Abstract
Endoplasmic reticulum–plasma membrane contact sites (ER–PM CS) play fundamental roles in all eukaryotic cells. Arabidopsis thaliana mutants lacking the ER–PM protein tether synaptotagmin1 (SYT1) exhibit decreased PM integrity under multiple abiotic stresses, such as freezing, high salt, osmotic stress, and mechanical damage. Here, we show that, together with SYT1, the stress-induced SYT3 is an ER–PM tether that also functions in maintaining PM integrity. The ER–PM CS localization of SYT1 and SYT3 is dependent on PM phosphatidylinositol-4-phosphate and is regulated by abiotic stress. Lipidomic analysis revealed that cold stress increased the accumulation of diacylglycerol at the PM in a syt1/3 double mutant relative to wild-type while the levels of most glycerolipid species remain unchanged. In addition, the SYT1-green fluorescent protein fusion preferentially binds diacylglycerol in vivo with little affinity for polar glycerolipids. Our work uncovers a SYT-dependent mechanism of stress adaptation counteracting the detrimental accumulation of diacylglycerol at the PM produced during episodes of abiotic stress.
Arabidopsis synaptotagmins 1 and 3 are localized at endoplasmic reticulum–plasma membrane contact sites and during abiotic stress episodes control diacylglycerol homeostasis at the plasma membrane
Introduction
As sessile organisms, plants are continuously exposed to changes in environmental conditions (Botella et al., 2007; Zhu, 2016; Amorim-Silva, 2019). In addition to forming a selective permeable barrier, the plasma membrane (PM) senses environmental cues and transforms them into highly regulated signaling outputs (Hou et al., 2016). Among these outputs, there are several lipids such as phosphatidic acid (PA) and phosphatidylinositol (PI) phosphates (also known as phosphoinositides, PIPs), which are synthetized at the PM and play important regulatory roles in cellular plasticity (Testerink and Munnik, 2005; Munnik and Testerink, 2009; Munnik and Nielsen, 2011; Hou et al., 2016; Colin and Jaillais, 2020). In particular, PA is rapidly generated at the PM in response to abiotic stresses, such as dehydration (Hong et al., 2008), salinity (Munnik et al., 2000; Yu et al., 2010), temperature stress (Ruelland, 2002; Arisz et al., 2013), and treatment with the stress-related hormone abscisic acid (Katagiri et al., 2005; Uraji et al., 2012; Li et al., 2019).
The production of PA at the PM can be triggered by the hydrolysis of structural phospholipids, such as phosphatidylcholine (PC) and phosphatidylethanolamine (PE), either directly by phospholipases D (PLD) or through the consecutive actions of nonspecific phospholipases C (NPC), that produce diacylglycerol (DAG), which is transformed into PA by DAG kinases (DGKs; Pokotylo et al., 2018). In addition, hydrolysis of the PIPs, for example, PI-4-phosphate (PI4P) and PI-4,5-biphosphate, by phospholipase C (PLC) also produces DAG, that can be transformed into PA by DGKs (Arisz et al., 2009, 2013; Testerink and Munnik, 2011). However, local accumulation of DAG at the PM could cause the disruption of the lamellar phase of lipid membranes (Campomanes et al., 2019), thus DAG concentration must be tightly controlled within cell membranes.
The PM forms extensive contacts with the endoplasmic reticulum (ER) at ER–PM contact sites (ER–PM CS), which can be defined as tight and not fused membrane junctions tethered by specific protein complexes (Bayer et al., 2017; Wu et al., 2018; Scorrano et al., 2019). These ER–PM CS are essential for communication between the ER and PM in mammalian, fungal, and plant cells, enabling lipid transport (Saheki et al., 2016), regulating calcium influx (Saheki and De Camilli, 2017), and maintaining the cortical ER (cER) morphology (Siao et al., 2016). The plant synaptotagmins (SYTs) and their ortholog counterparts, the mammalian extended SYTs (E-Syts) and the yeast tricalbins (Tcbs) are ER–PM CS tethers that share a common modular structure, comprising a N-terminal transmembrane (TM) domain that anchors them to the ER, a SYT-like mitochondrial-lipid binding (SMP) domain, and a variable number of the Ca2+- and phospholipid-binding C2 domains (Manford et al., 2012; Giordano et al., 2013; Perez Sancho et al., 2015). SMP domains are members of the tubular lipid-binding protein superfamily, with a common folding structure that can harbor lipids in a hydrophobic cavity (Kopec et al., 2010). The modular structure of these proteins implies two likely inter-related functions, namely the establishment of Ca2+ regulated ER–PM tethering by their C2 domains (Manford et al., 2012; Giordano et al., 2013; Perez Sancho et al., 2015; Saheki et al., 2016), and the SMP-dependent transport of lipids between the PM and the ER (Schauder et al., 2014; Saheki et al., 2016).
The yeast Tcbs and the mammalian E-Syts contain a variable number of C2 domains, which can bind membrane phospholipids either dependently or independently of Ca2+. In yeast, the C2C domains of Tcb1 and Tcb3 bind anionic phospholipids in a Ca2+-dependent manner, while all the C2 domains in Tcb2 are insensitive to Ca2+ (Schulz and Creutz, 2004). In mammals, E-Syt2 and E-Syt3 are localized at ER–PM CS due to the constitutive interaction of their C2 domains with PM PI(4,5)P2; meanwhile, E-Syt1 is localized throughout the ER at resting state (Giordano et al., 2013; Fernández-Busnadiego et al., 2015; Idevall-Hagren et al., 2015). However, elevation of cytosolic Ca2+ promotes its accumulation at the ER–PM CS and reduces the distance between ER and PM (Giordano et al., 2013; Fernández-Busnadiego et al., 2015; Idevall-Hagren et al., 2015; Bian et al., 2018). Interestingly, loss of all three Tcbs does not result in a substantial reduction of ER–PM CS in yeast (Manford et al., 2012), and suppression of all three E-Syts in mice neither affect their normal development, viability or fertility, nor their ER morphology (Sclip et al., 2016; Tremblay and Moss, 2016), indicating the presence of additional tethers.
A recent study has shown that Tcbs are necessary for the generation of peaks of extreme curvature at cER membrane and the maintenance of PM integrity in yeast. Although this study proposed that these processes were facilitated by the transport of lipids from the highly curved, less packaged, cER to the PM (Collado et al., 2019), the nature of the lipids transported remains elusive. Important insights about the function of E-Syts were provided by the structural analyses of the SMP domain of E-Syt2 (Schauder et al., 2014). The SMP-domain of E-Syt2 consists of six beta strands and two alpha helices arranged in a barrel that homodimerizes to form a cylinder whose interior is lined almost exclusively by the hydrophobic residues. Notably, the crystal structure of the SMP dimer revealed the presence of glycerophospholipids in the hydrophobic channel without the preference for any particular headgroup (Schauder et al., 2014). Further studies indicated that E-Syt1 also transfers glycerolipids in vitro without any preference for the head group and that an E-Syt1 mutant lacking the SMP domain lacks the lipid transfer ability (Saheki et al., 2016; Yu et al., 2016). Despite the difficulty of identifying the specific lipids preferentially transported by the SMP domain of E-Syts, analysis of cells lacking all three E-Syts supports the hypothesis that the E-Syts function in vivo is to clear DAG formed at the PM after receptor-triggered activation of PLC (Saheki et al., 2016; Bian et al., 2018). This proposed role of E-Syts in DAG homeostasis has been further supported by the finding that E-Syt1 regulates insulin secretion in pancreatic islets by clearing DAG from the PM (Xie et al., 2019).
The Arabidopsis thaliana genome encodes five SYTs (SYT1–SYT5), all of them containing a TM region, an SMP domain and two C2 domains (Perez Sancho et al., 2016). The C2 domains of SYT1 are targeted to the PM through their binding to negatively charged PIP, resulting in constitutive localization of SYT1 at the ER–PM CS (Perez Sancho et al., 2015). Originally, Arabidopsis SYT1 was identified using a forward genetic screen based on salt hypersensitivity (Schapire et al., 2008) and as a cold-induced protein required for Ca2+-dependent freezing tolerance (Yamazaki et al., 2008). Interestingly, salt stress causes an increase of SYT1 localization at ER–PM CS as well as an expansion of these ER–PM CS (Lee et al., 2019). In the last few years, additional roles for SYT1 have been uncovered based on the analysis of loss-of-function syt1 mutants, including the stability of the cER network (Siao et al., 2016; Ishikawa et al., 2018), wounding responses (Perez Sancho et al., 2015), and the resistance to several biotic factors, such as viruses (Lewis and Lazarowitz, 2010; Uchiyama et al., 2014; Levy et al., 2015) and fungi (Kim et al., 2016). Recent studies have shown that SYT5, a major interactor of SYT1, also localizes at ER–PM CS (Lee et al., 2020; Ishikawa et al., 2020) and displays similar changes in localization in response to salt stress as SYT1 (Lee et al., 2020).
Despite the defective responses reported for syt1 under multiple biotic and abiotic stresses, a mechanistic understanding is still lacking. It has been long established that abiotic stresses produce lipid changes at the PM; therefore, we hypothesized that SYT1 may function in the homeostasis of PM lipid composition by transporting lipids at ER–PM CS and therefore, in the tolerance for multiple and apparently unrelated stresses. Here, we reveal that SYT1 and SYT3 are ER–PM tethers that play an important role in PM lipid homeostasis by transporting DAG produced under stress. In addition, the dynamics of SYT1 and SYT3 in response to stress are consistent with their role in lipid transport in order to maintain PM integrity under adverse environmental conditions.
Results
SYT1 and SYT3 are important for PM stability under abiotic stress
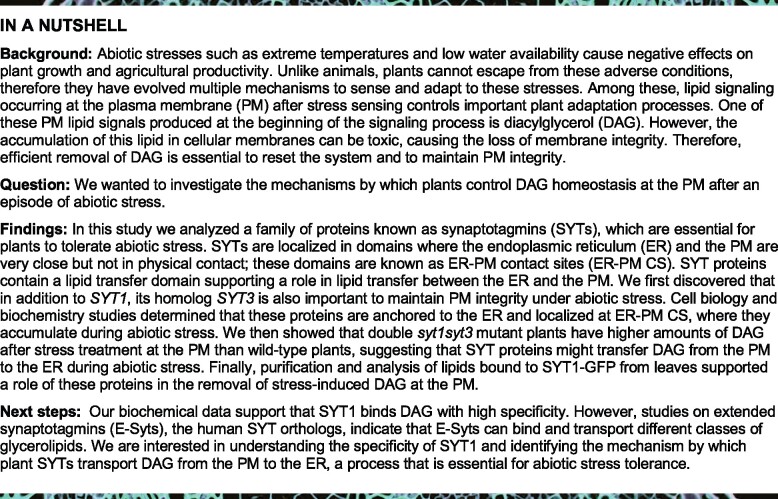
Earlier work has established that SYT1 gene is an important determinant of cell viability under various abiotic stresses (Schapire et al., 2008; Yamazaki et al., 2008; Perez Sancho et al., 2015); therefore, we investigated whether other SYTs play a role in stress resistance. The A. thaliana genome encodes five SYT homologs that can be divided in two clades (Supplemental Figure S1A). In silico expression analysis of SYT genes in vegetative tissues using available RNA sequencing data, for example, eFP Browser and TRAVA, showed that SYT1, SYT3, and SYT5 are ubiquitously expressed with SYT1 transcript levels being about 6 and 10 times higher than those of SYT5 and SYT3, respectively. SYT4 and SYT2, however, have low expression in vegetative tissues (Figure 1A;Supplemental Figure S1B). A similar analysis indicated that the expression of SYT1 and SYT3 but not SYT5 is regulated by abiotic stress (Supplemental Figure S1, C and D). This is consistent with our real-time quantitative PCR (RT-qPCR) data showing that SYT1 and SYT3 but not SYT2, SYT4, or SYT5 were induced after 24 h of cold stress (Figure 1B). Thus, we focused on SYT1 and SYT3 to further characterize their involvement in cell viability under abiotic stress conditions.
Figure 1.
SYT1 and SYT3 are important for PM stability under abiotic stress. A, RNAseq data of SYT1, SYT2, SYT3, SYT4, and SYT5 obtained from vegetative tissues at different developmental stages from eFP-seq Browser (https://bar.utoronto.ca/eFP-Seq_Browser/). Each dot represents a value of RPKM and the bar represent the median. B, SYT1 and SYT3 transcripts are induced by cold. Arabidopsis WT seedlings were grown under long-day photoperiod and 23°C for 7 d and then transferred to 4°C for 24 h or kept under control conditions. The relative expression level of the SYT genes was measured by RT-qPRC and normalized to the expression of ACTIN2. Columns are showing mean values of fold induction; error bars indicate sd (n = 3 pools of 20 seedlings per experiment). C–F, Confocal images and cell viability quantification after cold treatment in 6-d-old Arabidopsis roots. Seedlings grown in half-strength MS agar solidified medium under long-day photoperiod and 23°C. Cell viability was determined by FDA staining and quantified as the percentage of root area with fluorescence above an automatic threshold stablished by the “Moments” algorithm of Fiji (see “Methods” for details). Each dot represents a measurement of an individual root. Horizontal lines represent mean values. C, Increased sensitivity of syt1 compared to WT and syt3 roots. D, The cell viability of syt1 roots is complemented by a SYT1-GFP fusion protein driven by the SYT1 promoter. E, syt1/3 double mutant shows decreased cell viability compared to the single syt1 mutant. F, Cell viability of syt1 roots is complemented by a SYT3-GFP fusion protein driven by the 35S promoter. Significant different values are indicated by letters (P < 0.001; one-wayANOVA uncorrected Fisher’sLSD). Scale bars 50 µm. G, Three-week-old WT, syt1, syt3, and syt1/3 Arabidopsis plants were grown under long-day photoperiod at 20°C and then cold-acclimated for 7 d at 4°C. After that, plants were exposed to different freezing temperatures for 6 h and freezing tolerance was estimated as the percentage of plants surviving each specific temperature after 7 d of recovery under control conditions. Asterisks indicate statistical differences between mutant versus WT determined for each freezing temperature by one-way ANOVA and Tukey’s multiple comparison test. Data represent mean values, error bars are sd, n ≥ 30 plants per genotype for each freezing temperature.
