Abstract
Herein, we describe the development of a paper-based device to detect nucleic acids of pathogens of interest in complex samples using loop-mediated isothermal amplification (LAMP) by producing a colorimetric response visible to the human eye. To demonstrate the utility of this device in emerging public health emergencies, we developed and optimized our device to detect SARS-CoV-2 in human saliva without preprocessing. The resulting device was capable of detecting the virus within 60 min and had an analytical sensitivity of 97% and a specificity of 100% with a limit of detection of 200 genomic copies/μL of patient saliva using image analysis. The device consists of a configurable number of reaction zones constructed of Grade 222 chromatography paper separated by 20 mil polystyrene spacers attached to a Melinex® backing via an ARclean® double-sided adhesive. The resulting device is easily configurable to detect multiple targets and has the potential to detect a variety of pathogens simply by changing the LAMP primer sets.
Keywords: Paper-based diagnostics, Colorimetric LAMP, SARS-CoV-2, Saliva, Microfluidic paper-based analytical devices
1. Introduction
In 2020, the coronavirus disease 2019 (COVID-19) pandemic infected over 79 million individuals and claimed over 1.7 million lives worldwide (“Weekly epidemiological update - December 29, 2020,” 2020). Additionally, it is estimated that by 2030, the COVID-19 pandemic will have caused over $16 trillion in economic damages to the United States alone (Cutler and Summers, 2020). Most molecular tests for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, the virus responsible for COVID-19) are limited to the laboratory and thus have significant lag times (>24 h) to provide a result, preventing their widespread adoption in point-of-care settings (Yüce et al., 2021). Despite several attempts at developing a point-of-care test for SARS-CoV-2 (Tromberg et al., 2020), some key limitations remain: i) scalability (the demand for testing is in the order of millions per week, but manufacturing new tests at that scale is challenging), ii) the need for sample processing (many tests still require an extraction step when using saliva), and iii) readability (molecular tests often require the use of fluorescence and thus, a fluorescence reader to report the results) (Parupudi et al., 2021).
We overcome the limitations of current testing methods by developing a point-of-care test using paper-based devices and reverse-transcription loop-mediated isothermal amplification (RT-LAMP) that reports a color change in the presence of SARS-CoV-2 within 60 min using diluted saliva (5% v/v in water) as the sample. RT-LAMP is a nucleic acid amplification technique conducted at a constant temperature with diagnostic performance comparable to the current gold standard, reverse-transcription quantitative polymerase chain reaction (RT-qPCR), especially during the acute phase of infection (Inaba et al., 2021). Since RT-LAMP can be conducted at a constant temperature, there is no need for expensive thermal cycling equipment. Additionally, existing colorimetric reporters for LAMP products eliminate the need for fluorescence readers. Consequently, this test is suitable for use in point-of-care settings and is amenable to rapid development and scale-up, making it appropriate for use in public health emergencies.
RT-LAMP has been implemented on microfluidic paper-based analytical devices (μPADs) to detect the Zika virus (Kaarj et al., 2018), where image analysis was performed using a smartphone to distinguish between positive and negative responses. Here, we provide a high-contrast RT-LAMP reaction on paper that provides a color change that is visible to the naked eye. In addition, instead of using wax-printing—which would require precise alignment of printed areas and reagent addition—we used polystyrene spacers for preventing cross-talk between samples. The polystyrene spacers are amenable to roll-to-roll fabrication for scale up of production. There are no other reports yet demonstrating on-paper detection of SARS-CoV-2 using RT-LAMP with colorimetric reporters and saliva as the sample matrix.
Table 1 presents a selection of currently available nucleic-acid-based COVID-19 diagnostic methods. The novelty of our assay is the on-paper colorimetric detection of SARS-CoV-2 with minimal pre-processing. Our device has a sensitivity and specificity that is comparable to other RT-LAMP based assays; however, our assay is the only one that can detect SARS-CoV-2 on paper without pre-amplification. Other assays are conducted in solution which is not as scalable during manufacturing as paper-based assays. Additionally, our assay only requires a dilution step (requiring seconds to complete), whereas other assays require treatment with protease, heat-inactivation, and/or RNA extraction to detect SARS-CoV-2 (steps requiring at least 10 min to complete and additional equipment).
Table 1.
Performance and description of select nucleic-acid-based biosensors for COVID-19 detection.
| Test | Technology | Limit of detection (copies/μL) | Sensitivity (%) | Specificity (%) | Pre-processing needed | Format | Reference |
|---|---|---|---|---|---|---|---|
| EasyCOV | RT-LAMP | Not reported | 72.7 | 95.7 | Heat-inactivation at 65 °C for 30 min | Tube/Solution, Colorimetric | L'Helgouach et al. (2020) |
| SalivaDirect | RT-PCR | 6–12 | 92.7 | 99.9 | Proteinase K, Heat-inactivation at 95 °C for 5 min | Tube/Solution, Fluorometric | Vogels et al. (2021) |
| Un-named Point-of-care Test | RT-LAMP | 25 | 82.6 | 100 | Semi-alkaline protease for 15 min, heat treatment at 95 °C for 5 min | Tube/Solution, Colorimetric | Yamazaki et al. (2021) |
| Ambry COVID-19 RT-PCR Test | RT-PCR | 0.1 | 96.8 | 100 | RNA Extraction | Tube/Solution, Fluorometric | FDA (2021) |
| Rheonix COVID-19 MDx Assay | RT-PCR | 0.625 | 97.8 | 100.0 | RNA Extraction | Tube/Solution, Fluorometric | Rhoenix, Inc., 2021 |
| STOPCovid.v2 | CRISPR/SHERLOCK | 0.033 | 93.1 | 98.5 | RT-LAMP Amplification | Fluorescent or Lateral Flow Strip | Joung et al. (2020) |
| Unnamed extraction-free RT-LAMP | RT-LAMP | 59 | 85 | 100 | Heat-treatment at 65 C for 15 min and at 95 C for 5 min | Tube/Solution, Colorimetric | Lalli et al. (2021) |
| Unnamed extraction-free RT-LAMP on paper | RT-LAMP | 200 | 97 | 100 | Sample dilution with nuclease-free water | Colorimetric, Paper | This study |
We demonstrate the direct detection of SARS-CoV-2 in saliva via a distinct colorimetric response that can be read using the naked eye (Fig. 1 ). This format is amenable to roll-to-roll fabrication and is anticipated to cost ~$10/test (Table S1). The limit of detection (LoD) of our test is 200 copies/μL saliva. The analytical sensitivity (positive predictive value) is 76%, the specificity (negative predictive value) is 100%, and the accuracy is approximately 91% as determined using contrived samples (freshly collected saliva spiked with heat-inactivated SARS-CoV-2) when evaluated visually (Fig. 1F). Due to subjectivity in color perception of the control pad, respondents incorrectly identified a total of 20 out of the 80 devices presented as invalid, resulting in a false invalid rate of 25%. The sensitivity increases to 97% with an accuracy of 98% when the color change is quantified using image processing (Fig. 1C).
Fig. 1.
