Significance
Many amphipathic lipids, including cannabinoids, lysophospholipids, prostanoids, and leukotrienes, bind to G protein–coupled receptors (GPCRs) through the lipid bilayer. How these lipid ligands get anchored to their receptors and enter ligand-binding pockets is largely elusive. Our results revealed a distinctive lipid recognition mechanism for the prostaglandin D2 (PGD2) receptor CRTH2 that is not shared by other prostanoid receptors. Based on the structural analysis, we propose two representative binding modes of lipids in GPCRs. We also identified critical structural motifs and conformational changes of CRTH2 for capturing lipids from the lipidic environment and lipid transition to the binding pocket, thereby offering unprecedented molecular insight into the dynamic lipid-binding processes for GPCRs.
Keywords: CRTH2 (DP2), prostaglandin D2, lipid binding, crystal structure, MD simulations
Abstract
Prostaglandin D2 (PGD2) signals through the G protein–coupled receptor (GPCR) CRTH2 to mediate various inflammatory responses. CRTH2 is the only member of the prostanoid receptor family that is phylogenetically distant from others, implying a nonconserved mechanism of lipid action on CRTH2. Here, we report a crystal structure of human CRTH2 bound to a PGD2 derivative, 15R-methyl-PGD2 (15mPGD2), by serial femtosecond crystallography. The structure revealed a “polar group in”–binding mode of 15mPGD2 contrasting the “polar group out”–binding mode of PGE2 in its receptor EP3. Structural comparison analysis suggested that these two lipid-binding modes, associated with distinct charge distributions of ligand-binding pockets, may apply to other lipid GPCRs. Molecular dynamics simulations together with mutagenesis studies also identified charged residues at the ligand entry port that function to capture lipid ligands of CRTH2 from the lipid bilayer. Together, our studies suggest critical roles of charge environment in lipid recognition by GPCRs.
Eicosanoids constitute a group of signaling lipid mediators that are derived from arachidonic acid or other polyunsaturated fatty acids (1, 2). As the name indicates, they all possess a long hydrocarbon chain with 20 carbon units, which is usually attached to a carboxylic acid head group. Prostanoids including prostaglandins D2, E2, and F2 (PGD2, PGE2, and PGF2); prostacyclin (PGI2); thromboxane A2 (TXA2) and leukotrienes including leukotriene B4, C4, and D4 (LTB4, LTC4, and LTD4) are representative endogenous eicosanoids that can induce signaling through G protein–coupled receptors (GPCRs) to play critical roles in inflammation, immunity, hemostasis, and tissue repair (1, 3–5). In humans, nine prostanoid receptors have been identified: PGD2 receptors 1 and 2 (DP1 and DP2), PGE2 receptors 1 to 4 (EP1 to EP4), PGF2 receptor (FP), PGI2 receptor (IP), and TXA2 receptor (TP) (5). All of these receptors, except for DP2, belong to the α-branch of Class A GPCRs as close phylogenetic neighbors of aminergic receptors (5, 6). DP2, chemoattractant receptor–homologous molecule expressed on type 2 helper T cells (Th2) (also named CRTH2), is more closely related to a group of chemotactic GPCRs in the γ-branch of Class A GPCRs, including chemokine receptors and receptors for LTB4, formylpeptides, and complement peptides C3a and C5a (6, 7).
Certain types of immune cells, including eosinophils, innate lymphoid cells, and Th2 cells, express high levels of CRTH2 (8–11). PGD2 signaling through CRTH2 can induce chemotaxis of these immune cells, which is a major pathway that drives the onset of type 2 inflammation (11–15). Therefore, the roles of PGD2–CRTH2 signaling axis in type 2 inflammation-related diseases such as asthma and allergic rhinitis have attracted outstanding research interest (14, 16, 17). CRTH2 antagonists hold the potential of being a new class of anti-inflammatory drugs (16, 18–24). Although clinical trials have generated mixed results on different CRTH2 antagonists for asthma (17–20, 25), it is likely that a certain subpopulation of patients, such as those with a high baseline of Th2 cells or eosinophils, may benefit most from anti-CRTH2 therapy (17, 24). In addition, it has been proposed that insurmountable CRTH2 antagonists with prolonged receptor residence time can provide better, therapeutic efficacy, compared to reversible and fast, dissociating CRTH2 antagonists, which needs further investigation (26).
We have previously reported crystal structures of CRTH2 bound to two antagonists, CAY10471 and fevipiprant, which revealed a positively charged environment of the ligand-binding pocket and a potential ligand entry port (27). The structural analysis of antagonist-bound CRTH2 suggested that PGD2 may enter the ligand-binding pocket through the ligand entry port by opposite charge attraction (27). Recently, crystal structures of several other prostanoid receptors, EP3, EP4, and TP, and cryogenic electron microscopy (cryo-EM) structures of the EP2 and EP4 signaling complexes with PGE2 have also been reported (28–33). To further study the binding of lipid agonists to CRTH2 and investigate if CRTH2 differs from other prostanoid receptors in lipid recognition and receptor activation, we determined a 2.6-Å resolution, room temperature crystal structure of CRTH2 bound to a PGD2 derivative by serial femtosecond crystallography (SFX) using an X-ray free electron laser (XFEL) source. We performed computational simulation studies using the CRTH2–15R-methyl-PGD2 (15mPGD2) structure, which helped identify the roles of specific residues surrounding the ligand entry port and provided molecular insights into the events that could facilitate ligand binding. Results from our mutational analysis of the identified residues helped us further strengthen the proposed model for ligand capture and entry in CRTH2.
Results
Structure Determination of Lipid Agonist-Bound CRTH2 by Serial Femtosecond Crystallography.
We used the same construct of human CRTH2 that we previously used to solve crystal structures of antagonist-bound CRTH2 (27). To facilitate crystallogenesis, an engineered T4 lysozyme (mT4L) with an additional eight-residue linker was inserted into the intracellular loop 3, flexible C terminus was truncated at R340, and the potential glycosylation site N25 was mutated to alanine. The crystallization construct showed a comparable PGD2-binding ability as the wild-type CRTH2 (wtCRTH2) (27). We used 15mPGD2 instead of PGD2 in our structural studies because 15mPGD2 has a higher affinity for CRTH2 (SI Appendix, Fig. S1A), and it is a selective prostaglandin agonist for CRTH2 over the DP1 (34). The only difference between 15mPGD2 and PGD2 is a methyl group at the 15-position carbon (C-15) (Fig. 1A). The crystals of CRTH2 bound to 15mPGD2 were obtained in the lipidic cubic phase (LCP) (35). Because of the small sizes of crystals (maximal crystal size <10 μm), we used XFEL to collect SFX data and solved the crystal structure at an anisotropic resolution of 2.6 to 3.2 Å (Materials and Methods) (SI Appendix, Fig. S1B and Table 1). We observed clear and strong electron density of the ligand. Our calculated simulated-annealing ligand-omit map was continuous, even when contoured at 4 σ, which allowed unambiguous modeling of 15mPGD2 in the structure (SI Appendix, Fig. S1C).
