Abstract
KARRIKIN INSENSITIVE2 (KAI2) was first identified as a receptor of karrikins, smoke-derived germination stimulants. KAI2 is also considered a receptor of an unidentified endogenous molecule called the KAI2 ligand. Upon KAI2 activation, signals are transmitted through the degradation of D53/SMXL proteins via MAX2-dependent ubiquitination. Although components in the KAI2-dependent signaling pathway, namely MpKAI2A and MpKAI2B, MpMAX2, and MpSMXL, exist in the genome of the liverwort Marchantia polymorpha, their functions remain unknown. Here, we show that early thallus growth is retarded and gemma dormancy in the dark is suppressed in Mpkai2a and Mpmax2 loss-of-function mutants. These defects are counteracted in Mpkai2a Mpsmxl and Mpmax2 Mpsmxl double mutants indicating that MpKAI2A, MpMAX2, and MpSMXL act in the same genetic pathway. Introduction of MpSMXLd53, in which a domain required for degradation is mutated, into wild-type plants mimicks Mpkai2a and Mpmax2 plants. In addition, the detection of citrine fluorescence in Nicotiana benthamiana cells transiently expressing a SMXL-Citrine fusion protein requires treatment with MG132, a proteasome inhibitor. These findings imply that MpSMXL is subjected to degradation, and that the degradation of MpSMXL is crucial for MpKAI2A-dependent signaling in M. polymorpha. Therefore, we claim that the basic mechanisms in the KAI2-dependent signaling pathway are conserved in M. polymorpha.
Functions of genes in the KARRIKIN INSENSITIVE2-dependent signaling pathway are conserved in the liverwort Marchantia polymorpha and control early development of the thallus.
Introduction
Plants alter their developmental program to optimize their growth depending on environmental conditions. Plant hormones are crucial, linking environmental conditions with plant growth. Strigolactones (SLs), carotenoid-derived terpenoid lactones, are unique molecules that act as plant hormones in planta and control variable aspects of plant growth, including shoot branching, root growth, and senescence (Gomez-Roldan et al., 2008; Umehara et al., 2008; Ferguson and Beveridge, 2009). In addition, they act as rhizosphere-signaling molecules in the soil when exuded from roots (Akiyama et al., 2005). Here, they promote symbiosis with arbuscular mycorrhiza (AM), facilitating phosphate acquisition from soil. In seed plants, SL is perceived by the α/β-hydrolase superfamily protein, DWARF14 (D14; Arite et al., 2009; Nakamura et al., 2013; Kagiyama et al., 2013; Yao et al., 2016; Seto et al., 2019). D14 orthologs are known as DECREAED APICAL DOMINANCE 2 (DAD2) in petunia and RAMOSUS 3 (RMS3) in pea (Hamiaux et al., 2012; de Saint Germain et al., 2016). D14 is part of a small gene family containing a few to several genes that are divided into two major groups, one containing D14 and the other containing KARRIKIN INSENSITIVE2 (KAI2; Waters et al., 2012; Bythell-Douglas et al., 2017). The D14 clade includes D14-LIKE2 (DLK2). Although the function of DLK2 is not fully understood, the DLK2 expression depends on KAI2 signaling and may be involved in light signaling (Waters et al., 2012; Végh et al., 2017).
KAI2 was first identified in Arabidopsis as a receptor of karrikins, butenolide molecules found in smoke from burned vegetation (Waters et al., 2012). Karrikins play a role in the regulation of seed germination and seedling development. However, because the loss-of-function mutants of KAI2 showed smoke-independent phenotypes, KAI2 was proposed to be a receptor of an unidentified endogenous molecule called KAI2 ligand (KL; Waters et al., 2012; Scaffidi et al., 2013; Conn and Nelson, 2016; Stanga et al., 2016). Although the detailed mechanisms remain to be elucidated, it is widely accepted that the SL and KARRIKIN/KL signals are transduced by parallel proteolysis-dependent signaling pathways in which D14 and KAI2 work as their receptors, an F-box protein, which forms a Skp–Cullin–F-box (SCF) E3 ubiquitin ligase complex with Skp1 (ASK1) and Cullin (CUL1), and repressor proteins, are major components (Jiang et al., 2013; Zhou et al., 2013; Wang et al., 2015; Khosla et al., 2020). DWARF3 (D3) of rice and MAX2 of Arabidopsis are F-box proteins and are shared in both SL and KL signaling pathways (Stirnberg et al., 2002; Ishikawa et al., 2005; Nelson et al., 2011; Stanga et al., 2016). The D53 protein of rice was identified as a degradation target of SL signaling (Jiang et al., 2013; Zhou et al., 2013). SMAX1 and SMXLs of Arabidopsis, close homologs of D53, were also identified as likely targets of karrikin signaling-dependent proteolysis (Stanga et al., 2013; Wang et al., 2015; Liang et al., 2016). The D53 and SMAX1 genes form a small gene family (Bennett and Leyser 2014; Moturu et al., 2018; Walker et al., 2019). Among the eight SMXL genes of Arabidopsis, SMXL6, SMXL7, and SMXL8 are redundant orthologs of D53, and are involved in the control of shoot branching regulated by SL signaling (Wang et al., 2015; Liang et al., 2016). SMAX1 and SMXL2 are involved in the KAI2-dependent pathway controlling seed germination and hypocotyl elongation (Stanga et al., 2013; Soundappan et al., 2015). Recently, it was shown that the proteolysis of SMXL2 can be triggered by both SLs and KARs (Wang et al., 2020a). On the other hand, SMXL3, SMXL4, and SMXL5 have lost the sequence required for degradation during evolution and are thus not under proteolysis-dependent regulation (Wallner et al., 2017). It has been proposed that SMXL6, SMXL7, and SMXL8 repress gene expression through interaction with transcription factors and/or transcriptional corepressors. In addition, SMXL6 of Arabidopsis directly binds DNA and regulates the transcription of downstream genes (Wang et al., 2020b). These findings suggest that the expansion of the D53/SMXL protein families was likely to be important in conferring diversity in the function of SL signaling and KL signaling, enabling control of many aspects of growth and development.
Land plants evolved from an ancestral charophycean alga more than 450 million years ago (Delwiche and Cooper, 2015). Most land plants and green algae contain at least one KAI2 ortholog, while D14 orthologs exist only in seed plants (Delaux et al., 2012; Waters et al., 2012; Bythell-Douglas et al., 2017). This indicates that KAI2 is ancestral and D14 arose by gene duplication events before the evolution of seed plants. Among the components in the SL- and KAI2-dependent signaling pathways, the D3/MAX2 F-box proteins are highly conserved in all land plants and Coleochaete nitellarum but not identified in Chara braunii (Bythell-Douglas et al., 2017; Nishiyama et al., 2018). The D53/SMXL family proteins are also present only in land plant groups (Bennett and Leyser, 2014; Bythell-Douglas et al., 2017; Moturu et al., 2018; Nishiyama et al., 2018; Walker et al., 2019). Therefore, the major KAI2-dependent signaling components arose in the common ancestors of land plants.
The function of the SL signaling and KAI2-dependent signaling pathways in sister lineages has been studied using Physcomitrium (Physcomitrella) patens, whose genome contains at least 11 KAI2 (PpKAI2-LIKE (PpKAI2L)), 1 MAX2 (PpMAX2), and 5 SMXL (PpSMXL) homologs (Rensing et al., 2008; Lopez-Obando et al., 2016). A series of studies suggested that the PpKAI2L genes are involved in perception and signaling of SL-related and KL-related compounds via either PpMAX2 dependent or independent pathways (Lopez-Obando et al., 2016, 2018, 2020; Bürger et al., 2019). However, the genetic redundancy in PpKAI2L genes has hampered explicit elucidation of their functions. Marchantia polymorpha, the common liverwort, is suitable for molecular genetic studies because of its low genome redundancy and the availability of its whole genome sequence and essential tools for molecular genetic studies (Bowman et al., 2017). Marchantia, like Physcomitrium, is a nonvascular sister lineage to the vascular land plants. Here, toward understanding the ancestral function, prototype, and evolution of KAI2-dependent signaling pathway, we analyzed the function of genes putatively involved in this signaling pathway in M. polymorpha.
Results
Genes in the KAI2-dependent signaling pathways in M. polymorpha
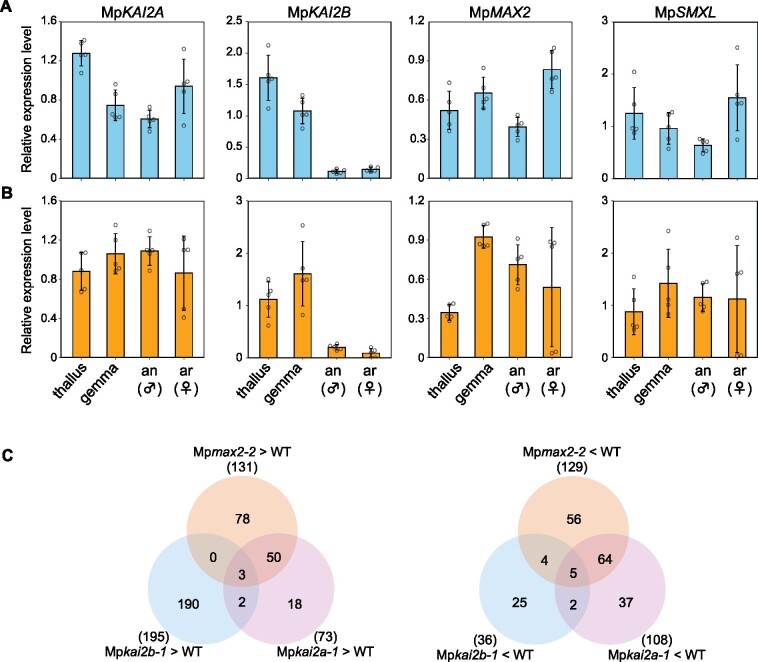
In this paper, we refer to the signaling pathway that presumably mediates signaling from the KL via the KAI2 receptor as the “KAI2-dependent signaling pathway.” Genetic nomenclature is as outlined in Bowman et al. (2016). Two KAI2 orthologs, one D3/MAX2 ortholog and one D53/SMXL ortholog (termed MpKAI2A and MpKAI2B, MpMAX2, and MpSMXL, respectively), have been identified in the genome of M. polymorpha (Waters et al., 2015; Bowman et al., 2017). The catalytic triad of KAI2 and D14, which is composed of three conserved amino acids, Ser, His, and Asp, is crucial to the hydrolase activity of these proteins (Hamiaux et al., 2012; Yao et al., 2016; Seto et al., 2019). The three amino acids in the catalytic triad are conserved in MpKAI2A and MpKAI2B (Supplemental Figure S1, A). The MpMAX2 amino acid sequence, including the last residue in the C-terminal LRR20 α-helix, is well conserved with MAX2 orthologs in other land plants (Supplemental Figure S1, B; Shabek et al., 2018). Amino acids conserved in SMXL proteins, including the RGKT motif required for degradation and the ethylene response factor-associated amphiphilic repression (EAR) motif required for repression of transcription in other species, are also conserved in MpSMXL (Supplemental Figure S1, C). We first examined the tissue specificity of the expression of MpKAI2A, MpKAI2B, MpMAX2, and MpSMXL and confirmed that all four genes are expressed in all organs tested, namely, the thallus, gemma, antheridiophore and archegoniophore, the male and female gametophores (Figure 1, A and B).
