Significance
This study identifies a previously undefined role for the inflammatory bowel disease–associated gene RNF186 in innate immunity. RNF186 is expressed in human macrophages and promotes outcomes downstream of receptors responding to microbial products. Upon stimulation of microbial response receptors, RNF186 contributes to assembly and ubiquitination of the signaling complex and subsequent cytokine secretion and antimicrobial pathway induction. Importantly, through different mechanisms, both rare (through impaired ubiquitination) and common (through reduced expression) disease-risk genetic variants in RNF186 lead to a loss in RNF186 function and impaired bacterial clearance in primary human macrophages. These studies highlight a key role for RNF186 in promoting essential innate immune functions contributing to intestinal immune homeostasis.
Keywords: innate immunity, ubiquitination, macrophage, inflammatory bowel disease
Abstract
Balancing microbial-induced cytokines and microbial clearance is critical at mucosal sites such as the intestine. How the inflammatory bowel disease (IBD)–associated gene RNF186 regulates this balance is unclear. We found that macrophages from IBD-risk rs6426833 carriers in the RNF186 region showed reduced cytokines to stimulation through multiple pattern recognition receptors (PRRs). Upon stimulation of PRRs, the E3-ubiquitin ligase RNF186 promoted ubiquitination of signaling complex molecules shared across PRRs and those unique to select PRRs. Furthermore, RNF186 was required for PRR-initiated signaling complex assembly and downstream signaling. RNF186, along with its intact E3-ubiquitin ligase activity, was required for optimal PRR-induced antimicrobial reactive oxygen species, reactive nitrogen species, and autophagy pathways and intracellular bacterial clearance in human macrophages and for bacterial clearance in intestinal myeloid cells. Cells transfected with the rare RNF186-A64T IBD-risk variant and macrophages from common rs6426833 RNF186 IBD-risk carriers demonstrated a reduction in these RNF186-dependent outcomes. These studies identify mechanisms through which RNF186 regulates innate immunity and show that RNF186 IBD-risk variants demonstrate a loss of function in PRR-initiated outcomes.
Inflammatory bowel disease (IBD) is characterized by dysregulated host–microbial responses and is composed of two subtypes: Crohn’s disease and ulcerative colitis (UC). Pattern recognition receptors (PRRs) are critical for recognition and responses to microbes. Both loss of function and gain of function in PRR-initiated outcomes can be associated with intestinal inflammation (1), thereby highlighting the importance of the balance in innate immune regulation in intestinal tissues. Despite the tremendous success in genetic studies identifying loci associated with susceptibility to IBD (2), altered functions for most of these loci are not well defined. One such region is on chromosome 1, encompassing the RNF186 gene (2, 3).
The UC rs6426833 A risk variant in the RNF186 region is observed at a 0.395 to 0.548 frequency in European ancestry healthy individuals (per the Single Nucleotide Polymorphism Database; accessed July 2019), and this common variant confers a 1.265 increased risk of developing UC (2). Importantly, a rare Ring Finger Protein 186 (RNF186) Ala64Thr mutation confers a 1.49-fold increased risk for developing UC (4). RNF186 is a member of the RING family E3 ubiquitin ligases and has a conserved C3HC4 type zinc finger (ZnF) motif in the RING domain consistent with its ability to catalyze ubiquitination of select downstream substrates (5, 6). E3 ubiquitin ligases are key mediators for posttranslational modifications of PRR-initiated signaling intermediates (7). Only a few reports have examined roles for RNF186, and these have focused on its functions in epithelial cells (4–6, 8). In HeLa cells, RNF186 can localize to the endoplasmic reticulum (ER) and enhance ER stress-associated apoptotic signaling (5). RNF186-deficient and rare variant RNF186 A64T knockin mice demonstrated more severe dextran sodium sulfate–induced colitis associated with intestinal epithelial dysregulation (6). RNF186 can regulate nutrient sensing in epithelial cells (8). Therefore, while some functions for RNF186 have been identified in epithelial cells, roles for RNF186 in mediating outcomes in innate immunity, including downstream of PRRs, have not been examined. Moreover, the consequences of UC-associated common genetic variants in the RNF186 region have not been defined.
We identify that RNF186 is expressed in human monocyte-derived and intestinal myeloid-derived cells. We establish that RNF186 is required for optimal PRR-induced signaling, cytokine secretion, and induction of a range of antimicrobial pathways. We define mechanisms and structural regions through which RNF186 regulates these PRR-initiated outcomes and identify that the rare and common RNF186 IBD-risk variants lead to a loss of function in these outcomes.
Results
Monocyte-Derived Macrophages from IBD-risk rs6426833 AA Carriers Demonstrate Decreased PRR-Induced Cytokine Secretion.
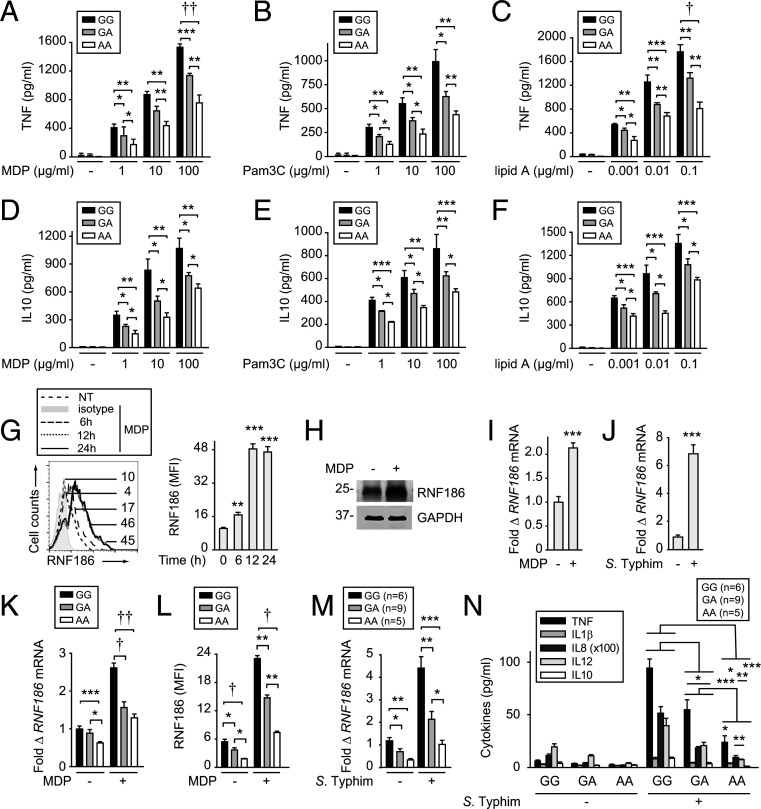
We first assessed if the rs6426833 genotype in the RNF186 region modulates PRR-induced cytokines in human monocyte-derived macrophages (MDMs). We examined the PRR NOD2 as a model innate receptor given the abundance of its ligand, muramyl dipeptide (MDP) (a component of peptidoglycan contained in the cell wall of most bacteria), in intestinal tissues (9). MDMs from rs6426833 AA disease-risk carriers showed decreased NOD2-induced tumor necrosis factor (TNF) secretion compared to GG carriers (Fig. 1A). These effects persisted across a range of MDP doses (Fig. 1A). IL1β, IL8, and IL12 showed similar results (SI Appendix, Fig. S1A). Rs6426833 genotype distribution did not change when stratified on NOD2 variants as assessed by high-density SNP mapping in the NOD2 region. As multiple microbial ligands are present in the intestine, we examined the additional PRRs, TLR2 and TLR4, and observed similar rs6426833 genotype–dependent regulation of cytokines (Fig. 1 B and C and SI Appendix, Fig. S1 B and C). The antiinflammatory cytokine IL10 demonstrated similar reduced PRR-induced secretion from rs6426833 AA risk MDMs (Fig. 1 D–F). Therefore, MDMs from rs6426833 AA UC-risk carriers show reduced PRR-induced pro- and antiinflammatory cytokines.