Cell viability after cold stress in roots of wild-type (WT), syt1 and syt3 (Supplemental Figure S1E), Arabidopsis seedlings were analyzed by confocal microscopy using fluorescein diacetate (FDA), a dye that only becomes fluorescent after its hydrolysis by esterases in living cells (Schapire et al., 2008). Thirty minutes of cold treatment (8°C) did not affect cell survival of WT nor syt3 roots, while it caused a dramatic ∼94% decrease in cell viability of syt1 roots (Figure 1C), quantified as the percentage of root area with visible FDA fluorescence above a threshold (see “Methods”). In addition, we showed that cold treatment causes the disruption of syt1 PM integrity by costaining the roots with FM4-64, a hydrophobic dye that is normally retained in the PM but rapidly enters and massively stains the cell interior when PM is disrupted. Thus, FDA/FM4-64 dual stain (Schapire et al., 2008) on WT and syt1 roots showed that all cells examined that underwent cell death (absence of FDA signal) also showed a loss of PM integrity (internal FM4-64 signal; Supplemental Figure S1F).
In order to investigate whether SYT3 has a role in cell viability which might be masked by the highly expressed SYT1, we generated a syt1 syt3 (syt1/3) double mutant. Remarkably, after 10 min of cold treatment, an early time point at which no cell viability defects were observed in syt1, around 80% of the cells in the syt1/3 double mutant were dead (Figure 1E). We then generated Arabidopsis lines transformed with a genomic SYT3 construct fused to green fluorescent protein (GFP) at the C-terminus driven by either the SYT3 promoter or the constitutive 35S promoter. The genomic SYT1 construct carrying GFP at the C-terminus (SYT1:SYT1-GFP) complemented the cell viability defects of syt1 (Figure 1D), indicating that SYT1-GFP is functional. Immunoblot analysis indicated that SYT3-GFP and SYT1-GFP proteins accumulated similarly in the 35S:SYT3-GFP and SYT1:SYT1-GFP lines, while no SYT3-GFP protein was detected in the SYT3:SYT3-GFP line (Supplemental Figure S1G). Next, we investigated whether SYT3-GFP could act redundantly with SYT1 by introducing the 35S:SYT3-GFP construct in the syt1 mutant background (syt1/35S:SYT3-GFP). As shown in Figure 1F, SYT3-GFP driven by a strong promoter complemented the syt1 cold hypersensitive phenotype (Figure 1F). Taken together, these results indicate that SYT1 and SYT3 function redundantly in maintaining cell viability under cold stress.
In silico expression data also indicated that, in addition to cold, SYT1 and particularly SYT3 are regulated by different environmental stresses (Supplemental Figure S1, C and D), possibly reflecting a broad functionality of both proteins in abiotic stress tolerance. Therefore, we investigated the sensitivity of WT, syt1, syt3, and syt1/3 plants to other abiotic stresses such as salt and osmotic stress. Although syt1 mutants showed increased damage compared with WT, cell viability of syt3 was very similar to WT. Similar to cold stress, syt1/3 roots showed significantly higher cellular damage than syt1 under salt stress (Supplemental Figure S1H). We next analyzed the responses of whole seedlings to osmotic stress by treating them with 20% of polyethylene glycol (PEG) and monitoring the cellular damage over time by measuring ion leakage. As shown in Supplemental Figure S1I, we observed similar damage in WT and syt3 seedlings, and an increased damage in syt1 that was further enhanced in syt1/3.
Cold acclimation is an adaptive response by which certain plants increase their freezing tolerance after being exposed for some days to low nonfreezing temperatures (Thomashow, 1999) and previous reports have shown a role of SYT1 in cold-acclimated freezing tolerance (Yamazaki et al., 2008). In contrast with syt1 and syt1/3 seedlings, adult syt1 and syt1/3 plants did not show obvious phenotypic differences when growing in soil during cold stress. Therefore, we evaluated the involvement of SYT1 and SYT3 in cold-acclimated freezing tolerance in adult plants by measuring the survival rate of cold-acclimated (7 d, 4°C) WT, syt1, syt3, and syt1/3 plants after exposure to freezing temperatures. As for other stresses, syt3 plants exhibited a survival rate after freezing similar to WT (Figure 1G), syt1 plants showed decreased freezing tolerance than WT plants, while the syt1/3 double mutant showed a decreased freezing tolerance than syt1 (Figure 1G). Taken together, our data indicate that SYT1 and SYT3 play a redundant role in the maintenance of PM stability in response to diverse abiotic stresses in both seedlings and adult plants.
SYT3 localizes at ER–PM CS
SYT3 is annotated as a pseudogene based on the presence of a stop codon in a cDNA sequence deposited in The Arabidopsis Information Resource Database (Yamazaki et al., 2008). The predicted protein lacks the C2 domains and is therefore expected to be nonfunctional. However, this observation is in clear disagreement with the role identified for SYT3 in abiotic stress (Figure 1; Supplemental Figure S1). To investigate this inconsistency, we performed a detailed sequence analysis using available RNAseq databases and RT-PCR amplification followed by sequencing of the cDNAs fragments (Supplemental Figure S2A). This allowed the identification of five splice variants (Supplemental Figure S2B). Four of these variants encode nonfunctional proteins while a transcript, the one coinciding with the sequence used for the SYT3-GFP constructs (AT5G04220.2), encodes a full SYT3 protein with similar domains to SYT1: a predicted TM region, an SMP domain and two C2 domains (Supplemental Figure S2C).
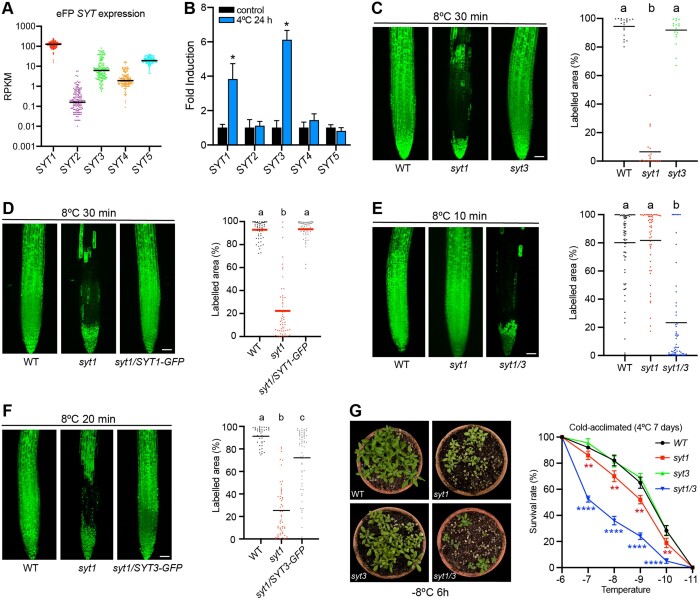
The analogous modular structure of SYT1 and SYT3 and their redundant function in stress resistance suggests that, like SYT1, SYT3 is also an ER–PM CS tether. SYT3-GFP expressed in Nicotiana benthamiana leaves showed a reticulated beads-and-string pattern at the cortical plane of the cell (Figure 2A) and formed focused puncta around the cell periphery at the equatorial plane (Figure 2B), a localization pattern reminiscent of the ER–PM tethers SYT1-GFP (Supplemental Figure S3A) and GFP-MAPPER an artificial ER–PM CS marker that localizes at ER–PM CS (Supplemental Figure S3B; Wang et al., 2014; Perez Sancho et al., 2015; Lee et al., 2019). Coexpression of different tagged constructs of SYT3, SYT1, and MAPPER produced a highly overlapping pattern (Figure 2C;Supplemental Figure S3, C and D). When we coexpressed SYT3-GFP with the ER localized proteins DGK2-mCherry (Angkawijaya et al., 2020) or FaFAH1-mCherry (Sánchez-Sevilla et al., 2014), we found that SYT3-GFP was mainly located in ER tubules and at the edges of ER sheets (Figure 2D;Supplemental Figure S3, E and F). This localization is similar to that of Arabidopsis SYT1 (Ishikawa et al., 2018) and yeast Tcbs (Collado et al., 2019; Hoffmann et al., 2019), which have been reported to have important implications in the formation of tubular ER network and peaks of extreme curvature.
Figure 2.
SYT3 localizes at ER–PM CS. A and B, SYT3-GFP shows a beads-and-string pattern in N. benthamiana leaves. Confocal images of lower epidermis cells transiently expressing SYT3-GFP and schematic representations of the cell planes that are showed in the images, either at the cortical region (A) or at the equatorial plane (B). Boxed regions are magnified in the insets (close up). See also Supplemental Figure S3, A and B. C, SYT3-mCherry colocalizes with ER–PM CS marker. Confocal images of N. benthamiana leaves transiently co-expressing SYT3-mCherry and the ER–PM CS marker MAPPER, showing the cortical region as in (A). Images of the individual channels as well as the merged are shown. See also Supplemental Figure S3, C and D. D, SYT3-GFP localizes at ER tubules and the edges of ER sheets. Confocal images of the lower epidermis cells of N. benthamiana leaves transiently co-expressing SYT3-GFP with the ER marker DGK2-mCherry. Images show the cortical region of the cells as in (A). Images of the individual channels as well as the merged images are shown. See also Supplemental Figure S3, E, F, and G. E and F, SYT3-GFP shows a beads-and-string pattern in Arabidopsis. Confocal images showing the localization of SYT3-GFP either at the cortical region of the cells (E) or at the equatorial plane (F) in epidermal cells from the cotyledon of 6-d-old Arabidopsis seedlings expressing 35S:SYT3-GFP grown in half-strength MS agar under long-day photoperiod and 23°C. Boxed regions are magnified in the insets (close up) G, SYT3-GFP colocalizes with endogenous SYT1 in Arabidopsis roots. Five-day-old Arabidopsis seedlings expressing 35S:SYT3-GFP grown in half-strength MS agar solidified medium under long-day photoperiod and 23°C were fixed with 4% PFA and permeabilized. The endogenous SYT1 protein was detected in the root tip with an anti-SYT1 primary antibody and a TRITC conjugated secondary antibody. Images of the individual channels as well as the merged image are shown. Scale bar 5 µm. See also Supplemental Figure S2G. A–F, Scale bars, 10 µm and scale bars for the close-up insets, 2 µm. A–G, Cortical plane images are a maximum Z-projection of several planes from the cell surface until a plane where cells are close but still not touching the neighbors. Equatorial plane images are single plane images Intensity plots along the white lines in close up views are shown for each co-localization pattern.
In the SYT3:SYT3-GFP Arabidopsis line, the GFP signal was only observed in stomata and root epidermis (Supplemental Figure 2D), indicating low expression of SYT3-GFP under its own promoter. Consistent with this, a transgenic line with the reporter β-glucuronidase (GUS) gene under control of a 1.9 kb SYT3 cis-regulatory region showed a similar expression pattern (Supplemental Figure S2E). Because of low expression of SYT3 in cotyledon pavement cells (the cell type that has been classically used to study ER–PM CS morphology in Arabidopsis (Perez Sancho et al., 2015; Ishikawa et al. 2018; Lee et al. 2019; Lee et al. 2020), we characterized the subcellular localization of SYT3 using the functional 35S:SYT3-GFP line (Figure 1E). Similar to SYT1-GFP localization, SYT3-GFP showed a beads-and-string pattern at the cortical plane of the cell (Figure 2E) and focused puncta around the cell periphery at the equatorial plane (Figure 2F). In addition, using specific antibodies against SYT1 to immunolocalize endogenous SYT1 in the SYT3:SYT3-GFP line, we obtained highly overlapping patterns (Figure 2G;Supplemental Figure S2G; Perez Sancho et al., 2015). Altogether, the experiments described above indicate that SYT3 localizes together with SYT1 at ER–PM CS.
SYT1 and SYT3 tether the PM through interaction with PI4P
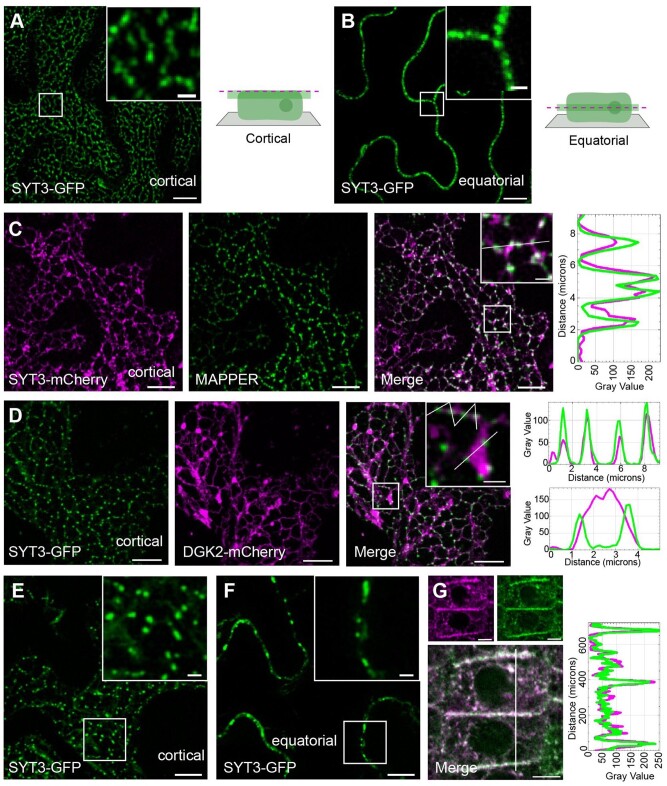
The expansion of ER–PM CS tethering is a defining property of ER–PM tether proteins when overexpressed (Eisenberg-Bord et al., 2016). Interestingly, N. benthamiana epidermal cells expressing GFP-MAPPER showed a characteristic punctate pattern (Figure 3A; cell on the left and close up 1), but when GFP-MAPPER was coexpressed with SYT3-mCherry, both MAPPER and SYT3 signals spread and colocalized throughout the entire cER (Figure 3A; cell on the right and close up 2), indicating the expansion of ER–PM CS. A similar effect was observed when GFP-MAPPER and SYT1-mCherry were coexpressed, occasionally causing an almost complete attachment of the ER to the PM (Supplemental Figure S4A), a phenomenon that was associated to leaf necrosis (Supplemental Figure S4B).
