Abstract
Light functions as the primary environmental stimulus and brassinosteroids (BRs) as important endogenous growth regulators throughout the plant lifecycle. Photomorphogenesis involves a series of vital developmental processes that require the suppression of BR-mediated seedling growth, but the mechanism underlying the light-controlled regulation of the BR pathway remains unclear. Here, we reveal that nuclear factor YC proteins (NF-YCs) function as essential repressors of the BR pathway during light-controlled hypocotyl growth in Arabidopsis thaliana. In the light, NF-YCs inhibit BR biosynthesis by directly targeting the promoter of the BR biosynthesis gene BR6ox2 and repressing its transcription. NF-YCs also interact with BIN2, a critical repressor of BR signaling, and facilitate its stabilization by promoting its Tyr200 autophosphorylation, thus inhibiting the BR signaling pathway. Consistently, loss-of-function mutants of NF-YCs show etiolated growth and constitutive BR responses, even in the light. Our findings uncover a dual role of NF-YCs in repressing BR biosynthesis and signaling, providing mechanistic insights into how light antagonizes the BR pathway to ensure photomorphogenic growth in Arabidopsis.
NF-YC transcription factors function as essential repressors of the brassinosteroid biosynthesis and signaling pathways to mediate photomorphogenic growth in Arabidopsis.
Introduction
Photomorphogenesis includes a series of vital developmental processes that are controlled by light signals throughout the plant lifecycle. Following germination, seedlings emerging from dark to light conditions undergo photomorphogenic growth, which leads to de-etiolation phenotypes characterized by open and expanded cotyledons, green chloroplasts, and short hypocotyls (de Wit et al., 2016). When confronting shade conditions, neighboring plants usually alter their architecture to capture light by accelerating internode and petiole elongation, reducing shoot branching and leaf expansion, and promoting the formation of horizontal leaf angles, which are collectively referred to as shade avoidance syndrome (Casal, 2012). Light-regulated growth helps plants adapt to various environmental challenges. To trigger such dramatic morphological changes, thousands of target genes involved in plant growth and development are regulated by the light signaling pathway by interacting with phytohormones such as brassinosteroids (BRs) (Li et al., 1996; Wang et al., 2012).
BRs are plant steroidal hormones that play important roles in multiple aspects of plant development and physiological responses. The BR biosynthetic pathway has been largely elucidated (Zhao and Li, 2012). Campesterol, the first precursor of the BR biosynthetic pathway, is converted to castasterone (CS) via an early or late C-6 oxidation pathway by a series of BR biosynthetic enzymes, including DE-ETIOLATED2 (DET2), CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM (CPD), ROTUNDIFOLIA3 (ROT3), CYP90D1, DWARF4 (DWF4), and BRASSINOSTEROID-6-OXIDASE1 (BR6ox1). BR6ox2 catalyzes the final biosynthetic step, converting CS into brassinolide (BL) (Zhao and Li, 2012).
In recent decades, multiple genetic and biochemical studies have greatly advanced our understanding of the BR signaling pathway in plants (Wang et al., 2012). The Shaggy/glycogen synthase kinase 3 (GSK3)-like protein kinase BRASSINOSTEROID INSENSITIVE2 (BIN2) acts as a critical negative regulator of BR signaling in Arabidopsis thaliana by phosphorylating and inactivating two transcription factors, BRASSINAZOLE (BRZ) RESISTANT 1 (BZR1) and BRI1-EMS-SUPPRESSOR1 (BES1), to repress BR-responsive gene expression (He et al., 2002; Yin et al., 2002). BR directly binds to the extracellular domain of the plasma membrane-localized receptor kinase BRASSINOSTERIOD INSENSITIVE1 (BRI1), resulting in the rapid activation of the intracellular kinase domain of BRI1 (Li and Chory, 1997). The activated BRI1 triggers a series of downstream phosphorylation events, including the phosphorylation of BRI1 SUPPRESSOR1 (BSU1). The phosphorylated BSU1 protein dephosphorylates BIN2, which triggers the degradation of BIN2 by the ubiquitin 26S proteasome, thereby releasing the repression of the BR pathway (Li et al., 2001; Kim et al., 2009; 2011).
Studies have revealed significant crosstalk between the BR and light signaling pathways in plants (Luo et al., 2010; Oh et al., 2012; Li and He, 2016). Both loss-of-function mutants of BR biosynthesis and signaling display similar de-etiolated morphology characterized by short hypocotyls, open cotyledons, and increased expression of light-induced genes, even in the dark (Chory et al., 1991; Li et al., 1996; Szekeres et al., 1996; Wang et al., 2012). Light inhibits BR production and signal transduction for de-etiolation. The BR signaling regulator BZR1 directly interacts with several key transcription factors in the light signaling pathway, including GATA2, PHYTOCHROME INTERACTING FACTOR4 (PIF4), and ELONGATED HYPOCOTYL 5 (HY5) to mediate photomorphogenesis (Luo et al., 2010; Oh et al., 2012; Li and He, 2016). In addition, BIN2 interacts with and phosphorylates PIF3 and PIF4, leading to their degradation by the ubiquitin 26S proteasome, thus modulating skotomorphogenesis and diurnal hypocotyl growth (Bernardo-Garcia et al., 2014; Ling et al., 2017). Light represses BR signaling by inhibiting the DNA binding activity of BZR1/BES1 and promoting BZR1 degradation in a photoreceptors-dependent manner (Li et al., 2017a, 2017b; Liang et al., 2018; Wang et al., 2018; Dong et al., 2019; He et al., 2019). Light may also inhibit BR biosynthesis via PIFs-mediated regulation (Wei et al., 2017; Martínez et al., 2018). However, the detailed molecular mechanism underlying how light mediates the BR biosynthesis and signaling pathways to control photomorphogenic processes remains to be further investigated.
NUCLEAR FACTOR-Y C proteins (NF-YCs) are subunits of the NF-Y heterotrimeric complex that specifically recognize the CCAAT-box-containing promoters of their target genes in eukaryotes (Petroni et al., 2012; Nardini et al., 2013). In plants, NF-YCs function as major regulators of phytohormone-regulated growth and developmental processes, including flowering and abscisic acid (ABA)/gibberellin (GA)-mediated seed germination (Hou et al., 2014; Liu et al., 2016a, 2016b; Hwang et al., 2019). There are 10 NF-YC genes in Arabidopsis, among which the closest homologs, NF-YC1, NF-YC3, NF-YC4, and NF-YC9, are expressed in light- and dark-grown seedlings (Siefers et al., 2009). These four NF-YC homologs positively regulate photomorphogenesis via histone deacetylation and an HY5-independent light signaling pathway in Arabidopsis (Myers et al., 2016; Tang et al., 2016). The diverse roles of NF-YCs together with the two other NF-Y subunits NF-YA/B might be derived from the flexible formation of NF-Y complexes, which is spatially and temporally regulated by intrinsic and external cues (Myers and Holt 3rd, 2018).
In this study, we demonstrate that the NF-YC homologs NF-YC1, NF-YC3, NF-YC4, and NF-YC9 play redundant and essential roles in the light-mediated inhibition of the BR biosynthesis and signaling pathways in Arabidopsis. In the light, NF-YCs directly repress the expression of the BR biosynthesis gene BR6ox2 to inhibit BR biosynthesis. On the other hand, NF-YCs physically interact with the BR signaling repressor BIN2 to mediate its autophosphorylation, thereby protecting BIN2 from degradation by the 26S proteasome pathway. Our findings indicate that NF-YCs function as essential repressors of the BR biosynthesis and signaling pathways to regulate light-controlled hypocotyl growth.
