Abstract
In cultivated tetraploid potato (Solanum tuberosum), reduction to diploidy (dihaploidy) allows for hybridization to diploids and introgression breeding and may facilitate the production of inbreds. Pollination with haploid inducers (HIs) yields maternal dihaploids, as well as triploid and tetraploid hybrids. Dihaploids may result from parthenogenesis, entailing the development of embryos from unfertilized eggs, or genome elimination, entailing missegregation and the loss of paternal chromosomes. A sign of genome elimination is the occasional persistence of HI DNA in some dihaploids. We characterized the genomes of 919 putative dihaploids and 134 hybrids produced by pollinating tetraploid clones with three HIs: IVP35, IVP101, and PL-4. Whole-chromosome or segmental aneuploidy was observed in 76 dihaploids, with karyotypes ranging from 2n = 2x − 1 = 23 to 2n = 2x + 3 = 27. Of the additional chromosomes in 74 aneuploids, 66 were from the non-inducer parent and 8 from the inducer parent. Overall, we detected full or partial chromosomes from the HI parent in 0.87% of the dihaploids, irrespective of parental genotypes. Chromosomal breaks commonly affected the paternal genome in the dihaploid and tetraploid progeny, but not in the triploid progeny, correlating instability to sperm ploidy and to haploid induction. The residual HI DNA discovered in the progeny is consistent with genome elimination as the mechanism of haploid induction.
A large potato progeny population produced by crossing tetraploid cultivated clones to diploid Phureja lines displays rare instances of haploid inducer chromosomes, which are frequently damaged.
Introduction
Cultivated potato (Solanum tuberosum) is predominantly autotetraploid (2n = 4x = 48), vegetatively propagated, highly heterozygous, and can be severely affected by inbreeding depression. These attributes make potato improvement through conventional breeding slow and difficult. Thus, there have been renewed efforts to reinvent potato as a diploid and inbred-based crop based on true seed in order to keep pace with a rapidly changing market and climate (Jansky et al., 2016). The first step is to capture useful genetic diversity of elite tetraploid cultivars at the diploid level (Lindhout et al., 2011; Jansky et al., 2016). This can be routinely achieved through pollination of a tetraploid of interest with select clones that act as a haploid inducer (HI). In a 4× by 2× HI cross, a fraction of the progeny are 2n = 2x = 24 primary dihaploids lacking chromosomes from the HI parent. Several diploid accessions of Andigenum group potato (formerly Solanum phureja [Spooner et al., 2014]) were demonstrated to act as efficient HIs over half a century ago (Gabert, 1963). Subsequent breeding efforts were successful in obtaining more efficient HIs, as well as incorporating a dominant marker that aids in distinguishing dihaploids from hybrids, which are usually discarded (Hermsen and Verdenius, 1973; Hutten et al., 1993). In practice, most hybrids from 4× by 2× crosses of potato are tetraploid rather than triploid, presumably because of a triploid block (Marks, 1966; Hanneman and Peloquin, 1968; Hanneman and Ruhde, 1978; Jackson et al., 1978).
Relatively little is known about the molecular basis of haploid induction in potato, but cytological evidence provides some clues. In a 4× wild-type (WT) by 2× HI cross, dihaploids originate from seeds with hexaploid (6×) endosperm, which is the expected outcome of a 4× central cell fertilization by a 2× sperm (Wangenheim et al., 1960). In different HI clones, 30%–40% of pollen fails to complete the second mitosis, resulting in a single, larger restitution sperm (RS; single male gamete) or occasionally two sperm cells that potentially have unbalanced chromosome sets (Montezuma-de-Carvalho, 1967; Montelongo-Escobedo and Rowe, 1969). Furthermore, colchicine-treated pollen, but not untreated pollen of wild potato species Solanum tarijense develops RS and induces potato dihaploids, and colchicine treatment of HI pollen further increases the haploid induction rate (Montelongo-Escobedo and Rowe, 1969). Unreduced 2× sperm cells are also produced from restitution of the first or second meiotic divisions, but this increased rate of meiotic restitution is not associated with increased haploid induction efficiency (Hermsen and Verdenius, 1973; Peloquin et al., 1996). From these results, it was concluded that the 2× sperm fertilizes the central cell, leaving no sperm to fertilize the egg, which then develops parthenogenetically. However, HI-specific DNA markers in dihaploids or near-dihaploid aneuploids were reported in some studies (Clulow et al., 1991; Waugh et al., 1992; Clulow et al., 1993; Wilkinson et al., 1995; Allainguillaume et al., 1997; Clulow and Rousselle-Bourgeois, 1997; Straadt and Rasmussen, 2003; Ercolano et al., 2004; Bartkiewicz et al., 2018; Pham et al., 2018), but not others (Samitsu and Hosaka, 2002; Amundson et al., 2020). This suggests that DNA of the HI parent is retained either as chromosomes or segments thereof in an otherwise haploid plant, a diagnostic feature of uniparental chromosome elimination in plants (Laurie and Bennett, 1986; Riera-Lizarazu et al., 1996; Kynast et al., 2001; Gernand et al., 2005; Ishii et al., 2010; Zhao et al., 2013; Kuppu et al., 2015; Maheshwari et al., 2015; Tan et al., 2015; Li et al., 2017). In Arabidopsis thaliana haploid induction systems, selective instability of the HI chromosomes is observed among the hybrid byproducts (Kuppu et al., 2015; Maheshwari et al., 2015; Tan et al., 2015).
Given the role of dihaploids in efforts to convert potatoes into a diploid inbred crop, the incidental transfer of HI DNA remains a concern, as it has been reported to influence the phenotypes of dihaploids (Allainguillaume et al., 1997). The relatively small sample sizes of previous studies, including our previous evaluation of 167 dihaploids that did not identify instances of HI DNA transfer (Amundson et al., 2020), warrant a robust characterization of the frequency and molecular state of incidental HI DNA transfer in primary dihaploids. Furthermore, it is important to investigate the genomic stability of triploid and tetraploid hybrids obtained from potato haploid induction crosses. Toward these ends, we asked two questions: (1) How often, if ever, do potato HIs transmit chromosomes or chromosome fragments to dihaploids? (2) Do dihaploids or hybrid byproducts of potato haploid induction exhibit evidence of genome instability?
In this study, we used genome resequencing to search for DNA from the HI parent and determine its molecular state in 919 dihaploids and 134 hybrids obtained from potato haploid induction crosses. We found that 8.27% of primary dihaploids were aneuploid, which in ∼90% of cases was due to additional chromosomes from the non-inducer parent. Eight primary dihaploids exhibited one to three additional chromosomes from the HI parent, some of which appeared fragmented due to genome instability. Chromosome breakage in dihaploids and hybrids suggests an association between haploidization and genome instability. However, this instability appears to be ploidy-dependent: chromosomes from the HI were fragmented much less often in triploid hybrids than in either dihaploids or tetraploid hybrids. Comparison with tetraploid self-pollinated progeny suggested that the HI genome instability observed in tetraploid hybrids could not be attributed to pollen, sperm, or embryo ploidy per se. In summary, our results indicate that, although rarely, HI can transmit whole or partial chromosomes during the production of dihaploids, and they suggest a role for ploidy of the HI gamete in potato haploid induction.
Results
Widespread aneuploidy among primary dihaploids
Potato haploid induction crosses can yield dihaploids, triploid hybrids, and tetraploid hybrids, with possible aneuploidy from either parent at any ploidy level (Figure 1). We pollinated 19 tetraploid clones with HIs IVP35, IVP101, or PL-4, recorded presence or absence of the inducer-specific embryo spot, and evaluated the ploidy of each plantlet by counting guard cell chloroplasts or by flow cytometry (Supplemental Table S1 and Supplemental Data Set S1). Next, we selected 919 putative dihaploids and 134 hybrids for chromosome dosage analysis by low-coverage whole genome sequencing as previously described (Supplemental Data Set S2; Amundson et al., 2020). For each dihaploid, read depth per chromosome was standardized to that dihaploid’s tetraploid parent such that values near 1, 2, or 3 corresponded to monosomy, disomy, or trisomy, respectively. Aneuploids were then identified as individuals with one or more outlier chromosomes. The analysis was carried out for all 919 dihaploids; representative results are shown for all 229 dihaploids extracted from clone WA.077 (Figure 2, A). In this cohort, we identified 27 aneuploids: 25 had one additional chromosome (2n = 2x + 1 = 25), one had two additional chromosomes (2n = 2x + 2 = 26), and one had three chromosomes (2n = 2x + 3 = 27).
