Significance
Modeling cancer formation requires introduction of relevant oncogenic perturbations into normal cells in a tissue/cell-type–specific manner. Genetically engineered mouse models are powerful but require significant time and cost to generate and maintain. The ability to edit primary epithelial cells ex vivo followed by orthotopic transplantation provides an alternative strategy for cancer modeling but requires efficient gene editing, typically in a multiplex fashion. Here we successfully engineer multigenic perturbations or chromosomal rearrangements in primary prostate organoids through single-step Cas9–sgRNA ribonucleoprotein electroporation. This approach can also address cell-of-origin questions by directly editing and transplanting freshly isolated subpopulations without the intermediate step of organoid culture, providing a rapid complement to traditional lineage tracing approaches.
Keywords: CRISPR, organoids, editing, cancer modeling
Abstract
The increasing complexity of different cell types revealed by single-cell analysis of tissues presents challenges in efficiently elucidating their functions. Here we show, using prostate as a model tissue, that primary organoids and freshly isolated epithelial cells can be CRISPR edited ex vivo using Cas9–sgRNA (guide RNA) ribotnucleoprotein complex technology, then orthotopically transferred in vivo into immunocompetent or immunodeficient mice to generate cancer models with phenotypes resembling those seen in traditional genetically engineered mouse models. Large intrachromosomal (∼2 Mb) or multigenic deletions can be engineered efficiently without the need for selection, including in isolated subpopulations to address cell-of-origin questions.
Models to study the earliest stages in cancer progression must, by definition, start with normal cells to assess the consequences of a suspected oncogenic perturbation. Inbred mouse strains and genetically engineered mouse models (GEMMs) have, for decades, served as gold standards for such studies but require significant time (years) to generate, breed, and age mice. CRISPR technology has greatly accelerated the pace of generating GEMMs through delivery of sgRNAs (guide RNAs) and/or Cas9 to tissues such as lung and liver (1–3). The ability to grow primary tissues ex vivo as organoids and introduce precise genetic changes into these cultures provides an alternative platform to model genomic alterations with speed and efficiency, as reported for intestine and prostate (4–7). Here we demonstrate highly efficient (50 to 90%) editing of primary prostate epithelial organoid cultures, including multigenic or intrachromosomal (>2 Mb) deletions, through transient electroporation of Cas9–sgRNA ribonucleoprotein (cRNP) complexes. We also show cRNP-based CRISPR editing can be performed on freshly isolated prostate epithelial cells and then transplanted orthotopically into the prostates of recipient mice in a single day, enabling extremely rapid generation of in vivo cancer models in immunodeficient as well as immunocompetent settings. Finally, we show that this approach can be used to address cancer cell-of-origin questions by multigenic editing selectively in luminal versus basal epithelial cells.
Results and Discussion
As a first step to optimize CRISPR editing in primary organoids, we compared the efficiency of a conventional virus-based approach, in which vectors expressing Cas9 and sgRNAs are stably introduced into cells (4), to the cRNP approach, in which Cas9 protein and sgRNAs are transiently electroporated into cells (8–10). Prostate organoids derived from a GEM model of ERG-positive human cancers (R26LSL-ERG/LSL-ERG;Ptenfl/fl;Pb-Cre, EPC) (11) were used as a model in which the ERG transgene is activated via a Cre-mediated removal of the preceding Lox-STOP-Lox (LSL) cassette. There is no endogenous Erg expression in these cells. We quantified editing efficiency by flow cytometric assessment of loss of ERG expression, an assay sensitive enough to distinguish between biallelic (R26+/+), heterozygous (R26LSL-ERG/+), or absence of ERG expression (R26KO, created by CRISPR knockout of the R26+/+ allele) (Fig. 1A).
Fig. 1.
Efficient one-step organoid population editing by cRNP. (A) (Top) Schematic describing the R26LSL-ERG allele. LSL, LoxP-STOP-LoxP cassette. (Bottom) ERG detection in organoid cells by intracellular flow cytometry. (B) Cas9 protein detection in bulk EPC organoid cells after Cas9 lentiviral transduction. (C) ERG loss in mCherry-labeled sgRNA-infected R26LSL-ERG/LSL-ERG;Ptenfl/fl;Pb-Cre (EPC) organoids as measured by intracellular flow cytometry. EV, empty vector control. (D) sgERG–cRNP dosage titration in EPC organoids with corresponding editing outcomes as measured by ERG intracellular flow cytometry (n = 1). Unt, untreated. (E) Western blot comparing efficiency of editing the Ptenfl/fl allele by Adeno-Cre and sgPten–cRNP in R26LSL-ERG/LSL-ERG;Ptenfl/fl organoids. (F) Pten loss in wild-type (WT) organoids by cRNP as measured by intracellular flow cytometry. (G) Representative images showing a hyperplastic morphology displayed by sgPten–cRNP-treated organoids. (Scale bar, 100 µm.)
