This review discusses and illustrates nine extracellular strategies that plant pathogens and symbionts use to avoid the recognition by cell surface receptors.
Abstract
Recognition of microbe-associated molecular patterns (MAMPs) by cell-surface receptors is pivotal in host-microbe interactions. Both pathogens and symbionts establish plant-microbe interactions using fascinating intricate extracellular strategies to avoid recognition. Here we distinguish nine different extracellular strategies to avoid recognition by the host, acting at three different levels. To avoid the accumulation of MAMP precursors (Level 1), microbes take advantage of polymorphisms in both MAMP proteins and glycans, or downregulate MAMP production. To reduce hydrolytic MAMP release (Level 2), microbes shield MAMP precursors with proteins or glycans and inhibit or degrade host-derived hydrolases. And to prevent MAMP perception directly (Level 3), microbes degrade or sequester MAMPs before they are perceived. We discuss examples of these nine strategies and envisage three additional extracellular strategies to avoid MAMP perception in plants.
Introduction
Plants are a rich source of nutrients for many organisms (Cardinale et al., 2011). Their root and aerial systems are exposed to a wide range of microbes including bacteria, fungi, and oomycetes. Some of these microbes are plant pathogens that cause detrimental effects on plant fitness (Dangl and Jones, 2001; Dodds and Rathjen, 2010), while symbiotic microbes can promote plant growth, such as rhizobacteria and arbuscular mycorrhizal fungi (Pérez-de-Luque et al., 2017).
When microbes enter the extracellular space within plant tissues (the apoplast), they interact with host cells that carry surface receptors that recognize conserved molecules, conventionally called microbe-associated molecular patterns (MAMPs, Jones and Dangl, 2006). MAMPs are fragments of proteins or glycans that are essential for the biology of microbes and are absent in the host plant. Examples are fragments of flagellin and peptidoglycans (PGN) from bacteria and fragments of chitin and β-glucans from fungi and oomycetes. These MAMPs are recognized on the host cell surface by pattern recognition receptors (PRRs). PRRs are transmembrane receptor-like kinases (RLKs) or receptor-like proteins (RLPs) that often carry extracellular leucine-rich repeat (LRR) or lysine motif (LysM) domains to confer MAMP recognition (Couto and Zipfel 2016; Tang et al., 2017; Boutrot and Zipfel, 2017; Schellenberger et al., 2019). PRR activation triggers a series of immune responses (Bigeard et al., 2015), resulting in pattern-triggered immunity (PTI) and preventing colonization by nonadapted microbes (Bigeard et al., 2015; Andersen et al., 2018). Importantly, many MAMPs require hydrolytic release from their precursors before they can be perceived by PRRs. For instance, hydrolytic release from precursors is required for the recognition of flagellin and elongation factor Tu (EF-Tu), and for the recognition of chitin and β-glucan.
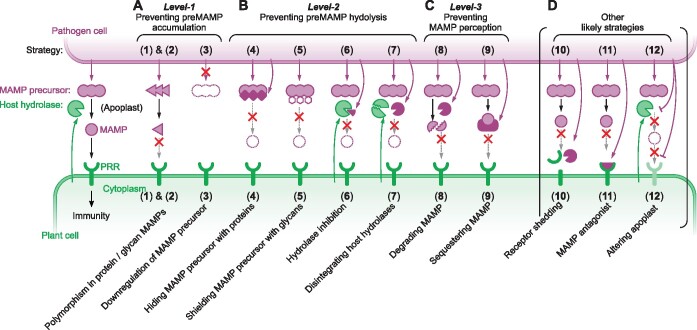
To colonize plants, adapted pathogens and symbionts have adopted strategies to avoid recognition by the immune system. Many effector proteins that act inside the host cell interfere in signaling downstream of PRRs (Toruño et al., 2016). In this review, however, we describe nine extracellular strategies that plant pathogens and symbionts use to avoid recognition by PRRs (Table 1). These strategies occur at three levels: MAMP production (Figure 1A), MAMP release (Figure 1B), and MAMP perception (Figure 1C).
Table 1.
Overview of strategies employed by microbes to evade MAMP recognition in plants
| Microbe | MAMP or MAMP precursor | Microbial protein | Plant host | Host target (PRR) | References | ||
|---|---|---|---|---|---|---|---|
| Level 1: Preventing MAMP production | |||||||
| Strategy 1: Polymorphisms in protein MAMPs | |||||||
| Bacteria | Agrobacterium tumefaciens | flg22 | N/A | Arabidopsis | (FLS2) | Felix et al. (1999) | |
| Ralstonia solanacearum | Arabidopsis | Pfund et al. (2004); Wei et al. (2018) | |||||
| Xanthomonas campestris pv. campestris | Arabidopsis | Sun et al. (2006) | |||||
| Xanthomonas oryzae pv. and pv. oryzicola | rice | Wang et al. (2015) | |||||
| Xanthomonas arboricola pv. juglandis | Juglans regia | Cesbron et al. (2015) | |||||
| Sinorhizobium meliloti | tomato, rice | Felix et al. (1999) | |||||
| Burkholderia phytofirmans | grapevine | Trdá et al. (2014) | |||||
| Pseudomonas syringae pv. tomato Col338 | flgII-28 | tomato | (FLS3) | Cai et al. (2011) | |||
| Xanthomonas campestris pv. campestris and Pseudomonas syringae pv. tomato | elf18 | N/A | Arabidopsis | (EFR) | Lacombe et al. (2010) | ||
| Xanthomonas oryzae pv. oryzae | RaxX | N/A | rice | (Xa21) | Pruitt et al. (2015) | ||
| Strategy 2: Polymorphisms in glycan MAMPs | |||||||
| Fungi | Verticillium dahliae | Chitin | VdPDA1 | cotton | (CERK1) | Gao et al. (2019) | |
| Puccinia striiformis f. sp. tritici | Pst13661 | wheat | Xu et al. (2020) | ||||
| Puccinia graminis f. sp. tritici | N/A | wheat | El Gueddari et al. (2002) | ||||
| Uromyces fabae | N/A | broad bean | |||||
| Colletotrichum graminicola | N/A | maize | |||||
| Pestalotiopsis sp. | PesCDA | rice | Cord-Landwehr et al. (2016) | ||||
| Strategy 3: Downregulating MAMP production | |||||||
| Bacteria | Pseudomonas | Flagellin | cdG | N/A | (FLS2) | Hickman and Harwood (2008) | |
| Pseudomonas syringae |
N. benthamiana and Arabidopsis |
Pfeilmeier et al. (2016) | |||||
| Pseudomonas aeruginosa | |||||||
| Pseudomonas fluorescens | |||||||
| Pseudomonas syringae pv. tomato | AlgU | N. benthamiana | Bao et al. (2020) | ||||
| Pseudomonas syringae pv. syringae B728a | fliC | bean | Yu et al. (2013) | ||||
| Fungi | Colletotrichum graminicola | β-glucan | KRE5 and KRE6 | maize | N/A | Oliveira‐Garcia and Deising (2016) | |
| Strategy 4: Hiding MAMP precursors with proteins | |||||||
| Fungi | Cladosporium fulvum | Chitin | Avr4 | tomato | (CERK1) | Van den Burg et al. (2006) | |
| Verticillium nonalfalfae | VnaChtBP | hop (Humulus lupulus L.) | Volk et al. (2019) | ||||
| Verticillium dahliae strain VdLs17 | Vd2LysM | tomato | Kombrink et al. (2017) | ||||
| Colletotrichum higginsianum | ChELP1 and ChELP2 | monocot and dicot crops | Takahara et al. (2016) | ||||
| Parastagonospora nodorum | SnTox1 | wheat | Liu et al. (2016) | ||||
| Clonostachys rosea | Cell wall | LysM1 and LysM2 | wheat | Dubey et al. (2020) | |||
| Serendipita indica | β-glucan | FGB1 | Arabidopsis, barley, and N. benthamiana | Wawra et al. (2016) | |||
| Strategy 5: Shielding MAMP precursors with glycans | |||||||
| Bacteria | Pseudomonas syringae pv. tabaci 6605 | Flagellin |
flagellin glycan |
tobacco | N/A | Taguchi et al. (2006) | |
| Pseudomonas syringae pv. glycinea race 4 | soybean | Takeuchi et al. (2003) | |||||
| Xanthomonas campestris pv. campestris XcA | cabbage | Ichinose et al. (2013) | |||||
| Acidovorax avenae K1 strain | rice | Hirai et al. (2011) | |||||
| Pseudomonas syringae pv. syringae B728a | N. benthamiana | Buscaill et al. (2019) | |||||
| Xylella fastidiosa | LPS | LPS glycan | grapevine | N/A | Rapicavoli et al. (2018) | ||
| Pseudomonas syringae pv. syringae B728a | bean | Helmann et al. (2019) | |||||
| Fungi | Magnaporthe oryzae | Chitin | α-1,3-glucan | rice | N/A | Fujikawa et al. (2009) | |
| Cochlioborus miyabeanus | Fujikawa et al. (2012) | ||||||
| Rhizoctonia solani | |||||||
| Strategy 6: Blocking MAMP release by inhibiting the activity of host hydrolases | |||||||
| Oomycete | Phytophthora sojae | β-glucan | GIP1 | soybean | EGaseA | Rose et al. (2002) | |
| Phytophthora infestans | GIPs | tomato | EGases | Damasceno et al. (2008) | |||
| Phytophthora infestans | PC2 | EPIs | tomato | P69B | Wang et al. (2021) | ||
| Bacteria | Pseudomonas syringae pv. tomato | Flagellin | galactosyrin | N. benthamiana |
BGAL1 (FLS2) |
Buscaill et al. (2019) | |
| Strategy 7: Disintegrating host-derived hydrolases | |||||||
| Fungi | Verticillium dahlia | Chitin | SSEP1 | cotton | Chi28 | Han et al. (2019) | |
| Fusarium oxysporum f. sp. lycopersici | FoMep1/Sep1 | tomato | Chi1 and Chi13 | Jashni et al. (2015) | |||
| Fusarium verticillioides | fungalysin | maize | ChitA and ChitB | Naumann et al. (2011) | |||
| Colletotrichum graminicola | Cgfl | maize | chitinases | Sanz‐Martín et al. (2016) | |||
| Colletotrichum cinereus | fungalysin | N/A | N/A | Lilly et al. (2008) | |||
| Ustilago maydis | UmFly1 | maize | ZmChiA | Ökmen et al. (2018) | |||
| Level 3: Preventing MAMP perception | |||||||
| Strategy 8: Degrading MAMPs | |||||||
| Bacteria |
Pseudomonas aeruginosa and Pseudomonas syringae pv. tomato |
Flagellin/flg22 | AprA | Arabidopsis | (FLS2) | Bardoel et al. (2011) | |
| Fungi | Podosphaera xanthii | Chitin | EWCAs | melon | (CERK1) | Martínez-Cruz et al. (2021) | |
| Strategy 9: Sequestering released MAMPs | |||||||
| Fungi | Cladosporium fulvum | Chitin | Ecp6 | tomato | (CERK1) | De Jonge et al. (2010) | |
| Mycosphaerella graminicola | Mg3LysM | wheat | Marshall et al. (2011) | ||||
| Magnaporthe oryzae | Slp1 | rice | Mentlak et al. (2012) | ||||
| Verticillium dahliae strain VdLs17 | Vd2LysM | tomato | Kombrink et al. (2017) | ||||
| Colletotrichum higginsianum | ChELP1 and ChELP2 | monocot and dicot crops | Takahara et al. (2016) | ||||
| Rhizoctonia solani | RsLysM | sugar beet | Dölfors et al. (2019) | ||||
| Trichoderma atroviride | Tal6 | Arabidopsis and tomato | Romero-Contreras et al. (2019) | ||||
| Rhizophagus irregularis | RiSLM | Medicago truncatula | Zeng et al. (2020) | ||||
| Moniliophthora perniciosa | MpChi | cacao | Fiorin et al. (2018) | ||||
| Magnaporthe oryzae | MoChia1 | rice | Yang et al. (2019) | ||||
| Bacteria | Bacillus subtilis | Flagellin | BSn5 | Arabidopsis and voodoo lily | (FLS2) | Deng et al. (2019) | |
See Supplemental Data Set S1 for a version of this table in excel format.
