Abstract
The neutralization assays are considered the gold-standard being capable of evaluating and detecting, functional antibodies. To date, many different protocols exist for micro-neutralization (MN) assay which varies in several steps: cell number and seeding conditions, virus amount used in the infection step, virus-serum-cells incubation time and read out.
The aim of the present preliminary study was to carry out SARS-CoV-2 wild type MN assay in order to investigate which optimal tissue culture infective dose 50 (TCID50) infective dose in use is the most adequate choice for implementation in terms of reproducibility, standardization possibilities and comparability of results. Therefore, we assessed the MN by using two viral infective doses: the “standard” dose of 100 TCID50/well and a reduced dose of 25 TCID50/well. The results obtained, yielded by MN on using the lower infective dose (25 TCID50), were higher respect to those obtained with the standard infective dose. This suggests that the lower dose can potentially have a positive impact on the detection and estimation of real amount of neutralizing antibodies present in a given sample, showing higher sensitivity maintaining high specificity.
Keywords: SARS-CoV-2, Infective dose, Live virus, Micro-neutralisation, Immunological responses
The detection and quantitation of serum antibodies to different viral antigens, after natural infection and/or immunization, has long been used to assess the likelihood of protection against a specific pathogen (Petherick, 2020). The Enzyme Linked-immunosorbent assay (ELISA) is one of the most used method for total antibodies detection. This method is able to detect all the immunoglobulins (class and subclass) present in a given sample able to bind the specific antigen of interest coated in a dedicated plate. It is fast, cheap and safe because it does not require the handling of live pathogens. Another classical way of measuring antibody response for agglutinating viruses such as Influenza, is the Haemagglutination Inhibition assay (HAI). This method is considered as the gold standard in Influenza field (Hirst, 1942; Salk, 1944) and correlates of protection have been established. It is based upon the principle that antibody able to bind the globular head of the haemagglutinin (HA) can inhibit the HA’s ability to agglutinate red blood cells (RBCs) by prevent the binding between the head domain (HA1) and the sialic acids (SA) present on the RBC surface. Both, ELISA and HAI suffer from the fact that they are not able to give a precise indication about the functionality of the antibodies detected. Given these limitations, the neutralization assays are an attractive alternative for the assessment of baseline sero-status and the evaluation of the humoral responses following natural infection and/or vaccination (Klimov et al., 2012). MN assays were developed in 1990 (Okuno et al., 1990; Bachmann et al., 1999). This is a functional assay, and it is able to detect neutralizing antibodies capable of prevent the virus infection of different mammalian cell lines and the neutralization activity is measured as the ability of the sera to reduce the cytopathic effect (CPE) due to inhibition of viral entry and subsequent replication (WHO, 2011). Compared to the ELISA-based methods, the results derived by the MN represent a more precise and relevant estimation of antibody-mediated protection in-vitro (Sicca et al., 2020).
On the other hand, MN is more complex to manage due to some requirements: the need of live viruses and biosecurity level 4, 3 or 2+ laboratories (in case of class IV, III or II pathogens), the costs associated with the assay and the difficulties in protocol standardization across laboratories (e.g. cell lines, infective dose, days of incubation and read-out).
In the present small and investigative study, we focused our attention on the performance of the MN assay with SARS-CoV-2 wild type virus using two different input of viral dose: the standard 100 Tissue Culture Infective Dose 50 % (TCID50) and the 25 TCID50 infective dose. As it is well known in the field of enzymology and enzyme kinetics (Adamczyk et al., 2011), there is a close bond between the half maximum inhibitory concentration (IC50) value and the chosen concentration of the enzyme/molecule in a given system. In this case, by lowering the SARS−COV-2 viral input we expect to observe a general improve in antibody titers and, the focus of this work was to try to evaluate what is the most appropriate value of viral dose to perform the MN in order to have strong sensitivity and specificity as well. Regarding this, a total of 102 human serum samples, anonymously collected in compliance with Italian ethics law, were collected as part of an epidemiological study performed at the University of Siena, Italy (Marchi et al., 2019). The human monoclonal antibody (mAb) IgG1 SAD-S35 (Acrobiosystem) was tested along with the serum samples in the MN assay and ELISA Kit (Euroimmun) as positive control. Human serum minus IgA/IgM/IgG (S5393−1VL) (Sigma, St. Louis, MO, USA) was used as a negative control. SARS-CoV 2 Italy-INMI1, Clade V - wild type virus was purchased from the European Virus Archive goes Global (EVAg, Spallanzani Institute, Rome). The virus was propagated and titrated as previously reported (Manenti et al., 2020). The plates were observed daily for a total of four days for the presence of CPE by means of an inverted optical microscope. The 102 human serum samples were heat-inactivated for 30 min at 56 °C then tested in MN as already reported (Manenti et al., 2020).