Results
NF-YCs are required for BR-mediated hypocotyl elongation in the light
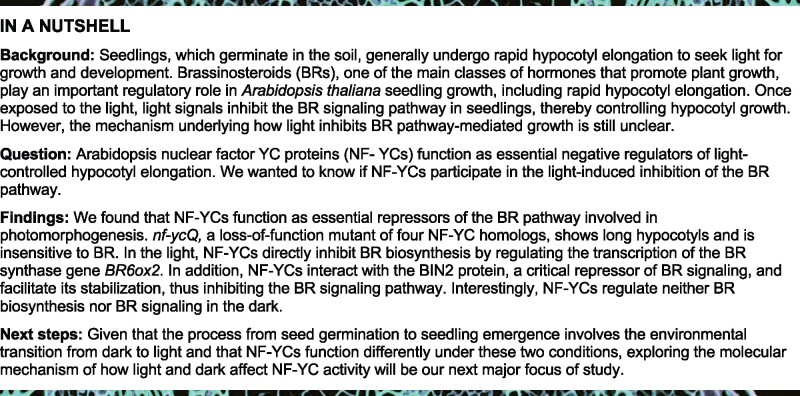
During post-germination, seedlings that germinated underground must undergo etiolated growth (i.e. hypocotyl elongation) to seek light in a BR-dependent manner (Chory et al., 1991; Clouse, 2001; Minami et al., 2019). Once that have emerged from the soil, seedlings require the rapid light-triggered inhibition of BR responses in order to undergo photomorphogenesis (Li et al., 1996; Neff et al., 2005; Wang et al., 2012). To confirm this notion, we examined BR biosynthesis and signaling in Arabidopsis during the early seedling stage. Real-time qPCR (RT-qPCR) analysis showed that the expression levels of key BR biosynthesis genes were significantly lower in light-grown seedlings than in seedlings grown in the dark (Supplemental Figure S1; Figure 1A). The BR-responsive genes exhibited similar expression pattern to BR biosynthesis genes (Supplemental Figure S2). Correspondingly, the contents of the active BRs CS (the precursor of BL) and BL, but not 6-DeoxoCS (the precursor of CS), were dramatically lower or not detected in light-grown seedlings compared with dark-grown plants (Figure 1B). bri1-301, a weak BR-insensitive allele, was used as a control to measure BR levels. BR levels were significantly higher in bri1-301 than Col-0, confirming the reliability of our BR measurement system (Supplemental Figure S3; Chung and Choe, 2013). These results suggest that light represses the BR pathways.
Figure 1.
NF-YCs are involved in BR-mediated hypocotyl growth in the light. A, RT-qPCR analysis of BR biosynthesis gene expression in Col-0 seedlings grown in the light or dark for 3 d. Relative gene expression was calculated by comparing the value with that of seedlings grown in the dark. Data represent mean ± sd of three biological replicates. PP2A was amplified as an internal control. B, Endogenous BR levels of Col-0 seedlings grown in the light or dark for 3 d. Data represent mean ± sd of two biological replicates. Asterisks in (A) and (B) indicate significant differences between dark and light conditions (P < 0.01, Student’s t test). NS indicates no significant difference. ND indicates no detection. C, Hypocotyl phenotypes of Col-0, nf-ycT, and nf-ycQ grown in the light with BL (20 nM), PIC (10 µM), and GA3 (10 µM) treatment for 5 d. Scale bar, 1 mm. D, Hypocotyl lengths of the seedlings shown in (C). Data represent means ± sd of at least 20 seedlings with three biological replicates. Asterisks indicate significant differences in seedlings treated with BL, PIC, and GA3 compared with the mock (P < 0.01, Student’s t test). E, Hypocotyl length of Col-0 and nf-ycQ seedlings treated with different concentrations of BL in the light for 5 d. Data represent means ± sd of at least 20 seedlings with three biological replicates. F, Relative hypocotyl length of Col-0 and nf-ycQ seedlings treated with different concentrations of BRZ in the light for 5 d. Relative hypocotyl length was calculated by comparing the value of BRZ treatment with that of the mock. Data represent means ± sd of at least 20 seedlings with three biological replicates. Asterisks indicate significant differences between Col-0 and nf-ycQ (P < 0.01, Student’s t test).
Phytohormones such as GA, auxin, and BR also mediate photomorphogenic growth (de Wit et al., 2016; Nolan et al., 2020). As we previously demonstrated that NF-YCs are involved in light-controlled hypocotyl elongation via epigenetic regulation (Tang et al., 2016), we wondered whether NF-YCs function in early seedling development via these phytohormone pathways. We treated 5-d-old seedlings of various nf-yc mutants and the wild-type (WT) with different phytohormones and measured hypocotyl length. The nf-yc3 nf-yc4 nf-yc9 triple mutant (nf-ycT) and nf-yc1 nf-yc3 nf-yc4 nf-yc9 (nf-ycQ) quadruple mutant exhibited markedly longer hypocotyls than single and double nf-yc mutants and WT in the light, while all seedlings showed less of a difference in the dark, as previously described (Figure 1, C and D; Supplemental Figure S4; Tang et al., 2016). Interestingly, loss-of-function of the four NF-YCs (nf-ycQ) disabled the response to BL, but the response to GA and picloram (PIC; an auxin analog) treatment remained normal (Figure 1, C and D), while other nf-yc mutants showed enhanced hypocotyl growth in response to all hormones in the light, as did WT seedlings (Supplemental Figure S5). In contrast, there was no difference in hypocotyl length between the nf-yc mutants and WT following hormone treatment in the dark (Supplemental Figure S4). In addition to promoting hypocotyl elongation, BR also accelerates the petiole elongation of adult seedlings for shade avoidance (Kozuka et al., 2010; Liu et al., 2016a, 2016b). nf-ycQ petioles exhibited significantly lower sensitivity (13.7%) to BL than WT (38.9%; Supplemental Figure S6). These results suggest that NF-YCs might function redundantly in the light-mediated control of the BR pathway.
We then examined the response of nf-ycQ seedlings to different concentrations of BL. The hypocotyl length of WT markedly increased with increasing BL concentration, whereas nf-ycQ seedlings exhibited no difference in hypocotyl length, even when high concentrations of BL were applied (Figure 1E). We then reduced the endogenous BR levels in nf-ycQ and WT seedlings by treating the plants with different concentrations of BRZ, a BR biosynthesis inhibitor that inhibits DWF4 enzyme activity. The hypocotyl length of nf-ycQ was more dramatically reduced by higher concentrations of BRZ, indicating a higher sensitivity to BRZ than the WT (Figure 1F;Supplemental Figure S7). Together, these results suggest that NF-YCs mediate the light-induced inhibition of BR responses during photomorphogenic growth.
NF-YCs directly inhibit BR biosynthesis in the light
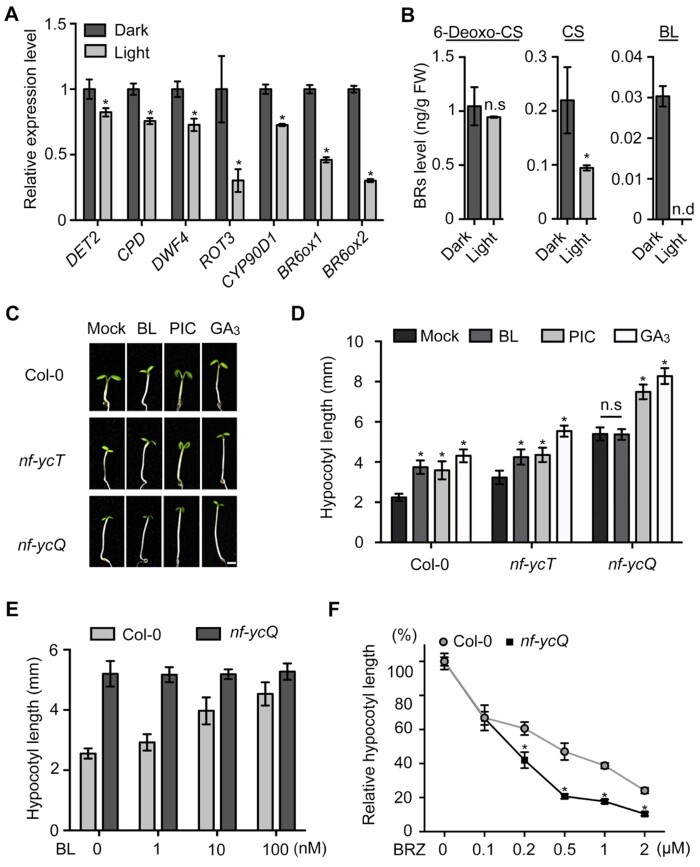
Even though the hypocotyl growth of nf-ycQ did not respond to exogenous BR treatment, hypocotyl growth was suppressed by inhibiting BR biosynthesis. It is therefore possible that NF-YCs are involved in regulating BR biosynthesis. To test this notion, we monitored the transcript levels of genes encoding key enzymes that catalyze different steps of BR biosynthesis (Supplemental Figure S1). The expression levels of DWF4, ROT3, BR6ox1, and BR6ox2 were significantly higher in nf-ycQ seedlings compared with WT in the light (Figure 2A). The BR-responsive genes also presented a similar expression pattern (Supplemental Figure S8). These observations prompted us to measure endogenous BR levels in nf-ycQ seedlings. In the light, 6-Deoxo-CS and CS level in nf-ycQ seedlings increased to 150% that of the WT (Figure 2B). Notably, the level of BL, the endpoint of BR biosynthesis catalyzed by BR6ox2, was much higher in nf-ycQ seedlings compared to the WT, in which BL was not detected (Figure 2B). This might explain why nf-ycQ had little response to exogenous BL in the light. In contrast, there were no significant differences in the expression levels of BR biosynthesis or BR-responsive genes or endogenous BR levels between dark-grown nf-ycQ and WT seedlings (Supplemental Figure S9, A–C). These results suggest that NF-YCs affect BR contents in light-grown seedlings primarily by regulating the final step of BR biosynthesis.
