Abstract
Flowering is the developmental transition from the vegetative to the reproductive phase. FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), and LEAFY (LFY) are floral integrators. These genes are repressed by several floral repressors including EARLY FLOWERING3 (ELF3), SHORT VEGETATIVE PHASE (SVP), TEMPRANILLO1 (TEM1), and TEM2. Although gibberellin (GA) promotes flowering by activating the floral integrator genes, the exact molecular mechanism remains unclear. DELLAs are negative regulators in GA signaling and act as coactivators of the transcription factor GAI ASSOCIATED FACTOR 1 (GAF1). GAs convert the GAF1 complex from a transcriptional activator to a repressor. Here, we show that GAF1 functions in the GA-dependent flowering pathway by regulating FT and SOC1 expression in Arabidopsis thaliana. We identified four flowering repressors, ELF3, SVP, TEM1, and TEM2, as GAF1-target genes. In response to GAs, GAF1 forms a transcriptional repressor complex and promotes the expression of FT and SOC1 through the repression of four flowering repressor genes, ELF3, SVP, TEM1, and TEM2.
Gibberellins change the function of a transcription factor complex and promote flowering via the activation of floral integrator genes including florigen through the inhibition of floral repressors.
Introduction
The induction of flowering is a key developmental decision in a plant’s life cycle, and its timing is controlled by a complex combination of developmental and environmental signals. In Arabidopsis thaliana, the autonomous, photoperiod, vernalization, thermosensory, and gibberellin (GA) pathways (Simpson and Dean, 2002; Blazquez et al., 2003; Halliday et al., 2003; Amasino, 2004) promote flowering by activating the floral integrator genes FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), and LEAFY (LFY). FT, expressed in leaves, functions as a florigen that is transported to the shoot apex to induce flowering. Several transcription factors regulate FT expression. CONSTANS (CO) expression in the phloem is circadian-dependent; it induces FT expression under long day (LD) conditions. By contrast, FT expression is repressed by TEM1 and TEM2, which are RAV subfamily transcription factors containing an AP2/ERF and a B3 DNA-binding domain (Matias-Hernandez et al., 2014). Thus, CO and TEMs act as an FT activator and repressors, respectively. The binding sites of CO and TEM1 within the FT promoter are closely aligned, and their balance controls the FT expression level (Castillejo and Pelaz, 2008). The circadian clock component EARLY FLOWERING3 (ELF3) and MADS-box transcription factor SHORT VEGETATIVE PHASE (SVP) are other negative regulators of flowering in Arabidopsis (Hartmann et al., 2000), which are involved in FT regulation in the leaves. In addition, SVP represses the expression of SOC1 in the shoot apex and leaves and plays a role in the autonomous and GA pathways of flowering (Li et al., 2008). GAs promote FT expression in the leaves and SOC1 expression in the shoot apex (Moon et al., 2003; Hisamatsu and King, 2008), but it is unclear how GAs regulate these genes.
GAs are critical for flowering in Arabidopsis under short day (SD) conditions (Wilson et al., 1992). The GA-deficient mutant ga1-3 flowers moderately late under LD conditions but does not flower under SD conditions. Before floral initiation, the levels of bioactive GAs increase in the shoot apex via the activation of GA20ox1, encoding the GA-biosynthetic enzyme GA 20-oxidase, which promotes the expression of the floral integrators SOC1 and LFY (Eriksson et al., 2006). DELLA proteins act as negative regulators of GA signaling (Sasaki et al., 2003; Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Sun, 2011; Hauvermale et al., 2012). Arabidopsis contains five DELLAs, including GIBBERELLIN INSENSITIVE (GAI) (Sun and Gubler, 2004). DELLAs are rapidly degraded in the presence of GAs. A DELLA quadruple mutant of Arabidopsis flowers early under SD conditions (Cheng et al., 2004). Recently, we identified a DELLA-interacting factor, GAI-ASSOCIATED FACTOR1 (GAF1), and revealed the role of DELLAs as transcriptional coactivators (Fukazawa et al., 2014). GAF1 also interacts with corepressor TOPLESS-RELATED (TPR). DELLAs and TPR are coactivators and a corepressor of GAF1, respectively (Fukazawa et al., 2014). GAs convert the GAF1 complex from a transcriptional activator to a repressor. GAF1 is a transcription factor with zinc-finger motifs, belonging to the INDETERMINATE1 (ID1) (Colasanti et al., 1998) domain (IDD) family. In maize (Zea mays), an id1 mutant flowers late (Singleton, 1946; Colasanti et al., 1998), indicating its role in floral transition in monocots. Among the Arabidopsis IDD family members, INDETERMINATE DOMAIN1 (IDD1)/ENHYDROUS was found to be most closely related to GAF1. In Arabidopsis, the gaf1 idd1 double mutant also flowers later than the wild-type under SD conditions, whereas GAF1 overexpressing lines flower earlier than the wild-type (Figure 1, A and previously shown [Fukazawa et al., 2014]), suggesting that the GAF1 complex is involved in the GA-mediated regulation of flowering.
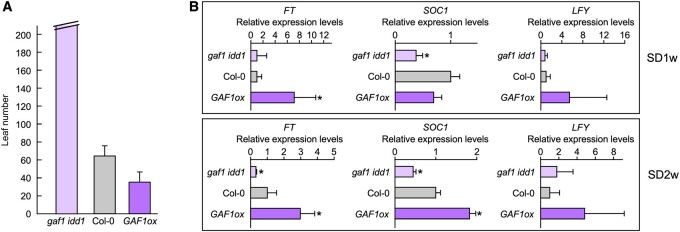
Figure 1.
The GAF1 overexpressor and gaf1 idd1 mutant exhibit early and late-flowering phenotypes, respectively, via alterations to the expression levels of flowering integrator genes. A, Flowering time analysis (rosette leaf number) under SD conditions. The GAF1 overexpressor exhibited an early flowering phenotype under SD conditions. The gaf1 idd1 mutant exhibited an extremely delayed flowering phenotype under SD conditions Error bars indicate sd of the mean (n > 8). Asterisks represent Student’s t test significance compared with Col-0 (*P < 0.05). B, Plants were grown under SD conditions for 1 or 2 weeks. Plants were harvested at zeitgeber (ZT) 8. Relative expression levels of FT, SOC1, and LFY in Col-0, gaf1 idd1, and GAF1 overexpressor plants are shown. The results are shown as relative expression levels to UBQ11. Error bars indicate the sd of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with Col-0 (*P < 0.05).
In this study, we identified four flowering repressors, ELF3, TEM1, TEM2, and SVP, as GAF1-target genes. In response to GAs, GAF1 forms a transcriptional repressor complex and promotes the expression of FT and SOC1 by repressing genes encoding negative regulators of flowering including ELF3, TEM1, TEM2, and SVP. We demonstrated that ELF3 represses the expression of FT in combination with LUX and ELF4 as part of the Evening Complex (EC). We provide a model of the GA flowering pathway in which GAs promote flowering via GAF1-TPR-dependent repression of flowering repressors.
Results
GAF1 regulates flowering via altering the expression levels of GA-controlled flowering genes
To gain insight into the role of the GAF1 complex in the regulation of flowering, we investigated the expression levels of genes encoding the floral pathway integrators FT, SOC1, and LFY in the gaf1 idd1 double mutant and the GAF1 overexpressor under SD conditions (Figure 1, A). The expression levels of FT, SOC1, and LFY were increased in the GAF1 overexpressor, whereas the expression levels of FT and SOC1 were decreased in the gaf1 idd1 double mutant (Figure 1, B). The induction of SOC1 in the GAF1 overexpressor was later, by approximately 1 week, than the induction of FT under SD conditions (Figure 1, B). Because GAF1 forms a transcriptional repressor complex under GA-sufficient conditions (Fukazawa et al., 2014), it cannot directly promote the expression of FT, SOC1, and LFY in GA-controlled flowering. Rather, the expression of FT, SOC1, and LFY might be induced by the GAF1-dependent downregulation of flowering repressor genes.