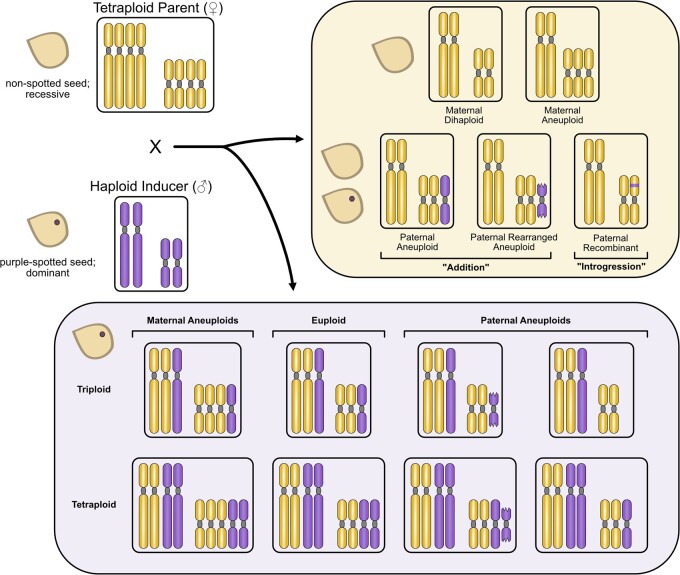
Figure 1.
Possible outcomes of potato haploid induction crosses. The haploid-inducing pollinator genotypes used in this study were homozygous for the dominant embryo spot trait. Presence or absence of HI DNA in the ensuing progeny is expected to manifest as presence or absence of the embryo spot. Progeny from spotted seeds include hybrids that may be aneuploid as well. If potato haploid induction is due to post-zygotic elimination of paternal chromosomes, then occasional failure to eliminate all paternal DNA is expected to result in additional intact or rearranged (addition) paternal chromosomes and/or the integration of paternal DNA segments into maternal chromosomes (introgression). Spotted seeds presumably contain the HI genome and may be triploid or tetraploid, depending on the ploidy of the sperm that took part in fertilization. Diploid sperm may result from unreduced pollen or the blockage of generative cell division (Montelongo-Escobedo and Rowe, 1969).
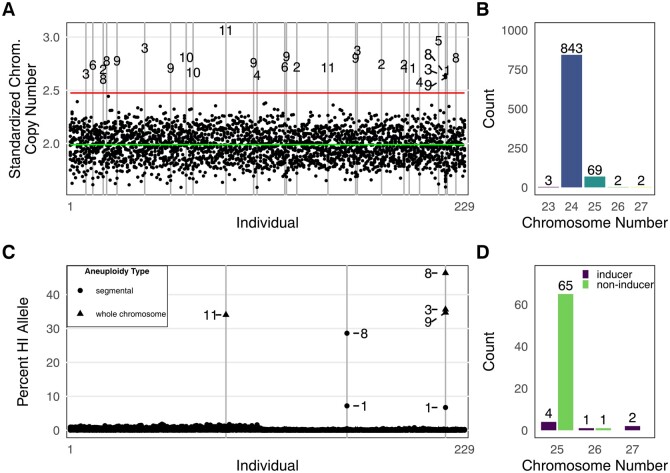
Figure 2.
Incidence and parental origin of aneuploidy among putative dihaploids. A, Standardized chromosome coverage of 229 dihaploids (inferred from flow cytometry) that were extracted from CIP315047 (WA.077). Each individual is displayed along the X-axis, with the stack of 12 points at each coordinate along the X-axis corresponding to the estimated copy number of the 12 potato chromosomes. The green (lower) line corresponds to the population all-chromosome mean, and the red (upper) line corresponds to a cutoff of three standard deviations greater than the mean, which was our criterion for calling whole-chromosome aneuploidy. Outliers in this distribution correspond to additional chromosomes, all of which are numbered by homolog. B, Count of dihaploids by chromosome number inferred from low pass sequencing for all dihaploids evaluated for chromosome dosage (n = 919) in this study. C, Per-chromosome contributions of the HI alleles of the 229 flow-cytometry confirmed dihaploids shown in panel A. Each individual is displayed along the X-axis as a stack of 12 points, with each point corresponding to the HI allele contribution of 1 of the 12 chromosomes. Outliers are numbered by chromosome. Chromosomes identified as outliers in panel A are labeled as whole-chromosome aneuploids; those not identified as outliers are labeled as segmental aneuploids. D, Parental origin of chromosomal deficiencies and excesses for all near-dihaploid aneuploids analyzed for parental origin in this study (n = 73). All compound trisomics resulted in the inheritance of multiple additional chromosomes from the same parent, that is, a 26-chromosome individual exhibited additional chromosomes from either the maternal or paternal parent, but not both.
Among all sequenced dihaploids, 8.3% were aneuploid. Primary trisomics (i.e. single chromosome aneuploids) comprised 91% of the aneuploid class, with the remaining aneuploids consisting of monosomics (2n = 2x−1 = 23) and primary trisomics for multiple chromosomes (2n = 2x + 2 = 26 or 2n = 2x + 3 = 27; Figure 2, B). Each of the 12 homologous chromosomes was recovered as a trisomic. Differences among chromosomes were not significant for either aneuploidy of any type (gains and losses pooled, P = 0.07296; df = 11; χ2 test) or chromosome gains only (P = 0.06726; df = 11; χ2 test; Supplemental Figure S1). It is worth noting that flow cytometric analyses did not readily detect aneuploidy in primary dihaploids.
To evaluate the effect of parental genotype on aneuploidy frequency, we grouped dihaploids based on the genotypes of the parents. When grouped by maternal genotype, aneuploidy frequency ranged from 6.4% to 11.3%, and differences between maternal genotypes were not significant (P = 0.5987; df = 6; χ2 test; Supplemental Figure S2). When grouped by paternal genotype, aneuploidy frequency ranged from 7.6% to 10.3%, and differences between inducer genotypes were not significant either (P = 0.3626; df = 2; χ2 test; Supplemental Figure S3). Taken together, our data show that ∼8% of presumed dihaploids were aneuploid, without detectable aneuploidy bias for parental genotype in this material.
Retention of HI chromosomes
To determine the parental origin of the additional chromosomes in the aneuploid dihaploid progeny, we identified homozygous single-nucleotide polymorphisms (SNPs) between each pair of tetraploid seed parent and HI. We used these SNPs to calculate the percentage of reads that originated from the HI across the genome of every dihaploid. If the additional chromosome originated from the HI, this percentage was expected to be ∼ 33%, while it was expected to be close to 0% if all copies originated from the tetraploid parent. Representative SNP dosage plots are shown in Figure 2, C. In this population, two of the aneuploids identified in Figure 2, A carried chromosomes from the HI parent. One of these two individuals also exhibiting HI alleles above background levels on chromosome 1 (Figure 2, C). A third individual was not aneuploid according to dosage analysis but showed HI alleles above background levels on chromosomes 1 and 8 (Figure 2, C).
Among all dihaploids, we found 66 aneuploids with additional chromosomes exclusively from the tetraploid parent and 8 with additional chromosomes from the HI (Figure 2, D). Two aneuploid lines, MM247 and MM890, were segmental aneuploids with additional HI chromosome segments; the six others showed HI-derived aneuploidy of entire chromosomes (Figure 3 and Supplemental Figure S4). We refer to these eight lines as HI addition dihaploids hereafter. All of the HI genotypes contributed genetic material to at least one dihaploid, and the frequencies at which they did so were not significantly different (P = 0.7747; Fisher’s exact test). The frequency of aneuploidy was consistent with the results of previous analyses of primary dihaploid populations (Frandsen, 1967; Hermsen, 1969; Hermsen et al., 1970; Wagenvoort and Lange, 1975; Samitsu and Hosaka, 2002; Pham et al., 2018; Amundson et al., 2020), and the low frequency (8/919; 0.87%) of aneuploidy due to additional HI chromosomes agrees with previous results in which chromosomes from the inducer parent were not detected in cohorts of ˂200 individuals (Samitsu and Hosaka, 2002; Pham et al., 2018; Amundson et al., 2020).
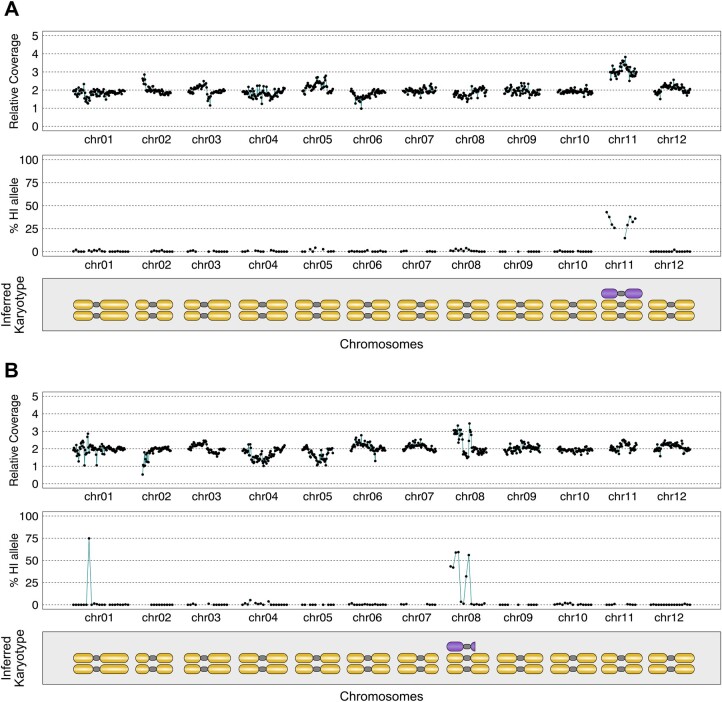
Figure 3.
