Significance
Blueprints of in-cell protein interaction landscapes are essential for our understanding of cellular structures and functions, which have been challenging to study at the systems level. Cross-linking–mass spectrometry (XL-MS) represents a high-throughput method for global profiling of interaction networks and can determine the identity and connectivity of native PPIs simultaneously without cell engineering. While in vivo XL-MS experiments are feasible, in-depth analyses remain difficult due to technical limitations on sample preparation. Here, we have developed a new Alkyne-A-DSBSO–based in vivo XL-MS platform that enabled us to obtain the most comprehensive PPI maps in cells. This approach can be adopted for proteome-wide XL-MS studies in any organisms and sample origins, thus advancing interactome biology beyond proteome identity and abundance.
Keywords: in vivo cross-linking mass spectrometry, protein–protein interactions, Alkyne-A-DSBSO, click chemistry enrichment, proteome-wide XL-MS
Abstract
Defining protein–protein interactions (PPIs) in their native environment is crucial to understanding protein structure and function. Cross-linking–mass spectrometry (XL-MS) has proven effective in capturing PPIs in living cells; however, the proteome coverage remains limited. Here, we have developed a robust in vivo XL-MS platform to facilitate in-depth PPI mapping by integrating a multifunctional MS-cleavable cross-linker with sample preparation strategies and high-resolution MS. The advancement of click chemistry–based enrichment significantly enhanced the detection of cross-linked peptides for proteome-wide analyses. This platform enabled the identification of 13,904 unique lysine–lysine linkages from in vivo cross-linked HEK 293 cells, permitting construction of the largest in vivo PPI network to date, comprising 6,439 interactions among 2,484 proteins. These results allowed us to generate a highly detailed yet panoramic portrait of human interactomes associated with diverse cellular pathways. The strategy presented here signifies a technological advancement for in vivo PPI mapping at the systems level and can be generalized for charting protein interaction landscapes in any organisms.
Protein–protein interactions (PPIs) are fundamental to the formation of protein complexes and crucial for regulating cellular activities required for cell viability and homeostasis. As a result, aberrant PPIs have been implicated in a variety of human diseases, including cancers (1). Recent studies have demonstrated the potential of modulating PPIs through their interaction interfaces, representing a therapeutic paradigm for mechanism-driven drug discovery (2). Therefore, detailed analysis of PPIs at the systems level will not only advance our understanding of protein sociology and their cellular functions but also facilitate the exploration of novel interaction-based therapeutics. Affinity purification–mass spectrometry (AP-MS) has been proven powerful in profiling PPIs at the global scale due to its versatility, speed, and scope (3–6). However, PPIs can reorganize upon cell lysis, and weak/transient interactions can be lost during APs (2, 7–11), preventing the full characterization of native PPIs. Recently, AP-MS has been coupled with protein proximity labeling (i.e., APEX [ascorbate peroxidase] and BioID) to improve the detection of PPIs within vicinity (∼20 nm) (12, 13). Although successful, these approaches only provide a list of putative interactors and often result in high background due to label diffusion and/or nonspecific binding (11, 14). Additionally, cell engineering associated with AP-based methods could potentially affect protein localization, interaction, and function (2, 10, 11). Thus, reciprocal purification is often needed for further validation, and iterative tagging of a large number of baits is required to dissect proteome-wide PPI networks (3–6), leading to limited analysis throughput. Recent developments in cofractionation-based MS approaches and complex-centric data analysis have allowed global characterization of protein complexes from cell lysates with increased accuracy and throughput (15, 16). While these technologies have been effective in defining cellular PPIs and complexes, it remains challenging to differentiate between direct and indirect interactors, as well as determine PPI interfaces of proteome networks in their native environments. To advance PPI studies, cross-linking–mass spectrometry (XL-MS) has been coupled with AP to facilitate the capture and identification of interactions of protein complexes (8, 9, 17–19). The unique capability of XL-MS technologies to covalently tether proximal amino acids (AA) of interacting proteins (∼30 Å) enables preservation of native PPIs in living cells, as well as simultaneous identification of PPI identities and contact sites (20–24). Thus, XL-MS represents a high-throughput method for defining interaction network topologies at the systems level. Apart from PPI mapping, the identified cross-links can be used not only for validating and fine-tuning existing structures but also for assisting de novo structural modeling to elucidate architectures of protein complexes (19, 24–30).
While in vivo PPI profiling is most attractive for unraveling protein interactions and structures in their native cellular environments, most XL-MS studies have been restricted to in vitro experiments. This is largely due to the difficulty in detecting low-abundance and heterogeneous cross-link products from complex peptide mixtures, especially at the proteome scale. Various efforts have been made to address the limitations of XL-MS analysis, resulting in technological advances in bioinformatics tools, XL reagents, and enrichment strategies (24–29). Most notably, recent innovations in MS-cleavable XL reagents have made a significant impact on XL-MS studies, enabling their applications in mapping PPIs at the systems level (21, 22, 31–34). To advance XL-MS studies, we have developed a class of sulfoxide-containing MS-cleavable XL reagents (e.g., disuccinimidyl sulfoxide [DSSO]), which allow fast and accurate identification of cross-linked peptides using multistage MS (MSn) and conventional database searching tools (22, 35–38). Importantly, our MS-cleavable reagents have demonstrated effectiveness in defining PPIs and elucidating architectures of protein complexes in vitro and in vivo (22, 24, 30, 39, 40). In addition, they have been successfully employed to delineate proteome-wide interaction networks by in vitro XL of cell lysates (31, 34). While these results highlight the potential of MS-cleavable cross-linkers in PPI studies, reagents with enrichment capabilities are ultimately advantageous for enhancing MS detectability of low-abundance cross-links in complex mixtures. This has been exemplified by the success of proteome-wide in vivo XL-MS analyses using Protein Interaction Reporter, resulting in the most representative in vivo PPI network in human cells composed of 893 nodes and 1,042 edges to date (21). While impressive, the scope of the uncovered PPI networks was limited, describing only a fraction of the human interactome. To enhance in vivo XL-MS studies, we have developed two multifunctional sulfoxide-containing XL reagents, that is, Azide-A-DSBSO and Alkyne-A-DSBSO (Azide/Alkyne-tagged, acid-cleavable disuccinimidyl bissulfoxide) (22, 41), which are membrane permeable, enrichable, and MS cleavable. Both reagents carry a small bio-orthogonal handle that allows click chemistry–based biotin conjugation for enrichment of cross-linked peptides. In addition, they possess two symmetric sulfoxide-containing MS-cleavable bonds for robust cross-link separation and identification. Moreover, incorporation of an acid-cleavable site improves cross-link recovery and identification through removal of the biotin handle. Finally, the spacer length (∼14 Å) is well suited for mapping PPIs. Collectively, the combined features of Azide-A-DSBSO and Alkyne-A-DSBSO make them ideal reagents for studying PPIs in intact cells. While we have previously demonstrated the feasibility of in vivo XL-MS analysis using Azide-A-DSBSO (22), the proteome content remains underrepresented. To further in vivo XL-MS studies, we have developed an Alkyne-A-DSBSO–based in vivo XL-MS analytical platform by coupling sample preparation strategies with advanced instrumentation to maximize the yield and identification of cross-linked peptides at the global scale. These developments have enabled us to generate an expansive in vivo XL-MS dataset consisting of 13,904 unique lysine–lysine linkages (9,289 intersubunit and 4,615 intrasubunit), permitting construction of the largest in vivo human interactome to date. This network comprises 6,439 edges within 2,484 nodes, uncovering PPIs involving 795 CORUM (comprehensive resource of mammalian) protein complexes (with at least 50% subunits identified) associated with various cellular pathways. Importantly, the methodologies presented here will help facilitate future XL-MS studies of various sample origins at the systems level.
Results
Establishing a Robust In Vivo XL-MS Platform for Proteome-Wide Analysis.