Identification of novel GAF1-target genes involved in the regulation of flowering
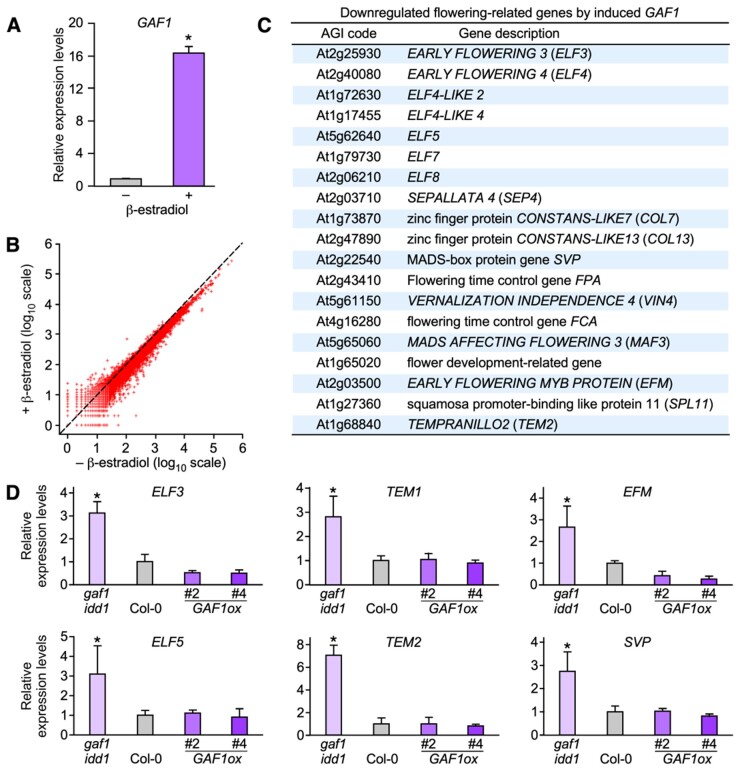
To identify GAF1-target genes involved in flowering, RNA-seq analysis was conducted using transgenic plants harboring estradiol-inducible GAF1 (Figure 2, A; Zuo et al., 2000). Flowering-related genes including ELF3, ELF5, TEM1, TEM2, EARLY FLOWERING MYB PROTEIN (EFM), and SVP were selected as candidates of novel GAF1-target genes among genes downregulated after the induction of GAF1 (Figure 2, B and C and Supplemental Data set S1). Their expression levels increased in the gaf1 idd1 double mutant but the expression levels of ELF3 and EFM decreased and other genes were slightly decreased in the GAF1 overexpressor (Figure 2, D).
Figure 2.
Search for GAF1 target genes using the inducible GAF1 transgenic plant. A, Transgenic plants carrying pER10-GAF1 grown under LD conditions for 1 week. Whole plants were treated with or without β-estradiol for 24 h and were harvested at ZT8 under LD conditions. Relative expression levels of GAF1 of the pER10-GAF1 transgenic plant with or without β-estradiol treatment are shown. Error bars indicate the SD of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with Col-0 (*P < 0.05). B, RNA-Seq analysis using an inducible GAF1 transgenic line in which the XVE system induced the expression of GAF1 in 7-d-old pER10-GAF1 Arabidopsis seedlings treated with β-estradiol or mock treated for 24 h. C, A list of flowering-related genes downregulated by induced GAF1. D, Plants were grown under SD conditions for 2 weeks. Whole seedlings were harvested at ZT8. Relative expression levels of ELF3, TEM1, EFM, ELF5, TEM2, and SVP in Col-0, gaf1 idd1, and GAF1 overexpressor plants. Error bars indicate the SD of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with Col-0 (*P < 0.05).
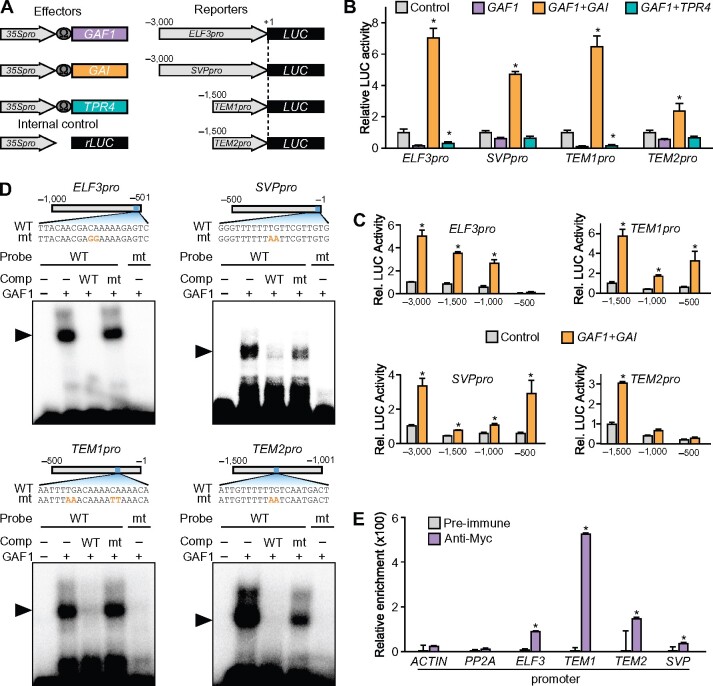
To investigate whether the GAF1 complex was involved in the regulation of these genes, we performed a transient assay using Arabidopsis protoplasts. The promoter of each gene was fused with the LUC reporter gene (Figure 3, A). The ELF3, SVP, TEM1, and TEM2 gene promoters were activated by the DELLA-GAF1 complex but repressed by the TPR4-GAF1 complex (Figure 3, B). However, the DELLA-GAF1 complex did not affect the activity of the EFM and ELF5 promoters (Supplemental Figure S1). These results suggested that ELF3, SVP, TEM1, and TEM2 are directly regulated by the GAF1 complex. To define the cis-acting elements for the DELLA-GAF1 complex in each promoter of the GAF1 target genes, we generated a series of 5′-deletions in each promoter—ELF3Δ-3000, ELF3Δ-1500, ELF3Δ-1000, ELF3Δ-500; SVPΔ-3000, SVPΔ-1500, SVPΔ-1000, SVPΔ-500; TEM1Δ-1500, TEM1Δ-1000, TEM1Δ-500; and TEM2Δ-1500, TEM2Δ-1000, TEM2Δ-500—fused with the LUC reporter gene and determined the promoter activities in a transactivation assay (Figure 3, C). The DELLA-GAF1 complex activated ELF3 (ELF3Δ-1500 and ELF3Δ-1000), SVP (SVPΔ-500), TEM1 (TEM1Δ-1000, TEM1Δ-500), and TEM2 (TEM2Δ-1500), suggesting that each promoter contains a cis-acting element for DELLA-GAF1 in the 500-bp region between −1000 and −501 of the ELF3 promoter, −500 to −1 of the SVP promoter, −500 to −1 of the TEM1 promoter, and −1500 to −1001 of the TEM2 promoter. The consensus binding motif for IDD family transcription factors is TTTTGTCG and GAF1 also binds to this sequence and closely related motifs (Fukazawa et al., 2014, 2017). Putative binding motifs for DELLA-GAF1 were found in each of the experimentally identified 500-bp regions of the ELF3, SVP, TEM1, and TEM2 promoters. The electrophoretic mobility shift assay (EMSA) indicated that recombinant GAF1 directly binds to these sequences, which are ELF3 (−529 to −510), SVP (−20 to −1), TEM1 (−152 to −133), and TEM2 (−1,288 to −1,269), in vitro (Figure 3, D). To investigate the binding of GAF1 to each gene promoter in vivo, we used transgenic plants expressing Myc-tagged GAF1 under the control of the CaMV 35S promoter. ChIP assays showed that GAF1 binds to each promoter (ELF3, −620 to −420; SVP, −103 to +97; TEM1, −241 to −41; TEM2, −1,382 to −1,182; Figure 3, E), indicating that GAF1 interacts directly with each target gene promoter in vivo.
Figure 3.