Paternal genomic contributions to maternal dihaploids. In silico karyotypes of trisomic dihaploids demonstrate the presence of HI DNA. A, Dihaploid MM246, with additional chromosome 11 from HI IVP35. B, Dihaploid MM247, with segmental aneuploidy of chromosome 8 from HI IVP35. The HI segmental addition on chromosome 1 cosegregates with chromosome 8 in each of seven dihaploid populations we evaluated in this study, suggesting that the two loci are physically linked and that potato is either polymorphic for Chr8-1 translocation or that this is an assembly error in the reference genome.
Detection of HI-derived DNA segments in dihaploids
The appearance of inducer DNA fragments shorter than entire chromosomes has also been reported among potato dihaploids (Wilkinson et al., 1995; Pham et al., 2018). Our low coverage sequencing cannot detect segments of this size, but they may be detected with higher coverage sequence. To test whether this type of transfer occurred in our material, we sequenced three HI addition lines to 27–30× coverage and searched for secondary introgressions, that is, segments of chromosomes other than the trisomic chromosome showing HI-specific SNP alleles. Overall, few markers (0.39%–0.69%) were consistent with HI introgression, and HI alleles were underrepresented in allele-specific read depth (Supplemental Figure S5). When considering only putative introgressions covering ≥3 adjacent markers, we found that each of the three lines exhibited putative introgressions, with 3–13 events per line and a total of 21 events (Table 1). Of these putative introgressions, 8 showed identical coordinates in MM247 and MM1114, both of which carried part or all of chromosome 8 from IVP35. One of these putative introgressions, a 1.8-Mb region of chromosome 1, showed linkage to chromosome 8 in seven of our uniparental dihaploid populations (Supplemental Figure S6) and at least three independent mapping populations (Bourke et al., 2015; Endelman and Jansky, 2016), suggesting they are located on the trisomic-HI derived chromosome and only appear as introgressions due to errors in the genome assembly used for analysis (DM1-3 version 4.04). Consistent with this prediction, when we aligned each putative introgression to the updated long-read genome assembly (version 6.1; Pham et al., 2020), we found that eight had an unambiguous top hit to the chromosome corresponding to the HI-derived addition chromosome (Table 1). For the remaining 13, we used short read alignments to the version 4.04 assemblies to locate possible breakpoint junctions. Short read alignments of DM1-3 to itself indicated that these regions were matched uniformly by short reads, but that the putative boundaries were not matched by reads from IVP35, WA.077, and the dihaploid (Table 2 and Supplemental Data Set S3). In conclusion, our analysis did not provide evidence of true introgressions of short HI DNA segments, indicating instead that they can be attributed to genome assembly artifacts or structural variation that occurred prior to haploid induction.
Table 1.
Putative introgressions of HI DNA segments in dihaploid potatoes
| HI addition dihaploid | Tetraploid parent | HI parent | Trisomic chromosome(s) | Segments of HI alleles | Seen in multiple dihaploidsa | Best hit in version 6.1 assembly (if different from original chromosome)b |
|---|---|---|---|---|---|---|
| MM246 | WA.077 | IVP35 | Chr11 | Chr01:45392940-45400039 | No | |
| Chr05:15510198-15510533 | No | |||||
| Chr07:7833610-7844546 | No | |||||
| Chr07:7880027-7880413 | No | |||||
| MM247 | WA.077 | IVP35 | Chr08 | Chr01:24228918-25308116 | Yes | Chr08 |
| Chr01:25309455-26003606 | Yes | Chr08 | ||||
| Chr01:38172625-38174004 | Yes | Chr08 | ||||
| Chr07:9467446-9467874 | Yes | Chr08 | ||||
| MM1114 | WA.077 | IVP101 | Chr03, Chr08, Chr09 | Chr01:24228918-26003606 | Yes | Chr08 |
| Chr01:38172625-38174004 | Yes | Chr08 | ||||
| Chr01:41494766-41499334 | No | |||||
| Chr01:67944376-67945914 | No | Chr08 | ||||
| Chr01:84519844-84521775 | No | |||||
| Chr05:280661-281197 | No | |||||
| Chr05:901127-901352 | No | |||||
| Chr05:15812350-15814025 | No | |||||
| Chr07:5953200-5953784 | No | |||||
| Chr07:9467446-9467874 | Yes | Chr08 | ||||
| Chr10:52856295-52859220 | No | |||||
| Chr10:54984736-54988488 | No | |||||
| Chr12:55689534-55690262 | No |
For each HI addition dihaploid, the trisomic chromosome and reference genome coordinates of putative segmental introgressions are shown.
Coordinates of putative introgression common to two HI addition dihaploids.
Top hit to version 6.1 assembly was to a trisomic HI-derived chromosome with full query coverage and >99.5% nucleotide identity.
Table 2.
Frequency of chromosome breakage among progeny of potato haploid induction crosses
| Chromosome origin | HI cross progeny | Individuals scored | Chromosomes |
P Fisher exact | |
|---|---|---|---|---|---|
| Intact | Fragmented (%) | ||||
| Paternal chromosomes | Dihaploids | 8 | 11 | 2 (15.38) | A |
| 3× hybrids | 30 | 360 | 1 (0.28) | B | |
| 4× hybrids | 104 | 2,366 | 109 (4.40) | C | |
| Maternal chromosomes | Dihaploids | 918 | 22,091 | 5 (0.02) | D |
| 3× hybrids | 30 | 714 | 2 (0.28) | E | |
| 4× hybrids | 104 | 2,470 | 3 (0.12) | F | |
| Putative tetraploid selfs | 14 | 673 | 0 (0) |
A-B: 0.0033. B-C: 0.00001. A-C: 0.1117. D-E: 0.0186. E-F: 0.3135. D-F: 0.0387.
For each progeny class, maternal and paternal chromosomes were recorded as appearing in an intact or fragmented state. The number and frequency of chromosomes of each type (intact versus fragmented) were then grouped by progeny ploidy and parental origin.
Selective instability of the HI genome in dihaploids and tetraploid hybrids
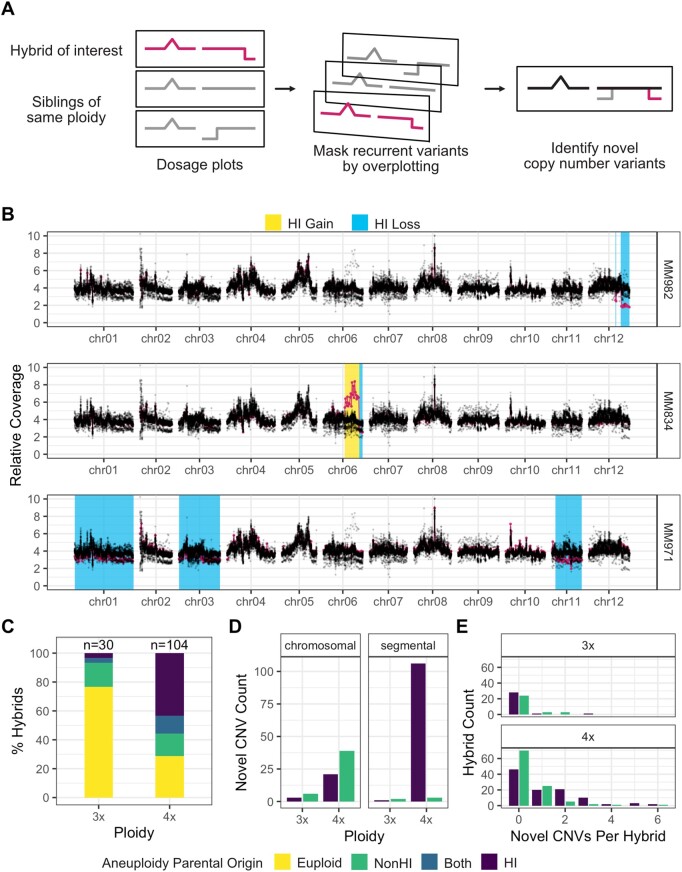
Next, we asked whether the hybrid byproducts of potato haploid induction showed signs of genome instability. Seeds with the dominant, inducer-specific embryo spot marker were germinated and analyzed by flow cytometry, yielding 30 triploids and 104 tetraploids. As a control, we included 14 progeny that did not show the nodal banding phenotype, were tetraploid based on flow cytometry, and lacked HI alleles in the low coverage sequencing; these are likely self-pollinated progeny of the tetraploid clones. To distinguish novel dosage variants attributable to genome instability from recurring variants likely due to pre-existing structural variation in the parents, each offspring was evaluated in the context of its siblings of the same ploidy (Figure 4, A and B). Aneuploids made up a greater proportion of tetraploid hybrids (>70% versus 22% of triploid hybrids), with the frequency of maternally and paternally derived aneuploidy both increasing (Figure 4, C). The per-chromosome rate of HI-derived segmental aneuploidy was significantly lower in the triploid hybrids than the corresponding rate in either dihaploids or tetraploid hybrids, suggesting a greater degree of HI genome instability in dihaploids and tetraploid hybrids (Table 2). Chromosome breakage was not observed in tetraploid selfs, suggesting that the instability of HI-derived chromosomes seen in tetraploid hybrids was not a consequence of 2× pollen and/or sperm per se (Table 2). Relative to triploid hybrids, tetraploid hybrids showed a strong and highly significant increase in the incidence of HI-derived genome instability (P < 0.001; log odds 95% confidence interval 1.51–4.54; Figure 4, C), a difference driven by more frequent segmental aneuploidy of HI-derived chromosomes (Figure 4, D). Most tetraploid hybrids exhibited no more than two novel copy number variations (CNVs) of each parental genome, indicating that genome instability is not restricted to a few exceptional tetraploid hybrids but is pervasive (Figure 4, E). In conclusion, our data suggest that HI-derived chromosomes are selectively unstable in dihaploids and tetraploids, suggesting a specific role of 2× HI sperm in potato haploid induction.
