (homologous data) = 0.39. Second, we provided another set of kmax values (Dataset S7) estimated using unbiased flux random sampling (SI Appendix), which correlate strongly (R2 = 0.97) with the FBA-based kmax values in log scale. To evaluate the effect of a single high outlier value due to protein measurement we also correlated in log scale the second largest kapp across all conditions with in vitro kcat but found similar R2 values, i.e., R2 (all data) = 0.25, R2 (heterologous data) = 0.1, and R2 (homologous data) = 0.39. Moreover, by correlating kapp of each condition with in vitro kcat we found most correlations to be poor in log scale (Dataset S8). A recent study, using another proteomic dataset and a different GEM, also showed a poor correlation in log scale of R2 = 0.27 between yeast kapp and in vitro kcat at one condition (12).
We analyzed the yeast kapp under various conditions based on the ratio of condition-specific kapp over kmax for each reaction. By plotting the average ratio for each condition versus the corresponding growth rate (μ), we observed an increasing trend (Fig. 1D), which is in line with findings for E. coli (13). Furthermore, we compared the ratio of individual reactions between slow (μ ≤ 0.2/h) and fast (μ > 0.2/h) growth and found that kapp is significantly higher in faster-growing cells (Fig. 1E). We therefore conclude that kapp of yeast enzymes increase with growth rate. This suggests that proteome is more efficiently used at faster growth (13) and also indicates that growth could be controlled by efficiency of specific enzymes independent of conditions.
As turnover numbers are essential parameters in enzyme-constrained GEMs (ecGEMs) (9), we tested the use of kapp and in vitro kcat in a yeast ecGEM ecYeast8 (8). We parameterized the model with 1) an assumed same kcat for all enzymes, 2) default in vitro kcat in the original ecYeast8, 3) general kmax, and 4) μ-dependent kmax. Note that μ-dependent kmax is defined as the maximum kapp across the conditions under which growth rate is not greater than the given μ. To compare model performance, we used the model to predict proteomics data for growth on various carbon sources (14, 15), and we compared with the data not used to estimate our kapp dataset. We found that kmax outperforms the assumed kcat and default in vitro kcat (Fig. 2), confirming our estimation of kmax to be reliable. Notably, μ-dependent kmax can further improve the predictions (Fig. 2), meaning that it is more effective to use μ-dependent kmax values than condition-independent maximum values, which are adopted in most published ecGEMs (9). In addition, we found that default in vitro kcat in the original ecYeast8 outperforms the assumed same kcat for all enzymes (Fig. 2), meaning that it is still acceptable to use in vitro kcat when kapp values are unavailable.
Fig. 2.
Predictions of proteomics data on various carbon sources by ecYeast8 parameterized with an assumed same kcat, default in vitro kcat, general kmax, and μ-dependent kmax. Model performance is evaluated by root-mean-square error (RMSE) between predicted and measured protein levels on a log10 scale. N is the number of proteins with predicted nonzero concentrations by four parameterization strategies.
Overall, we present a kapp dataset of S. cerevisiae under various conditions, which can be used by ecGEMs for simulating the corresponding conditions. As kapp depends generally on growth rates rather than conditions (Fig. 1D), we believe that our μ-dependent kmax can be used to parameterize ecGEMs for predicting other conditions that are not involved in our dataset.
Materials and Methods
The metabolic fluxes were simulated using Yeast8 constrained by measurements. The absolute proteomics data were processed, i.e., the units of protein abundances were converted to millimoles per gram cell dry weight (gCDW). Details of all the materials and methods are provided in SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge funding from the European Union’s Horizon 2020 research and innovation program under grant agreement 686070. We also acknowledge funding from the Novo Nordisk Foundation (grant NNF10CC1016517).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108391118/-/DCSupplemental.
Data Availability
The data and codes are available at https://github.com/SysBioChalmers/Yeast_kapp. All other study data are included in the article and/or supporting information.
References
- 1.Davidi D., Milo R., Lessons on enzyme kinetics from quantitative proteomics. Curr. Opin. Biotechnol. 46, 81–89 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Davidi D., et al., Global characterization of in vivo enzyme catalytic rates and their correspondence to in vitro kcat measurements. Proc. Natl. Acad. Sci. U.S.A. 113, 3401–3406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heckmann D., et al., Kinetic profiling of metabolic specialists demonstrates stability and consistency of in vivo enzyme turnover numbers. Proc. Natl. Acad. Sci. U.S.A. 117, 23182–23190 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lahtvee P.-J., et al., Absolute quantification of protein and mRNA abundances demonstrate variability in gene-specific translation efficiency in yeast. Cell Syst. 4, 495–504.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Di Bartolomeo F., et al., Absolute yeast mitochondrial proteome quantification reveals trade-off between biosynthesis and energy generation during diauxic shift. Proc. Natl. Acad. Sci. U.S.A. 117, 7524–7535 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu R., et al., Nitrogen limitation reveals large reserves in metabolic and translational capacities of yeast. Nat. Commun. 11, 1881 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu R., Vorontsov E., Sihlbom C., Nielsen J., Quantifying absolute gene expression profiles reveals distinct regulation of central carbon metabolism genes in yeast. eLife 10, e65722 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu H., et al., A consensus S. cerevisiae metabolic model Yeast8 and its ecosystem for comprehensively probing cellular metabolism. Nat. Commun. 10, 3586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y., Nielsen J., Mathematical modelling of proteome constraints within metabolism. Curr. Opin. Syst. Biol. 25, 50–56 (2021). [Google Scholar]
- 10.Küken A., Gennermann K., Nikoloski Z., Characterization of maximal enzyme catalytic rates in central metabolism of Arabidopsis thaliana. Plant J. 103, 2168–2177 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Chen Y., Nielsen J., Flux control through protein phosphorylation in yeast. FEMS Yeast Res. 16, fow096 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Hanson A. D., et al., The number of catalytic cycles in an enzyme’s lifetime and why it matters to metabolic engineering. Proc. Natl. Acad. Sci. U.S.A. 118, e2023348118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Brien E. J., Utrilla J., Palsson B. O., Quantification and classification of E. coli proteome utilization and unused protein costs across environments. PLOS Comput. Biol. 12, e1004998 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulo J. A., O’Connell J. D., Gaun A., Gygi S. P., Proteome-wide quantitative multiplexed profiling of protein expression: Carbon-source dependency in Saccharomyces cerevisiae. Mol. Biol. Cell 26, 4063–4074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulo J. A., et al., Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. J. Proteomics 148, 85–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and codes are available at https://github.com/SysBioChalmers/Yeast_kapp. All other study data are included in the article and/or supporting information.