Identification of novel GAF1 target genes involved in the regulation of flowering. A, Schematic representation of the reporter and effector. Fragments (3,000 bp) of the ELF3 and SVP promoters and 1,500-bp fragments of the TEM1 and TEM2 promoters were fused with the LUC gene. The effector plasmid expressed full-length GAF1, GAI, or TPR4 under control of the CaMV 35S promoter with a viral translation enhancer (Ω). B, Transient expression assay showed that the DELLA-GAF1 complex activated and the TPR4-GAF1 complex repressed the ELF3, SVP, TME1, and TEM2 promoters. The reporter plasmids consisted of the 3,000-bp promoter regions of ELF3 and SVP and the 1,500-bp promoter regions of TEM1 and TEM2 fused with the LUC reporter gene. The results are shown as LUC/rLUC activity. Error bars indicate the sd of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with mock treated control (*P < 0.05). C, Transactivation assay of GAF1 and GAI. The effector, reporter, and internal control constructs were co-transfected into Arabidopsis protoplasts. The transfected cells were incubated for 20 h, and Luc and rLUC activities were measured. The results are shown as LUC/rLUC activity. Error bars indicate sd of the mean (n = 3). Asterisks represent Student’s t test significance compared with mock treated control (*P < 0.05). D, Identification of GAF1-binding regions in the ELF3, SVP, TEM1, and TEM2 promoters in vitro. EMSA analysis using recombinant GAF1 protein. Oligonucleotides containing ELF3pro (−529 to −510, wild-type; lanes 1–4) or mutated (mt) ELF3pro (mt; lane 5), SVPpro (−20 to −1, wild-type; lanes 1–4) or mtSVPpro (mt; lane 5), TEM1pro (−152 to −133, wild-type; lanes 1–4) or mtTEM1pro (mt; lane 5), and TEM2pro (−1,288 to −1,269, wild-type; lanes 1–4) or mtTEM2pro (mt; lane 5) were used as probes. Orange letters indicate mutated bases. Wild-type and mt indicate competition with a 100-fold to 500-fold excess of unlabeled wild-type and mutated probe, respectively. The specific GAF1–DNA complexes are indicated by arrowheads. +, Addition to the reaction mixtures; –, omission from the reaction mixtures. E, Plants were grown under SD conditions for 2 weeks. Whole seedlings were harvested at ZT8. GAF1 binds to a region of the ELF3, SVP, TEM1, and TEM2 promoters in vivo. ChIP assays were performed with pre-immune or anti-myc in CaMV 35S:myc-GAF1 transgenic plants. The co-precipitated level of each DNA fragment was quantified by real-time PCR and normalized to the input DNA. Error bars indicate sd of three technical replicates (n = 3). Asterisks represent Student’s t test significance compared with pre-immune as control (*P < 0.05). Experiments were repeated twice with independently grown plants, with similar results.
Tissue-specific expression profiles and GA sensitivity of flowering integrators and GAF1 target genes
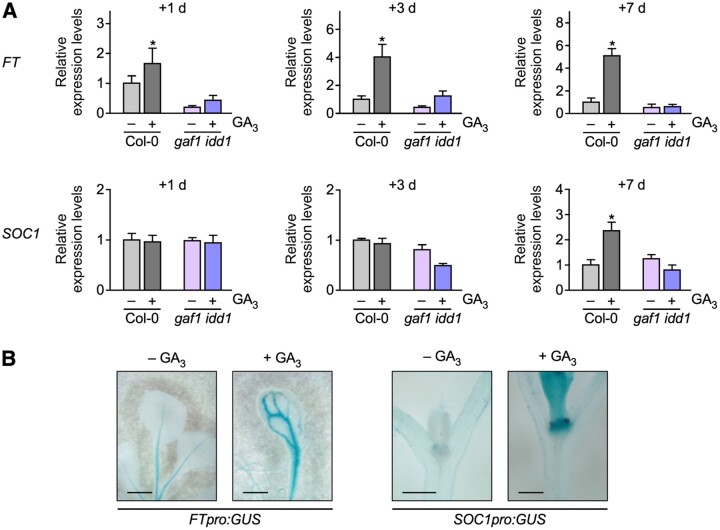
To investigate the temporal and spatial expression of flowering integrators and GAF1 target genes with or without GA, we carried out real-time PCR and histochemical analysis using transgenic plants. FT and SOC1 expression levels were increased by GAs within 1 d and 7 d under SD conditions, respectively. The induction of FT and SOC1 by GAs was not observed in the gaf1 idd1 mutant, indicating that GAF1 and IDD1 were involved in the induction of FT and SOC1 by GAs under SD conditions (Figure 4, A). Histochemical analysis using transgenic plants carrying the FT and SOC1 promoter fused with GUS showed that GAs promote FT and SOC1 expression in the leaves and the shoot apex, respectively (Figure 4, B).
Figure 4.
Tissue-specific expression profiles and GA sensitivity of FT and SOC1. A, Relative expression levels of FT and SOC1 in Col-0 and gaf1 idd1 with or without GA3. Plants were grown under SD conditions for 1 week and were treated for 1 d, 3 d, or 1 week with or without GA3. Whole seedlings were harvested at ZT8. Error bars indicate sd of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with control (*P < 0.05). B, GUS expression pattern in transgenic Arabidopsis plants carrying FT promoter:GUS and SOC1 promoter:GUS. Plants were grown under SD conditions for 1 week with or without GA3. Scale bar indicates 1 mm.
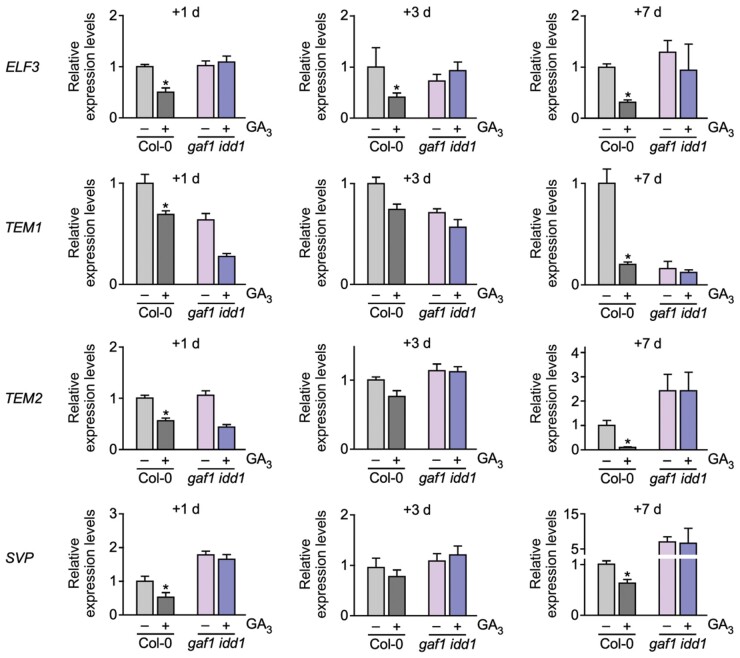
To investigate the response of the GAF1 target genes to GA, we examined the expression levels of these genes with or without GAs. We found that the expression levels of ELF3, TEM1, TEM2, and SVP were significantly reduced by GAs within 1 d (Figure 5). These reductions in ELF3, TEM1, TEM2, and SVP by GAs were not observed in the gaf1 idd1 mutant under SD conditions, indicating that GAs promote the expression of FT and SOC1 via the reduction of GAF1 target genes, including ELF3, TEM1, TEM2, and SVP (Figure 5).
Figure 5.
Tissue-specific expression profiles and GA sensitivity of GAF1 target genes. A, Relative expression levels of ELF3, TEM1, TEM2, and SVP in Col-0 and gaf1 idd1 with or without GA3 treatment. Plants were grown under SD conditions for 1 week and were treated with GA3 or mock-treated for 1 d, 3 d, or 1 week. Whole seedlings were harvested at ZT8. Error bars indicate sd of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with the mock-treated control (*P < 0.05).