Figure 4.
HI genome instability in tetraploid potato hybrids. Chromosomal variation was investigated by dosage analysis in triploid and tetraploid hybrids. A, Schematic of dosage plot generation for a hybrid cohort. An individual displaying novel dosage variation is identified as an outlier track (pink) compared with common structural variation (gray) in overlaying dosage plots obtained by plotting siblings of the same ploidy. The parental origin of each novel variant is inferred from allele-specific read depth at parent-informative SNP loci. B, Overlay plots of the same sibling hybrid family with novel CNVs highlighted. Regions corresponding to DNA gains or losses are shaded with yellow or blue backgrounds, respectively. In these examples, all novel CNV are attributable to gained or lost HI DNA. C, Increased paternal aneuploidy in near-tetraploid versus near-triploid offspring. Combined data from all cohorts in this study. D, Paternally derived segmental variation is preponderant in tetraploid hybrids. The bars display counts of aneuploidy according to the paternal origin and ploidy of individuals. They also display the counts of whole chromosome aneuploidy versus segmental aneuploidy. E, Number of novel CNV events per hybrid, subdivided by parental origin, showing that HI genome instability in hybrids is not restricted to few individuals.
Tetraploid hybrids produced by first meiotic division restitution of the HI
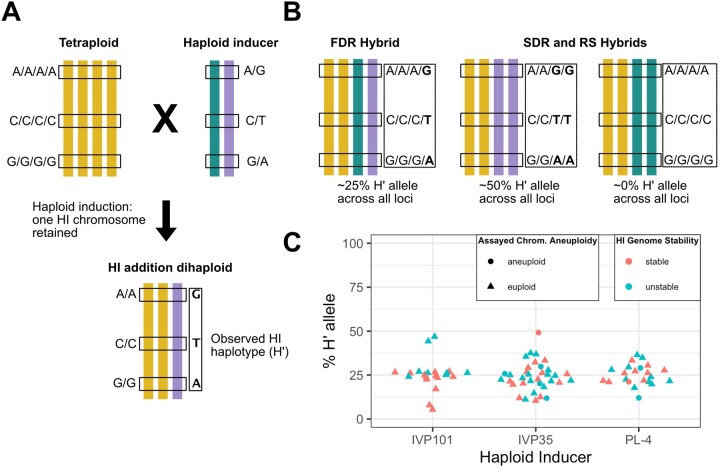
The rate of RS, but not of 2n pollen, appears to be associated with potato haploid induction (Montelongo-Escobedo and Rowe, 1969; Dongyu et al., 1995; Peloquin et al., 1996). We next asked how each 2× sperm was formed, and whether any one mechanism was more likely to be associated with HI genome instability. Based on the high-coverage sequencing of each dihaploid addition line, we derived the HI haplotypes of the additional chromosome(s) in each line (Figure 5, A), which we refer to as H′ hereafter. Using these haplotypes, we then used the low-pass sequencing of 134 hybrids to genotype the centromeres of the chromosomes contributed by the HI. As a control, we analyzed triploid hybrids and found the expected transmission of a single HI haplotype through the centromere and into the chromosome arms (Supplemental Figure S7), indicating that our centromeric HI haplotype phasing was robust. For tetraploid hybrids, the HI-contributed sequences at centromere-linked markers were expected to be heterozygous (show ∼25% H′ allele) if they were derived from 2n first division restitution (FDR) pollen, but homozygous (show either ∼0% or ∼50% HI allele) if they were derived from either 2n second division restitution (SDR) pollen or 2× RS (Figure 5, B). Among the 78 tetraploid hybrids, all but five showed HI heterozygosity at the centromeres, implicating FDR as the dominant mechanism of hybrid formation (Figure 5, C andSupplemental Figure S8). Among the five hybrids with the SDR or RS pattern, one showed ∼50% H′ allele of Cen11, but this individual was disomic for chromosome 11, with both maternal homologs missing (Supplemental Figure S9); together, these results are also consistent with FDR. Of the four remaining hybrids, all of which were derived from IVP101, two showed signs of HI genome instability and two did not. In conclusion, FDR hybrids predominated among tetraploids, with minor contributions possibly from SDR pollen or RS. On the other hand, no mechanism was uniquely associated with HI genome instability.
Figure 5.
Tetraploid hybrid formation during potato haploid induction. A, Biallelic SNP loci used for analysis are homozygous in the tetraploid parent and heterozygous but unphased in the HI. The retained HI chromosome(s) in each HI addition dihaploid represents a phased HI-derived haplotype. To avoid confounding effects from crossovers, only expectations and data for the non-recombining centromere are shown. B, Expected representation of HI haplotype alleles in a non-recombining region of tetraploid hybrids. For all tetraploid hybrids, read information for adjacent loci is binned and the percentage of reads with H′ alleles is calculated. If tetraploids are the product of FDR of the HI, then H′ alleles are expected to appear in 25% of all binned reads. If tetraploids are the product of SDR or RS, the expected percentage could be 50% or 0%, depending on which HI’s haplotype was inherited. C, Percentage of H′ alleles in the centromeres of tetraploid hybrids. Each point corresponds to the percentage of H′ alleles among reads spanning the non-recombining region of a chromosome of one tetraploid hybrid. Chromosome 8 was used to assess IVP101 hybrids. Chromosome 10 was used to assess PL-4 hybrids. Chromosome 11 was used to assess IVP35 hybrids.
Discussion
Multiple studies investigating the presence of HI genetic material in potato dihaploids have come to different conclusions (Clulow et al., 1991, 1993; Waugh et al., 1992; Wilkinson et al., 1995; Allainguillaume et al., 1997; Clulow and Rousselle-Bourgeois, 1997; Samitsu and Hosaka, 2002; Straadt and Rasmussen, 2003; Ercolano et al., 2004; Bartkiewicz et al., 2018; Pham et al., 2018; Amundson et al., 2020). Documenting the existence and extent to which this type of DNA transfer occurs is critical for basic understanding of haploid induction, as well as the use of primary dihaploids for diploid potato breeding. Here, we sequenced a cohort of 919 dihaploids and found that 0.87% of primary dihaploids contained one to three chromosomes or chromosome fragments from the HI. Introgressions of smaller HI DNA segments were also detected, but in approximately one-third of cases, this could be explained by errors in reference genome assembly, while the remaining putative introgressions could not be robustly confirmed. HI chromosomes were generally stable in triploid hybrids but unstable in tetraploid hybrids, most of which were products of FDR pollen.
We documented the occasional appearance of small HI segments (0.5 to few kb). These could represent small translocations or gene conversion derived from the HI genome before elimination. The evidence in support of their presence is robust because it is based on a continuous haplotype that encompasses multiple SNPs. However, in 8 out of 21 instances these segments were physically linked to another HI chromosome present in these samples. For example, identical HI segments were present in HI addition lines MM247 and MM1114, which were both trisomic for chromosome 8. Part of chromosome 8 was previously identified as translocated or misassembled in at least two genetic mapping populations (Bourke et al., 2015; Endelman and Jansky, 2016) as well as in our dihaploid populations. Therefore, these short introgressions are physically part of the trisomic chromosome and map to a different location of the reference assembly, possibly due to genome assembly artifacts or pre-existing translocations in the HI relative to the reference genome. The remaining 13 were flanked by poorly mapping sequences and could not be anchored to a chromosome. While introgression could not be ruled out in these cases, the probability that these are segmental recombination events is low.