Fig. 1.
Human MDMs from rs6426833 A risk carriers demonstrate reduced PRR-induced cytokines and RNF186 expression. (A–F) MDMs from rs6426833 GG, GA, or AA carriers (n = 10 donors/genotype) were treated for 24 h with 1, 10, or 100 µg/mL MDP (recognized by NOD2) (A and D); 1, 10, or 100 µg/mL Pam3Cys (recognized by TLR2) (B and E); or 0.001, 0.01, or 0.1 µg/mL lipid A (recognized by TLR4) (C and F). TNF (A–C) and IL10 secretion (D–F) stratified by rs6426833 genotype. (G–I) MDMs were treated with 100 μg/mL MDP. (G) RNF186 protein expression at the indicated times. (Left) Representative histogram with mean fluorescence intensity (MFI) values. Isotype control is from 24-h treated cells. (Right) Summary graph (n = 4 donors; similar results in additional n = 4). (H) RNF186 expression at 24 h assessed by Western blot. (I) RNF186 mRNA expression at 4 h (n = 4 donors; similar results for an additional n = 4). (J) MDMs (n = 5 donors; similar results in additional n = 4) were cocultured with S. Typhimurium. RNF186 mRNA at 4 h. (K and L) MDMs (n = 10 donors/genotype; similar results in additional n = 6/genotype) were treated with 100 μg/mL MDP. (K) RNF186 mRNA at 4 h. (L) RNF186 protein expression at 24 h. (M and N) Human intestinal myeloid cells were cocultured with S. Typhimurium. RNF186 mRNA at 2 h (M) and cytokine secretion at 12 h (N) stratified on rs6426833 genotype. Mean + SEM. NT, no treatment. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5.
MDMs from IBD-Risk rs6426833 A Carriers Demonstrate Reduced RNF186 Expression.
As the rs6426833 variant is located in a noncoding region ∼31 kb from RNF186, we hypothesized that RNF186 expression would be regulated in a genotype-dependent manner. We therefore first assessed whether PRR stimulation regulates RNF186 expression in human MDMs. Upon NOD2 stimulation, RNF186 protein expression increased within 6 h and peaked after 12 to 24 h as assessed by flow cytometry (Fig. 1G). We confirmed increased NOD2-induced RNF186 expression by Western blot (Fig. 1H). NOD2 regulated RNF186 expression at the transcriptional level (Fig. 1I). RNF186 expression also increased in MDMs upon coculture with live bacteria (Fig. 1J). Importantly, RNF186 expression was modulated by rs6426833 genotype, with MDMs from AA risk carriers demonstrating less baseline and particularly NOD2-induced RNF186 messenger RNA (mRNA) (Fig. 1K) and protein (Fig. 1L and SI Appendix, Fig. S1D) expression compared to GG carriers. We next assessed whether the rs6426833 genotype modulates human intestinal myeloid cells. Intestinal myeloid cells respond poorly to PRR ligands (10) but are responsive to live bacteria (11, 12). Rs6426833 modulated baseline and S. Typhimurium–induced RNF186 mRNA expression (Fig. 1M) and cytokines (Fig. 1N) in intestinal myeloid cells. Therefore, RNF186 increases with PRR and live bacterial stimulation, and RNF186 expression is modulated by IBD-risk rs6426833 genotype.
RNF186 Is Required for PRR-induced NFκB and MAPK Activation and Cytokine Secretion in MDMs.
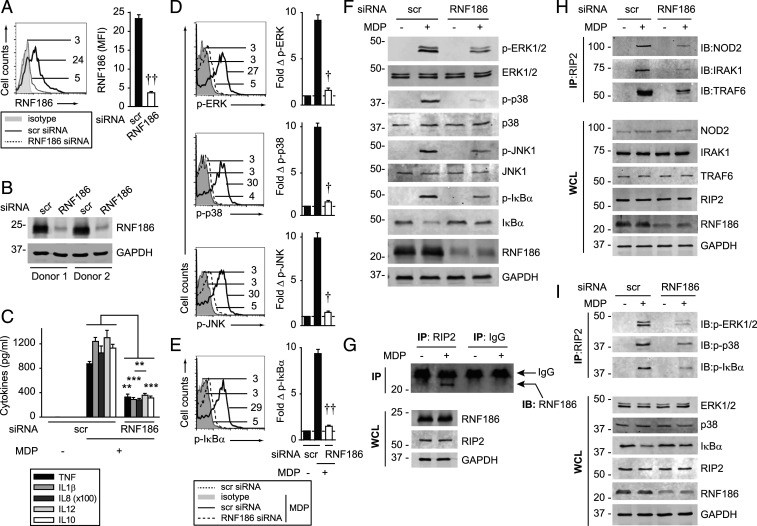
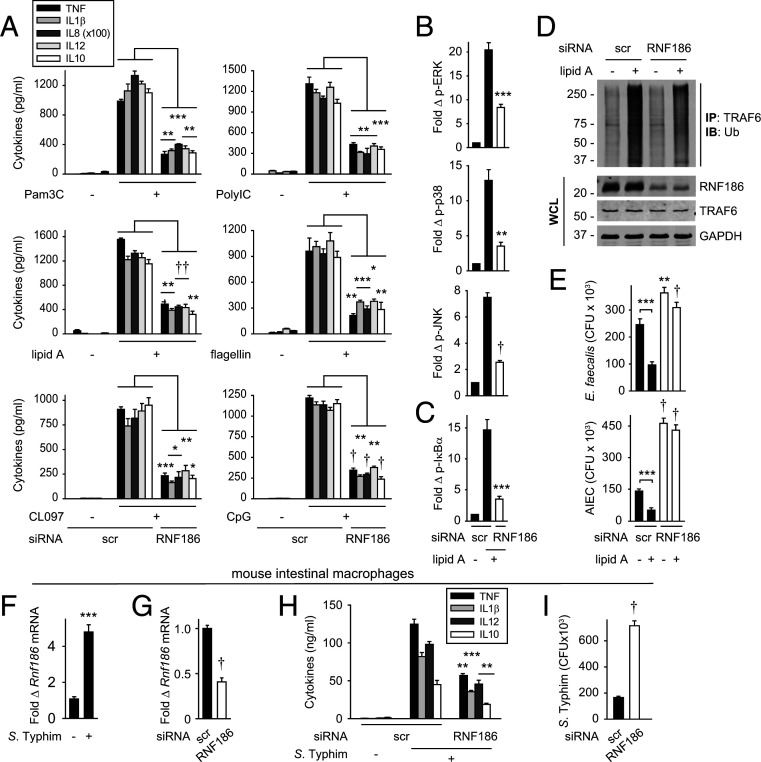
To clearly define RNF186 contributions to PRR-induced cytokines in MDMs, we used small interfering RNA (siRNA) to effectively knockdown RNF186 in MDMs as assessed by RNA (SI Appendix, Fig. S2A) and protein (Fig. 2 A and B) expression. RNF186 knockdown efficacy was confirmed with an additional RNF186 antibody (SI Appendix, Fig. S2 B and C). With reduced RNF186 expression, NOD2-induced pro- and antiinflammatory cytokines decreased (Fig. 2C). Cell viability was intact (SI Appendix, Fig. S2 D and E). Dectin-induced cytokine secretion also remained intact with RNF186 knockdown (SI Appendix, Fig. S2F), thereby demonstrating cell functionality through an alternative PRR. NOD2 stimulation activates mitogen-activated protein kinases (MAPKs) and NFκB pathways, which are critical for the induction of subsequent cytokines (12). With RNF186 knockdown, NOD2-induced MAPK and NFκB pathways were reduced per flow cytometry (Fig. 2 D and E) and Western blot (Fig. 2F). Macrophage differentiation remained intact (SI Appendix, Fig. S3 A–C). PRR pathways can be regulated similarly across myeloid-derived cells. Consistently, reducing RNF186 expression in monocyte-derived dendritic cells (SI Appendix, Fig. S3 D–F) reduced NOD2-induced signaling and cytokines (SI Appendix, Fig. S3 G–I). Therefore, RNF186 is required for PRR-induced MAPK and NFκB pathway activation and cytokine secretion in human myeloid cells.