Figure 3.
SYT1 and SYT3 tether the PM through interaction with PI4P. A, SYT3-mCherry expression expands ER–PM CS in N. benthamiana. Confocal images of the cortical region of lower epidermis cells of N. benthamiana leaves transiently co-expressing SYT3-mCherry and the ER–PM marker MAPPER. Two cells are shown, one cell expressing only MAPPER (left cell, close up 1) and one coexpressing SYT3-mCherry and MAPPER (right cell, close up 2). Images of the individual channels as well as the merged images are shown. Boxed regions are magnified in the close ups. The intensity plot along the white line in close up 2 is shown. Scale bar, 10 µm and scale bar for close-up images, 2 µm. B and C, Recombinant GST fusions of C2A domain (B) and C2B domain (C) of SYT3 containing the indicated residues were used in phospholipid binding assays. SYT3 domains were incubated with liposomes composed of 25% PS/75% PC in the indicated concentrations of free Ca2+ (clamped with Ca/EGTA buffers) in order to determine the half-maximal binding for free Ca2+. Liposomes were precipitated by centrifugation, and bound proteins were analyzed by SDS-PAGE. The EC50 for SYT3-C2A was 4.5 ± 0.6 µM and for SYT3 C2B was 5 ± 0.5 µM (paired t test, P < 0.05; n = 4 assays). D, SYT3-GFP, SYT1-GFP and MAPPER localization on ER–PM CS depends on PM PI4P. Confocal images of the cortical region of lower epidermis cells of N. benthamiana leaves transiently co-expressing SYT3-GFP (left row), SYT1-GFP (central row), or MAPPER (right row) either with an empty vector (top panels) or with the PM bound PI4P phosphatase MAP-mTU2-SAC1. Scale bars 10 µm.
Like SYT1 (Perez Sancho et al., 2015), SYT3 is expected to be anchored to the ER and to bind in trans to the PM via its C2 domains (Supplemental Figure S2C). Earlier work has shown that the C2A domain of SYT1 binds artificial liposomes containing phosphatidylserine (PS) and PC in a Ca2+-dependent manner, while binding of SYT1-C2B is independent of Ca2+ (Schapire et al., 2008). Calcium binding sites in C2 domains are characterized by acidic residues (Asp/Glu) confined in conserved loops that play crucial roles in coordinating Ca2+ ions (Fernández-Chacón et al., 2001). Sequence analysis of the C2 domains of SYT1, SYT3, and E-Syt1 predicted the presence of conserved Ca2+-binding sites in the C2A domains of SYT1 and SYT3 and in the C2A and C2C of E-Syt1 (Supplemental Figure S4C). In addition, 3D modeling of the independent C2A and C2B domains of SYT3 identified three Ca+2 coordination pockets with a high confidence score (Cs > 0.5) for C2A, while no Ca+2 coordination pockets were identified for C2B (Supplemental Figure S4D).
Nevertheless, since Ca2+ and phospholipid binding properties of C2 domains cannot be reliably predicted from sequence analysis (Dai et al., 2004), we investigated the Ca2+-dependent phospholipid binding properties of SYT3 C2A and C2B domains. As shown in Figure 3B, the purified SYT3-C2A domain fused to glutathione-S-transferase (SYT3-C2A-GST) bound negatively charged liposomes (25% PS/75% PC) in a Ca2+-dependent manner with an estimated half-maximal Ca2+-binding value of 4.5 ± 0.6 µM free Ca2+, which is slightly smaller than that of SYT1-C2A (6 ± 0.6 µM free Ca2+; Schapire et al., 2008) and very similar to the C2A of rat SYT 1 (4.1 ± 0.3 µM free Ca2+; Fernández-Chacón et al., 2001). However, SYT3-C2B-GST showed some Ca2+-independent binding to these liposomes and its binding increased in the presence of Ca2+ (EC = 5.0 ± 0.5 µM free Ca2+; Figure 3C). These results indicate that the C2B domain of SYT3 might contain additional Ca2+-binding sites that are different from the canonical ones.
In addition to PS, C2 domains can bind other negatively charged phospholipids (Schapire et al., 2008; Perez Sancho et al., 2015). Because PI4P is the main determinant generating negative charges at the inner surface of the PM (Simon et al., 2016), we investigated the role of PI4P in the localization of SYT3-GFP and SYT1-GFP at ER–PM CS in vivo. In order to do so, SYT3-GFP and SYT1-GFP were coexpressed in N. benthamiana with the PM anchored MAP-mTU2-SAC1 phosphatase which depletes PI4P from the PM (Simon et al., 2016). In addition, we used the artificial tether GFP-MAPPER, which binds the PM through the interaction of a polybasic domain with negatively charged phospholipids (Chang et al., 2013). Expression of individual SYT1-GFP, SYT3-GFP, and GFP-MAPPER resulted in a typical cortical ER–PM CS localization (Figure 3D, left). Nonetheless, their coexpression with MAPmTU2-SAC1 caused the redistribution of the GFP signals throughout the ER (Figure 3D, right).
In an alternative approach, we decreased the content of PI4P at the PM in Arabidopsis roots by the application of phenylarsine oxide (PAO), an inhibitor of the PI-4-kinase; (Simon et al., 2016). PAO treatment in a transgenic line expressing SYT1-GFP and the PI4P biosensor 1xPHFAPP1 triggered the dissociation of the biosensor from the PM into the cytosol (Supplemental Figure S4E, left) and caused a marked increase in perinuclear staining of SYT1-GFP (Supplemental Figure S4E, central and right), indicating a SYT1-GFP relocalization from ER–PM CS to a broader distribution in the ER. These results are consistent with a tethering role for SYT1 and SYT3 in which their N-terminal TM domain anchors them to the ER and their C-terminal C2 domains bind in trans to the negatively charged PM.
SYT1 and SYT3 show cold-dependent dynamics at ER–PM CS
SYT1 was originally identified as a PM protein that accumulates in the PM fractions in response to cold stress (Kawamura and Uemura, 2003). However, using specific SYT1 antibodies (Perez Sancho et al., 2015), we did not detect increased accumulation of total SYT1 protein after 1, 3, or 7 d of cold exposure in WT or in the syt3 mutant (Figure 4A). This indicates that the total amount of SYT1 protein is neither altered by cold treatment nor by the absence of SYT3. SYT3 expression is induced by several stresses, including cold (Figure 1B;Supplemental Figure S1, C and D); therefore, we investigated the level of SYT3-GFP protein after cold in the SYT3:SYT3-GFP line using immunoblot. However, the amount of SYT3-GFP remained below the detection level in control conditions or after cold treatment (Figure 4B;Supplemental Figure S1F). Therefore, we concentrated SYT3-GFP using GFP-trap columns, and showed that, in contrast to SYT1, cold treatment caused a ∼6-fold induction of SYT3-GFP (Figure 4B).
Figure 4.
SYT1 and SYT3 show cold-dependent dynamics at ER–PM CS A, Total SYT1 protein level does not change after cold treatment. Arabidopsis WT and syt3 plants were grown for 3 weeks on soil under long-day photoperiod at 20°C and then transferred to 4°C for 1, 3, or 7 d. Proteins were extracted from shoots and detected by immunoblot with anti-SYT1 antibody. Equal loading was confirmed by Coomassie blue (CBB) staining. Experiment was repeated three times with similar results. B, SYT3-GFP protein accumulation is induced by cold. Arabidopsis seedlings expressing SYT3:SYT3-GFP were grown under long-day photoperiod and 23°C for 7 d and then transferred to 4°C for 24 h or kept at 23°C. Proteins from whole seedlings were detected by immunoblot with anti-GFP antibody. No signal was detected in the crude extracts (INPUT). SYT3-GFP was concentrated using GFP-Trap beads (IP:αGFP), detected by immunoblot with anti-GFP antibody and quantified using FIJI. C–F, Representative confocal images showing SYT1-GFP (C and D) and SYT3-GFP (E and F) dynamics with cold. Arabidopsis seedlings expressing SYT1:SYT1-GFP or 35S:SYT3-GFP were grown under long-day photoperiod and 23°C for 7 d. Cold treatment plates were transferred to 4°C for 24 h (D and F), while control plates kept growing in control conditions (C and E). Mounting of the cold-treated seedlings in the coverlids was done under cold conditions and with prechilled water. Images show a maximum Z-projection of the cortical region of the epidermal cells from the cotyledon. Close ups are shown for detailed view. Scale bars 10 and 5 µm for the close-ups. G–H, Quantification of ER–PM CS labeled by SYT1-GFP (G) or SYT3-GFP (H) in cotyledons of 7-d-old Arabidopsis epidermal cells in control conditions or after 24 h at 4°C. Contacts sites were identified as intensity maxima in the cortical region using FIJI (see “Methods” for details). Dots represent individual measurements from independent ROI from at least five cotyledons. Box plots display the first and third quartiles, split by the median; whiskers extend to include the maximum and minimum values. Different lowercase letters indicate significant differences. Data were analyzed with one-way ANOVA and Tukey’s multiple comparison test; P < 0.05. I, Quantification of the amount of SYT1-GFP labeled ER–PM CS in the epidermal cells of the cotyledon of 7-d-old Arabidopsis seedlings in control conditions or after 3 h, 15 h, 24 h, and 3 d at 4°C. “Control + 3 days” is included to show that differences in the amount of CS are not due to cotyledon age. Contacts sites were identified as intensity maxima in the cortical region using FIJI (see “Methods” for details). Dots represent individual measurements from independent ROI from at least 5 cotyledons. Statistical analysis as in panels (G–H). J, Quantification of SYT1-GFP labeled contact sites in the epidermal cells of the cotyledon of 7-d-old Arabidopsis seedlings in mock conditions or after 10 and 30 min of cold treatment. Arabidopsis seedlings expressing SYT1:SYT1-GFP were grown under long-day photoperiod and 23°C for 7 d. Seedlings for cold treatment were covered with liquid half-strength MS prechilled at 4°C and the plates were transferred to a box with ice for the specified times. Mounting of the seedlings in the cover-slides was done under cold conditions and with prechilled water. Mock seedlings were covered with liquid half-strength MS at 23°C for the specified times. Contacts sites and statistical analysis as were quantified as described in panels (G–J). K, SYT1 coimmunoprecipitates with SYT3-GFP. Seven-day-old Arabidopsis WT and expressing SYT3:SYT3-GFP and 35S:GFP were subjected to 23°C and 4°C for 24 h. Total (input), IP, and CoIP proteins were analyzed by immunoblotting. Equal loading in the input was confirmed by CBB. Free GFP was used as a negative control for CoIP. SYT3-GFP and free GFP were detected with anti-GFP antibody and SYT1 was detected with anti-SYT1 antibody.
Next, we investigated the subcellular dynamics of SYT1-GFP and SYT3-GFP using the SYT1:SYT1-GFP and 35S:SYT3-GFP lines under cold stress. In control conditions, SYT1-GFP and SYT3-GFP were observed at cER, showing the characteristic beads-and-string pattern (Figure 4, Cand E). Although cold does not affect the total amount of endogenous SYT1 (Figure 4A) and SYT3-GFP (which is driven by the constitutive 35S promoter) proteins, we observed an increase of SYT1-GFP and SYT3-GFP signals at the cortical plane after 24 h of cold treatment (Figure 4, D and F). Image analysis confirmed significant changes in the localization of SYT1-GFP and SYT3-GFP after cold treatment (see “Methods”; Figure 4, G and H). Quantification of SYT1-GFP signal, 3 h, 15 h, 24 h, and 3 d after cold treatment, indicated an increased cortical accumulation at 3 h that remained up to 3 d of cold treatment (Figure 4I). Shorter cold treatments (10 and 30 min) did not produce any significant changes in SYT1-GFP localization (Figure 4J).
E-Syts form homodimers and heterodimers and, in fact, these interactions affect their respective localization and Ca2+-dependent lipid binding (Giordano et al., 2013). Arabidopsis SYT1 forms homodimers as well (Lee et al., 2020). To assess formation of heterodimers between SYT1 and SYT3, SYT3:SYT3-GFP plants in WT background were used and analyzed for their interaction using coimmunoprecipitation (CoIP) assays in control conditions (22°C) or after 24 h of cold treatment. As previously shown, cold treatment did not change the amount of SYT1 while increased the amount of SYT3-GFP (Figure 4K). In both conditions, SYT3-GFP Co-IP endogenous SYT1 proteins indicating an in vivo association (Figure 4K). The amount of SYT1 CoIP by SYT3-GFP was higher after cold treatment; however, the ratio of SYT1 interacting with SYT3 was similar in control of after cold (Figure 4K).