Figure 2.
NF-YCs directly target the BR6ox2 promoter to inhibit BR biosynthesis. A, RT-qPCR analysis of BR biosynthesis gene expression in Col-0 and nf-ycQ seedlings grown in the light for 3 d. Relative gene expression was calculated by comparing the value with that of Col-0. Data represent mean ± sd of three biological replicates. Asterisks indicate significant differences from Col-0 (P < 0.01, Student’s t test). PP2A was amplified as an internal control. B, Endogenous BR levels of Col-0 and nf-ycQ seedlings grown in the light for 3 d. Data represent mean ± sd of two biological replicates. Asterisks indicate significant differences between Col-0 and nf-ycQ seedlings in the light (P < 0.01, Student’s t test). Note: the BR contents in Figures 1, B and 2, B were measured together and thus the light-grown Col control is same in these two figures. C, Transient expression assay indicating that the expression of BR6ox2 is repressed by NF-YC9. Upper part shows a schematic diagram of the effector and reporter constructs used in the transient expression assay. Lower part shows the results of the transient expression assay. Either the reporter (BR6ox2pro) or the relevant empty vector (reporter control) was co-transformed with the effector and the relevant empty vector (effector control) into Arabidopsis mesophyll protoplasts. Asterisks indicate significant difference compared with the Effector control+BR6ox2pro (P < 0.01, Student’s t test). D, ChIP analysis of NF-YC9 binding to the BR6ox2 promoter regions. Upper part shows the 3.3-kb promoter regions of BR6ox2. Lower part shows that NF-YC9-FLAG was enriched in the P5 region of the BR6ox2 promoter. The nf-yc9-1 NF-YC9pro:NF-YC9-3FLAG (NF-YC9-FLAG) and Col-0 seedlings were grown in the light for 3 d and harvested for ChIP-qPCR analysis. Data represent mean ± sd of three biological replicates. Asterisks indicate significant differences in fold enrichment between NF-YC9-FLAG and Col-0 (P < 0.01, Student’s t test).
We then examined the effect of NF-YC on BR6ox2 expression by performing transient assays. A 3.2 kb fragment of the BR6ox2 promoter was fused to the luciferase (LUC) gene as the reporter. The effector construct 35Spro:NF-YC9-6HA was transformed together with the reporter or control into Arabidopsis mesophyll protoplasts (Figure 2C). After 2 h of culture in the dark to activate reporter expression, the protoplasts were transferred to the light and cultured for 12 h. NF-YC9, but not the effector control, significantly reduced LUC activity in the protoplasts, supporting the role of NF-YC9 in repressing BR6ox2 transcription (Figure 2C). Since NF-YCs regulate gene expression by associating with DNA in plants (Liu et al., 2016a, 2016b), we performed a chromatin immunoprecipitation assay to examine the binding ability of NF-YC9 to BR6ox2 in vivo. NF-YC9 strongly bound to the BR6ox2 promoter at the P5 region, which contains a CCAAT element recognized by NF-Y (Petroni et al., 2012), in light-grown seedlings (Figure 2D). No enrichment of NF-YC9 was observed in the BR6ox2 promoter in seedlings grown in the dark (Supplemental Figure S9D). These results support the notion that NF-YCs function as negative regulators of BR biosynthesis genes to inhibit hypocotyl elongation in the light.
NF-YCs mediate BR signaling independently of BR biosynthesis
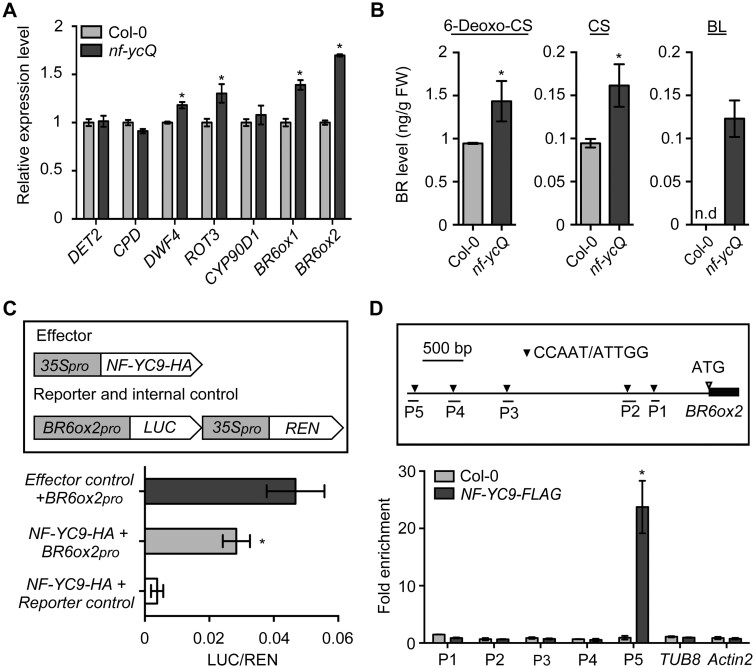
To investigate whether NF-YCs mediate the BR pathway by regulating BR biosynthesis, we abolished BR biosynthesis in seedlings via BRZ treatment. We treated seedlings with BL and BRZ and monitored seedling growth in the light. We expected that nf-ycQ and WT hypocotyls would exhibit a comparable length after treatment. Strikingly, although nf-ycQ and WT initially had the same degree of hypocotyl shortening caused by BRZ treatment, nf-ycQ seedlings exhibited significantly longer hypocotyls than WT when they were subsequently treated with up to 500 nM BL, indicating the nf-ycQ is more sensitive to BR than the WT (Figure 3A). This observation raised the possibility that, apart from BR biosynthesis, NF-YCs also play a role in regulating BR signaling.
Figure 3.
BR signaling is enhanced in nf-ycQ seedlings independent of BR biosynthesis. A, Hypocotyl length of Col-0 and nf-ycQ seedlings treated with BRZ (2 µM) and different concentrations of BL in the light for 5 d. Data represent means ± sd of at least 20 seedlings with three biological replicates. Asterisks indicate significant differences from Col-0 (P < 0.01, Student’s t test). B, Hypocotyl phenotypes of Col-0 and nf-ycQ seedlings treated with Bikinin (10 µM) in the light for 5 d. C, Hypocotyl lengths of the seedlings shown in (B). Data represent means ± sd of at least 20 seedlings with three biological replicates. Asterisks indicate significant differences between the treatment and the mock (P < 0.01, Student’s t test). D, Hypocotyl lengths of Col-0 and nf-ycQ seedlings treated with BRZ (2 µM) and different concentrations of Bikinin in the light for 5 d. Data represent means ± sd of at least 20 seedlings with three biological replicates. Asterisks indicate significant differences from Col-0 (P < 0.01, Student’s t test).