Fig. 2.
RNF186 is required for PRR-induced signaling complex assembly, MAPK and NFκB activation, and cytokine secretion. (A–F, H, and I) MDMs were transfected with scrambled or RNF186 siRNA. (A and B) RNF186 protein expression (Aviva System Biology). Flow cytometry with summary graph (n = 6 donors; similar results in additional n = 4) (A) and Western blot (B). (C) MDMs were treated with 100 μg/mL MDP for 24 h. Cytokine secretion (n = 4 donors; similar results in additional n = 4). (D–F, H, and I) MDMs were treated with 100 μg/mL MDP for 15 min. (D–F) Phospho-proteins. (D and E) (Left) Representative flow cytometry plots with MFI values. (Right) Fold phospho-protein induction (n = 6). (F) Western blot. (H and I) RIP2 was immunoprecipitated (IP), and protein recruitment was assessed by immunoblot (IB). (G) MDMs were treated with 100 μg/mL MDP for 15 min. RIP2 was IP followed by immunoblotting for RNF186. Representative of two to three independent experiments for F–I. The respective proteins and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in whole cell lysates (WCLs) served as loading controls. IP with IgG was used as a control (data shown specifically for G). Mean + SEM. Scr, scrambled. **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5.
RNF186 Interacts with RIP2 and Is Required For Assembly of the NOD2 Signaling Complex.
Molecules recruited to a complex assembled upon PRR stimulation can be both shared across PRRs (e.g., TRAF6) and unique to select PRRs. As RNF186 regulates both MAPK and NFκB pathways, we assessed whether RNF186 interacts with and contributes to the formation of the upstream PRR-induced signaling complex, with an initial focus on NOD2, and examination of additional PRRs in subsequent studies. Upon NOD2 stimulation in MDMs, RNF186 interacted with Receptor-Interacting Protein 2 (RIP2) (Fig. 2G), the adaptor protein downstream of NOD2, which then forms a signaling complex with IRAK1 and TRAF6 (13–15). Moreover, upon RNF186 knockdown, the assembly of NOD2, IRAK1, and TRAF6 with RIP2 (Fig. 2H), as well as the recruitment of phospho–extracellular signal-regulated kinase (ERK), phospho-p38, and phospho-IκBα to the RIP2 complex (Fig. 2I), was reduced. As RNF186 did not regulate Dectin-1–induced cytokines (SI Appendix, Fig. S2F), we examined the Dectin-1 signaling complex as a negative control. RNF186 did not associate with the curdlan-induced Dectin-1 and Syk2 complex (SI Appendix, Fig. S2G). Therefore, upon NOD2 stimulation, RNF186 associates with, and is required for optimal assembly of, the NOD2-signaling complex in human MDMs.
RNF186 Ubiquitinates RIP2 and RNF186-Dependent E3 Ubiquitin Ligase Activity Is Required For NOD2-Induced Signaling and Cytokine Secretion.
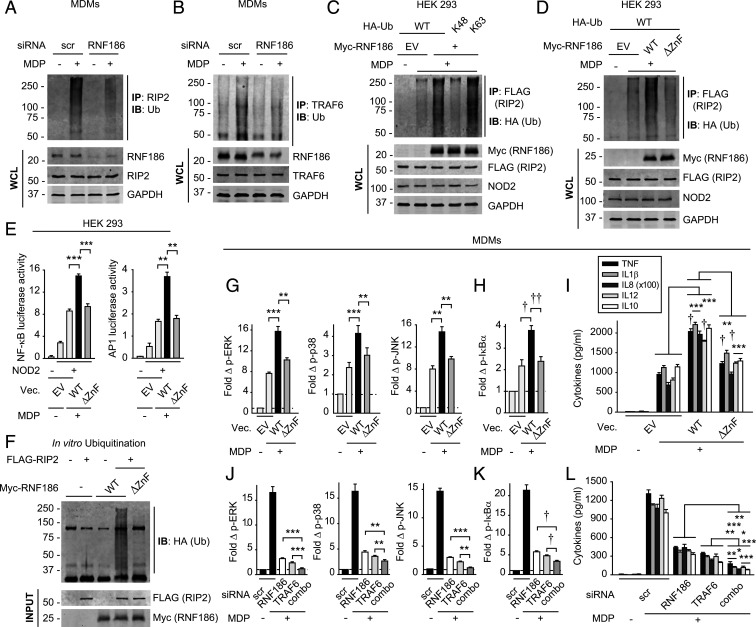
Ubiquitination of various molecules in the PRR-initiated complex is critical for downstream signaling; RIP2 ubiquitination promotes NOD2-induced signaling (16). We observed optimal RIP2-associated complex ubiquitination in MDMs 45 min after NOD2 stimulation (SI Appendix, Fig. S4A). Upon RNF186 knockdown, NOD2-induced ubiquitination of the RIP2-associated complex was reduced (Fig. 3A). We also assessed ubiquitination of the complex associated with TRAF6 as a signaling molecule shared across PRRs; NOD2-induced TRAF6-associated complex ubiquitination was also reduced with RNF186 knockdown (Fig. 3B). We next assessed whether RNF186 promotes K63- or K48-linked ubiquitination of the RIP2 complex; MDP-induced K63-linked polyubiquitination of RIP2 can activate NFκB signaling (17, 18). We coexpressed myc-RNF186 and FLAG-RIP2 together with wild type (WT) ubiquitin or K63- or K48-detecting constructs in human embryonic kidney 293 (HEK293) cells. Upon NOD2 stimulation, RNF186 led to K63-linked ubiquitination of the RIP2-associated complex (Fig. 3C).
Fig. 3.
