Significance
Toxin–antitoxin (TA) systems are proposed to protect bacteria from deadly stresses encountered in their environmental niches. Yet clear examples supporting this hypothesis are lacking. The abundant Mycobacterium tuberculosis (Mtb) TA systems are implicated in enabling this pathogen to evade killing by our immune system and persist inside macrophages as a latent tuberculosis infection. We used a multiomics approach to track the activity of one TA system toxin, VapC4, and the series of molecular events it triggers. VapC4 first depletes tRNACys to mimic a state of Cys starvation. This reprograms Mtb metabolism to specifically defend against oxidative and copper stresses. Thus, VapC4 plays a direct and vital role in mediating these critical stress pathways essential for Mtb viability during infection.
Keywords: RNA-seq, mycothiol, protein translation, mass spectrometry, sulfur assimilation
Abstract
The Mycobacterium tuberculosis (Mtb) VapBC4 toxin–antitoxin system is essential for the establishment of Mtb infection. Using a multitier, systems-level approach, we uncovered the sequential molecular events triggered by the VapC4 toxin that activate a circumscribed set of critical stress survival pathways which undoubtedly underlie Mtb virulence. VapC4 exclusively inactivated the sole transfer RNACys (tRNACys) through cleavage at a single site within the anticodon sequence. Depletion of the pool of tRNACys led to ribosome stalling at Cys codons within actively translating messenger RNAs. Genome mapping of these Cys-stalled ribosomes unexpectedly uncovered several unannotated Cys-containing open reading frames (ORFs). Four of these are small ORFs (sORFs) encoding Cys-rich proteins of fewer than 50 amino acids that function as Cys-responsive attenuators that engage ribosome stalling at tracts of Cys codons to control translation of downstream genes. Thus, VapC4 mimics a state of Cys starvation, which then activates Cys attenuation at sORFs to globally redirect metabolism toward the synthesis of free Cys. The resulting newly enriched pool of Cys feeds into the synthesis of mycothiol, the glutathione counterpart in this pathogen that is responsible for maintaining cellular redox homeostasis during oxidative stress, as well as into a circumscribed subset of cellular pathways that enable cells to defend against oxidative and copper stresses characteristically endured by Mtb within macrophages. Our ability to pinpoint activation or down-regulation of pathways that collectively align with Mtb virulence–associated stress responses and the nonreplicating persistent state brings to light a direct and vital role for the VapC4 toxin in mediating these critical pathways.
The vast majority of people infected with Mycobacterium tuberculosis (Mtb) do not develop active tuberculosis (TB). Instead, in most infected individuals, this bacterial pathogen reprograms its physiology to evade immune clearance and establish a latent TB infection. The Mtb genome harbors ∼90 Type II toxin–antitoxin (TA) systems (1, 2), the majority belonging to the 50-member virulence-associated protein VapBC family (VapB: antitoxin, VapC: toxin) unique to pathogenic bacteria. In general, Type II TA systems comprise adjacent genes encoding two small (∼10 kDa) proteins, a toxin and its cognate antitoxin that inhibits toxin activity through the formation of a stable TA protein–protein complex. In the prevailing model, stress conditions alter the toxin–antitoxin balance, resulting in a preponderance of free toxin, which exerts its growth regulating and/or other functions within the bacterial cells (3, 4). Because the activity of individual Mtb toxins typically leads to growth inhibition, they have been implicated in the establishment of latent TB infection (1–3, 5, 6). Consistent with this, a subset of the 50 VapBC TA systems are differentially expressed when subjected to stresses relevant to Mtb infection and nonrepicating persistence TB (7). More importantly, TA module deletion strains used for guinea pig and mouse infection models demonstrated that VapBC3, VapBC4, and VapBC22 are essential to establish Mtb infections (7, 8). VapC4, VapB5, VapC20, VapC26, and VapC45 are required for Mtb survival in macrophages and the lungs of mice and nonhuman primates (9).
VapC toxins are structure- and sequence-specific endoribonucleases (10). Only a few of the 50 Mtb VapC toxins have been studied in detail. In vitro cleavage studies demonstrated that VapC4 (also known as VapC-mt4, Rv0595c) and VapC11 (also known as VapC-mt11, Rv1561) require a cleavage consensus sequence and specifically target transfer RNA (tRNA) (11–13). Other Mtb VapC toxins with apparent tRNase activity were identified using an RNA–VapC interaction screen in a Mycobacterium smegmatis host (14). However, only recently did we learn that accurate tRNA target identification requires in vivo cleavage in the matched mycobacterial host cell (11).
Here, we applied physiologically relevant, low-level toxin expression studies in tandem with powerful genome-scale approaches to elucidate the mechanism underlying VapBC4 virulence. VapC4 cleaves and inactivates a single tRNACys in vivo. Its activity leads to widespread ribosome stalling at Cys codons, which enabled the discovery of unnannotated Cys codon-containing genes. We demonstrate that some are orthologous to small open reading frames (sORFs) that act as Cys-responsive attenuators as recently described in M. smegmatis (15). Ribosome stalling also unleashes a cascade of downstream events that precisely manipulates the transcriptome based on codon usage to down-regulate growth and redirect metabolism toward the synthesis of Cys, which in turn selectively activates genes involved in the oxidative stress and copper detoxification responses, endowing Mtb with the molecular defenses needed to withstand the deadly assaults of the host innate immune response.
Results
VapC4 Cleaves Many Mtb tRNAs within Their Anticodon Stem Loop In Vitro.
VapC4 expression in Mtb leads to growth arrest (12). To understand how this toxin down-regulates cell growth, we first determined how many Mtb tRNAs were cleaved by VapC4 in vitro. Eight of the complete set of 45 tRNAs (16, 17) were cleaved to completion: tRNAAla2UGC, tRNAAla11CGC, tRNAAla31GGC, tRNACys21GCA, tRNAGly20GCC, tRNAPhe10GAA, tRNASer24GGA, and tRNASer26GCU (SI Appendix, Fig. S1 A and B, shaded black). Surprisingly, five of these eight Mtb in vitro tRNA targets did not harbor an ACGC, ACUGC, or ACAGC VapC4 cleavage sequence identified in earlier reports (12, 13). In addition, 19 of the 45 tRNAs were partially cleaved (SI Appendix, Fig. S1 A and B, shaded gray). Therefore, 27 of the 45 tRNAs were fully or partially cleaved by VapC4. This data suggested that the growth inhibition associated with VapC4 toxin expression might be due to a depletion of multiple tRNAs.
VapC4 Does Not Completely Inhibit Protein Synthesis.
Since VapC4 appeared to disable so many tRNAs, we next tested if its role is to comprehensively shut down translation. In fact, others have concluded that tRNA-cleaving toxins generally act by globally inhibiting translation because their expression characteristically leads to bacterial cell growth arrest (14, 18). We used a click chemistry approach for metabolic labeling to monitor de novo protein synthesis in Mtb cells with and without toxin expression. The incorporation of the azide-containing Met mimetic azidohomoalanine (AHA) enabled fluorescent visualization of AHA-containing proteins upon coupling to the alkyne tetramethylrhodamine (TAMRA). Total proteins were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the newly synthesized population was visualized by fluorescent imaging (Fig. 1 A and B). Surprisingly, VapC4 did not completely inhibit protein synthesis even though its expression leads to growth arrest (Fig. 1C).
Fig. 1.
VapC4 inhibits growth but does not completely inhibit translation. (A) Newly synthesized (AHA-labeled) proteins from ±VapC4 Mtb H37Rv cells at 12, 24, and 48 h postinduction were visualized with an alkyne-TAMRA conjugate. Each lane was normalized to contain 10 μg total protein. (B) Whole lane fluorescence was measured by ImageJ software selecting equal areas. The average fluorescence signals from triplicates are shown. Error bars correspond to SEM. (C) Growth profiles obtained from Mtb H37Rv cultures with (blue) or without (orange) VapC4 expression for 0, 24, 48, and 72 h. Dotted lines are OD600 readings for each replicate, and solid lines represent the average of the triplicates. Error bars represent the SEM.
VapC4 Only Targets tRNACys in Mtb Cells.
Because the effects on translation were not consistent with the large number of tRNAs cleaved by VapC4 in vitro, we next expressed this toxin in Mtb to determine if the tRNAs cleaved by VapC4 in vitro were also targeted in vivo. RNAs cleaved by VapC toxins are marked with a specific chemical tag at their 5′ end that permits facile isolation, identification, and mapping of the toxin cleavage site. Therefore, we enlisted a specialized RNA sequencing (RNA-seq) approach developed in our laboratory, 5′ RNA-seq, that enabled genome-wide VapC4 target identification as well as high-resolution, single-nucleotide (nt) cleavage site mapping (19). The 5′ RNA-seq method captures and amplifies RNAs carrying the specific chemical moieties, a 5′-phosphate (5′-P) or 5′-hydroxyl (5′-OH), that are generated upon cleavage by VapC4 and other endoribonuclease toxins.