Figure 1.
Expression patterns of KAI2-dependent signaling genes. A and B, Expression levels of MpKAI2A, MpKAI2B, MpMAX2, and MpSMXL relative to the M. polymorpha ELONGATION FACTOR-1α gene (Mp3g23400.1; upper panel; blue, A) and Actin gene (Mp6g11010.1; lower panel; yellow, B) in the thallus, gemma, antheridiophore (an), and archegoniophore (ar) are shown. qPCR was used to measure expression levels. Data are the mean ± sd from five biological replications. B, Venn diagrams of genes upregulated (left) or downregulated (right) by more than two-fold in the Mpkai2a-1, Mpkai2b-1, and Mpmax2-2 compared with WT.
Then, the loss-of-function mutants of these genes were generated in M. polymorpha using a CRISPR/Cas9 genome editing system (Bortesi and Fischer, 2015). Mutations in the transgenic plants were confirmed by sequencing and at least two independent alleles for each gene, representing complete loss-of-function alleles, were selected for subsequent phenotypic analysis (Supplemental Figure S2).
We next analyzed transcriptomes in Mpkai2a, Mpkai2b, and Mpmax2 mutants by RNA seq analysis (Figure 1, C). Mature gemmae cultured on fresh medium for 24 h were used for this analysis. We found a high degree of overlap between genes up- and downregulated in Mpkai2a and Mpmax2. By contrast, there was much less of an overlap between up- and downregulated genes in Mpkai2b and the other two mutants, Mpkai2a or Mpmax2. Notably, MpKAI2B [Mapoly0031s0148 (Mp2g04930.1)] expression is extensively decreased in both the Mpkai2a and Mpmax2 mutants, suggesting that MpKAI2B expression is positively regulated via an MpKAI2A- and MpMAX2-dependent signaling pathway (Supplemental Data Sets S1, S2). The expression of MpSMXL [Mapoly0006s0101.1 (Mp3g06310.1)] also was decreased in Mpkai2a and Mpmax2.
Genetic and phenotypic analysis of MpSMXL, MpKAI2A, and MpMAX2
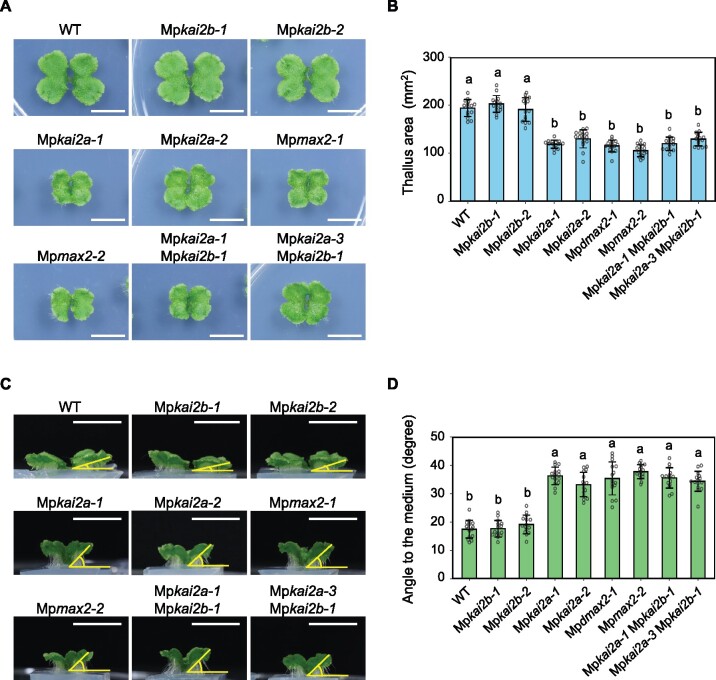
We observed thallus growth in loss-of-function mutants of three putative KAI2-dependent signaling genes (Figure 2). At least two independent alleles of each gene were analyzed. Thalli from mutant plants with either allele of Mpkai2b (Mpkai2b-1 or Mpkai2b-2) were morphologically indistinguishable from those of wild-type (WT) plants (Figure 2, A–D). By contrast, thalli from mutant plants with either allele of Mpkai2a (Mpkai2a-1 or Mpkai2a-2) or Mpmax2 (Mpmax2-1 or Mpmax2-2) showed similar abnormalities, in which early growth of the thalli was retarded and the thalli curved upward. The size of the thalli of Mpkai2a-1, Mpkai2a-2, Mpmax2-1, and Mpmax2-2 mutants was significantly smaller than that of the WT, whereas there was no significant difference between Mpkai2a and Mpmax2 (Figure 2, B). The angle of thallus curving also was larger in Mpkai2a-1, Mpkai2a-2, Mpmax2-1, and Mpmax2-2 plants compared with the WT, while no significant differences were observed between Mpkai2a and Mpmax2 (Figure 2, D). To test the possibility that the defects in the Mpkai2b mutants were masked by the function of the MpKAI2A gene, which contains a high sequence similarity to MpKAI2B, we produced double mutants of Mpkai2a and Mpkai2b. The Mpkai2a-1 Mpkai2b-1 and Mpkai2a-3 Mpkai2b-1 double mutants were not significantly different from Mpkai2a-1 and Mpkai2a-2, implying that MpKAI2B is not significantly involved in the control of thallus growth. These data indicate that MpKAI2A and MpMAX2 control thallus growth in a similar way.
Figure 2.
MpKAI2A and MpMAX2 influence thallus growth in a similar manner. A, Top view of 14-d-old thalli. Bars = 1 cm. B, Area of 14-day-old thalli. C, Side view of the thalli shown in (A). Bars = 1 cm. D, Angles between the 14-d-old thalli and the growth media. Data in (B) and (D) are the mean ± sd from 15 gemmae. Tukey’s honestly significant difference (HSD) test was used for multiple comparisons. Statistical differences (P < 0.05) are indicated by different letters.
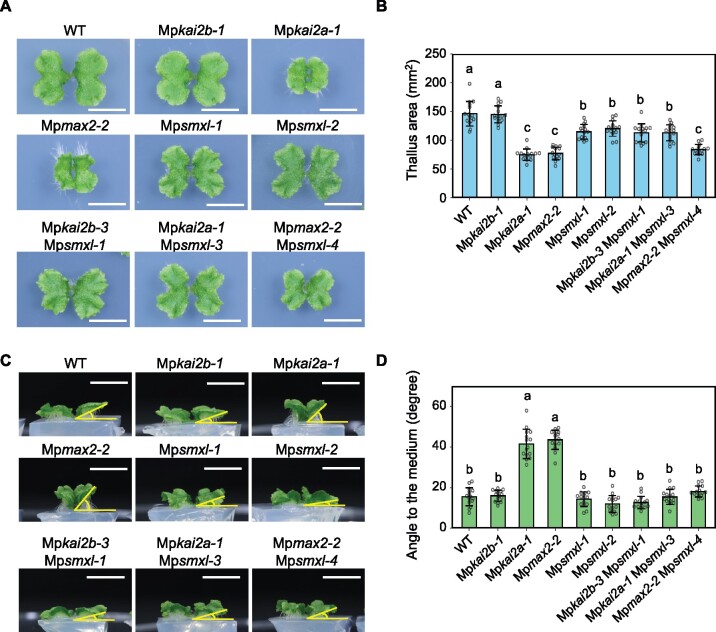
M. polymorpha contains a single SMXL/D53 family gene, MpSMXL. In Mpsmxl loss-of-function mutants, the size of the thallus was reduced to a size between that of the WT and Mpkai2a or Mpmax2 plants (Figure 3, A and B). In contrast to Mpkai2a and Mpmax2, Mpsmxl mutants did not show upward curving of the thalli (Figure 3, C and D). The morphology and the size of the thalli in the Mpsmxl Mpkai2a and Mpsmxl Mpkai2b double mutant plants were not significantly different from those of the Mpsmxl single mutant plants. The thallus-curving phenotype of Mpkai2a and Mpmax2 plants was suppressed in the double mutants of Mpsmxl Mpkai2a and Mpsmxl Mpmax2, suggesting that MpSMXL works downstream of MpKAI2A and MpMAX2 (Figure 3, C and D). However, the size of the thalli in Mpsmxl Mpmax2 double mutant plants was smaller than that of the Mpsmxl plants (Figure 3, A and B). This may indicate that SMXL works somewhat independently from MpMAX2, as reported in Arabidopsis (Khosla et al., 2020).