RNF186 promotes NOD2-induced K63-linked ubiquitination of RIP2 and RNF186-dependent E3 ligase activity is required for NOD2-induced signaling and cytokines. (A and B) MDMs were transfected with scrambled or RNF186 siRNA and then treated with 100 μg/mL MDP for 45 min. RIP2 (A) or TRAF6 (B) was IP, and ubiquitinated proteins (Ub) were assessed by IB. (C) HEK293 cells were transfected with empty vector (EV) or WT myc-RNF186, FLAG-RIP2, NOD2, and HA-tagged WT or K48- or K63-detecting constructs of ubiquitin. Cells were then treated with 100 μg/mL MDP for 45 min. RIP2 (FLAG) was IP, and the association of ubiquitinated proteins was assessed by α-HA (IB). (D) HEK293 cells were transfected with EV or myc-tagged WT or ΔZnF RNF186, FLAG-RIP2, and NOD2. Cells were then treated with 100 μg/mL MDP for 45 min. RIP2 (FLAG) was IP followed by IB with α-HA. WCLs were assessed for equal loading. (E) HEK293 cells were transfected with EV, WT, or ΔZnF RNF186 along with NOD2 ± NFκB or AP-1 luciferase and Renilla constructs. Cells were then treated with 100 μg/mL MDP (four replicates; representative of two independent experiments). NFκB and AP-1 luciferase activity at 6 h. (F) In vitro ubiquitination was assessed as per Materials and Methods with purified HA-ubiquitin ± purified FLAG-RIP2 ± purified myc-WT or −ΔZnF RNF186. Ubiquitin protein (α-HA) was detected by Western blot. Representative of two to three independent experiments for A, C, D, and F. (G–I) MDMs from rs6426833 AA risk carriers (low RNF186-expressors) were transfected with EV or FLAG-tagged WT or ΔZnF RNF186 (n = 6 donors; similar results in additional n = 6). (J–L) MDMs were transfected with scrambled or RNF186 or TRAF6 siRNA, alone or in combination (comb) (n = 6; similar results in additional n = 6). (G–L) Cells were then treated with 100 μg/mL MDP. (G, H, J, and K) Fold phospho-protein induction at 15 min. (I and L) Cytokines at 24 h. Mean + SEM. Scr, scrambled; Vec, vector. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5.
To assess whether the ZnF motif in the RING domain of RNF186 is required for RIP2-associated complex ubiquitination, we coexpressed either WT or a ZnF mutant of RNF186 (RNF186-ΔZnF) (mutant construct as per SI Appendix, Fig. S4B) together with RIP2 and NOD2 in HEK293 cells. The respective constructs expressed at similar levels (Fig. 3D). WT RNF186 enhanced NOD2-induced ubiquitination of the RIP2-associated complex, whereas RNF186-ΔZnF did not (Fig. 3D). Consistently, WT RNF186 enhanced NOD2-induced NFκB and AP-1 luciferase activity (Fig. 3E) and cytokine secretion (SI Appendix, Fig. S4C) in these transfected HEK293 cells, whereas these outcomes were impaired with RNF186-ΔZnF (Figs. 3E and SI Appendix, Fig. S4C). To establish if RNF186 directly ubiquitinates RIP2, we conducted in vitro ubiquitination assays with purified RNF186 and RIP2 (purity confirmations in SI Appendix, Fig. S4 D and E). Importantly, RNF186 directly catalyzed RIP2 ubiquitination (Fig. 3F). Moreover, this ubiquitination was impaired with RNF186-ΔZnF (Fig. 3F).
To assess the requirement for the E3 ligase activity of RNF186 in NOD2-induced outcomes in MDMs, we transfected either WT RNF186 or RNF186-ΔZnF into MDMs from low RNF186-expressing carriers (rs6426833 AA) so as to minimize endogenous RNF186 expression. We ensured equal levels of transfection (SI Appendix, Fig. S4 F and G). WT RNF186 increased NOD2-induced signaling (Fig. 3 G and H) and cytokines (Fig. 3I) in MDMs, thereby providing a complementary approach to the knockdown RNF186 studies. RNF186-ΔZnF did not as effectively increase these NOD2-induced outcomes (Fig. 3 G–I). Therefore, the E3 ligase activity of RNF186 is required for NOD2-induced RIP2 ubiquitination, signaling, and cytokines.
As TRAF6 is a well-described E3 ligase that ubiquitinates the signaling complex downstream of PRRs, including NOD2 (19), we compared RNF186 and TRAF6 contributions to NOD2-initiated outcomes in MDMs. We effectively reduced TRAF6 expression (SI Appendix, Fig. S5 A–C). NOD2-induced ubiquitination of the RIP2 complex was reduced with both RNF186 and TRAF6 knockdown (SI Appendix, Fig. S5D). NOD2-induced MAPK and NFκB signaling and cytokine secretion were also reduced to similar levels (Fig. 3 J–L). Furthermore, reduction in both RNF186 and TRAF6 expression led to a greater reduction in signaling and cytokines compared to reduction with each alone (Fig. 3 J–L). Therefore, RNF186 and TRAF6 function in a nonredundant manner to mediate NOD2-induced outcomes, and they further cooperate in these outcomes.
The focus to date on RNF186 functions has been in epithelial cells; however, its ability to mediate innate immune outcomes in epithelial cells in response to microbial products is poorly defined. RNF186 association with RIP2 increased in HeLa cells upon NOD2 stimulation (SI Appendix, Fig. S5E). Through complementary overexpression (SI Appendix, Fig. S5F) and knockdown (SI Appendix, Fig. S5G) approaches, we observed that RNF186 and TRAF6 mediated nonredundant and cooperative roles in NOD2-induced cytokines in HeLa cells.
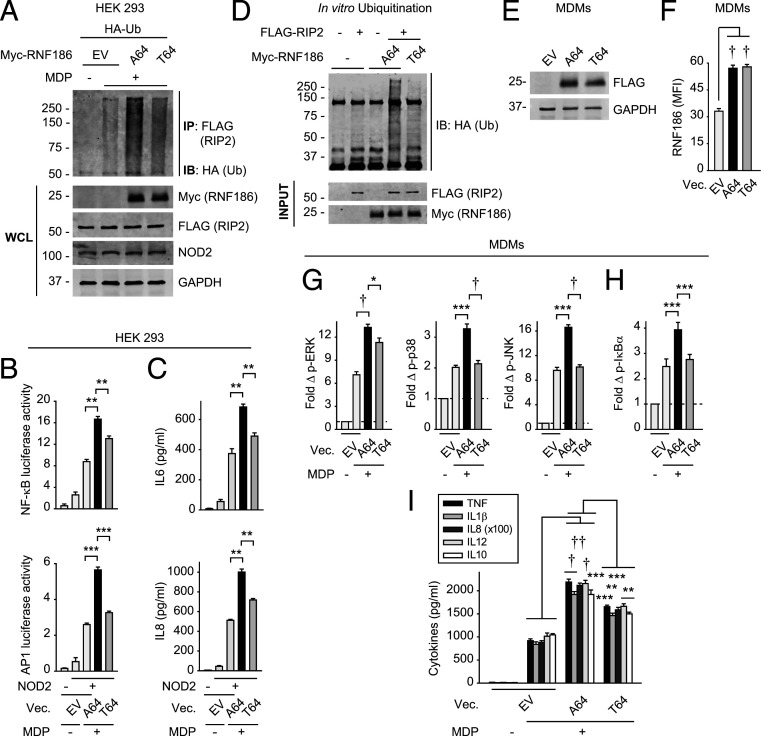
The Rare RNF186-A64T Disease-Risk Variant Shows Reduced NOD2-Induced Ubiquitination of RIP2 and Downstream Signaling.