After four days of incubation, the plates were inspected by an inverted optical microscope. The highest serum dilution protecting more than the 50 % of cells from CPE was taken as the neutralization titre.
The data obtained have been evaluated to investigate the optimal viral dose that could be effectively used for SARS-CoV-2 strain in the MN assay.
Among various serological tests, the MN is the only assay that can offer a high throughput in processing samples along with the information regarding the capability of the antibodies to prevent the attachment/entry of the virus into the target cells. To date, MN assay is considered the reference standard method for detection of neutralizing antibodies, which may be used as a correlate of protective immunity. Although alternative BSL2 protocols using SARS CoV-2 pseudotyped viruses are being developed to obviate culture of live SARS-CoV-2 virus (Hyseni et al., 2020; Crawford et al., 2020; Nie et al., 2020) these methods remain in the research area.
Historically, such as for Influenza virus, the MN assay is routinely carried out in 96-micro-well plates, by mixing different 2-fold serial dilutions of a serum-containing antibodies with a well-defined viral dose containing 100 TCID50/well. However, for newly emerging viruses such as SARS-CoV-2, the viral dose needs to be accurately evaluated necessitating agreement on a consensus assay protocol for future studies.
The viral load equal to 100 TCID50, in accordance with the empirical formula obtained by applying the Poisson distribution, should be equal to approximately 70 plaque-forming units (pfu), which represents the measure of the infectious viral particles in a certain volume of medium used in each well of the microplate. Clearly, this is valid if the same cell system is used and the virus is able to form plaques on the cells monolayer.
All the 102 serum samples screened have been assayed by Commercial ELISA test in order to assess more specifically the presence/absence of anti-SARS-CoV-2 binding antibodies. Among the ELISA positive sample 19.8–20 % of sera were found positive in MN assay with 100 TCID50 and 25 TCID50 of viral dose.
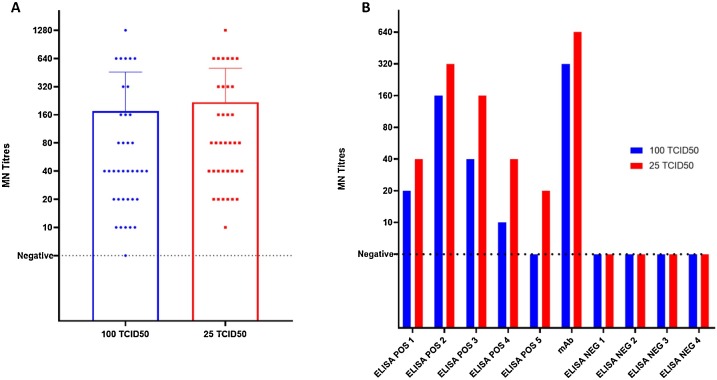
Our results show that, with the lower dose (25 TCID50) in the majority of the cases the MN titres are higher of one or two dilution steps (Fig. 1 A and B). This is also confirmed for the neutralizing mAb, used as a positive control sample for the assay, with a titre equal of 320 using 100 TCID50 and 640 using 25 TCID50. More interestingly, one sample (Fig. 1B; ELISA POS 5) with ELISA positive signal but tested negative in MN 100TCID50 resulted to be low positive for the presence of neutralizing antibodies with 25 TCID50 with a titre of 20. All the ELISA negative samples were also confirmed negative by MN 25TCID50.
Fig. 1.
A) ELISA and MN positive CPE- viral titres obtained when 102 samples were tested against 100 TCID50 and 25 TCID50 SARS-CoV-2 analysed by GraphPad using Kruskal-Wallis non-parametric test; B) impact of the viral load on the neutralization titre in different samples (5 ELISA positive, the neutralizing mAb, 4 ELISA negative sample).
Although it has already been studied by others (Magnus, 2013; Klasse, 2014), these results are of considerable importance supporting the evidence that even if a lower infective dose is used, the possibility to have false positives in ELISA and MN 100 TCID50 confirmed-negative samples is low. Indeed, the sensitivity of the assay to detect functional antibodies could be improved by reducing the viral dose.
Thus, confirming that even with a lower infective dose the cell monolayer is able to results in high percentage of CPE after 4 days (128 h) of incubation, avoiding the possibility to have false positive outcomes due to non-specific inhibition of the viral infection by the high serum concentration at the first sample dilution.