N/A, nonapplicable.
Figure 1.
Nine known and three possible strategies to evade extracellular recognition in plants. Illustration of microbial strategies that evade extracellular recognition (A) by preventing the accumulation of MAMP precursors (Level 1, Strategies 1–3); (B) by preventing the hydrolysis of MAMP precursors (Level 2, Strategies 4–7); and (C) by preventing MAMP perception (Level 3, Strategies 8 and 9). (D) Other possible but as yet unreported strategies. MAMP precursors, MAMPs, and other microbial molecules are colored in purple, and host hydrolases and receptors in green. Secretion by microbe and plant are represented by arrows in purple and green, respectively.
Level 1: Three strategies Preventing MAMP production
We distinguish three different strategies that microbes use to prevent the accumulation of MAMP precursors (Level 1, Figure 1A). The first strategy is to accumulate mutations in protein-based MAMPs to avoid recognition of the MAMP fragment. The second strategy is a similar genetic adaptation to alter glycan-based MAMPs beyond recognition. The third strategy is to downregulate the accumulation of MAMP precursors upon infection. The latter can involve transcription factors, epigenetic regulation, and posttranscriptional control. Specific examples of the three strategies are illustrated in Figure 2.
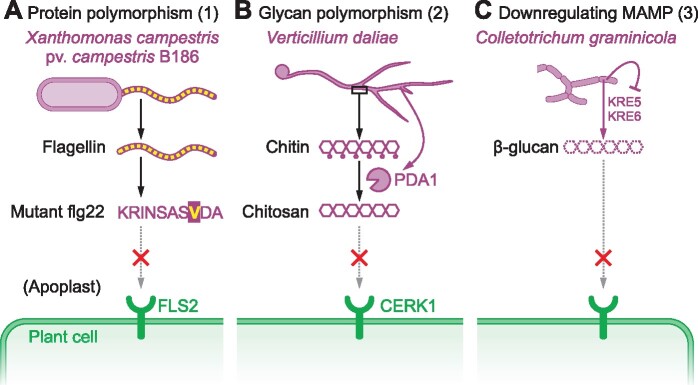
Figure 2.
Three mechanisms to avoid MAMP accumulation (Level 1). A, Example of Strategy 1: The bacterial rice pathogen X. campestris pv. campestris B186 avoids the recognition of its flagellin through an amino acid substitution in the flg22 sequence. B, Example of Strategy 2: the fungal cotton pathogen V. dahliae avoids recognition of chitin with secreted polysaccharide deacetylase-1 (PDA1), which converts chitin into chitosan. C, Example of Strategy 3: The fungal maize pathogen C. graminicola downregulates the expression of KRE5 and KRE6, which encodes two biosynthesis enzymes required for β-glucan biosynthesis.
Strategy 1: Polymorphisms in protein MAMPs
Polymorphisms in MAMP protein sequences are a frequently used strategy to avoid detection. Sequence polymorphisms have been described for MAMPs from bacterial flagellin, EF-Tu, and RaxX.
In most angiosperms, recognition of bacterial flagellin is mediated by Flagellin Sensing 2 (FLS2), a PRR with extracellular LRRs (Gómez-Gómez and Boller, 2000). FLS2 recognizes peptides from a highly conserved 22-amino acids region in the N-terminal domain of flagellin, called flg22 (Felix et al., 1999; Zipfel et al., 2004). However, some flagellated bacteria carry flg22 sequences that are not recognized by FLS2. flg22 peptides from most ε-, δ-, and α-proteobacteria induce moderate, weak, or no response, respectively, in contrast to flg22 peptides from the majority of γ- and β-proteobacteria, which trigger strong responses (Cheng et al., 2020). For instance, flagellin of the crown gall disease pathogen Agrobacterium (Agrobacterium tumefaciens, an α-proteobacterium) possesses a different flg22 sequence that does not trigger FLS2-mediated immune responses in most plant species including Arabidopsis (Arabidopsis thaliana; Felix et al., 1999). However, some γ- and β-proteobacteria also evade FLS2 recognition with sequence polymorphism. For example, the bacterial wilt pathogen Ralstonia solanacearum (a β-proteobacterium) and some strains of Xanthomonas campestris pathovar (pv.) campestris (Xcc; a γ-proteobacterium) have nonrecognizable versions of the flg22 sequence that evade FLS2-mediated defenses in their respective hosts (Pfund et al., 2004; Sun et al., 2006; Wei et al., 2018). Specifically, an aspartate (D) to valine (V) substitution at amino acid position 14 within the flg22 sequence of flagellin from Xcc results in reduced immune responses associated with increased virulence of Xcc on Arabidopsis (Figure 2A;Sun et al., 2006). Similarly, X. oryzae pv. oryzae (Xoo) and pv. oryzicola (Xoc) evade rice (Oryza sativa) FLS2-mediated recognition with substitutions in the flg22 sequence (Wang et al., 2015). Consistent with selection for immune evasion, nonpathogenic strains of X. arboricola pv. juglandis carry the conserved flg22 sequence whereas pathogenic strains carry polymorphisms within the flg22 sequence that evades recognition by FLS2 (Cesbron et al., 2015). Evasion of flagellin recognition by flg22 polymorphisms is also observed with symbiotic bacteria. For instance, the plant beneficial endophytic bacterium Burkholderia phytofirmans (a β-proteobacterium) and the plant beneficial bacterium Sinorhizobium meliloti (an α-proteobacterium) carry flg22 sequences with weak elicitor activity in grapevine (Vitis vinifera) and with no elicitor activity, respectively (Felix et al., 1999; Trdá et al., 2014).
Flagellin is also recognized by FLAGELLIN-SENSING 3 (FLS3), which is present only in some solanaceous plant species including some cultivars of tomato (Solanum lycopersicum), potato (S. tuberosum), and pepper (Capsicum annuum; Hind et al., 2016). FLS3 recognizes flgII-28, a 28-amino acid peptide from the central region of the flagellin protein. Interestingly, polymorphisms within flgII-28 sequences are observed between strains of Pseudomonas syringae pv. tomato (Pto; a γ-proteobacterium). Flagellin of the Col338 strain of Pto contains a valine (V) residue at position 13 in the flgII-28 sequence, and this peptide triggers a weaker immune response in tomato than the flgII‐28 peptide from the PtoT1 strain, which carries an alanine (A) residue at this position (Cai et al., 2011).
Evasion of recognition caused by flagellin polymorphisms has also been described for animal pathogens. For instance, the human pathogenic bacteria Campylobacter jejuni, Helicobacter pylori, and Bartonella bacilliformis evade flagellin recognition by Toll-Like Receptor 5 (TLR5) with mutations within the key recognition sites of the flagellin N-terminal D1 domain (Andersen-Nissen et al., 2005).
Besides flagellin, evasion with protein polymorphism has also been reported for peptides containing the first 18 amino acids of bacterial Elongation Factor Thermal unstable (EF-Tu), called elf18, which is recognized in Arabidopsis by the EF-Tu Receptor (EFR; Kunze et al., 2004; Zipfel et al., 2006). Polymorphism within the elf18 sequence correlates with different elicitation activity. For instance, elf18 peptides from Xcc and PtoDC3000 trigger only 0.8%–3.2% of the immune activity as compared to peptides from Agrobacterium, Ralstonia, and other Xanthomonas and Pseudomonas strains (Lacombe et al., 2010).