We next assessed the consequences of the rare RNF186 A64T UC-risk variant; this mutation is located in the RING domain (required for E3 ubiquitin ligase activity) of RNF186 (SI Appendix, Fig. S6A). As this is a rare variant (0.8% allele frequency), we utilized a transfection approach to assess this disease-risk coding mutation. We coexpressed myc-RNF186-A64 (WT) or myc-RNF186-T64 (risk variant) with FLAG-RIP2 and NOD2 in HEK293 cells and ensured equivalent expression (Fig. 4A). Whereas RNF186-A64 enhanced NOD2-induced ubiquitination of the RIP2-associated complex, RNF186-T64 demonstrated a less effective increase in this ubiquitination (Fig. 4A) and, in turn, less effective NFκB and AP-1 activation (Fig. 4B) and cytokine secretion (Fig. 4C and SI Appendix, Fig. S6B). Importantly, the ability of purified RNF186-T64 (SI Appendix, Fig. S4D) to directly ubiquitinate RIP2 in vitro was reduced compared to RNF186-A64 (Fig. 4D). We further assessed the effects of the rare RNF186-T64 variant on NOD2-induced outcomes in MDMs. Upon transfection into low RNF186-expressor (rs6426833 AA) MDMs so as to minimize endogenous RNF186 expression, both variants expressed at similar levels (Fig. 4 E and F). RNF186-T64 demonstrated a reduced ability to enhance NOD2-induced signaling (Fig. 4 G and H) and cytokines (Fig. 4I) compared to RNF186-A64 (WT). Therefore, the disease-associated A64T mutation located in the RING domain of RNF186 results in lower levels of NOD2-induced RIP2 ubiquitination, signaling, and cytokine secretion.
Fig. 4.
The rare RNF186 A64T IBD-risk variant results in decreased NOD2-induced outcomes compared to WT RNF186. (A) HEK293 cells were transfected with EV, myc-RNF186-A64 (WT) or myc-RNF186-T64 (rare risk variant), and FLAG-RIP2, NOD2, and HA-ubiquitin. Cells were then treated with 100 μg/mL MDP for 45 min. RIP2 (FLAG) was IP and the association of ubiquitinated proteins was assessed by α-HA (IB). WCL were assessed for equal loading. Representative of three independent experiments. (B and C) HEK293 cells were transfected with EV, RNF186-A64 (WT), or RNF186-T64 (risk variant) along with NOD2 ± AP-1 or NFκB luciferase and Renilla constructs. Cells were then treated with 100 μg/mL MDP (representative of two independent experiments). NFκB and AP-1 luciferase activity at 6 h (four replicates) (B) and cytokine secretion at 24 h (five replicates) (C). (D) In vitro ubiquitination was assessed with purified HA-ubiquitin ± purified FLAG-RIP2 ± purified myc-RNF186-A64 or myc-RNF186-T64. Ubiquitin protein (anti-HA) was detected by Western blot. Representative of three independent experiments. (E–I) MDMs from rs6426833 AA risk carriers (low RNF186-expressors) were transfected with EV, FLAG-tagged RNF186-A64 (WT), or RNF186-T64 (risk variant). (E and F) RNF186 protein expression by Western blot (E) or flow cytometry (F) (n = 6 donors). (G–I) Cells were treated with 100 μg/mL MDP (n = 6). (G and H) Fold phospho-proteins at 15 min. (I) Cytokines at 24 h. Mean + SEM. Vec, vector. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5.
RNF186 Is Required for Optimal Induction of Antimicrobial Pathways and Intracellular Bacterial Clearance.
Given that both the rare RNF186 A64T UC-risk variant and common RNF186 rs6426833 A UC-risk variant result in reduced PRR-induced outcomes, we sought to define mechanisms through which a loss of function in RNF186 might confer IBD risk. Reduced PRR-induced signaling and impaired antimicrobial responses can increase risk for IBD (20–27). PRR-initiated signaling promotes antimicrobial pathways (1) and, consistent with residence in an environment of ongoing PRR signaling, intestinal macrophages more effectively clear microbes compared to peripheral macrophages (10). As such, the chronic NOD2 stimulation observed in intestinal tissues enhances microbial clearance (1, 28). We therefore hypothesized that RNF186 might enhance bacterial clearance mechanisms in macrophages and that loss of function in RNF186 would impair antimicrobial pathways. Upon knockdown of RNF186 in human MDMs, cells less effectively cleared the gut resident Enterococcus faecalis (E. faecalis) in untreated macrophages and particularly in chronic NOD2-stimulated macrophages (Fig. 5A). We observed similar results with adherent invasive Escherichia coli, which is enhanced in the ilea of Crohn’s disease patients (29), and with S. Typhimurium (Fig. 5A).
Fig. 5.
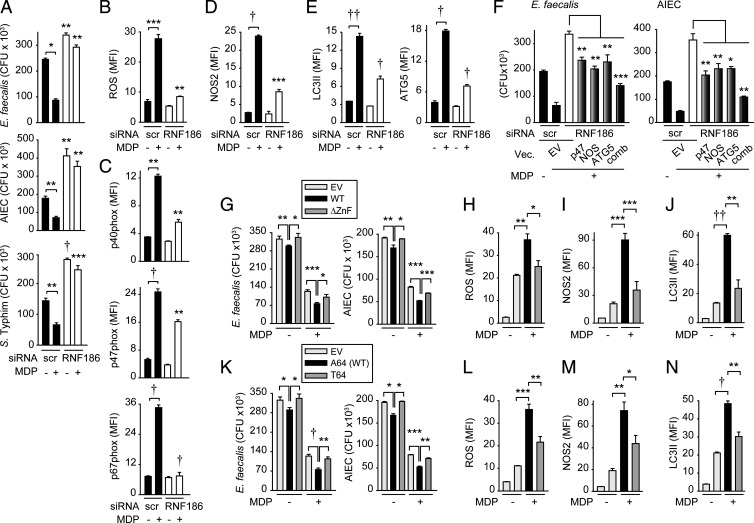
RNF186 is required for NOD2-induced antimicrobial pathways and intracellular bacterial clearance. (A–E) MDMs (n = 4 donors; similar results in additional n = 4) were transfected with scrambled or RNF186 siRNA and then treated with 100 µg/mL MDP for 48 h. (A) Intracellular bacterial clearance (for S. Typhimurium n = 6 and similar results in additional n = 4). (B) ROS production. (C) p40phox, p47phox, and p67phox expression. (D) NOS2 expression. (E) LC3II and ATG5 expression. (F) MDMs (n = 6 donors) were transfected with scrambled or RNF186 siRNA ± EV, p47phox-, NOS2-, or ATG5-expressing vectors, alone or in combination (comb), then left untreated or treated with 100 μg/mL MDP for 48 h. Intracellular bacterial clearance. (G–N) MDMs (n = 6; similar results in additional n = 6) were transfected with EV, WT RNF186, or ∆ZnF RNF186 (G–J) or RNF186-T64 (K–N) and then treated with 100 µg/mL MDP for 48 h. (G and K) Intracellular bacterial clearance. (H and L) ROS. (I and M) NOS2. (J and N) LC3II. Mean + SEM. Significance is compared to the respective scrambled-transfected condition or as indicated in A–E. Scr, scrambled; Vec, vector. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5.