To evaluate the viral approach, we stably introduced a Cas9-expressing vector into organoids, confirmed protein expression in the bulk population (Fig. 1B), then introduced a second vector containing a sgRNA targeting ERG and a cis-linked mCherry cassette. Flow cytometric analysis confirmed loss of ERG expression in ∼34% of mCherry-positive cells (Fig. 1C). Using the cRNP method, we observed a dose-dependent increase in editing efficiency after electroporation of sgERG–cRNP, with up to 90% of cells showing loss of ERG expression (Fig. 1D). Although it may be possible to achieve greater deletion efficiency using the viral approach by increasing the level of Cas9 expression, we were struck by the remarkable efficiency and simplicity of the cRNP method, as well as the speed, since fully edited cells were obtained within 3 d following electroporation. In addition to its simplicity, the cRNP approach offers the advantage of transient Cas9 expression and potentially reduced off-target editing as a consequence (8, 12, 13).
Given the efficiency of cRNP editing in the ERG model system, we extended this approach to tumor suppressor genes by first targeting Pten in primary mouse prostate organoids. Pten knockout (KO) populations were readily generated following sgPten–cRNP electroporation with >90% efficiency as measured by flow cytometry and Western blot, comparable to that observed after Pten deletion in Ptenfl/fl organoids infected with Cre-expressing adenovirus (Fig. 1 E and F). In addition to loss of Pten protein expression, the edited organoids acquired known morphologic features of Pten loss such as a hyperplastic morphology with a filled-in lumen and loss of basal/luminal epithelial polarity (14) (Fig. 1G).
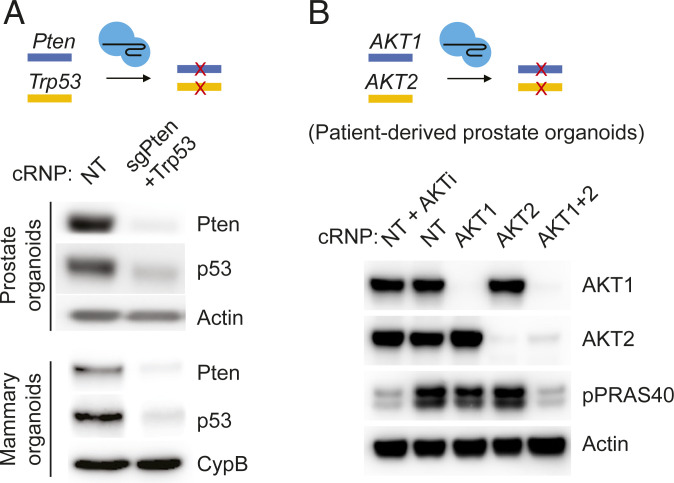
Having documented rapid, highly efficient single-gene editing in organoids (ERG, Pten), we considered the possibility of editing two genes simultaneously. We selected Pten and Trp53, which are commonly deleted in human metastatic prostate cancer and cooperatively induce prostate cancer in GEM models (15–17). Indeed, codelivery of two cRNP complexes, sgPten–cRNP and sgTrp53–cRNP, resulted in substantial ablation of both targets as measured by Western blot within 7 d following electroporation (Fig. 2A). Efficient multigenic editing is not unique to prostate, as similar results were obtained by targeting Pten and Trp53 in mouse mammary organoids (Fig. 2A). We also observed robust multigenic cRNP editing in human prostate organoids, as shown by deletion of AKT1 and AKT2 in an organoid line derived from a patient with metastatic prostate cancer (Fig. 2B). The near complete loss of phosphorylation of the AKT substrate PRAS40, as seen following treatment with a pan-AKT inhibitor, was observed only in dual sgAKT1–cRNP/sgAKT2–cRNP electroporated organoids. In addition to confirming highly efficient multigene deletion in a human organoid model, this result demonstrates the importance of dual rather than isoform-specific AKT inhibition to block downstream signaling.
Fig. 2.
Multiplexed cRNP editing in organoids across different tissues and species. (A) Western blot showing codisruption of Pten and p53 proteins in WT (C57BL/6J albino) mouse prostate organoids and mouse mammary organoids by cRNP. (B) Western blot showing codisruption of AKT1 and AKT2 proteins in a patient-derived prostate organoid line by cRNP. AKTi, AKT inhibitor ipatasertib treatment (500 nM for 4 h). Unt, untreated; NT, nontargeting sgRNA control.