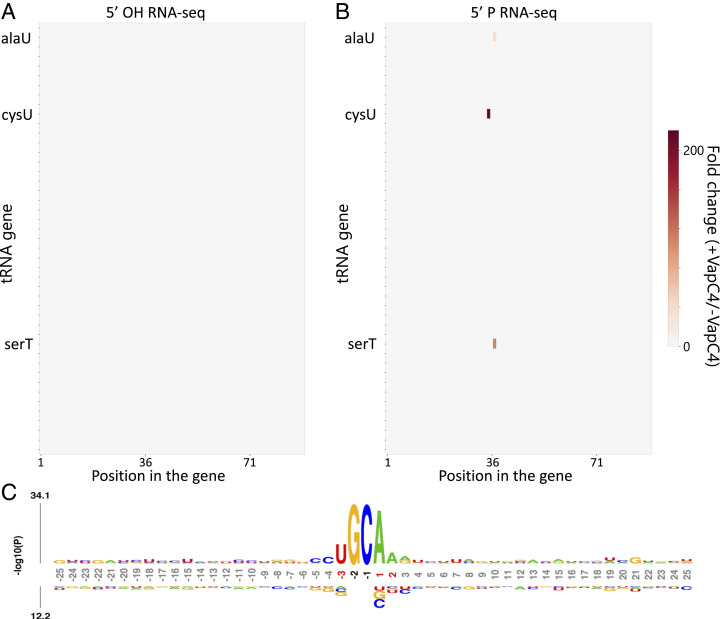
Total RNA was isolated from VapC4 induced and uninduced cells. We created 5′-P and 5′-OH RNA-seq libraries and identified a single tRNA, tRNACysGCA, as the sole target of VapC4 (Fig. 2 A and B). This is the only Cys tRNA in Mtb, and it services both the UGC and the UGU Cys codons. The site of toxin cleavage was within the Cys anticodon (Fig. 2C), therefore functionally disabling this tRNA for protein synthesis. This tRNACysGCA target is also consistent with that identified upon ectopic expression of VapC4 in M. smegmatis (14). Interestingly, in contrast to the MazF family of toxins, which generate a 5′-OH upon RNA cleavage, VapC4 instead appeared to generate a 5′-P upon RNA cleavage since there were no tRNA targets detected in the 5′-OH RNA-seq libraries. To prove that VapC4 directly generated a 5′-P upon RNA cleavage, instead of downstream phosphorylation of a 5′ end that was initially hydroxylated, we incubated VapC4 with total Mtb RNA and again created 5′-P and 5′-OH RNA-seq libraries. tRNACysGCA was cleaved in the 5′-P library and not the 5′-OH library (Fig. 3 A and B and Dataset S1). This toxin has an absolute requirement for the sequence U G34 C35 A36 A in the proper structural context to recognize and cleave only tRNACysGCA (Fig. 3C). Therefore, VapC4, and likely all VapC family toxins, are distinct from other ribonuclease toxins in that they generate 5′-P ends upon RNA cleavage.
Fig. 2.
tRNACysGCA is the sole VapC4 target in vivo. (A) Heat map obtained from 5′-P RNA-seq libraries (represented in fold change of internal 5′ monophosphate ends in induced versus uninduced samples) indicating internal cleavage at a single position in only one (cysU) of the Mtb 45 tRNA genes 24 h post VapC4 induction in Mtb H37Rv cells. (B) Bar graph showing the fold change of 5′ monophosphate ends (induced versus uninduced) in the only Cys tRNA gene after 24 h of VapC4 induction. (C) Representation of tRNACysGCA (D, D-loop; T, TψC loop; ASL, anticodon stem loop). Cleavage site, yellow arrow. Anticodon positions are numbered according to standardized tRNA guidelines.
Fig. 3.
VapC4 cleavage generates 5′ monophosphate ends and lacks specificity in in vitro assays. Heat map showing fold changes obtained in 5′ RNA-seq for internal 5′-OH (A) or 5′-P (B) ends in all 45 tRNA genes after incubating purified VapC4 with total RNA extracted from Mtb mc2 6206. (C) Probability logo (obtained using kpLogo) showing the consensus sequence observed in 100 RNA hits with highest fold change (induced versus uninduced) in the in vitro cleavage 5′-P libraries. The RNA sequence flanking the cleavage site (25 nucleotides up- and downstream) is shown. Nucleotide positions (shown below the kpLogo) are numbered relative to the cleavage site and colored red if the nucleotide is statistically enriched at the position or black if the nucleotide is statistically enriched and its frequency is above 75%.
tRNACysGCA Cleavage Leads to Ribosome Stalling at Cys Codons.
We recently reported that MazF-mt9 toxin-mediated cleavage of a single species of tRNA, tRNALysUUU, leads to selective ribosome stalling at the Lys codon (AAA) requiring that depleted tRNA (20). This highly specific genome-wide ribosome stalling leads to preferential depletion of proteins containing the Lys AAA codon. Serendipitously, these ribosome stalling events were first suggested within the MazF-mt9 5′-OH RNA-seq dataset and then rigorously proven using ribosome profiling (Ribo-seq) (20). In perfect agreement, we documented the same two effects for VapC4 following cleavage of tRNACysGCA.
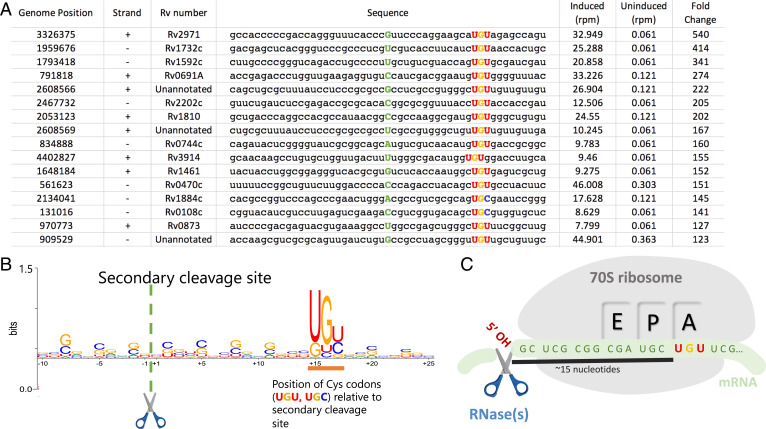
First, within our VapC4 5′-OH RNA-seq datasets, we saw the hallmarks of ribosome stalling at several hundred Mtb transcripts. Each stalling event is readily detected because there is a conspicuous ∼15 nt distance in this 5′-OH RNA-seq dataset to an in-frame “hungry” Cys codon requiring the depleted tRNA (Fig. 4 A and B). In Barth et al., we used Ribo-seq to demonstrate that this 15-nt distance corresponds to the footprint of the stalled ribosome spanning its 5′ edge to the position of the hungry codon at the A-site (Fig. 4C) (20). We fortuitously detect these stalled ribosomes because some transcripts that harbor them are recycled by an RNase distinct from VapC4 that happens to leave a 5′OH upon cleavage. Second, consistent with our earlier study, we found that the proteome in VapC4-expressing cells contained fewer Cys-containing proteins relative to control cells (SI Appendix, Fig. S2). Therefore, VapC4 expression leads to inactivation of the primary target of the toxin, tRNACysGCA, followed by ribosome stalling at Cys codons because the only Cys tRNA in Mtb is now in deficit. These events impact the proteome, resulting in an overall reduction in the steady-state level of Cys-containing proteins.
Fig. 4.
The 5′-OH RNA-seq method reveals ribosome stalling at Cys codons with single nucleotide resolution. (A) Top 16 mRNA hits in 5′-OH RNA-seq libraries constructed from Mtb mc2 6206 RNA extracted after 24 h of VapC4 induction. Cysteine codons are highlighted, ∼15 nucleotides downstream of the 5′-OH cleavage site (5′ of the green capitalized letter). The genome position and strand where the secondary cleavage occurs is shown as well as the Rv number of the gene containing the Cys codon. (B) Web logo showing the consensus sequence from the top 100 mRNA hits found by 5′-OH RNA-seq. Positions are numbered relative to 5′-OH cleavage site (scissor). Cysteine codons are predominantly positioned at +15 to +17 (orange underline). (C) Following VapC4-mediated cleavage of tRNACys, ribosome stalling occurs when a transcript containing a Cys codon (UGU or UGC) reaches the A-site because the pool of tRNACys is depleted by VapC4 cleavage. The 5’-OH cleavage site (scissor) represents a secondary cleavage event on the mRNA by an unknown RNase that generates a 5′-OH, enabling accurate detection Cys stalling events within our 5′-OH RNA-seq dataset.
An Mtb sORF Is the Ortholog of a Cys-Responsive Attenuator in M. smegmatis.