We next examined mechanisms through which RNF186 contributes to intracellular bacterial clearance. Reactive oxygen species (ROS) and reactive nitrogen species (RNS) can contribute to bacterial clearance, and they cooperate to maintain homeostasis in environments heavily populated by microbes, such as the intestine (30). Chronic PRR stimulation induces these pathways. Upon RNF186 knockdown in MDMs, chronic NOD2-induced ROS was reduced (Fig. 5B and SI Appendix, Fig. S7A), and accordingly, induction of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase subunits p40phox, p47phox, and p67phox was reduced (Fig. 5C and SI Appendix, Fig. S7B). Chronic NOD2 stimulation also enhanced expression of the RNS-producing enzyme NOS2; however, this was similarly less effective in RNF186-deficient MDMs (Fig. 5D and SI Appendix, Fig. S7C). Autophagy is yet another key antimicrobial pathway. With chronic NOD2 stimulation, autophagy increased as assessed by the marker LC3II (Fig. 5E and SI Appendix, Fig. S7D). In contrast, RNF186-deficient MDMs less effectively induced autophagy (Fig. 5E). To define mechanisms wherein RNF186 enhances autophagy, we examined the expression of the autophagy-associated molecule ATG5. ATG5 protein expression increased with chronic NOD2 stimulation, and optimal ATG5 induction required RNF186 (Fig. 5E and SI Appendix, Fig. S7D).
We ensured that upon effective knockdown of each p40phox, p47phox, and p67phox (SI Appendix, Fig. S7E), intracellular bacterial clearance was less effective, with p47phox contributing to the greatest degree (SI Appendix, Fig. S7F). While the NADPH oxidase (p47phox), NOS2, and autophagy (ATG5) pathways each contributed to NOD2-induced intracellular bacterial clearance, they particularly cooperated in this clearance, as assessed by combined knockdown of the pathways (SI Appendix, Fig. S7 G–I). To clearly establish that each of the RNF186-dependent antimicrobial pathways was contributing to intracellular bacterial clearance in MDMs, we restored ROS production through transfection of p47phox (SI Appendix, Fig. S7 J and K), NOS2 induction (SI Appendix, Fig. S7L), and autophagy induction through transfection of ATG5 (SI Appendix, Fig. S7M) in RNF186-deficient cells. Restoring each pathway in RNF186-deficient cells partially rescued intracellular bacterial clearance (Fig. 5F). Combined restoration of ROS (p47phox), RNS (NOS2), and autophagy (ATG5) pathways in RNF186-deficient cells more effectively restored intracellular bacterial clearance, highlighting cooperation between these pathways (Fig. 5F). These data indicate that RNF186 is required for optimal induction of ROS, RNS, and autophagy pathways and, in turn, intracellular bacterial clearance in macrophages.
We next sought to establish whether the ubiquitin activity of RNF186 is required for the RNF186-dependent antimicrobial pathways we had defined. Transfection of WT RNF186 into MDMs enhanced NOD2-induced bacterial clearance (Fig. 5G), as well as NOD2-induced ROS, RNS, and autophagy pathways (Fig. 5 H–J), thereby providing evidence for RNF186 in promoting these pathways in a complementary approach to the knockdown studies above. Importantly, the RNF186 ∆ZnF mutant demonstrated less effective NOD2-induced bacterial clearance and ROS, RNS, and autophagy pathway induction compared to WT RNF186-transfected MDMs (Fig. 5 G–J).
We next assessed how the rare RNF186-A64T IBD-risk variant regulates bacterial clearance and the implicated antimicrobial pathways. MDMs transfected with RNF186-T64 (UC-risk variant) demonstrated reduced NOD2-induced bacterial clearance (Fig. 5K) along with reduced induction of ROS, RNS, and autophagy pathways compared to RNF186-A64 (Fig. 5 L–N). Taken together, these data indicate that RNF186 promotes NOD2-induced antimicrobial pathways in human MDMs; this regulation requires an intact RNF186 RING domain and is impaired with the RNF186-T64 IBD-risk variant.
RNF186 Promotes Outcomes Downstream of a Broad Range of PRRs.
Intestinal cells are exposed to a broad range of microbial components, such that we sought to define RNF186 contributions to additional PRRs. Reduced RNF186 expression led to decreased cytokine secretion upon stimulation of TLR2, TLR3, TLR4, TLR5, TLR7, and TLR9 (Fig. 6A). Further, RNF186 knockdown led to reduced TLR4-induced MAPK and NFκB signaling (Fig. 6 B and C), ubiquitination of the TRAF6-associated complex (Fig. 6D), and intracellular bacterial clearance (Fig. 6E). In a complementary approach, transfection of WT RNF186 into MDMs increased TLR2- and TLR4-induced signaling, cytokines, and antimicrobial pathways, and importantly, the rare RNF186 A64T risk variant did not as effectively increase these TLR2- and TLR4-induced outcomes (SI Appendix, Fig. S8). A prior report described contamination of media/serum with peptidoglycan fragments (31). Peptidoglycan recognition proteins detected in media from MDMs was negligible (SI Appendix, Fig. S9 A and B). Importantly, responses to the TLR2 and TLR4 ligands were specific to their respective receptors and not attributable to peptidoglycan contamination leading to NOD2 activation. In particular, upon NOD2 and RIP2 (adapter molecule for NOD2) knockdown, MDP-induced cytokines were dramatically impaired, while treatment with TLR2 and TLR4 ligands remained intact (SI Appendix, Fig. S9 C–F), thereby indicating that the TLR2 and TLR4 responses were not attributable to peptidoglycan/MDP contamination. In contrast, knockdown of TLR2 and TLR4 selectively reduced responses to their respective ligands (SI Appendix, Fig. S9 G–J). Taken together, RNF186 promotes signaling, signaling complex ubiquitination, cytokines, and intracellular bacterial clearance across a range of PRRs.
Fig. 6.
RNF186 is required for cytokine secretion downstream of multiple PRRs and promotes TLR4-induced TRAF6-associated complex ubiquitination, signaling, and bacterial clearance. (A–E) MDMs were transfected with scrambled or RNF186 siRNA. (A) Cells were treated with 10 μg/mL Pam3Cys (TLR2), 100 μg/mL polyI:C (TLR3), 0.1 μg/mL lipid A (TLR4), 5 ng/mL flagellin (TLR5), 1 μg/mL CL097 (TLR7), or 10 μg/mL CpG (TLR9) for 24 h. Cytokine secretion (n = 4 donors; similar results in additional n = 4). (B–E) Cells were treated with 0.1 μg/mL lipid A. (B and C) Fold phospho-proteins at 15 min (n = 6 donors). (D) TRAF6 was IP and the association of ubiquitinated proteins was assessed by α-HA (IB) at 45 min. WCLs were assessed for equal loading. (E) After 48 h, intracellular bacterial clearance was assessed (n = 6 donors). (F) Mouse intestinal macrophages were cocultured with S. Typhimurium. Rnf186 mRNA at 4 h (n = 5; representative of two independent experiments). (G–I) Scrambled or RNF186 siRNA was administered to mice (n = 5; n = 6 in additional independent dose–response experiment). Colonic macrophages were then isolated. (G) Rnf186 mRNA. (H and I) Cells were then cocultured with S. Typhimurium. (H) Cytokine secretion at 24 h. (I) Intracellular bacterial clearance. Mean + SEM. Significance is compared to the respective scrambled siRNA-transfected condition or as indicated. Scr, scrambled. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5.
RNF186 Promotes Intracellular Bacterial Clearance in Intestinal Macrophages.
We next assessed whether RNF186 promotes both cytokine secretion and bacterial clearance in intestinal myeloid cells. To this end, we utilized mouse systems and first ensured that RNF186 regulated outcomes in mouse bone marrow–derived macrophages (SI Appendix, Fig. S10 A–D) similar to that observed in human MDMs. Upon coculture of intestinal macrophages (SI Appendix, Fig. S10E) with S. Typhimurium, Rnf186 mRNA expression increased (Fig. 6F). We then delivered RNF186 siRNA to mice in vivo and confirmed reduced Rnf186 expression in the colon (SI Appendix, Fig. S10F), as well as in colonic macrophages (Fig. 6G). Similar to observations in peripheral macrophages, in Rnf186-deficient intestinal macrophages, S. Typhimurium–induced cytokines (Fig. 6H) and intracellular bacterial clearance (Fig. 6I) were reduced.