Figure 3.
MpSMXL acts downstream of MpKAI2A and MpMAX2. A, Top view of 14-d-old thalli. Bars = 1 cm. B, Area of 14-d-old thalli. C, Side view of the thalli shown in (A). Bars = 1 cm. D, Angles between the 14-d-old thalli and the growth media. Data shown in (B) and (D) are the mean ± sd from 15 gemmae. The HSD test was used for multiple comparisons. Statistical differences (P < 0.05) are indicated by different letters.
Phenotypic data on some of the genotypes were collected in more than one set of experiments and is shown in more than one figure. For example, both show WT, Mpkai2b-1, Mpkai2a-1, and Mpmax2-2 images and data in comparison to other Mpkai2 mutants (Figure 2) or to various Mpmax2 and Mpsmxl mutants (Figure 3). In all cases, data for each figure were collected from independent experiments, with all genotypes grown on the same media at the same time for each experiment (i.e. no images or data are duplicated in more than one figure). Similar results were obtained on the same genotype under the same conditions in the separate experiments; however, some experimental variation is noticeable. For example, the mean WT thallus area of 14-d-old thalli was roughly 200, 150, or 130 mm2 in experiments shown in Figures 2, 3, and 6, respectively.
Figure 6.
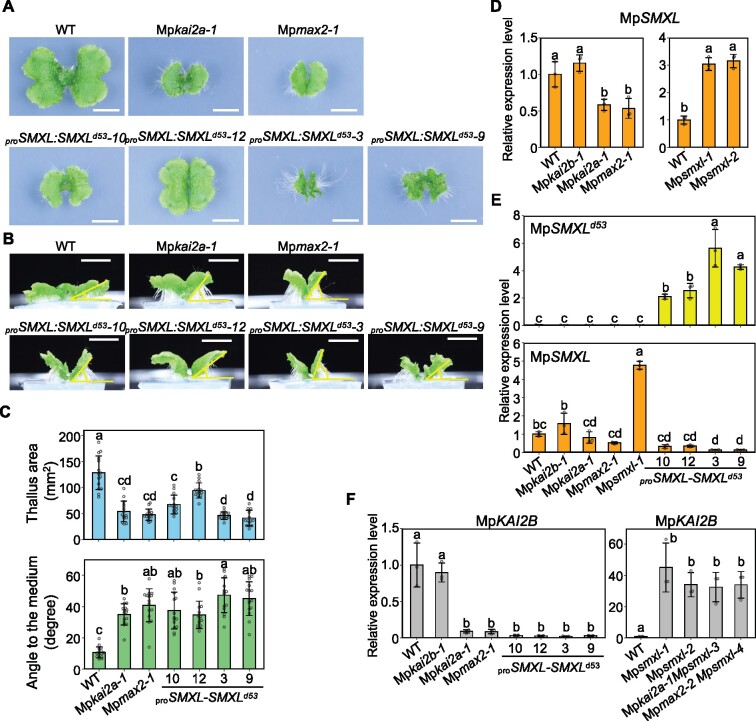
Functional analysis of the conserved RGKT motif of MpSMXL: MpSMXL functions as a repressor in the MpSMXL degradation-dependent KAI2-dependent signaling pathway. A–C, Effects of the expression of the MpSMXL gene containing the same mutation as rice d53 (proSMXL:SMXLd53) on thallus growth. Top (A) and side (B) view of 14-d-old thalli. Bars = 1 cm. C, Thallus area and angle to the growth medium. Data are the mean ± sd from 15 gemmae used in each analysis. D, The expression of MpSMXL relative to M. polymorpha ELONGATION FACTOR-1α in the thallus. E, The expression of the introduced proSMXL:SMXLd53 and endogenous MpSMXL genes relative to MpEF in the thallus. F, The expression of MpKAI2B relative to MpEF in the thallus. Data in (D)–(F) are the mean ± sd from three biological replications. The HSD test was used for multiple comparisons in (C)–(F). Statistical differences (P < 0.05) are indicated by different letters.
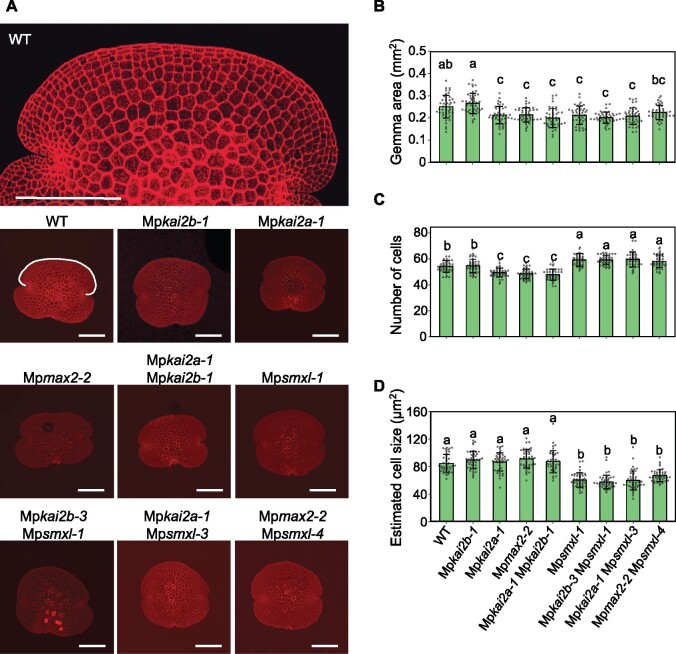
To investigate the cause of the reduced thallus size, we examined the gemmae in more detail. Gemmae are propagules produced in a cup-like structure called a gemma cup, and they directly give rise to new plants containing the thallus. Many gemmae of a uniform developmental stage can be obtained easily and it is possible to count the number of cells due to their small size and flat shape (Figure 4, A). We measured the size of the gemmae and counted the total number of their epidermal cells. Gemmae in Mpkai2a, Mpmax2, and Mpsmxl mutant plants were smaller than those of the WT plants at the mature gemma stage (Figure 4, B). Consistent with this, the number of cells in the gemma was reduced in Mpkai2a and Mpmax2 plants (Figure 4, C). On the other hand, despite the decrease in gemmae size, the number of cells in the gemma was increased in Mpsmxl plants (Figure 4, C). Mpkai2a Mpsmxl and Mpmax2 Mpsmxl double mutants also contained more cells in the gemma than in the WT plants. Cell size estimated from the gemma area and the number of cells was not significantly different between WT, Mpkai2a, Mpkai2b, and Mpmax2 mutants (Figure 4, D). In contrast, the estimated cell size was smaller in Mpsmxl and double mutants containing Mpsmxl compared with that of the WT plants. These results indicated that MpKAI2A and MpMAX2 act to increase the cell number in the gemmae without affecting the cell size, while MpSMXL functions to suppress cell proliferation and to promote cell growth. In addition, these results support the hypothesis that MpSMXL works downstream of MpKAI2A and MpMAX2 in the genetic pathway, which regulates gemma development.
Figure 4.
MpKAI2A and MpMAX2 act to increase cell number in the gemmae without affecting the cell size, while MpSMXL acts to suppress cell proliferation and to promote the cell growth. A, Gemmae of WT and mutant plants stained with propidium iodide. Bars = 200 µm. B, Area of gemmae. C, Number of cells in the outermost cell layer of the dorsal side of gemmae as shown in white in the WT plant in (A). D, Relative size of a cell estimated from the gemma area shown in (B) and the number of cells shown in (C). Data shown in (B)–(D) are the mean ± sd from 45 gemmae. The HSD test was used for multiple comparisons. Statistical differences (P < 0.05) are indicated by different letters.
Degradation-resistant mutation in MpSMXL mimics Mpkai2a and Mpmax2 mutants
We next analyzed whether the degradation of MpSMXL is required in the KAI2-dependent signaling pathway in M. polymorpha as is the case in the SMXL/D53 family proteins in rice and Arabidopsis (Jiang et al., 2013; Stanga et al., 2013; Zhou et al., 2013; Wang et al., 2015; Liang et al., 2016; Moturu et al., 2018). In rice and Arabidopsis, D53/SMXL proteins containing the RGKT motif are ubiquitinated via the SCF E3 (SCFD3/MAX2) complex and degraded through the 26S proteasome.