To confirm GA responsibility of novel GAF1 target genes including ELF3, TEM1, TEM2, and SVP, we investigated the expression levels of these gene following shorter exposures to GA. ELF3 and TEM1 are decreased by GA within 3 h and TEM2 and SVP was decreased by GA within 6 h (Supplemental Figure S2). Thus, GAF1 target genes respond to GA and decreased at least within 6 h.
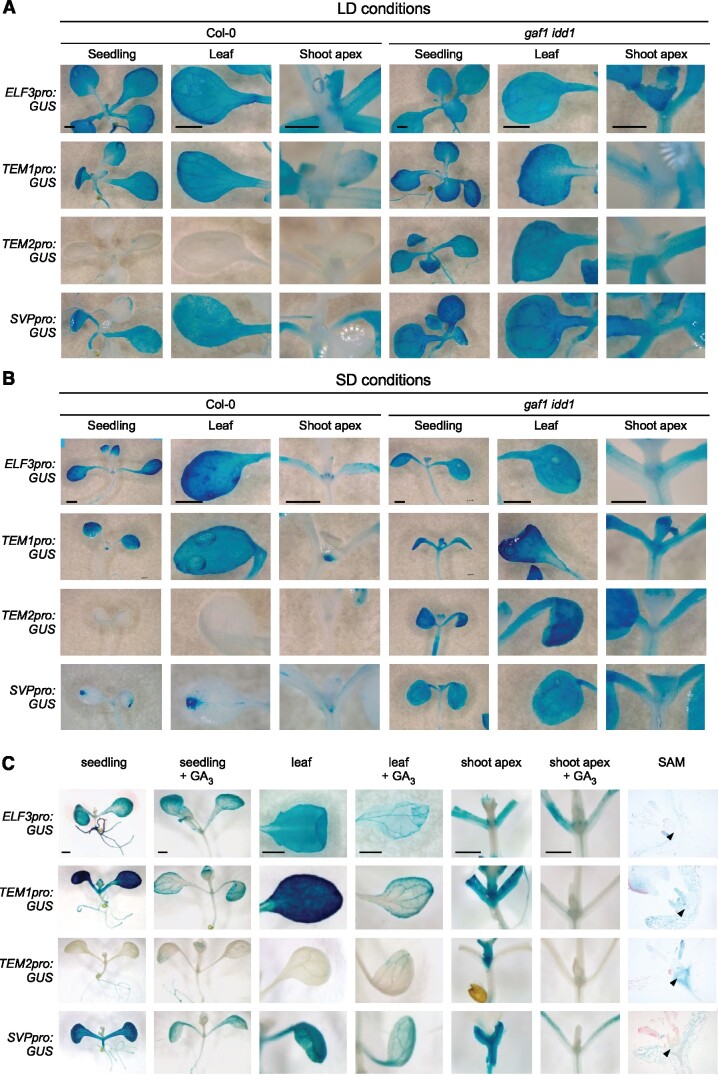
Next, we examined the effects of GA on the tissue-specific expression patterns of GAF1 target genes using transgenic plants carrying each GAF1 target gene promoter fused with GUS. The GUS staining assay showed expression of ELF3, TEM1, and SVP in the leaves and TEM1, TEM2, and SVP in the shoot apex under both LD and SD conditions (Figure 6, A and B). Furthermore, GUS activity in all transgenic lines decreased with GA treatment in the seedling, leaf, and the shoot apex (Figure 6, C). To investigate the contribution of GAF1 and IDD1 to the spatial expression of TEM1, TEM2, ELF3, and SVP, we generated transgenic plants carrying the TEM1, TEM2, ELF3, and SVP promoters fused with GUS in the gaf1 idd1 mutant background. The GUS activity of the transgenic plant in the gaf1 idd1-background was relatively high compared with that in GUS transgenic plants in the Col-0 background (Figure 6, A and B), indicating that GAF1 and IDD1 were involved in the repression of TEM1, TEM2, ELF3, and SVP.
Figure 6.
Spatial expression pattern and GA sensitivity of GAF1 target genes. A and B, GUS expression patterns in transgenic Arabidopsis plants carrying ELF3 promoter:GUS, TEM1 promoter:GUS, TEM2 promoter:GUS, and SVP promoter:GUS in Col-0 (left side nine panels) and gaf1 idd1 (right side nine panels). Plants were grown under LD conditions for 2 weeks (A) and SD conditions for 1 week (B). C, GUS expression patterns in transgenic Arabidopsis plants carrying ELF3 promoter:GUS, TEM1 promoter:GUS, TEM2 promoter:GUS, and SVP promoter:GUS. Seedling (first and second colums), leaves (third and fourth colums) and meristematic regions (fifth and sixth colums), and SAM (arrow head in seventh colums) in transgenic plants were analyzed. Plants were grown for 1 week with or without GA3. Scale bar indicates 1 mm.
The expression of FT and SOC1 is repressed by TEM1 and SVP
We next investigated how the floral integrator genes SOC1 and FT are affected by the GAF1 target genes and if this potential effect is GA-dependent. Although LFY is also a floral integrator, it was excluded from this analysis because SOC1 directly activates LFY in the shoot apex (Lee and Lee, 2010), indicating that LFY acts downstream of SOC1 in flowering. Previous reports showed that SVP directly represses SOC1 and FT by binding to their promoters (Li et al., 2008) and that TEM1 and TEM2 also act as direct FT repressors (Castillejo and Pelaz, 2008; Osnato et al., 2012). To confirm whether SVP, TEM1, and TEM2 regulate the expression of SOC1 and FT, we conducted a transient expression assay (Figure 7, A). We observed that SVP represses the SOC1 promoter and that TEM1 and SVP repress the FT promoter. TEM2 also repressed the FT promoter, but this repression activity of TEM2 was weaker than that of TEM1 (Figure 7, B and C).
Figure 7.
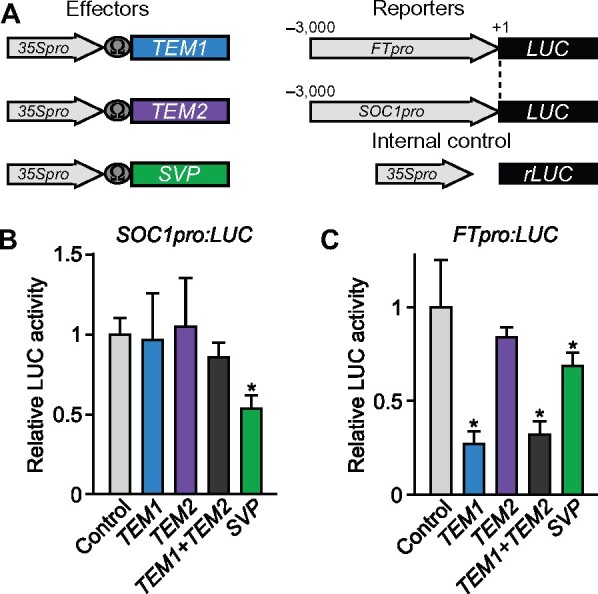
The expression of FT and SOC1 is repressed by TEM1 and SVP. A, Schematic representation of the reporter and effector. A 3,000-bp fragment of the FT and SOC1 promoter was fused with the LUC gene. The effector plasmid expressed the full-length TEM1, TEM2, and SVP under the control of the CaMV 35S promoter with a viral translation enhancer (Ω). B and C, The SOC1 promoter is regulated by SVP; the FT promoter is regulated by TEM1. The effector, reporter, and internal control constructs were co-transfected into Arabidopsis protoplast cells. The transfected cells were incubated for 20 h and then Luc and rLUC activities were measured. The results are shown as LUC/rLUC activity. Error bars in (B) and (C) indicate sd of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with mock treated control (*P < 0.05).