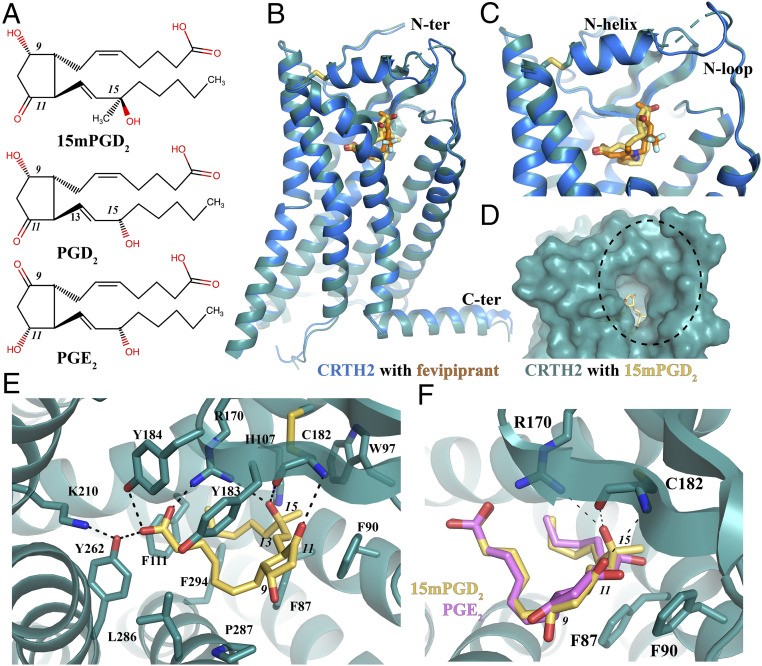
Fig. 1.
Overall structure of CRTH2 bound to 15mPGD2 and its ligand-binding pocket. (A) Chemical structures of 15mPGD2, PGD2, and PGE2. C-9, C-11, and C-15 positions are indicated with italic numbers. (B) Overall structure of CRTH2 bound to 15mPGD2 and structure alignment with CRTH2 bound to fevipiprant. (C) Overlapping, binding positions of 15mPGD2 and fevipiprant. (D) Potential lipid entry port. (E) Binding pocket of 15mPGD2. (F) Docking of PGE2 (pink) into the 15mPGD2- (yellow) binding pocket. Hydrogen bonds are shown as dashed lines.
Table 1.
Data collection and refinement statistics of CRTH2–15mPGD2 complex
| Number of crystals | 7,061 |
| Space group | P212121 |
| Cell dimensions, Å | |
| a | 51.4 |
| b | 67.8 |
| c | 265.4 |
| Resolution, Å | 32.4–2.60 (2.69–2.60)* |
| No. reflections | 1,809,804 (12,462) |
| No. unique reflections | 29,269 (2,540) |
| Rsplit, % | 26.4 (513.5) |
| Mean I/σ(I) | 2.4 (0.2) |
| CC*, % | 99.1 (51.2) |
| Completeness, % | 98.9 (90.6) |
| Multiplicity | 61.8 (4.9) |
| Refinement | |
| Resolution, Å | 32.4–2.6† |
| No. reflections/free set | 15,632/781 |
| Rwork/Rfree | 0.229/0.267 |
| No. atoms | |
| Protein | 3,424 |
| Ligand | 26 |
| Water and ions | 51 |
| B-factors, Å2 | |
| Overall | 52.3 |
| Protein | 52.1 |
| Ligand | 36.2 |
| Water and ions | 77.2 |
| rmsds | |
| Bond lengths, Å | 0.002 |
| Bond angle, ° | 0.48 |
| Ramachandran stats, % | |
| Favored | 95.12 |
| Allowed | 4.88 |
| Disallowed | 0 |
Values in parentheses are for highest-resolution shell.
Data were anisotropically truncated by the STARANISO web server at 3.2-, 3.2-, and 2.6-Å resolution along the reciprocal unit cell vectors a*, b*, and c*, respectively.
Overall Structure and Lipid-Binding Mode.
The overall structure of 15mPGD2-bound CRTH2 is nearly identical to the antagonist fevipiprant-bound CRTH2 (27), with a rmsd of 0.39 Å for all atoms of CRTH2 in both structures (Fig. 1B), suggesting that we captured an agonist-bound, inactive conformation of the receptor. Specifically, important structural motifs for receptor activation can be well aligned in both structures (SI Appendix, Fig. S1D). It is likely that agonists alone cannot stabilize CRTH2 in an active conformation, similarly to that observed for the β2-adrenergic receptor (36) and melatonin receptors (37, 38). Additionally, the inactive receptor conformation may be constrained by the fusion partner.
The lipid agonist 15mPGD2 occupies the same ligand-binding pocket in CRTH2 as the antagonist fevipiprant (Fig. 1C). The N-terminal region of CRTH2 bound to 15mPGD2 forms a well-folded structure with an α-helix (N-helix) and a loop (N-loop). The gap formed between the N-loop and the extracellular part of TM7 has been proposed to serve as the potential ligand entry port (27). The N-helix and the extracellular loop 2 (ECL2) constitute a lid domain to cover the binding pocket for 15mPGD2, leaving the ligand entry port as the only open end of the ligand-binding pocket (Fig. 1D). 15mPGD2 adopts a “U” shape–binding pose with the two hydrocarbon chains extending from the central hydroxycyclopentanone ring almost parallel to each other. The carboxyl head group of 15mPGD2 can be well superimposed to the carboxyl group of fevipiprant, which forms a similar polar interaction network with nearby residues R170ECL2, Y184ECL2, K2105.42, and Y2626.51 [superscripts indicate Ballesteros–Weinstein numbering (39)] (Fig. 1E). The alkyl ω-chain of 15mPGD2 is surrounded by mostly aromatic residues, including F872.60, F902.63, W97ECL1, H1073.28, F1113.32, Y183ECL2, and F2947.43. These residues together with P2877.36 and L2867.35 form extensive hydrophobic interactions with the ω-chain of 15mPGD2 (Fig. 1E). Supporting such a binding mode, previous mutagenesis studies from our group and others showed that mutations of R170ECL2A in human CRTH2 (27) and mutations of H3.28A, F3.32A, K5.42A, and Y6.51A in mouse CRTH2 (40) significantly reduced or abolished PGD2 binding. In addition, the hydroxyl group at C-15 of 15mPGD2 forms hydrogen bonds with the side chain of R170ECL2 and the main chain carbonyl group of C182ECL2; the carbonyl group at C-11 forms a hydrogen bond with the main chain amine group of C182ECL2 (Fig. 1E). The ∆13 carbon–carbon double bond in 15mPGD2 is close to the aromatic side chain of F902.63, engaging in potential 𝜋–𝜋 interactions (Fig. 1E).
It has been reported that 15mPGD2 is over five times more potent than PGD2 in inducing cellular signaling events (34), and the inhibitory constant (Ki) value of 15mPGD2 in competing antagonist ramatroban is about 20-fold lower than that of PGD2 (41). CRTH2 also prefers PGD2 over other endogenous prostaglandins such as PGE2 (>2,000-fold selectivity) (42). In our structure, the 15-hydroxyl group extends toward R170ECL2, because of the R-configuration at the C-15 position, to form a hydrogen bond, while in PGD2 the reversed stereochemical configuration at C-15 position would place the 15-hydroxyl group away from R170ECL2, thus breaking the hydrogen-bonding interaction (Fig. 1E). Similarly, 15S-methyl-PGD2 with the S-configuration at C-15 also exhibited a much lower affinity for CRTH2 compared to 15mPGD2 (34). We further docked PGE2 into the 15mPGD2-binding pocket by superposing a three-dimensional conformer of PGE2 on top of the 15mPGD2 molecule. In the docked structure, both the C-11 and C-15 hydroxyl groups in PGE2 point toward F902.63 of CRTH2, causing steric hindrance and breaking polar interactions with R170ECL2 and C182ECL2 of CRTH2 (Fig. 1F). This may potentially lead to the lower affinity of PGE2 for CRTH2 compared to 15mPGD2.
Distinctive Lipid Recognition by CRTH2.
Recently, structures of several prostaglandin receptors have been reported, including crystal structures of EP3 bound to two prostaglandin agonists, misoprostol and PGE2, antagonist-bound EP4 and TP, and cryo-EM structures of the EP2–Gs and EP4–Gi complexes with PGE2 (28–33). The structural comparison of 15mPGD2-bound CRTH2 to those structures of other prostanoid receptors reveals significant differences in lipid recognition and receptor activation.