Another bacterial MAMP with protein polymorphism is the tyrosine-sulfated peptide RaxX, which is perceived by the rice immune receptor XA21 (Pruitt et al., 2015; Luu et al., 2019). XA21 confers resistance to most strains of X. oryzae pv. oryzae (Xoo; Wang et al., 1996). However, Xoo isolates carrying nonsynonymous substitutions of tyrosine residue Y41 in RaxX evade XA21-mediated immunity (Pruitt et al., 2015).
In conclusion, protein MAMP polymorphism is an efficient and frequently used strategy to evade host immunity, employed by both pathogenic and symbiotic bacteria. The polymorphisms in MAMPs highlight the exceptional genetic plasticity associated with host adaptation of bacteria. However, amino acid sequence polymorphism is of course restricted by protein function. Flagellin, for instance, cannot accept many substitutions in the flg22 sequence without affecting flagellin function because this region acts as a hinge that facilitates important conformational changes in the flagellin structure when reversing the spin of flagellar rotation (Fliegmann and Felix, 2016; Wang et al., 2017). And obviously, while evasion of immunity by MAMP polymorphism is relevant for protein-based MAMPs, different strategies are needed for nonproteinaceous MAMPs.
Strategy 2: Polymorphisms in glycan MAMPs
Glycan polymorphism is the second strategy used by microorganisms to evade host immunity. This strategy is illustrated here by the deacetylation of chitin by fungi.
Many fungi evade host immunity by deacetylating chitin into chitosan. Chitin is a structural element of fungal cell walls and chitin fragments are conserved MAMPs that are almost universally recognized in the plant kingdom by Chitin Elicitor Receptor Kinase 1 (CERK1) as a signature of fungal invasion (Pusztahelyi, 2018). Chitosan, however, induces a weaker immune response than chitin, so the deacetylation of chitin is frequently used by plant-associated fungi to avoid recognition (Vander et al., 1998). The soil-borne pathogenic fungus Verticillium dahliae, for example, secretes Polysaccharide Deacetylase 1 (VdPDA1) to deacetylate chitin oligomers and prevent chitin-triggered immunity in cotton (Gossypium hirsutum) plants (Figure 2B; Gao et al., 2019). Fusarium oxysporum f. sp. vasinfectum PDA1 is also required for virulence during wilt disease in cotton (Gao et al., 2019). Similarly, the wheat stripe rust fungus Puccinia striiformis f. sp. tritici suppresses chitin‐induced plant defense by secreting the Polysaccharide Deacetylase Pst13661 (Xu et al., 2020). The wheat (Triticum aestivum) stem rust fungus P. graminis f. sp. tritici, the broad bean (Vicia faba) rust fungus Uromyces fabae, and the maize (Zea mays) anthracnose fungus Colletotrichum graminicola also use chitosan instead of chitin in their hyphae (El Gueddari et al., 2002). Likewise, the endophytic fungus Pestalotiopsis sp. avoids recognition by secreting chitin deacetylases (PesCDA; Cord-Landwehr et al., 2016). Chitosan is also produced by human pathogens to evade immunity. The human fungal pathogen Cryptococcus neoformans, for instance, secretes chitin deacetylase CnCda4 to further deacetylate chitosans that are already partially deacetylated by other chitin deacetylases to evade host immunity (Hembach et al., 2020).
In conclusion, the evolution of glycoforms on the exposed portion of microbes is an efficient strategy to dampen the host immune response. More examples of microbial glycan polymorphisms that evade immunity remain to be discovered. For instance, bacterial pathogens of animals modify the structure of their peptidoglycans (PGNs) through deacetylation and thus evade the antibacterial activity of lysozyme and delay pro-inflammatory immune responses (Boneca et al., 2007; Shimada et al., 2010; Wolf et al., 2011; Grifoll-Romero et al., 2019). However, PGN modification remains to be described for plant pathogens.
Strategy 3: Downregulating MAMP production
Another microbial strategy to evade host detection is to reduce the amount of MAMPs by downregulating their biosynthesis during infection. This strategy has been described for both bacteria and fungi.
Pathogenic, opportunistic, and commensal bacteria downregulate flagellin biosynthesis during infection. Biosynthesis of flagella in Pseudomonas is downregulated by the second messenger cyclic‐di‐GMP (cdG; Hickman and Harwood, 2008). Elevated cdG levels in the plant pathogen P. syringae, the plant opportunist P. aeruginosa and the plant commensal P. fluorescens reduce flagellin levels, and thus contribute to the evasion of FLS2‐mediated immune response in Nicotiana benthamiana and Arabidopsis (Pfeilmeier et al., 2016). Flagellin protein levels are also downregulated in PtoDC3000 via reduction in flagellin expression by the gene expression regulator AlgU to avoid host immune responses (Bao et al., 2020).
Downregulation of flagellin genes is also observed upon entry of P. syringae pv. syringae B728a (PsyB728a) into the leaf of bean (Phaseolus vulgaris) plants (Yu et al., 2013). Comparison of the global transcriptome profiling of PsyB728a in epiphytic and apoplastic sites reveals that the mean induction level of genes related to flagellar biosynthesis and motility was >4.5-fold greater on leaf surface than in planta (Yu et al., 2013). Regarding the underlying sensory mechanism to downregulate flagellin expression, Escherichia coli downregulates flagellar genes in response to immobilization with anti-flagellar antibodies (Cullender et al., 2013), indicating that flagellin downregulation occurs when bacteria are immobilized.
In fungal pathogens, downregulation of β-glucan biosynthesis reduces exposure to glycan MAMPs. For instance, during the biotrophic phase of infection, the fungal maize pathogen C. graminicola downregulates the expression of genes encoding Killer toxin resistant 5 (KRE5) and KRE6, which are key enzymes for the biosynthesis of β‐glucan (Figure 2C;Oliveira‐Garcia and Deising, 2016). However, KRE5 and KRE6 expressions are indispensable for the formation of appressoria and necrotrophic hyphae. Consistent with a need for β-glucan, KRE genes are also required for full virulence of fungal human pathogens C. neoformans and Candida albicans (Herrero et al., 2004; Gilbert et al., 2010).
In conclusion, downregulating MAMP precursor levels is a common strategy used by both fungi and bacteria to avoid host detection. Obviously, this strategy is only beneficial for the microbe when production of the MAMP precursor is not required for full virulence. For instance, bacterial proliferation and spread do not rely on flagellin after host entry and the altered fungal cell wall composition may no longer need β-glucans upon host entry.
Level 2: Preventing MAMP release
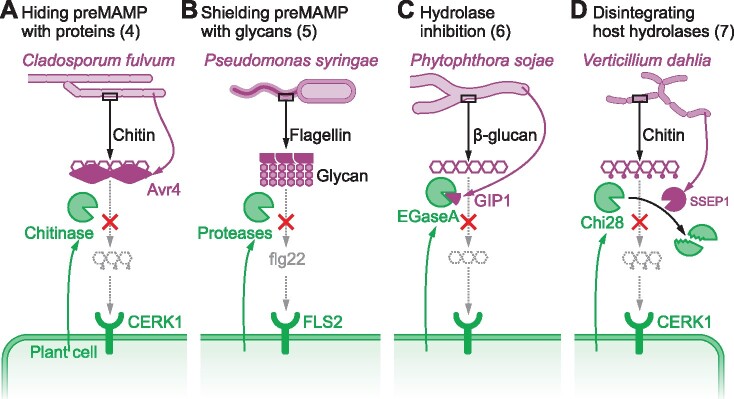
MAMPs discussed in this section are released from microbes by host-secreted hydrolases, such as chitinases and proteases. We distinguish four strategies to block MAMP release from microbes (Figure 1B). MAMP precursors are protected against host hydrolases by microbial proteins and glycans, and host hydrolases are also inhibited and disintegrated. Specific examples of the four strategies are illustrated in Figure 3.
Figure 3.
Four mechanisms to avoid hydrolytic MAMP release (Level 2). A, Example of Strategy 4: the fungal tomato pathogen C. fulvum prevents the release of chitin fragments by secreting Avirulence protein-4 (Avr4) to hide chitin in the cell wall from hydrolysis by plant-secreted chitinases. B, Example of Strategy 5: the bacterial tobacco (N. tabacum) pathogen Pseudomonas syringae pv. tabaci 6605 prevents the proteolytic release of the flagellin elicitor flg22 by carrying a glycan covering the flagellin polymer. C, Example of Strategy 6: The oomycete soybean pathogen Phytophthora sojae prevents the release of β-glucan-based elicitors by secreting glucanase inhibiting protein-1 (GIP1), which inhibits the plant-secreted endoglucanase EGaseA. D, Example of Strategy 7: the fungal cotton pathogen V. dahlia prevents the release of immunogenic chitin fragments by Secreting Serine Protease (SSEP1), which inactivates host-secreted chitinase Chi28.
Strategy 4: Hiding MAMP precursors with proteins
Microbial organisms can evade host recognition by secreting proteins that cover MAMP precursors to prevent hydrolytic release of MAMPs. There are seven examples of this strategy, involving structurally unrelated secreted proteins used by both pathogenic and symbiotic fungi.
The tomato leaf mold fungus (Cladosporium fulvum syn. Passalora fulva) produces Avr4, a member of Carbohydrate-binding module family 14 (CBM14). Avr4 specifically binds to chitin in the fungal cell wall to protect it against plant chitinases, which release chitin elicitors (Van den Burg et al., 2006; Figure 3A). Homologs of Avr4 are present in other pathogenic fungi, indicating a similar protection of chitin cell walls by other fungi (Stergiopoulos et al., 2010).
Likewise, the hemibiotrophic xylem-invading fungus V. nonalfalfae prevents chitin hydrolysis by secreting VnaChtBP, a CBM18 protein that binds chitin and suppresses chitin-triggered host immunity (Volk et al., 2019). VnaChtBP is present in 28 V. nonalfalfae isolates, suggesting a high evolutionary stability and testifying its importance for the fungal lifestyle.