The presence of HI DNA in some dihaploids could be explained if, after the formation of a hybrid genome, the HI genome was imperfectly eliminated. Acceptance of this hypothesis would imply that uniparental genome elimination is the mechanism that generates haploids and that the dihaploids with no HI DNA contamination underwent perfect elimination of the HI genome. There are, however, at least two alternative explanations. First, all or a fraction of the perfect dihaploids could result from parthenogenesis. This would require two independent mechanisms of haploid induction to function in the potato system, which seems implausible. Second, the mechanism of haploid induction could entail incomplete gamete fusion. A defective sperm may thus deliver both egg-activating factors and, occasionally, chromosomes but fail to carry out proper karyogamy. This mechanism has not been explicitly proposed, to our knowledge, but may explain observations from the maize (Zea mays) haploid induction system, where in addition to mutations of phospholipases, mutations of fusogenic proteins both enhance the haploid induction rate and induce haploid formation in the WT phospholipase background (Zhong et al., 2019, 2020; Jacquier et al., 2020).
What fates are possible for the HI genome? The genomes delivered by sperm formed by the tetraploid selfs were stable. We could assess HI genome integrity in HI-contaminated dihaploids and in triploid and tetraploid hybrids. Dihaploids that had inherited and maintained HI chromosomes shared high instability of the HI genome with tetraploid hybrids. Triploids, on the other hand, did not. This demonstrates that the HI genome can be inherited and maintained with fidelity, and that instability is not intrinsic to the formation of hybrid zygotes. The tetraploids result from hybridization of a 1n(=2x) egg cell with 2n(=2x) sperm. We infer that either during formation of the 2n sperm or upon fertilization, the HI genome becomes unstable. The instability displayed by tetraploid hybrids could be related to that displayed by HI-containing dihaploids. This provides a potential explanation for the long-standing proposal that 2n sperm triggers haploid induction (Wangenheim et al., 1960; Montezuma-de-Carvalho, 1967; Montelongo-Escobedo and Rowe, 1969). These studies documented the formation of 2x sperm from restitution of the generative cell mitosis in pollen of HIs and suggested a connection to haploid induction. Genome maintenance may become compromised during the formation or growth of 2n pollen, resulting in a fragmented genome that is incompetent for replication and subject to elimination.
It is also possible that this instability is unrelated to genome elimination. We explored the nature of 2n sperm cells in the haploid induction crosses by analyzing the HI contribution in tetraploid hybrids. Restitution of second mitosis predicts homozygosity of 2n sperm. Instead, we found heterozygosity indicative of meiotic FDR in most cases. This agrees with a previous study of microsporogenesis in IVP35 that reported moderate frequencies of parallel spindles (16.88%–26.13%) and fused spindles (1.29%–17.65%) that resulted in ∼29% dyads (Ramanna, 1979). Both parallel and fused spindles are associated with FDR (Peloquin et al., 1999; d’Erfurth et al., 2009; Li et al., 2010; De Storme and Geelen, 2011). However, this leaves unanswered the role of 2x sperm in haploid induction. It demonstrates, however, that the instability observed in the tetraploid hybrids may be connected to FDR. This instability could result from missegregation during meiosis (Ly et al., 2019, 2017; Umbreit et al., 2020), but its relationship to haploid induction is mysterious.
The frequency of aneuploids among primary dihaploids (8.27%) was within the range of previously studied dihaploid populations (1.5%–11.4%; Frandsen, 1967; Hermsen, 1969; Hermsen et al., 1970; Wagenvoort and Lange, 1975; Samitsu and Hosaka, 2002; Pham et al., 2018; Amundson et al., 2020). Based on the availability of robust polymorphic markers, we identified HI DNA in 8 individuals out of 919, including 6 with whole chromosomes and 2 with large fragments. That aneuploids with HI chromosomes make up ˂1% of primary dihaploids is good news for diploid potato breeding, as our results suggest that most primary dihaploids will be free of residual HI DNA. More often, primary dihaploids carry additional maternal chromosomes, possibly due to meiotic non-disjunction in the autotetraploid, female parent. Little evidence is available on the cytology of female meiosis in potato and other autopolyploids (Ramsey and Schemske, 2002), but in male meiosis, high frequencies of univalents and multivalents at metaphase I (Swaminathan, 1954b; He et al., 2018) and unbalanced chromosome sets at metaphase II (Swaminathan, 1954a) have been reported. Regardless of origin, the immediate and potentially lasting impacts of aneuploidy (Henry et al., 2010) are likely adverse and best left avoided. Unlike the primary trisomics of Datura, Arabidopsis, and tomato (Solanum lycopersicum), which show diagnostic phenotypes (Blakeslee, 1922; Rick and Barton, 1954; Steinitz-Sears, 1963; Koornneef and Van der Veen, 1983), primary trisomics could not be readily detected by phenotype alone in primary dihaploids of potato (Hermsen et al., 1970; Wagenvoort and Lange, 1975), indicating that cytological or genetic assays would be required to detect it.
In conclusion, using a large-scale approach, we examined the genomes of 1,053 progeny from haploid induction crosses and determined that, regardless of parental genotype, a small but definite fraction of dihaploids display paternal HI contribution. This large-scale study provides the solid evidence needed to interpret previous studies, and calibrates the expectations for potato haploid induction crosses. In addition, we made an unexpected observation. The HI genome, which is stable when inherited by triploid hybrids, displays selective instability in both tetraploid hybrids and in dihaploids. The interpretation and meaning of these findings are still unclear. At a minimum, they indicate that genome stability is compromised in the HI 2n pollen. They also reinforce the hypothesis that 2n pollen may be required for haploid induction, suggesting future lines of investigation to elucidate mechanisms contributing to this unusual, but highly relevant phenomenon.
Materials and methods
Plant material
Primary dihaploids and hybrids of potato (S. tuberosum) were obtained from 19 tetraploid clones (Supplemental Table S2) via pollination with HIs IVP101 (Hutten et al., 1993), IVP35 (Hermsen and Verdenius, 1973), or PL-4 (also known as CIP596131.4) in greenhouses located at the International Potato Center's (CIP) experimental station in the Peruvian Andes (3,216 meters above sea level, −12.01039°, −75.22411°). Haploid induction crosses were performed during 2015, 2016, and 2017 in greenhouses (average temperature: 19.5°C day and 11.6°C night; relative humidity range: 56.8%–87.4%) located at the CIP’s experimental station. An extended photoperiod of 16 h was arranged by adding one supplementary white lightbulb/m2. Flower buds of the pistillate parents were emasculated and pollinated with HI pollen the following day. All HIs are homozygous for a dominant embryo spot that facilitates the detection of hybrids (Hermsen and Verdenius, 1973; Hutten et al., 1993). Seeds were extracted from mature fruit, recorded for presence or absence of the embryo spot, and germinated in soil. The ploidy of each established seedling was determined by either chloroplast counting as described in Amundson et al. (2020) or flow cytometric measurement of nuclear DNA content against maternal and paternal parents as standards. Refer to Supplemental Data Set S1 for an expanded description of the plant material.
Flow cytometry
Approximately 50–60 mg of greenhouse-grown leaf tissue was harvested from each sample and homogenized in 500 µL of LB01 buffer (Doležel et al., 1989) and left to rest for 1 min. About 250 µL of homogenate was passed through a 20-µm filter (Partec 04-0042-2315) into tubes containing 12 µL of 1 mg/mL propidium iodide and 2.5 µL of 5 mg/mL RNase. Samples were incubated in the dark for 5 min and analyzed in an Accuri C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) with the following filter configurations: (1) FL-1 530/14-nm bandpass filter, (2) FL-2 585-20-nm bandpass filter, and (3) FL-3 670-nm longpass filter. Threshold levels were set to 10,000 for forward scatter with a secondary threshold of 1,000 for FL-2 (Galbraith et al., 2011).
Whole genome resequencing
Genomic DNA was extracted from young leaflets as previously described (Ghislain et al., 1999). For each sample, ∼750 ng of genomic DNA was sheared to an average size of 300 bp as previously described (Amundson et al., 2020). Libraries were constructed using all sheared input DNA with a KAPA Hyper Prep kit (KAPA Biosystems KK8504) with half-scale reactions used throughout the protocol, custom 8 bp index adapters, and amplification cycles as described in Supplemental Data Set S2. Libraries were sequenced on the Illumina HiSeq 4000 or NovaSeq 6000 platform at the University of California, Davis DNA Technologies Core, Vincent Coates Genome Sequencing Laboratory, or University of California San Francisco Center for Advanced Technologies, as specified in Supplemental Data Set S2. Libraries were demultiplexed using custom Python scripts available on our laboratory website (allprep-12.py; http://comailab.genomecenter.ucdavis.edu/index.php/Barcoded_data_preparation_tools). Publicly available sequencing reads from Pham et al. (2017), Hardigan et al. (2017), and Amundson et al. (2020) were retrieved from NCBI Sequence Read Archive (SRA) and incorporated in subsequent analyses.