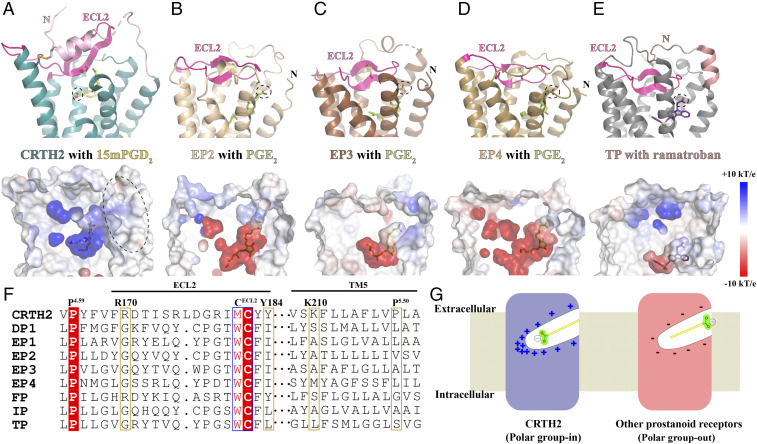
As mentioned in the introductory section, CRTH2 belongs to a different branch of Class A GPCRs compared to other prostanoid receptors. Consequently, the extracellular region of CRTH2 for ligand binding is distinct from that in other prostanoid receptors. In CRTH2, the ECL2 with a β-hairpin structure protrudes from the 7-transmembrane (7-TM) domain and forms a lid domain together with the well-folded N-terminal region to cover the ligand-binding pocket (Fig. 2A). In EP2, EP3, EP4, and TP, the ECL2 is buried inside the 7-TM domain, projecting toward TM1 to form the lid domain by itself, which prevents ligand access from the extracellular side (Fig. 2 B–E). As a result, PGE2 binds to its receptors more deeply inside the 7-TM domain compared to 15mPGD2 in CRTH2. Interestingly, the orientations of 15mPGD2 and PGE2, relative to their receptors, are also different (Fig. 2). The carboxyl group of 15mPGD2 is buried in the distal end of the positively charged ligand-binding pocket of CRTH2, away from TM1 and TM7 (Fig. 2A). The positive charge environment is important for PGD2 binding. Mutations of individual CRTH2 residues R170ECL2, Y184ECL2, K2105.42, and Y2626.51 at the distal end of the ligand-binding pocket to the negatively charged glutamic acid led to undetectable PGD2 binding (SI Appendix, Fig. S2). In contrast, in the structures of PGE2-bound EP2, EP3, and EP4, the carboxyl group of PGE2 is close to TM1 and TM7, facing the extracellular side of the receptors with the ω-chain extending toward the inside of the 7-TM domain (Fig. 2 B–D). Such a binding mode is associated with a predominantly negatively charged environment of the ligand-binding pockets in EP2, EP3, and EP4, which can help to position the carboxyl group of PGE2 at the top of the ligand-binding pockets through an electrostatic repulsion mechanism (Fig. 2 B–D). It is likely that the binding pose of TXA2 in TP is similar to PGE2, considering the structural similarity between TP and PGE2 receptors and the fact that the ligand-binding pocket of TP is also negatively charged (Fig. 2E). Supporting distinct lipid recognition mechanisms for CRTH2 and other prostanoid receptors, EP2 residues R7.40, Y2.65, and TECL2, that interact with the carboxyl group of PGE2 are highly conserved in all prostanoid receptors except for CRTH2 (33), while CRTH2 residues, R170ECL2, Y184ECL2, and K2105.42, that form a polar interaction network with the carboxyl group of PGD2 are not conserved in most of other prostanoid receptors (Fig. 2F). Therefore, we propose two lipid recognition modes for the prostanoid receptors, the “polar group in” mode for CRTH2 and the “polar group out” mode for the other prostanoid receptors (Fig. 2G). These two lipid-binding modes for the prostanoid receptor family may also apply to other lipid GPCRs as well (see Discussion).
Fig. 2.
Structural comparison analysis. (A) CRTH2 with 15mPGD2. The N-terminal region preceding TM1 is colored in light pink. (B–D) EP2, EP3, and EP4 with PGE2 (Protein Data Bank [PDB] IDs: 7CX2, 6AK3, and 7D7M, respectively). (E) TP with ramatroban (PDB ID: 6IIU). The N-terminal region preceding TM1 is colored in salmon. ECL2 in each receptor is colored in magenta. 15mPGD2, PGE2, and ramatroban are colored in yellow, green, and purple, respectively. The carboxyl group in each ligand is circled. The charge distributions of ligand-binding pockets of these prostanoid receptors are shown in the lower panels. (F) Sequence alignment of all prostanoid receptors. The alignment of ECL2 sequences was based on the alignments of two highly conserved residues, P4.50 and the cysteine residue, in ECL2, forming the conserved extracellular disulfide bond. The alignment of TM5 sequences was based on the alignment of 5.50 residues in the receptors. (G) Two lipid-binding modes.
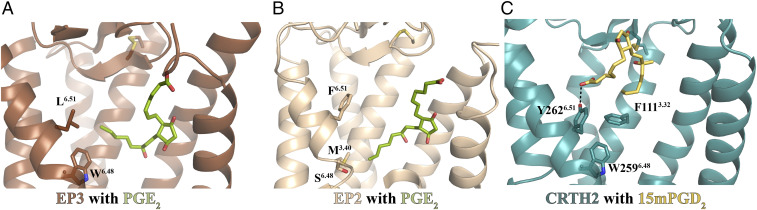
The different binding poses of lipid agonists in CRTH2 and other prostanoid receptors also suggest nonconserved mechanisms of receptor activation. The structure of EP3 bound to PGE2 represents an active-like conformational state, characterized by an outward displacement of the cytoplasmic end of TM6 starting at W6.48 in comparison to antagonist-bound EP4 (30). In the EP3–PGE2 structure, the end of the ω-chain of PGE2 extends toward TM6, forming hydrophobic interactions with residues L2986.51 and W2956.48 (Fig. 3A). It has been proposed that the direct contact of the ω-chain of PGE2 with W6.48 and surrounding residues is important for the activation of the receptor (30). Indeed, previous studies on other GPCRs suggested that W6.48 functions as a “toggle switch” or a part of a “transmission switch” with F6.44 that links the ligand binding at the extracellular region to the activation of the receptor at the cytoplasmic region (43, 44). For EP2 and EP4, the conserved W6.48 is replaced by S6.48 (31–33). Structural studies on the EP2 signaling complex with PGE2 suggested that a direct interaction between the ω-chain of PGE2 and M1243.40 instead of W6.48, which packs against F2736.44, may stabilize the active conformation of EP2 (33) (Fig. 3B). In CRTH2, the lipid agonist 15mPGD2 binds to a much more superficial binding pocket compared to PGE2, and it does not directly interact with the F6.44xxCW6.48 motif (Fig. 3C). In the U shape–binding pose of 15mPGD2, the carboxyl group forms a hydrogen bond with Y2626.51, and the ω-chain points toward TM3, forming a hydrophobic interaction with F1113.32. Both Y2626.51 and F1113.32, in turn, pack against W2596.48 (Fig. 3C). This implies a distinct activation mechanism of CRTH2 compared to the PGE2 receptors, which may involve residues Y2626.51, F1113.32, and W2596.48.
Fig. 3.
PGE2 and PGD2 interact with different sets of residues to activate their receptors. (A) EP3 (brown) with PGE2 (green) (PDB ID: 6AK3). (B) EP2 (light brown) with PGE2 (green) (PDB ID: 7CX2). The ω-chain of PGE2 extends below the 6.51 residue to interact with W2956.48 in EP3 or M1243.40 in EP2. (C) CRTH2 (cyan) with 15mPGD2 (yellow). PGD2 binds to a superficial binding pocket in CRTH2 so that the ω-chain of PGD2 is above the residue Y6.51. The hydrogen bond is shown as a black dashed line.