ELF3 controls flowering through the regulation of FT expression
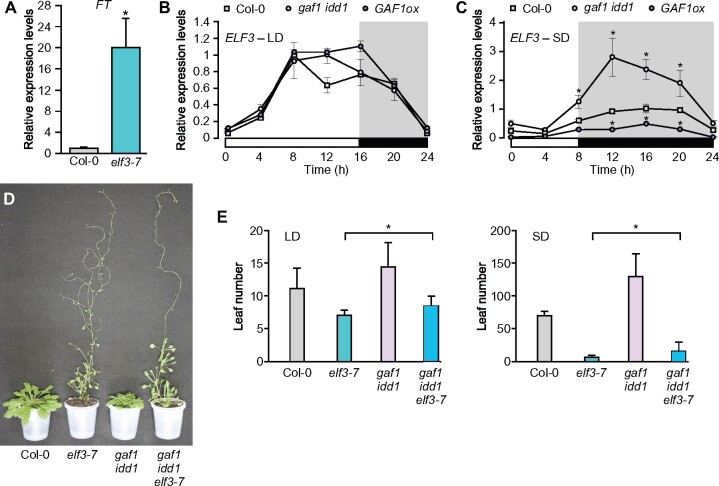
Although Boden et al. (2014) reported that the expression level of FT is increased in a barley (Hordeum vulgare) elf3 mutant, it was unclear how ELF3 regulates flowering through the regulation of FT expression. We confirmed that FT expression is also increased in the Arabidopsis elf3 mutant (Figure 8, A). The phenotype of elf3 was more pronounced than that of the other GAF1 target gene mutants. Therefore, we focused our analysis on ELF3. Since GAs promote the conversion of the GAF1 complex from an activator to a repressor, the GAF1-TPR complex appears to function as the repressor of ELF3 under GA-sufficient conditions. We found that ELF3 expression levels decreased with GAs (Figure 5). Although gaf1 or idd1 single mutant did not exhibit obvious phenotypes, the gaf1 idd1 double mutant exhibited an extremely late-flowering phenotype under SD conditions (Figure 1, A;Fukazawa et al., 2014), indicating that both GAF1 and IDD1 are involved in the GA flowering pathway. ELF3 is a circadian clock gene that contributes to photoperiod-dependent flowering, and the early flowering phenotype of elf3 was clearer under SD conditions than under LD conditions (Figure 8, D and E). We further investigated ELF3 expression under LD and SD conditions. We found that ELF3 expression is higher in gaf1 idd1 plants than in Col-0 plants under SD conditions; whereas ELF3 expression was lower in the GAF1 overexpressor than in Col-0 under SD conditions (Figure 8, B and C).
Figure 8.
ELF3 acts downstream of GAF1 and IDD1 in flowering. A, Relative expression levels of FT in Col-0 or elf3-7. Plants were grown under SD conditions for 2 weeks. Whole seedlings were harvested at ZT8. Error bars indicate sd of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with Col-0 as control (*P < 0.05). B, Diurnal expression of ELF3 in Col-0, gaf1 idd1, and GAF1ox under LD conditions. Plants were grown under LD conditions for 2 weeks. Whole seedlings were harvested at ZT0, 4, 8, 12, 16, 20, and 24. Error bars indicate sd of three biological replicates (n = 3). C, Diurnal expression of ELF3 in Col-0, gaf1 idd1, and GAF1ox under SD conditions. Seedlings were grown in SD conditions for 2 weeks. Whole seedlings were harvested at ZT0, 4, 8, 12, 16, 20, and 24. Error bars indicate sd of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with Col-0 as control (*P < 0.05). D, Col-0, elf3-7, gaf1 idd1, and gaf1 idd1 elf3-7 plants were analyzed at 104 d after germination under SD conditions. All mutants had a Col-0 background. E, Flowering time of Col-0, elf3-7, gaf1 idd1, and gaf1 idd1 elf3-7 plants under LD or SD conditions. Data are means of at least eight plants. Error bars indicate the sd of total leaf number. Asterisks represent Student’s t test significance compared between elf3-7 and gaf1 idd1 elf3-7 (*P < 0.05).
ELF3 acts downstream of GAF1 and IDD1 in flowering
The elf3-7 mutant exhibited the early flowering phenotype, and the gaf1 idd1 double mutant had the late-flowering phenotype under both LD and SD conditions. If ELF3 acts as a flowering repressor downstream of GAF1 and IDD1 in the GA flowering pathway, the gaf1 idd1 elf3 triple mutant should exhibit an early flowering phenotype similar to that of the elf3 mutant. To investigate flowering phenotype-associated epistasis between the elf3 and gaf1 idd1 mutations, we determined the flowering time of elf3, gaf1 idd1, and gaf1 idd1 elf3-7. As expected, the gaf1 idd1 elf3-7 triple mutant flowered earlier than the wild-type Col-0 or gaf1 idd1 mutant (Figure 8, D and E), indicating that elf3-7 is epistatic to gaf1 idd1. This result suggests that ELF3 acted downstream of GAF1 and IDD1 in flowering. The gaf1 idd1 elf3-7 triple mutant flowered slightly later than the elf3-7 mutant (Figure 8, D and E), implying that other factors including SVP, TEM1, and TEM2 might act downstream of GAF1 and IDD1 in flowering.
ELF3 represses the expression of FT in combination with LUX and ELF4 as part of the EC
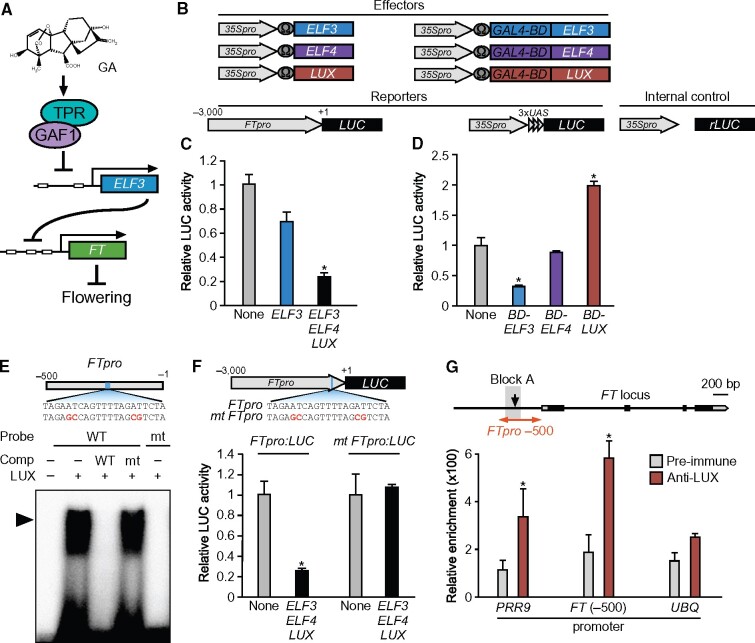
Our data indicated the possibility that ELF3 inhibits the expression of FT and that GAs activate FT via ELF3 repression induced by the GAF1-TPR complex (Figure 9, A). However, ELF3 does not have a conventional DNA-binding domain, suggesting that a DNA-binding transcription factor was involved in the ELF3-mediated transcriptional regulation of FT. ELF3 is a component of the trimeric EC, formed by interactions among ELF3, EARLY FLOWERING4 (ELF4), and LUX ARRHYTHMO (LUX) (Nusinow et al., 2011). Both elf4 and lux mutants also exhibit an early flowering phenotype (Doyle et al., 2002; Hazen et al., 2005). Because the EC acts as the transcriptional repressor complex that represses the expression of PIF4 and PIF5 (Nusinow et al., 2011), it might also repress FT. To test this possibility, we performed a transient expression assay using T87 Arabidopsis cultured cells, suggesting that the EC repressed the FT promoter (Figure 9, B and C). We further investigated the transcriptional repression activity of each EC protein in the transient expression assay. Only ELF3 possessed transcriptional repression activity, whereas ELF4 and LUX did not exhibit any repression activity in this assay (Figure 9, B and D). The LUX protein belongs to the MYB transcription factor family that binds to the GATACG consensus sequence (Helfer et al., 2011). EMSA showed that LUX binds to a GATACG-related motif in the FT promoter (−207 to −187; Figure 9, E). To determine whether the GATACG-related sequence is required for the repression of the FT promoter by the EC in vivo, we constructed a mutant FT promoter-LUC fusion, in which the LUX binding sequence was removed. The mutation in the LUX binding sequence in the FT promoter completely abolished transcriptional repression by the EC (Figure 9, F). The binding site of the EC including LUX is located within Block A in the proximal FT promoter (Figure 9, G). Since the binding site is conserved among Brassicaceae species, this FT promoter region might be required for the transcriptional repression of FT and the response to GA-dependent FT expression (Adrian et al., 2010). To test the binding of LUX protein to the FT promoter in vivo, we produced an anti-LUX antibody and performed a ChIP assay. The ChIP assay demonstrated that LUX binds to the FT promoter (−300 to −100) in plants (Figure 9, G). These data indicated that ELF3 functions as a co-repressor and that the EC interacts with the FT promoter via the LUX transcription factor to repress FT. Therefore, our results suggest that GAF1 promotes the expression of FT via transcriptional repression of ELF3, a component of the EC in GA-controlled flowering.