Our ability to definitively map the presence and precise position of stalled ribosomes on messenger RNAs (mRNAs) within the VapC4 5′-OH RNA-seq dataset was exploited as a powerful tool to identify transcripts actively undergoing translation and whose expression is thus impacted by these stalling events. We detected hundreds of stalled ribosomes on mRNAs, corresponding to 444 distinct transcripts within the VapC4 5′-OH RNA-seq dataset (Dataset S2). The ribosomes were stalled at either of the two Cys codons, UGC and the UGU, serviced by the single Cys tRNA (tRNACysGCA) in Mtb (Fig. 4 A and B).
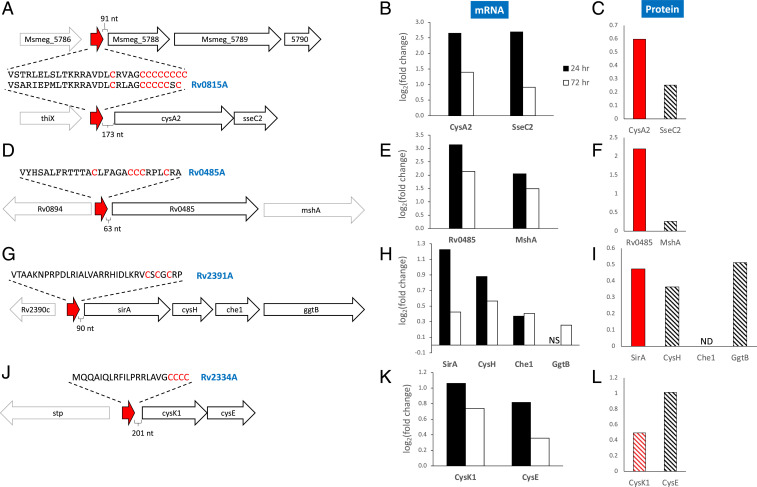
In addition, we identified four leaderless sORFs (lacking a 5′ untranslated region) whose genome location and sequence revealed compelling functional clues. One of the four sORFs encoded a 29 amino acid small protein with an extremely Cys-rich carboxy terminus: VSARIEPMLTKRRAVDLCRLAGCCCCCSC. We discovered that it is the ortholog of the M. smegmatis sORF (Ms5788A) first described as one of the many unannotated small leaderless transcripts that are unusually common in mycobacteria (Fig. 5A) (21). Moreover, recent follow-up studies on the function of Ms5788A support its role in a novel mode of Cys attenuation (15). Ms5788A is so named because it exerts translational control of the downstream three-gene operon beginning with the Ms5788 (unknown function) followed by the M. smegmatis CysA2 sulfotransferase and SseC2 putative sulfotransferase, both of whose up-regulation is logical under Cys-limiting conditions. When Cys availability is low, ribosomes are proposed to stall at the Cys codons of Ms5788A and preclude formation of a secondary structure that would otherwise block the availability of the Shine–Dalgarno (SD) ribosome binding site needed to recruit the ribosome and translate the downstream mRNA (Fig. 5B) (15). Distinct from the classic tryptophan attenuation in Escherichia coli, where the absence or presence of ribosome stalling at tryptophan codons in a small leader peptide dictates the formation of alternate secondary structures that either permit or prevent transcription termination, this M. smegmatis Cys-rich small protein acts as a sensor for Cys concentration and dictates whether transcripts that follow are translated.
Fig. 5.
Downstream genes of identified Cys-containing sORFs are generally up-regulated. (A, D, G, and J) Genomic organization of the region surrounding the Cys-containing sORFs that occur upstream of annotated genes. Unannotated putative sORFs (red arrow) and their amino acid sequence (Cys residues highlighted in red) are shown. The Cys-containing sORFs in A, D, and G have orthologs in M. smegmatis (15, 21). VapC4 up-regulates the transcription of genes downstream of these sORFs in Mtb mc2 6206 (B, E, H, and K; all adjusted P values ≤ 0.05; NS, not statistically significant) and translation of their corresponding proteins (C, F, I , and L; solid red, Strimmer q value ≤ 0.05; striped red, Strimmer q value ≤ 0.1; striped black, Strimmer q value ≥ 0.1; ND, not detected).
Based on the function of the Ms5788A sORF in M. smegmatis, we predicted that its leaderless Mtb counterpart in front of an operon with similar architecture would also serve as a Cys-responsive attenuator. We designated this Mtb sORF as Rv0815A, following the convention of newly annotated genes in Mycobrowser [also used by Canestrari et al. (15)] to add an A to the name of the gene it precedes, since it resides 173 nt in front of the cysA2 (Rv0815c) and sseC2 genes (Fig. 5A). We also already knew that ribosomes stall within Rv0815A because that was the basis of its detection within our 5′ RNA-seq dataset. Therefore, stalling at its Cys-rich carboxyl terminus would be expected to alter the secondary structure and thus influence expression of cysA2 and sseC2. We performed conventional RNA-seq coupled with quantitative mass spectrometry to determine how expression of VapC4 influenced the abundance of the transcripts and proteins corresponding to these two genes and additional genes downstream of other Cys-rich sORFs.
We expected up-regulation of cysA2 and sseC2 in cells expressing VapC4, consistent with the luciferase reporter experiments for M. smegmatis Ms5788A (15). Indeed, RNA-seq performed on RNA harvested 24 and 72 h after VapC4 induction showed significant (all P values < 0.0006) increases in both cysA2 and sseC2 transcripts relative to the uninduced controls (Fig. 5B). In support of the RNA-seq data, quantitative mass spectrometry of newly synthesized AHA-labeled proteins from ±VapC4 cells also demonstrated that translation of the CysA2 and SseC2 proteins was up-regulated (Fig. 5C).
VapC4-Triggered Ribosome Stalling at Leaderless sORFs Identifies Additional Cys Attenuation-Responsive Regulons.
We documented VapC4-dependent ribosome stalling on three more unannotated Mtb leaderless sORF mRNAs that are also implicated as Cys-responsive attenuators. Two, Rv0485A and Rv2391A, were previously recognized as orthologs of M. smegmatis leaderless sORFs encoding small proteins with the telltale Cys-rich tract (21) (Fig. 5 D and G). In agreement with a role in attenuation, the abundance of both of these M. smegmatis small proteins increased under low Cys conditions (15). We identified the third Mtb sORF based on its distinctive Cys-rich carboxy terminus MQQAIQLRFILPRRLAVGCCCC and characteristic genome position (sORF starts 270 base pairs [bp] upstream of the cysK1 cysteine synthase—cysE serine acetyltransferase operon) (Fig. 5J) (15). In support of their role as Mtb Cys-responsive attenuators, our H37Rv RNA-seq data 24 h after VapC4 induction showed significant increases in the abundance of all transcripts immediately downstream of each of the three leaderless mRNAs, consistent with active translation of these transcripts (Fig. 5 E, H, and K). All genes were still up-regulated 72 h postinduction but generally showed log2 fold change values lower than the 24 h samples (Fig. 5 E, H, and K). We observed a polar effect on the polycistronic sirA-cysH-che1-ggtB operon (Fig. 5 G and H), that is, a decrease in translation as the ribosomes traverse the polycistronic mRNA, in alignment with the findings reported by Canestrari et al. (15). Finally, the transcriptional up-regulation we documented for each operon downstream of these three sORFs was again supported by an increase in translation, especially for the first protein encoded by each operon (Fig. 5 F, I, and L).
VapC4 Regulates Growth through Targeted Manipulation of Discrete Pathways.
In addition to the functional insights obtained through identification of the four sORFs described above, we used the annotation tool “Database for Annotation, Visualization and Integrated Discovery” (DAVID) (22, 23) to obtain a comprehensive summary of biological themes from RNA-seq datasets derived from total RNA harvested 24 h after VapC4 induction in Mtb H37Rv (Datasets S3 and S4). These biological themes were consistent when DAVID was performed on RNA-seq datasets from different induction times (72 h) in H37Rv or 24 and 72 h postinduction in the attenuated strain mc26206. This DAVID analysis also reflects the global impact of the 444 mRNAs for which we identified stalled ribosomes at one or more Cys codons (96 unannotated and 348 annotated).