Schematics and colorimetric characterization of the paper-based device. A) Schematic illustration of the workflow for device use. The control zone indicates the no-primer control. B) Colorimetric LoD on paper using heat-inactivated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) at the indicated concentration (copies/μL undiluted saliva) in 5% saliva. The negative replicates are RT-LAMP reactions using nuclease-free water in lieu of heat-inactivated SARS-CoV-2. Data was taken from Fig. S27. C) Schematic layout of the paper device. D) Typical colorimetric results of a negative and positive run. Controls are RT-LAMP reactions without LAMP primers included. Positive reactions have 800 copies/μL inactivated virus spiked into saliva. E) Color gradient of possible results derived from the colorimetric results of panel B. F) Summary table of observations used to calculate the analytical sensitivity and specificity of the paper device based on survey responses. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2. Methods
2.1. Primer design and screening
RT-LAMP primer sets (Table S2) were designed using Primer Explorer v5 (http://primerexplorer.jp/lampv5e/index.html) with parameters found in Table S3. Primer sets were designed using portions of the SARS-CoV-2 genome (NCBI accession number: NC_045512). Primer sets for RegX were designed by choosing three random 2000 nt regions. In-silico analyses were used to the predict sensitivity and specificity of each primer set. Optimal primer sets underwent experimental cross-reactivity studies to ensure specificity to SARS-CoV-2.
2.2. Saliva collection device
Three commercial saliva collection devices were selected to evaluate their effect on the RT-LAMP reaction in saliva. The three devices were “Saliva Sampler™” (StatSure Diagnostic Systems, Inc.), “Pure·SAL™” (Oasis Diagnostics®), and “Super·SAL™” (Oasis Diagnostics®).
2.3. Cost of goods table with source, price, and product details
Heat-inactivated SARS-CoV-2 was obtained from ATCC (ATCC VR-1986HK) or BEI (NR-52286). Pooled human saliva (991-05-P-50) and saliva from individual donors (991-05-S) was purchased from Lee Biosolutions. All oligonucleotides (desalted) were purchased from Life Technologies. Materials used in the colorimetric RT-LAMP reaction and their prices can be found in Table S1.
2.4. Optimization of composition of assay
Final optimized colorimetric RT-LAMP master mix consisted of KCl (50 mM), MgSO4 (8 mM), dNTP mixture (1.4 mM each dNTP), Bst 2.0 WarmStart® DNA Polymerase (0.32 U/μL), WarmStart® RTx Reverse Transcriptase (0.3 U/μL), Phenol red (0.25 mM), dUTP (0.14 mM), Antarctic Thermolabile UDG (0.0004 U/μL), Tween® 20 (1% v/v), betaine (20 mM), BSA (500 μg/mL), and trehalose (10% w/v).
2.5. Fabrication and optimization of devices
The final device (Fig. 1C) had dimensions of 6 mm × 20 mm and consisted of: a reading layer, two reaction strips, and spacers to prevent crosstalk. The reading area consisted of an optically clear 3 mil MELINEX (Tekra MELINEX® 454 Polyester (PET)) backer for support. This was attached to two reaction strips of 5 mm × 6 mm chromatography paper (Ahlstrom-Munksjö Grade 222) using a double-sided adhesive (Adhesives Research acid-free ARclean® 90178). The strips were separated by 2.5 × 6 mm 20 mil polystyrene spacers (Tekra Double White Opaque High Impact Polystyrene (HIPS) Litho Grade). 25 μL of sample was added to saturate the strips when rehydrating.
2.6. Determining LoD, sensitivity, specificity
Serial dilutions of heat-inactivated virus in water (100–105 copies/reaction for 25 μL reactions) were used as template in liquid reactions to establish a baseline LoD for candidate primer sets. Reactions were run in triplicate on a qPCR plate (Thermo Scientific AB-0800W) for each viral concentration and heated to 65 °C in a standard 75 L biological incubator (Fisherbrand Isotemp Microbiological Indicator, 15-103-0513) for 60 min. The color of the reaction mixtures at different time points was recorded by scanning the plate on a tabletop scanner (Epson Perfection V800 Photo Color). The LoD for a primer set was determined by the lowest viral concentration that resulted in a distinctive color change in all three replicates.
The sensitivity and specificity were determined using 30 contrived positive samples at varying multiples of the LoD (1x, 2x, 4x, 40x, and 400x with 10, 10, 4, 3, and 3 replicates, respectively) and 30 non-template control (NTC) negative saliva samples. Saliva was diluted to 5% v/v in nuclease-free water before spiking in heat-inactivated SARS-CoV-2 at the appropriate concentrations. The reported concentrations are copies/µL of undiluted saliva (i.e., expected concentration in a patient sample). The colorimetric response intensity was determined using ImageJ to extract the average green channel intensity of each reaction zone. A receiver-operating characteristic (ROC) curve was generated by varying the threshold cutoff between positive and negative reactions and calculating the sensitivity and specificity at each threshold value. The sensitivity was calculated as a ratio of the number of true positives to total positives, including false positives. The specificity was calculated as a ratio of true negatives to total negatives, including false negatives.
A color bar was created by averaging RGB values of phenol red on Grade 222 chromatography paper over a range of pH values. A linear gradient was created using these average RGB values and the optimal threshold value determined from the ROC curve was annotated on the color bar (Fig. 1C).
2.7. Human subjects
Fresh saliva was collected from participants who enrolled in the study in accordance with Purdue University IRB Protocol # IRB-2020-527. Participants who reported a COVID-19 diagnosis within the past 60 days were not permitted to donate. Saliva was collected from participants after receiving informed consent. Samples were assigned an ID based on the donor, date of sample collection, and method used for collection. No identifying information on participants was recorded.
Colorimetric perception surveys (Additional File 1) were collected from participants enrolled in the study in accordance with Purdue University IRB Protocol # IRB-2021-375. Participants were given a color bar with a threshold annotated (Fig. 1E). Participants were given multiple paper device scans and were asked to classify the control pad (left reaction zone) as valid or invalid and the SARS-CoV-2 reaction (right reaction zone) as positive or negative. Observations classified as invalid were discarded from assay performance analysis and the proportion of incorrectly identified invalid assays was reported as the false invalid rate.
3. Results
The workflow of our assay is illustrated in Fig. 1A: collect saliva, transfer sample to paper-based device, incubate at 65 °C, and read a result; a typical result is shown in Fig. 1D.
3.1. Design and screening of primers
We designed at least 3 primer sets for each of the following SARS-CoV-2 genes: N gene, RdRp gene, and orf1ab (Table S2). We screened primer sets using a fluorescent RT-LAMP kit with: i) in-vitro transcribed SARS-CoV-2 RNA for the gene in water or ii) heat-inactivated SARS-CoV-2 in 18% saliva (final reaction concentration). After screening the primer sets (Fig. S1–S3), we decided that orf1ab.II was our optimal primer set due to it possessing the best LoD of 200 copies/μL of reaction (reaction volume 25 μL).
In case downstream testing using orf1ab.II failed, we designed primers for random portions of the SARS-CoV-2 genome, labeled RegX. These primers were screened using a fluorometric RT-LAMP kit in 18% saliva (final reaction concentration), and RegX.III was chosen due to its fast reaction time and lack of false-positives (Table 2 and Fig. S4).
Table 2.
Sequence of optimal primers used for detection of SARS-CoV-2.
| Primer | Sequence (5’ - 3′) |
|---|---|
| orf1ab.II_F3 | ACTTAAAAACACAGTCTGTACC |
| orf1ab.II_B3 | TCAAAAGCCCTGTATACGA |
| orf1ab.II_FIP | TGACTGAAGCATGGGTTCGCGTCTGCGGTATGTGGAAAG |
| orf1ab.II_BIP | GCTGATGCACAATCGTTTTTAAACGCATCAGTACTAGTGCCTGT |
| orf1ab.II_LF | GAGTTGATCACAACTACAGCCATA |
| orf1ab.II_LB | TTGCGGTGTAAGTGCAGCC |
| orf7ab.I_F3 | CGGCGTAAAACACGTCTA |
| orf7ab.I_B3 | GCTAAAAAGCACAAATAGAAGTC |
| orf7ab.I_FIP | GGAGAGTAAAGTTCTTGAACTTCCTAGTTACGTGCCAGATCAG |
| orf7ab.I_BIP | TGCGGCAATAGTGTTTATAACACTATGAAAGTTCAATCATTCTGTCT |
| orf7ab.I_LF | TGTCTGATGAACAGTTTAGGTGAAA |
| orf7ab.I_LB | TTGCTTCACACTCAAAAGAA |
In-silico inclusivity studies and sequence identity studies indicated over 99% conservation of orf1ab.II and orf7ab.I in available SARS-CoV-2 sequences and did not indicate cross-reactivity with other microorganisms found in human saliva or the human respiratory tract (Table S3 and Table S4). We experimentally verified that the orf1ab.II and orf7ab.I primer sets were not cross-reactive using genomic extracts of the analyzed microorganisms (Table S7 and Table S8).