SYT1 and SYT3 maintain DAG homeostasis at the PM upon cold stress
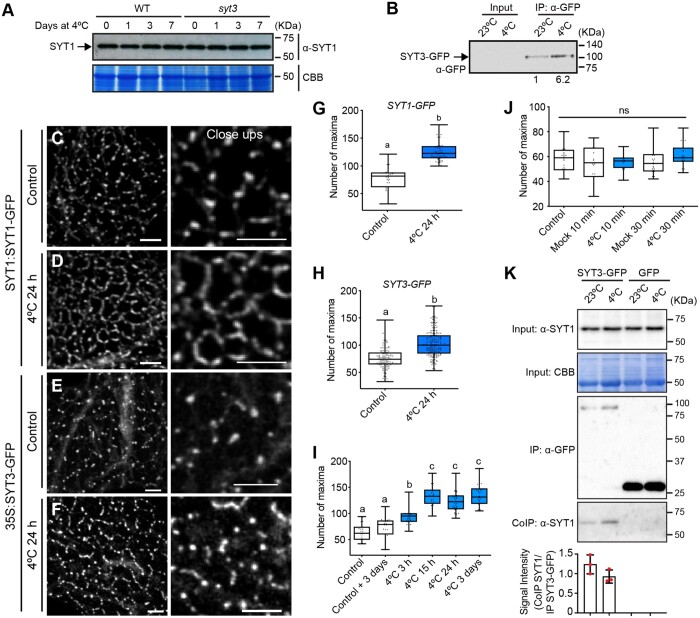
Human E-Syts contain a SMP domain with a hydrophobic groove that harbors glycerolipids (Schauder et al., 2014). Therefore, to evaluate a possible role of SYT1 and SYT3 proteins in glycerolipid homeostasis, we analyzed the leaf glycerolipidome of WT, syt1, syt3, and syt1/3 plants in control conditions and after 7 d at 4°C (a timepoints in which the lipidomic remodeling associated to cold acclimation has already occurred). Using ultraperformance liquid chromatography (UPLC) coupled to Fourier-transform mass spectrometry -based high-resolution lipidome analyses, we profiled 181 molecular species from nine glycerolipid classes (Supplemental Data Set S1). In WT plants, cold stress caused an increase in the amount of unsaturation in most lipid classes (Supplemental Figure S5A), a decrease of monogalactosyldiacylglycerols (MGDG) and the accumulation of unsaturated triacylglycerols (TAG), which is consistent with previous reports (Degenkolbe et al., 2012; Tarazona et al., 2015). As shown in Figure 5A, the total glycerolipid composition of syt1, syt3, and syt1/3 was similar to that of WT plants in both control conditions and after a cold treatment, ruling out a role for SYT1 and SYT3 proteins in the substantial lipid remodeling that takes place after cold stress.
Figure 5.
SYT1 and SYT3 maintain DAG homeostasis at the PM. A, Heatmap panels representing the fold change in the peak area of each glycerolipid class and unsaturation grade of the acyl chains when comparing WT against syt1, syt3, or syt1/3. Four-week-old rosettes were analyzed after growing under control conditions and after a 7 d cold treatment (4°C). See also Supplemental Figure S5A. Color scale code expresses mean fold changes of the corresponding glycerolipids (n = 3 pools of five independently grown plants). B, ESI–MS/MS analysis of the molecular species of PM glycerolipids from 4-week-old WT and syt1/3 rosettes grown at 23°C followed by 3 d of cold treatment (4°C). PM samples were purified by two phase partitioning protocol and lipids were extracted following as described in “Methods.” Column bars represent mean values of three biological replicates (pools of >50 plants) and error bars are showing SD. See also Supplemental Figure S5B. C–F, Distribution of the identified PC (C), PE (D), PA (E), and DAG (F) molecular species in PM of WT and syt1/3. Acyl chains are expressed as number of acyl carbons: number of acyl double bonds. Data of lipids in PM are represented as column bars (mean values of three biological replicates; pools of >50 plants) and error bars are showing sd. B–F, Asterisks indicate statistically significant differences between PM in WT and syt1/3 plants as determined by the Fisher LSD test; ****P < 0,0001; ***P < 0,0002; **P < 0,0021; *P < 0,0332.
The localization and the increased accumulation of SYT1 and SYT3 at ER–PM CS after cold treatment point toward a lipid redistribution between ER and PM more than to a general lipid remodeling. Thus, we investigated lipid changes that take place specifically in the PM of WT and syt1/3 plants after 3 d of cold treatment, a time point in which SYT1 showed a strong relocalization at ER–PM CS. To achieve a good resolution, we utilized a two-phase partitioning protocol to obtain enriched PM fractions minimizing ER contamination. Immunoblot analysis (Supplemental Figure S5B) showed that the PM fraction was highly enriched in AHA3, a H+-ATPase localized at the PM, while the luminal binding proteins BiP (a common ER marker covering BiP1, BiP2, and BiP3), and SYT1 were depleted. In contrast, BiP and SYT1 were enriched in the intracellular membrane fractions , while AHA3 was diminished. This analysis confirmed a high enrichment of PM fractions and corroborated the ER anchoring of SYT1 via its TM domain. Electrospray ionization tandem mass spectrometry (ESI–MS/MS) of the PM fractions allowed the quantification of 78 glycerolipids species belonging to nine lipid classes. The low amount of chloroplastic MGDG and digalactosildiacylglycerol (DGDG) lipids in the PM samples further supported the purity of the isolated membranes (Supplemental Data Set S2). The analysis showed no significant changes in the composition of major lipids, such as PE, PS, PG, and PA between WT and syt1/3 PM, while significant differences were found in PC and DAG (Figure 5B). Examination of single lipid species revealed a complex pattern of changes in syt1/3 related to WT in PC (Figure 5C) and minor changes in PE or PA (Figure 5, D–E). Remarkably, most DAG molecular species displayed a tendency to accumulate in syt1/3 PM (Figure 5F), with DAG32:2 and DAG32:4 being the most enriched species.
SYT1 preferentially binds to DAGs in vivo
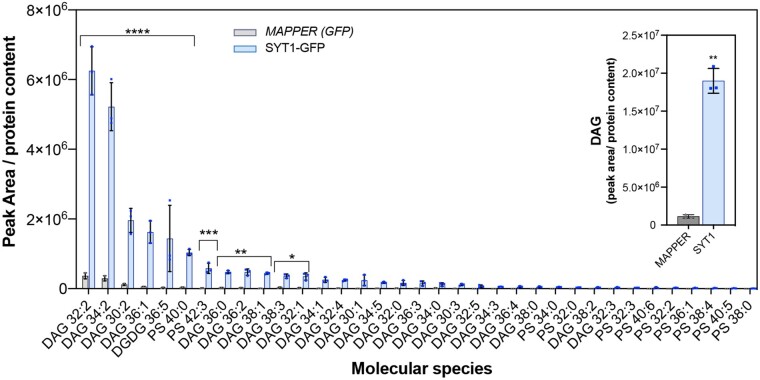
SMP domain of E-Syts forms a hydrophobic groove that can transfer glycerolipids between membranes, but the nature of the transported glycerolipids and their directionality is not unequivocally resolved (Schauder et al., 2014; Saheki et al 2016). Therefore, we sought to identify glycerolipids bound to the SYT1 SMP domain in vivo by analyzing the glycerolipids that CoIP with the functional SYT1-GFP (syt1/SYT1:SYT1-GFP Arabidopsis line). The artificial ER–PM tether GFP-MAPPER was used as negative control because it colocalizes with SYT1 at ER–PM CS but does not contain a SMP domain. We first established that GFP-MAPPER does not coimmunoprecipitate SYT1 (Supplemental Figure S6), as this would render lipids in the GFP-MAPPER sample that would be bound to the copurified SYT1. Then, SYT1-GFP and MAPPER were purified using CHAPS as a detergent, which has been reported to not displace lipids bound to the SMP domain of Mdm12 (AhYoung et al., 2015). The lipids bound to the CoIP proteins were subsequently extracted and analyzed by ESI–MS/MS (Supplemental Table S1). Surprisingly, the analysis did not detect the presence of species of highly abundant lipid classes, such as PC, PE, or phosphatidylglycerol (PG). Remarkably, out of the 35 lipid species identified in SYT1-GFP samples, 24 were DAGs (Figure 6). The two most abundant DAG species enriched in SYT1-GFP were DAG32:2 and DAG34:2. In addition to DAGs, a single species of DGDG (36:5), PS40:0, and minor amounts of some PS species were also detected in SYT1-GFP. These results are consistent with the finding that most lipids differentially accumulated in syt1/3 PM are molecular species of DAG and support a role for SYT1 and SYT3 in transporting DAGs from the PM to the ER.
Figure 6.
SYT1 preferentially binds DAGs in vivo. A, Glycerolipid molecular species determined by ESI–MS/MS analysis and identified in the immunoprecipitated SYT1-GFP and MAPPER proteins from 4-week-old Arabidopsis plants grown at 23°C followed by a 3 d cold treatment (4°C). Leaves of Arabidopsis plants expressing SYT1-GFP or MAPPER proteins were homogenized, and proteins were affinity purified using beads coated with anti-GFP antibody. Lipids were extracted using Bligh and Dyer protocol. Acyl chains are expressed as (number of acyl carbons: number of acyl double bonds). The inset graph shows the comparison of total DAG in SYT1-GFP and MAPPER immunoprecipitated proteins. Verification that SYT1-GFP did not CoIP with MAPPER is in Figure S6A. Lipid species were expressed as mean peak area (mean values of three biological replicates; pools of >50 plants). Error bars indicate SD. Asterisks indicate statistically significant differences between SYT1-GFP and MAPPER lipids as determined by the Fisher LSD test; ****P < 0.0001; ***P < 0.0002; **P < 0.0021; *P < 0.0332.
Discussion
Our results provide direct evidence that SYT1 and likely SYT3 are implicated in the clearance of PM DAG that is produced during stress. The lack of these proteins in syt1/3 plants results in the accumulation of DAG at the PM, that conceivably causes the defective PM integrity exhibited in response to multiple stresses. SYT1 and SYT3 localization at ER–PM CS are directed by their TM anchor to the ER and the binding of their C2 domains in trans to PM PI4P and follows a stress-dependent dynamics. Thus, the efficiency of DAG removal is likely enhanced by the SYT1 and SYT3 accumulation at ER–PM CS after abiotic stress.
SYT1 and SYT3 function as dynamic ER–PM CS tethers in response to abiotic stress
SYT1 and SYT3 proteins function as tethers in the formation of ER–PM CS. This activity is dependent on their ER anchoring by their N-TM domain and the interaction of their C2 domains with PI4P at the PM. Consistent with this, SYT1 purifies within the ER fraction and the depletion of PI4P in the cytosolic leaflet of the PM by MAP-SAC1 triggers the loss of SYT1-GFP and SYT3-GFP focal localization at ER–PM CS, resulting in a distribution of these proteins throughout the ER. As it is anticipated for proteins that function as ER–PM tethers (Eisenberg-Bord et al., 2016), overexpression of SYT1 and SYT3 causes a drastic expansion of ER–PM CS that can eventually results in cell death. Interestingly, SYT3 shows an enrichment in ER domains that display high curvature, like tubules and sheet edges. This localization to high ER curvature areas has been previously reported for SYT1 (Ishikawa et al., 2018) and Tcb proteins (Collado et al., 2019; Hoffmann et al., 2019), and may have important functional implications in lipid transport as we discuss below. Cold stress increases the ER–PM connectivity by promoting the cortical expansion of ER–PM CS containing SYT1 and SYT3 without changes in the total amount of SYT1 nor SYT3, which is driven by the constitutive 35S promoter. A similar increase of ER–PM CS marked by SYT1 has been previously reported by salt stress (Lee et al., 2019), indicating that SYT1 (and likely SYT3) dynamics are not specific to cold and establishing SYT1 and SYT3 as general abiotic stress-regulated tethers between the ER and the PM.
An obvious question is what the molecular events that trigger SYT1 and SYT3 relocalization and ER–PM CS expansion are. Based on previous data from E-Syt1, one evident candidate that triggers SYT1 and SYT3 stress-dependent dynamics is cytosolic Ca2+. First, it is known that cytosolic Ca2+ increases as a response to multiple abiotic stresses (Knight et al., 1997, 1991). Second, the C2 domains of SYT1 (Schapire et al., 2008) and SYT3 (this work) show phospholipid binding that is dependent on micromolar increase of Ca2+. However, the temporal dynamics of Arabidopsis SYT1 and SYT3 make it difficult to reconcile a Ca2+ increase as being the molecular trigger for this relocalization. The relocation of plant SYTs to ER–PM CS after cold is slow (∼3 h) and persistent (3 d), while cold stress is known to cause very fast and transient increases in cytosolic Ca2+ (Knight and Knight, 2000). This slow relocation is in contrast to the fast (within seconds/minutes) and transient Ca2+-dependent recruitment to of E-Syt1 ER–PM CS (Giordano et al., 2013; Saheki et al., 2016), revealing striking differences between the dynamics of animal E-Syts and plant SYTs. Although we cannot rule out that the cold ER–PM CS expansion is a downstream response to the transient Ca2+, changes in the content of PI(4,5)P2 could also regulate SYT1 and SYT3 dynamics. This lipid species is induced by different stresses at the PM (Heilmann, 2016) and salt stress causes a similar SYT1 relocalization at ER–PM CS that is associated with changes in the content of PI(4,5)P2, but not PI4P (Lee et al., 2019).
A role of SYT1 and SYT3 in DAG homeostasis during abiotic stress
SYT1 is not required for the expansion of ER–PM CS after stress, suggesting additional functions for SYT1 other than ER–PM CS tethering (Lee et al., 2019). Thus, it is conceivable that a defective lipid transport associated to their SMP domains could be the cause underlying the sensitivity of syt1/3 to stress. Consistent with this, PM fractions from cold treated syt1/3 plants showed a significant accumulation of DAG compared with the WT. Importantly, most DAG species were increased, while significant changes were found in DAG32:2, DAG32:4, DAG34:2, and DAG34:3. Moreover, IP of SYT1-GFP followed by lipidomic analysis revealed the presence of many DAG species, suggesting a lack of specificity with respect to any particular DAG species.
Upon stress, PLCs and NPCs (which hydrolyze PIPs and membrane structural phospholipids, such as PC or PE, respectively) trigger the production of DAG at the PM. Although both NPCs and PLCs can induce DAG formation, the DAG molecular species they produce are different due to the substrates they hydrolyze. Although DAG produced via the PLC pathway is mainly composed of C32 and C34 species, NPC generates DAG species with longer (C34 and C36) and more unsaturated fatty acids (Peters et al., 2010). When analyzing the PM of syt1/3, we did not detect any significant changes in DAG molecular species with 36 carbons, and the biggest changes were found in shorter DAG with 32 carbons. These results suggest that DAG species in syt1/3 PM are likely produced due to the activation of PLCs.