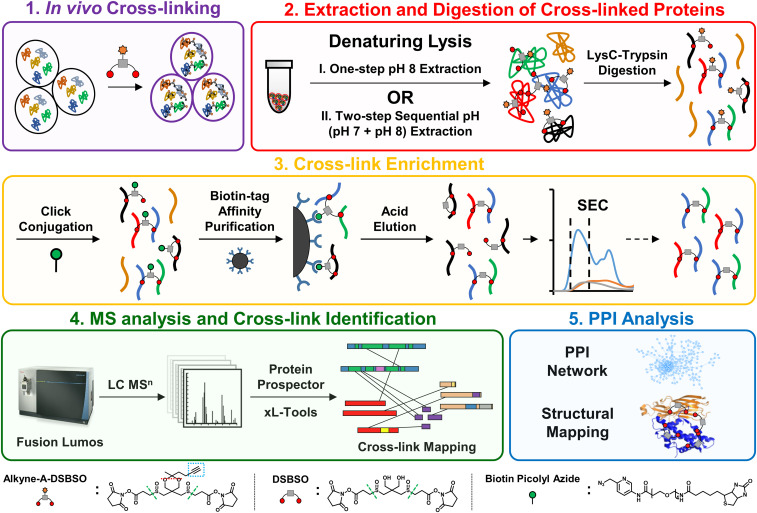
Successful proteome-wide XL-MS analyses of living cells rely on four major steps: 1) XL of intact cells; 2) extraction of cross-linked proteins; 3) enrichment of cross-linked peptides; and 4) MS analysis and identification of cross-linked peptides. The performance of these steps determines the scope and comprehensiveness of in vivo PPI profiling. Through the years, innovations in MS have continuously enhanced the MS analysis of cross-linked peptides. However, improvements in sample preparation required for in-depth in vivo PPI mapping have been limited. The remaining challenges are mostly associated with efficient protein extraction and cross-link enrichment, the bottlenecks of existing methods (22). To address these limitations, we have established an Alkyne-A-DSBSO–based in vivo XL-MS platform by developing sample preparation strategies and integrating them with high-performance Lumos Orbitrap instrumentation. Immunoblotting analysis revealed that Alkyne-A-DSBSO cross-linked intact cells effectively (Fig. 1, step 1) (SI Appendix, Fig. S1).
Fig. 1.
The Alkyne-A-DSBSO–based in vivo XL-MS platform for PPI mapping at the proteome scale. The overall workflow is divided into five major steps as displayed. The chemical structures of Alkyne-A-DSBSO, DSBSO, and BPA are illustrated, in which green dashed lines represent MS-cleavable sites, red dashed lines represent the acid cleavage site, and the blue dashed box represents the enrichable alkyne handle.
In vivo cross-linked proteins are often extracted from cells with a commonly used denaturing lysis buffer (8 M urea, pH 8) (Fig. 1, step 2) (22). While effective, the resulting sample typically contains cross-linked histones in excessive abundance. Previous analysis has shown that such overrepresentation prevents the effective detection and identification of cross-links from nonhistone proteins, thus hindering deeper proteome-wide PPI profiling (22). In order to better delineate protein interaction landscapes in living cells, we intended to first separate cross-linked histones from nonhistone proteins. It has been shown that native histones in cells can be extracted at acidic pH due to their highly positive charges (42). However, it was unclear that a similar strategy could be used for cross-linked cells. To test whether and how pH modulates the solubility of cross-linked histones, we lysed cross-linked cells at pH 6, 7, and 8 under fully denaturing conditions (8 M urea). Immunoblotting analysis showed that denaturing lysis at pH 7 considerably decreased the solubility of in vivo cross-linked histones, while predominantly maintaining the solubility of cross-linked nonhistone proteins compared to the commonly used pH 8 lysis (SI Appendix, Fig. S2A). These results suggest that lysine XL decreased the overall positive charges of histones, making them less soluble in acidic pH, opposite of native histones. This observation is significant as it allows us to establish a two-step sequential pH extraction procedure to separate cross-linked histones from nonhistone proteins, by lysing cross-linked cells at pH 7 first, followed by extraction of the residual pellet at pH 8 (Fig. 1, step 2). The two extracts were then collected and processed separately for downstream experiments, significantly improving the identification of nonhistone cross-linked peptides (SI Appendix, Fig. S2B) and yielding an ∼2.5-fold increase in the total number of identified cross-linked proteins in a single XL-MS experiment (SI Appendix, Fig. S2C). These results indicate that separation of cross-linked products at the protein level is beneficial to expanding the range of MS analysis.
Developing a Cross-Link Enrichment Strategy.
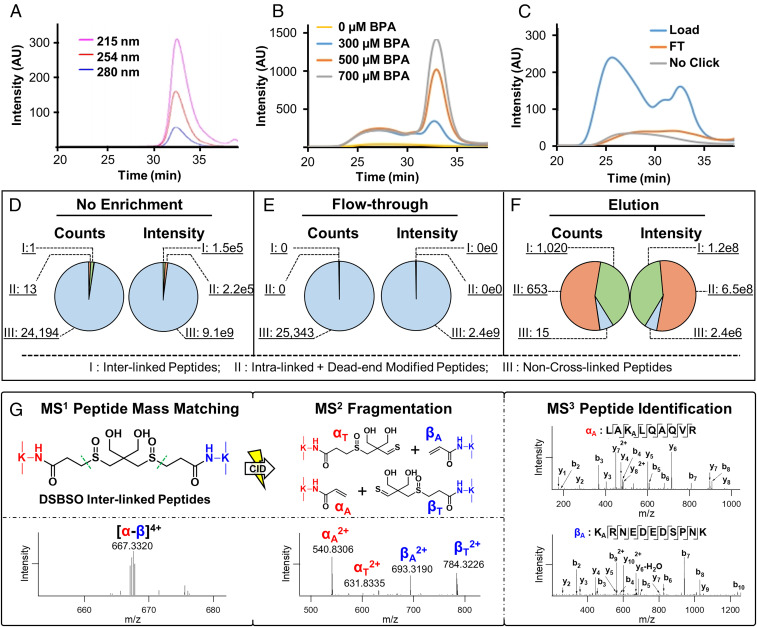
The next critical step in in vivo proteome-wide XL-MS experiments is efficient cross-link enrichment to reduce sample complexity and enhance MS detection of low-abundance cross-linked peptides (Fig. 1, step 3). Click chemistry–based enrichment strategies have been widely used in chemical proteomics due to the bio-orthogonality, specificity, and advantageously small sizes of affinity handles (i.e., azide or alkyne tags) (43, 44). However, its use in XL-MS studies has been limited due to the low-abundance and heterogeneity of cross-linked products, often leading to difficulty in achieving high efficiency for both click conjugation and subsequent peptide purification (22). To address current limitations, we developed a cross-link enrichment workflow by implementing three features to advance the analysis of Alkyne-A-DSBSO cross-linked peptides. First, instead of biotin conjugation at the protein level (22), we performed click reactions directly on Alkyne-A-DSBSO cross-linked peptides to increase their accessibility for more effective and thorough labeling (Fig. 1, step 3). Second, we utilized a water-soluble biotin conjugating reagent (biotin picolyl azide, also known as BPA) to achieve a highly sensitive and efficient click reaction. This implementation was based on the reported capability of BPA to label proteins and cells with dramatically higher efficiency and reduced dependence of Cu(I) compared to water-insoluble reagents (45). In addition, due to the presence of the picolyl moiety, BPA has ultraviolet (UV) absorbance at multiple wavelengths, including the commonly used 215 nm for peptide backbone absorbance (Fig. 2A). Interestingly, our result showed that BPA-labeled peptides exhibit much higher absorbance at 254 nm compared to unlabeled peptides, making it the ideal wavelength for monitoring the presence of BPA-conjugated cross-linked peptides. The unique UV detectability of BPA and BPA-labeled peptides is important and advantageous as it enables their chromatographic tracking and thus permits the optimization of each step of click chemistry–based enrichment, which has been difficult to monitor in the past. Specifically, these features permit effective examination and optimization of click-based biotin conjugation (Fig. 2B) and subsequent enrichment of cross-linked peptides by binding to streptavidin beads (Fig. 2C). The bound BPA-labeled cross-linked peptides were then acid eluted, yielding highly purified cross-linked peptides (Fig. 1, step 3). The acid cleavage is beneficial as it enables the removal of BPA and converts Alkyne-A-DSBSO cross-linked peptides into DSBSO cross-linked peptides, significantly reducing mass addition and thereby simplifying subsequent MS analysis and identification.
Fig. 2.
Enrichment and analysis of Alkyne-A-DSBSO cross-linked peptides. (A) SEC separation and detection of free BPA at three wavelengths: 215, 254, and 280 nm. (B) Assessment of biotin conjugation efficiency of Alkyne-A-DSBSO cross-linked peptides with increasing amounts of BPA monitored at 254 nm by SEC. (C) Evaluation of purification efficiency of BPA-labeled cross-linked peptides before binding to streptavidin beads (load) and flow through (FT) after binding through SEC. This process was monitored by comparing peptide UV absorbance at 254 nm. Unlabeled cross-linked peptides were used as control. (D–F) Benchmarking of cross-link enrichment performance using cross-linked cell lysates. Comparative analysis was performed among nonenriched (original), FT after binding of BPA-labeled cross-linked peptides to streptavidin beads, and elution (enriched cross-linked peptides) samples. Note: counts describe the total number of identified peptides, and intensity represents the total ion intensity of each defined group. Three groups of peptides in each sample were identified and compared: I, interlinked peptides; II, intralinked + dead-end modified peptides; and III, non–cross-linked peptides. (G) MSn analysis of a representative DSBSO interlinked peptide (α-β). The parent ion mass (m/z 667.33204+) was detected in MS1, which produced two characteristic fragment ion pairs modified with complementary DSBSO remnants, that is, αA/βT (m/z 540.83062+/784.32262+) and αT/βA (m/z 631.83352+/693.31902+) using collision-induced dissociation (CID) during MS2 analysis. Subsequent MS3 analyses of individual fragment ions αA (m/z 540.83062+) and βA (m/z 693.31902+) yielded series of b and y ions that identified them as 55LAKALQAQVR63 of BTF3 and 91KARNEDEDSPNK101 of RPL31, respectively. The integration of MS1, MS2, and MS3 data accurately determined a cross-link between BTF3:K57 and RPL31:K91. Note: KA represents alkene modified lysine.