3.2. Screening of colorimetric dyes
We evaluated several metal-ion-chelating, DNA intercalating, and pH-sensitive colorimetric reporters for their ability to produce a robust and discernible colorimetric response during the RT-LAMP reaction using primer sets our lab had previously developed to target Histophilus somni (Mohan et al., 2021). Some of the investigated metal-ion-chelating and DNA intercalating indicators were able to produce a colorimetric response in solution but were unable to produce a colorimetric response on paper (Fig. S5–S11).
We investigated several pH-sensitive colorimetric indicators for use in our assay (Fig. S12-S16). Of the indicators screened, phenol red had the most apparent and contrasting colorimetric response (Fig. S16).
3.3. Optimization of colorimetric assay
We deemed an initial pH of 8.5 in solution to be optimal with respect to reported color consistency across replicates and contrast of color between positive and negative reactions (Fig. S16). On paper, an initial pH of 8.0 is optimal (Fig. S17).
We found that diluting saliva to 25% before adding to the RT-LAMP reaction (which further dilutes to a final concentration of 5% saliva) was sufficient to reduce the buffering capacity of saliva as well as the concentration of interferents (e.g., RNases) and produce a discernible colorimetric result in saliva within 60 min (data not shown).
Inclusion of both carrier DNA and guanidine hydrochloride as seen in the literature (Hardinge and Murray, 2019; Kiddle et al., 2012; Zhang et al., 2020) provided a colorimetric response and LoD that was comparable in both water and 5% saliva (Fig. S18 and Fig. S19). These components could not be added to paper, however, because they caused a significant color change upon drying the reagents (data not shown). The addition of RNase inhibitors appeared to worsen the LoD (Fig. S20).
3.4. Selection of saliva collection device
We evaluated several commercially available saliva processing devices to eliminate particles in collected saliva (Fig. S21). We selected a sponge-based collection device (Oasis Diagnostics® Pure·SAL™) due to its ability to standardize starting color and to provide the most consistent and best LoD of the observed devices.
3.5. Design of paper-based devices
Fig. 1C provides a schematic of our paper device's structure. The paper device consisted of two Grade 222 cellulose reaction pads separated by 20 mil polystyrene spacers to prevent crosstalk between reaction zones. These components were attached to a transparent backing for structural support via a double-sided adhesive. Reagents to conduct RT-LAMP are dried onto the reaction pad during fabrication. These reagents are rehydrated when the user adds the sample to the reaction zones.
Inclusion of ammonium sulfate caused a color from red to yellow upon drying of RT-LAMP reagents when no template was present. We prevented this color change by increasing the phenol red concentration and replacing ammonium sulfate with betaine (Fig. S24). Furthermore, the addition of trehalose and bovine serum albumin (BSA) increased the reaction speed and improved LoD (Fig. S25).
After loading the sample onto the paper-based device, we placed the device into a 1″ x 1” re-sealable plastic bag to prevent contamination during the RT-LAMP reaction. The plastic bag containing the paper device was then placed into an incubator at 65 °C for 60 min. The bag was removed, scanned with a flatbed scanner (Fig. 1D), and compared against a color chart (Fig. 1E) created by averaging RGB values from the phenol red response on grade 222 pads in buffers at known pH values from 6 to 9 (Fig. S26). The threshold value in the color chart corresponds to the threshold determined from ROC analysis (Fig. 2 B).
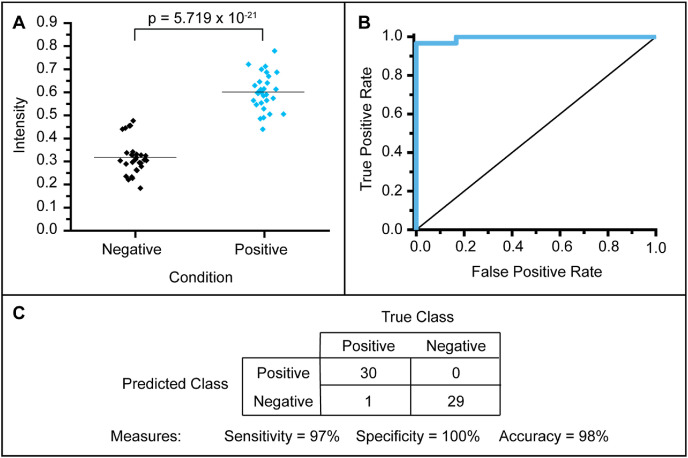
Fig. 2.
Digital analysis of colorimetric responses on paper. A) Box plot for the green channel intensity of 30 positive and 30 negative results of RT-LAMP on paper. B) Receiver-operating characteristic (ROC) curve for 30 positive and 30 negative results of RT-LAMP on paper. C) Summary table for the observations based on image analysis. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Validation of contrived samples
After determining an LoD of 200 copies/μL in saliva (Fig. 1B), we created contrived samples of 1x, 2x, 4x, 40x, and 400x LoD. We used 30 aliquots of freshly collected saliva as negative samples (Fig. S27). We quantified the results using image processing (Fig. 2A and Fig. S28). We found the difference between the green channel intensity of the negative and positive colorimetric reaction pads to be significant (p < 0.001) using a two-tailed student's t-test. The specificity using image analysis was 100%, the sensitivity was 97%, and the accuracy was 98% (Fig. 2C).
3.7. Colorimetric interpretation survey
We asked four participants to classify 10 positive and 10 negative reactions presented in Fig. S27 as valid or invalid (according to the left control zone) and positive or negative for SARS-CoV-2 (according to the right reaction zone) using a color bar (Fig. 1C).
Observations deemed invalid by the participant were discarded. Of the 40 true positive observations (all valid using image analysis), participants incorrectly classified 19 as invalid. This contrasts with the 40 true negative observations (36 of which were valid using image analysis) where only 1 observation was incorrectly classified as invalid. Thus, the calculated specificity and sensitivity of our device while accounting for colorimetric interpretation were 100% and 76%, respectively, with an accuracy of 91% (Fig. 1F) and a false invalid rate of 25%.
4. Discussion
4.1. Overview of device design and procedure
We designed each step of the assay (Fig. 1A) to reduce complexity and mitigate user errors in point-of-care settings. The sample collection step uses a sponge-based collection device which allows for self-collection by the patient, removes particulates from the collected saliva, and minimizes variance in results from patient to patient. The transfer step requires placing 25 μL of diluted saliva (5% v/v in water) onto each of the two reaction zones on the device. For the incubation step, the paper device with the sample loaded is sealed in a resealable plastic bag and placed in an incubator set at 65 °C for 60 min. Finally, for the read step, the user compares the color of the control and reaction zones to the color bar in Fig. 1C to determine if the results are valid and if the pathogen of interest is present.