Potential Lipid-Binding Process Revealed by MD Simulations.
To explore the conformational dynamics associated with the lipid-bound CRTH2, we carried out equilibrium molecular dynamics (MD) simulations of CRTH2 in its ligand-free (apo) and 15mPGD2-bound (holo) states. We generated four independent 1-μs trajectories of CRTH2 embedded in TIP3P-solvated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayers, two in apo (apo-1 and apo-2) and two in holo states (holo-1 and holo-2). The system preparation and MD simulation protocol are described in the Materials and Methods section. All four trajectories remained stable over the 1-μs simulation, with the receptor in the holo-1 system exhibiting relatively larger conformational deviations, as compared to the other three systems (SI Appendix, Fig. S3). In all four simulation systems, the receptor mainly stayed in the inactive conformation, as in the crystal structure. We analyzed the tilt of the seven transmembrane helices TM1 to TM7 and helix 8 with respect to the normal to the plane of the membrane (z-axis) and observed no major conformational changes of the transmembrane helices. The average tilt angles remained close to those observed in the crystal structure in all four systems during the course of the simulation (SI Appendix, Fig. S4). The SD values for the tilt angles of all seven transmembrane helices were in the range of 0.8 to 2.3° only, suggesting that there were no significant shifts in their relative orientations. In contrast, H8 in all four trajectories showed significant shifts in the tilt angle (SI Appendix, Fig. S4).
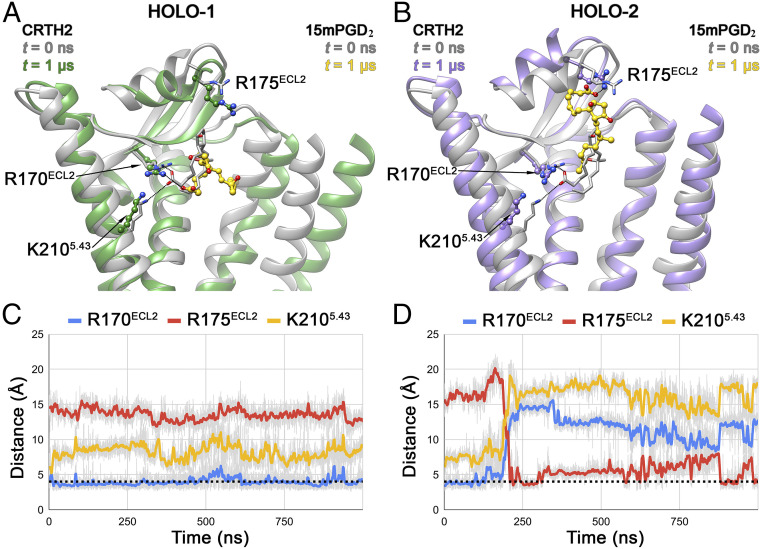
In contrast to the subtle conformational changes of the transmembrane helices, the 15mPGD2 molecule in both holo simulations showed significant rearrangements during the simulation compared to its initial conformation (Fig. 4 A and B). In holo-1, the conformational changes associated with the bound 15mPGD2 molecule were relatively subtle compared to those observed in holo-2, with its anionic headgroup rearranged but still interacting directly with R170ECL2 and its aliphatic ω-chain wedged between TM2 and TM3 (Fig. 4 A and C). In contrast to the subtle changes of 15mPGD2 in holo-1, in holo-2, 15mPGD2 exhibited dramatic changes with respect to its initial conformation. The molecule adopted a more linear conformation, in contrast to the U-shaped conformation in the crystal structure. The anionic headgroup of 15mPGD2 moved away from both R170ECL2 and K2105.42 toward the extracellular side to form new interactions with R175ECL2, which was accompanied by a displacement of the N-helix toward the extracellular milieu (Fig. 4 B and D). This partial dissociation of 15mPGD2 may represent an intermediate state in the ligand-binding/unbinding processes.
Fig. 4.
Conformational dynamics of 15mPGD2 in CRTH2 holo-1 and holo-2 systems. The CRTH2 structures at t = 0 ns (translucent gray ribbon) are superposed with structures at t = 1 µs in holo-1 (A) (translucent green ribbon) and t = 1 µs in holo-2 (B) (translucent purple ribbon). In both panels, conformations of bound 15mPGD2 at t = 0 ns (translucent gray) and t = 1 µs (gold) are shown in stick and ball-and-stick representation, respectively. Salt–bridge interactions formed by the carboxyl group of 15mPGD2 with R170ECL2, R175ECL2, and K2105.42 are shown as black dotted lines in A and B. Evolution of the salt bridges during the 1-µs simulation in holo-1 (C) and holo-2 (D) systems are presented as blue, red, and yellow lines (50-point moving average) corresponding to 15mPGD2-R170ECL2, 15mPGD2-R175ECL2, and 15mPGD2-K2105.42 interactions, respectively, with the dotted black line indicating the cutoff distance of 4.0 Å.
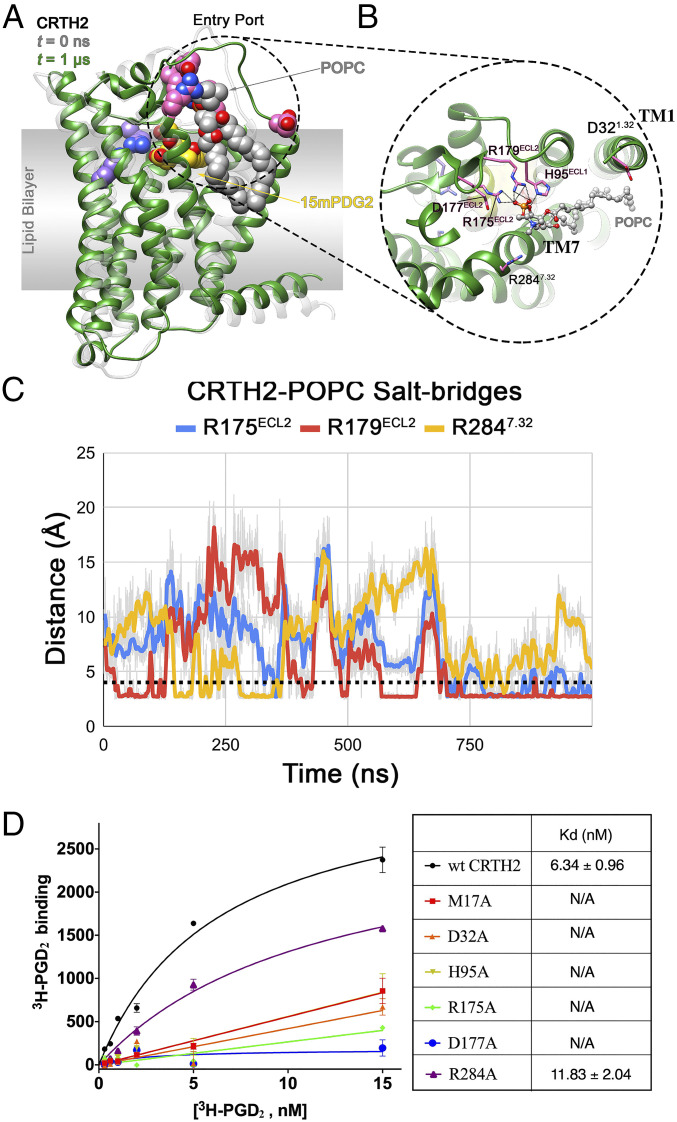
The subtle movement of 15mPGD2 in holo-1 was associated with binding of a POPC molecule from the lipid bilayer to CRTH2, which potentially hindered the partial dissociation of 15mPGD2, as that observed in holo-2. In holo-1, the POPC molecule, POPC330, moved toward ECL2 through the putative entry port between TM1 and TM7. At t = 0 ns, POPC330 was positioned near the entry port with one of its acyl chains wedged between TM1 and TM7 (SI Appendix, Fig. S5A). In this position, the phosphate group of POPC330 was in close proximity of charged/polar residues lining the entry port, including D321.32, R2847.32, and H95ECL1 at the water–bilayer interface, while the acyl chains were flanked by hydrophobic residues from TM1 and TM7. The cleft formed by TM1 and TM7 has been shown to be a high-occupancy site for phospholipids in atomistic and coarse-grained simulations of β2-adrenergic (45) and serotonin1A receptors (46), along with the role of membrane cholesterol in regulating the occupancy of the TM1 to TM7 cleft (47). Over the course of the simulation in holo-1, the phosphate group of POPC330 formed salt bridges and hydrogen bonds with H95ECL1, R2847.32, and R179ECL2, causing the POPC headgroup to be laterally pulled toward ECL2 from the bilayer by ∼7 Å along the membrane plane (Fig. 5 A and B and SI Appendix, Fig. S5B). The phosphate group initially formed transient salt–bridge interactions with R179ECL2 and/or R2847.32. Subsequently, the interaction with R179ECL2 was stabilized, allowing the POPC headgroup to move further inward and, in turn, establish an interaction with R175ECL2 (Fig. 5C). No such inward movement of a bilayer POPC molecule was observed in the other three trajectories, despite the presence of a POPC molecule near the entry port at t = 0 ns in those systems (SI Appendix, Fig. S5). In order to accommodate the inward movement of the headgroup and acyl chains of the captured POPC, TM1 moved away from TM7, increasing the space within the entry port.