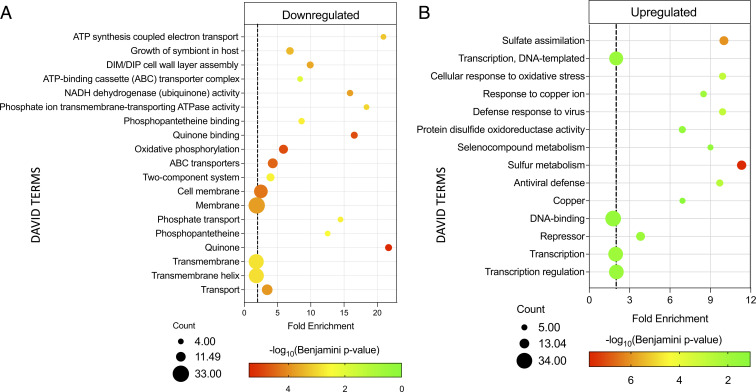
Among down-regulated gene categories, those specifically involved in respiratory ATP synthesis—“NADH dehydrogenase (ubiquinone) activity,” “quinone binding,” “quinone,” “ATP synthesis coupled electron transport,” “oxidative phosphorylation”—were the most affected (Fig. 6A and SI Appendix, Fig. S3). The “cell wall/cell membrane” as well as “ABC transporters” gene groups were down-regulated (Fig. 6A). ABC transporters are substrate-specific couriers of molecules such as amino acids, peptides, polysaccharides, proteins, ions, and antibiotics into or out of the cell. “Phosphate transport” was also identified as a down-regulated category (Fig. 6A) based on the decreased expression of the majority of ABC phosphate transporter subunit transcripts. Although inorganic phosphate is essential for many cellular processes, it is often limiting in the cellular environment so must be transported into cells. Consequently, phosphate limitation restricts growth, and importantly, phosphate depletion triggers Mtb persistence (24). Therefore, rather than broadly shutting down translation as previously proposed for tRNA-cleaving VapC family members (14), our data instead suggest that VapC4 controls growth and alters Mtb physiology by specifically down-regulating discrete pathways that modulate growth, the mycobacterial envelope, energy generation, transport of essential factors/nutrients, and synthesis of essential metabolites.
Fig. 6.
DAVID analysis of transcripts down- and up-regulated by VapC4. (A) VapC4 engages multiple mechanisms for growth control. DAVID Functional Analysis Tool terms (38, 39) associated with genes that were significantly (twofold) down-regulated after 24 h of VapC4 induction by total RNA-seq analysis. (B) VapC4 mimics Cys starvation to activate the oxidative and Cu stress responses. DAVID terms associated with genes that were significantly (twofold) up-regulated after 24 h of VapC4 induction by RNA-seq analysis. The dotted lines in both A and B represent a twofold enrichment between the proportion of the term in the observed genes compared to the expected proportion of the term when considering all genes in the genome. The area of each circle in both A and B is proportional to the number of observed genes in the corresponding category.
VapC4 Up-Regulates Sulfur Metabolism and Oxidative Stress Pathways.
Overall, both RNA-seq and supporting proteomic data exposed selective up-regulation of a subset of key stress survival pathways (Fig. 6B). The “sulfate assimilation/sulfur metabolism” categories had the highest statistical significance and are consistent with a toxin whose primary target is tRNACys. Sulfur is an essential element that possesses a variety of chemical properties—redox capacity, metal binding proficiency, nucleophilicity, and the ability to form strong disulfide bonds—that support its critical role in Mtb metabolism. In its reduced form, sulfur is used for the biosynthesis of both Cys and methionine. Cys is particularly important, as it is incorporated into proteins, coenzymes, and the glutathione counterpart in Actinobacteria called mycothiol (25).
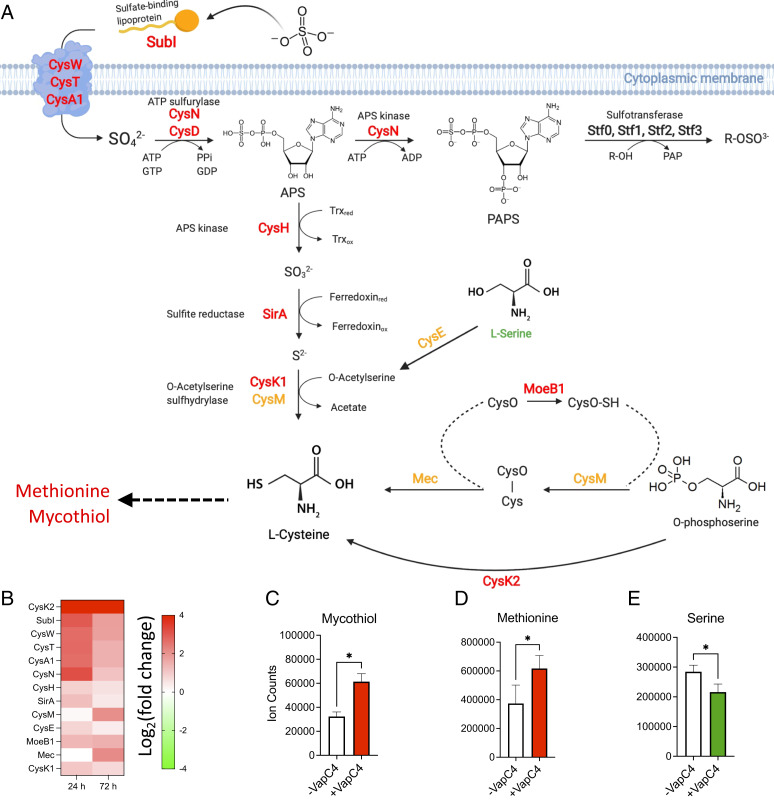
Notably, we found that all of the enzymes in the Mtb sulfur import and sulfur assimilation pathway to Cys synthesis (26) were up-regulated when VapC4 was expressed (Fig. 7 A and B). In addition, all enzymes in two alternate synthetic pathways to Cys (27, 28) were up-regulated: CysE and CysK1 that catalyze synthesis of Cys from serine (Fig. 7 A, Middle), CysM, Mec and MoeB1, or CysK2 that catalyze the synthesis of Cys from O-phosphoserine (Fig. 7 A, Bottom Right). Overall, 16 of the 19 the “cys”-assigned genes in Mycobrowser were up-regulated (Dataset S4, “VapC4 DAVID_upregulated” tab).
Fig. 7.
VapC4 mimics Cys starvation to activate the oxidative and Cu stress responses. (A) Transcripts involved in sulfate assimilation are up-regulated in Mtb cells expressing VapC4. Red, up-regulated at both 24 and 72 h in Mtb H37Rv cells (≥1.5 fold, adjusted P value ≤ 0.05); orange, up-regulated at 72 h only (≥1.5 fold, adjusted P value ≤ 0.05). CysN is sometimes referred to as CysC. Data from refs. 27, 28, 53, and 70. Sufur assimilation illustration created with https://Biorender.com. (B) Heat map of transcripts corresponding to up-regulated enzymes shown in A. (C) Mass spectrometry of mycothiol (asterisk represents P value of 0.02), (D) methionine (asterisk represents P value of 0.02), and (E) serine (asterisk represents P value of 0.05); all P values for C–E used the paired Wilcoxon signed-rank test.
Another up-regulated DAVID category “oxidative stress/disulfide oxidoreductase” represents pathways that are functionally related to “sulfate assimilation/sulfur metabolism” (see Fig. 6B). The ability to sense and respond to oxidative stress is critical for Mtb survival upon infection. We found that VapC4 expression results in up-regulation of other oxidative stress/disulfide oxidoreductase pathways that are consistent with an increase in sulfur metabolism shown in Fig. 7A. A major end product of sulfur uptake and metabolism pathways, Cys, is required for multiple pathways in Mtb, including intra and intermolecular disulfide bond formation, coordination of metal ions, and in oxidoreductase proteins. Cys also supports the production of mycothiol (Fig. 7A). Mycothiol, present at millimolar levels in the cell, is a Cys-ligated disaccharide that is essential for Mtb growth and is responsible for maintaining an intracellular reducing environment and minimizing the damaging effects of oxidizing agents such as hydrogen peroxide and NO (25). Increased production of mycothiol in response to oxidative stress would consume large amounts of Cys. In support of mycothiol up-regulation, transcription of the first enzyme in the pathway for its synthesis, MshA, is also strongly increased 24 and 72 h post VapC4 induction (Fig. 5E), and newly synthesized MshA protein is also up-regulated (Fig. 5F). The gene encoding MshA appears to be regulated by the Rv0485A sORF because it directly follows the first gene, Rv0485, in this Cys attenuation-responsive operon (Fig. 5 D–F). More definitively, we confirmed that mycothiol and methionine production increases upon VapC4 expression (Fig. 7 C and D), while serine levels decrease as expected because it is a substrate for Cys (Fig. 7E). Cys is not detectable by mass spectrometry because it has a very short half-life. Instead, Mtb mycothiol is the storage form of Cys; it is present at levels >600 times that of Cys (29).