Our platform comprises three main components: primer sets imposing specificity to the assay, a paper device containing two reaction zones (one control and one reaction), and a heating source used to heat the paper device to reaction temperatures. The primer sets determine what pathogen the assay targets. Therefore, our platform can be reconfigured to target a different pathogen by redesigning the primer sets while keeping all other aspects of our device and formulation the same. Additionally, the paper-based device can be configured to accommodate numerous reaction zones allowing for multiple targets to be detected simultaneously.
4.2. Design and screening of primers
We designed primer sets targeting the N gene (Corman et al., 2020), RNA-dependent RNA Polymerase (nsp12) gene (Lu et al., 2020), and portions of the orf1ab genes (Huang et al., 2020). Our assay utilized a no-primer control to ensure that the reaction zones do not change color when heating. Other assays utilize a Human RNaseP mRNA control which increases diagnostic power (Yang et al., 2021; Yaren et al., 2021). We screened primer sets in water using in-vitro transcribed RNA of the gene to assess performance and ability to dimerize. We further screened primer sets in saliva (18% reaction concentration) to assess performance in complex samples and off-target interactions. Since only one primer set – with a relatively poor LoD – passed both stages of screening, primer sets targeting random regions of the SARS-CoV-2 genome were designed and screened in 18% saliva. We selected primer set RegX.III (renamed to orf7ab.I) for further assay development alongside orf1ab.II. For further discussion on primer screening, see Supplemental Information.
4.3. Screening of colorimetric reporters
Although fluorescent reporters work well for quantitative RT-LAMP in a real-time thermocycler, they require an additional ultraviolet (UV) light source to be read by the naked eye which adds technical complexity to our assay. Thus, we developed a colorimetric RT-LAMP assay using indicators previously reported in the literature for both LAMP and PCR (Tanner et al., 2015). We evaluated three classes of indicators: i) metal ion indicators (Rodriguez-Manzano et al., 2016), ii) DNA intercalating indicators (Roy et al., 2017), and iii) pH indicators (Tanner et al., 2015). Whereas some of the screened metal-ion and DNA-intercalating indicators produced a colorimetric response in solution, none were able to produce a colorimetric response on paper (Fig. S5–S11).
Therefore, we screened pH dyes used as indicators in literature (Fig. S12–S16). Of the pH indicators we examined, cresol red and phenol red (Fig. S16) produced the most vibrant colorimetric contrast between positive and negative LAMP reactions. We used phenol red as our indicator since it was used frequently as a reporter in literature (Ahn et al., 2019; Tanner et al., 2015; Thi et al., 2020).
The initial pH also has a drastic effect on the 60-min end-point values. For initial pH values of 8.1, all negative reactions are false positives. For pH values of 8.3, some negative replicates at the 60-min time point also appear to be changing color (Fig. S16). Consequently, an initial pH value of around 8.5 is necessary to achieve a consistent, distinguishable colorimetric response, despite the contrast between positive and negative reactions being lower at this pH. Previous studies have found that the dependence of the final colorimetric response on initial pH is likely due to the diversity of compositions and buffering capacities of complex samples (Uribe-Alvarez et al., 2021). We speculate a similar explanation is responsible for the observed dependence of the final colorimetric response on initial pH.
4.4. Selection of saliva processing device
To standardize initial pH and to remove particulates from collected saliva, we evaluated three commercially available saliva collection devices to pre-process saliva. We chose Pure·SAL™ due to consistent initial starting pH, the best LoD, and the highest contrast between positive and negative reactions at the end of the reaction using our optimized LAMP formulation (Fig. S21). We found reports of Pure·SAL™ used in lateral flow strips and other investigations (Dalirirad et al., 2020; Sa et al., 2020), but not in isothermal amplification assays. Literature investigations of Pure·SAL™ suggest that it removes greater than 70% of mucinous material from saliva, which would act as an inhibitor to our amplification assay (Khurshid et al., 2017). When using our optimized LAMP formulation, processing saliva with Pure·SAL™ provides a four-fold better LoD when compared to unprocessed saliva accompanied by a more robust colorimetric response. (Fig. S23).
4.5. Optimization of colorimetric assay
We determined that diluting the saliva to 25% with nuclease-free water and further diluting to a final concentration of 5% saliva upon addition to the RT-LAMP reaction was needed to obtain results within 60 min. Dilution reduces the buffering capacity of saliva and decreases the concentration of inhibitory components, both of which would delay colorimetric reporting. We found dilution preferable to more commonly used pre-treatment steps found in a variety of studies, such as pre-treatment with proteases (Janíková et al., 2021, Wei et al., 2021a), Chelex® 100 (Yaren et al., 2021) or RNA extraction steps to inactivate inhibitory components of saliva (Garneret et al., 2021; Yamazaki et al., 2021), as it is less complex on the end-user.
The LoD of the unoptimized colorimetric assay in 5% saliva that has been processed using Pure·SAL™ is 1000 copies/reaction (reaction volume 25 μL), which corresponds to 800 copies/μL of patient saliva after accounting for dilution (Fig. S21). This LoD is several times worse than other LoDs found in the literature for other RT-LAMP based reactions on whole saliva (generally ~50 copies/reaction, but some reported values are as poor as 500 copies/reaction) and is several orders of magnitude higher than RT-PCR assays or other assays utilizing RNA extraction (on the order of 1 copy/reaction) (Oliveira et al., 2021). However, almost all of these studies are accompanied by pretreatment protocols and/or RNA extraction steps to achieve the reported LoD (Chomczynski et al., 2021; Ganguli et al., 2020; Lalli et al., 2021; Reynés et al., 2021; Yang et al., 2021). To improve this LoD, we investigated the use of RNase inhibitors (Lalli et al., 2021), Guanidine HCl (Zhang et al., 2020), and carrier DNA which had previously been reported to increase the sensitivity of RT-LAMP and RT-PCR (Hardinge and Murray, 2019; Kiddle et al., 2012). The addition of RNase inhibitors seems to worsen the LoD in 5% saliva (Fig. S20). This result directly contradicts literature reports where RT-LAMP assays in saliva utilize RNase inhibitors improve the LoD (Janíková et al., 2021; Wei et al., 2021b); this discrepancy may be due to the type of RNase inhibitor used. Both Guanidine HCl and carrier DNA improved the LoD (Fig. S18 and Fig. S19) and was added to our RT-LAMP reaction formulation for colorimetric solution reactions (Lalli et al., 2021; Wei et al., 2021b). We did not include these components in our paper RT-LAMP formulation, however, as they resulted in a color change when drying on paper (due to unexplained reasons). Finally, uracil-DNA glycosylase (UDG) and deoxyuridine triphosphate (dUTP; Fig. S22) were included to reduce carryover contamination (Hsieh et al., 2014; Tang et al., 2016; Wei et al., 2021b; Yaren et al., 2017; Zeng et al., 2020). When including Guanidine HCl, carrier DNA, and UDG, the LoD of our optimized RT-LAMP colorimetric assay in 5% processed saliva in solution improves to 250 copies/reaction (Fig. S23).
4.6. Design and optimization of paper-based device
Following optimization of the colorimetric RT-LAMP in solution, we transferred our assay onto paper. Paper is widely used in pH indicators and urine strips primarily due to the inexpensive cost, low technical complexity, and ease of production using roll-to-roll manufacturing (Yetisen et al., 2013). For our devices, we evaluated the use of several types of papers and selected chromatography paper (Fig. S7-S9). Chromatography paper is typically used in paper-based biosensors due to its improved wicking ability compared to other papers (Gutiérrez-Capitán et al., 2020; Singh et al., 2018; Verma et al., 2018). Most paper-based devices in literature use Grade 1 chromatography paper (Busa et al., 2016); however, due to the large area (5 mm × 20 mm) of the Grade 1 chromatography paper needed, we noticed that the distribution of solution across the paper was uneven. Thus, we selected a different type of chromatography paper, Grade 222 (0.83 mm thick), which is approximately 4.6 times thicker than Grade 1 (0.18 mm thick). Due to its increased thickness, the same volume of liquid can be loaded onto a 70% smaller reaction area by reducing the size of our device (5 mm × 6 mm), allowing for even distribution.