Recurrent multigenic chromosomal deletions are among the most common alterations in cancer genomes, particularly in epithelial tumors such as prostate cancer (18, 19). The high efficiency of cRNP-based gene disruption we observed in primary mouse prostate organoids led us to test whether we could efficiently engineer larger deletions. As an initial test we returned to the R26-ERG organoid model, which contains a 2.7-kb LSL cassette between the Rosa26 promoter and the ERG cDNA cassette (R26LSL-ERG/LSL-ERG), with the goal of deleting this region using cRNP (instead of Cre). Over 50% of cells electroporated with cRNPs targeting both sides of the STOP cassette displayed robust ERG expression within 3 d following electroporation, indicative of effective deletion (Fig. 3A). Importantly, ERG expression was not observed in cells electroporated with each RNP alone, ruling out large deletions created by single sgRNAs (20).
Fig. 3.
cRNP generates large chromosomal deletions in mouse prostate organoids. (A) (Left) Schematic showing a paired-cRNP strategy to induce ERG expression by disrupting the preceding LSL cassette. (Right) ERG expression from the cRNP-treated R26LSL-ERG/LSL-ERG organoid population was measured by intracellular flow cytometry. (B) (Top) Schematic showing Tmprss2-Erg fusion by paired cRNPs with different sgRNA combinations in WT organoids. (Bottom) Genomic fusion detection from the electroporated organoid populations by fusion-specific genomic PCRs. (C) ERG expression of the paired cRNP-treated WT organoid population from B was measured by intracellular flow cytometry. (D) Fusion-specific genomic PCRs of organoid clones treated with indicated cRNP pairs. DNA electrophoresis results of representative clones are shown. (E) Paired CRISPR-mediated deletion of the Foxp1-Shq1 region (schematic shown on the Left). WT organoids were either electroporated with cRNP or infected with lentiGuide (with preengineered Cas9 expression). Fusion-specific genomic PCR of resulting organoid clones and quantification of deletion-positive clones are shown on the Right.
Having established that multikilobase deletions can be generated quickly and efficiently in organoids using sgRNA–cRNPs flanking the region of interest, we next asked whether we could model larger (megabase scale) deletions found in human prostate cancer. Complex translocations or deletions have been previously generated with CRISPR tools but suffer from limited efficiency, require template construct engineering, or rely on the strong selective advantage from the edited cells (e.g., tumor formation) to be successful (3, 21, 22). To address the efficiency question directly, we designed cRNPs to generate the 2.1-Mb intrachromosomal deletion that results in aberrant expression of the ETS family transcription factor ERG in human prostate cancer (23). Using a single sgRNA targeting TMPRSS2 intron 1 and two different sgRNAs targeting Erg intron 2, we detected the expected fusion products by PCR of genomic DNA from the bulk population within 3 d of electroporation (Fig. 3B and SI Appendix, Fig. S1A). We subsequently documented an efficiency of ∼20% by flow cytometry analysis of ERG protein expression (Fig. 3C) and PCR screening of single-cell clones (Fig. 3D). These results replicate earlier work using CRISPR-directed homologous recombination to generate the Tmprss2-Erg translocation in mouse prostate organoids (24) but eliminate the time and effort needed to generate a genomic targeting construct containing the relevant 5′ and 3′ homology arms of Tmprss2 and Erg.
As a second example, we chose to delete the multigenic 2.0-Mb tumor suppressor locus at human 3p13-14, found in ∼15% of localized prostate cancers (18, 25). This region is of particular interest because it is almost exclusively associated with prostate cancer and lacks a canonical tumor suppressor gene within the locus. Our group previously documented that this region is, indeed, tumor suppressive in a GEM model, but this approach required a generation of a complex array of LoxP sites and extensive breeding (26). To simplify this process and simultaneously create a platform that enables rapid functional interrogation of specific genes within the locus, we tested whether a syntenic 3p13-14 deletion could be generated directly in mouse organoids, using the conventional lentiviral approach with stable Cas9 and sgRNA expression as well as cRNP, followed by screening of individual subclones. As with ERG deletion experiments discussed earlier, cRNP-based CRISPR editing was fast and efficient (20 of 42 positive clones, 48%) compared to the lentiviral Cas9/sgRNA expression (3 of 37 positive clones, 5%); albeit higher lentiviral efficiencies might be achievable by isolation of high Cas9-expressing subclones (Fig. 3E and SI Appendix, Fig. S1B). Importantly, all alterations above were created without any exogenous selection. Thus, cRNP technology is a versatile and convenient tool for multigenic and long-range genome editing in organoids.