Similarly, the fungal vascular wilt pathogen V. dahliae strain VdLs17 secretes the lineage‐specific LysM effector Vd2LysM, a CBM50 protein (Akcapinar et al., 2015), which binds chitin, suppresses chitin‐induced immune responses, and protects fungal hyphae against hydrolysis by plant hydrolytic enzymes (Kombrink et al., 2017). Likewise, the causative fungus of anthracnose diseases, C. higginsianum, produces the extracellular LysM proteins 1 and 2 (ChELP1 and ChELP2), which bind chitin and prevent chitin-triggered immunity (Takahara et al., 2016).
The wheat Septoria nodorum blotch (SNB) pathogen Parastagonospora nodorum secretes SnTox1, a protein that also binds chitin and protects the pathogen from wheat chitinases (Liu et al., 2016). Interestingly, SnTox1 also induces host cell death, supporting the necrotrophic lifestyle of this pathogen (Liu et al., 2012).
Plant beneficial fungi that are parasites of pathogenic fungi also avoid plant immune response by covering MAMPs with proteins. For example, the mycoparasite Clonostachys rosea (sin. Gliocladium roseum) produces CBM50 members LysM1 and LysM2 to protect hyphae against chitinases to prevent MAMP-induced defenses during wheat root colonization by its host F. graminearum (Dubey et al., 2020).
Fungi also prevent MAMP release by hiding β-glucans with proteins. The root endophyte Serendipita indica (Si, syn. Piriformospora indica), avoids recognition of its β‐glucan by secreting a fungal-specific β-glucan-binding lectin, Fungal Glucan-Binding 1 (FGB1). SiFGB1 binds β-glucan to reduce β-glucan-triggered immunity in several host plants, including Arabidopsis, barley (Hordeum vulgare), and N. benthamiana (Wawra et al., 2016).
In conclusion, covering MAMP precursors with proteins to prevent their hydrolysis is a mechanism used by many fungal pathogens and symbionts. Remarkably, several structurally unrelated carbohydrate-binding proteins (Avr4, ChtBP, Vd2LysM, ChELP1 and ChELP2, SnTox1, LysM2, and FGB1) have convergently evolved to protect fungal hyphae from hydrolysis by plant chitinases and glucanases, which would otherwise release MAMP from their precursors. In addition to preventing MAMP release, these proteins can also promote virulence by strengthening the cell wall.
Strategy 5: Shielding MAMP precursors with glycans
Glycosylation of MAMP precursors to shield them from hydrolytic release of MAMPs is the fifth strategy to evade host recognition. For instance, glycosylation of bacterial flagellin and fungal chitin suppress MAMP release.
O-glycosylation of flagellin is very common in bacteria, including important plant pathogens from the genera Xanthomonas, Pseudomonas, Burkholderia, Dickeya, Erwinia, Pantoea, and Pectobacterium (Taguchi et al., 2010; Ichinose et al., 2013; De Maayer and Cowan, 2016). This flagellin glycosylation is currently thought to prevent the hydrolytic release of the flagellin MAMP (Figure 3B). For instance, glycosylation mutants of P. syringae pv. tabaci 6605, P. syringae pv. glycinea race 4 and X. campestris pv. campestris XcA lacking the flagellin glycosyltransferase (FGT1) are less virulent on tobacco, soybean (Glycine max), and cabbage (Brassica oleracea), respectively (Takeuchi et al., 2003; Taguchi et al., 2006; Ichinose et al., 2013). Flagellin glycosylation is also important for Acidovorax avenae, a Gram-negative bacterial pathogen causing leaf blight in rice. Flagellin isolated from the avirulent N1141 strain induces immune responses, whereas flagellin from the virulent K1 strain does not. These flagellin protein sequences are identical but their glycosylation pattern is different: strain K1 carries a 2,150-Da O-glucan while strain N1141 harbors a 1,600-Da O-glycan (Hirai et al., 2011). Thus, glycosylation avoids flagellin recognition, presumably by preventing the hydrolytic release of immunogenic flagellin fragments.
Consistent with shielding glycans, plant‐secreted β-galactosidase 1 (BGAL1) acts in immunity by promoting the release of immunogenic peptides from glycosylated flagellin of PtoDC3000 and P. syringae pv. tabaci 6605 (Pta6605), which both carry a terminal-modified viosamine (mVio) sugar on flagellin O-glycans (Buscaill et al., 2019). BGAL1 is not required to release the flagellin MAMP from the Δfgt1 mutant of Pta6605, which produces nonglycosylated flagellin. Interestingly, pv. syringae B728a (PsyB728a) evades host immunity by having O-glycans on flagellin that are resistant to hydrolysis by BGAL1 (Buscaill et al., 2019), even though PsyB728a has an flg22 sequence recognized by FLS2 (Segonzac et al., 2011). Importantly, mVio biosynthesis genes are absent from PsyB728a, which instead carries a putative (1,2)-linked terminal GlcNac (N-acetylglucosamine) on its O-glycan (Yamamoto et al., 2011; Chiku et al., 2013). This suggests that different glycoforms on flagellin are required for the colonization of different hosts and that hosts may use different glycosidases to release flagellin MAMPs.
Bacteria also use polymorphic lipopolysaccharides (LPSs) to evade immunity. LPSs are the major component of the outer membrane of Gram-negative bacteria and consist of lipid A, a di-glucosamine carrying four to seven fatty acids, and an oligosaccharide core region that carries an O-polysaccharide (OPS) comprising a variable number of oligosaccharide repeats. OPS composition is highly diverse among bacterial species and strains. Glycans covering bacterial LPS may alter host responses. For instance, the plant pathogenic bacterium Xylella fastidiosa possesses a long chain O-antigen that delays recognition by the host plant (Rapicavoli et al., 2018). Mutant X. fastidiosa lacking these O-antigens induces faster immune responses (Rapicavoli et al., 2018). In addition, genes that encode glycosyltransferase domains and cause strong virulence phenotypes when disrupted in PsyB728a are suspected to be involved in the biosynthesis of O-antigens that decorates LPS (Helmann et al., 2019).
Glycosylation of fungal cell walls is also used to prevent MAMP release. For instance, the rice blast fungus Magnaporthe oryzae, the rice brown spot fungus Cochlioborus miyabeanus, and the rice sheath blight fungus Rhizoctonia solani accumulate α-1,3-glucan on the surface of infectious hyphae (Fujikawa et al., 2009, 2012). Fungal mutants with reduced α-1,3-glucan levels have reduced virulence, indicating that α-1,3-glucan may mask chitin and β-glucans in the fungal cell wall to shield it against hydrolytic MAMP release (Fujikawa et al., 2012). The large phylogenetic distance between these ascomycete and basidiomycete rice pathogens indicates that this strategy is a widespread mechanism that may be used by fungal pathogens to evade host innate immunity.
Glycan shielding of MAMP precursors has also been described for human pathogens, including viruses, fungi, and bacteria, as a strategy to evade recognition by the host immune system (Walls et al., 2016; Hernández-Chávez et al., 2017; Poole et al., 2018; PradHan et al., 2019). For instance, flagellin glycosylation reduces recognition of human bacterial pathogens, providing an evasive strategy for P. aeruginosa (Arora et al., 2005) and B. cenocepacia (Hanuszkiewicz et al., 2014). In conclusion, glycan shielding of MAMP precursors is a common strategy used by many pathogens to enhance infection. These findings predict that many more examples of glycan shielding will be discovered for plant pathogens and symbionts.
Strategy 6: Blocking MAMP release by inhibiting the activity of host hydrolases
The inhibition of MAMP-releasing hydrolases is another strategy used by plant pathogens. Examples of this strategy have been identified in oomycetes and bacterial pathogens.
Phytophthora species secrete glucanase inhibitor proteins (GIPs) during invasion of their hosts, which themselves inhibit MAMP release through extracellular Endo-β-1,3-Glucanases (EGases). For example, P. sojae secretes GIP1 to inhibit soybean EGaseA, thereby preventing EGaseA-mediated release of elicitor-active glucan oligosaccharides (Figure 3C; Rose et al., 2002). Analysis of tomato leaves inoculated with P. infestans showed that P. infestans GIPs and tomato EGases form stable complexes in the apoplast (Damasceno et al., 2008), indicating that GIPs-mediated inhibition of EGases to prevent MAMP release is a common strategy used by Phytophthora in different hosts.
Phytophthora species also produce Kazal-like Extracellular Protease Inhibitors (EPIs) during infection. P. infestans EPI1 and EPI10 inhibit the secreted subtilisin-like protease P69B of tomato (Tian et al., 2004; Tian et al., 2005). P69B releases a fragment from the apoplastic effector PC2 of P. infestans that then triggers immune responses and a hypersensitive response (HR) in solanaceous plants (Wang et al., 2021). Thus, EPI1 might suppress PC2-elicited host immunity by inhibiting the protease that releases the elicitor.
Bacterial pathogens also produce hydrolase inhibitors to prevent MAMP release. For instance, PtoDC3000 produces the small molecule BGAL1 inhibitor galactosyrin (Buscaill et al., 2019). BGAL1 promotes the release of MAMPs from glycosylated flagellin carrying mVio on their O-glycan (see Strategy 5). BGAL1 is suppressed by galactosyrin during infection to prevent the release of immunogenic peptide from flagellin (Buscaill et al., 2019).
GIP1, EPI1, and galactosyrin are just the first examples of pathogen-derived inhibitors targeting MAMP-releasing host hydrolases. Further studies on widespread pathogen-derived inhibitors will probably uncover many more examples. However, in addition to preventing MAMP release, these inhibitors also protect the physical integrity of flagellin and the microbial cell wall by preventing their degradation.
Strategy 7: Disintegrating host-derived hydrolases
Destruction of MAMP-releasing host hydrolases is the seventh strategy used by invading microorganisms to evade immunity. We currently know three unrelated classes of proteases from fungal pathogens that disintegrate host chitinases to prevent MAMP release.