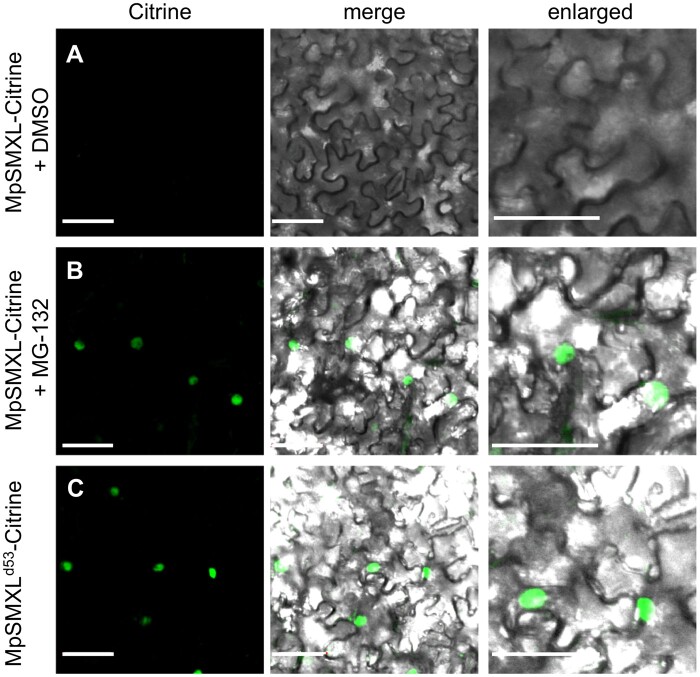
To observe the MpSMXL protein, MpSMXL was fused with Citrine, and driven by its own promoter in M. polymorpha. We produced more than 10 independent transgenic lines and confirmed the expression of the introduced MpSMXL–Citrine mRNA; however, we did not detect Citrine fluorescence under the microscope, nor did we detect MpSMXL–Citrine protein by protein immunoblotting in any of the transgenic lines. Similar difficulty to observe SMAX1 was recently reported (Khosla et al., 2020). This suggested the possibility of proteasome-dependent degradation of the MpSMXL–Citrine protein. To test this, we used a transient expression system in N. benthamiana epidermis cells (Figure 5). The MpSMXL gene fused with the Citrine gene and the CaMV35S promoter was introduced into N. benthamiana cells. Citrine fluorescence was detected in the N. benthamiana cells after treatment with MG132, a proteasome inhibitor, supporting our hypothesis that MpSMXL is degraded (Figure 5, A and B). In the dominant d53 mutant of rice, an arginine in the RGKT motif of the D53 protein is replaced with threonine and a subsequent GKTGI amino acid sequence is deleted (Jiang et al., 2013; Zhou et al., 2013). To further confirm the degradation of the MpSMXL, we replaced the MpSMXL gene with the -MpSMXLd53, in which the RGKT domain was mutated to the same sequence as the d53 mutant allele of rice, fused with the Citrine gene, and introduced into N. benthamiana cells. Citrine fluorescence was observed without MG132 treatment (Figure 5, C). These results indicate that MpSMXL is subjected to proteosome-dependent degradation.
Figure 5.
MpSMXL is subject to proteosome-dependent degradation. A–C, Subcellular localization of MpSMXL–Citrine in N. benthamiana epidermal cells treated with 0.25% (v/v) DMSO (A), or 100-μM MG-132 (B). Subcellular localization of MpSMXLd53–Citrine in N. benthamiana epidermal cells (C). Micrographs showing cells expressing MpSMXL–Citrine or MpSMXLd53–Citrine were examined under fluorescence (left), bright field by confocal microscopy. Merged (middle) and enlarged (right) images are shown for each analysis. Bar = 50 μm.
To understand the consequence(s) of degradation for the function of MpSMXL, the MpSMXLd53 gene was fused with the promoter region of MpSMXL and introduced into WT M. polymorpha. We generated more than 20 independent transgenic lines (proSMXL:SMXLd53). Although the severity of the phenotype varied among the transgenic lines, most showed defects resembling Mpkai2a and Mpmax2, in which the thallus is small and curved upward. We selected transgenic lines showing moderate (proSMXL:SMXLd53-10, -12) and strong (proSMXL:SMXLd53-3, -9) phenotypes for further analysis (Figure 6). The lines with strong phenotypes showed equivalent defects to those of the Mpkai2a and Mpmax2 mutant plants (Figure 6, A–C). These data suggest that the degradation of MpSMXL is required in KAI2-dependent signaling.
We next examined feedback regulation in the expression of genes in the KAI2-dependent signaling. In rice and Arabidopsis, the accumulation of D53, SMXL6, SMXL7, and SMXL8 transcripts is subjected to negative feedback control in the SL signaling pathway. In rice, D53 expression is suppressed in SL signaling and biosynthesis mutants, while it is upregulated by the addition of GR24, a synthetic SL (Jiang et al., 2013; Zhou et al., 2013). In M. polymorpha, the expression level of MpSMXL mRNA was decreased in the Mpkai2a and Mpmax2 mutants, while it was enhanced in Mpsmxl loss-of-function mutants (Figure 6, D andSupplemental Data Set S1). This implies that MpSMXL expression was subjected to conserved feedback suppression by KAI2-dependent signaling. In the transgenic lines, the level of introduced proSMXL:SMXLd53 expression varied, possibly due to the different integration sites, and was roughly consistent with the severity of the defects (Figure 6, E). The amount of endogenous MpSMXL transcript, however, appeared to decrease depending on the severity of the defects (Figure 6, E).
Consistent with the RNA seq data (Supplemental Data Set S1), MpKAI2B expression was dramatically reduced in Mpkai2a and Mpmax2 plants but was unchanged in Mpkai2b plants (Figure 6, F). This implies that MpKAI2B expression depends on KAI2-dependent signaling, and therefore is suitable as a marker for the signaling. We observed that MpKAI2B expression was severely suppressed in line with moderate (-10 and -12) and strong (-3 and -9) phenotypes, indicating that KAI2-dependent signaling was dampened by the suppression of MpSMXL protein degradation in the transgenic lines. On the other hand, MpKAI2B expression was dramatically enhanced in Mpsmxl-1 and Mpsmxl-2, the loss-of-function mutants of MpSMXL (Figure 6, F). The addition of Mpkai2a or Mpmax2 mutation to the Mpsmxl mutants did not affect the enhanced expression of MpKAI2B (Figure 6, F). These findings further support the hypothesis that MpKAI2A, MpMAX2, and MpSMXL work in the same pathway. All these results suggest that MpSMXL functions as the repressor in the MpSMXL degradation-dependent KAI2-dependent signaling pathway, in which MpKAI2A and MpMAX2 are involved.
KAI2-dependent signaling pathway suppresses gemma growth in the dark
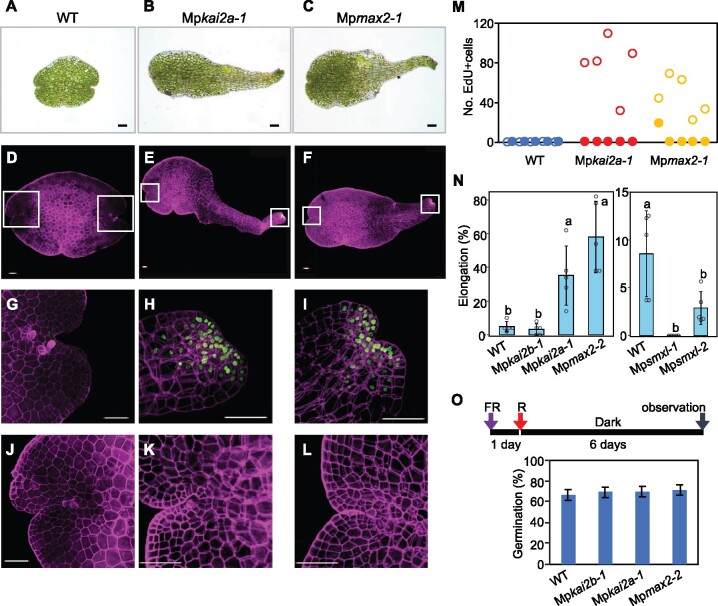
The KAI2-dependent pathway is involved in the control of seed germination in the light and the expression of light-responsive genes in Arabidopsis (Nelson et al., 2010, 2011; Sun and Ni, 2011; Shen et al., 2012; Waters et al., 2012; Waters and Smith, 2013). Therefore, we analyzed light-related phenotypes in M. polymorpha. We found that gemmae of WT plants were maintained in a dormant state and retained their green color in dark conditions for 2 weeks (Figure 7, A). In contrast, the elongation of one side of the gemma was often observed in Mpkai2a and Mpmax2 mutants under these conditions (Figure 7, B and C). Staining the gemmae with 5-ethynyl-2′-deoxyuridine (EdU) showed that cell proliferation occurred in the gemmae of Mpkai2a-1and Mpmax2-1 mutants (Figure 7, D–L). Interestingly, cell proliferation occurred at one side of the gemma (Figure 7, M). In contrast to Mpkai2a and Mpmax2, gemma dormancy was strengthened in Mpsmxl mutants (Figure 7, N). These results indicate that KAI2-dependent signaling is required to inhibit the proliferation of cells around the apical region of the gemma and maintain the gemma in a dormant state in the dark. Since gemmae of M. polymorpha need light to germinate (Inoue et al., 2016), we tested whether KAI2-dependent signaling is involved in germination itself or in post-germination growth. A germination assay with a single pulse of light showed that the germination was not affected in the mutants (Figure 7, O). Taken together, these results suggest that KAI2-dependent signaling inhibits post-germination growth in the dark.
Figure 7.
KAI2-dependent signaling suppresses gemma growth in the dark. A–C, WT (A), Mpkai2a-1 (B), and Mpmax2-1 (C) gemmae grown in the dark for 2 weeks. Bars = 100 μm. D–L, Gemma stained with 5-ethynyl-2′-deoxyuridine (EdU) after 2 weeks’ incubation in the dark. (G)–(L) are enlargements of the meristem regions of (D)–(F), respectively. (G)–(I) are enlargements of the apical region at right side, and (J)–(L) are enlargements of the apical region at left side. (D), (G), and (J) are WT, (E), (H), and (K) are Mpkai2a-1, and (F), (I), and (L) are Mpmax2-1. Bars = 100 μm. M, The number of cells stained with EdU in each meristem. Open and solid circles indicate the results for elongated and nonelongated meristems, respectively. Five gemmae were used. N, frequency of gemma elongation. Data are the mean ± sd from five independent experiments. Over 50 gemmae were used in each experiment. The HSD test was used for multiple comparisons. Statistical differences (P < 0.05) are indicated by different letters. O, Germination frequencies of WT, Mpkai2b-1, Mpkai2a-1, and Mpmax2-1. Gemmae imbibed on the medium in the dark for 1 d were irradiated with a pulse of Red light (3,000 µmol photons m−2) followed by incubation in the dark for 6 d. Fifty gemmae were used in each experiment. Data are the mean ± sd from three independent experiments. A schematic illustration of the experimental procedure is shown above.