While click chemistry–based purification permits separation of cross-linked peptides from non–cross-linked peptides, the resulting enriched mixture contains all types of cross-linked products including interlinked, intralinked, and dead-end modified peptides (46). Among them, interlinked peptides are most informative about protein interaction contacts. Therefore, lastly, the enriched cross-linked peptides were further separated by size-exclusion chromatography (SEC) (Fig. 1, Step 3) to maximize the detection and identification of interlinked peptides (47). Free BPA in the cross-linked peptide mixture was also concurrently removed during SEC, eliminating its interference in MS analysis (Fig. 2A). The final enriched DSBSO cross-linked peptides were analyzed and identified by liquid chromatography (LC)-MSn, the most effective data acquisition strategy for accurate identification of sulfoxide-containing MS-cleavable cross-linked peptides (Fig. 2G) (22, 24, 35–38). Together, these developments enable the establishment of a robust and traceable click chemistry–based enrichment strategy for cross-linked peptides, significantly improving their yield and analysis at the proteome scale, as shown in this study.
Benchmarking Cross-Link Enrichment.
To illustrate the effectiveness and robustness of our cross-link enrichment procedure and its benefits for proteome-wide XL-MS studies, we have performed benchmarking studies by two types of comparative analyses. In order to properly assess enrichment efficiency and specificity, resulting samples were analyzed directly by MS without SEC fractionation unless stated otherwise. The initial benchmarking was accomplished by spiking Alkyne-A-DSBSO cross-linked BSA peptides into a HEK 293 cell lysate matrix in two different ways. First, a fixed amount (1 µg) of cross-linked BSA peptides was mixed with increasing amounts (i.e., 1, 10, and 100 µg) of non–cross-linked cell lysate digests, respectively, prior to click chemistry–based cross-link enrichment. LC-MSn analysis of the purified samples identified 31, 27, and 24 cross-linked peptides of BSA (including dead-end, intralinked, and interlinked peptides), of which 14, 12, and 10 are interlinks from the 1:1, 1:10, and 1:100 samples, respectively (Dataset S1A). The results indicate that the enrichment is efficient as no significant loss in cross-linked peptide identification was observed when increasing amounts of complex matrix backgrounds were present. In comparison, cross-linked peptides of BSA were only identified in the 1:1 mixed sample without enrichment, in which only one interlinked peptide was found, signifying the importance and effectiveness of cross-link enrichment.
Second, we added increasing amounts (i.e., 1, 10, or 100 µg) of cross-linked BSA in a fixed amount (100 µg) of non–cross-linked cell lysate digests and normalized peptide concentration before injection. Using this approach, we identified 38, 31, and 28 cross-linked peptides from the 1:1, 1:10, and 1:100 samples, respectively, of which, 19, 17, and 13 were interlinked peptides (Dataset S1B). These results further demonstrate the sensitivity and efficiency of our cross-link enrichment as 100-fold differences in the amount of cross-linked peptide spike only marginally affected the peptide recovery. It is noted that the number of identified interlinks were further increased by ∼2.3- to 5.5-fold for the three sample conditions when coupled with SEC fractionation, with the 1:100 sample showing the most significant improvement (Dataset S1C). This indicates that incorporating SEC fractionation is beneficial to further enhance the detection of low-abundance cross-links after enrichment.
The second type of benchmarking was carried out using cross-linked cell lysates to evaluate whether the enrichment would significantly augment the identification of cross-linked peptides at the proteome scale. To better understand the efficiency of cross-link enrichment, we have compared the number of identified non–cross-linked and cross-linked peptides in samples with and without enrichment. Without enrichment, a total of 24,208 peptides were identified in the mixture by tandem MS (MS2) and triple-stage MS (MS3), of which more than 99.94% of identifications were linear non–cross-linked peptides (Fig. 2D), and only 0.06% (i.e., 14 peptides) represented cross-linked products. In contrast, a total of 1,688 peptides were identified after enrichment, among which 99.0% were cross-linked peptides and only 1.0% belonged to non–cross-linked peptides (Fig. 2F). As expected, almost all of the non–cross-linked peptides (a total of 25,343 identified) remained in the flow through during AP of cross-linked peptides (Fig. 2E). Additionally, no cross-linked peptides were found in the flow through by MS3. These results demonstrate high binding specificity and purification efficiency. Apart from the total number of peptide identifications, more than 99.99% of the ion intensities observed were attributed to non–cross-linked peptides in both unenriched and flow-through samples (Fig. 2 E and F). In contrast, 99.99% of the total ion intensities were represented by cross-linked peptides in the enriched sample (Fig. 2F). Based on the total identifications, these numbers indicate a roughly 1,000-fold enrichment efficiency and 99% specificity for cross-linked cell lysates. In comparison to immobilized metal ion affinity chromatography (IMAC)-based enrichment of PhoX cross-linked peptides (48), click chemistry–based enrichment appears to have a comparable specificity (i.e., 99% versus 97% [PhoX]) but a significantly higher enrichment efficiency (i.e., 1,000- versus 300-fold [PhoX]). Collectively, these benchmarking results establish the robustness and effectiveness of the cross-link enrichment strategy developed here.
Defining the XL Proteome in Mammalian Cells.
Identification of cross-linked peptides was performed by LC-MSn (22, 35) and exemplified by the MSn analysis of a representative DSBSO interlinked peptide (α-β) from in vivo cross-linked HEK293 cells (Fig. 2G). In this work, we have performed two biological replicates for both one-step (pH 8 only) and two-step (pH 7 + 8) extraction procedures. In total, 5,316 proteins were identified with at least one cross-linked peptide (i.e., interlinked, intralinked, and/or dead-end modified peptide), describing the breadth of the proteome targeted by the cross-linker. To illustrate PPIs, we next derived an XL proteome that only contains proteins identified with at least one interlinked peptide. The resulting XL proteome was composed of 2,484 proteins that were identified with a total of 17,063 unique interlinked peptides from 181,207 cross-linked peptide spectrum matches (CSMs) at <1% false discovery rate (FDR), representing 13,904 unique K–K linkages (9,289 intersubunit and 4,615 intrasubunit) (Dataset S2 A–D). These signified 5,401 interprotein and 1,038 intraprotein interactions. The XL proteome obtained here consists of a significantly (∼6- to 10-fold) higher number of protein interactions and cross-links than any previously reported in vitro and in vivo XL proteomes (21, 31) (Dataset S3), thus representing the largest XL-PPI dataset at the systems level.
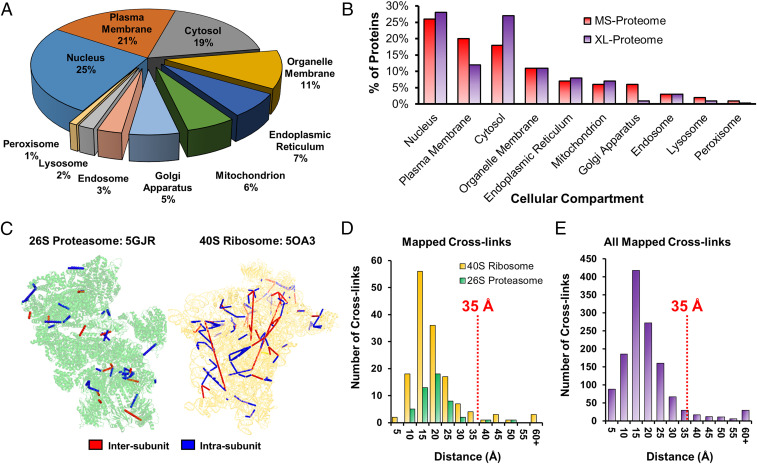
To understand the extent of in vivo XL, we compared our XL proteome to the MS proteome of HEK 293 cells containing 7,201 proteins determined by shotgun proteomics in ProteomicsDB (49). As a result, ∼95% of our XL proteome was covered, reaffirming the validity of the identified cross-linked proteins. In addition, Gene Ontology (GO) analysis has revealed that all major compartments (i.e., cytosol, nucleus, and plasma membrane) have been mapped by in vivo XL-MS analysis (Dataset S4), with an overall distribution similar to that of the MS proteome (Fig. 3 A and B). Noticeably, plasma membrane and the Golgi apparatus were better represented in the MS proteome, whereas the cytosol was better described in the XL proteome. The observable differences are likely attributed to multiple factors including protein solubility and abundance, as well as protein lysine content, accessibility, and cross-linkability. As in vivo XL preserves native PPIs in cells, the interactions captured are expected to be within the same cellular compartments. Indeed, 92% of the PPIs identified represent intracompartmental interactions, confirming the preservation of cellular interactions by in vivo XL. The small number of observed intercompartmental PPIs are most likely ascribed to proteins that can be present in different subcellular locations.