The root-infecting fungal pathogen V. dahlia produces Secreted Serine Protease 1 (SSEP1, family S8) during the invasion of cotton root cells to hydrolyze cotton Chitinase 28 (Chi28; Figure 3D; Han et al., 2019). Likewise, the vascular wilt pathogen of tomato, F. oxysporum f. sp. lycopersici, secretes family-M36 metalloprotease fungalysin FoMep1 and family-S8 subtilisin-like protease FoSep1 to remove the extracellular chitin-binding domain (CBD) from tomato chitinases SlChi1 and SlChi13 (Jashni et al., 2015). Removal of the CBD significantly reduces chitinase and antifungal activity, thereby playing a pivotal role in virulence of F. oxysporum. Similarly, the fungal pathogens F. verticillioides, C. graminicola, Coprinopsis cinerea, and Ustilago maydis also secrete family-M36 metalloprotease fungalysins to cleave plant chitinases (Lilly et al., 2008; Naumann et al., 2011; Sanz‐Martín et al., 2016; Ökmen et al., 2018). Interestingly, animal fungal pathogens also produce fungalysin during infection, presumably with the same objective (Li and Zhang, 2014).
In conclusion, SSEP1, FoSep1, FoMep1, and other fungalysins represent different protease families that cleave plant chitinases to prevent MAMP release. The phylogenetic distance between these fungi and proteases indicates that the inactivation of chitinases evolved convergently. Besides preventing MAMP release, these proteases also protect the physical integrity of the cell wall by preserving chitin.
Level 3: Preventing MAMP perception
Once MAMPs are released, we know two effective strategies that prevent the perception of MAMPs by PRRs (Figure 1C). One strategy degrades MAMPs before they reach their receptors, the other strategy sequesters MAMPs before they are perceived. Specific examples of the two strategies are illustrated in Figure 4.
Figure 4.
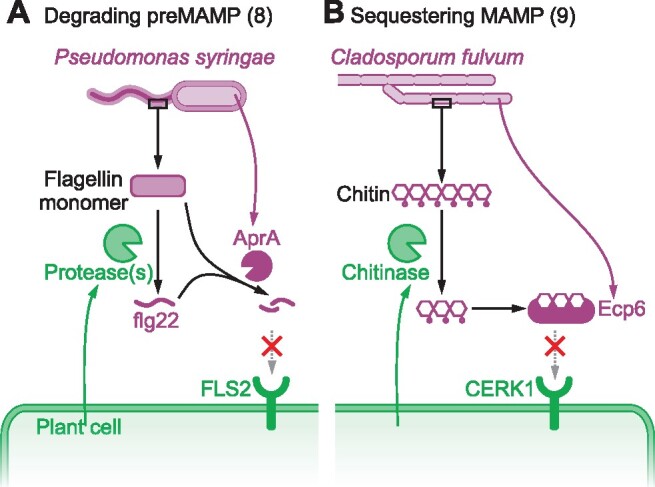
Two mechanisms to prevent MAMP perception (Level 3). A, Example of Strategy 8: the bacterial plant pathogen Pseudomonas syringae prevents the accumulation of immunogenic flagellin fragments by secreting the metalloprotease AprA, which can cleave both the flagellin monomer and the flg22 elicitor peptide. B, Example of Strategy 9: The fungal tomato pathogen C. fulvum prevents the accumulation of immunogenic chitin fragments by secreting Extracellular cysteine-rich protein-6 (Ecp6), which sequesters chitin fragments.
Strategy 8: Degrading MAMPs
The eighth strategy used by pathogens is based on the targeted degradation of released MAMPs by pathogen-derived proteases. Examples are the bacterial effectors AprA and LasB and fungal effectors with chitinase activity (EWCAs), explained below.
Pseudomonas species secrete alkaline protease ArpA, a 50-kD zinc metalloprotease (MEROPS family M10 of clan MA), which prevents flagellin-triggered immune responses by degrading flagellin monomers and flg22 (Figure 4A; Bardoel et al., 2011). Flagellin polymers resist degradation by AprA and this preserves the integrity of the flagellum (Bardoel et al., 2011). In Arabidopsis, AprA prevents flg22- and flagellin-induced immune responses and delays stomatal closure. AprA of PtoDC3000 is required for its full virulence on both Arabidopsis and tomato. Interestingly, AprA is widespread among human- and plant-pathogenic bacteria, including the bacterial plant pathogen P. syringae and human pathogen P. aeruginosa, suggesting a conserved infection mechanism among bacteria.
In addition to AprA, P. aeruginosa secretes a second zinc metalloprotease, the 33-kD pseudolysin LasB (MEROPS family M4 of clan MA), which also degrades flagellin and acts in concert with AprA to prevent flagellin-mediated immune recognition (Casilag et al., 2016). The production of two different proteases targeting flagellin monomers likely provides P. aeruginosa with robust immune suppression mechanisms.
Fungal pathogens also degrade MAMPs during infection. The cucurbit powdery mildew fungus Podosphaera xanthii releases effectors with chitinase activity (EWCAs) when penetrating melon (Cucumis melo) plant cells to degrade immunogenic chitin oligomers and thereby prevents the activation of chitin-triggered immunity (Martínez-Cruz et al., 2021). Remarkably, EWCA homologs are also widely distributed in plant fungal pathogens but also in fungi that are pathogens of insects, nematodes, and animals, suggesting a conserved infection mechanism among fungi (Martínez-Cruz et al., 2021).
In conclusion, the degradation of MAMPs is an efficient mechanism to avoid recognition but only a few of these proteases have been identified. Additional pathogen-produced proteases that promote virulence (Chandrasekaran et al., 2016; Figaj et al., 2019) may also act by degrading MAMPs. Likewise, pathogen-secreted glycosidases may degrade glycan-based MAMPs. For instance, the human pathogen Histoplasma capsulatum secretes endo-β-1,3-glucanase Eng1 to evade host detection (Garfoot et al., 2016) and many plant pathogens having glycan-based MAMPs secrete glycosidases (Ospina-Giraldo et al., 2010; Murphy et al., 2011; Vermassen et al., 2019; McGowan et al., 2020). Notably, MAMP degradation must require fine regulation of these hydrolases to avoid unspecific or premature degradation of MAMP precursors or inadvertent MAMP release.
Strategy 9: Sequestering released MAMPs
Elicitor sequestration is the ninth strategy used by plant pathogens to evade recognition. In this strategy, pathogen-secreted proteins bind released elicitors to prevent them from binding host receptors. Many fungi secrete proteins to sequester chitin elicitors.
During infection, C. fulvum secretes Extracellular protein 6 (Ecp6), a LysM-containing protein of the CBM50 family that binds chitin fragments. Ecp6 binds these elicitors with greater affinity than the chitin receptor, so C. fulvum evades chitin recognition by using Ecp6 to sequester chitin fragments, (Figure 4B; De Jonge et al., 2010). The widespread presence of Ecp6 orthologs suggests that this is a common strategy of many pathogenic fungi to avoid host recognition (De Jonge and Thomma, 2009). Indeed, the fungal wheat pathogen Mycosphaerella graminicola produces Mg3LysM, an Ecp6 homolog, that plays a major role in pathogen virulence on wheat plants by preventing the elicitation of chitin-induced immunity (Marshall et al., 2011). Similarly, the rice blast fungus M. oryzae produces Secreted LysM protein-1 (Slp1), which binds chitin fragments and prevents chitin-triggered immunity (Mentlak et al., 2012). Interestingly, Vd2LysM from V. dahlia and ChELP1 and ChELP2 from C. higginsianum, also sequester chitin oligomers and additionally protect chitin polymers against chitinases (Strategy 4; Takahara et al., 2016; Kombrink et al., 2017).
Symbiotic fungi also use LysM proteins to establish compatible interactions. The endophytic fungus Trichoderma atroviride and the arbuscular mycorrhiza fungus Rhizophagus irregularis, for instance, produce the LysM proteins Tal6 and RiSLM, respectively, to evade extracellular recognition (Romero-Contreras et al., 2019; Zeng et al., 2020). Surprisingly, even necrotrophic fungi use LysM proteins to evade immunity. For instance, the necrotrophic fungus R. solani, which kills seedlings and causes root rot in a broad range of plant species, secretes RsLysM, which associates with chitin oligomers to prevent early chitin perception during sugar beet (Beta vulgaris) colonization (Dölfors et al., 2019).
LysM proteins are also used by animal fungal pathogens and contribute to fungal virulence during host invasion. The insecticidal fungus Beauveria bassiana secretes the two LysM effectors Blys2 and Blys5 that bind chitin and prevent the activation of immunity in insects (Cen et al., 2017).
Sequestration of chitin fragments is also achieved through different evolutionary paths. The cacao (Theobroma cacao) witches broom disease Moniliophthora perniciosa produces an enzymatically inactive chitinase (MpChi) that prevents chitin-triggered immunity by sequestering chitin fragments (Fiorin et al., 2018). Remarkably, its sister species M. roreri encodes a second, nonorthologous catalytically inactive chitinase (MrChi). MpChi and MrChi are both highly expressed during the biotrophic phase of infection. Despite lacking chitinolytic activity, both proteins sequester immunogenic chitin fragments. Similarly, the fungal rice pathogen M. oryzae secretes Chitinase 1 (MoChia1) that binds chitin and can prevent immune responses (Yang et al., 2019).
Bacteria also use the sequestration strategy by targeting MAMP precursors. For instance, the endophyte bacterium Bacillus subtilis BSn5 enhances its colonization of Arabidopsis and voodoo lily (Amorphophallus konjac) through minimizing the stimulation of flg22-induced defense by producing the antimicrobial peptide (lantibiotic) subtilomycin, which binds to its own flagellin (Deng et al., 2019). The presence of subtilomycin biosynthesis genes in genomes of other bacteria suggests that flagellin sequestration is a common strategy of endophytic bacteria to adapt to endosphere niches (Deng et al., 2019).
MAMP sequestration is common to fungal and bacterial microbes, but more details remain to be discovered in other invading microorganisms. The success of this strategy depends on the competition between the microbial-derived sequestering protein and the host PRR.