Fig. 5.
CRTH2–POPC interaction in holo-1 simulation and mutagenesis data. (A) Interaction of a bilayer POPC molecule, with the residues lining the entry port in holo-1 simulation. The structures of the receptor at t = 0 ns (translucent gray) and t = 1 μs (green) are shown in ribbon representation. The entry port residues (pink), bound 15mPGD2 (yellow), POPC (gray), and binding site residues R170ECL2 and K2105.42 (purple) are shown in sphere representation. (B) Close up of the CRTH2 entry port–POPC interaction, with the bound POPC (gray) and entry port residues (pink) shown in ball-and-stick and stick representation, respectively. The interactions between the POPC and the protein residues are shown as black dotted lines. (C) Evolution of CRTH2–POPC intermolecular salt bridges during the 1-µs holo-1 simulation, with the dotted black line indicating the cutoff distance of 4.0 Å. (D) Saturation-binding assays on different CRTH2 constructs using 3H-PGD2. All CRTH2 constructs were transiently expressed in human embryonic kidney 293 cells, and cell membranes were prepared and used in the ligand-binding assays. Expression of each construct was confirmed by cell surface staining. Nonspecific binding measured in the presence of access amount of PGD2 was subtracted. The dissociation equilibrium constants (Kds) of PGD2 for the wtCRTH2 and the R284A mutant are listed in the table shown on the right of panel D. For other CRTH2 mutants, no saturable 3H-PGD2 binding was observed, indicating mostly nonspecific binding. Each data point in the left panel is shown as mean ± SEM and n = 3.
The binding of 15mPGD2 and POPC330 near the bilayer–water interface observed in the two holo simulations may be suggestive of how CRTH2 captures anionic lipid ligands. We propose that the “cationic tetrad” of R2847.32, H95ECL1, R179ECL2, and R175ECL2, which line at the ligand entry port and do not directly interact with CRTH2 antagonists (27) or 15mPGD2 in the crystal structures, constitute a lipid trap to enable the initial capture of anionic eicosanoid lipids from the cell membrane. Previous studies showed that mutation of R178ECL2 to Ala in mouse CRTH2, which corresponds to R179ECL2 in human CRTH2, significantly decreased the affinity of PGD2 for CRTH2 (40). We also performed 3H-PGD2 saturation–binding assays on CRTH2 mutants R175ECL2A, H95ECL1A, and R2847.32A (Fig. 5D). All mutants showed comparable expression levels as the wtCRTH2 (SI Appendix, Fig. S6), with compromised PGD2 binding. The results suggested the important roles of these residues in PGD2 binding, supporting our proposed, lipid-capturing mechanism. Interestingly, the side chains of R175ECL2 and R179ECL2, despite being cationic, maintained their proximity and almost parallel orientation to each other in all four trajectories without any electrostatic repulsion. This was due to the presence of an anionic residue, D177ECL2, between R175ECL2 and R179ECL2 (Fig. 5B and SI Appendix, Fig. S7), potentially circumventing any repulsion between the two residues. Therefore, we further propose that D177ECL2 functions as the foil to help maintain the proper orientation of the two cationic residues, which in turn allows the effective capturing of anionic eicosanoid lipids. The vital role of D177ECL2 in PGD2 binding was confirmed by mutagenesis study (Fig. 5D and SI Appendix, Fig. S6).
Conformational Dynamics of CRTH2 in MD Simulations.
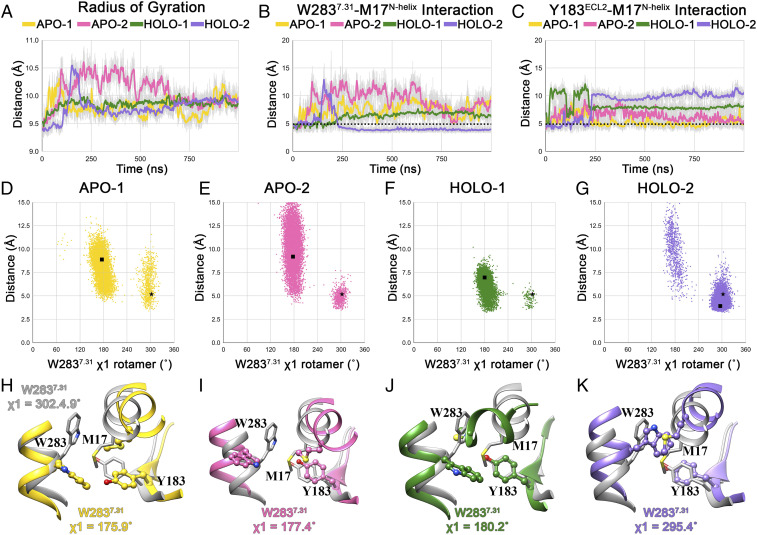
The tetrad of R2847.32, H95ECL1, R179ECL2, and R175ECL2 is centered around the entry port at the water–bilayer interface, ∼19 Å away from R170ECL2 and K2105.42 in the binding pocket. Once captured, how the anionic group of the ligand is translocated across such a distance toward R170ECL2 and K2105.42 is unclear. Our simulation studies show that the binding pocket of CRTH2 undergoes structural rearrangements, leading to significant changes in residue packing. We characterized the residue packing changes within the binding site in the simulations by calculating the radius of gyration of the binding pocket (Fig. 6A). We defined the binding pocket by considering those residues that are within 5.0 Å of the 15mPGD2 molecule in the crystal structure. The analysis shows that the removal of the bound 15mPGD2 in apo simulations causes the packing within the pocket to relax, causing it to open up (Fig. 6A). Similar relaxation of the binding site residue packing is also observed in holo simulations but to a lesser magnitude, relative to the apo simulations. In holo-2, the brief spike in the radius of gyration at t ∼150 ns corresponds to the 15mPGD2-unbinding event. The changes in residue packing can be attributed to conformational changes associated with M17N-helix, W2837.31, and Y183ECL2 and the interactions between these residues (Fig. 6A). In the crystal structure, the N-helix is nestled between the extracellular end of TM7 and ECL2, with M17N-helix forming numerous contacts with the surrounding residues. In particular, the side chain of M17N-helix is flanked by W2837.31 and Y183ECL2 and is buried within the upper half of the binding pocket (SI Appendix, Fig. S8). Such an arrangement of these three residues is reminiscent of an Aro-Met-Aro “bridging” motif and is likely mediated by S-π/CH-π–type interactions (48). Such Met-aromatic interactions are abundant in protein structures (48–51).