Independent of DAVID analysis, we also found significant up-regulation of transcripts encoded by 19 previously identified genes in the alternate sigma factor SigH regulon, at 24 and/or 72 h post VapC4 induction (30, 31). SigH is required for virulence and regulates a network of genes that respond to oxidative and heat stress, both to limit cellular damage and re-establish redox equilibrium in response to these stresses (30, 32). These regulon members include two other stress-response alternate sigma factor genes (sigB and sigE), a thioredoxin/thioredoxin reductase pair (trxB1/B2) involved in maintaining redox homeostasis, and a mycothiol-dependent reductase (Rv2466c) that is required for Mtb viability in the presence of hydrogen peroxide stress (33). The 18 SigH-regulated genes that show increased expression in response to VapC4 induction are: Rv1221, sigE; Rv1471, trxB1; Rv1528c, papA4; Rv1875; RV2466c; Rv2707; Rv2710, sigB; Rv3206c, moeB1; Rv3223c, sigH; Rv3913, trxB2; Rv0140; Rv0384c, clpB; Rv0991c; Rv1039c, ppe15; Rv1259, udgB; Rv1801, ppe29; Rv3054c; and Rv3463 (31) (Dataset S3, “SigH regulon” tab). ClpB is part of a multichaperone protein damage repair system that is induced by cell stress, and Rv0991c is an oxidation-activated molecular chaperone (34). UdgB functions in DNA excision repair by removing promutagenic uracil bases created by cytosine deamination to uracil. The high guanine/cytosine (GC) content of Mtb makes it especially susceptible to cytosine deamination upon exposure to reactive nitrogen and oxygen species (35). PPE15 is required for triacylglycerol accumulation (36), the dominant Mtb carbon source under nonreplicating conditions (37).
VapC4 Up-Regulates the CsoR and RicR Copper-Resistance Pathways.
Although copper (Cu) is an essential micronutrient, it is deadly at high concentrations. Macrophages increase the phagosomal concentration of Cu upon Mtb infection. Therefore, Cu resistance mechanisms are thought to help Mtb evade macrophage-mediated killing. DAVID analysis identified up-regulation of “Copper” and “Copper ion” functional categories (Fig. 6B).
There are currently three independently regulated Mtb copper-response pathways that function in concert (38), although Cu treatment itself induces many more genes beyond these defined pathways (39). In the first pathway, the copper-sensitive operon repressor CsoR controls the expression of a four-gene operon (Fig. 8A). The CsoR dimer lacking bound Cu binds DNA and functions as the repressor. Cu binding to CsoR induces a conformational change, release from DNA, and derepression. The second Cu resistance pathway engages the RicR (regulated in copper) repressor, which binds to a palindrome in the promoter region of several genes/operons (40) (Fig. 8A). Its mechanism of derepression is presumed to parallel that of CsoR. The third Cu resistance pathway involves the action of a single membrane protein, MctB, a mycomembrane protein thought to block the entry of copper into the cell (41) (Fig. 8A).
Fig. 8.
VapC4 up-regulates key components of Cu homeostasis. (A) CsoR and RicR regulon proteins with known functions are illustrated; all were up-regulated by VapC4 (labeled in black text). MctB was not affected by VapC4 expression (labeled with gray text). (B) Heat map of up-regulated CsoR and RicR regulon transcripts 24 and 72 h post VapC4 induction in Mtb H37Rv cells (≥1.5 fold, adjusted P value ≤ 0.05). The proteins encoded by the mmcO and cysK2 (white text, boxed in red) genes were also identified in quantitative mass spectrometry datasets (adjusted P values ≤ 0.05 [RNA-seq, Strimmer q values ≤ 0.05 (QMS). Since socA and B were not annotated in the NCBI Mtb H37Rv database, we used their genome positions to identify read counts within the raw RNA-seq datasets (log2FC at 24/72 h: socA 1.17/2.23, socB 1.17/3.59; all adjusted P values ≤ 0.05). The illustration of Mtb Cu pathways (A) was created with https://Biorender.com. (C) VapC4 transcript levels are elevated in M. tuberculosis cultures exposed to copper at 5 μM and 50 μM [data from Ward. et al (39)]. Error bars represent the SEM from microarray data obtained from two replicates.
We found that all of the 14 genes in the CsoR and RicR regulons were up-regulated upon VapC4 expression, but we did not observe up-regulation of MctB, the sole component of the third Cu resistance pathway (Fig. 8B and Dataset S3, “CsoR-RicR regulon” tab). In addition to the dramatically up-regulated, divergently transcribed RicR-regulated mmcO and cysK2 transcripts, we identified marked increases in newly synthesized MmcO and CysK2 protein by quantitative mass spectrometry (Fig. 8B, white text in red box). Interestingly, three of the transcripts in the RicR regulon, mmcO, lpqS, and cysK2, were consistently up-regulated during the Wayne model of nonreplicating persistence in independent microarray experiments (42–44).
Although the mechanisms of action of many of the genes controlled by CsoR and RicR are not known, mymT, mmcO, cysK2, and ctpV encode proteins whose roles have been identified (38). Mycobacterium metallothionein MymT is central to Cu homeostasis because it is able to bind and sequester up to six Cu ions (45), the mycobacterium multicopper oxidase MmcO oxidizes toxic Cu+ to less toxic Cu2+ (46), and the CtpV ATPase appears to export Cu (Fig. 8A). CysK2 is an S-sulfocysteine synthase which catalyzes the synthesis of Cys from O-phosphoserine (28) (Fig. 7 A, Bottom Right), Therefore, the transcription of cysK2 is dependent on the presence of Cu, and the activity of the metabolic CysK2 enzyme leads to synthesis of mycothiol, suggesting that the redox homeostasis and Cu homeostasis pathways are interconnected in Mtb.
Many Cu-Response Genes Are Metabolic Enzymes Involved in Sulfate Assimilation and Oxidative Stress.
Among the genes identified in the proteasome mutant screen by the Darwin laboratory that fortuitously identified members of the RicR regulon were three “cys” genes, cysK2, cysNC, and cysD (40); all three were up-regulated upon VapC4 expression. CysNC, CysD, and CysK2 each catalyze reactions in the sulfate assimilation pathway (Fig. 7A). Four “cys” genes were identified in a screen for Cu-responsive genes: cysA2, cysA3, cysK2, and cysW (39). CysA2 and CysA3 are “thiosulfate sulfurtransferases.” CysW is a component of an ABC transporter for sulfate that shuttles sulfate into the cell for the sulfate assimilation pathway as illustrated in Fig. 7A. Each of these “cys” annotated genes were up-regulated in response to VapC4 expression as well. Curiously, cysA2 resides downstream of one of the sORFs we identified by Cys ribosome stalling (Fig. 5 A–C). As detailed above, CysA2 is the Mtb ortholog of MSMEG_5788 that resides within the well-characterized M. smegmatis operon regulated by Cys attenuation through the Cys codon-rich sORF MSMEG_5788A (Fig. 5 A–C) (15). Taken together, the common themes between sulfur assimilation and Cu-resistance pathways are reflected by the six “cys” annotated genes represented in the two pathways—cysA2, cysK2, cysA3, cysNC, cysD, and cysW—and all are up-regulated by VapC4 expression.
Finally, Ward et al. found that 13 of the 24 genes up-regulated during the Mtb Cu response are involved in the response to oxidative stress as well (39). Since many were transcription regulators for oxidative stress, they concluded that the Cu response is not specific to Cu exposure but falls within the generalized response to oxidative stress. We found that all 24 genes up-regulated in response to Cu (including the 13 also involved in the oxidate stress response) were also up-regulated upon VapC4 expression (Dataset S3, “Oxidative & Cu stress response” tab). Therefore, expression of VapC4 produces an expression profile that integrates Mtb sulfur assimilation, the response to Cu, and oxidative stress pathways. In alignment with this integrated theme, we found the up-regulation of VapC4 itself within the Cu-response microarray data in Ward et al. (39) (Fig. 8C). Therefore, Cu may be a physiological trigger for VapC4.
Discussion
The abundant Mtb TA systems have been suggested to be involved in the establishment of latent TB infection based on the observation that the expression of E. coli toxin components typically leads to a reversible state of growth arrest and a “quasidormant” metabolic state, a phenotype that has been observed with many but not all Mtb toxins (3, 4). Early studies of TA toxins in E. coli—most of which are endoribonucleases—almost exclusively identified mRNA as the toxin target (4, 47, 48). Therefore, for many years, the prevailing view was that all endoribonuclease TA toxins in bacteria were sequence-specific “mRNA interferases” that imparted reversible growth arrest through widespread mRNA degradation that then leads to the inhibition of protein synthesis. However, once our laboratory and others began to characterize the targets of TA toxins in Mtb, it was clear that this pathogen was quite different from E. coli and the gram-positive pathogen Staphylococcus aureus. Not only does Mtb employ endoribonuclease toxins for the inactivation of 23S ribosomal RNA (rRNA) at helix loop 70 (19, 49, 50) or the highly conserved sarcin–ricin loop (51), it likely engages a large proportion of its TA systems for highly selective isoacceptor-specific tRNA cleavage (11, 12, 20, 52).