M. polymorpha responds to rac-GR24 possibly through MpKAI2A but does not respond to karrikins
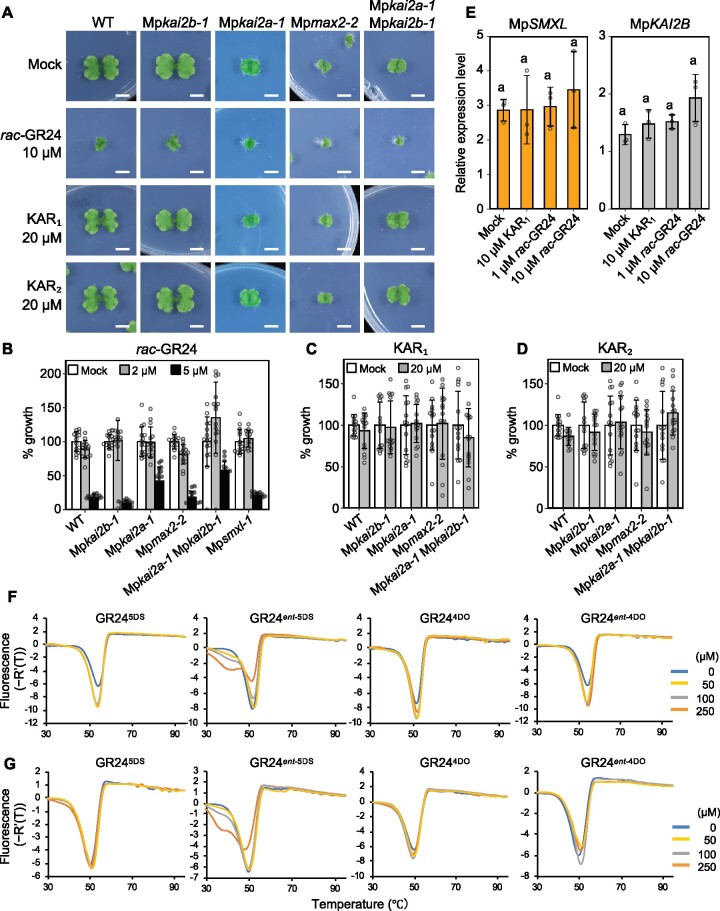
To obtain insights into the ligands involved in the KAI2-dependent signaling pathway in M. polymorpha, we examined the responses of the mutants to karrikins (KAR1 and KAR2) and the synthetic SL, rac-GR24. Gemmae were grown on media containing rac-GR24, KAR1, or KAR2 and the growth of thalli was measured after 14 d culture. Growth of thalli was not affected by the addition of 2 μM rac-GR24, while it was severely suppressed by the addition of higher concentration of rac-GR24 (Figure 8, A and B). The suppression was weakened in Mpkai2a-1 and Mpkai2a-1 Mpmax2-1 double mutants, while it was not affected in the Mpkai2b-1 or Mpmax2 plants (Figure 8, A and B). This suggests that MpKAI2A is partially involved in the growth suppression by high concentration of rac-GR24. On the other hand, addition of neither KAR1 nor KAR2 affected the growth of thalli in any of the genotypes examined (Figure 8, A, C, and D). We asked whether the expression of MpKAI2B and MpSMXL, markers that are positively regulated by the KAI2-dependent signaling pathway in M. polymorpha (Figure 6, D and F), is changed by addition of the chemicals (Figure 8, E). Neither MpKAI2B nor MpSMXL expression was affected by the addition of a high concentration (10 μM) of rac-GR24. This suggests that the growth retardation caused by the addition of a high concentration (higher than 5 μM) of rac-GR24 is partially dependent on MpKAI2A but independent of KAI2-dependent signaling. Finally, we investigated the physical interaction between MpKAI2A or MpKAI2B with four stereoisomers of GR24 by differential scanning fluorimetry (DSF) analysis. Only GR24ent-5DS, that has an unnatural d-ring configuration, interacted weakly with MpKAI2A and MpKAI2B (Figure 8, F and G).
Figure 8.
Effects of exogenously supplied rac-GR24, KAR1, and KAR2 on thallus growth. A, Top view of the thalli grown on the media containing rac-GR24, KAR1, or KAR2. Bars = 1 cm. B–D, Growth of thalli exposed to 2- or 5-µM rac-GR24 (B), 20-µM KAR1 (C), and 20-µM KAR2 (D) relative to a mock-treated control. Data are the mean ± sd from 15 gemmae used in each experiment. E, The expression of MpSMXL and MpKAI2B relative to M. polymorpha ELONGATION FACTOR-1α gene in the thalli exposed to 10 μM KAR1 and 1- or 10-μM rac-GR24. Data are the mean ± SD from three biological replications. The Tukey’s HSD test was used for multiple comparisons. Statistical differences (P < 0.05) are indicated by different letters. F and G, Differential Scanning Fluorimetry (DSF) analysis with 4 stereoisomers of GR24 and MpKAI2A (F) and MpKAI2B (G) proteins. A slight change of melting points was detected when the GR24ent-5DS was incubated with MpKAI2A and MpKAI2B. The enantiomers ent-5DS and ent-4DO have not been reported to be produced in nature.
Discussion
Components in the KAI2-dependent signaling pathway are conserved in M. polymorpha
The M. polymorpha genome contains one D3/MAX2, one SMXL, and two KAI2 homologs (Bowman et al., 2017). In this study, we showed that the loss-of-function mutants of MpKAI2A, one of the two KAI2 homologs, and MpMAX2, showed indistinguishable defects in gemma and thallus growth and that Mpsmxl loss-of-function mutants were epistatic to Mpkai2a and Mpmax2. These results indicated that MpKAI2A, MpMAX2, and MpSMXL are likely function in the same genetic pathway.
In the dominant d53 mutant of rice, the mutation in the RGKT motif confers resistance to degradation of the D53 protein upon SL perception and leads to more branched phenotypes (Jiang et al., 2013; Zhou et al., 2013; Wang et al., 2015). The deletion of the RGKT sequence from Arabidopsis SMXL6 also confers resistance to degradation and causes more shoot branching phenotypes (Wang et al., 2015). The RGKT motif and EAR motif, which are involved in transcriptional suppression of downstream genes, are conserved in MpSMXL (Soundappan et al., 2015; Wang, 2015; Ma et al., 2017; Moturu et al., 2018; Wang et al., 2020a), suggesting that the MpSMXL protein may be involved in proteolysis-dependent regulation and function as a repressor of the signaling pathway. We showed that introduction of the mutant version of the MpSMXL gene containing the same amino acid sequence as the d53 mutant of rice into WT M. polymorpha mimicked the phenotypes of the Mpkai2a and Mpmax2 mutants. We also showed that the introduction of the Citrine gene fused with MpSMXL into N. benthamiana epidermis cells resulted in Citrine fluorescence when the introduced MpSMXL contained the d53 mutation or the cells were treated with proteolysis inhibitor. These findings support our hypothesis that degradation of MpSMXL is crucial in the signaling of MpKAI2A.
F-box protein-mediated protein degradation is used for signaling of multiple plant hormones and is present in the nonvascular as well as vascular land plants and is therefore presumed to have been established early in the evolution of land plants (Blázquez et al., 2020). In M. polymorpha and P. patens, auxin is perceived by TIR1, an F-box protein, and the signal is transduced through the degradation of AUX/IAA transcription factors, as is the case in seed plants (Prigge et al., 2010; Flores-Sandoval et al., 2015; Kato et al., 2015; Lavy et al., 2016). The F-box protein coronatine-insensitive 1 (COI1), the receptor of jasmonate in seed plants, works as a receptor of dinor-12-oxo-phytodienoic acid (dinor-OPDA), a molecule related to jasmonate, in the proteolysis-dependent signaling cascade in M. polymorpha (Monte et al., 2018). Although the F-box protein with homology to TIR1 and COI1 exists in charophytes, the sequences necessary for interaction with ligands and repressor proteins are lacking, suggesting that TIR1 and COI1 established their function by neofunctionalization during the early evolution of land plants (Mutte et al., 2018). D3/MAX2 orthologs are found in Coleochaetales but SMXL are present only in land plants (Bythell-Douglas et al., 2017). Therefore, the function of D3/MAX2 was also recruited to the KAI2-dependent signaling pathway in early land plants. Taking all these observations into account, it appears that the proteolysis-dependent signaling pathway through KAI2 and D3/MAX2 was established in the common ancestor of land plants. Notably, many genes whose expression is MAX2 dependent but independent from KAI2A or KAI2B were identified by RNAseq analysis. This suggests that MpMAX2 may be involved in other pathways in M. polymorpha. Conversely, SL-related molecules are perceived by a distinctive subset of PpKAI2Ls and their signaling does not require PpMAX2 in P. patens, suggesting that D3/MAX2 was recruited in SL signaling later in the common ancestor of seed plants (Lopez-Obando et al., 2018). Future studies of D3/MAX2 function in more species including hornworts as well as more diverse plant lineages will help to improve our understanding of the evolution of D3/MAX2 and F-box protein-mediated proteolysis-dependent hormone signaling in plants.
The number of SMXL genes has increased during evolution (Walker et al., 2019; Machin et al., 2020). The multiple copies of SMXL genes in seed plants are classified into subgroups, each regulating different aspects of growth and development (Stanga et al., 2013; Soundappan et al., 2015; Liang et al., 2016; Wallner et al., 2017; Villaécija-Aguilar et al., 2019). In contrast, M. polymorpha contains only a single SMXL gene, which functions in the MpKAI2A signaling pathway. We cannot exclude the possibility that the MpKAI2A and MpKAI2B pathways share MpSMXL as a repressor; however, the observation that MpSMXL expression is not affected in Mpkai2b indicates that MpSMXL is unlikely to be involved in the signaling of MpKAI2B. It is of interest to know whether the function of MpSMXL fully depends on MpKAI2A and MpMAX2 or whether MpSMXL acts partially independently from them.