Fig. 3.
Annotation of the XL proteome and validation of identified cross-links. (A) Cellular compartmental distribution of the in vivo Alkyne-A-DSBSO XL proteome. (B) Comparison of cellular compartmental distribution between our XL proteome and the MS proteome of HEK293 cells determined by shotgun proteomics in ProteomicsDB (49). (C) Structural mapping of the 26S proteasome (PDB: 5GJR) and the 40S ribosome (PDB: 5OA3) using the identified cross-links. Intersubunit cross-links shown in red and intrasubunit cross-links shown in blue. (D) Respective distance distributions of cross-links mapped to the 26S proteasome (green) and 40S ribosome (yellow) complexes as in C. In total, 94.8% of cross-links from both complexes satisfy the distance restraint of 35 Å. (E) The distance distribution of all in vivo cross-links that could be mapped to the known high-resolution structures of the 286 protein complexes (SI Appendix, Dataset S6). A total of 91.8% of cross-links fall below the distance constraint of 35 Å.
To estimate the dynamic range of the XL proteome, we plotted the abundance distribution of the identified cross-linked proteins based on the intensity-based absolute quantification (iBAQ) values of the HEK 293 MS proteome (49) (SI Appendix, Fig. S3). While the abundance distributions of XL and MS proteomes were similar, the former is shifted toward higher abundance proteins. This is not surprising as XL reaction kinetics are dependent on protein concentrations and that the resulting products are often substoichiometric, representing only a fraction of proteins involved. Nevertheless, our results demonstrate that the Alkyne-A-DSBSO–based in vivo XL-MS platform is capable of targeting cellular proteins localized across all cellular compartments and with abundances spanning several orders of magnitude.
Validation of Cross-Links by Structural Mapping.
To evaluate cross-link validity, we mapped all identified K–K linkages onto available high-resolution structures of protein complexes. We first categorized the XL proteome into 1,552 CORUM protein complexes (50), in which 795 complexes were identified with at least 50% composition, and 148 complexes were recovered with full composition (Dataset S5). Among them, only 286 protein complexes have high-resolution structures available in Protein Data Bank (PDB) (Dataset S6). Considering the length of the Alkyne-A-DSBSO spacer (∼14 Å), lysine side chains, and backbone dynamics, lysine residues with Cα-Cα distance <35 Å were expected to be preferentially cross-linked. To illustrate, we first mapped cross-links identified from 26S proteasome and 40S ribosome complexes onto their respective structures (PDB: 5GJR and 5OA3) (Fig. 3C). In total, 48 of 61 proteasome cross-links and 145 of 264 ribosome cross-links were successfully mapped, with 96% and 94% of cross-links <35 Å, respectively (Fig. 3D). In total, 812 K–K linkages were covered by the high-resolution structures of 286 protein complexes with a median Cα-Cα distance of 14.5 Å and 95% satisfaction (Fig. 3E and Dataset S6), confirming the validity of the identified cross-links.
Analysis and Validation of In Vivo XL PPIs.
To further analyze XL-MS data, we have assessed the reproducibility of unique K–K linkages for the two extraction strategies. As a result, pH 8–only extraction yielded 47.3%, and sequential pH extraction produced 45.7% overlaps between their two biological replicates, respectively (SI Appendix, Fig. S4). This level of reproducibility in K–K linkages is comparable to those of published proteome-wide XL-MS analyses, ranging from 23.5% to 43.5% (20, 32, 33) (Dataset S7). In addition, we have evaluated the reproducibility at the PPI level; both extraction protocols resulted in similar overlaps not only for PPIs (33% versus 34%) but also for K–K linkages (54% versus 54%) in the shared PPIs between their respective two biological replicates (SI Appendix, Fig. S5). These results further support the robustness of the in vivo XL-MS workflow established here.
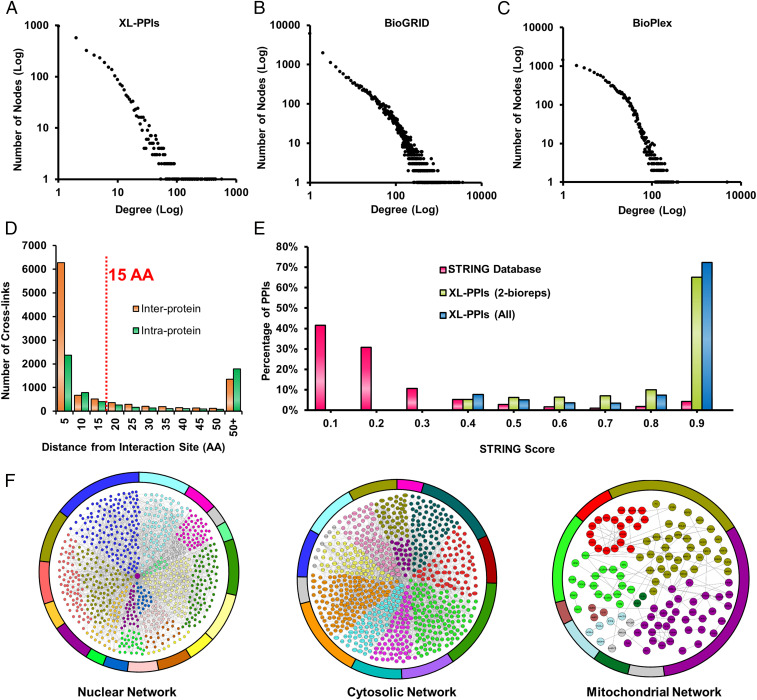
To illustrate how proteins connect in cells, we generated an XL-PPI network map comprising 2,484 nodes and 6,439 edges, denoting 5,401 interprotein and 1,038 intraprotein interactions (SI Appendix, Fig. S6). Interestingly, more than 80% of the nodes were interconnected in a single major network, demonstrating extensive communication among cellular components and activities. It is noted that 3,204 of the total interprotein interactions involve homologous proteins identified with at least one unique peptide, thus yielding 4,198 protein group–protein group interactions. To exclude the possibility of the XL-PPI network being random, we have plotted its degree distribution and discovered that it follows a power law log-log fit, representing a typical scale-free network topology where a few hub nodes support a large portion of the connections (51) (Fig. 4A). This feature corroborates very well with degree distribution characteristics of existing large-scale PPI networks derived by other methods in BioGRID (biological general repository for interaction datasets) and BioPlex (Fig. 4 B and C), substantiating the legitimacy of the identified XL PPIs. In comparison to the three selected PPI databases (i.e., STRING [search tool for the retrieval of interacting genes/proteins], BioGRID, BioPlex) containing a total of 769,748 pair-wise interactions, 41% of our in vivo XL PPIs were considered as known interactions and 59% as PPIs (SI Appendix, Fig. S7). The observed differences are most likely attributed to experimental variance in PPI profiling methods, and the information obtained here is complementary to existing PPI data.
Fig. 4.
Validation and analysis of in vivo XL PPIs. (A–C) Degree distribution plots of XL PPIs (A), BioGRID PPIs (B), and BioPlex PPIs (C). (D) AA distance distribution of cross-linked lysines from interaction interfaces for interprotein and intraprotein interactions predicted by Interactome INSIDER. (E) STRING score distributions for STRING PPIs (pink), XL PPIs detected in at least two biological replicates (yellow green), and all XL PPIs (aqua blue). (F) Three major cellular compartmental XL-PPI networks (nuclear, cytosolic, and mitochondrial) generated from intersubunit interactions captured by in vivo XL for proteins belonging to each compartment. Each network was organized into different colors based on biological processes associated with each protein node (SI Appendix, Dataset S4), in which a total of 1,093 biological processes was included.
XL-PPI contact sites are defined by residue-to-residue linkages (<35Å) and expected to be within close proximity of interaction interfaces (30, 40). To test this, we investigated the distribution of cross-linked lysines to the predicted interaction interfaces by Interactome INSIDER (52). As a result, 82.4% of the interprotein and 58.5% intraprotein cross-linked lysines were found to be within 15 AA of predicted interaction interfaces (Fig. 4D), of which the majority were localized within 5 AA. These results augment the validity of the identified XL PPIs and suggest that interprotein cross-linked sites occur more frequently in the predicted PPI interaction regions.