To reduce the complexity of our device, we dried the components of the RT-LAMP reaction (without the template) on the paper. Drying is one of the hallmark advantages of using paper-based devices as it allows for stable distribution of the device and easy operation without compromising diagnostic performance (Ratajczak and Stobiecka, 2020); the user simply adds the sample to rehydrate the reagents. Upon drying our reagents on paper, however, we noticed that the papers changed color from red to yellow over time without any template being present, indicating a decrease in the pH of the paper. Through a series of leave-one-out experiments, we were able to determine that ammonium sulfate was responsible for this change (Fig. S24). We speculate this color change could result from the oxidation of cellulose caused by heating and the oxidizing nature of ammonium sulfate or the acidification of reagents by degassing of ammonia from the RT-LAMP mixture. To prevent the color change in the absence of amplification, we replaced ammonium sulfate with betaine and increased the concentration of phenol red which acts as an antioxidant (Fig. S25). Literature reports vary concerning the effectiveness of betaine in LAMP reactions (Foo et al., 2020; Zhou et al., 2014); however, evidence exists indicating that betaine reduces oxidative damage, and we, therefore, included it in our formulation (Willingham et al., 2020). Trehalose has been reported to act as a protein stabilizer by resisting dehydration when drying (García-Bernalt Diego et al., 2019; Hayashida et al., 2015). Furthermore, bovine serum albumin (BSA) has been reported to act as a protein stabilizer when freeze-drying (Shimizu et al., 2017). As both trehalose and BSA have been shown to improve the performance of isothermal amplification methods (Mok et al., 2016), we added them to our formulation on paper (Fig. S25).
For our device, we elected to use two reaction zones; one targeting SARS-CoV-2 and one providing a no-primer control to determine the stability of our reagents on paper (which should not react with any sample). A schematic of our final device is shown in Fig. 1C and results from our device with and without heat-inactivated SARS-CoV-2 spiked into 5% saliva are shown in Fig. 1D. As can be seen in Fig. 1B, the LoD of our assay on paper (250 copies/reaction) is comparable to the LoD observed in solution for our optimized colorimetric RT-LAMP formulation (Fig. S23).
For the construction of our device, we used a Melinex® backing to provide structural support. Using a double-sided adhesive, we were able to attach two reaction pads to the backing without altering their pH. The number of reaction zones can be arbitrarily increased to allow for multiplexed detection without altering the design of our device. We found similar designs in literature where a solid substrate provided structural support to the reaction sites and utilized an adhesive in some cases (Choi et al., 2016; Reboud et al., 2019; Verma et al., 2018). Finally, we added 20 mil polystyrene spacers between reaction pads to provide a physical barrier inhibiting leakage from one reaction zone to an adjacent reaction zone, thus eliminating crosstalk during both reagent and sample addition. Typically for paper-based devices, the reaction zones are separated by hydrophobic barriers resulting from wax printing preventing sample from crossing (Altundemir et al., 2017; Carrilho et al., 2009; Martinez et al., 2007); however, to enable roll-to-roll manufacturing, we wanted to eliminate the usage of wax and thus settled on the incorporation of spacers (Kaarj et al., 2018).
4.7. Validation of contrived samples
To evaluate the analytical sensitivity and specificity of our assay on paper, we utilized contrived samples with heat-inactivated SARS-CoV-2 at multiples of the LoD for orf7ab.I to run our RT-LAMP assay on paper. We used a total of 30 positive samples and 30 corresponding negative samples which is the minimum number required for emergency use authorization (EUA) in the United States (Ravi et al., 2020). To determine the optimal colorimetric threshold to differentiate positive from negative reactions, we constructed an ROC curve by calculating the sensitivity and specificity at varying green channel intensity threshold values (Fig. 2). At the optimum threshold, our assay has the following analytical metrics: sensitivity of 97%, specificity of 100%, and accuracy of 98% (Fig. 2A and B). We found the difference between the positive and negative groups to be significantly different (p < 0.001). The paper strips were cut by hand and small differences in the size of the paper can cause differences in the colorimetric response. Therefore, we speculate large-scale fabrication and quality control will further improve the consistency within the two groups (positive and negative). This sensitivity is comparable to assays using RNA extracts (sensitivity ~95%), and better than that reported for crude samples where significant decreases in sensitivity (to ~80%) are common (Subsoontorn et al., 2020). Due to restrictions imposed by the Purdue University Institutional Biosafety Committee, we were unable to test samples obtained from COVID-19 positive and negative patients in a timely manner, and thus, we were unable to establish diagnostic sensitivity, specificity, and accuracy for our assay. We aim to perform this characterization in future studies.
4.8. Colorimetric interpretation of paper-based device
To observe the effect of color perception on the performance of our device, we surveyed four participants and asked them to interpret the results of our device. We provided each participant with a color bar and scans of our device after 60 min (Additional File 1) and asked them to classify the result as valid or invalid (using the control pad) and positive or negative based on the threshold marked on the color bar. The sensitivity and accuracy of our device decreased to 76% and 91%, respectively when we introduced user interpretation to our analysis, which is lower than other assays using crude samples (Fig. 1F). This low sensitivity stems from respondents identifying many positive reactions as invalid based on the control pad, leading to a false invalid rate of 25%, which may be attributed to contamination of the control pad with amplicons during the reaction. Additionally, user interpretation of the pads where regions of both yellow and red exist could introduce ambiguity, resulting in an increased false-positive rate or false invalid rate. Recent findings suggest this ambiguity is due to a third, intermediate color cluster (along with positive/negative clusters), that is not adequately addressed in colorimetric assays. (Aoki et al., 2021; de Oliveira Coelho et al., 2021). Since invalid results were discarded from further analysis, this elevated false invalid rate may artificially influence the specificity and accuracy metrics of our device.
5. Conclusion
We developed a platform capable of on-paper detection of SARS-CoV-2 from saliva using colorimetric reporters that produce responses visible to the naked eye. Our platform has the following eight advantages: i) it uses saliva, ii) it requires minimal operator training, iii) it can be fabricated using roll-to-roll methods to achieve millions of tests, iv) it performs similar to a RT-qPCR assay in terms of analytical sensitivity and specificity, v) it provides a colorimetric response visible to the naked eye, vi) it is amenable to point-of-care use, vii) it provides results in less than 60 min, and viii) it is estimated to cost ~$10/test.
Four limitations of the current approach are: i) LAMP-based assays are prone to false-positives due to the large amount of DNA product produced and carryover contamination in the form of aerosols and thus, the reaction needs to be sealed well externally to avoid cross-contamination, ii) an external heater is still needed for conducting the assay (although we envision that an integrated battery-operated heater could be developed for each test strip), iii) although red-yellow color transitions are vivid, variations in perception of color could affect interpretation of results and their validity (this could be overcome using a cellphone or camera to capture images and quantify color), and iv) sample needs to be added directly to each reaction zone (development and incorporation of a spreading layer would enable the user to add sample to a single site and the spreading layer would distribute the sample evenly to all reaction zones).