To verify this hypothesis, we treated nf-ycQ and WT seedlings with bikinin, a specific inhibitor of GSK3-like protein kinase activity that could deactivate the BR signaling repressor BIN2 to release BR signaling. Like BL treatment (Figure 1E), bikinin had no effect on the hypocotyl growth of nf-ycQ (Figure 3, B and C). In contrast, bikinin promoted the growth of WT hypocotyls to the same length as nf-ycQ, which BL treatment failed to achieve (Figure 3, A–C), supporting the possibility that NF-YC is involved in regulating both BR biosynthesis and signaling. Given that BR activates the downstream response in the BR signaling pathway by triggering the degradation of BIN2 (Wang et al., 2012), we applied BRZ together with different concentrations of bikinin to seedlings and examined the effect of BR signaling in the absence of BR biosynthesis. nf-ycQ seedlings were still more sensitive to bikinin than WT and 35Spro:BIN2-MYC even though BR biosynthesis had been abolished in these plants (Figure 3D). These results suggest that NF-YCs play an important role in regulating the BR signaling pathway.
NF-YCs interact with BIN2
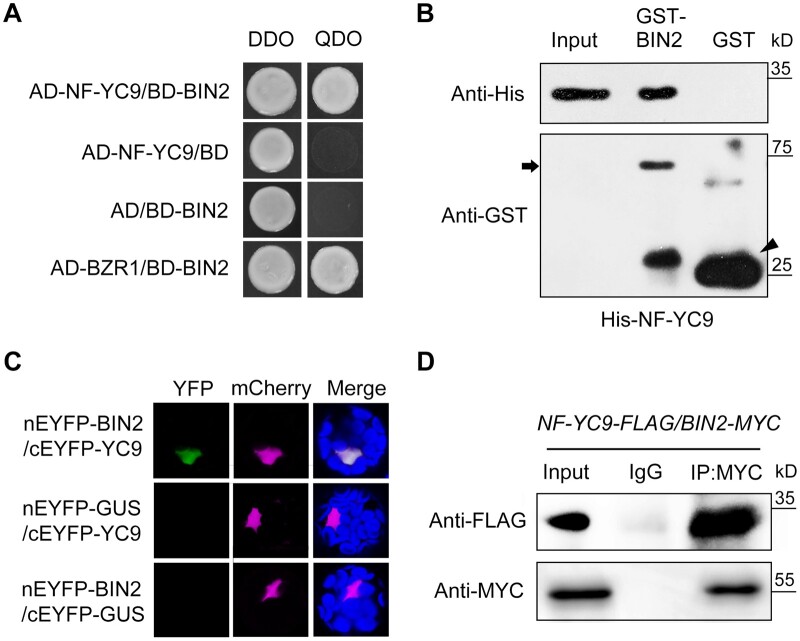
NF-YCs regulate diverse developmental processes in Arabidopsis via direct protein–protein interactions with other key regulatory factors (Hou et al., 2014; Liu et al., 2016a, 2016b; Hwang et al., 2019). To examine whether NF-YCs function with BR signaling regulators in BR-mediated photomorphogenic growth, we performed a yeast two-hybrid assay and found that, interestingly, NF-YC homologs interacted with BIN2 but not with BZR1, a positive regulator of BR signaling, in yeast (Figure 4A;Supplemental Figure S10). Pull-down assays showed that GST-BIN2, but not GST, interacted with His-NF-YC1, 3, 4, and 9 in vitro (Figure 4B;Supplemental Figure S11). Due to the functional redundancy of the four NF-YC homologs in light-controlled hypocotyl elongation (Tang et al., 2016; Supplemental Figure S4), we selected NF-YC9 as the representative for further investigation. Bimolecular fluorescence complementation (BiFC) assays revealed the interaction between NF-YC9 and BIN2 in Arabidopsis protoplasts (Figure 4C). In addition, co-immunoprecipitation (Co-IP) assays using nf-yc9-1 NF-YC9pro:NF-YC9-3FLAG/35Spro:BIN2-6MYC seedlings revealed that the interaction between NF-YC9 and BIN2 in plants occurs in the light but not in the dark and is not regulated by exogenous BL (Figure 4D;Supplemental Figure S12). These results suggest that NF-YCs function in light-mediated regulation of the BR signaling pathway together with BIN2.
Figure 4.
NF-YCs interact with BIN2 in vitro and in vivo. A, Yeast two-hybrid assays showing the interaction between NF-YC9 and BIN2. Transformed yeast cells were grown on SD/-Trp/-Leu/-His/-Ade and SD/-Trp/-Leu media. The interaction between BIN2 and BZR1 was used as a positive control. B, In vitro pull-down assay showing the direct interaction between His-NF-YC9 and GST-BIN2 recombinant proteins. His-tagged proteins were incubated with GST-BIN2 or GST proteins and the immunoprecipitated fractions were detected by anti-His and anti-GST antibodies. Arrow and arrowhead indicate GST-BIN2 and GST protein bands, respectively. C, BiFC analysis showing the interaction between NF-YC9 and BIN2 in Arabidopsis protoplasts. YFP, fluorescence of enhanced yellow fluorescent protein; mCherry, mCherry was fused with a nucleus localization signal (VirD2NLS-mCherry) as the nucleus indicator; Merge, merged of YFP and mCherry signals. The GUS gene served as a negative control. D, Co-IP assay of the interaction between NF-YC9 and BIN2 in planta. Nuclear extracts from 3-d-old nf-yc9-1 NF-YC9pro:NF-YC9-3FLAG/35Spro:BIN2-6MYC seedlings grown in the light were immunoprecipitated by anti-MYC antibody or preiummune serum Immunoglobulin G (IgG). The co-immunoprecipitated proteins were detected by anti-MYC or anti-FLAG antibody.
NF-YCs repress the BR signaling pathway by inhibiting BIN2 degradation
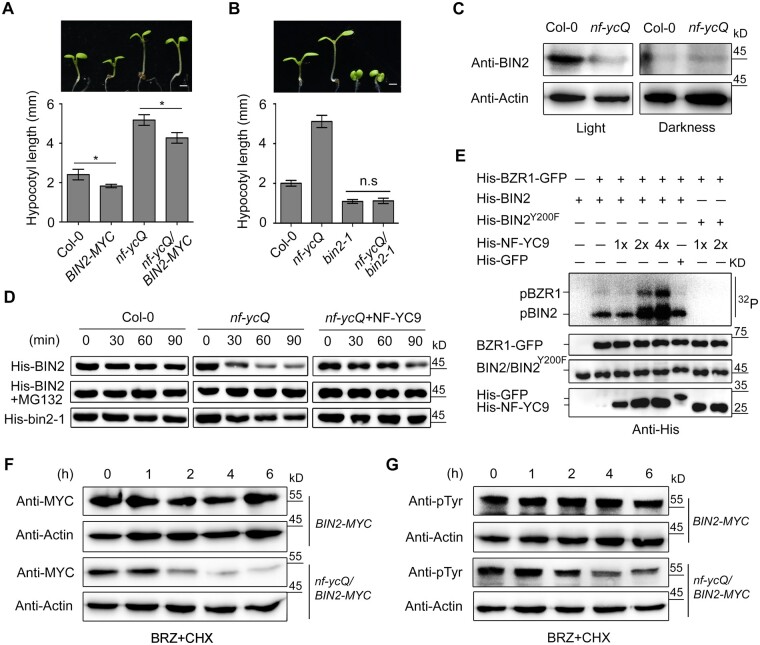
We then investigated the biological role of the interaction between NF-YCs and BIN2 in plants by performing genetic analysis. Overexpressing BIN2 (35Spro:BIN2-6MYC) reduced the hypocotyl length of both nf-ycQ and WT seedlings (Figure 5A;Supplemental Figure S13). Since BIN2 is unstable in the presence of BR even in the overexpression plants (Zhu et al., 2017), it is difficult to determine the genetic relationship of NF-YC and BIN2 using 35Spro:BIN2-6MYC. Thus, we employed bin2-1, a gain-of-function mutant of BIN2 in which a point-mutated form of BIN2 is present in a constitutively phosphorylated form and is barely degraded (Li et al., 2001; Kim et al., 2009). Like NF-YC9 overexpression lines (Tang, et al., 2016), bin2-1 exhibited photomorphogenic growth with severely inhibited hypocotyl elongation in both the light and dark (Figure 5B;Supplemental Figure S13). Notably, loss-of-function of NF-YCs had little effect on the short hypocotyl phenotype of bin2-1, indicating that BIN2 is epistatic to NF-YC (Figure 5B;Supplemental Figure S13).