Figure 9.
ELF3 represses the expression of FT in combination with LUX and ELF4 as part of the EC. A, GA promotes FT expression through repression of ELF3 by the GAF1-TPR complex. B, Schematic representation of the reporter and effector. A 3,000-bp fragment of the FT promoter was fused with the LUC gene. The effector plasmid expressed full-length ELF3, ELF4, LUX, GAL4BD-ELF3 (GAL4 DNA-binding domain fused ELF3), GAL4BD-ELF4, and GAL4BD-LUX under the control of the CaMV 35S promoter with a viral translation enhancer (Ω). C, Transient expression assay of ELF3, ELF4, and LUX. The effector, reporter, and internal control constructs were co-transfected into Arabidopsis protoplasts. The transfected cells were incubated for 20 h and then Luc and rLUC activities were measured. The results are shown as LUC/rLUC activity. Error bars indicate sd of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with mock treated control (*P < 0.05). D, Transient expression assay of GAL4BD-ELF3, GAL4BD-ELF4, and GAL4BD-LUX. The effector, reporter, and internal control constructs were co-transfected into Arabidopsis protoplasts. The transfected cells were incubated for 20 h and then Luc and rLUC activities were measured. The results are shown as LUC/rLUC activity. Error bars indicate sd of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with mock treated control (*P < 0.05). E, Identification of GAF1-binding regions in the FT promoter in vitro. An EMSA was performed using the recombinant LUX protein. Oligonucleotides containing FTpro (−216 to −196, wild-type; lanes 1–4) or mutated (mt) FTpro (mt; lane5) were used as probes. Red letters indicate mutated bases. Wild-type and mt indicate competition with a 1,000-fold excess of the unlabeled wild-type and mutated probe, respectively. The specific GAF1-DNA complexes are indicated by an arrowhead. +, Addition to the reaction mixtures; –, omission from the reaction mixtures. F, Transient expression assay of ELF3, ELF4, and LUX. The effector, reporter, and internal control constructs were co-transfected into Arabidopsis protoplasts. The transfected cells were incubated for 20 h and then Luc and rLUC activities were measured. The results are shown as LUC/rLUC activity. Error bars indicate sd of three biological replicates (n = 3). Asterisks represent Student’s t test significance compared with mock treated control (*P < 0.05). G, A triangle in the schematic map indicates the LUX-binding site in the FT promoter. The 200-bp region of the FT promoter used in the ChIP assay is indicated on the right. The PRR9 promoter region was used as a positive control and UBQ11 was used as a negative control. LUX binds to a region of the FT promoter in vivo. ChIP assays were performed with pre-immune serum or anti-LUX in Col-0. The level of each co-precipitated DNA fragment was quantified by real-time PCR and normalized to the input DNA. Error bars indicate sd of three technical replicates (n = 3). Asterisks represent Student’s t test significance compared with mock treated or pre-immune control (*P < 0.05). Experiments were repeated twice with independently grown plants, with similar results.
Discussion
In Arabidopsis, GA deficient mutants exhibited non-flowering phenotypes under SD conditions, suggesting that the GA flowering pathway is important under SD conditions (Wilson et al., 1992). Endogenous GA levels increase substantially before floral initiation under SD conditions and promote flowering via the induction of two floral integrator genes, FT in the leaves and SOC1 in the shoot apex. However, initiating the GA-dependent flowering pathway requires the suppression of several transcriptional repressors that inhibit the expression of FT and SOC1. Our study demonstrated that GA promotes the expression of FT and SOC1 by suppressing a group of flowering repressors, ELF3, SVP, TEM1, and TEM2, via the GAF1-TPR complex. GUS staining analysis revealed that ELF3, SVP, and TEM1 are expressed in the leaves and that SVP, TEM1, and TEM2 are expressed in the shoot apex (Figure 6). The transition from SD to LD promotes the expression of FT by suppressing EC, TEM1, and TEM2, leading to flowering. However, to promote flowering under SD conditions, another mechanism is needed to suppress EC, TEM1, and TEM2, which maintain low FT expression levels. Our data indicate that GAs activate the expression of FT in the leaves by inducing the formation of the GAF1-TPR complex to repress ELF3, which encodes an EC component, along with TEM1 and SVP. This process ultimately facilitates the translocation of FT from the leaves to the shoot apex. In the shoot apex, GAs decreased the expression of SVP, which acts as a repressor of SOC1 (Andres et al., 2014), and promotes SOC1 expression by mediating the translocation of FT from leaves (Figure 10).
Figure 10.
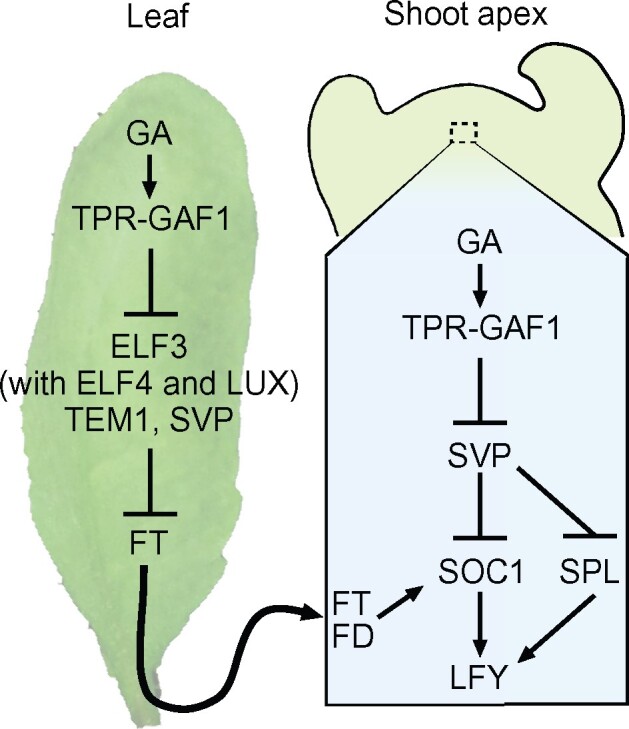
Regulation model of the GA-dependent flowering pathway by the GAF1 complex. GA promotes the induction of FT in the leaves via the suppression of ELF3, and the FT protein translocates from the leaves to the shoot apex. In the shoot apex, GA represses SVP, and SOC1 expression is induced by the translocated FT protein.
The expression level of ELF3 in the gaf1 idd1 mutant is higher than that in Col-0 under SD conditions but did not differ from that in Col-0 under LD conditions (Figure 8, B and C). These results are consistent with the phenomenon that the GA-dependent flowering pathway became more apparent under SD conditions. ELF3 overexpression represses the expression of FT (Nieto et al., 2015). In this study, we showed that ELF3 acts as a transcriptional repressor of FT in association with ELF4 and LUX. The PIF4 transcription factor mediates the thermal induction of flowering via the direct activation of FT under SD conditions (Kumar et al., 2012). ELF3 also suppresses PIF4 activity through transcriptional repression and sequestration. The transcription of PIF4 is repressed by EC during the early part of the night, and the binding of ELF3 to the PIF4 protein prevents PIF4-binding to the FT promoter (Nusinow et al., 2011; Nieto et al., 2015). Thus, ELF3 might regulate FT expression via the suppression of PIF4 activity and EC-dependent transcriptional repression under SD conditions (Figure 9, C).