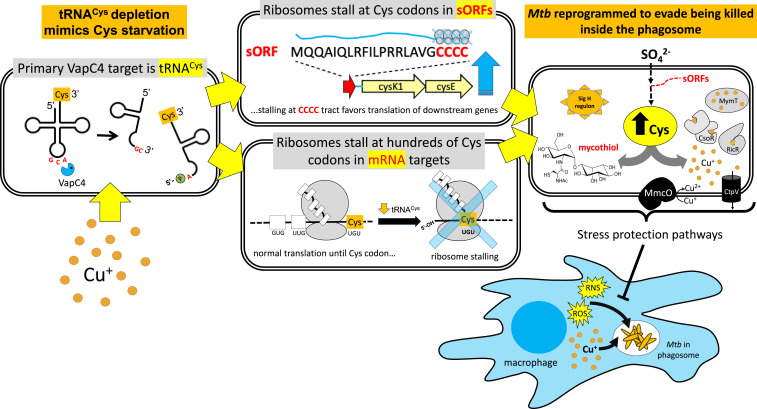
In this study, we induced low-level expression of VapC4 to recapitulate the physiologic effects of VapC4 produced by the not yet identified natural in vivo trigger(s) of toxin activation, thus enabling us to isolate the effects of this toxin on Mtb physiology and separate these effects from other broad stress responses. We then used a spectrum of powerful systems-level approaches to pinpoint the cellular pathways regulated by VapC4 expression (illustrated in Fig. 9) in order to elucidate the molecular mechanism underlying the essential role for VapBC4 in virulence (7). We discovered an extensive but highly specific Mtb response based on the selection of tRNACys as the primary toxin target (Fig. 9, Top Left). This toxin utilizes tRNACys inactivation to mimic a state of Cys starvation, leading the cell to retool its metabolism toward Cys synthesis. This shift is mediated in part by ribosome stalling at tracts of Cys codons within sORFs (Fig. 9, Middle Top) that are projected to block the formation of secondary structures that sequester the ribosome-binding site for the downstream operon, thus allowing translation of downstream genes. In conjunction with the increased production of proteins regulated by these sORFs, we identified highly specific proteome reprogramming via selective depletion of transcripts based on their Cys codon content (Fig. 9, Middle Bottom). The effects we observed with VapC4 are consistent with the importance of sulfur in Mtb, in which inorganic sulfur is assimilated into Cys, which then serves as a precursor to a variety of key metabolites such as mycothiol and Fe–sulfur enzymes to maintain protein synthesis and a redox balance (Fig. 9, Right) (26, 53, 54). Cys biosynthesis in Mtb is up-regulated in response to oxidative stress (55), is protective for the cell survival of oxidative stress (56), and protects cells during the persistent phase of infection in mice (56).
Fig. 9.
VapC4 reprograms Mtb physiology to protect against oxidative and Cu stress. Cu up-regulates VapC4 transcription, and VapC4 toxin precisely cuts the sole tRNACys at its anticodon GC↓A to create tRNA halves. These tRNA halves are nonfunctional in protein synthesis, resulting in ribosome stalling at Cys codons (UGU, UGC) requiring this depleted tRNA. Stalled ribosomes at Cys codons globally reduce new synthesis of Cys-containing proteins (SI Appendix, Fig. S2). A subset of transcripts with stalled ribosomes were Cys codon-containing unannotated small ORFs in which ribosome stalling at tracts of Cys codons within these sORFs is predicted to be a novel mode of Cys attenuation to regulate translation of downstream genes (as shown for sORF Rv2334A). Many of these sORFs up-regulate translation of enzymes for sulfate assimilation to Cys. As macrophages attempt to eliminate Mtb with ROS/RNS and toxic Cu+, the enriched pool of Cys can be incorporated into molecules needed to defend against these assaults. Key components of these two major stress response pathways are up-regulated by VapC4. For oxidative stress, these include mycothiol, the abundant, essential, glutathione counterpart in Mtb that regulates cellular redox status and multiple genes in the SigH regulon. For copper stress, these include multiple mechanisms controlled by copper-responsive regulators RicR and CsoR to mitigate copper toxicity.
While mining the themes in up-regulated genes led us to valuable insights into the metabolic changes that are expected to lead to protection against certain stresses, our data also suggested that certain features of the oxidative stress response and the response to Cu are coordinated, as they both share several Cys biosynthetic genes. An equally careful analysis of down-regulated genes led to many surprising modes of growth control engaged by this toxin, especially at the level of energy generation.
With over 90 Mtb TA systems, many of which are thought to be activated by stress, and a finite number of infection-associated stresses (e.g., hypoxia, oxidative stress, nutrient limitation, nitrosative stress, low pH), it is likely that more than one toxin is activated in response to infection. We demonstrated that the VapC4 mechanism of action begins with cleavage of a single tRNA and transitions to codon-dependent proteome remodeling. These findings suggest a model in which comprehensive defense against host immune responses would enlist the coordinated activation of multiple tRNA-cleaving toxins that each target a single, discrete tRNA isoacceptor. Based on our findings with VapC4, each of these toxins would have unique effects on remodeling the proteome to activate or repress complementary pathways that would allow Mtb to collectively attain comprehensive protection against host-mediated stresses. We hypothesize that VapC4 engages the integrated sulfur assimilation/oxidative stress and Cu-response pathways as a survival strategy early in infection. In fact, since VapC4 expression is up-regulated when Mtb cells are exposed to Cu in the growth medium (Fig. 8C), Cu may be the environmental trigger for the activation of VapC4 during infection (Fig. 9, Bottom Left). However, VapC4-mediated protection from these stresses would not fully protect this pathogen from the combined assaults of the immune response. We speculate that each toxin engages unique arms of the multifaceted Mtb stress response to infection. These toxin-induced arms of defense differ because each is defined by the codon dependence dictated by the toxin tRNA target. A coordinated defense system engaging an armamentarium of tRNA-cleaving toxins would provide a sophisticated, multitier evolutionary adaptation for survival upon Mtb infection of the human host, one that could adapt to each stage of infection and respond to both innate and adaptive immune mechanisms.
Materials and Methods
Strains, Plasmids, and Reagents.
All experiments were performed using either Mtb strain H37Rv (American Type Culture Collection 25618), Mtb mc2 6206 (ΔpanCD ΔleuCD, generously provided by the William Jacobs Laboratory, Albert Einstein College of Medicine, Bronx, NY), or Mtb mc2 6206 ΔVapBC4. VapC4 (Rv0595c locus) was amplified by PCR from Mtb H37Rv genomic DNA. The amplified gene was cloned under the control of an anhydrotetracycline (ATC)-inducible promoter in the pMC1s plasmid. Induction of VapC4 was obtained by adding ATC to the media to a final concentration of 200 ng/mL and replenishing it every 48 h. The average induction levels of the VapC4 transcript from the pMC1s plasmid expressed in H37Rv is 6.5-fold (Dataset S3).
Mtb cells were grown under constant shaking at 200 rpm at 37 °C in 7H9 Middlebrook media containing 1× oleic acid, albumin, dextrose, catalase (OADC) supplement (BD Biosciences), 0.05% of tyloxapol, and kanamycin at 25 μg/mL for plasmid selection. The media was supplemented to 50 µg/mL pantothenic acid and 100 µg/mL leucine for growth of the attenuated strain Mtb mc2 6206.
Construction of VapBC4 Deletion Strain.
To accurately assess whether VapC4 preferentially down-regulates translation of Cys-rich Mtb proteins (SI Appendix, Fig. S2), we created a strain with the VapBC4 (ΔVapBC4 or ΔRv0596c-Rv0595c) module deleted from the Mtb mc2 6206 parental strain using the oligonucleotide-mediated recombineering followed by Bxb1 integrase targeting (ORBIT) recombineering method (57). The Bxb1 attP-containing oligonucleotide was designed to contain the first and last 60 bp of the Rv0596c-Rv0595c toxin–antitoxin module (SI Appendix, Table S1). To confirm successful deletion, we performed PCR using oligonucleotides targeting the module flanking regions in combination with internal pKM464 plasmid oligonucleotides (SI Appendix, Table S1).
RNA Isolation.
Mycobacterial cells were grown in the absence or presence of an inducer for 24 or 72 h. Cells were centrifuged at 2,000 × g for 10 min at 4 °C, and supernatants were removed. The cell pellets were resuspended in 1 mL TRIzol and transferred to lysing tubes (Bertin Corp.) containing 0.1 mm glass beads. Cells were lysed in four cycles of 30 s at 9,000 rpm using a Precellys Evolution homogenizer (Bertin Corp.) with 1-min cooldown periods on ice in between each cycle. The lysate was centrifuged at 14,000 rpm at 4 °C, and the supernatant was used to extract total RNA using the Direct-zol RNA MiniPrep Plus Kit (Zymo Research). RNA was treated with an additional step of genomic DNA removal in a TURBO DNase (Thermo Fisher) digestion reaction for 30 min at 37 °C and repurified using the Zymo RNA Clean and Concentrator Kit. The extracted RNA was quantified by spectrophotometry using a µCuvette in a BioSpectrometer (Eppendorf).
In Vitro Cleavage Assays with Purified VapC4.