Figure 5.
NF-YCs repress BIN2 degradation by mediating i phosphorylation of BIN2. A and B, Hypocotyl phenotypes and lengths of Col-0, 35Spro:BIN2-6MYC, nf-ycQ, bin2-1, nf-ycQ/35Spro:BIN2-6MYC, and nf-ycQ/bin2-1 seedlings grown in the light for 5 d. Scale bar, 1 mm. Data represent means ± sd of at least 20 seedlings with three biological replicates. Asterisks indicate significant differences between Col-0 and 35Spro:BIN2-6MYC, or nf-ycQ and nf-ycQ/35Spro:BIN2-6MYC seedlings (P < 0.01, Student’s t test). C, BIN2 levels in Col-0 and nf-ycQ seedlings grown in the light and dark for 3 d detected by immunoblot. Actin was used as the internal control. BIN2 was detected by anti-BIN2 polyclonal antibody (AS163203; Agrisera Vännäs, Sweden). D, Cell-free degradation assay of BIN2. His-BIN2 and His-bin2-1, with or without His-NF-YC9, were incubated with equal amounts of plant cell crude extracts from Col-0 and nf-ycQ seedlings containing CHX (1 µM) at room temperature for 0–90 min. MG132 (50 μM) was added as indicated. The proteins were detected by anti-His antibody. E, In vitro kinase assay of the phosphorylation activity of BIN2 and BZR1. His-BIN2 was used as the autophosphorylation substrate, and His-BZR1-GFP as the phosphorylation substrate of BIN2. His-BIN2 and His-BIN2Y200F were incubated with different concentrations of His-NF-YC9 or His-GFP and His-BZR1-GFP at 30°C for 1 h in [γ-32P] ATP kinase buffer. Autophosphorylated BIN2 and phosphorylated BZR1 were detected by 32P autoradiography. His-BIN2, His-BIN2Y200F, His-GFP, His-BZR1-GFP, and His-NF-YC9 were detected by anti-His antibody. His-GFP was used as a negative control of His-NF-YC9. pBZR1, phosphorylated BZR1; pBIN2, autophosphorylated BIN2. 32P, autoradiography of [γ-32P] ATP-labeled proteins. F, BIN2 levels in 35Spro:BIN2-6MYC and nf-ycQ/35Spro:BIN2-6MYC plants. Seedlings were treated with BRZ (2 µM) in the light for 3 d and transferred to MS plates containing CHX (1 µM) for time-course treatment (0–6 h). BIN2 was detected with anti-MYC antibody. Actin was used as a loading control. G, Immunoblot analysis of BIN2 phosphorylation in plants as described in (F). Phosphorylated BIN2 was detected with anti-pTyr279/Tyr216 (pTyr) antibody. Actin was used as a loading control.
Genetic analysis suggested that NF-YCs mediate photomorphogenic growth by interacting with and facilitating BIN2 activity. RT-qPCR analysis showed that NF-YC does not regulate the transcription of BIN2 (Supplemental Figure S14). We then examined BIN2 accumulation in nf-ycQ, WT, 35Spro:BIN2-6MYC, and nf-ycQ/35Spro:BIN2-6MYC seedlings via immunoblotting using anti-BIN2 and anti-MYC antibodies. Interestingly, the abundance of BIN2 was markedly lower in nf-ycQ than in WT seedlings in the light, while it was maintained at a similarly low level in dark-grown plants (Figure 5C;Supplemental Figure S15), indicating that NF-YCs are involved in the light-mediated regulation of BIN2 stabilization. A cell-free degradation assay showed that the loss-of-function of NF-YCs promoted the degradation of His-BIN2 when expressed in Escherichia coli, which was inhibited by treatment with MG132, a 26S proteasome inhibitor (Figure 5D). The addition of His-NF-YC9 strongly attenuated the degradation of His-BIN2 in nf-ycQ (Figure 5D). These results, together with the observation that the level of ubiquitinated BIN2 was markedly higher in nf-ycQ/35Spro:BIN2-6MYC than in 35Spro:BIN2-6MYC (Supplemental Figure S16), support the notion that NF-YCs prevent BIN2 from undergoing 26S proteasome-mediated degradation.
NF-YCs mediate the autophosphorylation of BIN2
BIN2 functions as a critical negative regulator in the BR signaling pathway and an important component linking many other signaling pathways. Although BIN2 activity is regulated by several chemical modifications, the phosphorylation of BIN2 Tyr 200 (Y200), which is catalyzed by BIN2 itself or by other kinases, is an essential modification for the stabilization and full activity of BIN2 (Kim et al., 2009; Nolan et al., 2020). As NF-YCs were not required for the stabilization of constitutively phosphorylated BIN2 in bin2-1 (Figure 5, B and D), we reasoned that NF-YCs might contribute to the phosphorylation of BIN2. To test this hypothesis, we performed an in vitro kinase assay and found that His-NF-YC9, but not His-GFP, directly promoted the autophosphorylation of GST-BIN2 (Supplemental Figure S17A). In addition, the autophosphorylation level of BIN2 significantly increased with increasing amounts of His-NF-YC9 added to the kinase reactions (Supplemental Figure S17B). A radioactive in vitro kinase assay also verified that NF-YC9 promoted the 32P-labeled autophosphorylation of BIN2 (Supplemental Figure S17C).
To further confirm this observation, we employed a dephosphorylated BIN2Y200F protein, a mutation of Tyr 200 to Phe (Y200F) that largely impairs the phosphorylation activity of BIN2. As expected, NF-YC9 markedly promoted the 32P autophosphorylation of BIN2, but not BIN2Y200F (Figure 5E). These results suggest that NF-YCs regulates BIN2 phosphorylation in a dosage-dependent manner. We then examined the stabilization and phosphorylation status of BIN2 in planta. In this experiment, seedlings were pretreated with BRZ to suppress BR biosynthesis for 3 d, and cycloheximide (CHX), an inhibitor of protein synthesis, was then applied to the seedlings in a time course to block de novo biosynthesis of BIN2. Similar to the cell-free degradation assays, BIN2 remained stable in 35Spro:BIN2-MYC transgenic plants but was rapidly degraded in the nf-ycQ/35Spro:BIN2-MYC background (Figure 5F). Consistently, the phosphorylated form of BIN2-MYC was present at lower levels in nf-ycQ compared to the WT background (Figure 5G). These results indicate that NF-YCs help maintain the phosphorylation status of BIN2 to ensure its stabilization.
Since BZR1 is the substrate of BIN2 kinase in the BR signaling pathway (He et al., 2002), we examined whether NF-YCs mediate the phosphorylation status of BZR1 via BIN2. As expected, the de-phosphorylated form of BZR1 strongly accumulated in nf-ycQ seedlings compared to the WT in the light, while its levels in both genotypes were comparable in the dark (Supplemental Figure S18). These results are in line with the observations of BIN2 abundance (Figure 5C). In addition, NF-YC9-induced BIN2 autophosphorylation resulted in the subsequent increase in the level of the phosphorylated form of BZR1 in vitro. In contrast, NF-YC9 neither facilitated the autophosphorylation of BIN2Y200F nor the phosphorylation of BZR1 in the absence of BIN2 (Figure 5E). These results confirm the notion that NF-YCs regulate BR signaling via the BIN2–BZR1 phosphorylation cascade.