It has been shown that STRING scores of PPIs can be used to infer the likelihood of an interaction being true (53). To further evaluate the validity of the captured interactions, we obtained the STRING scores for 1,120 XL PPIs and compared their distribution to that of 687,000 human PPIs present in the STRING database (Fig. 4E). As shown, 89% of STRING human PPIs carry scores below 0.4, indicating that the majority of them display only low to medium confidence. In contrast, all of the 1,120 XL PPIs have STRING scores above 0.4, and more than 70% of them possess scores ≥0.9, illustrating their high probability and confidence in being true interactions. Importantly, the STRING score distribution remains similar for XL PPIs found in one or more biological replicates, further demonstrating the reliability of in vivo XL PPIs and suggesting that the confidence levels are comparable across the entire dataset regardless of their occurrence in detection. Therefore, we have included all of the identified PPIs for subsequent analyses.
Compartmental XL-PPI Networks.
To understand the content of the XL-PPI networks, we next categorized them into three main compartmental networks (i.e., nucleus, cytosol and mitochondria), which were constructed and color coded based on protein-associated cellular processes (Fig. 4F and Dataset S4). The nuclear network comprises 2,753 interprotein interactions among 1,118 proteins and is involved in a wide range of nucleus-related biological processes including gene expression, cell-cycle control, and cell death (Fig. 4 F, Left). The cytosolic PPI network contains 944 proteins that are intertwined through 1,821 interprotein interactions (Fig. 4 F, Middle) involved in various biological processes ranging from protein translation by ribosomal machinery to developmental processes orchestrated by kinases and actin-associated machinery. The mitochondrial PPI network was composed of 141 nodes connected by 138 pair-wise interactions (Fig. 4 F, Right). The majority of these proteins are involved in metabolic processes such as ATP synthesis, mitochondrial organization, and transport. Although our in vivo XL-MS analysis covered only one-third of an ex vivo XL-PPI map of isolated mitochondria (54), it did capture core PPIs in mitochondria—including those among ATP synthase subunits and mitochondrial ribosomal subunits. The results indicate that global in vivo XL-MS analysis is capable of providing a glimpse of mitochondrial PPIs even without subcellular fractionation. Collectively, within each compartmental PPI network, a large degree of connectivity within and between biological pathways were observed, suggesting extensive cross talk among cellular machineries to coordinate their functions in various biological processes.
The Histone PPI Subnetwork.
One of the most well-represented PPI subnetworks determined in this work involved histone interactions identified with 4,970 unique histone-containing K–K linkages. This resulted in an extensive in vivo histone PPI subnetwork with 684 nodes and 1,689 pair-wise interactions. High-density interactions within core histones constituted 2,975 unique cross-links (2,264 interprotein and 711 intraprotein), illustrating broad connectivity across their folded and tail regions. A significant portion of histone cross-links were localized to unstructured N-terminal regions that remain unresolved in human core nucleosome structures. These results thus reveal structural information in regions that cannot be easily assessed by high-resolution structural tools. In addition, extensive interactions between core histones (H2A, H2B, H3, H4) and linker histone variants (H1.1 to H1.5) were also identified (Dataset S2A). For example, 14 cross-links exhibited proximal interactions between the globular domains of H1 variants with H2A and H3, respectively, half of which involved the H1.3 variant (SI Appendix, Fig. S8). Interestingly, these interactions were only captured here by our in vivo and an ex vivo XL studies (32), and not by in vitro XL-MS analyses (31), suggesting that they are most likely weak/transient and are lost during native lysis prior to in vitro XL. Thus, this exemplifies the importance of in vivo XL for preserving physiological PPIs in their native environments.
In addition to core and linker histones, 623 proteins were identified as histone interactors, primarily categorized into five major biological processes (i.e., localization, gene expression, metabolism, chromosomal rearrangement, and process regulation) (SI Appendix, Fig. S9A). Of all histones, H2B was found to be involved in the most interprotein cross-links, whereas H4 had the least nonhistone interactions. Histone interactors were predominantly cross-linked to the C termini of H2A/H2B, or the N terminus of H3. The disparity in the number and location of cross-links between histone variants and interacting proteins is more likely due to differences in their structural flexibility and accessibility to cross-linkers as well as the availability of cross-linkable lysines (32). Among histone interactors, ∼33% of them were involved in regulating gene expression (SI Appendix, Fig. S9A), which is illustrated by extensive connectivity between histones and several known regulators (i.e., nucleophosmin [NPM1], high mobility group proteins [HMGA1, HMGN1, HMGN2, HMGN3], and MCM complex) involved in chromatin remodeling (SI Appendix, Fig. S9B). Their proximities to histones inferred by identified cross-links support their function through disruption of chromatin-DNA binding. Collectively, these results demonstrate that in vivo XL-MS analysis permits the generation of comprehensive histone PPI networks, which can be useful to understanding the importance of histone PPI networks during epigenetic regulation and chromatin remodeling in the future.
The Chaperonin PPI Subnetwork.
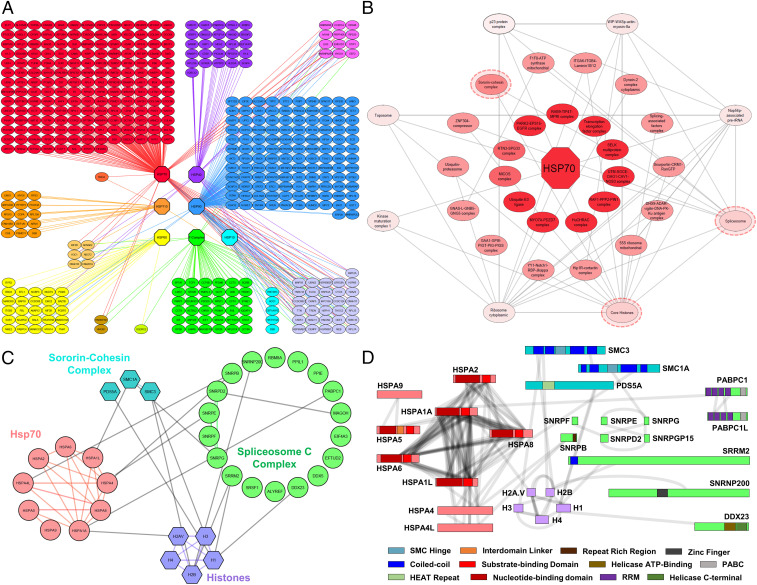
Another comprehensive PPI subnetwork captured in this study is the heat shock protein (HSP)-associated chaperonin network, composed of 467 nodes and 518 interprotein interactions. To illustrate this PPI subnetwork, HSP interactors were first clustered into groups according to their direct physical contacts with members of seven major chaperone families (i.e., Hsp70, Hsp40, Hsp110, Hsp90, Hsp60, Hsp10, and T complexes) (Fig. 5A). Members of each chaperone family were also grouped together and collapsed as individual nodes. In total, ∼80% of the 518 PPIs were interactions between HSPs and a wide range of proteins including chaperon regulators, adaptors, and clients. Among the seven HSP families, Hsp70 interactors were found to be the most diverse, including transcription factors, transport proteins, and metabolic proteins. Based on 1,421 unique Hsp70-containing K–K linkages, an Hsp70 subnetwork was constructed with 151 nodes and 231 edges—of which, only 35 edges were previously known. These Hsp70 interactors were further collapsed into 24 CORUM-defined protein complexes involved in an array of biological functions ranging from chromatin structural modeling to cytoskeletal attachment, reinforcing the importance of the Hsp70 family as ubiquitous housekeeping chaperons.
Fig. 5.
The chaperone XL-PPI networks. (A) The proteins identified with direct interactions with at least one of the chaperones were illustrated as individual nodes. The chaperones were grouped by seven major families shown as octagons. Chaperone-interacting proteins were color coded based on which chaperones they interacted with. Blending of colors was used when proteins interacted with multiple chaperones. (B) Hsp70-associated CORUM protein complex network defined by Hsp70 XL PPIs, illustrating connectivity among protein complexes. Complexes at the innermost ring only had direct interactions with Hsp70, while complexes at outer rings interacted with both Hsp70 and its associated complexes. (C) The interaction network of the three circled complexes from B arranged into subunits that compose these complexes, that is, sororin-cohesin, histones, and spliceosome C complexes. (D) 2D XL maps of the interactions seen between the subunits shown in C. Domains of interest were colored according to the scheme shown.