Organoids can also be used for in vivo cancer modeling through orthotopic injection studies, and the transient presence of Cas9 protein using the cRNP method may have advantages since it eliminates any potential risk of Cas9 immunogenicity that can occur using viral methods (27–29). To explore this, we injected the sgPten+Trp53–cRNP-deleted organoids orthotopically into the prostate dorsal lobes of syngeneic recipient mice (Fig. 4A). Remarkably, 100% (six of six) B6 recipient mice developed locally invasive adenocarcinomas with median survival of ∼17 wk, comparable to that observed in Pb-Cre;Ptenfl/fl;Trp53fl/fl GEM models (16). In contrast, all mice injected orthotopically with sgPten–cRNP and/or control cRNP-edited organoids survived beyond 30 wk with no evidence of disease (Fig. 4 B and C).
Fig. 4.
Organoid editing by cRNP enables cancer modeling in immunocompetent hosts. (A and B) Orthotopic transplantation of sgPten+Trp53–cRNP organoids (from Fig. 2A) into syngeneic immunocompetent mice (schematic shown in A). (B) Representative images of the urogenital systems at harvest (Left) and histological analysis (Right) of a prostate tumor harvested at 15 wk posttransplantation. Dashed circles highlight the prostate tumor regions. (Scale bar, 100 µm.) (C) Survival analysis of mice with orthotopic transplants of indicated prostate organoids. ****P < 0.0001 (log-rank Mantel–Cox test). Ctrl, control sgRNAs targeting intergenic regions.
The high efficiency of cRNP-based editing in organoids led us to ask whether this technology could be applied directly to prostate epithelial cells freshly isolated from a donor mouse and then reintroduced as an orthograft into a recipient mouse, thereby avoiding the need for organoid culture. In addition to speed, this approach could potentially be used to address cancer cell-of-origin questions by comparing the outcome of edited cells in different subpopulations isolated by flow cytometry. Toward that end, we sorted freshly isolated mouse prostate epithelial cells into basal (Cd49f-high) and luminal (Cd24-high) subpopulations (30), electroporated each with sgPten–cRNP and sgTrp53–cRNP as described earlier for organoids, and separately injected each subpopulation orthotopically into the dorsal prostate lobes of recipient mice (Fig. 5A). Substantially reduced p53 and Pten protein expression was confirmed in these postedited primary cells (Fig. 5B). At the 17-wk endpoint (selected based on the organoid orthograft data described earlier; Fig. 4C), two of six mice engrafted with cRNP-edited luminal cells developed large prostate tumors (one at 14 wk due to early death), whereas no tumors were observed in four mice engrafted with basal cells (Fig. 5C). Immunohistochemical analysis confirmed the presence of glandular orthografted cells in the dorsal lobes of all mice, based on phospho-Akt staining (indicative of Pten loss) and absence of p53 staining (Fig. 5D), as well as PCR analysis of the sgRNA-targeted region from bulk prostate tissue (SI Appendix, Fig. S2). Although additional studies are required to define the efficiency, these results document the feasibility of same-day cRNP editing of freshly isolated prostate epithelial cells for cancer modeling. The fact that tumors were observed only in mice engrafted with cRNP-edited luminal cells is consistent with prior evidence from GEM models implicating luminal cells as the cell of origin for prostate cancers initiated by Pten loss (31, 32).
Fig. 5.
Rapid cancer modeling with cRNP-edited freshly isolated prostate epithelial cells. (A) Workflow of prostate cancer modeling by cRNP editing of freshly isolated prostate epithelial cells followed by orthotopic transplantation. (B) Western blot showing codisruption of Pten and p53 proteins in freshly isolated prostate epithelial cells. Cells from indicated lineage origin were analyzed 7 d post-cRNP treatment. NT, nontargeting sgRNA control. PP, sgPten+Trp53. (C) Images showing prostate tumor development from mice engrafted with cRNP-edited luminal or basal cells. The dorsal prostate lobes of NSG mice were transplanted with sgPten+Trp53–cRNP-treated luminal or basal cells and harvested at indicated weeks (w) posttransplantation. The urogenital systems or dorsal lobes at harvest are displayed, with an H&E staining showing tumor histology. Dashed circles highlight the prostate tumor regions. (Scale bar, 200 µm.) (D) Histological analysis confirming the engraftment of edited cells in host prostates. The 17-wk endpoint prostate tissues carrying the sgPten+Trp53–cRNP-treated orthografts from C were stained as indicated, with high power magnifications shown in Inset. The lineage origins of the grafted cells are as indicated. A normal gland from the host dorsal lobe is shown in Top. Arrows highlight a gland with a pAkt-positive and p53-negative staining, indicative of Pten Trp53 double knockout. Asterisk, a pAkt/p53 double positive gland, indicative of Pten single knockout. (Scale bar, 200 µm.)