Due to the simplicity and scalability of this test, we envision that it could be used in a wide variety of settings, potentially including in-home diagnostics. Our platform can be readily reconfigured to target different pathogens simply by screening primer sets in solution and multiplexing is enabled by adding additional reaction sites to the device. The reconfigurable nature of our platform makes it an ideal tool for deployment and detection of emerging pathogens in future public health emergencies.
Funding sources
This work was funded partially by a contract with Raytheon BBN Technologies and by the Disease Diagnostics INventors Challenge created by the Purdue Institute of Inflammation, Immunology, and Infectious Diseases in partnership with the Department of Comparative Pathobiology.
CRediT authorship contribution statement
Josiah Levi Davidson: contributed the following, design and screening (using fluorometric methods) of all primers, Formal analysis, all in-silico analysis, Writing – original draft, (lead), writing – formatting, Writing – review & editing, (lead), Project administration, development and administration of saliva collection IRB protocol (lead), Methodology, (equal), and project coordination (support). Jiangshan Wang: contributed the following: development of paper-based devices (equal), optimization of formulation for RT-LAMP on paper (equal), characterization of paper-based device (lead), development and administration of colorimetric interpretation survey IRB protocol (lead), Writing – original draft, (support), and, Methodology, (equal). Murali Kannan Maruthamuthu: contributed the following: screening (colorimetric methods) of primers (equal), development and optimization of formulation for RT-LAMP in solution (lead), screening and selection of saliva processing and devices, and, Writing – original draft, (support). Andres Dextre: contributed the following: development of paper-based devices (equal), optimization of formulation for RT-LAMP on paper (equal), and, Writing – original draft, (support). Ana Pascual-Garrigos: contributed the following: screening of colorimetric indicators (equal) and, Writing – original draft, (support). Suraj Mohan: contributed the following: screening (colorimetric methods) of primers (equal), screening of colorimetric indicators (equal), Data curation, development of protocol for colorimetric data collection on BMG, troubleshooting the heating and sealing of RT-LAMP solutions reactions (equal), and, Writing – original draft, (support). Sai Venkata Sravan Putikam: contributed the characterization of heating devices. Fujr Osman Ibrahim Osman: contributed by troubleshooting the heating and sealing of RT-LAMP solution reactions (equal) and, Writing – original draft, (support). Darby McChesney: contributed by screening and selection of paper materials and pH indicators (equal). Jordan Seville: contributed by screening and selection paper materials and pH indicators (equal) and, Writing – review & editing, (support). Mohit S. Verma: contributed the following, Conceptualization, Writing – original draft, (support), writing – formatting, Writing – review & editing, (support), development of saliva collection IRB protocol (support), development of colorimetric interpretation survey IRB protocol (support), Project administration, project coordination (lead), and, Methodology, (lead).
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: M.S.V. has interests in Krishi LLC, a company interested in licensing the technology developed here. The work performed here was not funded by Krishi LLC. Employees of Raytheon BBN (sponsor) were involved in the conceptualization and analysis of the data presented here. The contents of this manuscript have been filed for provisional patent protection by Purdue University and Raytheon BBN.
Acknowledgments
We are grateful for collaborations with Raytheon BBN Technologies (Bryan Bartley, Aaron Adler, Allison Taggart, Jake Beal, Miles Rogers), PortaScience (Mike Gavin, Hanna Richman), LaDuca LLC (Frank LaDuca), and Cortex Design. We are also grateful to Jacqueline Linnes from Purdue University for her help with troubleshooting. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, Heat-inactivated, NR-52286. Other reagents obtained from BEI are located in Table S7 and Table S8.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biosx.2021.100076.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ahn S.J., Baek Y.H., Lloren K.K.S., Choi W.-S., Jeong J.H., Antigua K.J.C., Kwon H., Park S.-J., Kim E.-H., Kim Y., Si Y.-J., Hong S.B., Shin K.S., Chun S., Choi Y.K., Song M.-S. Rapid and simple colorimetric detection of multiple influenza viruses infecting humans using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. BMC Infect. Dis. 2019;19:676. doi: 10.1186/s12879-019-4277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altundemir S., Uguz A.K., Ulgen K. A review on wax printed microfluidic paper-based devices for international health. Biomicrofluidics. 2017;11 doi: 10.1063/1.4991504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki M.N., de Oliveira Coelho B., Góes L.G.B., Minoprio P., Durigon E.L., Morello L.G., Marchini F.K., Riediger I.N., do Carmo Debur M., Nakaya H.I., Blanes L. Colorimetric RT-LAMP SARS-CoV-2 diagnostic sensitivity relies on color interpretation and viral load. Sci. Rep. 2021;11:9026. doi: 10.1038/s41598-021-88506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busa L.S.A., Mohammadi S., Maeki M., Ishida A., Tani H., Tokeshi M. Advances in microfluidic paper-based analytical devices for food and water analysis. Micromachines. 2016;7:86. doi: 10.3390/mi7050086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrilho E., Martinez A.W., Whitesides G.M. Understanding wax printing: a simple micropatterning process for paper-based microfluidics. Anal. Chem. 2009;81:7091–7095. doi: 10.1021/ac901071p. [DOI] [PubMed] [Google Scholar]
- Choi J.R., Hu J., Gong Y., Feng S., Abas W.A.B.W., Pingguan-Murphy B., Xu F. An integrated lateral flow assay for effective DNA amplification and detection at the point of care. Analyst. 2016;141:2930–2939. doi: 10.1039/C5AN02532J. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Chomczynski P.W., Kennedy A., Rymaszewski M., Wilfinger W.W., Heiny J.A., Mackey K. Rapid processing of SARS-CoV-2 containing specimens for direct RT-PCR. PloS One. 2021;16 doi: 10.1371/journal.pone.0246867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler D.M., Summers L.H. The COVID-19 pandemic and the $16 trillion virus. J. Am. Med. Assoc. 2020;324:1495. doi: 10.1001/jama.2020.19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalirirad S., Han D., Steckl A.J. Aptamer-based lateral flow biosensor for rapid detection of salivary cortisol. ACS Omega. 2020;5:32890–32898. doi: 10.1021/acsomega.0c03223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Coelho B., Sanchuki H.B.S., Zanette D.L., Nardin J.M., Morales H.M.P., Fornazari B., Aoki M.N., Blanes L. Essential properties and pitfalls of colorimetric Reverse Transcription Loop-mediated Isothermal Amplification as a point-of-care test for SARS-CoV-2 diagnosis. Mol. Med. 2021;27:30. doi: 10.1186/s10020-021-00289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA Emergency use authorization (EUA) summary for the ambry COVID-19 RT-PCR test. 2021. https://www.fda.gov/media/145410/download