Possible ligands of MpKAI2A and MpKAI2B
The D14 clade, including core D14 genes and D14-Like2 (DLK2), arose through gene duplication of KAI2 paralogs prior to the emergence of seed plants (Bythell-Douglas et al., 2017). In Arabidopsis, D14 specifically recognizes SLs and KAI2 does not respond to natural SLs (Scaffidi et al., 2014). Based on this, it is hypothesized that the generation of D14 led to the establishment of the SL perception system. The existence of canonical SLs in sister lineages to the land plants is currently under debate (Delaux et al., 2012; Decker et al., 2017; Yoneyama et al., 2018; Walker et al., 2019). In the SL biosynthesis pathway, all-trans-β-carotene is converted to carlactone by actions of DWARF27 (D27) and two carotenoid cleavage dioxygenases (CCD7 and CCD8; Alder et al., 2012). Carlactone is converted to carlactonoic acid by the cytochrome P450 encoded by MORE AXILLARY GROWTH 1 (MAX1; Abe et al., 2014; Yoneyama et al., 2018) and further converted to canonical SLs. Although the MAX1 gene is absent in P. patens, the development of caulonema and protonema is affected in Ppccd8 and Ppccd7 mutants and the defects were complemented by the application of GR24, indicating that the products of CCD8 or their derivatives are active (Proust et al., 2011; Hoffmann et al., 2014; Decker et al., 2017; Lopez-Obando et al., 2018, 2020). If sister lineages to land plants produce SLs, how they are perceived without D14 remains unknown. Recently, it was found that KAI2 genes are extensively amplified in parasitic plants and some of them act as highly sensitive SL receptors (Toh et al., 2015; Tsuchiya et al., 2015). This implies that the machinery for SL perception can be generated through the modification of ligand reception specificity of KAI2 so that they can respond to SLs. These findings raise the possibility that a subset of KAI2s may work as SL receptors in sister lineages to land plants, while their function as SL receptors was substituted by D14 in seed plants. Consistent with this idea, biochemical and molecular characteristics of PpKAI2L proteins are divergent, including ligand preferences, enzymatic activities, and the size of the predicted ligand binding pocket (Lopez-Obando et al., 2016, 2020; Bürger et al., 2019).
In M. polymorpha, it is unlikely that canonical SLs are produced, at least through a canonical biosynthesis pathway, due to the absence of CCD8 and MAX1 (Bowman et al., 2017). This suggests that the ligand of MpKAI2A is unlikely to be an SL. Indeed, MpKAI2A driven by the Arabidopsis KAI2 promoter did not complement the shoot branching phenotype of Arabidopsis d14 mutants, indicating that MpKAI2A does not respond to Arabidopsis endogenous SLs (Waters et al., 2015). We showed that thallus responded to rac-GR24 in a manner partially dependent on MpKAI2A. It is known that AtKAI2 responds to GR24ent-5DS, a non-natural SL isomer (Scaffidi et al., 2014; Waters et al., 2015). Results of DSF analysis in this study indicated that MpKAI2A and MpKAI2B probably interact with GR24ent-5DS. The closest homologs of MpKAI2A in P. patens, PpKAI2L-C, -D, and -E, also show a shift in the protein melting point with high specificity to GR24ent-5DS (Bürger et al., 2019; Lopez-Obando et al., 2020). Considering these data, the ligand specificity of MpKAI2A may be similar to that of AtKAI2 and the natural ligand of MpKAI2A may be KL or a KL-related molecule. However, MpKAI2A expressed by the Arabidopsis KAI2 promoter did not rescue the seedling phenotype of kai2-2, suggesting the possibility that KLs have diverged between Arabidopsis and M. polymorpha (Waters et al., 2015). The absence of a response to exogenously supplied KAR1 or KAR2 observed in this study also supports the idea that the specificity of ligand perception has also diverged in seed plants. However, in cross-species complementation experiments, the possibility that MpKAI2A does not physically interact with Arabidopsis SMAX1, SMXLs, or MAX2 cannot be ruled out.
Function of MpKAI2B
We did not find any defects in growth of loss-of-function Mpkai2b mutants even though MpKAI2B is expressed in all organs examined at a comparable level to that of MpKAI2A. A possible explanation for the lack of an Mpkai2b phenotype is that MpKAI2B works as an SL receptor in Marchantia, but due to the loss of SL biosynthesis ability in M. polymorpha, the function of MpKAI2B is masked. However, the lack of physical interaction between GR245DS or GR244DO, synthetic analogs of natural SL compounds, and MpKAI2A or MpKAI2B does not support this possibility. Alternatively, MpKAI2B may function under special conditions or in specific cells, thus defects are not apparent under normal growth conditions. RNA-seq analysis showed that the expression of many genes is changed in Mpkai2b mutant plants compared with WT plants, and the expression of most of the differentially expressed genes is independent of the function of MpKAI2A and MpMAX2. These imply that MpKAI2B is a functional protein, which plays roles in unknown pathways. To reveal the function of MpKAI2B, a more detailed and comprehensive analysis of loss-of-function mutants under variable growth conditions using various experimental tools, such as transcriptomics, proteomics, and metabolomics, is required.
A recent phylogenetic study revealed that the duplication of KAI2 occurred at the base of the land plants (Bythell-Douglas et al., 2017). Bythell-Douglas et al. (2017) also show that MpKAI2A is in the KAI2 lineage while MpKAI2B is located at the base of the DDK lineage, which also contains D14 and DLK2 lineages. The overexpression of DLK2 results in more elongated hypocotyls independently from D14 or KAI2 function, whereas no clear defects were observed in dlk2 loss-of-function mutants in Arabidopsis (Waters et al., 2015; Végh et al., 2017). Although the lack of clear phenotypes in the loss-of-function mutants has hampered our understanding of its function, DLK2 is used as a positive marker of KL signaling because the normal expression of DLK2, as well as induction by SL or karrikin treatment, depends on MAX2 and KAI2 function (Scaffidi et al., 2013; Stanga et al., 2013; Waters and Smith, 2013). We showed that the expression of MpKAI2B is positively regulated by the signaling cascade of KAI2A and MpMAX2. We also showed that the MpKAI2B expression was extensively induced in Mpsmxl loss-of-function mutants. The expression of DLK2 is increased in smxl1,2 double mutants in Arabidopsis (Stanga et al., 2016). It was shown that DLK2 neither perceives nor hydrolyzes the natural ligand of D14 and KAI2 (Végh et al., 2017). Although the regulation of Arabidopsis DLK2 expression mostly depends on MAX2 function, DLK2 function does not depend on MAX2. This is also the case for MpKAI2B. These similarities between DLK2 and MpKAI2B suggest the possibility that MpKAI2B may play an equivalent role to DLK2 in Arabidopsis. MAX2- and KAI2-dependent regulation of MpKAI2B expression may play a significant role in the fine tuning of this signaling.
In conclusion, we showed that the basic regulatory mechanisms of the proteolysis-dependent signaling pathway containing KAI2, MAX2, and SMXL are conserved in M. polymorpha. This implies that this signaling pathway arose in the common ancestors of land plants. In near future, the elucidation of the nature of KL, as well as the identification of SL or SL-related compounds in sister lineages to the vascular land plants and seed plants, is expected to dramatically improve our understanding of the ancestral roles and the evolution of SL- and KAI2-dependent signaling systems.
Materials and methods
Plant materials and growth conditions
Marchantia polymorpha strains Takaragaike-1 (Tak-1; Japanese male line) and Takaragaike-2 (Tak-2; Japanese female line; Ishizaki et al., 2015) were used as the WTs in this study. Growth phenotypes were observed using Tak-1. Plants were grown on half-strength Gamborg’s B5 medium with 1.0% agar under continuous light (50–60 μmol photons m−2s−1) at 22°C.
Mutagenesis by CRISPR
pMpGE_En01 (GenBank LC090754; Sugano et al., 2018), digested with SacI and PstI, was separated by electrophoresis and purified using the GE Healthcare illustra GFX PCR DNA and Gel Band Purification Kit. Primers were treated at 96°C for 5 min to make them double-stranded (ds). The ds primers were cloned into pMpGE_En01 (Sugano et al., 2018; GenBank LC090756) using the Takara In-Fusion HD Cloning Kit. The LR reaction was performed using the resultant plasmid (Entry clone) and pMpGE010 (Destination vector, Sugano et al., 2018) to make a plasmid containing gRNA and Cas9. Each construct was independently transformed into M. polymorpha.
Transformation of M. polymorpha
Plasmids were transformed into Agrobacterium tumefaciens strain GV2260 by electroporation using the GenePulser Xcell (BIO-RAD). The transformed Agrobacterium was introduced into cut thallus according to Kubota et al. (2013). Selection was performed using hygromycin or chlorsulfuron.
RNA extraction and expression analysis
RNA was extracted from frozen plant samples using the NucleoSpin RNA Plant kit (MACHEREY-NAGEL). cDNAs were synthesized using SuperScriptIII VILO (Invitrogen). Quantitative PCR was performed using the SYBR Green I with Light Cycler 480 system (Roche Applied Science). Primers used to amplify the cDNAs are shown in Supplemental Table S1. The expression of the endogenous MpSMXL and the introduced MpSMXLd53 genes were amplified with primer pairs endoMpSMXL-d53_qPCR-F and endoMpSMXL_qPCR-R for MpSMXL and endoMpSMXL-d53_qPCR-F MpSMXL and d53_qPCR-R for the MpSMXLd53. The elongation factor gene M. polymorpha ELONGATION FACTOR-1α (Mapoly0024s0116.1) and Actin (Mapoly0016s0139.1) were used as standards.