Variant calling
Adapter and low-quality sequences were trimmed from raw reads using Cutadapt version 1.15 (Martin, 2011), retaining reads ≥40 nt in length. Trimmed reads were aligned to the DM1-3 version 4.04 reference assembly, including DM1-3 chloroplast and mitochondrion sequences, using BWA mem (version 0.7.12r1039) and default settings (Li, 2013). Alignments were further processed to remove PCR duplicates, soft clip one mate of overlapping read pairs, remove read pairs with mates aligning to different chromosomes, and locally realign indels, as previously described (Amundson et al., 2020). Processed alignments were then used as input for joint variant calling and genotyping with FreeBayes version 1.3.2 (Garrison and Marth, 2012) with minimum mapping quality 20, base quality 20, Hardy–Weinberg priors off, and up to 4 alleles considered per variant, and all other parameters left at the default setting.
Initially, we genotyped a subset of parental clones with deep whole genome sequencing available from this study or from previous studies (Hardigan et al., 2017; Pham et al., 2017), which we designated “Cohort A” in Supplemental Data Set S2. Raw variants were filtered as follows: NUMALT == 1, CIGAR == 1X, QUAL ≥ 20, MQM ≥ 50, MQMR ≥ 50, |MQM-MQMR| < 10, RPPR ≤ 20, RPP ≤ 20, EPP ≤ 20, EPPR ≤ 20, SAP ≤ 20, SRP ≤ 20. For each pair of tetraploid parent and HI represented in the offspring of Cohort A, we identified loci with read depth within 1.5 times the genome-wide median of each parent, at least 10 supporting reads, and homozygous genotype calls for different alleles in the two parents. Each list of parental SNPs was used to provisionally determine chromosome dosage (see the “Chromosome dosage analysis” section) for all offspring of Cohort A. To determine parental origin of aneuploidy in offspring for which sequencing reads from the tetraploid parent were not available, we tested the possibility of using pooled reads from multiple dihaploids produced from the same parent instead. Specifically, we tested the effect of substituting pooled low-coverage sequencing from dihaploids for that same tetraploid parent at the SNP calling step, and tested if we could recapitulate the observations obtained using SNPs taken directly from the tetraploid parent. As a proof of concept, we pooled low-coverage alignments from 205 dihaploids of WA.077 at the variant calling step and repeated all downstream analysis of Cohort A samples. Upon obtaining acceptable results, pooled alignments from low coverage dihaploids extracted from C93.154 (n = 237), 93.003 (n = 73), C91.640 (n = 79), LR00.014 (n = 110), LR00.022 (n = 51), LR00.026 (n = 51), WA.077 (n = 205), and all deeply sequenced HI addition lines were included along with Cohort A samples for the variant calling and genotyping reported in the manuscript.
Chromosome dosage analysis
Read alignments from low coverage dihaploids and hybrids were filtered for mapping quality ≥10 and counted in non-overlapping 1-Mb bins using bedtools version 2.27.1. We calculated the fraction of all aligned reads that mapped to a chromosome, normalized this fraction to the corresponding fraction of a family-specific control sample (controls specified in Supplemental Data Set S1), and scaled the standardized coverage values to the expected ploidy state based on flow cytometry results. Putative aneuploids were identified as outliers with a standardized coverage value of ≥3 standard deviations from the within-family all-chromosome mean. In some families, segregation of pre-existing deletions on chromosome 12 resulted in a high rate of false positive trisomy and monosomy calls. False positives of this nature are listed in Supplemental Data Set S1. Individuals exhibiting a false positive signal on chromosome 12 were not recorded as aneuploid, unless they also exhibited aneuploidy of another chromosome type. To infer parental origin of numerical and structural aneuploidies, parental SNPs were identified for each combination of tetraploid parent and HI as described above. For each low-coverage dihaploid or hybrid, allele-specific read depth was then tallied at homozygous parent-informative SNP loci in non-overlapping 4-Mb bins, and bins with >30 reads covering all informative loci within a bin were withheld from analysis.
High resolution analysis of parental DNA contribution
For each HI addition line, genotype data were recorded as T = tetraploid parent H = HI. For high stringency filtering, loci were removed from consideration if any of the following criteria were met: (1) one or more reads matched the HI allele in the tetraploid parent, (2) three or more reads matched the HI allele in the dihaploid pool, (3) excessive read depth (greater than the mean depth plus four standard deviations greater than the mean depth) was observed in either parent or the dihaploid at hand (Li, 2014), (4) HI allele depth was <6 in the dihaploid at hand, or (5) the HI allele represented <15% of the total reads at a locus in the dihaploid at hand. For regions of interest, we viewed short read alignments to the reference genome DM1-3 using the Integrative Genome Viewer (Thorvaldsdottir et al., 2013).
Low-pass haplotype analysis
Biallelic SNP loci with homozygous genotype calls for either allele in WA.077, heterozygous genotype calls in IVP35, and heterozygous with a single dose of the HI-specific allele (i.e. 0/0/1 if the called tetraploid genotype was 0/0/0/0 and 0/1/1 if the called genotype was 1/1/1/1) were used to define phased alleles of H′. For each tetraploid hybrid, we then calculated the depth of reads with H′ and non-H′ alleles at all retained loci, aggregated counts across the DM1-3 coordinates defined as recombination-suppressed centromeres by Bourke et al. (2015) or by non-overlapping 4-Mb bins, and reported the ratio of reads matching H′ reads to H′ + non-H′ reads.
Statistical analyses
Proportion of aneuploids among dihaploids by female parent
Euploidy and aneuploidy were treated as discrete outcomes, and counts of each category were evaluated for statistical significance using the prop.test() function in R version 3.6.2 (R Core Team 2019). Only families with 30 or more dihaploids were included in the analysis.
Fisher exact counts of HI dihaploid introgression events by HI
Appearance of HI-derived chromosomes in an otherwise dihaploid plant was treated as a binary outcome and used to construct a 2 × 3 contingency table with each HI genotype. Only dihaploids for which we had sufficient SNP information to determine chromosome parental origin were considered. This included dihaploids from the following tetraploid parents: 93.003 (CIP390637.1), Atlantic (CIP800827), C01.020 (CIP301023.15), C91.640 (CIP3888615.22), C93.154 (CIP392820.1), Desiree (CIP800048), LR00.014 (CIP300056.33), LR00.022 (CIP300072.1), LR00.026 (CIP300093.14), and WA.077 (CIP397077.16). This table was used to conduct a Fisher exact test in R using the function fisher.test().
Linkage disequilibrium from dosage variable states
For each dihaploid, standardized coverage values and bin dosage states were derived for non-overlapping 1-Mb bins of the reference genome as previously described (Amundson et al., 2020). Fisher exact tests were then carried out between pairs of dosage states to assess linkage disequilibrium (LD) between bins. For example, assume that both Bin1 and Bin100 have three dosage states: standardized coverages 1, 2, and 3. To test whether Bin1-State1 was correlated with Bin100-CN3, the following four dihaploid sets were compared in a 2 × 2 contingency table: observed in Bin1-State1: observed not in Bin1-State1, expected in Bin1-State1: expected not in Bin1-State1, where the expectation was derived from the assumption of complete independence. Self-comparison and reciprocal comparisons were removed, and the remaining comparisons were controlled at false discovery rate = 0.05 unless otherwise noted. Chromosomal bins in statistically significant LD with one another for any state at that pair of bins were displayed in an LD matrix.
Logistic regression model for incidence of paternal (maternal) genome instability in triploid and tetraploid hybrids
For each hybrid, we determined the incidence and parental origin of whole-chromosome and segmental aneuploidy tetraploid hybrids as previously described for potato dihaploids (Amundson et al., 2020). Instability of the paternal genome was treated as a binary outcome and used in a logistic regression model with ploidy, genotype of the HI parent, and aneuploidy of maternal chromosomes included as predictor variables (Supplemental Tables S3–S5). Contribution of the maternal genome to aneuploidy of HI-derived chromosomes was determined from the maternal aneuploidy term in the model. Pairwise differences between ploidy levels were evaluated with Tukey’s multiple test correction. To evaluate the stability of maternal chromosomes, maternal aneuploidy was used as the binary response variable and paternal aneuploidy was incorporated as a predictor variable. Effects of paternal aneuploidy on maternal aneuploidy, as well as ploidy-dependent effects were evaluated as described above.