To further dissect the interconnectivity among Hsp70-interacting protein complexes, we created an interaction map concentrically centered at Hsp70 and expanding outward with increasing an number of protein complex interactions (Fig. 5B). While the innermost complexes had only direct associations with Hsp70, those in the second and third layers interact not only with Hsp70 but with other Hsp70-associated complexes (1 to 2 or 3+, respectively). One exemplary set of interactions was identified between Hsp70 and three nuclear complexes, that is, spliceosome C (20/32 subunits), sororin-cohesin (3/6 subunits), and histone family (5/5 subunits) complexes (Fig. 5C). These complexes were identified not only with direct binding to Hsp70 but also to one another. While the sororin-cohesion complex regulates sister chromatid cohesin and separation and DNA damage and repair, the spliceosome is responsible for removing introns from primary transcripts to generate mature messenger RNA. Although Hsp70 has been implicated in these coregulatory processes, the molecular details underlying its role are unclear. Our network analysis indicates that Hsp70 directly associates with these complexes, implicating its involvement in gene expression, cell-cycle processes, and DNA repair. To better define their interactions, we have generated a two-dimensional (2D) XL map comprising 123 unique K–K linkages to display their physical contact regions and associated domains (Fig. 5D). Clearly, a high degree of intersubunit connectivity was observed within the Hsp70 family members, indicative of their intricate relationships for coordinated activities. Among the selected three complexes, Hsp70 proteins were found to interact with the spliceosome C complex the most—specifically, through two RNA-binding subunits PABPC1 and SNRPG. Interestingly, PABPC1 is a known Hsp70 client, indicating chaperon–substrate interactions. Apart from Hsp70 interactions, 38 cross-links were identified among the three complexes (Fig. 5D). As shown, SMC3-containing cross-links (SMC3:K185 to H2B:K109 and SMC:K238 to H3:K80) have confirmed its interactions with histones and suggest that the docking of SMC3 on chromosomes may involve both H2B and H3, instead of H3 alone as previously reported. The coiled-coil domain of SMC3 (AA no. 179 to 350) represents a strong candidate for facilitating this interaction. In addition, histones H1, H2B, and H3 were found interacting with spliceosome C subunits (SNRNP200, DDX23, and SRRM2) responsible for early spliceosome formation. Collectively, our results have demonstrated that chaperonins form high-density interaction networks, physically linking them to various cellular pathways and underlying the importance of chaperon mediation in PPI regulation.
The PPI Subnetwork of the Ubiquitin-Proteasome System.
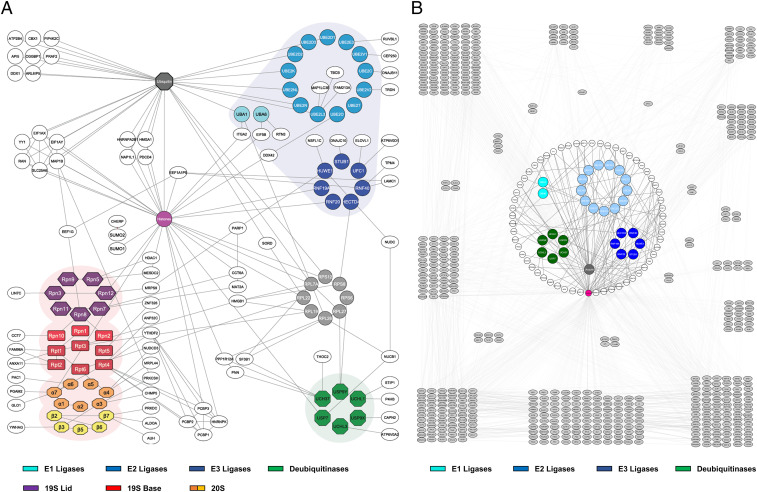
One of the interesting PPI subnetworks revealed here involves components within the ubiquitin-proteasome system (UPS), the pathway for proteasome-dependent degradation of ubiquitinated substrates in eukaryotes (55). Based on 1,198 unique K–K linkages, we have constructed the largest in vivo PPI map of the UPS obtained by XL-MS studies thus far, containing 185 interprotein interactions among 146 proteins directly associating with known UPS machinery (Fig. 6A). Of the 185 edges delineated in this network, 85 represent known interactions. Protein ubiquitination embodies multiple reactions through a cascade of enzymes involving E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin ligases), while proteasomal degradation entails an array of biochemical events including substrate recognition, delivery, deubiquitination, unfolding, and translocation. All of these processes are carried out by a variety of proteins and protein complexes, which interact stably and/or dynamically to fulfill their functions. Of the core UPS proteins, in vivo XL-MS analysis has identified intersubunit interactions involving 2 E1s, 13 E2s, 6 E3 ubiquitin ligases, 28 proteasome subunits, 6 deubiquitinases (DUBs), and ubiquitin.
Fig. 6.
The UPS XL-PPI networks. (A) An interaction network composed of the identified UPS components and their direct interactors. The UPS components were grouped and color coded based on complexes and functions, including 26S proteasome (19S lid, purple; 19S base, red; 20S α ring, orange; 20S β ring, yellow), E1 (cyan), E2 (light blue), E3 ubiquitin enzymes (royal blue), and DUBs (green). (B) Extended XL-PPI network of proteins found to interact with ubiquitin, DUBs, and ubiquitin ligases. Core ubiquitination machinery was color coded and shown in the center of the network. They were encircled by a ring of directly interacting proteins, shown in white. Proteins found to be cross-linked to direct interactors are shown in gray and categorized into groups based on their PPI similarities.
The 26S proteasome is the degradation machine composed of 19S regulatory and 20S core subcomplexes. In total, 85 unique proteasome-containing interprotein K–K linkages were identified, representing 54 pair-wise interactions within proteasome subunits and with their interacting proteins. Here, we identified 28 of the 33 subunits in the 26S proteasome, recovering 84.2% (16/19) and 85.7% (12/14) of the 19S and 20S subcomplexes, respectively. Of the 21 unique K–K linkages captured that describe interconnectivity within the proteasome, 13 were identified in our previous work on purified proteasomes (39). These results attest to the capability of the methodology in providing structural information of protein complexes even without purification. Among the 37 proteasome-interacting proteins, 24 were determined to have direct physical contacts with the 19S, whereas 12 were closely connected to the 20S, representing interactions covering a broad range of proteins. Although some proteins were considered as putative proteasome interactors through AP-MS experiments (http://www.BioGRID.org), our results yielded interaction contacts with specific subunits to demonstrate their direct associations with proteasomes. Interestingly, 34 out of 35 proteasome-interacting proteins can be ubiquitinated, and some of them (e.g., ALDOA, CCT7, EEF1G, PCBP1, etc.) have been previously linked to ubiquitin and ubiquitin ligases (http://www.BioGRID.org). Therefore, we suspect that these proteins may be ubiquitinated substrates docked on the proteasome prior to their degradation. However, it does not exclude the possibility of them being UPS regulators and/or facilitators, as the 26S proteasome is a dynamic entity with extensive protein interactions (55). Here, two subunits (CCT6A and CCT7) of CCT/TRiC complex were found to directly associate with the 26S proteasome through the 19S subunits (Rpn7 and Rpn10), suggesting a potential docking site for the TRiC complex that may be important for assisting proteasomal degradation of protein aggregates.
In addition to proteasomes, we have captured a total of 56 interactors associated with ubiquitin and four major groups of UPS enzymes (i.e., E1, E2, E3, and DUBs). Intriguingly, all of them can be modified by ubiquitin (http://www.BioGRID.org), confirming that their interactions are related to protein ubiquitination either as potential substrates and/or regulators. The identified interactions may play other functional roles in ubiquitination-related biological pathways (SI Appendix, Supplementary Results). To illustrate the influence of ubiquitin machinery over cellular functions, we have also generated an extended ubiquitin subnetwork (Fig. 6B) based on the connectivity of its directly interacting proteins. The innermost colored clusters represent E1s, E2s, E3s, DUBs, and ubiquitin, while their direct physical interactors are shown in a surrounding ring. We then grouped the next layer of proteins, which were found to be cross-linked to the direct interactors of ubiquitinating enzymes. In its entirety, this network consists of 759 nodes and 1,710 edges, indirectly connecting ubiquitination enzymes to a diverse array of cellular activities, including general housekeeping and protein translation. This is expected due to the role of proteasomal degradation in the regulation of almost all cellular processes required for cell maintenance and viability. One of the most highly enriched biological processes within this network is posttranslational protein folding, which is described by interactions of UPS enzymes to 10 chaperonin proteins and 3 chaperone-associated proteins, demonstrating extensive physical interactions among ubiquitin ligases and chaperones to underlie functional cross talk between protein degradation and folding.