MDMs from IBD-risk rs6426833 A Carriers Show Reduced NOD2-Induced Complex Ubiquitination, Signaling, and Antimicrobial Pathways.
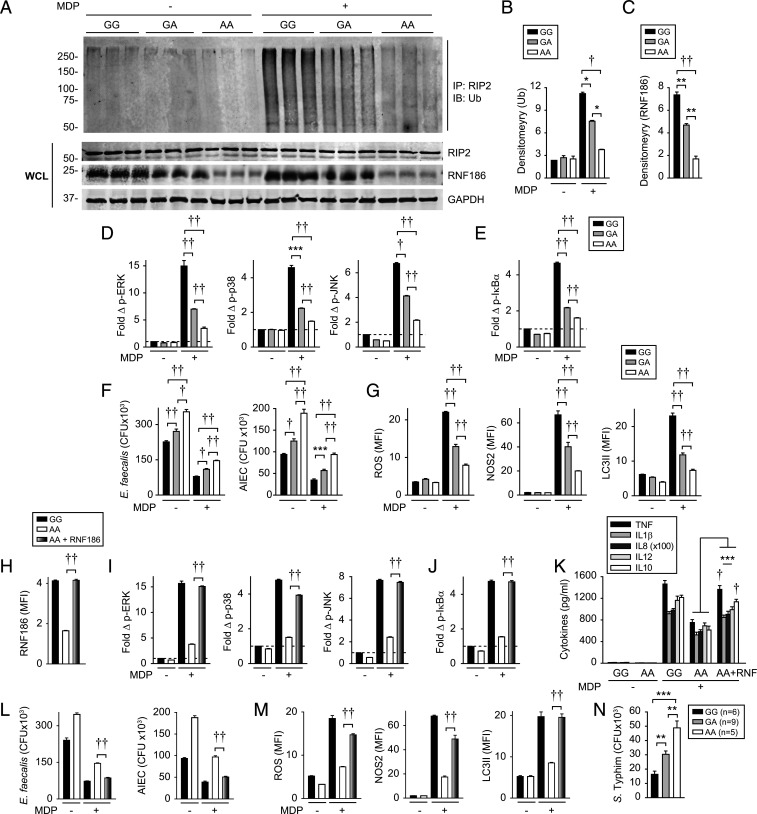
We next assessed whether the RNF186-dependent, PRR-initiated outcomes we had defined were regulated in an rs6426833 genotype–dependent manner; we had shown that the rs6426833 AA risk variant leads to reduced RNF186 expression and cytokines in Fig. 1. Relative to rs6426833 G carrier MDMs, A risk carrier MDMs demonstrated reduced NOD2-induced ubiquitination of RIP2-associated proteins (Fig. 7 A and B). These studies also confirmed reduced RNF186 expression in rs6426833 A risk carrier MDMs using Western blot as an independent approach (Fig. 7 A and C). Consistent with the decreased NOD2-induced ubiquitination (Fig. 7A) and cytokine secretion (Fig. 1), MDMs from rs6426833 A risk carriers demonstrated reduced NOD2-induced MAPK (Fig. 7D) and NFκB (Fig. 7E) activation compared to G carriers. Furthermore, MDMs from rs6426833 A risk carriers demonstrated a reduced baseline and NOD2-enhanced intracellular bacterial clearance compared to G carrier cells (Fig. 7F). Consistently, MDMs from rs6426833 A risk carriers demonstrated reduced NOD2-induced ROS, NOS2, and autophagy pathways compared G carriers (Fig. 7G). Rs6426833 is in a region that also contains the OTUD3 gene, and rs6426833 A risk carriers also demonstrated reduced OTUD3 expression (SI Appendix, Fig. S11). Therefore, to clearly establish the role for RNF186 in regulating the NOD2-induced outcomes above, we expressed RNF186 in AA carriers to the levels observed in GG carriers (Fig. 7H) and found that this increased NOD2-induced signaling, cytokines, and antimicrobial pathways to the levels observed in rs6426833 GG carriers (Fig. 7 I–M). Finally, we examined human intestinal myeloid cells and found intestinal myeloid cells from rs6426833 A carriers demonstrated less effective intracellular bacterial clearance compared to G carriers (Fig. 7N). Taken together, MDMs from rs6426833 A risk carriers demonstrate reduced NOD2-induced RIP2-complex ubiquitination, signaling, and antimicrobial pathways compared to rs6426833 G carriers.
Fig. 7.
IBD-risk rs6426833 A carriers show reduced NOD2-induced ubiquitination, signaling, and antimicrobial pathways. (A–C) MDMs from rs6426833 GG, GA, or AA carriers were left untreated or treated with 100 μg/mL MDP. (A) RIP2 was immunoprecipitated (IP) and ubiquitinated proteins (Ub) were assessed by Western blot at 45 min (shown is n = 3 donors/genotype). WCLs were assessed for loading controls. (B) Densitometric analysis of ubiquitinated complex from top 50 kDa from A (n = 6 donors/genotype). (C) Densitometric analysis of RNF186 expression from A. (D–G) MDMs from rs6426833 GG, GA, or AA carriers (n = 10 donors/genotype; similar results in additional n = 16 donors/genotype) were treated with 100 μg/mL MDP. (D and E) Fold phospho-proteins at 15 min. At 48 h (F and G), cells were assessed for intracellular bacterial clearance (F) and antimicrobial pathways (G). (H–M) MDMs from rs6426833 GG or AA carriers (n = 10 donors/genotype) were transfected with EV or RNF186-expressing vector. (H) RNF186 expression. Cells were then treated with 100 μg/mL MDP. (I and J) Fold phospho-proteins at 15 min. (K) Cytokines at 24 h. At 48 h (L and M), cells were assessed for intracellular bacterial clearance (L) and antimicrobial pathways (M). (N) Intracellular bacterial clearance in human intestinal myeloid cells stratified on rs6426833 genotype. Mean + SEM. *P < 0.05; **P < 0.01; ***P < 0.001; †P < 1 × 10−4; ††P < 1 × 10−5.
Discussion
In this study, we identify a previously undefined role for the IBD-associated gene RNF186 in innate immunity. We find that RNF186 is expressed in human MDMs and is required for PRR-induced signaling, cytokine secretion, and intracellular bacterial clearance. RNF186 contributes to optimal assembly of the PRR-initiated signaling complex and promotes PRR-induced ubiquitination of key molecules in the complex. Intact E3 ligase activity is required for RNF186 to regulate the identified RNF186-dependent PRR-induced outcomes. The rare RNF186 A64T IBD-risk variant located within the RING domain of RNF186 and the common rs6426833 AA risk variant in the RNF186 region leading to reduced RNF186 expression demonstrate reduced NOD2-initiated RIP2 ubiquitination, signaling, cytokine secretion, and intracellular bacterial clearance mechanisms. RNF186 bacterial clearance mechanisms are mediated through the cooperation of ROS, RNS, and autophagy pathways. RNF186 also promotes cytokine secretion and bacterial clearance in intestinal macrophages. Reduced PRR-induced signaling and antimicrobial responses can confer an increased risk for both common and early onset IBD (20, 23–27). Therefore, RNF186 enhances innate immune outcomes in human macrophages, and both the rare A64T and common rs6426833 A RNF186 UC-risk variants demonstrate a loss of function in RNF186-dependent outcomes, thereby providing insight into RNF186 contributions to pathways relevant to IBD pathogenesis (SI Appendix, Fig. S12).