VapC4 purification and cleavage reactions were performed as described in Sharp et al. and Cruz et al., respectively (12, 13). Briefly, 10 pmol of purified VapC4 was individually incubated with 2 pmol of each of the 45 in vitro–synthesized tRNAs from the Mtb H37Rv genome or 2 µg total RNA extracted from Mtb mc2 6206 for 3 h at 37 °C. Cleavage reactions with in vitro–synthesized tRNAs were run on a 15% Urea-PAGE gel, stained with SYBR Gold, and visualized in an ultraviolet transilluminator. Cleavage reactions using total RNA were purified using the Zymo RNA Clean and Concentrator Kit following the manufacturer’s protocol and were used for constructing 5′ RNA-seq libraries.
5′ RNA-Seq.
The preparation and analysis of 5′-OH libraries was performed as described in Barth et al. (20). For 5′ monophosphate libraries, the same procedure was followed except that the steps of digestion with Terminator 5′-Phosphate-Dependent Exonuclease (Epicentre) and phosphorylation with T4 PNK (New England Biolabs) were omitted. The libraries were sequenced in an Illumina HiSeq 2500/4000 platform at Genewiz Corp or New York University’s Genome Technology Center.
For data analysis, we only considered reads that had at least one read per million of mapped reads (rpm) for mRNAs and 5 rpm for tRNAs in the induced sample and a fold change of at least 20. Frequency logos were generated with kpLogo (58) or weblogo (59).
To identify unannotated Cys codon-containing ORFs, we searched for unannotated reads with either of the two Cys codons (UGC and UGU) ∼15 nt from the RNA cleavage site (corresponding to stalled ribosome at the “hungry” codon) in the 5′-OH RNA-seq dataset. We then determined if the stalled Cys codon was in frame and fit within our parameters for a potential ORF: at least 10 amino acids with a start (ATG, GTG, or TTG) and a stop (TGA, TAA, or TAG) codon. We selected the longest possible ORF since transcription start sites for these ORFs are not known.
Total RNA-Seq.
In order to remove 16S/23S ribosomal RNA from the total RNA, the samples were treated using the NEBNext Bacterial rRNA Depletion Kit (New England Biolabs). Approximately 100 ng rRNA-depleted RNA was used to generate the libraries using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs) and sequenced on an Illumina HiSeq 2500 or similar. Reads were mapped against the Mtb H37Rv reference genome (GenBank accession: AL123456.3) in a local Galaxy instance (60) using the default parameters of Bowtie 2.3.4.3 (61), featureCounts 1.6.4 (62), and limma 3.38.3 (63). Significantly up- or down-regulated genes (± 1.5-fold) with an adjusted P value < 0.05) were analyzed using the Functional Annotation tool in the DAVID platform using an Benjamini P value cutoff of 0.1 (23).
Labeling of Newly Synthesized Mtb proteins.
To assess the levels of newly synthesized proteins in Mtb after toxin induction, triplicate VapC4 samples were induced for 12, 24, and 48 h, and AHA (Anaspec Inc.) was added to the media to a final concentration of 50 µM and incubated for 6 h. AHA is an azide-containing methionine mimetic that is incorporated into proteins, allowing the capture or visualization of the newly synthesized proteins by a copper-catalyzed azide–alkyne cycloaddition reaction. To extract the AHA-labeled proteins, cells were pelleted, resuspended in lysis buffer (2% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 8 M Urea), and lysed in a Precellys Evolution homogenizer as described in RNA Isolation. The lysate was centrifuged, and the proteins from the supernatant were linked to an alkyne-containing fluorophore (TAMRA) using the Click-IT Protein Reaction Buffer Kit (Thermo Fisher). A total of 10 μg of protein from each sample were resolved on a 9% SDS-PAGE gel and imaged with a Typhoon FLA 9500 (GE Healthcare) image system. Fluorescence intensities from each lane were quantified using ImageJ software.
Proteomics of Newly Synthesized Proteins.
To identify newly synthesized proteins by quantitative mass spectrometry, mc2 6206 Mtb cells were harvested after VapC4 induced for 24 or 48 h. For SI Appendix, Fig. S1, ΔVapBC4 containing the vapC4-pMC1s plasmid were grown to an optical density at 600 nm (OD600) of 0.1 and induced for 48 h along with uninduced cultures, in triplicate. A total of 50 ml cultures were centrifuged at 2,000 × g at 4 °C for 5 min and washed with 1× phosphate-buffered saline two times to remove traces of the albumin-containing 7H9 media. The cell pellets were resuspended in lysis buffer (2% CHAPS, 8 M Urea) and lysed using a Precellys Evolution homogenizer as described in RNA Isolation. The lysates were pelleted at 12,000 × g at 4 °C for 10 min, and the AHA-labeled proteins contained in the supernatant were selectively captured using alkyne-coated agarose beads from the Click-iT Protein Enrichment Kit (Thermo Fisher) following the manufacturer’s protocol.
Tryptic digests were analyzed using an Orbitrap Tribrid mass spectrometer and nanoflow LC system (Thermo Scientific) as described in Barth et al. (20). The raw liquid chromatography–mass spectrometry data were converted into MASCOT Generic Format using Proteome Discover 2.1 (Thermo Fisher) and searched against either the National Center for Biotechnology Information (NCBI) Mtb database (accession: AL123456) together with a database of common laboratory contaminants (https://www.thegpm.org/crap/) using a local implementation of the global proteome machine (64).
Spectral counts were analyzed using the QuasiSeq package (https://cran.r-project.org/web/packages/QuasiSeq/index.html) for proteins containing 15 or more spectral counts total (65). Q-values are calculated using the fdrtool package of Strimmer (66) and considered significant if below 0.05. The Cys codon content of up- and down-regulated proteins was measured by estimating the codon content from the Mtb H37Rv annotated genes (GenBank accession: AL123456).
Metabolomics—Sulfur Assimilation Pathway.
To analyze metabolites by mass spectrometry, mc2 6206 Mtb cells containing the vapC4-pMC1s plasmid were grown to an OD600 of 0.1 and induced for 48 h along with uninduced cultures. Equal numbers of cells (normalized by OD600) for each sample were centrifuged at 2,000 × g at 4 °C for 10 min. The pellets were then transferred to microcentrifuge tubes and centrifuged at 10,000 × g at 4 °C followed by the removal of as much media as possible. The cell pellets were resuspended in 1.2 mL extracting solution (acetonitrile:methanol:water [40:40:20], 0.5% formic acid). Samples were mixed on ice, and 86 μL neutralization solvent (15% NH4HCO3) was added and mixed thoroughly. Samples were then centrifuged at 13,000 × g at 4 °C for 10 min. The supernatants were transferred to fresh tubes and analyzed as previously described (67). Briefly, the sample analysis was performed on a Thermo Q Exactive PLUS (Thermo Fisher Scientific) coupled with hydrophilic interaction chromatography (HILIC). HILIC separation was performed on a Vanquish Horizon UHPLC system (Thermo Fisher Scientific) with an XBridge BEH Amide column (Waters). The mass/charge (m/z) scan range was set to 72 to 1,000 m/z in negative ionization mode. The MS1 data were processed using Maven (68). The confirmation of the mycothiol retention time was aided by analysis of a panel of M. smegmatis mycothiol mutants (69) kindly provided by the Yossef Av-Gay Laboratory, University of British Columbia, Vancouver, BC, Canada.
Supplementary Material
Acknowledgments
We thank the William Jacobs laboratory (Albert Einstein College of Medicine) for providing the attenuated H37Rv strain mc2 6206 (ΔpanCD ΔleuCD), Jonathan Cruz for performing early 5′ RNA-seq experiments that helped inform some conclusions in this paper, and Michael DeCandia for assisting with uncovering ribosome stalling trends in the original datasets. We especially appreciate the considerable efforts of Mary Ko and the Yossef Av-Gay laboratory (University of British Columbia) for providing multiple mycothiol-deficient M. smegmatis strains to aid in an accurate identification of the mycothiol retention time. Quantitative mass spectrometry was performed by the Biological Mass Spectrometry Facility of Robert Wood Johnson Medical School supported by National Center for Research Resources Grant S10OD025140. Metabolomics experiments were performed by the Rutgers Cancer Institute of New Jersey Metabolomics Shared Resource, supported, in part, with funding from National Cancer Institute–Cancer Center Support Grant P30 CA072720-5923. This work was funded in part by a fellowship to V.C.B. from the Brazilian Federal Agency for Support and Evaluation of Graduate Education within the Ministry of Education of Brazil and New Jersey Commission on Cancer Research Fellowship DFHS18PPC045 to U.C., as well as by NIH Grants R21 AI135461 to N.A.W. and R.N.H. and RO1 AI154464 to N.A.W.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022136118/-/DCSupplemental.
Data Availability
The sequencing datasets generated in this study were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA662430. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD025774.