DAG molecules are conical in shape within the membrane and thus pack poorly in planar bilayers and generate areas of negative curvature leading to a disruption of the lamellar phase of lipid membranes (Campomanes et al., 2019). Consequently, it is predicted that an uncontrolled accumulation of DAG caused by mutations in SYT1 and SYT3 will lead to a local accumulation of these molecules and a decrease of PM integrity, highlighting the importance of DAG removal during stress episodes. Interestingly, although no significant alterations in steady-state PM glycerolipids were detected in genome-edited cells lacking all E-Syts, a transient accumulation of DAG after PLC activation was observed, supporting their role in DAG transport (Saheki et al., 2016). This function was further reinforced from the analysis of pancreatic islets, where a decreased expression of E-Syt1 resulted in prolonged accumulation of DAG in the PM causing an increase in insulin secretion (Xie et al., 2019).
Recently, it has been shown that Tcbs create peaks of extreme curvature on the cER and these peaks are important to maintain PM integrity under stress, likely by facilitating lipid transport (Collado et al., 2019). Like Tcbs, SYT1 and SYT3 are localized in regions of the ER that display high curvature (Ishikawa et al., 2018). Accordingly, it is conceivable that Tcbs, like SYTs and E-Syts may also operate in the regulation of DAG levels at the PM and their specific localization play a role in the mechanism of lipid transfer. In pancreatic β-cells, DAG formation occurs in microdomains that coincide with sites of insulin granule exocytosis and are accompanied by recruitment of E-Syt1 to the same sites. It is tempting to speculate that the formation of DAG by PLC in plants might occur in specific ER–PM CS microdomains that overlap with the localization of SYT1 and SYT3, increasing the efficiency of transfer.
The large evolutionary distance between eukaryotes, such as yeast, mammals, and plants has resulted in the generation of independent mechanisms to deal with developmental and environmental challenges. However, they all have in common the presence of ER–PM CS that plays important metabolic functions and express a class of closely related proteins. Mammal E-Syts, yeast Tcbs and plant SYTs localize in these microdomains, functioning as tethers that connect the ER with the PM. Still, whether or not these proteins have common roles in lipid transfer through their SMP domains remains elusive. This report provides insights into the role of the SMP domain containing proteins SYT1 and SYT3 that probably can be translated to yeast and mammals.
In summary, our studies support a role for ER–PM CS tethers SYT1 and SYT3 in the regulation of PM DAG homeostasis under abiotic stress. We propose a model in which activation of PLC induces rapid and transient formation of DAG during stress episodes. Because of its structural characteristics, DAG does not form lipid bilayers and therefore must be actively removed. Part of this pool of DAG is transformed into PA by action of DAG kinases, while the remaining DAG is efficiently transferred to the ER by SYT1 and SYT3 at ER–PM CS in order to avoid membrane damage. This removal of DAG from the PM is further boosted by the stress-induced expansion of ER–PM CS marked by SYT1 and SYT3.
Materials and methods
Plant material
All Arabidopsis (A. thaliana) plants in this study are Col-0 ecotype. Arabidopsis mutant lines are: syt1 (AT2G20990) SAIL_775_A08 (previously described in Perez Sancho et al., 2015), syt3 (AT5G04220) SALK_037933 (obtained from the Arabidopsis Biological Resource Center, Ohio State University) and syt1/3 double mutant obtained by crossing syt1 with syt3. The transgenic lines SYT1:SYT1-GFP (Perez Sancho et al., 2015), syt1/SYT1:SYT1-GFP (Lee et al, 2019), and 35S:GFP (Wang et al., 2019) were previously described. Line syt1/SYT1:SYT1-GFP/1xPHFAPP1 was generated by crossing syt1:SYT1-SYT1-GFP with the PI4P marker P1xPHFAPP1 (Simon et al., 2016). Generation of transgenic lines SYT1:MAPPER, SYT3:SYT3-GFP, 35S:SYT3-GFP, and SYT3:GUS is described in the section “Generation of transgenic plants.” Line syt1/35S:SYT3-GFP was obtained by crossing syt1 with the 35S:SYT3-GFP line.
Plant manipulation and growth conditions
Arabidopsis standard handling procedures and conditions were used to promote seed germination and growth. First, Arabidopsis seeds were surface-sterilized and cold treated for 3 d at 4°C. Next, seeds were sowed onto half-strength Murashige and Skoog (MS) agar solidified medium (0.6% [w/v] agar for horizontal growth and 1% [w/v] for vertical growth) containing 1.5% [w/v] sucrose, unless otherwise stated. Plates were placed either vertically or horizontally in a culture chamber at 23°C ± 1°C, under cool-white light (at 120 µmol photon m−2 s−1) with a long-day photoperiod (at 16-h light/8-h dark cycle) unless otherwise stated. When required, seedlings were transferred to soil after 7 d of in vitro growth and watered every 2 d. In soil, plants were grown in a mixture of organic substrate and vermiculite (4:1 [v/v]) under controlled conditions at 23°C ± 1°C, 16-h light/8-h dark cycle (at 120 µmol photon m−2 s−1). Freshly harvested seeds were used for all phenotypic analyses.
Plasmid constructs
SYT3:SYT3-GFP, 35S:SYT3-GFP, and SYT3:GUS constructs were generated using Gateway. A 2,210-bp SYT3 promoter fragment was cloned into pDONR-P1-P4 via a BP reaction (Invitrogen) to generate pEN-L4-proSYT3-R1. The genomic fragment of the SYT3 coding sequence without stop codon was amplified from Col-0 DNA using primers listed in Supplemental Table S2 and recombined into pDONR221 via a BP reaction (Invitrogen) to generate pEN-L1-SYT3g-L2. For the SYT3:SYT3-GFP and 35S:SYT3-GFP, we recombined pEN-L4-proSYT3-R1 or pEN-L4-2R1 (proCaMV35S) from Karimi et al. (2007) with pEN-L1-SYT3g-L2 and pEN-R2-F-L3,0 (GFP) from Karimi et al. (2007) into pKm43GW (Karimi et al., 2005). For the SYT3:GUS construct, pEN-L4-proSYT3-R1 was recombined with pEN-L1-S-L2 (GUS) into pKm42GW (Karimi et al., 2005). The SYT1:MAPPER construct was generated by recombination of the previously described constructs pEN-L4-proSYT1-R1 and pEN-L1-MAPPER-L2 (Perez Sancho et al., 2015) with pEN-R2-empty-L3 and the destination vector pKm42GW.
Multisite Gateway cloning was used in the preparation of constructs for transient expression in N. benthamiana. The coding DNA sequence (CDS) without the stop codon of SYT1, SYT3, and DGK2 (AT5G63770) was PCR-amplified using the primers listed in Supplemental Table S2 and cloned into the pENTR/ZEO vector using BP cloning kit (Invitrogen). All the pENTR clones were verified by sequencing. The pENTR vector with the CDS of FaFAH1 (Sánchez-Sevilla et al., 2014) was a gift from Iraida Amaya. These pENTR clones in combination with the appropriate destination vectors (pDEST) were used to create the final Gateway-expression constructs by LR-reaction (Invitrogen). The pGWB5 was used to generate 35S:SYT1-GFP and 35S:SYT3-GFP constructs. The pENTR vectors, pEN-L4-pUBQ10-R1 and pEN L2-mCherry-R3 were used with the pDEST vector pH7m34GW and the generated pEN-L1-L2 vectors to produce UBQ10:SYT1-mCherry, UBQ10:SYT3-mCherry, UBQ10:DGK2-mCherry, and UBQ10:FaFAH1-mCherry constructs. The SYT1:MAPPER used for N. benthamiana experiments was the same as in the generation of A. thaliana lines. The construct MAP-mTU2-SAC1 is previously described (Simon et al., 2016).
Generation of Arabidopsis transgenic lines
Expression constructs were transformed into Agrobacterium tumefaciens strain GVG3101::pMP90 through electroporation. The SYT1:MAPPER, SYT3:SYT3-GFP, 35S:SYT3-GFP, and SYT3:GUS constructs were transformed into Arabidopsis Col-0 plants by floral dip to generate stable transgenic plants. Homozygous transgenic plants were used in this study.
Transient expression in N. benthamiana
Transient expression in N. benthamiana was performed using Agrobacterium strain (GV3101::pMP90) carrying the different constructs, together with the Agrobacterium expressing p19. Agrobacteria cultures were grown overnight in Luria-Bertani medium containing rifampicin (50 mg mL−1), gentamycin (25 mg mL−1), and the construct-specific antibiotic. Cells were then harvested by centrifugation and pellets were resuspended in agroinfiltration solution (10 mM MES, pH 5.6, 10 mM MgCl2, and 1 mM acetosyringone), and incubated for 2 h in dark conditions at room temperature. Agrobacterium strains were infiltrated at OD600 of 0.70 for the constructs and 0.25 for the p19 strain. For double infiltration experiments, Agrobacterium strains were infiltrated at OD600 of 0.40 for the constructs and 0.15 for the p19 strain. Four to 5-week-old N. benthamiana leaves were infiltrated with Agrobacterium into at the abaxial side of the leaf lamina. After infiltration, all plants were kept in the growth chamber and analyzed 2 d later.
Arabidopsis eFP Browser Data Analysis
Gene expression level data from abiotic stress responses were retrieved from the Arabidopsis eFP Browser (Abiotic Stress Series) website (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). Data used for the analysis were obtained from 18-d-old WT seedlings. Differential expression was calculated by dividing the expression value of each gene in a given abiotic stress by the corresponding control (fold change of abiotic stress relative to the mock). The abiotic stress gene expression response was calculated and the heatmap was created using Excel (Microsoft). In the heatmap, red represents induction and blue represents repression as response to the indicated hormone.
Phylogenenic analysis
Phylogenetic tree was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 3.26 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and are in the units of the number of amino acid differences per site. This analysis involved six amino acid sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 1,752 positions in the final dataset. Evolutionary analyses were conducted using MEGA X (Kumar et al., 2018).
Total RNA extraction and quantitative PCR analysis
Seven-day-old whole seedlings (20 seedlings per biological replicate) were used for total RNA extraction of 24-h cold treatment experiments. Plant tissue was grounded to a fine powder in liquid nitrogen. Approximately 100 mg of ground tissue per sample was homogenized in 1 mL of the commercial reagent TRIsure (Bioline), and total RNA was extracted following the manufacturer’s instructions. RNA samples (10 µg per sample) were treated with TurboDNA-free DNase (Ambion), and 1 µg of RNA per sample was run on a 1% agarose gel to confirm RNA integrity. First-strand cDNA was synthesized from 1 µg of RNA using the iScript cDNA synthesis kit (Bio-Rad), according to the manufacturer’s instructions. cDNAs were amplified in triplicate by quantitative PCR (qPCR) using SsoFast EvaGreen supermix (Bio-Rad) and the Rotor-Gene Q cycler (Qiagen). The amplification protocol includes an initial step at 95°C for 2 min, followed by 40 cycles of 5 s at 95°C, 15 s at 58°C, and 20 s at 72°C. The relative expression values were determined using ACTIN2 as a reference. Primers used for qPCR are listed in Supplemental Table S2.
Protein extraction
Total protein extractions were carried out as previously described (Amorim-Silva et al., 2019) with minor modifications. Briefly, Arabidopsis tissue was ground to a fine powder in liquid nitrogen. Approximately 100 mg of ground tissue per sample was used for total protein extraction. Denatured protein extracts were obtained by homogenizing and incubating plant material in 2X Laemmli buffer [125 mM Tris–HCl, pH 6.8, 4% (w/v) SDS, 20% (v/v) glycerol, 2% (v/v) b-mercaptoethanol, and 0.01% (w/v) bromophenol blue] for 20–30 min at 70°C. Extracts were centrifuged for 5 min at 20,000g at 10–15°C and supernatants were recovered. Total protein extracts from supernatant were separated in 10%SDS–PAGE gels and analyzed as described in the following section.
Immunoblot analyses
Proteins separated by SDS–PAGE polyacrylamide gel electrophoresis were electroblotted using Trans-blot Turbo Transfer System (Bio-Rad) onto polyvinylidene difluoride (PVDF) membranes (Immobilon-P, Millipore) following instructions by the manufacturer. PVDF membranes, containing electroblotted proteins, were then incubated with the appropriate primary antibody followed by the appropriate secondary peroxidase-conjugated antibody. The following primary antibodies were used for detection of epitope-tagged proteins: mouse monoclonal anti-GFP clone B-2 (1:1,000, catalog no. sc-9996, Santa Cruz Biotechnology) BiP lumenal-binding protein (rabbit antibody, catalog AS09 481, Agrisera), rabbit polyclonal AHA3 (a gift from Ramón Serrano) and rabbit polyclonal anti-SYT1 antibody (1:5,000). The secondary antibodies used in this study were as follows: anti-mouse IgG whole molecule-Peroxidase (1:80,000; catalog no. A9044, Sigma-Aldrich) and anti-rabbit IgG whole molecule-Peroxidase (1:80,000; catalog no. A0545, Sigma-Aldrich). Proteins and epitope-tagged proteins on immunoblots were detected using the Clarity ECL Western Blotting Substrate or SuperSignal West Femto Maximum Sensitivity Substrate according to the manufacturer’s instructions, and images of different time exposures were acquired using the Chemidoc XRS1System (Bio-Rad). Only images with no saturated pixels were used for protein quantification. SDS–PAGE and immunoblotted PVDF membranes were stained with Coomassie Brilliant Blue R 250 to confirm equal loading of the different samples in a given experiment.