RNA-seq analysis
Gemmae of WT, Mpkai2a-1, Mpkai2b-1, and Mpmax2-2 were incubated on half-strength Gamborg’s B5 medium for 24 h at 22ºC under continuous light. Three biological replicates were prepared for each genotype. Total RNAs were extracted from the gemmae using the NucleoSpin RNA Plant kit (MACHEREY-NAGEL, Germany) and used to isolate mRNAs with the NEB Next poly(A) mRNA Magnetic Isolation Module Kit (New England Biolabs, USA). The quality and quantity of the isolated mRNAs were checked using the NanoVue Plus (GE Healthcare, Japan) and Bioanalyzer 2100 microfluidics system (Agilent). The purified mRNAs were used for library construction using an NEB Next Ultra RNA Library Prep Kit for Illumina (New England Biolabs, USA). The quantity of each library was determined using the Library Quantification Kit (Takara Bio, Japan).
Sequencing was performed with the DNBseq platform (BGI) and 150-bp paired-end reads were generated using HiSeqXten (Illumina, USA). The Reads were mapped onto the Marchantia v3.1 (http://marchantia.info/) assembly using HISAT2 (Kim et al., 2015) for Galaxy (https://usegalaxy.org) with default parameters. Read counts were calculated by Feature Counts for Galaxy. Gene annotation data were obtained from MarpolBase. To identify differentially expressed genes, q-values were calculated by the TCC R package (Sun et al., 2013) and genes with q < 0.05 and log2-fold change >1 were selected as up- or downregulated genes. Summary statistics of RNA-seq analysis are available in Supplemental Data Set S1.
Analysis of thallus phenotype
Gemmae were grown on 1/2 B5 medium at 22ºC under continuous light for 14 d. Resultant plants were used for the analysis of thallus phenotypes. To accurately measure the area, plants were flattened with a cover slip on a glass slide and images were taken. The area of the thallus was analyzed using image processing software (ImageJ). To measure the angle between the thallus and the growth medium, images were taken from the side and analyzed using ImageJ. EdU uptake experiments were performed according to Naramoto et al. (2019).
Gemma germination assay
The germination assay was performed as described previously (Inoue et al., 2016). Gemmae were plated on half-strength Gamborg’s B5 medium containing 1% sucrose and 1.2% agar under green light, and then irradiated with far red (FR) light (30-µmol photons m−2 s−1) for 15 min to inactivate phytochromes. Gemmae imbibed in the dark for 1 d were irradiated with a pulse of red light (10-µmol photons m−2 s−1) for 5 min followed by incubation in the dark and scored for germination after 6 d.
Application of rac-GR24, KAR1, and KAR2
The karrikins KAR1 and KAR2 and the potent synthetic SL analog rac-GR24 were obtained from Chiralix B.V. (Netherlands). Chemicals were dissolved in acetone and added to half-strength Gamborg’s B5 medium. For mock treatment, acetone was added to the medium.
DSF analysis
The coding sequences for MpKAI2A and MpKAI2B were amplified from cDNA synthesized from total mRNA of the M. polymorpha thallus using primers described in Supplemental Table S1. The PCR products were cloned into the pHIS8 vector (Addgene) using the In-Fusion HD Cloning Kit (Clontech) to generate pHIS8-MpKAI2A and pHIS8-MpKAI2B. BL21 (DE3; Takara) was used for recombinant protein expression. Overnight cultures (3 mL) in LB liquid medium containing 100 µg/kanamycin-1 were inoculated into fresh LB medium (1 L) containing 100 µg/kanamycin-1 and incubated at 37°C until the OD600 value reached 0.6–0.8. Then, IPTG was added to 200 µM and the cells were further incubated at 25°C overnight. The cells were collected by centrifugation at 4,000 rpm for 15 min. The pellets were resuspended in 30 mL of buffer A [20-mM Tris–HCl (pH 8.0) and 200-mM NaCl]. The supernatants (30 mL) from the resulting lysates were crushed using a cell disruption device. The fractions were purified with Ni affinity beads pre-equilibrated with buffer A. MpKAI2A and MpKAI2B proteins were eluted with 5 mL of buffer A containing 125 mM of imidazole. The purified proteins were concentrated using an Amicon Ultra-4 10 K (Millipore).
For DSF analysis, 20 μL reaction mixtures containing 10 μg protein; 0.015 μL Sypro Orange; and GR24s with final concentrations of 250 µM, 100 µM, or 50 µM were prepared in 96-well plates. The final acetone concentration in the reaction mixture was 5%. 1× PBS buffer (pH 7.4) containing 5% acetone was used in the control reaction. DSF experiments were performed using a Light Cycler 480 II (Roche). Sypro Orange (Invitrogen) was used as the reporter dye. Samples were incubated at 25°C for 10 min, then the fluorescence wavelength was detected continuously from 30°C to 95°C. The denaturation curve was obtained using MxPro software (Agilent).
Accession numbers
RNA-seq is available at Sequence Read Archive (SRA) with the accession number DRA011767. Accession numbers of plant material analyzed in this study are MpKAI2A: Mapoly0023s0137.1, MpKAI2B: Mapoly0031s0148.1, MpMAX2: Mapoly0113s0006.1, MpSMXL: Mapoly0006s0101.1, rice D10: Os01g0746400. Accession numbers of genes analyzed in Supplemental Figure S1 are presented in Supplemental Table S2.
Supplemental data
Supplemental Figure S1. Alignment of amino acid sequences of KAI2A and D14 (A), MAX2 (B) and SMXL (C) proteins.
Supplemental Figure S2. Production of CRISPR mutants.
Supplemental Table S1. Primers used in this study.
Supplemental Table S2. Accession numbers of genes analyzed in Supplemental Figure S1.
Supplemental Data Set S1. Results of RNA-seq analysis.
Supplemental Data Set S2. List of genes differentially expressed in mutants.
Supplementary Material
Acknowledgments
The authors thank Yoshiya Seto for providing pHIS8-MpKAI2A and pHIS8-MpKAI2B plasmids.
Funding
This work was supported by a Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (20H05684, 18K19198, 17H06475, 16K14748) and The Canon Foundation to J.K.
Conflict of interest statement. None declared.
J.K. and T.K. designed the research and wrote the article. Y.M., A.K., S.S., S.N., Ke.I., X.X., and Ki.I. executed the experiments. Y.M., A.K., Ki.I., and S.N. produced plasmids and mutants. Y.M., A.K., and Ke.I. analyzed the growth phenotypes. X.X. prepared GR24 chemicals.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Junko Kyozuka (junko.kyozuka.e4@tohoku.ac.jp).
References
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-babili S (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335:1348–1351 [DOI] [PubMed] [Google Scholar]
- Abe S, Sado A, Tanaka K, Kisugi T, Asami K, Ota S, Kim HI, Yoneyama K, Xie X, Ohnishi T, et al. (2014) Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc Natl Acad Sci USA 111:18084–18089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435:824–827 [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50:1416–1424 [DOI] [PubMed] [Google Scholar]
- Bennett T, Leyser O (2014) Strigolactone signaling: standing on the shoulders of DWARFs. Curr Opin Plant Biol 22:7–13 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Nelson DC, Weijers D. (2020) Evolution of plant hormone response pathways. Annu Rev Plant Biol 71:327–353 [DOI] [PubMed] [Google Scholar]
- Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33:41–52 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Araki T, Arteaga-Vazquez MA, Berger F, Dolan L, Haseloff J, Ishizaki K, Kyozuka J, Lin SS, Nagasaki H, et al. (2016) The naming of names: guidelines for gene nomenclature in Marchantia. Plant Cell Physiol 57:257–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F,et al. (2017) Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171:287–304 [DOI] [PubMed] [Google Scholar]
- Bürger M, Mashiguchi K, Lee HJ, Nakano M, Takemoto K, Seto Y, Yamaguchi S, Chory J (2019) Structural basis of karrikin and non-natural strigolactone perception in Physcomitrella patens. Cell Rep 26:855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bythell-Douglas R, Rothfels CJ, Stevenson DWD, Graham SW, Wong GK, Nelson DC, Bennett T (2017) Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues. BMC Biol 15:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn CE, Nelson DC (2016) Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not Karrikin or Strigolactone. Front Plant Sci 6:1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, , Clavé G, , Badet-Denisot MA, , Pillot JP, , Cornu D, , Le Caer JP, , Burger M, , Pelissier F, , Retailleau P, , Turnbull C et al, . ( 2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol 12: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker EL, Alder A, Hunn S, Ferguson J, Lehtonen MT, Scheler B, Kerres KL, Wiedemann G, Safavi-Rizi V, Nordzieke S, et al. (2017) Strigolactone biosynthesis is evolutionarily conserved, regulated by phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss, Physcomitrella patens. New Phytol 216:455–468 [DOI] [PubMed] [Google Scholar]
- Delaux P-M, Xie X, Timme RE, Puech‐Pages VE, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Bécard G, Séjalon‐Delmas N (2012) Origin of strigolactones in the green lineage. New Phytrogist 195:857–871 [DOI] [PubMed] [Google Scholar]
- Delwiche CF, Cooper ED (2015). The evolutionary origin of a terrestrial flora. Curr Biol 25:R899–910 [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Beveridge CA (2009) Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiol 149:1929–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Sandoval E, Eklund DM, Bowman JL (2015). A simple auxin transcriptional response system regulates multiple morphogenetic processes in the liverwort Marchantia polymorpha. PLoS Genet 11:e1005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455:189–194 [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowdenet KC (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22:2032–2036 [DOI] [PubMed] [Google Scholar]
- Hoffmann B, Proust H, Belcram K, Labrune C, Boyer F-D, Rameau C, Bonhomme S (2014) Strigolactones inhibit caulonema elongation and cell division in the moss physcomitrella patens. PLoS ONE 9:e99206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Nishihama R, Kataoka H, Hosaka M, Manabe R, Nomoto M, Tada Y, Ishizaki K, Kohchi T (2016) Phytochrome signaling is mediated by PHYTOCHROME INTERACTING FACTOR in the liverwort Marchantia polymorpha. Plant Cell 28:1406–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46:79–86 [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Nishihama R, Ueda M, Inoue K, Ishida S, Nishimura Y, Shikanai T, Kohchi T (2015) Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha .PLoS ONE 10:e0138876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504:401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama M, Hirano Y, Mori T, Kim SY, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T (2013) Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18:147–160 [DOI] [PubMed] [Google Scholar]