The ectopic expression of GIBBERELLIN 2 OXIDASE 7 (GA2ox7), which catabolizes active GAs, in the vascular tissue and in the shoot apices using tissue-specific promoters revealed that GA-dependent flowering was regulated in two different tissues: the leaves and the shoot apices (Porri et al., 2012). Similar results were obtained when della-Δ17, the GA-insensitive constitutively active DELLA protein, was ectopically expressed in leaf or shoot apex tissues (Galvao et al., 2012). They showed that the degradation of DELLA by GAs promotes expression of FT and TWIN SISTER OF FT (TSF) in leaves and the expression of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) in the shoot apex. GAs also promote the expression of SOC1 in the shoot apex through the degradation of DELLA proteins under SD conditions (Moon et al., 2003). ELF3, TEM1, and TEM2 act as repressors of FT and SVP acts as a repressor of SOC1and SPL (Figures 7, 9; Castillejo and Pelaz, 2008; Li et al., 2008; Andres et al., 2014). These reports support the GAF1-dependent GA flowering model in which GA-induced DELLA degradation promotes flowering through the repression of GAF1 target genes, TEM1, TEM2, ELF3, and SVP, in leaves and/or shoot apices (Figure 10).
The ft-1 and soc1 mutants exhibit a low-GA sensitive phenotype rather than GA insensitive for flowering, indicating the existence of a GA-induced, FT-independent, and SOC1-independent flowering pathway (Moon et al., 2003; Hisamatsu and King, 2008). Moreover, TSF and FRUITFULL (FUL) are functionally redundant to FT and SOC1, respectively, in flowering. The ft tsf soc1 ful quadruple mutant exhibits a late-flowering phenotype, but GAs still promote flowering in this quadruple mutant, whereas the ft tsf soc1 ful svp quintuple mutant exhibits a GA-insensitive phenotype in flowering (Andres et al., 2014). These observations suggest that SVP regulates target genes other than FT, TSF, FUL, and SOC1 in GA-dependent flowering. SVP represses the expression level of SPL thorough the reduction of GAs (Andres et al., 2014). Some SPL transcription factors, including SPL3, SPL4, and SPL5, are activated in response to GAs (Galvao et al., 2012; Jung et al., 2012; Porri et al., 2012) and SPL3 binds to the LFY promoter directly (Yamaguchi et al., 2009). Our data provide a model of the GA flowering pathway by which flowering repressors are downregulated via the GAF1-TPR complex, promoting the expression of FT in leaves and that of SOC1 in the shoot apex. The expression of LFY was also increased in the GAF1 overexpressor line; GAs could induce LFY by repressing SVP via the GAF-TPR complex in the shoot apex. Previously we reported that the DELLA-GAF1 complex involved in GA feedback regulation (Fukazawa et al., 2017). Although endogenous GA levels are controlled by feedback regulation, why endogenous GAs are highly accumulated in the shoot apex before flowering under SD conditions remains unknown. Further molecular analysis is required to establish the regulation of GA accumulation in the shoot apex before flowering.
Materials and methods
Plant material and growth conditions
All transgenic lines were derived from A. thaliana ecotype Col-0 (wild-type). To generate transgenic plants overexpressing myc-tagged GAF1, a 4×myc tag was amplified and cloned into the NotI site of pBIJ4-GAF1. To generate transgenic plants, the FT, SOC1, ELF3, TEM1, TEM2, and SVP promoters were cloned into the SalI–BamHI or HindIII–BamHI site of the binary vector pBI101and pGBW3 using the Gibson Assembly system (New England Biolabs). Agrobacterium-mediated Arabidopsis transformation was carried out using the floral dip method (Clough and Bent, 1998). Kanamycin-resistant transgenic plants in the Col-0 background or Hygromycin-resistant transgenic plants in the gaf1 idd1 background were isolated and followed by selfing to obtain fully homozygous lines. The gaf1-1 (SALK_070916) and idd1 (SALK_022425) mutants were obtained from the ABRC (Ohio State University). gaf1 idd1 double mutants were generated by crossing the gaf1 and idd1 mutants, and the GAF1 overexpressor was described previously (Fukazawa et al., 2014). The primer sets used for cloning are listed in Supplemental Table S1. Plants were grown in a controlled growth chamber at 22°C under white light illumination (40 µmol m−2 s−1) and LD conditions (16 h light/8 h dark) or SD conditions (8 h light/16 h dark).
RNA-Seq library construction and sequencing
Transgenic Arabidopsis pER10:GAF1 was used for experiments. For β-estradiol treatment, 7-d-old seedlings were grown on MS agar medium with or without β-estradiol (10 µM) for 24 h under LD condition. Each 7-d-old plant was collected and then stored in −80°C until use. All RNA samples were isolated at ZT8 under LD conditions. Total RNA was isolated using the Total RNA Extraction Kit (RBC genomics) and sent to InfoBio for RNA-seq analysis. Library construction, sequencing, and basic data analysis were carried out by InfoBio. The cDNA library was prepared using the NEBNext mRNA Library Prep Master Mix Set for Illumina (New England Biolabs) in combination with a NEBNext single read run for Illumina (New England Biolabs). The quality of the RNA and fragmentation sizes was checked by InfoBio. For sequencing, two libraries were pooled for each lane of the Illumina Chip. High-throughput sequencing was performed on an Illumina HiSeq2000 platform following the manufacturer’s instructions. Mapping of the reads was performed using bowtie2 (version 2.2.2) onto the NCBI Reference sequence Database.
Short time GA treatment of plants
Fourteen-day-old seedlings of Col-0 grown on 1/2 MS agar medium were treated with 100 µM GA3 or water by spraying. Plants were grown under SD conditions. Total RNA was isolated at several time points after GA or water treatment.
Expression analysis
Total RNA was isolated using the Total RNA Extraction Kit (RBC genomics) and used for reverse transcription with Reve Tra Ace (TOYOBO) according to the manufacturer’s instructions. Quantitative real-time PCR was performed in triplicate on a CFX connect Real-Time PCR Detection System (BioRad) with GeneAce SYBR qPCR Mixα No Rox (NIPPON GENE). The relative expression levels were determined and normalized against UBQ11 or ACT2 expression. Primers used for gene expression analysis are listed in Supplemental Table S1.
GA treatment of GUS transgenic plants
Seven-day-old seedlings of transgenic plants grown on 1/2 MS agar medium were transferred to 1/2 MS agar plates with or without 10 µM GA3 to investigate GA sensitivity and grown for one additional week.
Transient transactivation assay
ELF3, TEM1, TEM2, and SVP promoters with 5′ deletions were cloned into the p-less LUC vector, which is a pUC18-based plasmid that contains the LUC gene (Takahashi et al., 1995). All primers used in this analysis are provided in Supplemental Table S1. The cDNA of GAF1, GAI, TPR4, ELF3, ELF4, TEM1, TEM2, SVP, and LUX was cloned into the pJ4 vector, which carries the CaMV 35S promoter with a viral translation enhancer, the Ω sequence (Fukazawa et al., 2000), to be used as the effector. Protoplasts were prepared from T87 Arabidopsis cultured cells, and protoplasts were transfected as described previously (Fukazawa et al., 2014). Relative LUC activity was calculated via normalization to rLUC activity. The data are presented as averages of three independent biological replicates.