In summary, we show that cRNP-based CRISPR editing offers several advantages for cancer modeling: high editing efficiency, ability to generate single/multiple perturbations and large chromosomal deletions in a single step, and ability to edit freshly isolated epithelial cells, in bulk or after sorting into luminal or basal subpopulations, followed by same day orthotopic transfer for tumorigenicity studies in vivo. In addition, by eliminating the risk of an immune response against Cas9, the cRNP method is a convenient strategy to study tumor–microenvironment interactions in an immunocompetent setting without having to resort to models with germline Cas9 expression. The ability to edit freshly isolated prostate epithelial cells after separation into luminal and basal subpopulations is particularly exciting because it provides a platform to address cancer cell-of-origin questions without having to generate new mouse strains expressing tissue- or cell-type–specific recombinases. This application is particularly compelling in light of the increased complexity of cell types in tissues revealed by single-cell analysis, evidenced recently by the identification of multiple luminal subtypes in the normal prostate (33–35).
Although canonical lineage tracing experiments remain important to shed light on cell-of-origin questions in the context of a native microenvironment, our approach allows for screening through various cell types for more rapid assessment of their tumorigenic potentials. It should be noted that this method provides insight in cell of origin only in the context of mouse tissues. Validation of human relevance is always warranted. It will also be important to extend the studies reported here in prostate and breast cells to other tissues, and our laboratory has ongoing studies in primary renal and intestinal organoid cultures.
In conclusion, the speed, convenience, and scalability of the cRNP-editing approach described here provides an opportunity to generate an expansive array of cancer models covering a broader number of complex genomic alterations linked with human cancers. These models can serve as a platform to address ongoing questions in cancer biology and to evaluate novel precision oncology medicines and immunotherapies in a genetically defined setting.
Materials and Methods
Constructs.
To create a lentiGuide-mCherry backbone, the sgRNA scaffold of LRG (Addgene plasmid #65656) (36) was first replaced with an enhanced scaffold (37) between BsmBI/EcoRI sites. The GFP sequence was then replaced with mCherry using BamHI and MluI. sgRNA sequences were cloned into either lentiGuide-mCherry (sgCtrl and sgERG) or LRG (F and S) using BsmBI.
GEM Models and Organoid Lines.
Mouse prostate organoid lines EPC and wild type (WT) (R26LSL-ERG/LSL-ERG;Ptenfl/fl or Ptenfl/fl) were derived from corresponding GEM models (11). EPC and WT (R26LSL-ERG/LSL-ERG;Ptenfl/fl) have been previously described (38). The WT line was derived from a C57BL/6 albino mouse at the age of 3 mo. For Cas9 expression, organoid cells were transduced with lentiCas9-blast (Addgene plasmid #52962) (39) that was 30× concentrated with Lenti-X Concentrator (Takara Bio 631231) and bulk selected with 10 µg/mL blasticidin for at least one additional week after all control cells died. Selection was not maintained afterward. To generate R26LSL-ERG/+;Pten−/− organoids, bulk R26LSL-ERG/LSL-ERG;Ptenfl/fl populations were transduced with adeno-Cre (Vector Biolabs), sorted for GFP expression, and cultured as a pool of clones with the expected genotype. To generate the ERG-KO control used in Fig. 1C, EPC cells were electroporated with lentiGuide-mCherry-sgERG1/2 plasmids, followed by a mCherry+GFP− sorting and a second GFP-negative sorting (ERG expression is linked with ires-GFP). In all cases, ERG and PTEN status was confirmed at the protein level by intracellular flow cytometry and/or Western blot. For ERG genotyping PCR, oligos 1 and 2 were used to detect the LSL allele (164 bp). Oligos 3 and 4 were used to detect the + allele (379 bp). Pten genotyping primers have been previously described (40).
Oligo 1: CATTCTGCACGCTTCAAAAG
Oligo 2: GCCCAGTCATAGCCGAATAG
Oligo 3: GCGGTTGAGGACAAACTCTTC
Oligo 4: CTCTGTCTTAGCCAGGTGTGGCG
Mouse Organoid Derivation and Culture.
Mouse prostates were isolated as described previously (14). Briefly, prostates were harvested and subsequently digested with collagenase type II (Gibco) for 2 h at 37 °C, followed by TrypLE (Gibco) digestion at 37 °C until a single-cell suspension was obtained. Digestions were supplemented with Y-27632 (10 μM) to inhibit anoikis. Cells were filtered before seeding for organoid culture. Organoid culture was maintained under standard conditions as described previously (14). Organoids were continuously grown in three-dimensional (3D) culture, with the exception of single-clone isolation and EPC culture, when the organoids were maintained as adherent culture. For morphology experiments, organoids were cultured in the absence of EGF before image acquisition under a brightfield microscope (ECHO). Mammary tumors from c-Myc–overexpressing mice (41) were dissected and cultured in the form of organoids as previously described (42).
Patient-Derived Prostate Organoids.