Accession numbers
All sequencing data generated in this study has been deposited into National Center for Biotechnology Information (NCBI) SRA under Project ID PRJNA699631. IVP101 whole genome sequencing was retrieved from NCBI SRA project ID PRJNA408137. Whole genome sequencing of cv “Atlantic” was retrieved from NCBI SRA project ID PRJNA287438. Code for read preprocessing, variant calling, and chromosome dosage analysis is available on https://github.com/kramundson/MM_manuscript. Custom Python scripts used to demultiplex the libraries are available on our laboratory website (allprep-12.py; http://comailab.genomecenter.ucdavis.edu/index.php/Barcoded_data_preparation_tools).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Histogram of aneuploidy by homologous chromosome affected and state loss or gain among 74 potato dihaploids.
Supplemental Figure S2. Aneuploidy frequency among putatively uniparental dihaploid potatoes.
Supplemental Figure S3. Aneuploidy frequency among putatively uniparental dihaploid potatoes.
Supplemental Figure S4. Chromosome dosage and parental allele dosage plots of eight HI addition dihaploids.
Supplemental Figure S5. Histograms of HI allele representation at putative introgression loci in eight HI addition dihaploids.
Supplemental Figure S6. Dosage variation linkage matrix of WA.077 dihaploids.
Supplemental Figure S7. Test of potato HI haplotype phasing.
Supplemental Figure S8. HI centromeric heterozygosity for tetraploid hybrids of the potato haploid induction cross.
Supplemental Figure S9. Dosage plot, SNP plot, and inferred karyotype of the WA.077 × IVP35 chromosome 11 disomic tetraploid potato hybrid.
Supplemental Table S1. Genomic content and embryo spot phenotype in progeny of potato haploid induction crosses.
Supplemental Table S2. Description of 19 tetraploid potato clones used for dihaploid extraction.
Supplemental Table S3. Coefficients of logistic regression model of HI genome instability of hybrids.
Supplemental Table S4. Analysis of deviance table for logistic regression model.
Supplemental Table S5. Confidence intervals (reported as log odds) for logistic regression model.
Supplemental Data Set S1. Description of plant material used in this study.
Supplemental Data Set S2. Summary of sequencing libraries constructed or analyzed in this study.
Supplemental Data Set S3. IGV screenshots of short read alignments to the version 4.04 reference genome showing ambiguous introgression junctions.
Supplementary Material
Acknowledgments
The authors thank Hannele Lindqvist-Kreuze for guidance and support of the CIP-based research and the CIP’s greenhouse technicians who performed most of the crossings and plant maintenance.
Funding
This work was supported by the National Science Foundation Plant Genome Integrative Organismal Systems Grant 1444612 (Rapid and Targeted Introgression of Traits via Genome Elimination) to L.C.
Conflict of interest statement. None declared.
L.C. and E.H.T. conceived the experiments. K.R.A., B.O., E.H.T., M.B., A.K., I.M.H., and L.C. designed the experiments. K.R.A., B.O., M.S., and L.C. performed the experiments and data analysis, K.R.A., M.L.N., E.H.T., I.M.H., and L.C. interpreted the data. B.O., M.S., M.B., and A.K. contributed the reagents and materials. K.R.A. and L.C. wrote the paper with input from all authors.
The author responsible for the distribution of materials integral to these findings presented in the article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Luca Comai (lcomai@ucdavis.edu).
References
- Allainguillaume J, Wilkinson MJ, Clulow SA, Barr SNR (1997) Evidence that genes from the male parent may influence the morphology of potato dihaploids. Theor Appl Genet 94: 241–248 [Google Scholar]
- Amundson KR, Ordoñez B, Santayana M, Tan EH, Henry IM, Mihovilovich E, Bonierbale M, Comai L (2020) Genomic outcomes of haploid induction crosses in potato (Solanum tuberosum L). Genetics 214: 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkiewicz AM, Chilla F, Terefe-Ayana D, Lübeck J, Strahwald J, Tacke E, Hofferbert HR, Linde M, Debener T (2018) Maximization of markers linked in coupling for tetraploid potatoes via monoparental haploids. Front Plant Sci 9: 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee AF (1922) Variations in datura due to changes in chromosome number. Am Nat 56: 16–31 [Google Scholar]
- Bourke PM, Voorrips RE, Visser RGF, Maliepaard C (2015) The double-reduction landscape in tetraploid potato as revealed by a high-density linkage map. Genetics 201: 853–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clulow SA, Rousselle-Bourgeois F (1997) Widespread introgression of Solatium phureja DNA in potato (S. tuberosum) dihaploids. Plant Breed 116: 347–351 [Google Scholar]
- Clulow SA, Wilkinson MJ, Burch LR (1993) Solanum phureja genes are expressed in the leaves and tubers of aneusomatic potato dihaploids. Euphytica 69: 1–6 [Google Scholar]
- Clulow SA, Wilkinson MJ, Waugh R, Baird E, Demaine MJ, Powell W (1991) Cytological and molecular observations on Solanum phureja-induced dihaploid potatoes. Theor Appl Genet 82: 545–551 [DOI] [PubMed] [Google Scholar]
- De Storme N, Geelen D (2011) The Arabidopsis mutant jason produces unreduced first division restitution male gametes through a parallel/fused spindle mechanism in meiosis II. Plant Physiol 155: 1403–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Binarová P, Lcretti S (1989) Analysis of nuclear DNA content in plant cells by flow cytometry. Biol Plant 31: 113–120 [Google Scholar]
- Dongyu Q, Dewei Z, Ramanna MS, Jacobsen E (1995) A comparison of progeny from diallel crosses of diploid potato with regard to the frequencies of 2n-pollen grains. Euphytica 92: 313–320 [Google Scholar]
- Endelman JB, Jansky SH (2016) Genetic mapping with an inbred line-derived F2 population in potato. Theor Appl Genet 129: 935–943 [DOI] [PubMed] [Google Scholar]
- Ercolano MR, Carputo D, Li J, Monti L, Barone A, Frusciante L (2004) Assessment of genetic variability of haploids extracted from tetraploid (2n = 4× = 48) Solanum tuberosum. Genome 47: 633–638 [DOI] [PubMed] [Google Scholar]
- d’Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Mercier R (2009) Turning meiosis into mitosis. PLoS Biol 7: e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen NO (1967) Haploid production in a potato breeding material with intensive back crossing to wild species. Zuchter 37: 120–134 [Google Scholar]
- Gabert AC (1963) Factors influencing the frequency of haploids in the common potato (Solanum tuberosum L.). PhD dissertation, University of Wisconsin
- Galbraith DW, Janda J, Lambert GM (2011) Multiparametric analysis, sorting, and transcriptional profiling of plant protoplasts and nuclei according to cell type. Methods Mol Biol 699: 407–429 [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing. arXiv [q-bio.GN]
- Gernand D, Rutten T, Varshney A, Rubtsova M, Prodanovic S, Brüß C, Kumlehn J, Matzk F, Houben A (2005) Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell 17: 2431–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Zhang DP, Herrera MR editors (1999) Molecular Biology Laboratory Protocols Plant Genotyping: Training Manual. International Potato Center, Lima, Peru [Google Scholar]
- Hanneman RE, Peloquin SJ (1968) Ploidy levels of progeny from diploid-tetraploid crosses in the potato. Am Potato J 45: 255–261 [Google Scholar]
- Hanneman RE, Ruhde RW (1978) Haploid extraction in Solanum tuberosum group Andigena. Am J Potato Res 55: 259–263 [Google Scholar]
- Hardigan MA, Laimbeer FPE, Newton L, Crisovan E, Hamilton JP, Vaillancourt B, Wiegert-Rininger K, Wood JC, Douches DS, Farré EM, et al. (2017) Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. Proc Natl Acad Sci USA 114: E9999–E10008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Braz GT, Torres GA, Jiang J (2018) Chromosome painting in meiosis reveals pairing of specific chromosomes in polyploid Solanum species. Chromosoma 127: 505–513 [DOI] [PubMed] [Google Scholar]
- Henry IM, Dilkes BP, Miller ES, Burkart-Waco D, Comai L (2010) Phenotypic consequences of aneuploidy in Arabidopsis thaliana. Genetics 186: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermsen JGT (1969) Induction of haploids and aneuhaploids in colchicine-induced tetraploid Solanum chacoense Bitt. Euphytica 18: 183–189 [Google Scholar]
- Hermsen JGT, Verdenius J (1973) Selection from Solanum tuberosum group phureja of genotypes combining high-frequency haploid induction with homozygosity for embryo-spot. Euphytica 22: 244–259 [Google Scholar]