Discussion
Under natural conditions, seedlings in soil undergo a series of developmental changes in order to seek light after germination. Once exposed to the light, plants must rapidly remove the photomorphogenic repressors that had accumulated in the dark and activate photomorphogenic promoters to ensure normal development (de Wit et al., 2016). The plant growth-promoting steroid hormone BR functions as a repressor of photomorphogenesis. The light signal-triggered developmental process requires the immediate inhibition of the BR pathway (Li et al., 1996, 2020; Wang et al., 2012). However, how plants precisely modulate the BR pathway to control growth in response to light remains largely unclear. Here, we revealed that the Arabidopsis NF-YC1/3/4/9 proteins function as essential repressors of BR-mediated photomorphogenic growth. In the light, NF-YCs directly associate with the promoter of the BR biosynthesis gene BR6ox2 and repress its transcription, thereby inhibiting BR biosynthesis. NF-YCs also interact with the BR signaling repressor BIN2 to induce its autophosphorylation, which facilitates the stabilization of BIN2 and enhances the suppression of BR signaling (Figure 6). The dual roles of NF-YCs in repressing BR biosynthesis and signaling allow plants to rapidly inhibit BR responses, which consequently maintain their steady photomorphogenic growth in the light. This model provides a mechanistic understanding of how the BR pathway is modulated for light-regulated plant growth.
Figure 6.
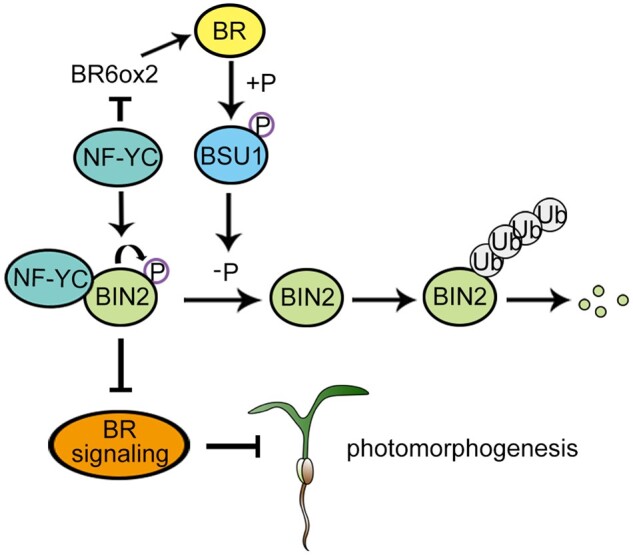
A working model of the role of NF-YCs in light-triggered repression of the BR pathway. In the light, NF-YCs inhibit BR biosynthesis by directly repressing the expression of BR6ox2. On the other hand, NF-YCs enhance BIN2 activity by promoting the autophosphorylation of BIN2 for its stabilization. The NF-YC-mediated inhibition of the BR pathway facilitates photomorphogenic growth by regulating both BR biosynthesis and signaling. The lines ending with arrows indicate promotion. The lines ending with bars indicate repression.
In the dark, the BR pathway is required for etiolated growth, such as long hypocotyls and unexpanded pale cotyledons (Szekeres et al., 1996; Clouse, 2001; Zhang et al., 2015). Consistently, dark-grown BR deficient and BR insensitive mutants display a photomorphogenic phenotype (Chory et al., 1991; Li et al., 1996; Szekeres et al., 1996; Wang et al., 2012). We observed no phenotypic difference between nf-ycQ and WT seedlings grown in the dark (Supplemental Figure S4; Tang et al., 2016), indicating that NF-YCs are not involved in BR-regulated etiolated growth. Unlike the changes that occurred in the light, the expression of BR biosynthesis genes was not altered and BIN2 accumulated to similarly low levels in nf-ycQ and WT seedlings in the dark (Supplemental Figure S9; Figure 5C). In addition, no interaction was observed between NF-YC and BIN2 in dark-grown plants (Supplemental Figure S12). Arabidopsis NF-YA2, an essential component for the formation of the NF-Y complex, is degraded in the dark (Myers et al., 2016). These findings support the conclusion that BR promotes skotomorphogenic growth independently of NF-YCs.
Light inhibits the BR signaling pathway by suppressing the DNA binding activity of BZR1/BES1 and accelerating BZR1 degradation in a photoreceptor-dependent manner (Li et al., 2017a, 2017b; Dong et al., 2019; He et al., 2019). The phosphorylated form of BZR1, which is induced by BIN2 for ubiquitin-mediated degradation, accumulates in light-grown seedlings (Li et al., 2017a, 2017b). In this study, we observed strong NF-YC-dependent accumulation of BIN2 in the light (Figure 5). Perhaps NF-YCs negatively regulate the BR signaling pathway via the BIN2–BZR1/BES1 regulatory cascade. PIFs interact with BZR1 to integrate the BR and light signaling pathways (Oh et al., 2012). Moreover, PIF4/5 directly bind to the promoters of DWF4 and BR6ox2 to activate their transcription by competing with BZR1/BES1, resulting in the de-repression of BR biosynthesis at dawn (Martínez et al., 2018). BIN2 interacts with and phosphorylates PIF4 for ubiquitin-mediated degradation, thereby inhibiting PIF-regulated hypocotyl elongation during the transition from dark to light (Bernardo-Garcia et al., 2014). Our study revealed a dual role for NF-YCs in repressing BR biosynthesis and BIN2 degradation. It would be worth investigating whether NF-YCs regulate light-mediated control of the BR pathway via BZR1/BES1, PIFs, or both.
NF-YB and NF-YC proteins (characterized by a histone-fold domain), together with the DNA binding protein NF-YA, function as the subunits of the heterotrimeric transcription factor NF-Y, which recognizes the CCAAT-boxes in its target genes in eukaryotes (Petroni et al., 2012). NF-YCs function in various developmental processes by interacting with other transcription factors or epigenetic factors. For instance, the NF-YC-RGL2-ABI5 module integrates ABA and GA signaling to regulate seed germination (Liu et al., 2016a, 2016b); NF-YCs interact with ABF3/4 and CONSTANS (CO) to mediate the expression of flowering genes during the floral transition (Hou et al., 2014; Hwang et al., 2019); NF-YCs recruit the histone deacetylase HDA15 to mediate light-regulated hypocotyl elongation (Tang et al., 2016); and so on. These studies illustrate the major roles of NF-YCs as co-transcription factors that regulate gene expression in plants, which is regarded as the current mode by which NF-YCs function.
Here, we revealed that NF-YCs directly regulate BR6ox2 expression to repress BR biosynthesis. In this scenario, NF-YCs might function together with other transcription factors, such as BZR1, which represses the transcription of BR biosynthesis genes (Wang et al., 2012), or with NF-YA/YB, as a CCAAT element is included in the NF-YC-associated promoter region (Figure 2D). CO forms a complex with NF-YB/C to recognize the CORE in the FT gene promoter during the floral transition (Gnesutta et al., 2017). Interestingly, a CORE motif also exists in the P5 region of the BR6ox2 promoter with which NF-YCs are associated, pointing to the possible role of CO-NF-YB/C in photomorphogenesis. In addition, since NF-YCs interact with HDA15 to regulate target gene expression (Tang et al., 2016), the role of NF-YCs in regulating BR6ox2 expression is thought to be dependent on HDA15. Unexpectedly, BR6ox2 had comparable transcript levels in hda15 and Col-0 seedlings (Supplemental Figure S19), indicating that it is not likely that HDA15 is recruited by NF-YCs to regulate BR6ox2 expression.