CoIP in Arabidopsis
CoIP experiments were carried out as in Amorim-Silva et al. (2019) with minor modifications. Eight-day-old Arabidopsis plants were ground to fine powder in liquid nitrogen. Approximated 0.5 g of ground tissue per sample were used and total proteins were then extracted with extraction buffer [50 mM Tris–HCl, pH 7.5; 150 mM NaCl; 10% glycerol; 10 mM EDTA, pH 8; 1 mM NaF; 1 mM Na2MoO4·2H2O; 10 mM DTT; 0.5 mMPMSF; 1% (v/v) P9599 protease inhibitor cocktail (Sigma); Nonidet P-40, CAS: 9036-19-5 (USB Amersham life science) 0.5% (v/v) added at 2 mL/g of powder using an end-over-end rocker for 30 min at 4°C. Samples were centrifuged 20 min at 4°C at 9,000g. Supernatants were filtered by gravity through Poly-Prep Chromatography Columns (#731-1550 Bio-Rad) and 100 μL was reserved for immunoblot analysis as input. The remaining supernatants were diluted (1:1 dilution) in extraction buffer without Nonidet P-40, so the final concentration of detergent (Nonidet P-40) was adjusted to 0.25% (v/v) to avoid unspecific binding to the matrix as recommended by the manufacturer. Protein samples were then incubated for 2 h at 4°C with 15-μL GFP-Trap coupled to agarose beads (Chromotek) in an end-over-end rocker. Beads were then collected and washed four times with the wash buffer (extraction buffer without detergent). Finally, beads were resuspended in 75 μL of 2× concentrated Laemmli Buffer and heated at 70°C for 30 min to dissociate immunocomplexes from the beads. Total (input), IP, and CoIP proteins were separated in a 10% SDS–PAGE and analyzed by immunoblot.
PAO treatment
Six-day-old seedlings of SYT1-GFP/1xPHFAPP1 were incubated in wells containing 60 µM of PAO (Sigma, PAO stock solution 60 mM in DMSO) in one half strength liquid MS for 4 h. The mock condition corresponds to the incubation of plants in wells supplemented with the same concentration of DMSO and treated for the same time. Roots were then subjected to imaging using confocal laser scanning microscopy.
Cold and salt treatments for cell viability in roots
Previous to cold treatment, 24-well plates loaded with liquid one tenth strength MS solution were placed on ice for 30 min. Six-day-old seedlings of the phenotypes in study (WT, syt1, syt3, syt1/3, syt1/SYT1:SYT1-GFP, and syt1/35S:SYT3-GFP) were incubated in the wells containing the mentioned liquid media at 8°C at different times between 0 and 30 min. Seedlings were then stained in 1/10 MS containing 10 µg mL−1 of FDA (Sigma, FDA stock solution 5 mg mL−1 in DMSO) for 5 min. Seedling were then washed once and imaging was performed. In all cases, roots were imaged within a 5-min time frame window around the indicated time. For salt treatment, 4-d-old seedlings of WT, syt1, syt3, and syt1/3 were incubated in 150 mM KCl in one-tenth MS strength for 1 h. Then, they were stained and imaged as described above. In both cases, at least 10 plants of each phenotype were used (and the experiment was repeated twice).
For double staining and after the cold treatment, seedlings were stained in 1/10 MS containing 10 µg mL−1 of FDA (stock solution as above) and 2-µM FM-4-64 (Invitrogen 2 mM stock solution in distilled water) for 5 min. Seedling were then washed once, mounted and imaging was performed as explained below.
Freezing tolerance assays
Two-week-old Arabidopsis plants growing in soil under standard conditions or under cold-acclimation conditions (for 7 additional days at 4°C) were transferred to a freezing chamber set to 4°C for 30 min in darkness. Subsequently, temperature was allowed to decrease at a rate of −1°C per 30 min until reaching the final desired freezing temperature which was maintained for 6 h. Then, the temperature was increased to 4°C at the same rate and thawing was allowed for 12 h before returning plants to control conditions for recovering. Tolerance to freezing was determined as the percentage of plants surviving after 2 weeks of recovery under control conditions.
Cold treatments for SYT1-GFP and SYT3-GFP dynamics
Arabidopsis SYT1:SYT1-GFP or 35S:SYT3-GFP was grown vertically in half-strength MS agar solidified medium under long-day photoperiod and 23°C for 7 d. For long exposure cold treatments (≳3 h), plates were transferred to a temperature programmable chamber and kept at 4°C for the specified times. Control plates kept growing in control conditions. For short-time cold exposure (<3 h), seedlings were covered with liquid half-strength MS prechilled at 4°C and the plates were transferred to a box with ice for the specified times. Mock seedlings were covered with liquid half-strength MS at 23°C for the specified times while control seedlings were undisturbed. Mounting of the cold-treated seedlings on the microscope slides was done under cold conditions and with prechilled water. In all cases, roots were imaged between 3 and 4-min time frame window after taken them out from the treatment condition.
Immunostaining of Arabidopsis roots
Five-day-old root tips were used, and all working solutions were prepared in microtubule stabilization buffer [MTSB, 50 mM PIPES, 5 mM EGTA, and 5 mM MgSO4 (pH 7.0)]. Samples were fixed in 4% paraformaldehyde and root tips were attached to polylisine coated slides following the basic protocol described in Sauer and Friml (2010). For immunodetection, a polyclonal rabbit anti-SYT1 anti-body (1:1,000) was incubated overnight at 4°C plus 2 additional hours at 37°C. Then, the root tips were incubated for 1 h with TRITC-conjugated AffiniPure Donkey anti-Rabbit IgG (1:200; Jackson Immunoresearch), and mounted on microscopy slides using the glycerol-based AF1 Mountant Solution (Citifluor).
Electrolyte leakage measurements
One-week-old seedlings grown in half-strength MS plates were transferred to tubes containing 5 mL of 20% PEG and incubated at 25°C for the indicated time points. Afterward, seedlings were carefully removed, washed with deionized water, and placed in new tubes containing 5 mL of deionized water. The tubes were shaken at 120 rpm for 3 h at 25°C and the conductivity of the solutions was measured. The tubes were then autoclaved and after cooling down to room temperature, the conductivities of the solutions were measured again. The percentage of electrolyte leakage was calculated as the percentage of the conductivity before autoclaving over that after autoclaving. Three independent experiments were performed.
Generation and purification of GST fusion proteins
RT-PCR of WT mRNA was used to produce GST fusion clones in pGEX-KG corresponding to the following SYT3 amino acid sequences: 244–402, corresponding to the SYT3-C2A domain, and 403–540, corresponding to the SYT3-C2B domain. Briefly, proteins expressed in BL21-RP were released by sonication and incubated with glutathione agarose beads (0.3 mL L−1 culture) overnight at 4°C. Proteins were washed on the agarose beads four times with 10 mL PBS, 1 mM EDTA, and 0.1 g L−1 PMSF. Purified protein was eluted from the agarose with 2.5 mL of 100 mM Tris, pH 8.0, and 40-mM glutathione and concentrated to 0.2 mL by Centricon centrifugation (Millipore). Protein concentrations were determined by the method of Bradford with protein dye reagent concentrate from Bio-Rad, using BSA as standard.
Phospholipid binding assays
Phospholipid binding to isolated C2A and C2B domains of SYT3 was assessed by a centrifugation assay as described previously (Schapire et al., 2008). Briefly, phospholipids (PS/PC = 25/75, w/w; Sigma-Aldrich) in chloroform were dried as a thin layer under a stream of nitrogen. Dried lipids were resuspended in buffer A (100 mM NaCl; 50 mM HEPES, pH 6.8; 4 mM EGTA) by vortexing for 20 min. Large multilamellar vesicles were collected by centrifugation for 20 min at 20,800g and resuspended in buffer A with various concentrations of free Ca2+ and used within 1 h. Calcium concentrations were calculated using the WEBMAXC STANDARD, RRID:SCR_003165). Purified soluble recombinant GST-C2 domains (6 μg) and vesicles (100 μg of phospholipids; total volume = 1 mL) were incubated for 15 min at 27°C with shaking at 250 rev/min on a platform shaker. Large multilamellar vesicles and bound protein were isolated by centrifugation for 10 min at 20,800g at 4°C. Pellets were washed three times with 0.5 mL of the corresponding incubation buffer, and the bound protein was analyzed by SDS–PAGE and densitometry using a Bio-Rad GS670 scanning densitometer of the Coomassie Brilliant Blue-stained gel.
Modeling of SYT3 C2 domains.
The 3D molecular modeling of SYT3 C2A and C2B domains were predicted using the Phyre2 multitemplate homology modeling tool (Kelley et al., 2015) coupled to the 3DLigandSite server for protein binding site prediction (Wass et al., 2010). The resulting PDB coordinates were refined using the Wincoot 3D toolkit (Emsley and Cowtan, 2004). The resulting 3D structure was validated by analysis of torsional angles using Rampage (Lovell et al., 2003) and Ramachandran plots were generated. As a result, very robust C2A244–402 and C2B403–540 3D models with a high confidence match (>99%) were generated.
Total lipid extraction and analysis by liquid chromatography–mass spectrometry
Lipid extraction was performed as previously described (Giavalisco et al., 2011) at Max Planck Institute of Molecular Plant Physiology. In brief, 100 mg of each biological replicate was extracted in 1 mL of a precooled mixture of methanol/methyl-tert-butyl-ether/water (1:3:1) and shaking in a cooled sonic bath for 10 min. Then, 500 µL of water: methanol (3:1) was added. This led to the formation of two phases. A fixed volume of lipid phases was transferred to an Eppendorf tube and vacuum-dried to dryness. Chromatographic separation was performed using a Waters Acquity UPLC system connected to an Exactive Orbitrap (Thermo Fischer Scientific) via a heated electrospray source (Thermo Fischer Scientific). Processing of chromatograms, peak detection and integrations were done with REFINER MS 7.5 (GeneData; http://www.genedata.com). The obtained features (m/z at a certain retention time) were queried against an in-house lipid database for further annotation.
PM isolation
PM purification was carried out as described previously (Bernfur et al., 2013) with some modifications. All steps in the preparation procedure were performed at 4°C or kept in ice. Fifteen grams of leaves were homogenized with a knife blender in 40 mL homogenization buffer: 0.33 M sucrose, 50 mM MOPS-KOH, pH 7.5, 5 mM EDTA, 5mM EGTA, 20mM NaF, and including 0.6% (w/v) polyvinylpoly–pyrrolidone, 5 mM ascorbate, 4 mM salicylhydroxamic acid (SHAM), 150 µM 4-(2-aminoethyl)-benzene-sulfonyl fluoride, and 5 mM dithiothreitol (DTT) that were added immediately before use. The homogenate was filtered through a nylon mesh (200 µm) and PMSF was added to a final concentration of 1 mM. The filtrate was centrifuged at 10,000g for 15 min, the pellet was discarded, and the supernatant was centrifuged at 100,000g for 2 h at 4°C. The microsomal pellet was resuspended in 1.5–2.0 mL resuspension buffer: 0.33 M sucrose, 5 mM K-phosphate, pH 7.8, 0.1 mM EDTA, and 1 mM DTT freshly added final weight of the resuspended pellet should be 3.0 g. To produce an aqueous polymer two-phase system with a final weight of 12 g, the resuspended pellet (3 g) was added to a 9.00 g phase mixture producing a 6.1% (w/w) dextran 500, 6.1% (w/w) PEG 3350 in 0.33 M sucrose, 5 mM K-phosphate, pH 7.8, 3 mM KCl phase system. Further purification of the PMs using the aqueous polymer two-phase system was performed according to (Larsson et al., 1994). The final upper phase, highly enriched in PMs, was diluted two-fold with 0.33 M sucrose, 5 mM K-phosphate, pH 7.8, and 0.1 mM EDTA before centrifugation at 100,000g for 2 h. The PM pellet was resuspended in 100 µL resuspension buffer plus 5 mM KCl and stored at −80° C until use.
Protein large-scale IP for lipidomics
Arabidopsis lines syt1/SYT1:SYT1-GFP and SYT1:MAPPER were used for this analysis. Four-week-old Arabidopsis rosettes were ground to fine powder in liquid nitrogen. Fifteen grams of ground leaves per sample were used and total proteins were then extracted with extraction buffer (50mM Tris–HCl pH 7.5, 150 mM NaCl, 10% glycerol; 16 mM CHAPS, 10 mM EDTA pH 8.0, 1 mM Na2MoO4·2H2O, 1 mM NaF, 10 mM DTT, 0.5 mM PMSF; and 1% [v/v] P9599 protease inhibitor cocktail [Sigma]) added at 2 mL/g of powder and mixed by pipetting and with an end-over-end rocker for 30 min at 4°C. Samples were centrifuged at 9,000g for 20 min at 4°C. Supernatants were filtered by gravity through Poly-Prep Chromatography Columns (#731-1550 Bio-Rad) and 100 μL was reserved for immunoblot analysis as input. The remaining supernatants were diluted (1:1 dilution) with extraction buffer without CHAPS. Protein samples were then incubated for 2 h at 4°C with 150-μL GFP-Trap coupled to agarose beads (Chromotek) in an end-over-end rocker. Following incubation, beads were collected and washed three times with wash buffer (extraction buffer without detergent) and one more time with wash buffer with 500-mM NaCl. An aliquot of the beads (approximately 1/10) was saved for CoIP analysis. For these samples, proteins were stripped from these beads by boiling in SDS loading buffer for 10 min. IP proteins were separated on SDS–PAGE acrylamide gels and detected by immunoblot. In addition, lipids of IP in the rest of the beads were extracted and analyzed. Lipids were extracted as previously described using the method of Bligh and Dyer (see below).
PM, SYT1-GFP, and MAPPER samples lipid extraction and analysis
Lipids were extracted as previously described using the method of Bligh and Dyer. Briefly, extraction of lipids from IP proteins in GFP-beads or PM samples was performed by the addition of 3.75 volumes of chloroform: methanol (1:2 v/v), followed by sonication for 1–2 min and vortex during 15–20 min. Then 1.25 volumes of chloroform were added, samples were vortexed for 1 min and 1.25 volumes of 0.5% acetic acid in 500 mM of chloroform were added, followed by 1 min of vortexing. The sample was centrifuged at low speed (300g) using a tabletop centrifuge for 5 min; then the bottom layer was gently recovered. Two upper phase re-extractions were done by adding 1.875 vol of chloroform, followed by vortex and centrifugation as above. Lipids were dried by adding gas N2.