References
- 1.Ramage H. R., Connolly L. E., Cox J. S., Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: Implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5, e1000767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sala A., Bordes P., Genevaux P., Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel) 6, 1002–1020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harms A., Brodersen D. E., Mitarai N., Gerdes K., Toxins, targets, and triggers: An overview of toxin-antitoxin biology. Mol. Cell 70, 768–784. (2018). [DOI] [PubMed] [Google Scholar]
- 4.Masuda H., Inouye M., Toxins of prokaryotic toxin-antitoxin systems with sequence-specific endoribonuclease activity. Toxins (Basel) 9, E140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arcus V. L., McKenzie J. L., Robson J., Cook G. M., The PIN-domain ribonucleases and the prokaryotic VapBC toxin-antitoxin array. Protein Eng. Des. Sel. 24, 33–40 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Arcus V. L., Rainey P. B., Turner S. J., The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 13, 360–365 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Agarwal S., et al., System-wide analysis unravels the differential regulation and in vivo essentiality of virulence-associated proteins B and C toxin-antitoxin systems of Mycobacterium tuberculosis. J. Infect. Dis. 217, 1809–1820 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Agarwal S., et al., VapBC22 toxin-antitoxin system from Mycobacterium tuberculosis is required for pathogenesis and modulation of host immune response. Sci. Adv. 6, eaba6944 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slayden R. A., Dawson C. C., Cummings J. E., Toxin-antitoxin systems and regulatory mechanisms in Mycobacterium tuberculosis. Pathog. Dis. 76fty039 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Cruz J. W., Woychik N. A., tRNAs taking charge. Pathog. Dis. 74, ftv117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cintrón M., et al., Accurate target identification for Mycobacterium tuberculosis endoribonuclease toxins requires expression in their native host. Sci. Rep. 9, 5949 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz J. W., et al., Growth-regulating Mycobacterium tuberculosis VapC-mt4 toxin is an isoacceptor-specific tRNase. Nat. Commun. 6, 7480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharp J. D., et al., Growth and translation inhibition through sequence-specific RNA binding by Mycobacterium tuberculosis VapC toxin. J. Biol. Chem. 287, 12835–12847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winther K., Tree J. J., Tollervey D., Gerdes K., VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res. 44, 9860–9871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canestrari J. G., et al., Polycysteine-encoding leaderless short ORFs function as cysteine-responsive attenuators of operonic gene expression in mycobacteria. Mol. Microbiol. 114, 93–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan P. P., Lowe T. M., GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 37, D93–D97 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan P. P., Lowe T. M., GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44 (D1), D184–D189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winther K. S., Gerdes K., Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 108, 7403–7407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schifano J. M., et al., An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat. Commun. 5, 3538 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barth V. C., et al., Toxin-mediated ribosome stalling reprograms the Mycobacterium tuberculosis proteome. Nat. Commun. 10, 3035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shell S. S., et al., Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 11, e1005641 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Huang W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rifat D., Bishai W. R., Karakousis P. C., Phosphate depletion: A novel trigger for Mycobacterium tuberculosis persistence. J. Infect. Dis. 200, 1126–1135 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Newton G. L., Buchmeier N., Fahey R. C., Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol. Mol. Biol. Rev. 72, 471–494 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatzios S. K., Bertozzi C. R., The regulation of sulfur metabolism in Mycobacterium tuberculosis. PLoS Pathog. 7, e1002036 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agren D., Schnell R., Oehlmann W., Singh M., Schneider G., Cysteine synthase (CysM) of Mycobacterium tuberculosis is an O-phosphoserine sulfhydrylase: Evidence for an alternative cysteine biosynthesis pathway in mycobacteria. J. Biol. Chem. 283, 31567–31574 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Steiner E. M., et al., CysK2 from Mycobacterium tuberculosis is an O-phospho-L-serine-dependent S-sulfocysteine synthase. J. Bacteriol. 196, 3410–3420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newton G. L., et al., Distribution of thiols in microorganisms: Mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178, 1990–1995 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raman S., et al., The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183, 6119–6125 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharp J. D., et al., Comprehensive definition of the SigH regulon of Mycobacterium tuberculosis reveals transcriptional control of diverse stress responses. PLoS One 11, e0152145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehra S., et al., The Mycobacterium tuberculosis stress response factor SigH is required for bacterial burden as well as immunopathology in primate lungs. J. Infect. Dis. 205, 1203–1213 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosado L. A., et al., The antibacterial prodrug activator Rv2466c is a mycothiol-dependent reductase in the oxidative stress response of Mycobacterium tuberculosis. J. Biol. Chem. 292, 13097–13110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker S. H., et al., Mycobacterium tuberculosis Rv0991c is a redox-regulated molecular chaperone. MBio 11, e01545–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar P., Bharti S. K., Varshney U., Uracil excision repair in Mycobacterium tuberculosis cell-free extracts. Tuberculosis (Edinb.) 91, 212–218 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Daniel J., Maamar H., Deb C., Sirakova T. D., Kolattukudy P. E., Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 7, e1002093 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maurya R. K., Bharti S., Krishnan M. Y., Triacylglycerols: Fuelling the hibernating Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol. 8, 450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darwin K. H., Mycobacterium tuberculosis and copper: A newly appreciated defense against an old foe? J. Biol. Chem. 290, 18962–18966 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward S. K., Hoye E. A., Talaat A. M., The global responses of Mycobacterium tuberculosis to physiological levels of copper. J. Bacteriol. 190, 2939–2946 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Festa R. A., et al., A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol. Microbiol. 79, 133–148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolschendorf F., et al., Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 108, 1621–1626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muttucumaru D. G., Roberts G., Hinds J., Stabler R. A., Parish T., Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinb.) 84, 239–246 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Voskuil M. I., Mycobacterium tuberculosis gene expression during environmental conditions associated with latency. Tuberculosis (Edinb.) 84, 138–143 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Voskuil M. I., Visconti K. C., Schoolnik G. K., Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb.) 84, 218–227 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Gold B., et al., Identification of a copper-binding metallothionein in pathogenic mycobacteria. Nat. Chem. Biol. 4, 609–616 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rowland J. L., Niederweis M., A multicopper oxidase is required for copper resistance in Mycobacterium tuberculosis. J. Bacteriol. 195, 3724–3733 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaguchi Y., Inouye M., Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 9, 779–790 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi Y., Park J. H., Inouye M., Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45, 61–79 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Schifano J. M., et al., Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc. Natl. Acad. Sci. U.S.A. 110, 8501–8506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schifano J. M., Woychik N. A., 23S rRNA as an a-Maz-ing new bacterial toxin target. RNA Biol. 11, 101–105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winther K. S., Brodersen D. E., Brown A. K., Gerdes K., VapC20 of Mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat. Commun. 4, 2796 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Schifano J. M., et al., tRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res. 44, 1256–1270 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhave D. P., Muse W. B. III, Carroll K. S., Drug targets in mycobacterial sulfur metabolism. Infect. Disord. Drug Targets 7, 140–158 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schelle M. W., Bertozzi C. R., Sulfate metabolism in mycobacteria. ChemBioChem 7, 1516–1524 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Pinto R., Tang Q. X., Britton W. J., Leyh T. S., Triccas J. A., The Mycobacterium tuberculosis cysD and cysNC genes form a stress-induced operon that encodes a tri-functional sulfate-activating complex. Microbiology (Reading) 150, 1681–1686 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Senaratne R. H., et al., 5′-Adenosinephosphosulphate reductase (CysH) protects Mycobacterium tuberculosis against free radicals during chronic infection phase in mice. Mol. Microbiol. 59, 1744–1753 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Murphy K. C., et al., ORBIT: A new paradigm for genetic engineering of mycobacterial chromosomes. MBio 9, e01467–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu X., Bartel D. P., kpLogo: Positional k-mer analysis reveals hidden specificity in biological sequences. Nucleic Acids Res. 45 (W1), W534–W538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Afgan E., et al., The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44 (W1), W3–W10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Ritchie M. E., et al., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beavis R. C., Using the global proteome machine for protein identification. Methods Mol. Biol. 328, 217–228 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Lund S. P., Nettleton D., McCarthy D. J., Smyth G. K., Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat. Appl. Genet. Mol. Biol. 11, 8 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Strimmer K., fdrtool: A versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24, 1461–1462 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Su X., et al., In-source CID ramping and covariant ion analysis of hydrophilic interaction chromatography metabolomics. Anal. Chem. 92, 4829–4837 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melamud E., Vastag L., Rabinowitz J. D., Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 82, 9818–9826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rawat M., et al., Mycothiol-deficient Mycobacterium smegmatis mutants are hypersensitive to alkylating agents, free radicals, and antibiotics. Antimicrob. Agents Chemother. 46, 3348–3355 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeng L., Shi T., Zhao Q., Xie J., Mycobacterium sulfur metabolism and implications for novel drug targets. Cell Biochem. Biophys. 65, 77–83 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing datasets generated in this study were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA662430. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD025774.