Interestingly, NF-YCs also contribute to the stabilization of BIN2 by interacting with this protein to negatively regulate the BR signaling pathway, prompting the question of whether NF-YCs repress BR biosynthesis via NF-YC-stabilized BIN2. The gain-of-function mutant dwf12-1d, in which BIN2 is stable like that in bin2-1, exhibits BR-deficient phenotypes such as dwarfism yet shows significant BR accumulation due to the negative feedback regulation of BIN2 (Choe et al., 2002). In this study, we observed higher BR biosynthesis and lower BIN2 abundance in nf-ycQ than the WT (Figures 2, C and 5, C). Therefore, it appears that NF-YC regulates BR biosynthesis and BIN2 stability via different mechanisms. NF-YB/C form a complex together with NF-YA or other factors to regulate gene expression. Here, we revealed that NF-YC alone regulates the stabilization and autophosphorylation of BIN2 in the absence of NF-YA/B. Although the diverse roles of NF-Y in development and stress responses in eukaryotes have been revealed, our finding uncovers a regulatory mode of NF-Y that might be involved in these processes, although the molecular details underlying how NF-YCs inhibit BIN2 degradation remain to be further elucidated.
GSK3 are highly conserved serine/threonine kinases found in all eukaryotes. In contrast to the two isoforms of GSK3s (GSK3α and GSK3β) in mammals, plants contain diverse GSK3-like kinases (Yoo et al., 2006). BIN2, the Arabidopsis ortholog of human GSK3β kinase, functions as a critical repressor of BR signaling and an important component linking many other signaling pathways (He et al., 2002; Yin et al., 2002; Nolan et al., 2020). The phosphorylation of Y216 (Tyr216) in GSK3β, which includes an intramolecular autophosphorylation event, is essential for the full kinase activity of GSK3s in mammalian cells (Hughes et al., 1993; Cole et al., 2004; Saidi et al., 2012). BIN2 Y200, the homolog of GSK3β Y216, was also detected as the autophosphorylation site of BIN2, which plays an essential role in the stabilization and full kinase activity of BIN2 (Kim et al., 2009). BIN2 is inactivated by BSU1 phosphatase through the direct dephosphorylation of pY200 and is then degraded. A gain-of-function mutation of BIN2 (bin2-1) carrying the E263K mutation contains BIN2 in a constitutively phosphorylated form that is barely degraded due to the block of Y200 dephosphorylation (Li et al., 2001; He et al., 2002; Kim et al., 2009). Thus, Y200 plays an essential role in BIN2 activity during BR-dependent plant growth.
HY5 plays a role in the autophosphorylation of BIN2 Y200. The interaction of HY5 with BIN2 allows the active catalytic sites of BIN2 to access its Y200 residue, promoting the phosphorylation of this site and increasing BIN2 activity (Li et al., 2020). Here we showed that NF-YC homologs redundantly interact with BIN2 and maintain BIN2 stabilization by promoting the autophosphorylation of Y200. Although the phosphorylation of Y200 of BIN2 was reduced in both hy5 and nf-ycQ (Li et al., 2020; Figure 5G), loss-of-function of HY5 did not markedly affect the role of NF-YC in regulating BIN2 abundance, or vice versa, in our cell-free assays (Supplemental Figure S20). These preliminary observations suggest that NF-YC and HY5 regulate BIN2 stabilization in at least a partially independent manner. This notion is consistent with the finding that NF-YC functions partially independently of HY5 in light perception, since the hypocotyls of nf-ycT hy5 mutants are considerably longer than those of either parental mutant line (Myers et al., 2016). Nonetheless, the possibility that NF-YC alters the protein structure of BIN2 to increase the catalytic efficiency of BIN2 kinase, as occurs for HY5, cannot be excluded. All NF-YC proteins contain a conserved histone fold domain. Whether this domain facilitates BIN2 autophosphorylation remains to be investigated.
Materials and methods
Plant materials and growth conditions
All A. thaliana NF-YC-related mutants and transgenic lines used in this study were obtained in our laboratory (Tang et al., 2016). The 35pro:BIN2-6MYC transgenic lines were described previously (Li et al., 2017a, 2017b). All plant materials used in this study are listed in Supplemental Table S1. About 75% (v/v) of ethanol with 0.05% (v/v) Triton X-100 were used to sterilize seeds. Plants were grown on MS plates with 1% sucrose and 0.8% agar. Following cold treatment for 3 d in the dark, the plates were incubated for 6 h in full-spectrum white fluorescent light (100 µmol/m−2/s−1) at 22°C for germination and grown vertically in white light under long-day conditions (16-h light/8-h dark) or in the dark for further investigation.
Phenotypic analysis of hypocotyls
Plants were grown in white light under long-day conditions (16-h light/8-h dark) or in the dark for 5 d after germination. Representative seedlings were photographed with a digital camera. The hypocotyls length of seedlings with different genetic backgrounds was measured using ImageJ software (http://imagej.nih.gov/ij/). At least 20 seedlings per genotype were measured as one biological replicate. Average hypocotyl length was calculated with three independent biological replicates that were cultured under the same growth conditions.
Measuring endogenous BR levels
Endogenous BR levels were quantified by Webiolotech (Nanjing, China) as previously reported with some modifications (Ding et al., 2013). Harvested seedlings were ground to a fine powder in liquid nitrogen. Two grams of ground tissue was incubated with 10 mL of 80% aqueous methanol (MeOH) in an ultrasonic bath at 4°C for 2 h and centrifuged at 13,000g at 4°C for 5 min. The supernatant was loaded onto an activated MCX cartridge (Poly-Sery MCX SPE, 500 mg/6 mL, CNW Technologies GmbH, Duesseldorf, Germany ) and eluted with 3 mL MeOH. After drying under an N2 stream, the residue was redissolved in 200 µL MeOH and the solution was filtered twice through a 0.22 µm nylon membrane (SLGNX13NL; Millipore, Burlington, MA, USA). Subsequently, 5 µL of sample was subjected to HPLC–tandem mass spectrometry (HPLC–MS/MS) analysis. BR analysis was performed on a quadruple linear ion trap hybrid MS (QTRAP 6500; AB SCIEX, Framingham, MA, USA) equipped with an electrospray ionization source coupled with an HPLC (Aglient1290; Aglient, Santa Clara, CA, USA). Poroshell 120 SB-C18 reversed-phase chromatography (2.1 × 150, 2.7 µm) was adopted for CS and BL analysis, and a Waters BEH reversed-phase chromatography column (2.1 × 150, 2.7 µm) was used for 6-deoxo-CS analysis. The concentration curve of BR standards was generated by analyzing different concentrations (0.2, 0.5, 1.0, 2.0, 5.0, 20, and 50 ng/mL) of BL (Sigma-Aldrich, St Louis, MO, USA), CS (OlchemIm, Olomouc, Czech Republic) and 6-deoxo-CS (OlchemIm, Czech Republic) standards respectively, by HPLC–MS/MS (see Supplemental File S1). To measure BL, CS, and 6-deoxo-CS levels, the multiple reaction monitoring transitions 481.2→445.3, 465.3→429.4, and 449.4→128.9 were used for quantification, and 481.2→315.3, 465.3→429.4, and 451.4→377.2 were used for qualification, respectively (Supplemental Table S2). The content of BRs was determined based on the concentration curve of BR standards (Supplemental File S1).
RNA extraction and RT-qPCR analysis
Total RNA was extracted from various 3-d-old seedlings using an E.Z.N.A. Total RNA Kit I (Omega, Bienne, Switzerland) and reverse transcribed to cDNA using MMLV-RTase (Promega, Madison, WI, USA). Gene expression levels were determined by RT-qPCR with KAPA SYBR Fast qPCR Kit Master Mix (KAPA BIO, Wilmington, MA, USA) and a Light Cycler 480 thermal cycler (Roche, Basel, Switzerland; Liu et al., 2016a, 2016b). PP2A was used as an internal control. Results were obtained from three biological replicates. All primers used are listed in Supplemental Table S3.
Transient expression assay
The promoter of the BR biosynthesis gene BR6ox2 was fused with the LUC gene and Renilla (REN) gene, and the internal control gene driven by the 35S promoter was used as the reporter construct. 35Spro:NF-YC9-6HA was used as the effector construct. Equal amounts of effector and reporter plasmids or the controls were co-transformed into Arabidopsis mesophyll protoplasts (Tang et al., 2016). After culturing in the dark for 2 h to express the reporter gene, the protoplasts were transferred to white light for 12 h and collected. The dual-LUC assay system (Promega) was used to measure LUC and REN activity with a Berthold Centro LB960 luminometer. The LUC/REN ratio was calculated as the final value.