Quantitative analyses of lipids (at Rothamsted Research, UK), including neutral (DAGs) and polar lipids (PC, PE, PI, PG, lysophosphatidylcholine [LPC], MGDG, and DGDG) were carried out using electrospray ionization tandem triple quadrupole mass spectrometry (API 4000 QTRAP; SCIEX; ESI–MS/MS) as described previously (Guo et al., 2019). The internal standards were supplied by Avanti (Alabama, USA), incorporated as 8 pmol 13:0-LPC, 0.086 nmol di24:1-PC, 0.080 nmol di14:0-PE, 0.05 nmol di18:0-PI, 0.080 di14:0-PG, 0.03 nmol di18:0-PS, and 0.03 nmol di14:0-PA. The standards were dissolved in chloroform and 25 μL of the samples were combined with chloroform/methanol/300 mM ammonium acetate (300:665:3.5 v/v/v) to make a final volume of 1 mL. The lipid extracts were infused at 15 μL min−1 with an autosampler (HTS‐xt PAL, CTC‐PAL Analytics AG). Data acquisition and acyl group identification of the polar lipids were carried out as described in Ruiz-Lopez et al. (2014) with modifications. For quantifying TAG and DAG, 25 μL of lipid extract, 0.857 nmol of tri15:0 and 0.043 nmol 18:0-20:4-DAG (Nu‐Chek‐Prep) were combined with chloroform/methanol/300 mM ammonium acetate (24:24:1.75 v/v/v), to final volumes of 1 mL for direct infusion into the mass spectrometer. DAGs were detected as [M + NH4+] ions by a series of different neutral loss scans, targeting losses of fatty acids. Full documentation of lipid profiling data is provided in Supplemental Data Set S2 and Supplemental Table S1.
Confocal imaging of Arabidopsis and N. benthamiana
Living cell images were obtained using two different inverted Leica TCS SP5 II confocal laser-scanning microscope, and a Nikon Eclipse Ti based Andor Revolution WD spinning-disk confocal microscope. One of the Leica TCS SP5 II laser-scanning microscopes was equipped with GaAsp HyD detectors. Both were equipped with HCX PL Apo 40x/NA1.3 and HCX PL Apo 63x/Na1.4 oil immersion objective lenses. The software Leica LAS X was used for image acquisition. The Andor Revolution WD microscope was equipped with a CSU-W1 spinning disk head (Yokogawa, Tokyo, Japan), an Andor iXon Ultra 888 (EMCCD) camera and a Nikon Apo TIRF 60x/NA 1.49 oil immersion objective lens. The software Leica LAS X was used for image acquisition. Andor iQ3.6. All microscopes were equipped with a 488 nm laser for GFP/Fluorescein excitation, and the Leica SP5 II were also equipped with a 561-nm He–Ne laser for mCherry/TRITC excitation.
For imaging on N. benthamiana leaves, 2 d after infiltration leaf-disks were excised from the leaves immediately before visualization under the Leica TCS SP5 II microscope with 40× lens and the lower epidermis of the leaf was 3D imaged from the equatorial plane until the cell surface with 600 nm spacing. GaAsp HyD detectors were used to improve the signal detection. For colocalization, sequential line scanning mode was used to separate signals. Cortical plane images are a maximum Z-projection of several planes from the cell surface until a plane where cells are close but still not touching the neighbors (to ease identification of individual cells). Equatorial plane images are single plane images.
Images of SYT1 and SYT3 patterns in Arabidopsis in control and cold conditions—long treatments—and immunostained Arabidopsis roots, were obtained with the Leica TCS SP5 II microscope. Images of SYTs patterns in Arabidopsis in mock and cold conditions—short treatments—were obtained with the Andor Revolution WD spinning-disk confocal microscope. Cotyledons from 7-d-old seedlings were excised immediately before visualization and the epidermal cells in the adaxial side were imaged as indicated for N. benthamiana. Immunostained roots in Arabidopsis were visualized under the 20× lens and sequential line scanning mode was used. Imaging of FDA and dual FM4-64/FDA stained roots was performed with the Leica TCS SP5 II microscope, using the sequential mode: excitation at 488 nm/emission 500–550 nm for FDA, and excitation at 561 nm, emission 620–700 for FM4-64. Z-stacks were taken starting from the surface of the roots and ending 25–30 µm deep inside the root (1 µm spacing). For the PAO treatment, single plane images were taken with the Leica TCS SP5 II microscope using the 40× objective. Sequential line scanning mode was used to capture the mCherry and GFP signals to avoid crosstalk.
Image analysis
All images were analyzed using FIJI.
For the analysis of cell damage, Z-projections (Max intensity) were obtained from FDA-stained confocal stacks of Arabidopsis roots. All images were selected to the same size using the root tip as reference. Then they were thresholder and converted into binary images (using Auto Threshold, method Moments) showing alive cells in white and dead cells in black. Five different regions of interests (ROIs, size 73.48 × 73.48 µm2) were chosen for each image and the area of the white signal was measured. As an example, a ROI where all cells are alive will have an area of 5,400 µm2, while other in which all cells are dead will score 0. Data were normalized to the ROIs where all the cells were alive (i.e. divided by 5,400) and expressed as a percentage.
For the analyzing the images of short-time cold treated and mock plants, images were first deconvolved using the Autoquant X3 Software (MediaCybernetics). In all cases, for the quantification of contact sites, z-projections were obtained according to the following macro:
run("Enhance Contrast…", "saturated=0.005 normalize process_all");
run("Subtract Background…", "rolling=20 stack");
run("Green");
waitForUser ("make a z projection");
run("Subtract Background…", "rolling = 20 stack");
run("Smooth");
waitForUser ("Do you like the result?")
saveAs("Tiff");
close();
Three different ROIs (size 150 × 150 px) were chosen for each image trying to avoid internal filamentous signals sometimes present. Intensity maxima (contact sites) were identified in each ROI using the following macro:
run("Duplicate…", " ");
run("8-bit");
run("Subtract Background…", "rolling = 20");
run("Smooth");
run("Smooth");
run("Enhance Contrast…", "saturated = 0.1 normalize");
run("Find Maxima…", "noise = 20 output=Count exclude");
GUS staining assays
GUS staining assays were performed as in (Laranjeira et al., 2015) with minor modifications. Briefly, whole 5-d-old seedlings were immersed in histochemical GUS staining buffer (100 mM NaPO4, pH 7; 0.5 mM K3[Fe(CN)6; 0.5 mM K4[Fe(CN)6]; 20% methanol; 0.3% Triton X-100; and 2-mM 5-bromo-4-chloro-3-indoxyl- b-D-glucuronide cyclohexylammonium) in 12-well plates (five seedlings per well), vacuum infiltrated (60 cm Hg) for 10 min, and then wrapped in aluminum foil and incubated at 37°C for 12 h. Samples were then washed with a mix of water and ethanol with increasing concentration of ethanol (25%, 50%, 75%, 95%) and finally several times with 95% ethanol until complete tissue clarification. Samples where then rehydrated by gradually reducing the ethanol concentration in the solution (95%, 75%, 50%, 25%). Samples were mounted in the microscopy slides in 50% glycerol and photographed using the Nikon AZ100 Multizoom microscope system.
Accession numbers
SYT1: AT2G20990; SYT2: AT1G20080; SYT3: AT5G04220; SYT4: AT5G11100; SYT5: AT1G05500.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. SYT1 and SYT3 are induced under abiotic stress (related to Figure 1).
Supplemental Figure S2. SYT3 alternative splicing, promotor expression and SYT3 subcellular localization (related to Figure 2).
Supplemental Figure S3. SYT1 and SYT3 localize at ER–PM CS (related to Figure 2).
Supplemental Figure S4. SYT1 and SYT3 function as ER–PM CS tethers and bind PI4P at the PM (related to Figure 3).
Supplemental Figure S5. Total lipid changes in WT plants after cold and analysis of PM fractions (related to Figure 5)
Supplemental Figure S6. SYT1-GFP and MAPPER do not associate at ER–PM CS (related to Figure 6).
Supplemental Table S1. ESI–MS/MS Analysis of Molecular Species (Peak Area/relative protein content; x106) in inmunoprecipitated SYT1-GFP and MAPPER.
Supplemental Table S1. ESI–MS/MS analysis of molecular species in inmunoprecipitated SYT1-GFP and MAPPER.
Supplemental Table S2. List of primers used in this work.
Supplemental Data Set 1. Descriptive details of the measured lipids.
Supplemental Data Set 2. ESI–MS/MS analysis of WT and syt1/syt3 lipid classes (MS signal peak area; arbitary unit).
Supplemental File 1. Arabidopsis SYT1, SYT2, SYT3, SYT4, and SYT5 and human E-Syt1 sequences alignment in FASTA format, related to Supplemental Figure S1A.
Supplemental File 2. Machine-readable tree file in Newick format related to Supplemental figure S1A.
Supplementary Material
Acknowledgments
We would also like to thank Lothar Willmitzer for the lipidomic analysis at the Max Planck Institute of Molecular Plant Physiology (Potsdam, Germany). We thank Manuela Vega from SCI for her technical assistance in image analysis. We thank John R. Pearson and the Bionand Nanoimaging Unit, F. David Navas Fernández and the SCAI Imaging Facility and The Plant Cell Biology facility at the Shanghai Center for Plant Stress Biology for assistance with confocal microscopy. The FaFAH1 clone was a gift from Iraida Amaya Saavedra (IFAPA-Centro de Churriana, Málaga, Spain). The AHA3 antibody against the H+-ATPase was a gift from Ramón Serrano Salom (Instituto de Biología Molecular y Celular de Plantas, Valencia, Spain). The MAP-mTU2-SAC1 construct was provided by Yvon Jaillais (Laboratoire Reproduction et Développement des Plantes, Univ Lyon, France). The pGWB5 from the pGWB vector series, was provided by Tsuyoshi Nakagawa (Department of Molecular and Functional Genomics, Shimane University). We thank Plan Propio from the University of Málaga for financial support.
Funding
This work was supported by the Ministerio de Economía y Competitividad, co-financed by the European Regional Development Fund (grant no. BIO2017-82609-R to M.A.B.), the Ministerio de Ciencia, Innovación y Universidades (grant no. PGC2018-098789-B-I00 to N.R.-L.) UMA-FEDER (grant UMA18-FEDERJA-154 to N.R.-L.), and the Marie Skłodowska-Curie actions (grant no. H2020-655366-IIF- PLICO to M.A.B. and N.R.-L.). N.R.L. was supported by the Ramón y Cajal program RYC-2013-12699 (MINECO, Spain). J.P.-S. and S.G.-H. were funded by the Ministerio de Economía y Competitividad in Formación del Personal Investigador Fellowship (grant no. BES-2012-052324) and (PRE2018-085284), respectively. R.P.H. and J.A.N. received support from the Biotechnology and Biological Sciences Research Council (BBSRC, UK) in the form of an Institute Strategic Programme Grant (grant no. BBS/E/C/000I0420). J.L. is supported by the Program of Introducing Talents of Discipline to Universities (111 Project, grant no. B13007). A.P.M. and J.P.-S. were supported by the Shanghai Center for Plant Stress Biology (Chinese Academy of Sciences), Chinese 1000 Talents Program. A.R. was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC-Discovery Grant no. RGPIN-2019-05568). Support was also provided by AEI/FEDER, UE (grant nos. BIO2016-79187-R and PID2019-106987RB-I00 to J.P.-S.) and by the European Research Council under the European Union's Seventh Framework Programme (grant no. FP7/2007-2013)/ERC grant agreement no. 742985 to J.F. and T-Rex (project number 682436 to D.V.D.).
Conflict of interest statement. The authors declare no competing interests.
These authors contributed equally to this work (N.R.-L. and J.P.-S.).
M.A.B., N.R.-L., and J.P.-S. conceived the project. All authors contributed to the experimental design. N.R.-L., J.P.-S., A.E.-d.V., R.P.H., S.V., R.C., C.P.-R., S.G.-H., J.V., and V.A.-S. performed the experiments and analyzed the data. M.A.B., N.R.-L., and J.P.-S. wrote the article. All authors reviewed and commented on the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Noemi Ruiz-Lopez (noemi.ruiz@uma.es) and Miguel A. Botella (mabotella@uma.es).