Discussion
In this work, we have established an Alkyne-A-DSBSO–based in vivo XL-MS analytical platform with advancements in sample preparation and data acquisition for comprehensive PPI profiling in living cells. Specifically, we have developed a pH-dependent sequential protein extraction protocol of cross-linked cells to minimize abundant histone interference on the identification of nonhistone proteins, allowing expanded coverage of in vivo PPIs. In addition, we have developed a robust and traceable click chemistry–based cross-link enrichment method that significantly outperforms previous enrichment procedures (22). The integration of a water-soluble, UV-traceable click reagent (i.e., BPA) enables biotin conjugation at the peptide level with high sensitivity and efficiency. The unique detectability of BPA and BPA-conjugated peptides permits quantitative assessment of cross-link enrichment to achieve maximum recovery. Additional peptide separation by SEC not only facilitates the detection of interlinked peptides but also eliminates the interference of the conjugating reagent in subsequent MS analysis. Due to its specificity and sensitivity, the click chemistry–based enrichment strategy developed here can be easily adopted to enable effective isolation and analysis of alkyne- and azide-tagged peptides for any types of proteomic studies. In combination with the advantages offered by Alkyne-A-DSBSO XL and advanced instrumentation, 13,904 unique K–K linkages were identified from cross-linked HEK293 cells. This allowed us to construct an in vivo interaction map composed of 2,484 proteins, providing a detailed yet panoramic view of cellular PPIs in their native environments.
In comparison to existing reports (21, 22), the scope and depth of the XL proteome and XL PPIs obtained here are much broader, illustrating the sensitivity and effectiveness of the established method. Among the identified 5,401 interactions, 60% of them have not been previously reported. Although 40% of PPIs are known, our results provide direct physical contacts to confirm their interactions. Importantly, almost all of the in vivo XL PPIs describe intracompartmental interactions with a degree distribution similar to those of existing PPIs. In addition, more than 70% of them have high-confidence STRING scores (≥0.9). Moreover, the majority of cross-linked sites are found in close proximity of the predicted interfaces of PPIs. Therefore, the identified XL PPIs most likely represent true interactions in cells. Collectively, the in vivo XL-MS platform presented here signifies a technological advancement in probing protein networks in cells at the systems level, which will enable us to dissect global interactomes from various sample origins to understand PPI-dependent function and regulation of cells.
In comparison to in vitro analysis, in vivo XL-MS analysis resulted in histone-rich data with much more extensive PPIs involved in biological functions ranging from gene expression to chromosome organization. This feature can be advantageous for studying nuclear networks as they are known to be dynamic and difficult to probe with conventional approaches. In addition, 51% of the identified in vivo XL PPIs represent interprotein interactions, much higher than what have been observed in in vitro analyses. These observations are most likely attributed to differences in protein environments prior to XL. In comparison to molecular crowding during in vivo XL, native cell lysis required for in vitro XL not only dissipates subcellular compartments and changes protein concentration but also reorganizes dynamic, transient, and/or weak protein assemblies and PPI networks. Because of these variabilities in in vitro analysis, in vivo XL is preferred for mapping PPIs for studying their function and regulation, especially for delineating dynamic proteomes under different physiological conditions.
Apart from compartmental interactions, the in vivo XL PPIs describe interactions of 795 protein complexes identified with at least 50% composition, including 148 complexes with full coverages. This result uncovers the largest number of protein complexes with identities and connectivity from proteome-wide XL-MS experiments, which provides complementary information to MS profiling of protein complexes separated chromatographically and biochemically. In addition to interactions within each protein complex, interprotein-complex PPIs were mapped that can be used to infer their functional links associated with specific cellular pathways. This was exemplified by extensive communications found among protein complexes in histone, chaperonin, and UPS PPI networks, respectively, establishing a molecular basis for future studies on their biological significance in related pathways. While direct XL of affinity purified complexes would be desired to delineate fine structural details in each complex, proteome-wide analysis could provide a global view of interaction networks of protein complexes to enable the assessment of their functions and dynamics in the context of a cell. Therefore, the method presented here provides a unique analytical platform for future studies on complex assembly and dynamics as well as protein interactions such as TF-associated and enzyme-substrate interactions that are inaccessible to conventional approaches. Importantly, it offers great potential in a wide range of applications ranging from defining disease-relevant PPIs to screening proteome-reshaping pharmacological compounds for better therapeutics.
Similar to any proteomic and XL-MS studies, our analysis has shown a preference for abundant proteins. While our strategy enabled the XL proteome to uncover ∼34% of the HEK 293 MS proteome (49) with considerably broader scope and depth than previous studies (21, 22, 31), membrane proteins remain underrepresented. Clearly, new developments would still be needed to improve sensitivity and dynamic range to further expand the coverage of XL proteomes at the level equivalent to MS proteomes defined by shotgun proteomics. Inclusion of multiple enzymatic digestions, new fragmentation techniques, and software tools would further increase the number of cross-link identifications. In addition, implementation of a combinatory XL-MS strategy with multiple XL chemistries (30) would enlarge the breadth and depth of PPI mapping and unravel interaction interfaces unapproachable by lysine-reactive reagents. Nonetheless, our established in vivo XL-MS platform has made a significant step forward and built a solid foundation for future studies toward the complete representation of PPI networks in cells.
Methods
In Vivo XL-MS Analysis of 293Rpn11-HTBH Cells.
The general procedures for cell culture and in vivo XL were similar as described (22). Briefly, for each in vivo XL-MS experiment, about 300 million cells stably expressing Rpn11-HTBH (i.e., 293Rpn11-HTBH) were grown to 80 to 90% confluency, washed, and cross-linked with 5 mM Alkyne-A-DSBSO in phosphate buffered saline (PBS) buffer (pH 7.4) for 1 h with rotation at 37 °C. XL reaction was quenched with addition of 50 mM glycine and incubated at 37 °C for 5 min. Then, cross-linked cells were spun down and washed with PBS buffer. For sequential two-step protein extraction, cells were first lysed with denaturing buffer no. 1 (8 M urea, 50 mM Tris ⋅ HCl pH 7.0). Supernatant was clarified by centrifugation at 21,000 g and collected. The remaining cell pellet was resuspended in denaturing buffer no. 2 (8 M urea, 50 mM Tris ⋅ HCl pH 8.0, 150 mM NaCl) and the extracted proteins collected. For single-step protein extraction, cross-linked cells were directly lysed using buffer no. 2. Cross-linked proteins solubilized by each lysis buffer were subsequently digested, enriched, and analyzed by LC-MSn and identified through database searching as previously described (SI Appendix, Supplementary Methods) (22, 35).
Click Chemistry–Based Cross-Link Enrichment.
To optimize click chemistry–based enrichment procedure, HEK 293 cell lysates were cross-linked in vitro by Alkyne-A-DSBSO similarly as described (31). Specifically, 293Rpn11-HTBH cells were grown to ∼90% confluency and lysed with native buffer (20 mM Hepes, 150 mM NaCl, 0.5 mM DTT, 1.5 mM MgCl2, pH 7.8) via sonication. Protein concentration of the clarified cell lysate was measured using a Nanodrop (Thermo Fisher) and adjusted to 1 mg/mL prior to XL. A total of 2 mg cell lysates generated from ∼15 million cells were cross-linked with 2 mM Alkyne-A-DSBSO (2% volume/volume DMSO) for 1 h at room temperature with rotation. The reaction was then quenched using 50 mM Tris ⋅ HCl pH 8.0 for 5 min. Samples were centrifuged over 30,000 nominal molecular weight limit (NMWL) Microcon centrifugal filter to remove excess XL reagent prior to protein digestion.
Click chemistry was performed by adding BPA (biotin picolyl azide), BTTAA, CuSO4, and sodium ascorbate to dried peptide digests to a final concentration of 300 µM, 500 µM, and 250 µM, and 2.5 mM, respectively (45). The resulting mixture was rotated in the dark at room temperature for 2 h and then incubated with 200 µL streptavidin beads in binding buffer (25 mM sodium phosphate, 150 mM NaCl, pH 7.5) for 2 h at room temperature. Streptavidin-bound peptides were washed extensively with binding buffer and eluted through acid cleavage using 10% formic acid overnight at 37 °C, collected, and dried. The enriched cross-linked peptides were either directly subjected to LC-MS analysis or separated by SEC as described (47) (SI Appendix, Supplementary Methods). Benchmarking was carried out to evaluate the specificity, sensitivity, and efficiency of the click chemistry–based cross-link enrichment (SI Appendix, Supplementary Methods).
PPI Network Mapping and Analysis.
XL-PPI networks were generated from the pair-wise interactions determined by in vivo XL. These interactions were visualized using Cytoscape version 3.6.0 (http://www.cytoscape.org). The latest known Homo sapien PPI database on BioGRID (https://thebiogrid.org/), STRING (https://string-db.org/), and BioPlex (https://bioplex.hms.harvard.edu/) were used for data comparison. CORUM (http://mips.helmholtz-muenchen.de/corum) was used to annotate and determine protein complexes. Functional enrichment was performed using the Gene Ontology Consortium (http://www.geneontology.org). Only high-confidence annotations (FDR < 0.01) for GO cellular compartment and biological process were reported. DisGeNET (https://www.disgenet.org/) was used to categorize disease associated proteins.