Patient-derived organoids (MSK-PCa12) were generated and cultured as previously described (43). The tissues were deidentified prior to use. The 2 × 106 cells of MSK-Pca12 were engrafted subcutaneously into 6- to 8-wk-old CB17-SCID (severe combined immunodeficiency disease) male mice (Taconic). Mice were castrated when the tumor reached ∼200 mm3 and treated with ipatasertib (50 mg/kg) for 4 wk. Resistant tumors were collagenase/trypsin-digested into single cells and cultured in standard human organoid culture with ipatasertib (500 nM). AKT inhibitor ipatasertib was provided by Genentech under an MTA (material transfer agreement) and was dissolved in 0.5% methylcellulose and 0.2% Tween-80 for in vivo study. Acquisition of human tissue for prostate cancer organoids was performed under MSKCC IRB (Institutional Review Board of Memorial Sloan Kettering Cancer Center)-approved protocols 06-107 and 12-001.
Preparation of Freshly Isolated Prostate Epithelial Cells for Gene Editing.
Prostates from six C57BL/6 mice (The Jackson Laboratory) were pooled together, dissociated into single cells, and stained for 1 h on ice with CD49f-PE (1:200; BD, 555736) and CD24-Alexa Fluor 647 (1:200; BioLegend, 101818). Sorted basal and luminal cells were centrifuged and directly used for electroporation and transplantation into four or six recipient NSG (NOD scid gamma) mice, respectively.
Electroporation.
Nucleofection was performed using a basic kit for primary mammalian epithelial cells (Lonza, VPI-1005) or kit R (Lonza, VVCA-1001) for prostate organoids or a home-made nucleofection solution (150 mM KH2PO4, 24 mM NaHCO3, 3.7 mM glucose, 7 mM ATP (adenosine triphosphate)-disodium salt, 12 mM MgCl2, pH 7.4) for mouse mammary organoids. The 0.5∼1 × 106 dissociated organoid cells or 350 k of freshly isolated basal/luminal cells were resuspended with nucleofection buffer, cRNP complexes, and electroporation enhancer (IDT 1075915, 1:1 molar ratio to cRNP) in a total volume of 100 µL. The cell suspension was transferred to a nucleofection cuvette and nucleofected using Lonza Amaxa Nucleofector II (program T-030 for prostate cells, X-005 for mammary cells). Cells were centrifuged and either seeded for culture (organoids) or transplanted into mouse prostates (primary lineage cells).
Genome Editing and Editing Outcome Verification.
For cRNP editing, Cas9 was first mixed with sgRNA (IDT) for 20 min to form the cRNP complex before nucleofection. A total of 1.2 µM cRNP was used per individual sgRNA. Editing outcome was analyzed within a week. For virus-based editing, Cas9-expressing organoids were transduced with sgRNA-expressing lentiviruses. Fusion-specific PCR was performed to detect Tmprss2-Erg fusion or Foxp1-Shq1 deletion: oligos 5 and 6 for CRISPRs T and E2, oligos 5 and 7 for CRISPRs T and E3, and oligos 8 and 9 for CRISPRs F and S. PCR products were subcloned using a TA CloningTM kit (Thermo Fisher K200001) and confirmed by Sanger sequencing. To detect indels from sgTrp53 and sgPten editing from mouse tissue, genomic DNA was recovered from formalin-fixed paraffin-embedded slides and PCRs were performed using oligos 10 and 11 (for sgp53) or oligos 12 and 13 (for sgPten). Indel frequency estimation was performed by Sanger peak decomposition using TIDE (44).
Oligo 5: GTGGGAGTGCACAGAGATTGAG
Oligo 6: CTCAGGCTAATCCAGCCGTTC
Oligo 7: ATTGTGCATCCACCTCGAAGAG
Oligo 8: TAACGGGCTTAACAACCCCT
Oligo 9: CCAGGCCCAAACCATTGAAA
Oligo 10: CTGATCGTTACTCGGCTTGTC
Oligo 11: TTCCACCCGGATAAGATGCTG
Oligo 12: GGTGGTATGATAGAAAGGGTGG
Oligo 13: GATAGGTCTTAAAGAACAACTCTAAC
sgRNA target sequences:
Ctrl (targeting intron): TCGTCGAGTTGGTCTACC
Ctrl-1 (targeting a nongenic region): TCGCGCTCGGCGGGTCACAG
Ctrl-2 (targeting a nongenic region): GTAGCGGTCTGGCTAGAACG
NT-1 (nontargeting, mouse) (39): CTTCACGCCTTGGACCGATA
NT-2 (mouse) (39): ACCCACGTATGTACTCGGGA
NT-1 (human) (39): ACGGAGGCTAAGCGTCGCAA
NT-2 (human) (39): CGCTTCCGCGGCCCGTTCAA
ERG-1: TAACTCTGCGCTCGTTCG
ERG-2: CTGCCGTAGTTCATCCCAA
ERG-3: TGCCGTAGTTCATCCCAA
Pten-1: ACCGCCAAATTTAACTGCAG
Pten-2: TGTGCATATTTATTGCATCG
Pten-3: TCACCTGGATTACAGACCCG
Trp53: AAGTCACAGCACATGACGG
AKT1-1: TGTGCCGCAAAAGGTCTTCA
AKT1-2: TCACGTTGGTCCACATCCTG
AKT2-1: TCTCGTCTGGAGAATCCACG
AKT2-2: CATCGAGAGGACCTTCCACG
LSL-1: TTCTAGGAATTCTACCGGGT
LSL-2: GTTACTAGTGGATCCGAGCT
T (Tmprss2 intron 1): ATCGTCGAGTTGGTCTACC
E2 (Erg intron 2): CACCACCATCCGATGACGA
E3 (Erg intron 2): ATTAGACCGACATTTACCG
F (Foxp1 vicinity): GACCAAACCGGGTGATACC
S (Shq1 vicinity): GTAGCGGTCTGGCTAGAACG
Intracellular Flow Cytometry.