Electrophoretic mobility shift assay
EMSA was performed following the procedure described previously (Fukazawa et al., 2000, 2010). GAF1 and LUX were cloned into pET30b vectors (Novagen, Madison, WI, USA). The recombinant proteins 6×His-GAF1 and 6×His-LUX were expressed and purified from Escherichia coli BL21 (DE3) pLysE using Ni+-resin (Novagen). The nucleotide sequences of the double-stranded oligonucleotides used for EMSAs are described in Supplemental Table S1. The oligonucleotides were annealed and then labeled using (α-32P) dCTP and the Klenow fragment of DNA polymerase I. Binding mixtures contained 50 fmol of the labeled probe, 1 μg of purified recombinant GAF1 or LUX, or 1 μg of control extract of E. coli, and 2 μg of poly (dI/dC). DNA competitor was used at a 100–500-fold excess molar concentration. The binding buffer for GAF1 consisted of 20 mM Tris–HCl pH 7.5, 3 mM MgCl2, 50 mM KCl, 1 mM EDTA, 10% (v/v) glycerol, and 2 μM ZnCl2. The binding buffer for LUX consisted of 10 mM Tris–HCl pH 7.0, 50 mM NaCl, 1 mM MgCl2, 1 mM TCEP, 3% glycerol, 20 µg mL−1 BSA, 2.5% CHAPS, and 1.27 mM spermidine. Reactions were incubated at 4°C for 30 min and loaded onto 4% polyacrylamide gels containing 6.7 mM Tris–HCl, pH 7.5, 1 mM EDTA, and 3.3 mM sodium acetate.
Histochemical staining
Kanamycin-resistant transgenic plants in the Col-0 background or Hygromycin-resistant transgenic plants in the gaf1 idd1 background were histochemically stained to detect GUS activity by immersing seedlings in a staining solution (100 mM sodium phosphate buffer, pH 7.0, with 50 mM NaCl, 1 mM potassium ferricyanide, 0.1% [v/v] Triton X-100, and 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide) overnight at 37°C. After staining, the samples were immersed in a fixing solution (5% [v/v] formaldehyde, 5% [v/v] acetic acid, 20% [v/v] ethanol) followed by dechlorophyllation in 70% (v/v) ethanol. Photographs of GUS-stained plants were taken using a DVM6 Digital Microscope (Leica). For sectioning, X-gluc-stained tissues were fixed and embedded in paraffin as described (Yamaguchi et al., 2001). Photographs of thin sections were taken using bright field microscopy (Nikon ECLIPSE Ni).
ChIP assay
The ChIP experiment was performed following the procedure described previously with some modifications (Fukazawa et al., 2010). Briefly, 2-week-old 4×myc-GAF1 transgenic or Col-0 plants were cross-linked in 1% (v/v) formaldehyde by vacuum filtration for 10 min and incubated at 4°C for 1 h. The aliquots of each protein sample were immunoprecipitated with anti-GST (Santa Cruz Biotechnology Inc. SC-138), anti-myc (MBL International Corporation 562/lot#055), and anti-LUX (generated in-house) for 12 h at 4°C. Chromatin–antibody complexes were precipitated with salmon sperm DNA/protein-G Dyna beads at 4°C for 2 h. The primers used for ChIP analysis are listed in Supplemental Table S1. The level of each co-precipitated DNA fragment was quantified by real-time PCR using specific primer sets and normalized to input DNA. The levels of co-precipitated pre-immune serum or anti-GST antibody (immunoprecipitated DNA/input DNA) were set to 1. The results are shown as relative DNA enrichment. Error bars indicate sd ranges (n = 3).
Statistical analysis
All data for quantification analyses are presented as means (±sd). The statistical analyses were performed by two-tailed Student’s t test (*P < 0.05). Statistical results are shown in Supplemental Data set S2.
Accession numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: GAF1 (At3g50700), IDD1 (At5g66730), GAI (At1g14920), AtGA20ox2 (At5g51810), ACTIN2 (At3g18780), TEM1 (At1G25560), TEM2 (At1G68840), ELF3 (At2g25930), ELF5 (At5g62640), SVP (At2g22540), FT (At1g65480), SOC1 (At2g45660), LFY (At5g61850), EFM (At2g03500), and UBQ11 (At4g05050).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of novel GAF1-target genes by transactivation assays with the DELLA-GAF1 complex.
Supplemental Figure S2. GA responsibility of GAF1 target genes by shorter time GA treatment.
Supplemental Table S1. Primer sequences used in this study
Supplemental Data set S1. RNA-seq data of GAF1-induced plant.
Supplemental Data set S2. Statistical analysis results.
Supplementary Material
Acknowledgments
The authors thank Dr. Tsuyoshi Nakagawa (Shimane University) for providing the pGBW3 vector and Dr. Nam-Hai Chua (Rockefeller University) for providing the pER10 vector. They also thank Dr. Belay T. Ayele (University of Manitoba, Canada) for helpful comments on the manuscript. Finally they thank Misaki Kohsaka and Yu Morimoto for technical assistance.
Funding
This study was supported in part by grants from the Japan Society for the Promotion of Science (JSPS) to J.F. (17K07449 and 20K06688) and Y.T. (15H04392) and by grants from the Sumitomo Foundation to J.F. (180382).
Conflict of interest statement. None declared.
J.F. and Y.T. designed the research and wrote the manuscript. J.F., Y.O., R.T., and K.N. performed the experiments.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the instructions for Authors (https://academic.oup.com/plcell) is: Jutarou Fukazawa (jutarouf@hirosima-u.ac.jp)
References
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F (2010) cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasino R (2004) Vernalization, competence, and the epigenetic memory of winter. Plant Cell 16: 2553–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres F, Porri A, Torti S, Mateos J, Romera-Branchat M, Garcia-Martinez JL, Fornara F, Gregis V, Kater MM, Coupland G (2014) SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc Natl Acad Sci USA 111: E2760–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Ahn JH, Weigel D (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33: 168–171 [DOI] [PubMed] [Google Scholar]
- Boden SA, Weiss D, Ross JJ, Davies NW, Trevaskis B, Chandler PM, Swain SM (2014) EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell 26: 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V (1998) The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Eriksson S, Bohlenius H, Moritz T, Nilsson O (2006) GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Nakata M, Ito T, Yamaguchi S, Takahashi Y (2010) The transcription factor RSG regulates negative feedback of NtGA20ox1 encoding GA 20-oxidase. Plant J 62: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Fukazawa J, Mori M, Watanabe S, Miyamoto C, Ito T, Takahashi Y (2017) DELLA-GAF1 complex is a main component in gibberellin feedback regulation of GA20 oxidase 2. Plant Physiol 175: 1395–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y (2000) Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Teramura H, Murakoshi S, Nasuno K, Nishida N, Ito T, Yoshida M, Kamiya Y, Yamaguchi S, Takahashi Y (2014) DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell 26: 2920–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvao VC, Horrer D, Kuttner F, Schmid M (2012) Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139: 4072–4082 [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Salter MG, Thingnaes E, Whitelam GC (2003) Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 33: 875–885 [DOI] [PubMed] [Google Scholar]
- Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, Huijser P (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Hauvermale AL, Ariizumi T, Steber CM (2012) Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol 160: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA (2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamatsu T, King RW (2008) The nature of floral signals in Arabidopsis. II. Roles for FLOWERING LOCUS T (FT) and gibberellin. J Exp Bot 59: 3821–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Ju Y, Seo PJ, Lee JH, Park CM (2012) The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J 69: 577–588 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee I (2010) Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot 61: 2247–2254 [DOI] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Matias-Hernandez L, Aguilar-Jaramillo AE, Marin-Gonzalez E, Suarez-Lopez P, Pelaz S (2014) RAV genes: regulation of floral induction and beyond. Ann Bot 114: 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Nieto C, Lopez-Salmeron V, Daviere JM, Prat S (2015) ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr Biol 25: 187–193 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, Kay SA (2011) The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osnato M, Castillejo C, Matias-Hernandez L, Pelaz S (2012) TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun 3: 808. [DOI] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G (2012) Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Singleton WR (1946) Inheritance of indeterminate growth in maize. J Heredity 37: 61–64 [DOI] [PubMed] [Google Scholar]
- Sun TP (2011) The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 21: R338–345 [DOI] [PubMed] [Google Scholar]
- Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Ann Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Sakai T, Ishida S, Nagata T (1995) Identification of auxin-responsive elements of parB and their expression in apices of shoot and root. Proc Natl Acad Sci USA 92: 6359–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D (2009) The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y, Sun T (2001) Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J 28: 443–453 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.