Cross-Link Mapping to High-Resolution Structures of Protein Complexes.
The 3.5-Å resolution structure of the human 26S proteasome (PDB: 5GJR) and the 4.2-Å resolution structure of the human 40S ribosome (PDB: 5OA3) were obtained from the RCSB PDB (http://www.rcsb.org). Cross-linked residues were mapped onto the structures and visualized using PyMOL. To automatically map remaining cross-links to published protein complexes, an in-house script was utilized to extract a list of protein complexes that were identified with cross-links from CORUM. We queried the UniProt knowledgebase to identify all published human structures annotated with a RCSB PDB entry determined with resolution better than 10 Å, using the BioPython package. We further mapped the identified cross-links onto the 10 best determined structures with highest resolutions. The distances between cross-linked residues were determined for validation.
Supplementary Material
Acknowledgments
We thank Prof. A. L. Burlingame and Robert Chalkley for support of the development version of Protein Prospector. This work was supported by NIH Grants R01GM074830 and R01GM130144 to L.H.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023360118/-/DCSupplemental.
Data Availability
Mass spec data have been deposited in PRIDE repository (PXD012788). All other data needed to evaluate the conclusions in the paper are present in the paper and/or supporting information.
References
- 1.Ryan D. P., Matthews J. M., Protein-protein interactions in human disease. Curr. Opin. Struct. Biol. 15, 441–446 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Budayeva H. G., Kirkpatrick D. S., Monitoring protein communities and their responses to therapeutics. Nat. Rev. Drug Discov. 19, 414–426 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Gavin A. C., et al., Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Huttlin E. L., et al., The BioPlex network: A systematic exploration of the human interactome. Cell 162, 425–440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hein M. Y., et al., A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Huttlin E. L., et al., Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasilescu J., Guo X., Kast J., Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics 4, 3845–3854 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Guerrero C., Milenkovic T., Przulj N., Kaiser P., Huang L., Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc. Natl. Acad. Sci. U.S.A. 105, 13333–13338 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu C., et al., Characterization of dynamic UbR-proteasome subcomplexes by in vivo cross-linking (X) assisted bimolecular tandem affinity purification (XBAP) and label-free quantitation. Mol. Cellular Proteomics 15, 2279–2292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snider J., et al., Fundamentals of protein interaction network mapping. Mol. Syst. Biol. 11, 848 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards A. L., Eckhardt M., Krogan N. J., Mass spectrometry-based protein-protein interaction networks for the study of human diseases. Mol. Syst. Biol. 17, e8792 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee H. W., et al., Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuzow N., et al., Mapping the mammalian ribosome quality control complex interactome using proximity labeling approaches. Mol. Biol. Cell 29, 1258–1269 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heusel M., et al., Complex-centric proteome profiling by SEC-SWATH-MS. Mol. Syst. Biol. 15, e8438 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fossati A., et al., PCprophet: A framework for protein complex prediction and differential analysis using proteomic data. Nat. Methods 18, 520–527 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Herzog F., et al., Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Shi Y., et al., A strategy for dissecting the architectures of native macromolecular assemblies. Nat. Methods 12, 1135–1138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S. J., et al., Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weisbrod C. R., et al., In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J. Proteome Res. 12, 1569–1579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chavez J. D., Schweppe D. K., Eng J. K., Bruce J. E., In vivo conformational dynamics of Hsp90 and its interactors. Cell Chem. Biol. 23, 716–726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaake R. M., et al., A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Mol. Cell. Proteomics 13, 3533–3543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang B., et al., Spontaneous and specific chemical cross-linking in live cells to capture and identify protein interactions. Nat. Commun. 8, 2240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu C., Huang L., Cross-linking mass spectrometry: An emerging technology for interactomics and structural biology. Anal. Chem. 90, 144–165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leitner A., Faini M., Stengel F., Aebersold R., Crosslinking and mass spectrometry: An integrated technology to understand the structure and function of molecular machines. Trends Biochem. Sci. 41, 20–32 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Sinz A., Cross-linking/mass spectrometry for studying protein structures and protein-protein interactions: Where are we now and where should we go from here? Angew. Chem. Int. Ed. Engl. 57, 6390–6396 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Chavez J. D., Bruce J. E., Chemical cross-linking with mass spectrometry: A tool for systems structural biology. Curr. Opin. Chem. Biol. 48, 8–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steigenberger B., Albanese P., Heck A. J. R., Scheltema R. A., To cleave or not to cleave in XL-MS? J. Am. Soc. Mass Spectrom. 31, 196–206 (2020). [DOI] [PubMed] [Google Scholar]
- 29.O’Reilly F. J., Rappsilber J., Cross-linking mass spectrometry: Methods and applications in structural, molecular and systems biology. Nat. Struct. Mol. Biol. 25, 1000–1008 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Gutierrez C., et al., Structural dynamics of the human COP9 signalosome revealed by cross-linking mass spectrometry and integrative modeling. Proc. Natl. Acad. Sci. U.S.A. 117, 4088–4098 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F., Lössl P., Scheltema R., Viner R., Heck A. J. R., Optimized fragmentation schemes and data analysis strategies for proteome-wide cross-link identification. Nat. Commun. 8, 15473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fasci D., van Ingen H., Scheltema R. A., Heck A. J. R., Histone interaction landscapes visualized by crosslinking mass spectrometry in intact cell nuclei. Mol. Cell. Proteomics 17, 2018–2033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Götze M., Iacobucci C., Ihling C. H., Sinz A., A simple cross-linking/mass spectrometry workflow for studying system-wide protein interactions. Anal. Chem. 91, 10236–10244 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Yugandhar K., et al., MaXLinker: Proteome-wide cross-link identifications with high specificity and sensitivity. Mol. Cell. Proteomics 19, 554–568 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kao A., et al., Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cellular Proteomics 10, M110.002212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu C., Kandur W., Kao A., Rychnovsky S., Huang L., Developing new isotope-coded mass spectrometry-cleavable cross-linkers for elucidating protein structures. Anal. Chem. 86, 2099–2106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutierrez C. B., et al., Developing an acidic residue reactive and sulfoxide-containing MS-cleavable homobifunctional cross-linker for probing protein-protein interactions. Anal. Chem. 88, 8315–8322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gutierrez C. B., et al., Development of a novel sulfoxide-containing MS-cleavable homobifunctional cysteine-reactive cross-linker for studying protein-protein interactions. Anal. Chem. 90, 7600–7607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., et al., Molecular details underlying dynamic structures and regulation of the human 26S proteasome. Mol. Cell. Proteomics 16, 840–854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez-Lozano M. A., et al., Stitching the synapse: Cross-linking mass spectrometry into resolving synaptic protein interactions. Sci. Adv. 6, eaax5783 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burke A. M., et al., Synthesis of two new enrichable and MS-cleavable cross-linkers to define protein-protein interactions by mass spectrometry. Org. Biomol. Chem. 13, 5030–5037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shechter D., Dormann H. L., Allis C. D., Hake S. B., Extraction, purification and analysis of histones. Nat. Protoc. 2, 1445–1457 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Yang J., et al., Global, in situ, site-specific analysis of protein S-sulfenylation. Nat. Protoc. 10, 1022–1037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weerapana E., Speers A. E., Cravatt B. F., Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)—A general method for mapping sites of probe modification in proteomes. Nat. Protoc. 2, 1414–1425 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Uttamapinant C., et al., Fast, cell-compatible click chemistry with copper-chelating azides for biomolecular labeling. Angew. Chem. Int. Ed. Engl. 51, 5852–5856 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schilling B., Row R. H., Gibson B. W., Guo X., Young M. M., MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 14, 834–850 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Leitner A., et al., Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cellular Proteomics 11, M111.014126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steigenberger B., Pieters R. J., Heck A. J. R., Scheltema R. A., Pho X., PhoX: An IMAC-enrichable cross-linking reagent. ACS Cent. Sci. 5, 1514–1522 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilhelm M., et al., Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Ruepp A., et al., CORUM: The comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 36, D646–D650 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rzhetsky A., Gomez S. M., Birth of scale-free molecular networks and the number of distinct DNA and protein domains per genome. Bioinformatics 17, 988–996 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Meyer M. J., et al., Interactome INSIDER: A structural interactome browser for genomic studies. Nat. Methods 15, 107–114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szklarczyk D., et al., STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47 (D1), D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schweppe D. K., et al., Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 114, 1732–1737 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finley D., Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spec data have been deposited in PRIDE repository (PXD012788). All other data needed to evaluate the conclusions in the paper are present in the paper and/or supporting information.