Intracellular flow cytometry was performed using a Fixation/Permeabilization Solution Kit (BD 554714). Dissociated organoid cells were fixed and permeabilized with the fixation/permeabilization buffer for 30 min on ice, followed by incubation with primary antibodies for at least 1 h at room temperature and secondary antibodies (for nonconjugated primary antibodies) for 30 min at room temperature with washes between the steps before analysis by flow cytometry (BD Fortessa). Primary antibodies used were ERG (1:500; Abcam ab92513), ERG-Alexa Fluor 647 (1:200; Abcam ab196149), ERG-Alexa Fluor 488 (1:200; Abcam ab196374), and Pten (1:500; Abcam ab170941). Secondary antibody was goat anti-rabbit Alexa Fluor 647 (1:1,000; Thermo Fisher Scientific).
In Vivo Experiments and Histology.
All animal work was done in compliance with the guidelines of Research Animal Resource Center of Memorial Sloan Kettering Cancer Center (Institutional Animal Care and Use Committee: 06-07-012). For orthotopic transplantation of organoid cells, cells were transduced with Lenti-luciferase-P2A-Neo (Addgene plasmid #105621) (45) and selected with 500 µg/mL G418. Dissociated organoid cells were resuspended in 40 μL of 50% Matrigel (Corning) and 50% growth medium before injection into prostate dorsal lobes of syngeneic C57BL/6 albino mice (The Jackson Laboratory) (3 × 106 cells/injection, six mice per group). For orthotopic transplantation of freshly isolated prostate lineage cells, electroporated cells were resuspended in 20 μL of 50% Matrigel (Corning) and 50% growth medium before injection into prostate dorsal lobes of immunodeficient NSG mice (The Jackson Laboratory) (90,000 cells/injection, four to six mice per group). Prostates were collected at autopsy, fixed using 4% paraformaldehyde, dehydrated with 70% ethanol, paraffin embedded, and sectioned on glass slides. Hematoxylin/eosin (H&E) and immunohistochemistry staining were carried out by the MSKCC Molecular Cytology Core or HistoWiz, Inc. The following antibodies were used for staining: pAkt-S473 (1:100; CST4060) and p53 (0.05 µg/mL; CST 2524).
Western Blot.
Boiled whole cell lysates were run on a precast Tris-acetate (Thermo Fisher Scientific) or Mini-PROTEAN TGX protein gel (Bio-Rad) and then transferred to a nitrocellulose membrane (Bio-Rad). The membrane was incubated overnight at 4 °C with primary antibodies against Cas9 (1:1,000; Active Motif 61577), Pten (1:1,000; Cell Signaling Technology 9188), ERG (1:1,000; Abcam ab92513), p53 (1:1,000; Cell Signaling Technology 2524), Apc (1:1,000; Millipore MABC202), AKT1 (1:2,000, Cell Signaling Technology 4060L), AKT2 (1:1,000, Cell Signaling Technology 5239S), pPRAS40-Thr246 (1:1,000, Cell Signaling Technology 2997S), Actin-HRP (horseradish peroxidase) (1:10,000; Abcam ab49900), Cyclophilin B (1:10000; Cell Signaling Technology 43603S) and Vinculin (1:10,000; Cell Signaling Technology 13901). Signal was visualized with secondary HRP-conjugated antibodies and chemiluminescent detection.
Supplementary Material
Acknowledgments
C.L.S. is supported by the Howard Hughes Medical Institute; NIH Grants CA193837, CA092629, CA224079, CA155169, and CA008748; and the Starr Cancer Consortium Grant I12-0007. W.F. is supported by Department of Defense Fellowship W81XWH.