Fig. 6.
Conformational dynamics of the ligand-binding pocket in MD simulations. (A) Evolution of radius of gyration (50-point moving average) of the binding site residues in 1-µs trajectories. Evolution of S-π–type W2837.31–M17N-helix (B) and Y183ECL2–M17N-helix (C) interactions in 1-µs trajectories. Scatter plot showing distribution of W2837.31 (ring centroid)–M17N-helix (S-atom) distances and W2837.31 χ1 rotamer angles in W2837.31 apo-1 (D), apo-2 (E), holo-1 (F), and holo-2 (G) trajectories. Structures with W2837.31 χ1 rotamer angle equal to the average W2837.31 χ1 rotamer angle in the last 500 ns of the simulation are selected as representatives from each trajectory. The distance and χ1 rotamer angle corresponding to the crystal structure and representative structure from each simulation are shown using a black star and square in the scatter plots. Organization of the W2837.31–M17N-helix–Y183ECL2 bridge motif in representative structures, along with their W2837.31 χ1 rotamer angle, from apo-1 (H) (yellow), apo-2 (I) (pink), holo-1 (J) (green), and holo-2 (K) (purple) trajectories. The bridge motif from the crystal structure (gray) is also shown superposed to the representative structure.
We observed the disruption of the bridging motif characterized by the breakage of W2837.31–M17N-helix and/or Y183ECL2–M17N-helix interactions (Fig. 6 B and C) in all four simulations, resulting in the disorganization of the tightly packed hydrophobic interaction network involving the extracellular end of TM7, N-helix, and ECL2. We characterized the bridge disruption by scrutinizing the distribution of W2837.31–M17N-helix distances and W2837.31 χ1 rotamer angles. In the crystal structure, the W2837.31 side chain is pointed inwards (χ1 = −57.6°), favoring the formation of the W2837.31–M17N-helix–Y183ECL2 bridge motif. However, in all simulations except holo-2, W2837.31 side chain flips outwards (χ1 ∼180°), breaking the W2837.31–M17N-helix interaction and leading to the bridge disruption (Fig. 6 D–G). As a consequence, the N-helix moves away from the TM core (Fig. 6 H–K), associated with an increase in the volume of the translocation channel. In selected representative conformations of CRTH2, the disruption of the bridge motif culminates in the increase of volume of the translocation pathway from 1,435 Å3 (apo-1 at t = 0 ns) to 2,626 Å3, 2,345 Å3, 1,773 Å3, and 1,786 Å3 in apo-1, apo-2, holo-1, and holo-2 simulations, respectively (SI Appendix, Fig. S9).
Based on this observation, we hypothesized that the bridging motif mediated by M17N-helix might be of critical importance for ligand binding. Abrogation of the W2837.31–M17N-helix–Y183ECL2 bridging motif by mutation M17N-helixA disrupts PGD2 binding (Fig. 5D), confirming the important role of M17N-helix in PGD2 binding. Considering the CRTH2–POPC interactions in holo-1, the sequence of events leading to the ligand unbinding in holo-2, and the associated changes in the bridging motif anchored by M17N-helix, we speculate that the holo simulations represent potential transitional states in the ligand-binding process: holo-1 gives glimpses into the initial ligand-binding events, while holo-2 shows stages of ligand unbinding. Collectively, our observations suggest that the translocation of the anionic lipid ligand toward the binding pocket is modulated by the dynamics of the N-helix mediated by M17N-helix.
Discussion
Previously reported structures of CRTH2 bound to antagonists revealed a nonuniform positive charge distribution in the ligand-binding pocket, which has been suggested to guide the carboxylate group of CRTH2 lipid ligands to reach the distal end by opposite charge attraction (27). Indeed, the structure of CRTH2 bound to 15mPGD2 confirms that the carboxylate group of 15mPGD2 resides at the distal end of the ligand-binding pocket, with the highest positive charge distribution, forming polar interactions with R170ECL2 and K2105.42, both of which have been shown to be critical for PGD2 binding (27, 40). The U shape lipid–binding mode explains well the selectivity of CRTH2 for different lipid ligands.
The polar group in–binding mode of PGD2 on CRTH2 contrasts the polar group out–binding modes of lipid ligands on other prostanoid receptors. These two distinct lipid-binding modes may also apply to other lipid GPCRs. For example, structures of sphingosine-1–phosphate receptor 1, lysophosphatidic acid receptor 1, and cannabinoid receptors revealed negatively charged, ligand-binding pockets associated with polar group out–binding modes of their lipid ligands (27). Interestingly, these GPCRs and all prostanoid receptors except CRTH2 belong to the α-branch of Class A GPCRs (6). On the other hands, crystal structures of cysteinyl leukotriene receptors (CLTRs) (52, 53) revealed positively charged, ligand-binding pockets with uneven charge distribution, similar to that of CRTH2 (SI Appendix, Fig. S10). In addition, the carboxylate group of cysteinyl LTD4 has been proposed to be buried inside the binding pocket of CLTR1 (53), adopting a polar group in–binding mode. Therefore, it is likely that cysteinyl leukotrienes enter the lipid-binding pockets of CLTRs through a similar opposite charge attraction-facilitated mechanism, although the potential ligand entrance in CLTRs is different from that in CRTH2 (53).
To study the conformational dynamics of the human CRTH2, we carried out equilibrium MD simulations of the receptor in its apo and holo states. In our simulations, the receptor structure in holo systems in the presence of the agonist 15mPGD2 did not exhibit or assume any active-like conformations with respect to the intracellular end of TM6 or other conserved GPCR structural motifs. However, conformations sampled by CRTH2 in our MD simulation studies, coupled with functional assays of receptor mutants, helped us identify the roles of critical residues that were hitherto unrecognized or merely speculated upon. Holo simulations provided glimpses into the nature of the ligand-binding/unbinding process and the associated, conformational changes there within. The observed interaction of a bilayer POPC molecule with CRTH2 led to the identification of the mechanistic role of the cationic tetrad in the initial ligand capture process. Our study highlights the cooperative effects of the cationic tetrad R2847.32, R175ECL2, R179ECL2, and H95ECL1, which act like fishhooks that aid in picking out anionic group-containing lipids from the membrane, with D321.32 potentially funneling the ligands toward the entry port. All of these residues at the ligand entry port do not directly interact with 15mPGD2 in the crystal structure; however, our mutagenesis studies confirmed their important roles in lipid recognition (Fig. 5D). In FPR2, the residue at position 7.32 (D2827.32) has been suggested to be important for interaction with short formylpeptides with cationic, terminal moieties (54, 55). Also, a similar fishhook-like role for a cationic residue in the capture of a lipid-like substrate has been suggested for the bacterial lysophospholipid transporter (56). Therefore, the role of the cationic tetrad and its interactions with anionic lipids in CRTH2 could potentially represent a common mechanism employed by certain GPCRs and other membrane proteins. Our studies also shed light on the mechanisms involved in the stabilization and orientation of the cationic tetrad, especially R175ECL2 and R179ECL2. One of the distinct features present in CRTH2 is the ordered N-terminal region, and our study addresses one of the potential roles of the N-helix in modulating the access to the binding pocket and its packing via the W2837.31–M17N-helix–Y183ECL2 bridge motif. Breaking of either or both W2837.31–M17N-helix and M17N-helix–Y183ECL2 S-π–type interactions displaces the W2837.31 and M17N-helix side chains from the binding pocket. Such a disruption increases the volume of the binding pocket in the simulations and could facilitate ligand binding/unbinding.