References
- AhYoung AP, Jiang J, Zhang J, Khoi Dang X, Loo JA, Zhou ZH, Egea PF (2015) Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc Natl Acad Sci USA 112: E3179–E3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim-Silva V, García-Moreno Á, Castillo AG, Lakhssassi N, Del Valle AE, Pérez-Sancho J, Li Y, Posé D, Pérez-Rodriguez J, Lin J, et al. (2019) TTL proteins scaffold brassinosteroid signaling components at the plasma membrane to optimize signal transduction in arabidopsis. Plant Cell 31: 1807–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angkawijaya AE, Nguyen VC, Gunawan F, Nakamura Y (2020) A pair of Arabidopsis diacylglycerol kinases essential for gametogenesis and er phospholipid metabolism in leaves and flowers. Plant Cell 32: 2602–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisz SA, Testerink C, Munnik T (2009) Plant PA signaling via diacylglycerol kinase. Biochim Biophys Acta 1791: 869–875 [DOI] [PubMed] [Google Scholar]
- Arisz SA, van Wijk R, Roels W, Zhu J-K., Haring MA, Munnik T (2013) Rapid phosphatidic acid accumulation in response to low temperature stress in Arabidopsis is generated through diacylglycerol kinase. Front Plant Sci 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EM, Sparkes I, Vanneste S, Rosado A (2017) From shaping organelles to signaling platforms: the emerging functions of plant ER-PM contact sites. Curr Opin Plant Biol 40: 89–96 [DOI] [PubMed] [Google Scholar]
- Bernfur K, Larsson O, Larsson C, Gustavsson N (2013) Relative abundance of integral plasma membrane proteins in Arabidopsis leaf and root tissue determined by metabolic labeling and mass spectrometry. PLoS One 8: e71206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X, Saheki Y, De Camilli P (2018) Ca2+ releases E-Syt1 autoinhibition to couple ER-plasma membrane tethering with lipid transport. EMBO J 37: 219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella MA, Rosado A, Bressan RA, Hasegawa PM (2007) Plant adaptive responses to salinity stress. Plant Abiotic Stress 3: 37–70 [Google Scholar]
- Campomanes P, Zoni V, Vanni S (2019) Local accumulation of diacylglycerol alters membrane properties nonlinearly due to its transbilayer activity. Commun Chem 2: 1–9 [Google Scholar]
- Chang C-L, Hsieh T-S, Yang TT, Rothberg KG, Azizoglu DB, Volk E, Liao J-C, Liou J (2013) Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep 5: 813–825 [DOI] [PubMed] [Google Scholar]
- Colin LA, Jaillais Y (2020) Phospholipids across scales: lipid patterns and plant development. Curr Opin Plant Biol 53: 1–9 [DOI] [PubMed] [Google Scholar]
- Collado J, Kalemanov M, Campelo F, Bourgoint C, Thomas F, Loewith R, Martínez-Sánchez A, Baumeister W, Stefan CJ, Fernández-Busnadiego R (2019) Tricalbin-mediated contact sites control ER curvature to maintain plasma membrane integrity. Dev Cell 51: 476–487.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Shin O-H, Machius M, Tomchick DR, Südhof TC, Rizo J (2004) Structural basis for the evolutionary inactivation of Ca2+ binding to synaptotagmin 4. Nat Struct Mol Biol 11: 844–849 [DOI] [PubMed] [Google Scholar]
- Degenkolbe T, Giavalisco P, Zuther E, Seiwert B, Hincha DK, Willmitzer L (2012) Differential remodeling of the lipidome during cold acclimation in natural accessions of Arabidopsis thaliana. Plant J 72: 972–82 [DOI] [PubMed] [Google Scholar]
- Eisenberg-Bord M, Shai N, Schuldiner M, Bohnert M (2016) A tether is a tether is a tether: tethering at membrane contact sites. Dev Cell 39: 395–409 [DOI] [PubMed] [Google Scholar]
- Emsley Paul, Cowtan Kevin (2004) Coot: model-building tools for molecular graphics. Acta Cryst D60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fernández-Busnadiego R, Saheki Y, De Camilli P (2015) Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proc Natl Acad Sci USA 112: E2004–E2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Chacón R, Königstorfer A, Gerber SH, García J, Matos MF, Stevens CF, Brose N, Rizo J, Rosenmund C, Sudhof TC (2001) Synaptotagmin I functions as a calcium regulator of release probability. Nature 410: 41–49 [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Li Y, Matthes A, Eckhardt A, Hubberten H-M., Hesse H, Segu S, Hummel J, Köhl K, Willmitzer L (2011) Elemental formula annotation of polar and lipophilic metabolites using (13) C, (15) N and (34) S isotope labeling, in combination with high-resolution mass spectrometry. Plant J 68: 364–376 [DOI] [PubMed] [Google Scholar]
- Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P. (2013). PI(4,5)P(2)-dependent and Ca2+-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153: 1494–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z-H, Ye Z-W, Haslam RP, Michaelson LV, Napier JA, Chye M-L (2019) Arabidopsis cytosolic acyl-CoA-binding proteins function in determining seed oil composition. Plant Direct 3: e00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann I (2016) Phosphoinositide signaling in plant development. Development 143: 2044–2055 [DOI] [PubMed] [Google Scholar]
- Hoffmann PC, Bharat TAM, Wozny MR, Boulanger J, Miller EA, Kukulski W (2019) Tricalbins contribute to cellular lipid flux and form curved ER-PM contacts that are bridged by rod-shaped structures. Dev Cell 51: 488–502.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Zheng S, Wang X (2008) Dual functions of phospholipase Dalpha1 in plant response to drought. Mol Plant 1: 262–269 [DOI] [PubMed] [Google Scholar]
- Hou Q, Ufer G, Bartels D (2016) Lipid signaling in plant responses to abiotic stress. Plant Cell Environ 39: 1029–1048 [DOI] [PubMed] [Google Scholar]
- Idevall-Hagren O, Lü A, Xie B, De Camilli P (2015) Triggered Ca2+ influx is required for extended synaptotagmin 1-induced ER-plasma membrane tethering. EMBO J 34: 2291–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Tamura K, Fukao Y, Shimada T (2020) Structural and functional relationships between plasmodesmata and plant endoplasmic reticulum-plasma membrane contact sites consisting of three synaptotagmins. New Phytol 226: 798–808 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Tamura K, Ueda H, Ito Y, Nakano A, Hara-Nishimura I, Shimada T (2018) Synaptotagmin-associated endoplasmic reticulum-plasma membrane contact sites are localized to immobile ER tubules. Plant Physiol 178: 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P (2007) Recombinational cloning with plant gateway vectors. Plant Physiol 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Ishiyama K, Kato T, Tabata S, Kobayashi M, Shinozaki K (2005) An important role of phosphatidic acid in ABA signaling during germination in Arabidopsis thaliana. Plant J 43: 107–117 [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Uemura M (2003) Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J 36: 141–154 [DOI] [PubMed] [Google Scholar]
- Kelley L, Mezulis S, Yates C, Wass Mark N, Sternberg Michael JE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kwon H, Kim S, Kim MK, Botella MA, Yun HS, Kwon C (2016) Synaptotagmin 1 negatively controls the two distinct immune secretory pathways to powdery mildew Fungi in Arabidopsis. Plant Cell Physiol 57: 1133–1141 [DOI] [PubMed] [Google Scholar]
- Knight H, Knight MR (2000) Imaging spatial and cellular characteristics of low temperature calcium signature after cold acclimation in Arabidopsis. J Exp Bot 51: 1679–1686 [DOI] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1997) Calcium signaling in Arabidopsis thaliana responding to drought and salinity. Plant J 12: 1067–1078 [DOI] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ (1991) Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Kopec KO, Alva V, Lupas AN (2010) Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics 26: 1927–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laranjeira S, Amorim-Silva V, Esteban A, Arró M, Ferrer A, Tavares RM, Botella MA, Rosado A, Azevedo H (2015) Arabidopsis Squalene Epoxidase 3 (SQE3) complements SQE1 and is important for embryo development and bulk squalene epoxidase activity. Molecular plant 8: 1090–1102 [DOI] [PubMed] [Google Scholar]
- Larsson C, Sommarin M, Widell S (1994) Isolation of highly purified plant plasma membranes and separation of inside-out and right-side-out vesicles. Methods Enzymol 228: 451–469 [Google Scholar]
- Lee E, Santana BVN, Samuels E, Benitez-Fuente F, Corsi E, Botella MA, Perez Sancho J, Vanneste S, Friml J, Macho A, et al. (2020) Rare earth elements induce cytoskeleton-dependent and PI4P-associated rearrangement of SYT1/SYT5 endoplasmic reticulum–plasma membrane contact site complexes in Arabidopsis. J Exp Bot 71: 3986–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Vanneste S, Perez Sancho J, Benitez-Fuente F, Strelau M, Macho AP, Botella MA, Friml J, Rosado A (2019) Ionic stress enhances ER-PM connectivity via phosphoinositide-associated SYT1 contact site expansion in Arabidopsis. Proc Natl Acad Sci USA 116: 1420–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Zheng JY, Lazarowitz SG (2015). Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr Biol 25: 2018–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Lazarowitz SG (2010) Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc Natl Acad Sci USA 107: 2491–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Song T, Wallrad L, Kudla J, Wang X, Zhang W (2019). Tissue-specific accumulation of pH-sensing phosphatidic acid determines plant stress tolerance. Nat Plants 5: 1012–1021 [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 50: 437–450 [DOI] [PubMed] [Google Scholar]
- Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD (2012) ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell 23: 1129–1140 [DOI] [PubMed] [Google Scholar]
- Munnik T, Nielsen E (2011) Green light for polyphosphoinositide signals in plants. Curr Opin Plant Biol 14: 489–497 [DOI] [PubMed] [Google Scholar]
- Munnik T, Testerink C (2009) Plant phospholipid signaling: "in a nutshell". J Lipid Res 50 Suppl: S260–S265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Meijer HJ, Riet Ter B, Hirt H, Frank W, Bartels D, Musgrave A (2000) Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J 22: 147–154 [DOI] [PubMed] [Google Scholar]
- Perez Sancho J, Tilsner J, Samuels AL, Botella MA, Bayer EM, Rosado A (2016) Stitching organelles: organization and function of specialized membrane contact sites in plants. Trends Cell Biol 26: 705–717 [DOI] [PubMed] [Google Scholar]
- Perez Sancho J, Vanneste S, Lee E, McFarlane HE, Esteban Del Valle A, Valpuesta V, Friml J, Botella MA, Rosado A (2015) The Arabidopsis synaptotagmin1 is enriched in endoplasmic reticulum-plasma membrane contact sites and confers cellular resistance to mechanical stresses. Plant Physiol 168: 132–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Li M, Narasimhan R, Roth M, Welti R, Wang X (2010) Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. The Plant Cell 22: 2642–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokotylo I, Kravets V, Martinec J, Ruelland E (2018) The phosphatidic acid paradox: too many actions for one molecule class? Lessons from plants. Prog Lipid Res 71: 43–53 [DOI] [PubMed] [Google Scholar]
- Ruelland E (2002) Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol 130: 999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Lopez N, Haslam RP, Napier JA, Sayanova O (2014) Successful high-level accumulation of fish oil omega-3 long-chain polyunsaturated fatty acids in a transgenic oilseed crop. Plant J 77: 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki Y, De Camilli P (2017). Endoplasmic reticulum—plasma membrane contact sites. Annu Rev Biochem 86: 659–684 [DOI] [PubMed] [Google Scholar]
- Saheki Y, Bian X, Schauder CM, Sawaki Y, Surma MA, Klose C, Pincet F, Reinisch KM, De Camilli P (2016) Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat Cell Biol 18: 504–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Sevilla JF, Cruz-Rus E, Valpuesta V, Botella MA, Amaya I (2014) Deciphering gamma-decalactone biosynthesis in strawberry fruit using a combination of genetic mapping, RNA-Seq and eQTL analyses. BMC Genomics 15: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Friml J (2010) Plant Developmental Biology, Methods and Protocols. Methods Mol Biol 655: 253–263 [DOI] [PubMed] [Google Scholar]
- Schapire AL, Voigt B, Jasik J, Rosado A, Lopez-Cobollo R, Menzel D, Mancuso S, Valpuesta V, Baluska F, Botella MA (2008) Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 20: 3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder CM, Wu X, Saheki Y, Narayanaswamy P, Torta F, Wenk MR, De Camilli P, Reinisch KM (2014) Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature 510: 552–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TA, Creutz CE (2004) The tricalbin C2 domains: lipid-binding properties of a novel, synaptotagmin-like yeast protein family. Biochemistry 43: 3987–3995 [DOI] [PubMed] [Google Scholar]
- Sclip A, Bacaj T, Giam LR, Südhof TC (2016). Extended synaptotagmin (ESyt) triple knock-out mice are viable and fertile without obvious endoplasmic reticulum dysfunction. PLoS One 11: e0158295–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano L, et al. (2019) Coming together to define membrane contact sites. Nat Commun 10: 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siao W, Wang P, Voigt B, Hussey PJ, Baluska F (2016) Arabidopsis SYT1 maintains stability of cortical endoplasmic reticulum networks and VAP27-1-enriched endoplasmic reticulum-plasma membrane contact sites. J Exp Bot 67: 6161–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MLA., Platre MP, Marquès-Bueno MM, Armengot L, Stanislas T, Bayle V, Caillaud M-C, Jaillais Y (2016) A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signaling in plants. Nature Plants 2: 16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona P, Feussner K, Feussner I (2015) An enhanced plant lipidomics method based on multiplexed liquid chromatography-mass spectrometry reveals additional insights into cold- and drought-induced membrane remodeling. Plant J 84: 621–633 [DOI] [PubMed] [Google Scholar]
- Testerink C, Munnik T (2011) Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J Exp Bot 62: 2349–2361 [DOI] [PubMed] [Google Scholar]
- Testerink C, Munnik T (2005) Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci 10: 368–375 [DOI] [PubMed] [Google Scholar]
- Thomashow MF (1999) PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Plant J 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Tremblay MG, Moss T (2016) Loss of all 3 extended synaptotagmins does not affect normal mouse development, viability or fertility. Cell Cycle 15: 2360–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama A, Shimada-Beltran H, Levy A, Zheng JY, Javia PA, Lazarowitz SG (2014) The Arabidopsis synaptotagmin SYTA regulates the cell-to-cell movement of diverse plant viruses. Front Plant Sci 5: 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraji M, Katagiri T, Okuma E, Ye W, Hossain MA, Masuda C, Miura A, Nakamura Y, Mori IC, Shinozaki K, Murata Y (2012) Cooperative function of PLDδ and PLDα1 in abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol 159: 450–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Hawkins TJ, Richardson C, Cummins I, Deeks MJ, Sparkes I, Hawes C, Hussey PJ (2014) The Plant Cytoskeleton, NET3C, and VAP27 mediate the link between the plasma membrane and endoplasmic reticulum. Curr Biol 24: 1397–1405 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li Y, Rosas-Diaz T, Caceres-Moreno C, Lozano-Duran R, Macho AP (2019) The IMMUNE-ASSOCIATED NUCLEOTIDE-BINDING 9 protein is a regulator of basal immunity in Arabidopsis thaliana. Mol Plant Microbe Interact 32: 65–75 [DOI] [PubMed] [Google Scholar]
- Wass MN, Kelley LA, Sternberg MJ (2010) 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res 38 Suppl: W469–W473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Carvalho P, Voeltz GK (2018) Here, there, and everywhere: the importance of ER membrane contact sites. Science 361: 6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B, Nguyen PM, Idevall-Hagren O (2019) Feedback regulation of insulin secretion by extended synaptotagmin-1. FASEB J 33: 4716–4728 [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Kawamura Y, Minami A, Uemura M (2008) Calcium-dependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1. Plant Cell 20: 3389–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Liu Y, Gulbranson DR, Paine A, Rathore SS, Shen J (2016) Extended synaptotagmins are Ca2+-dependent lipid transfer proteins at membrane contact sites. Proc Natl Acad Sci USA 113: 4362–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, Liu J, Xiao Y, Liang Y, Zhang W (2010) Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol 188: 762–773 [DOI] [PubMed] [Google Scholar]
- Zhu J-K (2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.