Footnotes
Competing interest statement: C.L.S. is on the board of directors of Novartis, is a cofounder of ORIC Pharmaceuticals, and serves on the scientific advisory boards of the following biotechnology companies: Agios, Arsenal, Beigene, Blueprint, Column Group, Foghorn, Housey Pharma, Nextech, KSQ Therapeutics, and PMV Pharma. He is a coinventor of the prostate cancer drugs enzalutamide and apalutamide, covered by US patents 7,709,517, 8,183,274, 9,126,941, 8,445,507, 8,802,689, and 9,388,159 filed by the University of California, Los Angeles.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110344118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Sánchez-Rivera F. J., et al., Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Platt R. J., et al., CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maddalo D., et al., In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516, 423–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zafra M. P., et al., Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 36, 888–893 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawasaki K., et al., Chromosome engineering of human colon-derived organoids to develop a model of traditional serrated adenoma. Gastroenterology 158, 638–651.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Leibold J., et al., Somatic tissue engineering in mouse models reveals an actionable role for WNT pathway alterations in prostate cancer metastasis. Cancer Discov. 10, 1038–1057 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roper J., et al., In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 35, 569–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S., Kim D., Cho S. W., Kim J., Kim J. S., Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin S., Staahl B. T., Alla R. K., Doudna J. A., Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riggan L., et al., CRISPR-Cas9 ribonucleoprotein-mediated genomic editing in mature primary innate immune cells. Cell Rep. 31, 107651 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., et al., ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat. Med. 19, 1023–1029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuris J. A., et al., Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J., et al., Efficient delivery of nuclease proteins for genome editing in human stem cells and primary cells. Nat. Protoc. 10, 1842–1859 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Karthaus W. R., et al., Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159, 163–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson D., et al., Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z., et al., Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436, 725–730 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou M., et al., Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 7, 736–749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor B. S., et al., Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hieronymus H., et al., Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. eLife 7, e37294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosicki M., Tomberg K., Bradley A., Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanoli F., et al., CRISPR-Cas9-guided oncogenic chromosomal translocations with conditional fusion protein expression in human mesenchymal cells. Proc. Natl. Acad. Sci. U.S.A. 114, 3696–3701 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi P. S., Meyerson M., Targeted genomic rearrangements using CRISPR/Cas technology. Nat. Commun. 5, 3728 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomlins S. A., et al., Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Driehuis E., Clevers H., CRISPR-Induced TMPRSS2-ERG gene fusions in mouse prostate organoids. JSM Biotechnol. Biomed. Eng. 4, 1076 (2017). [PMC free article] [PubMed] [Google Scholar]
- 25.Abeshouse A., et al., The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hieronymus H., et al., Deletion of 3p13-14 locus spanning FOXP1 to SHQ1 cooperates with PTEN loss in prostate oncogenesis. Nat. Commun. 8, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gough V., Gersbach C. A., Immunity to Cas9 as an obstacle to persistent genome editing. Mol. Ther. 28, 1389–1391 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Annunziato S., et al., Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev. 30, 1470–1480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D., et al., Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum. Gene Ther. 26, 432–442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawson D. A., Xin L., Lukacs R. U., Cheng D., Witte O. N., Isolation and functional characterization of murine prostate stem cells. Proc. Natl. Acad. Sci. U.S.A. 104, 181–186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi N., Zhang B., Zhang L., Ittmann M., Xin L., Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell 21, 253–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., et al., A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 461, 495–500 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karthaus W. R., et al., Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science 368, 497–505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crowley L., et al., A single-cell atlas of the mouse and human prostate reveals heterogeneity and conservation of epithelial progenitors. eLife 9, e59465 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo W., et al., Single-cell transcriptomics identifies a distinct luminal progenitor cell type in distal prostate invagination tips. Nat. Genet. 52, 908–918 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi J., et al., Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 33, 661–667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen B., et al., Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao N., et al., Aberrant expression of ERG promotes resistance to combined PI3K and AR pathway inhibition through maintenance of AR target genes. Mol. Cancer Ther. 18, 1577–1586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trotman L. C., et al., Pten dose dictates cancer progression in the prostate. PLoS Biol. 1, E59 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart T. A., Pattengale P. K., Leder P., Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell 38, 627–637 (1984). [DOI] [PubMed] [Google Scholar]
- 42.Duarte A. A., et al., BRCA-deficient mouse mammary tumor organoids to study cancer-drug resistance. Nat. Methods 15, 134–140 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Gao D., et al., Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brinkman E. K., Chen T., Amendola M., van Steensel B., Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y., et al., A TFIID-SAGA perturbation that targets MYB and suppresses acute myeloid leukemia. Cancer Cell 33, 13–28.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.