This aspect could be crucial in order to evaluate the immune response against new emerging viruses, such as the SARS-CoV-2, for which immunological and serological data need to be well interpreted. In fact, a variety of in vitro assays for the detection of SARS-CoV-2 neutralizing antibodies has been described but there is no doubt that the absence of oversight and standardisation of serologic tests is a concern. Given that, the available serologic assays are highly variable, differing in their format, the antibody class detected, the selected antigen, and the acceptable sample types (Laurie et al., 2015).
As evidenced before (Petherick, 1942; Theel et al., 2020) it is fundamental to note how serological assays able to detect neutralizing antibody responses could be crucial to provide the most accurate and precise results for vaccine immunogenicity trials. There are many topics of discussion involving antibody responses to SARS-CoV-2 (Chvatal-Medina et al., 2021), and plenty of research is yet to be done in some of these fields (e.g. kinetics and antibody-dependent enhancement mechanisms). However, confirming that the viral dose is not able to compromise the specificity of the neutralisation profiles it would definitely be of great importance for the successful development (design and pre-clinical stage) and assessment of new vaccines platform, such as RNA, DNA or nasal vaccine. Especially for the latter, it is extremely important to have tests able to detect even extremely low levels of Immunoglobulin A (IgA) with neutralizing capability generally present in high diluted human specimens such as nasal wash/swab or saliva (Gianchecchi et al., 2019). Noteworthy was the application of this MN method using 25TCID50 in the first phase of discovery of the extremely potent monoclonal antibody as reported and described in detail in the paper of (Andreano et al., 2021). The use of the lower infective dose allowed us to detect even very low concentration of neutralizing immunoglobulin after the sorting and culturing of single B cells.
As stated before, our observations are in line with the enzymology and competition kinetics laws: decrease in the viral titres lead to an increase in antibody titres, but we believe that the most important point is that the specificity of this assay remain higher. This highlights how the such viral input should be taken as the most appropriate one to perform the MN assay for SARS-CoV-2 virus, since no precise indications or protocols have been established yet.
Even if small and preliminary, this study aims to encourage further international collaborations towards the standardization of the SARS-CoV-2 neutralization assays, maximizing the yield in terms of sensitivity. Said that, albeit at present the ability of a give antibody to neutralize SARS-CoV-2 virus remains the main target for vaccine design and their subsequent approval, more studies are focusing the attention on some mechanisms that could be crucial in Covid-19 pathologies, such as the antibody-dependent cellular cytotoxicity (ADCC). Due to the countless functions of antibodies in immune responses, it is possible that they could mediate protection from disease though different more hidden effector mechanisms (Tso et al., 2021; Tauzin et al., 2021).
Author statement
Conceptualization: AM, EMO; Data curation; AM, EMO, MM, AT. Formal analysis: AM, EMO, MM; Funding acquisition: EM; Investigation; AM, MM, GL, AT Methodology: AM, MM, EMO; Resources: EM; Supervision: EM, AT; Validation:AM, MM; Visualization; AM, EMO; Roles/Writing - original draft: AM, EMO; Writing - review & editing: all authors have approved the final version of the manuscript.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
This publication was supported by the European Virus Archive goes Global (EVAg) project which has received founding from the European Union’s Horizon 2020 research and Innovation Programme under grant agreement No 653316. We would like to thank the Department of Molecular Epidemiology of the University of Siena for providing the human serum samples.