- Foo P.C., Nurul Najian A.B., Muhamad N.A., Ahamad M., Mohamed M., Yean Yean C., Lim B.H. Loop-mediated isothermal amplification (LAMP) reaction as viable PCR substitute for diagnostic applications: a comparative analysis study of LAMP, conventional PCR, nested PCR (nPCR) and real-time PCR (qPCR) based on Entamoeba histolytica DNA derived from faecal sample. BMC Biotechnol. 2020;20:34. doi: 10.1186/s12896-020-00629-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F., Ramirez S.A.S. de, Valera E., Cunningham B.T., King W.P., Bashir R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Bernalt Diego J., Fernández-Soto P., Crego-Vicente B., Alonso-Castrillejo S., Febrer-Sendra B., Gómez-Sánchez A., Vicente B., López-Abán J., Muro A. Progress in loop-mediated isothermal amplification assay for detection of Schistosoma mansoni DNA: towards a ready-to-use test. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-51342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneret P., Coz E., Martin E., Manuguerra J.-C., Brient-Litzler E., Enouf V., Obando D.F.G., Olivo-Marin J.-C., Monti F., van der Werf S., Vanhomwegen J., Tabeling P. Performing point-of-care molecular testing for SARS-CoV-2 with RNA extraction and isothermal amplification. PloS One. 2021;16 doi: 10.1371/journal.pone.0243712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Capitán M., Baldi A., Fernández-Sánchez C. Electrochemical paper-based biosensor devices for rapid detection of biomarkers. Sensors. 2020;20 doi: 10.3390/s20040967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardinge P., Murray J.A.H. Reduced false positives and improved reporting of loop-mediated isothermal amplification using quenched fluorescent primers. Sci. Rep. 2019;9:7400. doi: 10.1038/s41598-019-43817-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashida K., Kajino K., Hachaambwa L., Namangala B., Sugimoto C. Direct blood dry LAMP: a rapid, stable, and easy diagnostic tool for human african trypanosomiasis. PLoS Neglected Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K., Mage P.L., Csordas A.T., Eisenstein M., Soh H.T. Simultaneous elimination of carryover contamination and detection of DNA with uracil-DNA-glycosylase-supplemented loop-mediated isothermal amplification (UDG-LAMP) Chem. Commun. 2014;50:3747–3749. doi: 10.1039/C4CC00540F. [DOI] [PubMed] [Google Scholar]
- Huang W.E., Lim B., Hsu C.-C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M., Higashimoto Y., Toyama Y., Horiguchi T., Hibino M., Iwata M., Imaizumi K., Doi Y. Diagnostic accuracy of LAMP versus PCR over the course of SARS-CoV-2 infection. Int. J. Infect. Dis. 2021 doi: 10.1016/j.ijid.2021.04.018. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janíková M., Hodosy J., Boor P., Klempa B., Celec P. Loop‐mediated isothermal amplification for the detection of SARS‐CoV‐2 in saliva. Microb Biotechnol. 2021;14:307–316. doi: 10.1111/1751-7915.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Kim N.-G., Woolley A.E., Segel M., Barretto R.P.J., Ranu A., Macrae R.K., Faure G., Ioannidi E.I., Krajeski R.N., Bruneau R., Huang M.-L.W., Yu X.G., Li J.Z., Walker B.D., Hung D.T., Greninger A.L., Jerome K.R., Gootenberg J.S., Abudayyeh O.O., Zhang F. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. 2020;383:1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaarj K., Akarapipad P., Yoon J.-Y. Simpler, faster, and sensitive Zika virus assay using smartphone detection of loop-mediated isothermal amplification on paper microfluidic chips. Sci. Rep. 2018;8:12438. doi: 10.1038/s41598-018-30797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurshid Z., Moin S.F., Khan R.S., Agwan M.A.S., Alwadaani A.H., Zafar M.S. Human salivary protein extraction from RNAPro·SALTM, Pure·SALTM, and passive drooling method. Eur. J. Dermatol. 2017;11:385–389. doi: 10.4103/ejd.ejd_183_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiddle G., Hardinge P., Buttigieg N., Gandelman O., Pereira C., McElgunn C.J., Rizzoli M., Jackson R., Appleton N., Moore C., Tisi L.C., Murray J.A. GMO detection using a bioluminescent real time reporter (BART) of loop mediated isothermal amplification (LAMP) suitable for field use. BMC Biotechnol. 2012;12:15. doi: 10.1186/1472-6750-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli M.A., Langmade J.S., Chen X., Fronick C.C., Sawyer C.S., Burcea L.C., Wilkinson M.N., Fulton R.S., Heinz M., Buchser W.J., Head R.D., Mitra R.D., Milbrandt J. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin. Chem. 2021;67:415–424. doi: 10.1093/clinchem/hvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Wu X., Wan Z., Li Y., Jin X., Zhang C. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Helgouach N., Champigneux P., Schneider F.S., Molina L., Espeut J., Alali M., Baptiste J., Cardeur L., Dubuc B., Foulongne V., Galtier F., Makinson A., Marin G., Picot M.-C., Prieux-Lejeune A., Quenot M., Robles F.C., Salvetat N., Vetter D., Reynes J., Molina F. EasyCOV : LAMP based rapid detection of SARS-CoV-2 in saliva. medRxiv. 2020 doi: 10.1101/2020.05.30.20117291. 2020.05.30.20117291. [DOI] [Google Scholar]
- Martinez A.W., Phillips S.T., Butte M.J., Whitesides G.M. Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew. Chem. Int. Ed. 2007;46:1318–1320. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S., Pascual-Garrigos A., Brouwer H., Pillai D., Koziol J., Ault A., Schoonmaker J., Johnson T., Verma M.S. Loop-mediated isothermal amplification for the detection of pasteurella multocida, mannheimia haemolytica, and Histophilus somni in bovine nasal samples. ACS Agric. Sci. Technol. 2021;1:100–108. doi: 10.1021/acsagscitech.0c00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok E., Wee E., Wang Y., Trau M. Comprehensive evaluation of molecular enhancers of the isothermal exponential amplification reaction. Sci. Rep. 2016;6 doi: 10.1038/srep37837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira K.G. de, Estrela P.F.N., Mendes G. de M., Santos C.A. dos, Silveira-Lacerda E. de P., Duarte G.R.M. Rapid molecular diagnostics of COVID-19 by RT-LAMP in a centrifugal polystyrene-toner based microdevice with end-point visual detection. Analyst. 2021;146:1178–1187. doi: 10.1039/D0AN02066D. [DOI] [PubMed] [Google Scholar]
- Parupudi T., Panchagnula N., Muthukumar S., Prasad S. Evidence-based point-of-care technology development during the COVID-19 pandemic. Biotechniques. 2021;70:58–67. doi: 10.2144/btn-2020-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak K., Stobiecka M. High-performance modified cellulose paper-based biosensors for medical diagnostics and early cancer screening: a concise review. Carbohydr. Polym. 2020;229 doi: 10.1016/j.carbpol.2019.115463. [DOI] [PubMed] [Google Scholar]
- Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboud J., Xu G., Garrett A., Adriko M., Yang Z., Tukahebwa E.M., Rowell C., Cooper J.M. Paper-based microfluidics for DNA diagnostics of malaria in low resource underserved rural communities. Proc. Natl. Acad. Sci. Unit. States Am. 2019;116:4834–4842. doi: 10.1073/pnas.1812296116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynés B., Serra F., Palou A. Rapid visual detection of SARS-CoV-2 by colorimetric loop-mediated isothermal amplification. Biotechniques. 2021;70:218–225. doi: 10.2144/btn-2020-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoenix, Inc Rheonix COVID-19TM MDx assay: instructions for use. 2021. https://www.fda.gov/media/137489/download
- Rodriguez-Manzano J., Karymov M.A., Begolo S., Selck D.A., Zhukov D.V., Jue E., Ismagilov R.F. Reading out single-molecule digital RNA and DNA isothermal amplification in nanoliter volumes with unmodified camera phones. ACS Nano. 2016;10:3102–3113. doi: 10.1021/acsnano.5b07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Mohd-Naim N.F., Safavieh M., Ahmed M.U. Colorimetric nucleic acid detection on paper microchip using loop mediated isothermal amplification and crystal violet dye. ACS Sens. 2017;2:1713–1720. doi: 10.1021/acssensors.7b00671. [DOI] [PubMed] [Google Scholar]