Based on our computational and experimental findings, we propose the following mechanistic model for the anionic, lipid-binding process in CRTH2. We postulate that electrostatically driven interactions with residues of the cationic tetrad R2847.32, R175ECL2, R179ECL2, and H95ECL1, potentially assisted by D321.32, favor the initial capture of anionic, lipid-like ligands and their subsequent inward transition from the bilayer. Following the initial binding and lateral inward movement of the anionic group, the receptor could shift through transitional conformations, as observed in the holo simulations. Once the ligand anionic group is optimally positioned, the complete/partial reestablishment of the bridging motif W2837.31–M17N-helix–Y183ECL2 could facilitate the movement of the anionic group into the binding pocket of the receptor toward R170 and K210, establishing the ligand-bound conformation, as observed in the crystal structure.
Materials and Methods
Protein Expression, Purification, and Crystallization.
CRTH2 was engineered, expressed in insect Sf9 cells, and purified in complex with 15mPGD2, as previously described (27). Microcrystals (5 to 10 μm) for SFX were grown in LCP inside 100-μL gas-tight syringes, as previously described (57), and then transported to XFEL for data collection. During sample transportation to XFEL, the LCP in all syringes converted into a sponge phase. The sponge phase with crystals was carefully separated from the precipitant and converted back into LCP for injection in the XFEL beam. Details of protein production and sample preparation for the SFX are described in SI Appendix.
Diffraction Data Collection and Structure Determination.
SFX data were collected at the coherent X-ray imaging instrument (58) of the Linac Coherent Light Source (LCLS). A total of 850,000 images were recorded within ∼2 h using ∼50 μL sample. A total of 20,358 potential, single-crystal diffraction patterns (crystal hits) were identified using Cheetah (59). Indexing, integration, and merging of the crystal diffraction data were performed using CrystFEL (60). The final dataset was analyzed by the STARANISO webserver (61) and truncated to 3.2, 3.2, and 2.6 Å resolution along the reciprocal unit cell vectors a*, b*, and c*, respectively. The structure was solved by the molecular replacement method. Final data collection and refinement statistics are shown in Table 1. Details of data collection and structure determination are described in SI Appendix.
Radioligand-Binding Assay and Protein Surface Expression.
The expression levels of wtCRTH2 and mutants in human embryonic kidney 293 (HEK293) cells were determined by flow cytometry experiments (SI Appendix). Binding assays were performed using cell membranes expressing CRTH2 and mutants in the presence of the radioactive 3H-PGD2 and nonradioactive PGD2 or 15mPGD2 (SI Appendix).
MD Simulations.
Apo or holo configurations of CRTH2 were embedded into a TIP3P-solvated bilayer comprising 300 POPC molecules. The systems were neutralized by adding Na+ and Cl− ions. The protein, solvent, lipids, and ions were modeled using CHARMM36 force-field parameters (62). Initial parameters for the 15mPGD2 molecule were generated using CHARMM general force field [CGenFF (63)] implemented through the ParamChem (64, 65) web server and were further refined using the GAAMP (63) web server. Details are described in SI Appendix.
All simulations were performed using GROMACS version 5.1.2 (66). Long-range electrostatics were calculated with the particle mesh Ewald scheme, while a 12-Å cutoff was employed for estimating short-range, nonbonded interactions. After energy minimization, the systems were equilibrated by applying harmonic position restraints on nonsolvent heavy atoms, which were gradually reduced over six equilibration steps (details in SI Appendix). All restraints were removed postequilibration and 1 µs production runs were carried out at 303.15 K (Nośe–Hoover thermostat) and 1.0 bar pressure (Parrinello–Rahman barostat). Covalent bonds were constrained with linear constraint solver algorithm (67). Pressure coupling was applied semiisotropically during NPT equilibration steps and unrestrained production runs.
Analysis of MD Simulation Trajectories.
The analysis is described in SI Appendix.
Supplementary Material
Acknowledgments
R.N.V.K.D. and H.F. gratefully acknowledge the Biomedical Research Council, A*STAR for financial support. The computational work for this article was partially performed on resources of the National Supercomputing Centre, Singapore (https://www.nscc.sg). U.W., H.H., and V.C. acknowledge financial support from the Science and Technology Center Program of the NSF through BioXFEL under Agreement No. 1231306. Use of the LCLS, Stanford Linear Accelerator Center National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. C.Z. is supported by Grant No. R35GM128641 from the National Institute of General Medical Sciences, NIH of the United States.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102813118/-/DCSupplemental.
Data Availability
Atomic coordinates and structure factors for the structure of 15mPGD2–CRTH2 have been deposited in the Protein Data Bank with the accession code 7M8W. All other study data are included in the article and/or supporting information.
References
- 1.Boyce J. A., Eicosanoids in asthma, allergic inflammation, and host defense. Curr. Mol. Med. 8, 335–349 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Harizi H., Corcuff J.-B., Gualde N., Arachidonic-acid-derived eicosanoids: Roles in biology and immunopathology. Trends Mol. Med. 14, 461–469 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Funk C. D., Prostaglandins and leukotrienes: Advances in eicosanoid biology. Science 294, 1871–1875 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Bäck M., et al., International Union of Basic and Clinical Pharmacology. LXXXIV: Leukotriene receptor nomenclature, distribution, and pathophysiological functions. Pharmacol. Rev. 63, 539–584 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Woodward D. F., Jones R. L., Narumiya S., International Union of Basic and Clinical Pharmacology. LXXXIII: Classification of prostanoid receptors, updating 15 years of progress. Pharmacol. Rev. 63, 471–538 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Fredriksson R., Lagerström M. C., Lundin L.-G., Schiöth H. B., The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63, 1256–1272 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Nagata K., Hirai H., The second PGD(2) receptor CRTH2: Structure, properties, and functions in leukocytes. Prostaglandins Leukot. Essent. Fatty Acids 69, 169–177 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Nagata K., et al., Selective expression of a novel surface molecule by human Th2 cells in vivo. J. Immunol. 162, 1278–1286 (1999). [PubMed] [Google Scholar]
- 9.Cosmi L., et al., CRTH2 is the most reliable marker for the detection of circulating human type 2 Th and type 2 T cytotoxic cells in health and disease. Eur. J. Immunol. 30, 2972–2979 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Mjösberg J. M., et al., Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 12, 1055–1062 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Hirai H., et al., Prostaglandin D2 selectively induces chemotaxis in T helper type 2 cells, eosinophils, and basophils via seven-transmembrane receptor CRTH2. J. Exp. Med. 193, 255–261 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monneret G., Gravel S., Diamond M., Rokach J., Powell W. S., Prostaglandin D2 is a potent chemoattractant for human eosinophils that acts via a novel DP receptor. Blood 98, 1942–1948 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Xue L., et al., Prostaglandin D2 causes preferential induction of proinflammatory Th2 cytokine production through an action on chemoattractant receptor-like molecule expressed on Th2 cells. J. Immunol. 175, 6531–6536 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Kostenis E., Ulven T., Emerging roles of DP and CRTH2 in allergic inflammation. Trends Mol. Med. 12, 148–158 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Tait Wojno E. D., et al., The prostaglandin D2 receptor CRTH2 regulates accumulation of group 2 innate lymphoid cells in the inflamed lung. Mucosal Immunol. 