Concluding remarks and perspectives
Successful plant-associated microbes evade extracellular detection by the host immune system. While certain immune evasion mechanisms are used by microbes from diverse kingdoms, other mechanisms have so far only been described for certain microbes. However, it seems unlikely that these strategies are pathogen-specific, prompting us to expect that this is likely to change with further development of the field.
For most of the nine strategies described above, suppression MAMP perception also results in increased stability of the MAMP precursor. It has therefore not always been robustly demonstrated that the observed enhanced virulence associated with the strategy is caused by evading MAMP recognition or by increased stability of the MAMP precursor. A good way to investigate this experimentally would be to test for enhanced virulence in the absence of the PRR, as this would indicate an important role in the stabilization of the MAMP precursor.
We can think of at least three additional extracellular strategies that would prevent MAMP recognition (Figure 1D). First, MAMP recognition can be blocked by receptor shedding. This has been described for animal pathogens, but not for plant pathogens. For example, the fungal respiratory pathogen Coccidioides posadasii secretes the Metalloproteinase Mep1 during endospore differentiation. Mep1 digests the Spherule Outer Wall glycoprotein (SOWgp), resulting in the prevention of host recognition mediated by this receptor (Hung et al., 2005). Similarly, the opportunistic pathogen of human lungs P. aeruginosa secretes the metalloproteinase LasB, which cleaves the human urokinase-type Plasminogen Activator Receptor (uPAR) through domain-specific endoproteolysis (Leduc et al., 2007). There have been no reports of PRR inactivation by shedding in plant-microbe interactions yet. By contrast, ectodomain shedding of the legume Symbiosis Receptor Kinase (SYMRK) causes the formation of a signaling complex with Nod Factor Receptor 5 (NFR5, Antolín-Llovera et al., 2014).
A second possible strategy is the use of antagonists. MAMP antagonists would bind PRRs and prevent the binding of MAMPs to their respective receptors and therefore inhibit PRR function. For instance, C-terminal truncations of the flagellin flg22 elicitor can block flg22 perception by FLS2 in tomato (Meindl et al., 2000), but the existence and use of MAMP antagonists during infection remains to be reported.
A third possible strategy is to alter the apoplastic conditions such that MAMP release and/or perception is inhibited. Although this mechanism has not yet been demonstrated, the regulation of hydrolases by pH and redox status would offer opportunities for pathogens to interfere in MAMP release. Interestingly, several plant pathogens secrete homologs of plant regulatory peptides to suppress host immunity. For instance, the fungal wilt pathogen F. oxysporum secretes a functional homolog of rapid alkalinization factors (RALFs), peptide hormones that trigger an increase of apoplastic pH and enhances fungal colonization (Masachis et al., 2016; Thynne et al., 2017).
In conclusion, the extracellular detection of MAMPs by plants is an active and exciting research field. The presence of many MAMPs, hydrolytic enzymes, and hydrolase inhibitors implicate a large and mostly unexplored area of research, still holding most of its secrets. Increased understanding of this extracellular battlefield of both animal and plant pathogens will ultimately translate into new strategies for the prevention of infectious diseases.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Data Set S1. Overview of strategies employed by microbes to evade MAMP recognition in plants.
Supplementary Material
Acknowledgments
We thank our colleagues from the Plant Chemetics laboratory for useful comments and suggestions.
Funding
This work was financially supported by Consolidator grant 616447 ‘GreenProteases’ from the European Research Council and Biotechnology and Biological Sciences Research Council BBSRC grant BB/R017913/1.
Conlict of interest statement. The authors declare no competing interests.
The authors wrote the article together. The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Renier A. L. van der Hoorn (renier.vanderhoorn@plants.ox.ac.uk).
References
- Akcapinar GB, Kappel L, Sezerman OU, Seidl-Seiboth V (2015) Molecular diversity of LysM carbohydrate-binding motifs in fungi. Curr Genet 61:103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen EJ, Ali S, Byamukama E, Yen Y, Nepal MP (2018) Disease resistance mechanisms in plants. Genes 9:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, Aderem A (2005) Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci USA 102:9247–9252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antolín-Llovera M, Ried MK, Parniske M (2014) Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr Biol 24:422–427 [DOI] [PubMed] [Google Scholar]
- Arora SK, Neely AN, Blair B, Lory S, Ramphal R (2005) Role of motility and flagellin glycosylation in the pathogenesis of Pseudomonas aeruginosa burn wound infections. Infect Immun 73:4395–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Wei HL, Ma X, Swingle B (2020) Pseudomonas syringae AlgU downregulates flagellin gene expression, helping evade plant immunity. J Bacteriol 202:e00418–e00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardoel BW, van der Ent S, Pel MJ, Tommassen J, Pieterse CM, van Kessel KP, van Strijp JA (2011) Pseudomonas evades immune recognition of flagellin in both mammals and plants. PLoS Pathog 7:e1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8:521–539 [DOI] [PubMed] [Google Scholar]
- Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, Psylinakis E, Bouriotis V, Hugot JP, Giovannini M, et al . (2007) A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci USA 104:997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Zipfel C (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol 55:257–286 [DOI] [PubMed] [Google Scholar]
- Buscaill P, Chandrasekar B, Sanguankiattichai N, Kourelis J, Kaschani F, Thomas EL, Morimoto K, Kaiser M, Preston GM, Ichinose Y. et al. (2019) Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 364:145. [DOI] [PubMed] [Google Scholar]
- Cai R, Lewis J, Yan S, Liu H, Clarke CR, Campanile F, Almeida NF, Studholme DJ, Lindeberg M, Schneider D, et al . (2011) The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog 7:e1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale BJ, Matulich KL, Hooper DU, Byrnes JE, Duffy E, Gamfeldt L, Balvanera P, O'Connor MI, Gonzalez A (2011) The functional role of producer diversity in ecosystems. Am J Bot 98:572–592 [DOI] [PubMed] [Google Scholar]
- Casilag F, Lorenz A, Krueger J, Klawonn F, Weiss S, Häussler S (2016) The LasB Elastase of Pseudomonas aeruginosa acts in concert with alkaline protease AprA to prevent flagellin-mediated immune recognition. Infect Immun 84:162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen K, Li B, Lu Y, Zhang S, Wang C (2017) Divergent LysM effectors contribute to the virulence of Beauveria bassiana by evasion of insect immune defenses. PLoS Pathog 13:e1006604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesbron S, Briand M, Essakhi S, Gironde S, Boureau T, Manceau C, Fischer-Le Saux M, Jacques MA (2015) Comparative genomics of pathogenic and nonpathogenic strains of Xanthomonas arboricola unveil molecular and evolutionary events linked to pathoadaptation. Front. Plant Sci 6:1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran M, Thangavelu B, Chun SC, Sathiyabama M (2016) Proteases from phytopathogenic fungi and their importance in phytopathogenicity. JGeneral Plant Pathol 82: 233–239 [Google Scholar]
- Cheng JHT, Bredow M, Monaghan J, DiCenzo GC (2020) Proteobacteria encode diverse flg22 peptides that elicit varying immune responses in Arabidopsis thaliana. Mol Plant-Microbe Interact. 10.1094/MPMI-11-20-0314-SC [DOI] [PubMed] [Google Scholar]
- Chiku K, Yamamoto M, Ohnishi-Kameyama M, Ishii T, Yoshida M, Taguchi F, Ichinose Y, Ono H (2013) Comparative analysis of flagellin glycans among pathovars of phytopathogenic Pseudomonas syringae. Carbohydr Res 375:100–104 [DOI] [PubMed] [Google Scholar]
- Cord-Landwehr S, Melcher RL, Kolkenbrock S, Moerschbacher BM (2016) A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci Rep 6:38018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16:537–552 [DOI] [PubMed] [Google Scholar]
- Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA, et al . (2013) Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 14:571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damasceno CM, Bishop JG, Ripoll DR, Win J, Kamoun S, Rose JK (2008) Structure of the glucanase inhibitor protein (GIP) family from phytophthora species suggests coevolution with plant endo-beta-1,3-glucanases. Mol Plant Microbe Interact 21:820–830 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833 [DOI] [PubMed] [Google Scholar]
- De Jonge R, Thomma B.P.H.J. (2009) Fungal LysM effectors: extinguishers of host immunity? Trends Microbiol. 17:151–157 [DOI] [PubMed] [Google Scholar]
- De Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R, van der Krol S, Shibuya N, Joosten MH, Thomma BP (2010) Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329:953–955 [DOI] [PubMed] [Google Scholar]
- De Maayer P, Cowan DA (2016) Comparative genomic analysis of the flagellin glycosylation island of the Gram-positive thermophile Geobacillus. BMC Genomics 17:913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Chen H, Li C, Xu J, Qi Q, Xu Y, Zhu Y, Zheng J, Peng D, Ruan L. et al. (2019) Endophyte Bacillus subtilis evade plant defense by producing lantibiotic subtilomycin to mask self-produced flagellin. Commun Biol 2:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11:539–548 [DOI] [PubMed] [Google Scholar]
- Dubey M, Vélëz H, Broberg M, Jensen DF, Karlsson M (2020) LysM proteins regulate fungal development and contribute to hyphal protection and biocontrol traits in Clonostachys rosea. Front Microbiol 11:679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölfors F, Holmquist L, Dixelius C, Tzelepis G (2019) A LysM effector protein from the basidiomycete Rhizoctonia solani contributes to virulence through suppression of chitin-triggered immunity. Mol Genet Genomics 294:1211–1218 [DOI] [PubMed] [Google Scholar]