- Sa H., A W., N A., S A., Mf I., Z K., Sah B. Effect of sodium bicarbonate mouth wash on salivary pH and interleukin-1β levels among smokers. Eur. J. Dermatol. 2020;14:260–267. doi: 10.1055/s-0040-1709896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Korehisa T., Imanaka H., Ishida N., Imamura K. Characteristics of proteinaceous additives in stabilizing enzymes during freeze-thawing and -drying. Biosci. Biotechnol. Biochem. 2017;81:687–697. doi: 10.1080/09168451.2016.1274637. [DOI] [PubMed] [Google Scholar]
- Singh A.T., Lantigua D., Meka A., Taing S., Pandher M., Camci-Unal G. Paper-based sensors: emerging themes and applications. Sensors. 2018;18:2838. doi: 10.3390/s18092838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subsoontorn P., Lohitnavy M., Kongkaew C. The diagnostic accuracy of isothermal nucleic acid point-of-care tests for human coronaviruses: a systematic review and meta-analysis. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-79237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Chen H., Diao Y. Advanced uracil DNA glycosylase-supplemented real-time reverse transcription loop-mediated isothermal amplification (UDG-rRT-LAMP) method for universal and specific detection of Tembusu virus. Sci. Rep. 2016;6 doi: 10.1038/srep27605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N.A., Zhang Y., Evans T.C. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques. 2015;58:59–68. doi: 10.2144/000114253. [DOI] [PubMed] [Google Scholar]
- Thi V.L.D., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Miranda I.B., Schnitzler P., Kräusslich H.-G., Knop M., Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromberg B.J., Schwetz T.A., Pérez-Stable E.J., Hodes R.J., Woychik R.P., Bright R.A., Fleurence R.L., Collins F.S. Rapid scaling up of covid-19 diagnostic testing in the United States — the NIH RADx initiative. N. Engl. J. Med. 2020 doi: 10.1056/NEJMsr2022263. 0, null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe-Alvarez C., Lam Q., Baldwin D.A., Chernoff J. Low saliva pH can yield false positives results in simple RT-LAMP-based SARS-CoV-2 diagnostic tests. PloS One. 2021;16 doi: 10.1371/journal.pone.0250202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M.S., Tsaloglou M.-N., Sisley T., Christodouleas D., Chen A., Milette J., Whitesides G.M. Sliding-strip microfluidic device enables ELISA on paper. Biosens. Bioelectron. 2018;99:77–84. doi: 10.1016/j.bios.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C.B.F., Watkins A.E., Harden C.A., Brackney D.E., Shafer J., Wang J., Caraballo C., Kalinich C.C., Ott I.M., Fauver J.R., Kudo E., Lu P., Venkataraman A., Tokuyama M., Moore A.J., Muenker M.C., Casanovas-Massana A., Fournier J., Bermejo S., Campbell M., Datta R., Nelson A., Anastasio K., Askenase M.H., Batsu M., Bickerton S., Brower K., Bucklin M.L., Cahill S., Cao Y., Courchaine E., DeIuliis G., Earnest R., Geng B., Goldman-Israelow B., Handoko R., Khoury-Hanold W., Kim D., Knaggs L., Kuang M., Lapidus S., Lim J., Linehan M., Lu-Culligan A., Martin A., Matos I., McDonald D., Minasyan M., Nakahata M., Naushad N., Nouws J., Obaid A., Odio C., Oh J.E., Omer S., Park A., Park H.-J., Peng X., Petrone M., Prophet S., Rice T., Rose K.-A., Sewanan L., Sharma L., Shaw A.C., Shepard D., Smolgovsky M., Sonnert N., Strong Y., Todeasa C., Valdez J., Velazquez S., Vijayakumar P., White E.B., Yang Y., Dela Cruz C.S., Ko A.I., Iwasaki A., Krumholz H.M., Matheus J.D., Hui P., Liu C., Farhadian S.F., Sikka R., Wyllie A.L., Grubaugh N.D. SalivaDirect: a simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med. Plus. 2021;2:263–280. doi: 10.1016/j.medj.2020.12.010. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekly epidemiological update. 2020. https://www.who.int/publications/m/item/weekly-epidemiological-update---29-december-2020 29 December 2020 [WWW Document] (accessed 2.28.21)
- Wei S., Kohl E., Djandji A., Morgan S., Whittier S., Mansukhani M., Hod E., D'Alton M., Suh Y., Williams Z. Direct diagnostic testing of SARS-CoV-2 without the need for prior RNA extraction. Sci. Rep. 2021;11:2402. doi: 10.1038/s41598-021-81487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S., Suryawanshi H., Djandji A., Kohl E., Morgan S., Hod E.A., Whittier S., Roth K., Yeh R., Alejaldre J.C., Fleck E., Ferrara S., Hercz D., Andrews D., Lee L., Hendershot K.A., Goldstein J., Suh Y., Mansukhani M., Williams Z. Field-deployable, rapid diagnostic testing of saliva for SARS-CoV-2. Sci. Rep. 2021;11:5448. doi: 10.1038/s41598-021-84792-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham B.D., Ragland T.J., Ormsbee M.J. Betaine supplementation may improve heat tolerance: potential mechanisms in humans. Nutrients. 2020;12:2939. doi: 10.3390/nu12102939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki W., Matsumura Y., Thongchankaew-Seo U., Yamazaki Y., Nagao M. Development of a point-of-care test to detect SARS-CoV-2 from saliva which combines a simple RNA extraction method with colorimetric reverse transcription loop-mediated isothermal amplification detection. J. Clin. Virol. 2021;136 doi: 10.1016/j.jcv.2021.104760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Meyerson N.R., Clark S.K., Paige C.L., Fattor W.T., Gilchrist A.R., Barbachano-Guerrero A., Healy B.G., Worden-Sapper E.R., Wu S.S., Muhlrad D., Decker C.J., Saldi T.K., Lasda E., Gonzales P., Fink M.R., Tat K.L., Hager C.R., Davis J.C., Ozeroff C.D., Brisson G.R., McQueen M.B., Leinwand L.A., Parker R., Sawyer S.L. Saliva TwoStep for rapid detection of asymptomatic SARS-CoV-2 carriers. eLife. 2021;10 doi: 10.7554/eLife.65113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaren O., Alto B.W., Gangodkar P.V., Ranade S.R., Patil K.N., Bradley K.M., Yang Z., Phadke N., Benner S.A. Point of sampling detection of Zika virus within a multiplexed kit capable of detecting dengue and chikungunya. BMC Infect. Dis. 2017;17:293. doi: 10.1186/s12879-017-2382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaren O., McCarter J., Phadke N., Bradley K.M., Overton B., Yang Z., Ranade S., Patil K., Bangale R., Benner S.A. Ultra-rapid detection of SARS-CoV-2 in public workspace environments. PloS One. 2021;16 doi: 10.1371/journal.pone.0240524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yetisen A.K., Akram M.S., Lowe C.R. Paper-based microfluidic point-of-care diagnostic devices. Lab Chip. 2013;13:2210–2251. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- Yüce M., Filiztekin E., Özkaya K.G. COVID-19 diagnosis -A review of current methods. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Liu M., Xia Y., Jiang X. Uracil-DNA-glycosylase-assisted loop-mediated isothermal amplification for detection of bacteria from urine samples with reduced contamination. Analyst. 2020;145:7048–7055. doi: 10.1039/D0AN01001D. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ren G., Buss J., Barry A.J., Patton G.C., Tanner N.A. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques. 2020;69:178–185. doi: 10.2144/btn-2020-0078. [DOI] [PubMed] [Google Scholar]
- Zhou D., Guo J., Xu L., Gao S., Lin Q., Wu Q., Wu L., Que Y. Establishment and application of a loop-mediated isothermal amplification (LAMP) system for detection of cry1Ac transgenic sugarcane. Sci. Rep. 2014;4 doi: 10.1038/srep04912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.