8, 1313–1323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelaia C., et al., New treatments for asthma: From the pathogenic role of prostaglandin D2 to the therapeutic effects of fevipiprant. Pharmacol. Res. 155, 104490 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Kupczyk M., Kuna P., Targeting the PGD2/CRTH2/DP1 signaling pathway in asthma and allergic disease: Current status and future perspectives. Drugs 77, 1281–1294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barnes N., et al., A randomized, double-blind, placebo-controlled study of the CRTH2 antagonist OC000459 in moderate persistent asthma. Clin. Exp. Allergy 42, 38–48 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Erpenbeck V. J., et al., The oral CRTh2 antagonist QAW039 (fevipiprant): A phase II study in uncontrolled allergic asthma. Pulm. Pharmacol. Ther. 39, 54–63 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Kuna P., Bjermer L., Tornling G., Two phase II randomized trials on the CRTh2 antagonist AZD1981 in adults with asthma. Drug Des. Devel. Ther. 10, 2759–2770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bateman E. D., et al., Fevipiprant, an oral prostaglandin DP2 receptor (CRTh2) antagonist, in allergic asthma uncontrolled on low-dose inhaled corticosteroids. Eur. Respir. J. 50, 1700670 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Miller D., et al., A randomized study of BI 671800, a CRTH2 antagonist, as add-on therapy in poorly controlled asthma. Allergy Asthma Proc. 38, 157–164 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Ortega H., et al., A phase 2 study to evaluate the safety, efficacy and pharmacokinetics of DP2 antagonist GB001 and to explore biomarkers of airway inflammation in mild-to-moderate asthma. Clin. Exp. Allergy 50, 189–197 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asano K., et al., A phase 2a study of DP2 antagonist GB001 for asthma. J. Allergy Clin. Immunol. Pract. 8, 1275–1283.e1 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Santus P., Radovanovic D., Prostaglandin D2 receptor antagonists in early development as potential therapeutic options for asthma. Expert Opin. Investig. Drugs 25, 1083–1092 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Calbet M., et al., Pharmacological characterization of CRTh2 antagonist LAS191859: Long receptor residence time translates into long-lasting in vivo efficacy. Pharmacol. Res. 111, 208–216 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Wang L., et al., Structures of the human PGD2 receptor CRTH2 reveal novel mechanisms for ligand recognition. Mol. Cell 72, 48–59.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Audet M., et al., Crystal structure of misoprostol bound to the labor inducer prostaglandin E2 receptor. Nat. Chem. Biol. 15, 11–17 (2019). Correction in: Nat. Chem. Biol. 15, 206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan H., et al., Structural basis for ligand recognition of the human thromboxane A2 receptor. Nat. Chem. Biol. 15, 27–33 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Morimoto K., et al., Crystal structure of the endogenous agonist-bound prostanoid receptor EP3. Nat. Chem. Biol. 15, 8–10 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Toyoda Y., et al., Ligand binding to human prostaglandin E receptor EP4 at the lipid-bilayer interface. Nat. Chem. Biol. 15, 18–26 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Nojima S., et al., Cryo-EM structure of the prostaglandin E receptor EP4 coupled to G protein. Structure 29, 252–260.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Qu C., et al., Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E2 receptor EP2 subtype. Sci. Adv. 7, eabf1268 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monneret G., Cossette C., Gravel S., Rokach J., Powell W. S., 15R-methyl-prostaglandin D2 is a potent and selective CRTH2/DP2 receptor agonist in human eosinophils. J. Pharmacol. Exp. Ther. 304, 349–355 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Caffrey M., Cherezov V., Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenbaum D. M., et al., Structure and function of an irreversible agonist-β(2) adrenoceptor complex. Nature 469, 236–240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson L. C., et al., XFEL structures of the human MT2 melatonin receptor reveal the basis of subtype selectivity. Nature 569, 289–292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stauch B., et al., Structural basis of ligand recognition at the human MT1 melatonin receptor. Nature 569, 284–288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ballesteros J. A., Weinstein H., Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Method. Neurosci. 25, 366–428 (1995). [Google Scholar]
- 40.Hata A. N., Lybrand T. P., Breyer R. M., Identification of determinants of ligand binding affinity and selectivity in the prostaglandin D2 receptor CRTH2. J. Biol. Chem. 280, 32442–32451 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Sugimoto H., Shichijo M., Okano M., Bacon K. B., CRTH2-specific binding characteristics of [3H]ramatroban and its effects on PGD2-, 15-deoxy-Delta12, 14-PGJ2- and indomethacin-induced agonist responses. Eur. J. Pharmacol. 524, 30–37 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Sawyer N., et al., Molecular pharmacology of the human prostaglandin D2 receptor, CRTH2. Br. J. Pharmacol. 137, 1163–1172 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deupi X., Standfuss J., Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr. Opin. Struct. Biol. 21, 541–551 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Trzaskowski B., et al., Action of molecular switches in GPCRs–Theoretical and experimental studies. Curr. Med. Chem. 19, 1090–1109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prasanna X., Chattopadhyay A., Sengupta D., “Role of Lipid-Mediated Effects in β2 - Adrenergic Receptor Dimerization” in Biochemical Roles of Eukaryotic Cell Surface Macromolecules, Chakrabarti A., Surolia A., Eds. (Springer, 2015), pp. 247–261. [Google Scholar]
- 46.Patra S. M., et al., Differential dynamics of the serotonin1A receptor in membrane bilayers of varying cholesterol content revealed by all atom molecular dynamics simulation. Mol. Membr. Biol. 32, 127–137 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Prasanna X., Sengupta D., Chattopadhyay A., Cholesterol-dependent conformational plasticity in GPCR dimers. Sci. Rep. 6, 31858 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber D. S., Warren J. J., The interaction between methionine and two aromatic amino acids is an abundant and multifunctional motif in proteins. Arch. Biochem. Biophys. 672, 108053 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Valley C. C., et al., The methionine-aromatic motif plays a unique role in stabilizing protein structure. J. Biol. Chem. 287, 34979–34991 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orabi E. A., English A. M., Modeling protein S–aromatic motifs reveals their structural and redox flexibility. J. Phys. Chem. B 122, 3760–3770 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Krishna Deepak R. N. V., Chandrakar B., Sankararamakrishnan R., Comparison of metal-binding strength between methionine and cysteine residues: Implications for the design of metal-binding motifs in proteins. Biophys. Chem. 224, 32–39 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Gusach A., et al., Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors. Nat. Commun. 10, 5573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luginina A., et al., Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs. Sci. Adv. 5, eaax2518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhuang Y., et al., Structure of formylpeptide receptor 2-Gi complex reveals insights into ligand recognition and signaling. Nat. Commun. 11, 885 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen T., et al., Structural basis of ligand binding modes at the human formyl peptide receptor 2. Nat. Commun. 11, 1208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin Y., Deepak R. N. V. K., Zheng J. Z., Fan H., Zheng L., A dual substrate-accessing mechanism of a major facilitator superfamily protein facilitates lysophospholipid flipping across the cell membrane. J. Biol. Chem. 293, 19919–19931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu W., Ishchenko A., Cherezov V., Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nat. Protoc. 9, 2123–2134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boutet S., Williams G. J., The coherent X-ray imaging (CXI) instrument at the Linac Coherent Light Source (LCLS). New J. Phys. 12, 035024 (2010). [Google Scholar]
- 59.Barty A., et al., Cheetah: Software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Cryst. 47, 1118–1131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White T. A., et al., Recent developments in CrystFEL. J. Appl. Cryst. 49, 680–689 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tickle I. J., et al., STARANISO (Global Phasing Ltd., Cambridge, United Kingdom, 2018).
- 62.Klauda J. B., et al., Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanommeslaeghe K., et al., CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanommeslaeghe K., MacKerell A. D. Jr, Automation of the CHARMM General Force Field (CGenFF) I: Bond perception and atom typing. J. Chem. Inf. Model. 52, 3144–3154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vanommeslaeghe K., Raman E. P., MacKerell A. D. Jr, Automation of the CHARMM General Force Field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 52, 3155–3168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abraham M. J., et al., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015). [Google Scholar]
- 67.Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. E. M., LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the structure of 15mPGD2–CRTH2 have been deposited in the Protein Data Bank with the accession code 7M8W. All other study data are included in the article and/or supporting information.