- El Gueddari NE, Rauchhaus U, Moerschbacher BM, Deising HB (2002) Developmentally regulated conversion of surface‐exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol 156:103–112 [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18:265–276 [DOI] [PubMed] [Google Scholar]
- Figaj D, Ambroziak P, Przepiora T, Skorko-Glonek J (2019) The role of proteases in the virulence of plant pathogenic bacteria. Int J Mol Sci 20:672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorin GL, Sanchéz-Vallet A, Thomazella DPT, do Prado PFV, do Nascimento LC, Figueira AVO, Thomma BPHJ, Pereira GAG, Teixeira PJPL (2018) Suppression of plant immunity by fungal chitinase-like effectors. Curr Biol 28:3023–3030 [DOI] [PubMed] [Google Scholar]
- Fliegmann J, Felix G (2016) Immunity: flagellin seen from all sides. Nat Plants 2: 16136. [DOI] [PubMed] [Google Scholar]
- Fujikawa T, Kuga, Y, Yano S, Yoshimi A, Tachiki T, Abe K, Nishimura M (2009) Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol 73: 553–570 [DOI] [PubMed] [Google Scholar]
- Fujikawa T, Sakaguchi A, Nishizawa Y, Kouzai Y, Minami E, Yano S, Koga H, Meshi TNishimura M (2012) Surface α-1,3-glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathog 8: e1002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Zhang BS, Zhao JH, Huang JF, Jia PS, Wang S, Zhang J, Zhou JM, Guo HS (2019) Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat Plants 5:1167–1176 [DOI] [PubMed] [Google Scholar]
- Garfoot AL, Shen Q, Wüthrich M, Klein BS, Rappleye CA (2016) The Eng1 β-glucanase enhances histoplasma virulence by reducing β-glucan exposure. mBio 7:e01388–e–01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert NM, Donlin MJ, Gerik KJ, Specht CA, Djordjevic JT, Wilson CF, Sorrell TC, Lodge JK (2010) KRE genes are required for beta-1,6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans. Mol Microbiol 76:517–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoll-Romero L, Sainz-Polo MA, Albesa-Jové D, Guerin ME, Biarnés X, Planas A (2019) Structure-function relationships underlying the dual. J Biol Chem 294:19066–19080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5:1003–1011 [DOI] [PubMed] [Google Scholar]
- Han LB, Li YB, Wang FX, Wang WY, Liu J, Wu JH, Zhong NQ, Wu SJ, Jiao GL, Wang HY. et al. (2019) The cotton apoplastic protein CRR1 stabilizes Chitinase 28 to facilitate defense against the fungal pathogen. Plant Cell 31:520–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanuszkiewicz A, Pittock P, Humphries F, Moll H, Rosales AR, Molinaro A, Moynagh PN, Lajoie GA, Valvano MA (2014) Identification of the flagellin glycosylation system in Burkholderia cenocepacia and the contribution of glycosylated flagellin to evasion of human innate immune responses. J Biol Chem 289:19231–19244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann TC, Deutschbauer AM, Lindow SE (2019) Genome-wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast. Proc Natl Acad Sci USA 116:18900–18910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hembach L, Bonin M, Gorzelanny C, Moerschbacher BM (2020) Unique subsite specificity and potential natural function of a chitosan deacetylase from the human pathogen. Proc Natl Acad Sci USA 117:3551–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero AB, Magnelli P, Mansour MK, Levitz SM, Bussey H, Abeijon C (2004) KRE5 gene null mutant strains of Candida albicans are avirulent and have altered cell wall composition and hypha formation properties. Eukaryot Cell 3:1423–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Chávez MJ, Pérez-García LA, Niño-Vega GA, Mora-Montes HM (2017) Fungal Strategies to evade the host immune recognition. J Fungi 3:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JW, Harwood CS (2008) Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hind SR, Strickler SR, Boyle PC, Dunham DM, Bao Z, O'Doherty IM, Baccile JA, Hoki JS, Viox EG, Clarke CR, et al . (2016) Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat Plants 2:16128. [DOI] [PubMed] [Google Scholar]
- Hirai H, Takai R, Iwano M, Nakai M, Kondo M, Takayama S, Isogai A, Che FS (2011) Glycosylation regulates specific induction of rice immune responses by Acidovorax avenae flagellin. J Biol Chem 286:25519–25530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, Seshan KR, Yu JJ, Schaller R, Xue J, Basrur V, Gardner MJ, Cole GT (2005) A metalloproteinase of Coccidioides posadasii contributes to evasion of host detection. Infect Immun 73:6689–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose Y, Taguchi F, Yamamoto M, Ohnishi-Kameyama M, Atsumi T, Iwaki M, Manabe H, Kumagai M, Nguyen QT, Nguyen CL, et al . (2013) Flagellin glycosylation is ubiquitous in a broad range of phytopathogenic bacteria. J General Plant Pathol 79:359–365 [Google Scholar]
- Jashni MK, Dols IH, Iida Y, Boeren S, Beenen HG, Mehrabi R, Collemare J, De Wit PJGM (2015) Synergistic action of a metalloprotease and a serine protease from Fusarium oxysporum f. sp. lycopersici cleaves chitin-binding tomato chitinases, reduces their antifungal activity, and enhances fungal virulence. Mol Plant-Microbe Interact 28:996–1008 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329 [DOI] [PubMed] [Google Scholar]
- Kombrink A, Rovenich H, Shi‐Kunne X, Rojas‐Padilla E, Van den Berg GCM, Domazakis E, de Jonge R, Valkenburg DJ, Sánchez‐Vallet A, Seidl MF. et al. (2017) Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts. Mol Plant Pathol 18:596–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N-terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16:3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BP, Staskawicz B, et al . (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol 28:365–369 [DOI] [PubMed] [Google Scholar]
- Leduc D, Beaufort N, de Bentzmann S, Rousselle JC, Namane A, Chignard M, Pidard D (2007) The Pseudomonas aeruginosa LasB metalloproteinase regulates the human urokinase-type plasminogen activator receptor through domain-specific endoproteolysis. Infect Immun 75:3848–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang KQ (2014) Independent expansion of zincin metalloproteinases in Onygenales fungi may be associated with their pathogenicity. PLoS One 9:e90225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly WW, Stajich JE, Pukkila PJ, Wilke SK, Inoguchi N, Gathman AC (2008) An expanded family of fungalysin extracellular metallopeptidases of Coprinopsis cinerea. Mycol Res 112:389–398 [DOI] [PubMed] [Google Scholar]
- Liu Z, Gao Y, Kim YM, Faris JD, Shelver WL, De Wit PJGM, Xu SS, Friesen TL (2016) SnTox1, a Parastagonospora nodorum necrotrophic effector, is a dual-function protein that facilitates infection while protecting from wheat-produced chitinases. New Phytol 211:1052–1064 [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhang Z, Faris JD, Oliver RP, Syme R, McDonald MC, McDonald BA, Solomon PS, Lu S, Shelver WL, et al . (2012) The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harbouring Snn1. PLoS Pathog 8:e1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu DD, Joe A, Chen Y, Parys K, Bahar O, Pruitt R, Chan LJG, Petzold CJ, Long K, Adamchak C, et al . (2019) Biosynthesis and secretion of the microbial sulfated peptide RaxX and binding to the rice XA21 immune receptor. Proc Natl Acad Sci USA 116:8525–8534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R, Kombrink A, Motteram J, Loza-Reyes E, Lucas J, Hammond-Kosack KE, Thomma BP, Rudd JJ (2011) Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol 156:756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masachis S, Segorbe D, Turrà D, Leon-Ruiz M, Fürst U, El Ghalid M, Leonard G, López-Berges MS, Richards TA, Felix G, Di Pietro A (2016) A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat Microbiol 1:16043. [DOI] [PubMed] [Google Scholar]
- McGowan J, O'Hanlon R, Owens RA, Fitzpatrick DA (2020) comparative genomic and proteomic analyses of three widespread. Microorganisms 8:653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl T, Boller T, Felix G (2000) The bacterial elicitor flagellin activates its receptor in tomato cells according to the address-message concept. Plant Cell 12:1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentlak TA, Kombrink A, Shinya T, Ryder LS, Otomo I, Saitoh H, Terauchi R, Nishizawa Y, Shibuya N, Thomma BP. et al. (2012) Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 24:322–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cruz J, Romero D, Hierrezuelo J, Thon M, de Vicente A, Pérez-García A (2021) Effectors with chitinase activity (EWCAs), a family of conserved, secreted fungal chitinases that suppress chitin-triggered immunity. Plant Cell https://doi.org/10.1093/plcell/koab011 [DOI] [PubMed] [Google Scholar]
- Murphy C, Powlowski J, Wu M, Butler G, Tsang A (2011) Curation of characterized glycoside hydrolases of fungal origin. Database 2011:bar020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann TA, Wicklow DT, Price NP (2011) Identification of a chitinase-modifying protein from Fusarium verticillioides: truncation of a host resistance protein by a fungalysin metalloprotease. J Biol Chem 286:35358–35366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ökmen B, Kemmerich B, Hilbig D, Wemhöner R, Aschenbroich J, Perrar A, Huesgen PF, Schipper K, Doehlemann G (2018) Dual function of a secreted fungalysin metalloprotease in Ustilago maydis. New Phytol 220:249–261 [DOI] [PubMed] [Google Scholar]
- Oliveira-Garcia E, Deising HB (2016) Attenuation of PAMP-triggered immunity in maize requires down-regulation of the key β-1,6-glucan synthesis genes KRE5 and KRE6 in biotrophic hyphae of Colletotrichum graminicola. Plant J 87:355–375 [DOI] [PubMed] [Google Scholar]
- Ospina-Giraldo MD, Griffith JG, Laird EW, Mingora C (2010) The CAZyome of Phytophthora spp.: a comprehensive analysis of the gene complement coding for carbohydrate-active enzymes in species of the genus Phytophthora. BMC Genomics 11:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeilmeier S, Saur IM, Rathjen JP, Zipfel C, Malone JG (2016) High levels of cyclic-di-GMP in plant-associated Pseudomonas correlate with evasion of plant immunity. Mol Plant Pathol 17:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfund C, Tans-Kersten J, Dunning FM, Alonso JM, Ecker JR, Allen C, Bent AF (2004) Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol Plant-Microbe Interact 17: 696–706 [DOI] [PubMed] [Google Scholar]