- Hermsen JGT, Wagenvoort M, Ramanna MS (1970) Aneuploids from natural and colchicine-induced autotetraploids of solanum. Can J Genet Cytol 12: 601–613 [Google Scholar]
- Hutten RCB, Scholberg EJMM, Huigen DJ, Hermsen JGT, Jacobsen E (1993) Analysis of dihaploid induction and production ability and seed parent x pollinator interaction in potato. Euphytica 72: 61–64 [Google Scholar]
- Ishii T, Ueda T, Tanaka H, Tsujimoto H (2010) Chromosome elimination by wide hybridization between Triticeae or oat plant and pearl millet: pearl millet chromosome dynamics in hybrid embryo cells. Chromosome Res 18: 821–831 [DOI] [PubMed] [Google Scholar]
- Jackson MT, Rowe PR, Hawkes JG (1978) Crossability relationships of andean potato varieties of three ploidy levels. Euphytica 27: 541–551 [Google Scholar]
- Jacquier NMA, Gilles LM, Pyott DE, Martinant JP, Rogowsky PM, Widiez T (2020) Puzzling out plant reproduction by haploid induction for innovations in plant breeding. Nat Plants 6: 610–619 [DOI] [PubMed] [Google Scholar]
- Jansky SH, Charkowski AQ, Douches DS, Gusmini G, Richael C, Bethke PC, Spooner DM, Novy RG, De Jong H, De Jong WC, et al. (2016) Reinventing potato as a diploid inbred line–based crop. Crop Sci 56: 1412–1422 [Google Scholar]
- Koornneef M, Van der Veen JH (1983) Trisomics in Arabidopsis thaliana and the location of linkage groups. Genetica 61: 41–46 [Google Scholar]
- Kuppu S, Tan EH, Nguyen H, Rodgers A, Comai L, Chan SWL, Britt AB (2015) Point mutations in centromeric histone induce post-zygotic incompatibility and uniparental inheritance. PLoS Genet 11: e1005494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kynast RG, Riera-Lizarazu O, Vales MI, Okagaki RJ, Maquieira SB, Chen G, Ananiev EV, Odland WE, Russell CD, Stec AO, et al. (2001) A complete set of maize individual chromosome additions to the oat genome. Plant Physiol 125: 1216–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie DA, Bennett MD (1986) Wheat × maize hybridization. Can J Genet Cytol 28: 313–316 [Google Scholar]
- Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-bio.GN]
- Li H (2014) Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30: 2843–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhout P, Meijer D, Schotte T, Hutten RCB, Visser RGF, van Eck HJ (2011) Towards F1 hybrid seed potato breeding. Potato Res 54: 301–312 [Google Scholar]
- Li X, Meng D, Chen S, Luo H, Zhang Q, Jin W, Yan J (2017) Single nucleus sequencing reveals spermatid chromosome fragmentation as a possible cause of maize haploid induction. Nat Commun 8: 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Shen Y, Cai C, Zhong C, Zhu L, Yuan M, Ren H (2010) The type II Arabidopsis formin14 interacts with microtubules and microfilaments to regulate cell division. Plant Cell 22: 2710–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly P, Brunner SF, Shoshani O, Kim DH, Lan W, Pyntikova T, Flanagan AM, Behjati S, Page DC, Campbell PJ, et al. (2019) Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat Genet 51: 705–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly P, Teitz LS, Kim DH, Shoshani O, Skaletsky H, Fachinetti D, Page DC, Cleveland DW (2017) Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat Cell Biol 19: 68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Tan EH, West A, Franklin FCH, Comai L, Chan SWL (2015) Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet 11: e1004970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks GE (1966) The enigma of triploid potatoes. Euphytica 15: 285–290 [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12 [Google Scholar]
- Montelongo-Escobedo H, Rowe PR (1969) Haploid induction in potato: cytological basis for the pollinator effect. Euphytica 18: 116–123 [Google Scholar]
- Montezuma-de-Carvalho J (1967) The effect of N2O on pollen tube mitosis in styles and its potential significance for inducing haploidy in potato. Euphytica 16: 190–198 [Google Scholar]
- Peloquin SJ, Boiteux LS, Carputo D (1999) Meiotic mutants in potato. Valuable variants. Genetics 153: 1493–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloquin SJ, Gabert AC, Ortiz R (1996) Nature of “pollinator” effect in potato (Solanum tuberosumL.) haploid production. Ann Bot 77: 539–542 [Google Scholar]
- Pham GM, Braz GT, Conway M, Crisovan E, Hamilton JP, Laimbeer FPE, Manrique-Carpintero N, Newton L, Douches DS, Jiang J, et al. (2018) Genome-wide inference of somatic translocation events during potato dihaploid production. Plant Genome 12: 180079, doi: 10.3835/plantgenome2018.10.0079 [DOI] [PubMed] [Google Scholar]
- Pham GM, Hamilton JP, Wood JC, Burke JT, Zhao H, Vaillancourt B, Ou S, Jiang J, Buell CR (2020) Construction of a chromosome-scale long-read reference genome assembly for potato. Gigascience 9: giaa100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham GM, Newton L, Wiegert-Rininger K, Vaillancourt B, Douches DS, Buell CR (2017) Extensive genome heterogeneity leads to preferential allele expression and copy number-dependent expression in cultivated potato. Plant J 92: 624–637 [DOI] [PubMed] [Google Scholar]
- Ramanna MS (1979) A re-examination of the mechanisms of 2n gamete formation in potato and its implications for breeding. Euphytica 28: 537–561 [Google Scholar]
- Ramsey J, Schemske DW (2002) Neopolyploidy in flowering plants. Annu Rev Ecol Syst 33: 589–639 [Google Scholar]
- Rick CM, Barton DW (1954) Cytological and genetical identification of the primary trisomics of the tomato. Genetics 39: 640–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera-Lizarazu O, Rines HW, Phillips RL (1996) Cytological and molecular characterization of oat x maize partial hybrids. Theor Appl Genet 93: 123–135 [DOI] [PubMed] [Google Scholar]
- Samitsu Y, Hosaka K (2002) Molecular marker analysis of 24- and 25-chromosome plants obtained from Solanum tuberosum L. subsp. andigena (2n = 4× = 48) pollinated with a Solanum phureja haploid inducer. Genome 45: 577–583 [DOI] [PubMed] [Google Scholar]
- Spooner DM, Ghislain M, Simon R, Jansky SH, Gavrilenko T (2014) Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Bot Rev 80: 283–383 [Google Scholar]
- Steinitz-Sears LM (1963) Chromosome studies in Arabidopsis thaliana. Genetics 48: 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straadt IK, Rasmussen OS (2003) AFLP analysis of Solanum phureja DNA introgressed into potato dihaploids. Plant Breed 122: 352–356 [Google Scholar]
- Swaminathan MS (1954a) MICROSPOROGENESIS IN SOME COMMERCIAL POTATO VARIETIES. J Hered 45: 265–272 [Google Scholar]
- Swaminathan MS (1954b) Nature of polyploidy in some 48-chromosome species of the genus solanum, section tuberarium. Genetics 39: 59–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EH, Henry IM, Ravi M, Bradnam KR, Mandakova T, Marimuthu MP, Korf I, Lysak MA, Comai L, Chan SW (2015) Catastrophic chromosomal restructuring during genome elimination in plants. Elife 4: e06516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP (2013) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit NT, Zhang CZ, Lynch LD, Blaine LJ, Cheng AM, Tourdot R, Sun L, Almubarak HF, Judge K, Mitchell TJ, et al. (2020) Mechanisms generating cancer genome complexity from a single cell division error. Science 368: eaba0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenvoort M, Lange W (1975) The production of aneudihaploids in Solanum tuberosum L. group Tuberosum (the common potato). Euphytica 24: 731–741 [Google Scholar]
- Wangenheim KH, Peloquin SJ, Hougas RW (1960) Embryological investigations on the formation of haploids in the potato (Solanum tuberosum). Zh Vyssh Nerv Deiat Im I P Pavlova 91: 391–399 [DOI] [PubMed] [Google Scholar]
- Waugh R, Baird E, Powell W (1992) The use of RAPD markers for the detection of gene introgression in potato. Plant Cell Rep 11: 466–469 [DOI] [PubMed] [Google Scholar]
- Wilkinson MJ, Bennett ST, Clulow SA, Allainguillaume J, Harding K, Bennett MD (1995) Evidence for somatic translocation during potato dihaploid induction. Heredity 74(Pt 2): 146–151 [DOI] [PubMed] [Google Scholar]
- Zhao X, Xu X, Xie H, Chen S, Jin W (2013) Fertilization and uniparental chromosome elimination during crosses with maize haploid inducers. Plant Physiol 163: 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Chen B, Li M, Wang D, Jiao Y, Qi X, Wang M, Liu Z, Chen C, Wang Y, et al. (2020) A DMP-triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis. Nat Plants 6: 466–472 [DOI] [PubMed] [Google Scholar]
- Zhong Y, Liu C, Qi X, Jiao Y, Wang D, Wang Y, Liu Z, Chen C, Chen B, Tian X, et al. (2019) Mutation of ZmDMP enhances haploid induction in maize. Nat Plants 5: 575–580 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.