The importance of ubiquitination in promoting PRR-initiated pathways has been observed with additional E3 ubiquitin ligase members such as X-linked Inhibitor of Apoptosis (XIAP), cIAP1, cIAP2, and TRAF6 (18, 32, 33). Moreover, rare XIAP mutations are associated with early onset IBD (34). Rare variants in the E3 ubiquitin ligase TRIM22 are also associated with early onset IBD, with the focus on this variant to date being on its contributions to epithelial cell function (35). With respect to common IBD-associated variants that function in the ubiquitin pathway, A20 modulates the PRR-initiated ubiquitin cascade through both ubiquitin ligase and deubiquitinase activities (36). In contrast to RNF186, A20 down-regulates PRR-initiated signaling and downstream outcomes (36). In this study, we find RNF186 plays both a nonredundant and cooperative role with the well-described E3 ubiquitin ligase TRAF6. Similar to TRAF6 and consistent with the various shared signaling complex molecules between PRRs (e.g., IRAK4, IRAK1, TRAF6, and TAK1), RNF186 contributes to outcomes initiated across a broad range of PRRs. Of note is that RNF186 can also localize to the ER, and components of the PRR-initiated signaling complex (e.g., TRAF6 and RIP2) have been found either directly or indirectly to transiently localize to the ER (37, 38), raising another possible mechanism for RNF186-dependent outcomes. Therefore, this study now defines common IBD-associated genetic variants in RNF186 that promote PRR-induced signaling through modulating the ubiquitin cascade.
Our study highlights a previously undefined role for RNF186 in innate immunity and processes critical for intestinal immune homeostasis, including regulation of outcomes downstream of a broad range of PRRs in human macrophages. We further identify that both the rare RNF186 A64T (through reduced E3 ubiquitin ligase activity) and common rs6426833 A variant in the RNF186 region (through reduced expression) conferring risk for IBD demonstrate a loss of function in PRR-initiated ubiquitination, signaling, cytokines, and antimicrobial pathways, thereby highlighting that modulating either RNF186 levels or function may provide therapeutic benefit in intestinal inflammation and human IBD.
Materials and Methods
Myeloid Cell Isolation and Cell Culture.
Human cell studies in healthy donors were approved by the institutional review board at Yale University. Subjects provided informed consent as indicated. Genotyping was conducted by TaqMan (Life Technologies). Monocytes were purified from human peripheral blood mononuclear cells by adhesion and cultured with 10 ng/mL macrophage colony-stimulating factor (Shenandoah Biotechnology) in 10% fetal bovine serum–containing RPMI media for 7 d to generate MDMs. Please see SI Methods for detailed methods.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01DK099097). We thank Tony Eissa, Celine DerMardirossian, and Emiko Mizoguchi for reagents and Rosa Munoz Xicola for assistance with statistical genetics.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013500118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Abraham C., Medzhitov R., Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology 140, 1729–1737 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jostins L., et al., Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silverberg M. S., et al., Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat. Genet. 41, 216–220 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaudoin M., et al., Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 9, e1003723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang P., Wu Y., Li Y., Zheng J., Tang J., A novel RING finger E3 ligase RNF186 regulate ER stress-mediated apoptosis through interaction with BNip1. Cell. Signal. 25, 2320–2333 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Fujimoto K., et al., Regulation of intestinal homeostasis by the ulcerative colitis-associated gene RNF186. Mucosal Immunol. 10, 446–459 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Liu J., Qian C., Cao X., Post-translational modification control of innate immunity. Immunity 45, 15–30 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Lear T. B., et al., The RING-type E3 ligase RNF186 ubiquitinates Sestrin-2 and thereby controls nutrient sensing. J. Biol. Chem. 294, 16527–16534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng S., Abraham C., NF-κB1 inhibits NOD2-induced cytokine secretion through ATF3-dependent mechanisms. Mol. Cell. Biol. 33, 4857–4871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smythies L. E., et al., Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J. Clin. Invest. 115, 66–75 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franchi L., et al., NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat. Immunol. 13, 449–456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedl M., Abraham C., A TNFSF15 disease-risk polymorphism increases pattern-recognition receptor-induced signaling through caspase-8-induced IL-1. Proc. Natl. Acad. Sci. U.S.A. 111, 13451–13456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogura Y., et al., Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J. Biol. Chem. 276, 4812–4818 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Opitz B., et al., Nucleotide-binding oligomerization domain proteins are innate immune receptors for internalized Streptococcus pneumoniae. J. Biol. Chem. 279, 36426–36432 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Hedl M., Li J., Cho J. H., Abraham C., Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc. Natl. Acad. Sci. U.S.A. 104, 19440–19445 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez-Torres R. J., Chamaillard M., The ubiquitin code of NODs signaling pathways in health and disease. Front. Immunol. 10, 2648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasegawa M., et al., A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 27, 373–383 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y., et al., NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J. Biol. Chem. 282, 36223–36229 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Akira S., Takeda K., Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Uhlig H. H., et al., The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology 147, 990–1007.e3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muise A. M., et al., NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut 61, 1028–1035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Homer C. R., Richmond A. L., Rebert N. A., Achkar J. P., McDonald C., ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology 139, 1630–1641.e2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holland S. M., Chronic granulomatous disease. Clin. Rev. Allergy Immunol. 38, 3–10 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Hedl M., Lahiri A., Ning K., Cho J. H., Abraham C., Pattern recognition receptor signaling in human dendritic cells is enhanced by ICOS ligand and modulated by the Crohn’s disease ICOSLG risk allele. Immunity 40, 734–746 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamas B., et al., CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lahiri A., Hedl M., Yan J., Abraham C., Human LACC1 increases innate receptor-induced responses and a LACC1 disease-risk variant modulates these outcomes. Nat. Commun. 8, 15614 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Travassos L. H., et al., Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 11, 55–62 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Lahiri A., Abraham C., Activation of pattern recognition receptors up-regulates metallothioneins, thereby increasing intracellular accumulation of zinc, autophagy, and bacterial clearance by macrophages. Gastroenterology 147, 835–846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darfeuille-Michaud A., et al., High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127, 412–421 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Shiloh M. U., et al., Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10, 29–38 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Molinaro R., Mukherjee T., Flick R., Philpott D. J., Girardin S. E., Trace levels of peptidoglycan in serum underlie the NOD-dependent cytokine response to endoplasmic reticulum stress. J. Biol. Chem. 294, 9007–9015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertrand M. J., et al., Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity 30, 789–801 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Damgaard R. B., et al., The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol. Cell 46, 746–758 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Speckmann C., Ehl S., XIAP deficiency is a mendelian cause of late-onset IBD. Gut 63, 1031–1032 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Li Q., et al., Variants in TRIM22 That Affect NOD2 Signaling Are Associated With Very-Early-Onset Inflammatory Bowel Disease. Gastroenterology 150, 1196–1207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma A., Malynn B. A., A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat. Rev. Immunol. 12, 774–785 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu Q., et al., Toll-like receptor-mediated IRE1α activation as a therapeutic target for inflammatory arthritis. EMBO J. 32, 2477–2490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bist P., et al., E3 Ubiquitin ligase ZNRF4 negatively regulates NOD2 signalling and induces tolerance to MDP. Nat. Commun. 8, 15865 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.