References
- Adamczyk M., van Eunen K., Bakker B.M., Westerhoff H.V. Enzyme kinetics for systems biology when, why and how. Methods Enzymol. 2011;500:233–257. doi: 10.1016/B978-0-12-385118-5.00013-X. PMID: 21943901. [DOI] [PubMed] [Google Scholar]
- Andreano E., Nicastri E., Paciello I., et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell. 2021;184(7):1821–1835. doi: 10.1016/j.cell.2021.02.035. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann M.F., Ecabert B., Kopf M. Influenza virus: a novel method to assess viral and neutralizing antibody titers in vitro. J. Immunol. Methods. 1999;225(1):105–111. doi: 10.1016/S0022-1759(99)00034-4. [DOI] [PubMed] [Google Scholar]
- Chvatal-Medina M., Mendez-Cortina Y., Patiño P.J., Velilla P.A., Rugeles M.T. Antibody responses in COVID-19: a review. Front. Immunol. 2021;(April 12) doi: 10.3389/fimmu.2021.633184. PMID: 33936045; PMCID: PMC8081880.doi:10.1136/bmjopen-2019-032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12(5) doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianchecchi E., Manenti A., Kistner O., Trombetta C., Manini I., Montomoli E. How to assess the effectiveness of nasal influenza vaccines? Role and measurement of sIgA in mucosal secretions. Influenza Other Respir. Viruses. 2019;13(September (5)):429–437. doi: 10.1111/irv.12664. Epub 2019 Jun 21. PMID: 31225704; PMCID: PMC6692539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst G.K. The quantitative determination of influenza virus and antibodies by means of red cell agglutination. J. Exp. Med. 1942;75(1):49–64. doi: 10.1084/jem.75.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyseni I., Molesti E., Benincasa L., et al. Characterisation of SARS-CoV-2 lentiviral pseudotypes and correlation between pseudotype-based neutralisation assays and live virus-based Micro neutralisation assays. Viruses. 2020;12(9) doi: 10.3390/v12091011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse P.J. Neutralization of virus infectivity by antibodies: old problems in new perspectives. Adv. Biol. 2014;2014 doi: 10.1155/2014/157895. Epub 2014 Sep 9. PMID: 27099867; PMCID: PMC4835181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimov A., Balish A., Veguilla V., et al. In: Kawaoka Y., Neumann G., editors. Volume 865. Humana Press; 2012. Influenza virus titration, antigenic characterization, and serological methods for antibody detection. (Influenza Virus. Methods in Molecular Biology (Methods and Protocols)). [DOI] [PubMed] [Google Scholar]
- Laurie K.L., Engelhardt O.G., Wood J., et al. International laboratory comparison of influenza microneutralization assays for a(H1N1)pdm09, a(H3N2), and a(H5N1) influenza viruses by CONSISE. Clin Vaccine Immunol CVI. 2015;22(8):957–964. doi: 10.1128/CVI.00278-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus C. Virus neutralisation: new insights from kinetic neutralisation curves. PLoS Comput. Biol. 2013;9(2) doi: 10.1371/journal.pcbi.1002900. Epub 2013 Feb 28. PMID: 23468602; PMCID: PMC3585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manenti A., Maggetti M., Casa E., et al. Evaluation of SARS-CoV-2 neutralizing antibodies using a CPE-based colorimetric live virus micro-neutralization assay in human serum samples. J Med Virol. Published online May. 2020;8 doi: 10.1002/jmv.25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchi S., Montomoli E., Remarque E.J., et al. Pertussis over two decades: seroepidemiological study in a large population of the Siena Province, Tuscany Region, Central Italy. BMJ Open. 2019;9(10) doi: 10.1136/bmjopen-2019-032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Li Q., Wu J., et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9(1):680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y., Tanaka K., Baba K., Maeda A., Kunita N., Ueda S. Rapid focus reduction neutralization test of influenza A and B viruses in microtiter system. J. Clin. Microbiol. 1990;28(6):1308. doi: 10.1128/jcm.28.6.1308-1313.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petherick A. Developing Antibody Tests for SARS-CoV-2. 2020. Developing antibody tests for SARS-CoV-2; pp. 1101–1102. April 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salk A simplified procedure for titrating hemagglutinating capacity of influenza-virus and the corresponding antibody. J. Immunol. 1944;49(2):87. [Google Scholar]
- Sicca F., Martinuzzi D., Montomoli E., Hukriede A. 2020. Comparison of Influenza-specific Neutralizing Antibody Titers Determined Using Different Assay Readouts and Hemagglutination Inhibition Titers: Good Correlation but Poor Agreement; pp. 2527–2541. [DOI] [PubMed] [Google Scholar]
- Tauzin A., Nayrac M., Benlarbi M., et al. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T cell responses. Cell Host Microbe. 2021;29(July (7)):1137–1150. doi: 10.1016/j.chom.2021.06.001. e6 Epub 2021 Jun 4. PMID: 34133950; PMCID: PMC8175625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theel E.S., Slev Patricia, Wheeler S., Couturier Marc Roger, Wong S.J., Kadkhodaf Kamran. 2020. The Role of Antibody Testing for SARS-CoV-2: Is There One? The Role of Antibody Testing for SARS-CoV-2: Is There One? July 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso F.Y., Lidenge S.J., Poppe L.K., et al. Presence of antibody-dependent cellular cytotoxicity (ADCC) against SARS-CoV-2 in COVID-19 plasma. PLoS One. 2021;16(March (3)) doi: 10.1371/journal.pone.0247640. PMID: 33661923; PMCID: PMC7932539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza. WHO Press; 2011. Manual for the laboratory diagnosis and virological surveillance of influenza; pp. 63–77. [Google Scholar]