- Kato H, Ishizaki K, Kouno M, Shirakawa M, Bowman JL, Nishihama R, Kohchi T (2015) Auxin-mediated transcriptional system with a minimal set of components is critical for morphogenesis through the life cycle in Marchantia polymorpha. Pros Genet 11:e1005084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla A, Morffy N, Li Q, Faure L, Chang SH, Yao J, Zheng J, Cai ML, Stanga J, Flematti GR, et al. (2020) Structure–function analysis of SMAX1 reveals domains that mediate its karrikin-induced proteolysis and interaction with the receptor KAI2. Plant Cell 32:2639–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A, Ishizaki K, Hosaka M, Kohchi T (2013) Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalli. Biosci Biotechnol Biochem 77:167–172 [DOI] [PubMed] [Google Scholar]
- Lavy M, Prigge MJ, Tao S, Shain S, Kuo A, Kirchsteiger K, Estelle M (2016) Constitutive auxin response in Physcomitrella reveals complex interactions between Aux/IAA and ARF proteins. eLife 5:e13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ward S, Li P, Bennett T, Leyser O (2016) SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 28:1581–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Obando M, Conn CE, Hoffmann B, Bythell-Douglas R, Nelson DC, Rameau C, Bonhomme S (2016) Structural modelling and transcriptional responses highlight a clade of PpKAI2-LIKE genes as candidate receptors for strigolactones in Physcomitrella patens. Planta 243:1441–1453 [DOI] [PubMed] [Google Scholar]
- Lopez-Obando M, de Villiers R, Hoffmann B, Ma L, de Saint Germain A, Kossmann J, Coudert Y, Harrison CJ, Rameau C, Hills P, et al. (2018) Physcomitrella patens MAX2 characterization suggests an ancient role for this F-box protein in photomorphogenesis rather than strigolactone signalling. New Phytol 219:743–756 [DOI] [PubMed] [Google Scholar]
- Lopez-Obando M, Guillory A, Boyer FD, Cornu D, Hoffmann B, Le Bris P, Pouvreau J-B, Delavault P, Rameau C, de Saint Germain A, et al. (2020) The Physcomitrium (Physcomitrella) patens PpKAI2L receptors for strigolactones and related compounds highlight MAX2 dependent and independent pathways. bioRxiv (doi: 10.1101/2020.11.24.395954). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Duan J, Ke J, He Y, Gu X, Xu TH, Yu H, Wang Y, Brunzelle JS, Jiang Y, et al. (2017) A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex. Sci Adv 3:e1601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin DC, Hamon-Josse M, Bennett T (2020) Fellowship of the rings: a saga of strigolactones and other small signals. New Phytol 225:621–636 [DOI] [PubMed] [Google Scholar]
- Monte I, Ishida S, Zamarreño AM, Hamberg M, Franco-Zorrilla JM, García-Casado G, Gouhier-Darimont C, Reymond P, Takahashi K, García-Mina JM, et al. (2018) Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat Chem Biol 14:480–488 [DOI] [PubMed] [Google Scholar]
- Moturu TR, Thula S, Singh RK, Nodzynski T, Vareková RS, Friml J, Simon S (2018) Molecular evolution and diversification of the SMXL gene family. J Exp Bot 69:367–2378 [DOI] [PubMed] [Google Scholar]
- Mutte SK, Kato H, Rothfels C, Melkonian M, Wong GKS, Weijers D (2018) Origin and evolution of the nuclear auxin response system. eLife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Xue YL, Miyakawa T, Hou F, Qin HM, Fukui K, Shi X, Ito E, Ito S, Park SH, et al. (2013). Molecular mechanism of strigolactone perception by DWARF14. Nat Commun 4:2613. [DOI] [PubMed] [Google Scholar]
- Naramoto S, Jones VAS, Trozzi N, Sato M, Toyooka K, Shimamura M, Ishida S, Nishitani K, Ishizaki K, Nishihama R, et al. (2019) A conserved regulatory mechanism mediates the convergent evolution of plant shoot lateral organs. PLoS Biol 17:e3000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Flematti GR, Riseborough JA, Ghisalberti EL, Dixon KW, Smith SM (2010) Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc Natl Acad Sci USA 107:7095–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 108:8897–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Sakayama H, de Vries J, Buschmann H, Saint-Marcoux D, Ullrich KK, Haas FB, Vanderstraeten L, Becker D, Lang D, et al. (2018) The Chara genome: secondary complexity and implications for plant terrestrialization. Cell 174:448–464 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Lavy M, Ashton NW, Estelle M (2010) Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr Biol 9:1907–1912 [DOI] [PubMed] [Google Scholar]
- Proust H, Hoffmann B, Xie X, Yoneyama K, Schaefer DG, Yoneyama K, Nogué F, Rameau C (2011) Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138:1531–1539 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319:64–69 [DOI] [PubMed] [Google Scholar]
- Scaffidi A, Waters MT, Ghisalberti EL, Dixon KW, Flematti GR, Smith SM (2013) Carlactone-independent seedling morphogenesis in Arabidopsis. Plant J 76:1–9 [DOI] [PubMed] [Google Scholar]
- Scaffidi A, Waters MT, Sun YK, Skelton BW, Dixon KW, Ghisalberti EL, Flematti GR, Smith SM (2014) Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol 165:1221–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y, Yasui R, Kameoka H, Tamiru M, Cao M, Terauchi R, Sakurada A, Hirano R, Kisugi T, Hanada A, et al. (2019) Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat Commun 10:191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zhu L, Bu QY, Huq E (2012) MAX2 affects multiple hormones to promote photomorphogenesis. Mol Plant 5:750–762 [DOI] [PubMed] [Google Scholar]
- Shabek N, Ticchiarelli F, Mao H, Hinds TR, Leyser O, Zheng N (2018) Structural plasticity of D3–D14 ubiquitin ligase in strigolactone signalling. Nature 563:652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A, Leyser O, Nelson DC (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27:3143–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga JP, Morffy N, Nelson DC (2016) Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta 243:1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga JP, Smith SM, Briggs WR, Nelson DC (2013) SUPPRESSOR OF MORE AXILLARY GROWTH 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol 163:318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HM (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129:1131–1141 [DOI] [PubMed] [Google Scholar]
- Sugano SS, Nishihama R, Shirakawa M, Takagi J, Matsuda Y, Ishida S, Shimada T, Hara-Nishimura I, Osakabe K, Kohchi T (2018) Efficient CRISPR/Cas9-based genome editing and its application to conditional genetic analysis in Marchantia polymorpha .PLoS ONE 13:e0205117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Nishiyama T, Shimizu K, Kadota K (2013) TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinform 14:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XD, Ni M (2011) HYPOSENSITIVE TO LIGHT, an alpha/beta fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de-etiolation. Mol Plant 4:116–126 [DOI] [PubMed] [Google Scholar]
- Toh S, Holbrook-Smith D, Stogios PJ, Onopriyenko O, Lumba S, Tsuchiya Y, Savchenko A, McCourt P (2015) Structure–function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350:203–207 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Yoshimura M, Sato Y, Kuwata K, Toh S, Holbrook-Smith D, Zhang H, McCourt P, Itami K, Kinoshita T, et al. (2015) PARASITIC PLANTS. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349:864–868 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455:195–200 [DOI] [PubMed] [Google Scholar]
- Végh A, Incze N, Fábián A, Huo H, Bradford KJ, Balázs E, Soós V (2017) Comprehensive analysis of DWARF14-LIKE2 (DLK2) reveals its functional divergence from strigolactone-related paralogs. Front Plant Sci 8:1641–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaécija-Aguilar JA, Hamon-Josse M, Carbonnel S, Kretschmar A, Schmidt C, Dawid C, Bennett T, Gutjahr C (2019) SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLoS Genet 15:e1008327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CH, Siu-Ting K, Taylor A, O’Connell MJ, Bennett T (2019) Strigolactone synthesis is ancestral in land plants, but canonical strigolactone signalling is a flowering plant innovation. BMC Biol 17:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner ES, López-Salmerón V, Belevich I, Poschet G, Jung I, Grünwald K, Sevilem I, Jokitalo E, Hell R, Helariutta Y, et al. (2017) Strigolactone- and karrikin-independent SMXL proteins are central regulators of phloem formation. Curr Biol 27:1241–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu Q, Yu H, Ma H, Li X, Yang J, Chu J, Xie Q, Wang Y, Smith SM, et al. (2020a) Strigolactone and karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation in Arabidopsis. Plant Cell 32:2251–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang B, Yu H, Guo H, Lin T, Kou L, Wang A, Shao N, Ma H, Xiong G, et al. (2020b) Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 583:277–281 [DOI] [PubMed] [Google Scholar]
- Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J (2015) Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27:3128–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 39:1285–1295 [DOI] [PubMed] [Google Scholar]
- Waters MT, Scaffidi A, Moulin SLY, Sun YK, Flematti GR, Smith SM (2015) A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27:1925–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Smith SM (2013) KAI2And MAX2-mediated responses to karrikins and strigolactones are largely independent of HY5 in Arabidopsis seedlings. Mol Plant 6:63–75 [DOI] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L, et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536:469–473 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Mori N, Sato T, Yoda A, Xie X, Okamoto M, Iwanaga M, Ohnishi T, Nishiwaki H, Asami T, et al. (2018) Conversion of carlactone to carlactonoic acid is a conserved function of MAX1 homologs in strigolactone biosynthesis. New Phytol 218:1522–1533 [DOI] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signaling. Nature 504:406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.