- Poole J, Dayr CJ, von Itzstein M, Paton JC, Jennings MP (2018) Glycointeractions in bacterial pathogenesis. Nat Rev Microbiol 16: 440–452 [DOI] [PubMed] [Google Scholar]
- Pradhan A, Avelar GM, Bain JM, Childers D, Pelletier C, Larcombe DE, Shekhova E, Netea MG, Brown GD, Erwig L, Gow NAR, Brown AJP (2019) Non-canonical signalling mediates changes in fungal cell wall PAMPs that drive immune evasion. Nat. Commun 10:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RN, Schwessinger B, Joe A, Thomas N, Liu F, Albert M, Robinson MR, Chan LJ, Luu DD, Chen H, et al . (2015) The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci Adv 1:e1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusztahelyi T (2018) Chitin and chitin-related compounds in plant-fungal interactions. Mycology 9:189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-de-Luque A, Tille S, Johnson I, Pascual-Pardo D, Ton J, Cameron DD (2017) The interactive effects of arbuscular mycorrhiza and plant growth-promoting rhizobacteria synergistically enhance host plant defences against pathogens. Sci Rep 7:16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli JN, Blanco-Ulate B, Muszyński A, Figueroa-Balderas R, Morales-Cruz A, Azadi P, Dobruchowska JM, Castro C, Cantu D, Roper MC (2018) Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nat Commun 9:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Contreras YJ, Ramírez-Valdespino CA, Guzmán-Guzmán P, Macías-Segoviano JI, Villagómez-Castro JC, Olmedo-Monfil V (2019) Tal6 from Trichoderma atroviride is a LysM effector involved in mycoparasitism and plant association. Front Microbiol 10:2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Ham KS, Darvill AG, Albersheim P (2002) Molecular cloning and characterization of glucanase inhibitor proteins: coevolution of a counterdefense mechanism by plant pathogens. Plant Cell 14:1329–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Martín JM, Pacheco-Arjona JR, Bello-Rico V, Vargas WA, Monod M, Díaz-Mínguez JM, Thon MR, Sukno SA (2016) A highly conserved metalloprotease effector enhances virulence in the maize anthracnose fungus Colletotrichum graminicola. Mol Plant Pathol 17:1048–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger R, Touchard M, Clément C, Baillieul F, Cordelier S, Crouzet J, Dorey S (2019) Apoplastic invasion patterns triggering plant immunity: plasma membrane sensing at the frontline. Mol Plant Pathol 20:1602–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C, Feike D, Gimenez-Ibanez S, Hann DR, Zipfel C, Rathjen JP (2011) Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol 156:687–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Park BG, Wolf AJ, Brikos C, Goodridge HS, Becker CA, Reyes CN, Miao EA, Aderem A, Götz F, et al . (2010) Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe 7:38–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos I, Van den Burg HA, Okmen B, Beenen HG, van Liere S, Kema GH, De Wit PJGM (2010) Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc Natl Acad Sci USA 107:7610–7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Dunning FM, Pfund C, Weingarten R, Bent AF (2006) Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 18:764–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F, Yamamoto M, Ohnishi-Kameyama M, Iwaki M, Yoshida M, Ishii T, Konishi T, Ichinose Y (2010) Defects in flagellin glycosylation affect the virulence of Pseudomonas syringae pv. tabaci 6605. Microbiology 156:72–80 [DOI] [PubMed] [Google Scholar]
- Taguchi F, Takeuchi K, Katoh E, Murata K, Suzuki T, Marutani M, Kawasaki T, Eguchi M, Katoh S, Kaku H, et al . (2006) Identification of glycosylation genes and glycosylated amino acids of flagellin in Pseudomonas syringae pv. tabaci. Cell Microbiol 8:923–938 [DOI] [PubMed] [Google Scholar]
- Takahara H, Hacquard S, Kombrink A, Hughes HB, Halder V, Robin GP, Hiruma K, Neumann U, Shinya T, Kombrink E, et al . (2016) Colletotrichum higginsianum extracellular LysM proteins play dual roles in appressorial function and suppression of chitin-triggered plant immunity. New Phytol 211:1323–1337 [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Taguchi F, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y (2003) Flagellin glycosylation island in Pseudomonas syringae pv. glycinea and its role in host specificity. J Bacteriol 185:6658–6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou JM (2017) Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29:618–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thynne E, Saur IML, Simbaqueba J, Ogilvie HA, Gonzalez-Cendales Y, Mead O, Taranto A, Catanzariti AM, McDonald MC, Schwessinger B, et al . (2017) Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Mol Plant Pathol 18:811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Benedetti B, Kamoun S (2005) A Second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol 138:1785–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Huitema E, Da Cunha L, Torto-Alalibo T, Kamoun S (2004) A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J Biol Chem 279:26370–26377 [DOI] [PubMed] [Google Scholar]
- Toruño TY, Stergiopoulos I, Coaker G (2016) Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol 54:419–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trdá L, Fernandez O, Boutrot F, Héloir MC, Kelloniemi J, Daire X, Adrian M, Clément C, Zipfel C, Dorey S, Poinssot, B (2014) The grapevine flagellin receptor VvFLS2 differentially recognizes flagellin-derived epitopes from the endophytic growth-promoting bacterium Burkholderia phytofirmans and plant pathogenic bacteria. New Phytol 201:1371–1384 [DOI] [PubMed] [Google Scholar]
- Van den Burg HA, Harrison SJ, Joosten MHAJ, Vervoort J, De Wit PJGM (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant-Microbe Interact 19:1420–1430 [DOI] [PubMed] [Google Scholar]
- Vander P, V rum KM, Domard A, Eddine El Gueddari N, Moerschbacher BM (1998) Comparison of the ability of partially N-acetylated chitosans and chitooligosaccharides to elicit resistance reactions in wheat leaves. Plant Physiol 118:1353–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermassen A, Leroy S, Talon R, Provot C, Popowska M, Desvaux M (2019) Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front Microbiol 10: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk H, Marton K, Flajšman M, Radišek S, Tian H, Hein I, Podlipnik Č, Thomma BPHJ, Košmelj K, Javornik B. et al. (2019) Chitin-binding protein of Verticillium nonalfalfae disguises fungus from plant chitinases and suppresses chitin-triggered host immunity. Mol Plant Microbe Interact 32: 1378–1390 [DOI] [PubMed] [Google Scholar]
- Walls A, Tortorici M, Frenz B, Snijder J, Li W, Rey FA, DiMaio F, Bosch BJ, Veesle D(2016) Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol 23:899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Burrage AM, Postel S, Clark RE, Orlova A, Sundberg EJ, Kearns DB, Egelman EH (2017) A structural model of flagellar filament switching across multiple bacterial species. Nat Commun 8:960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Song WY, Ruan DL, Sideris S, Ronald PC (1996) The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Mol Plant Microbe Interact 9:850–855 [DOI] [PubMed] [Google Scholar]
- Wang S, Sun Z, Wang H, Liu L, Lu F, Yang J, Zhang M, Zhang S, Guo Z, Bent AF, Sun W (2015) Rice OsFLS2-mediated perception of bacterial flagellins is evaded by Xanthomonas oryzae pvs. oryzae and oryzicola. Mol Plant 8:1024–1037 [DOI] [PubMed] [Google Scholar]
- Wang S, Xing R, Wang Y, Shu H, Fu S, Paulus JK, Schuster M, Saunders DGO, Win J, Vleeshouwers V, et al . (2021) Cleavage of a pathogen apoplastic protein by plant subtilases activates immunity. New Phytol. doi: 10.1111/nph.17120 [DOI] [PubMed] [Google Scholar]
- Wawra S, Fesel P, Widmer H, Timm M, Seibel J, Leson L, Kesseler L, Nostadt R, Hilbert M, Langen G. , et al. (2016) The fungal-specific β-glucan-binding lectin FGB1 alters cell-wall composition and suppresses glucan-triggered immunity in plants. Nat Commun 7:13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Caceres-Moreno C, Jimenez-Gongora T, Wang K, Sang Y, Lozano-Duran R, Macho AP (2018) The Ralstonia solanacearum csp22 peptide, but not flagellin-derived peptides, is perceived by plants from the Solanaceae family. Plant Biotechnol J 16:1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AJ, Arruda A, Reyes CN, Kaplan AT, Shimada T, Shimada K, Arditi M, Liu G, Underhill DM (2011) Phagosomal degradation increases TLR access to bacterial ligands and enhances macrophage sensitivity to bacteria. J Immunol 187:6002–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Wang J, Zhao J, Xu J, Sun S, Zhang H, Wu J, Tang C, Kang Z, Wang X (2020) A polysaccharide deacetylase from Puccinia striiformis f. sp. tritici is an important pathogenicity gene that suppresses plant immunity. Plant Biotechnol J 18:1830–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Ohnishi-Kameyama M, Nguyen CL, Taguchi F, Chiku K, Ishii T, Ono H, Yoshida M, Ichinose Y (2011) Identification of genes involved in the glycosylation of modified viosamine of flagellins in Pseudomonas syringae by mass spectrometry. Genes 2:788–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Yu Y, Huang J, Meng F, Pang J, Zhao Q, Islam MA, Xu N, Tian Y, Liu J (2019) Binding of the Magnaporthe oryzae chitinase MoChia1 by a rice tetratricopeptide repeat protein allows free chitin to trigger immune responses. Plant Cell 31:172–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Lund SP, Scott RA, Greenwald JW, Records AH, Nettleton D, Lindow SE, Gross DC, Beattie GA (2013) Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites .Proc Natl Acad Sci USA 110:E425–E–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng T, Rodriguez-Moreno L, Mansurkhodzaev A, Wang P, Van den Berg W, Gasciolli V, Cottaz S, Fort S, Thomma BPHJ, et al . (2020) A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis .New Phytol 225:448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428:764–767 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125:749–760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.