ChIP-qPCR assay
ChIP assays were performed as previously described (Tang et al., 2016). Three-day-old nf-yc9-1NF-YC9pro:NF-YC9-3FLAG and Col-0 seedlings grown in white light and the dark were collected for fixation by 1% formaldehyde. Chromatin was extracted from the samples and sheared into DNA fragments with an average size of ∼500 bp by sonication. The sonicated chromatin was immunoprecipitated using Protein G PLUS/Protein A agarose (16-201; Millipore) with anti-FLAG antibody (F3165; Sigma-Aldrich), and the precipitated DNA fragments were recovered and analyzed by qPCR with KAPA SYBR Fast qPCR Kit Master Mix (KAPA BIO) using specific primers (Supplemental Table S3). Relative fold enrichment was calculated by normalizing the amount of a target DNA fragment against that of a PP2A genomic fragment and then against the respective input DNA samples. Actin2 and TUB8 were used as negative controls.
Yeast two-hybrid assay
Yeast constructs harboring NF-YC1, NF-YC3, NF-YC4, and NF-YC9 were described previously (Tang et al., 2016). The coding region of BIN2 was amplified and cloned into pGADT7 as AD-BIN2. The specific bait and prey constructs were co-transformed into yeast AH109 cells using the Yeastmaker Yeast Transformation System 2 (Clontech, Mountain View, CA, USA) according to the manufacturer’s instructions and grown on SD/-Trp/-Leu and SD/Trp/-Leu/-His/-Ade media for interaction tests.
Pull-down assay
The coding regions of NF-YC1, NF-YC3, NF-YC4, NF-YC9, and BIN2 were cloned into the pQE30 (Qiagen, Hilden, Germany) and pGEX-4T-1 (Pharmacia, Peapack, NJ, USA) vectors to produce His-NF-YC1, His-NF-YC3, His-NF-YC4, His-NF-YC9, and GST-BIN2, respectively. Primers used for vector construction are listed in Supplemental Table S3. The expression of GST and His recombinant proteins in E. coli Rosetta cells (DE3; Novagen, Madison, WI, USA) was induced by treatment with 0.5 mM IPTG for 4 h. The proteins were purified using Ni-NTA agarose beads (30210; Qiagen) or Glutathione Sepharose Beads (17-0756-01; Amersham Biosciences, Little Chalfont, UK). For the pull-down assay, His-NF-YCs were incubated in binding buffer (50 mM Tris–HCl, pH 8.0, 100 mM NaCl, and 1 mM EDTA) with immobilized GST or GST-BIN2 at 4°C for 4 h. After washing 3 times with binding buffer, proteins that were retained on the beads were separated by SDS–polyacrylamide gel electrophoresis (PAGE) and detected with anti-His antibody (AbM59012-18-PU, BGI) or anti-GST antibody (AB101-02; Tiangen, Beijing, China).
BiFC analysis
The coding regions of BIN2 and NF-YC9 were amplified and cloned into a pGreen binary vector containing the N-terminus of Enhanced Yellow Fluorescent Protein (EYFP) and the C-terminus of EYFP, respectively, as described previously (Hou et al., 2014). Equal amounts of the plasmids were co-transformed into Arabidopsis mesophyll protoplasts. The protoplasts were cultured overnight, and yellow fluorescent protein signals were detected under an inverted fluorescence microscope (M165C; Leica, Wetzlar, Germany). VirD2NLS-mCherry was used as the nucleus marker.
Co-IP assay
Three-day-old nf-yc9-1NF-YC9pro:NF-YC9-3FLAG/35Spro:BIN2-6MYC seedlings were grown in the light for the Co-IP assay. Total proteins were extracted from the samples with extraction buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM DTT, and 1% Triton X-100) and incubated with either anti-MYC antibody (Sigma-Aldrich) or pre-immune serum (IgG) immobilized onto Protein G PLUS/Protein A-Agarose Suspension (16-201, Millipore) in Co-IP buffer (50 mM HEPES, pH 7.5, 150 mM KCl, 10 mM ZnSO4, 5 mM MgCl2, 1% Triton X-100) at 4°C for 2 h. After incubation, the beads were washed 3 times before being eluted with 5× SDS loading buffer. The proteins that bound to the beads were resolved by SDS–PAGE and detected by anti-FLAG (F3165; Sigma-Aldrich) antibody and anti-MYC antibody (Sigma-Aldrich).
Cell-free protein degradation assay
The cell-free protein degradation assay was performed as described previously with some modifications (Sun et al., 2018). The cell extracts were generated by grinding plant tissue in degradation buffer (25 mM Tris–HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 5 mM DTT, 100 µM CHX, and 10 mM ATP) with equal amounts of plant grinding powder. Cell debris was removed by two rounds of centrifugation at 20,000g at 4°C for 15 min each time. Cell extracts (100 μL containing 500 μg total proteins) from different genotypes were incubated with equal amounts of His-BIN2 recombinant protein at 25°C for time course degradation assays.
In vitro kinase assay
The in vitro kinase assay was performed as described previously with some modifications (Kim et al., 2011). The expression of GST and His recombinant proteins in E. coli Rosetta cells (DE3; Novagen) was induced by adding IPTG (0.5 mM) and incubating at 28°C for 4–6 h. GST and His recombinant proteins were purified using Ni-NTA agarose beads (30210; Qiagen) and Glutathione Sepharose Beads (17-0756-01; Amersham Biosciences), respectively. GST or His recombinant proteins were eluted from the beads with elution buffer containing 20 mM glutathione or 250 µM imidazole. Glutathione or imidazole was removed from the proteins by ultrafiltration using a Centricon Plus-20 centrifuge filter unit. (Amicon Ultra; Millipore). The purified recombinant protein was separated on a gel and stained with Coomassie Blue (Supplemental Figure S21).
To prepare fully dephosphorylated BIN2 proteins, GST-BIN2 or His-BIN2 was incubated with CIP (calf intestine alkaline phosphatase; New England Biolabs, Ipswich, MA, USA) in buffer (20 mM Tris at pH 7.5, 1 mM MgCl2, 100 mM NaCl, and 1 mM DTT) at 37°C for 30 min. The protein mixture was incubated with Glutathione Sepharose beads or Ni-NTA agarose beads to purify GST-BIN2 and His-BIN2. For the autophosphorylation assay, GST-BIN2 (or His-BIN2) protein was incubated with equal amounts of His-NF-YC9 or His-GFP in kinase buffer (20 mM Tris at pH 7.5, 1 mM MgCl2, 100 mM NaCl, and 1 mM DTT) containing 100 µM ATP (Sigma-Aldrich) at 30°C for 1 h. Phosphorylated GST-BIN2 (or His-BIN2) protein was detected with anti-pTyr279/Tyr216 (pTyr) antibody (Sigma-Aldrich) and anti-GST antibody (or anti-His). 32P radioactive assays were performed as previously described with some modifications (Li et al., 2020). About 0.5 µg His-BIN2 was mixed with NF-YC9 (at a mole ratio of BIN2/NF-YC9 from 1:1 to 1:4, as indicated), and 1 µg His-BZR1-GFP or CIP-treated BIN2 was incubated with the mixture in 20 µL kinase buffer containing 0.2 µCi [γ-32P] ATP (PerkinElmer) at 30°C for 1 h. The reactions were terminated by adding 1× SDS loading buffer, and the samples were separated in 12% SDS–PAGE. 32P signals were detected with Typhoon FLA9500 (GE Healthcare, Chicago, IL, USA).
Statistical analysis
Student’s t tests were used to compare groups in the indicated experiments. P < 0.01 was considered to be statistically significant. In all comparisons, *P < 0.01 (Supplemental Data Set S1).
Accession numbers
Sequence data from this article can be found in the TAIR website under the following accession numbers: NF-YC1 (AT3G48590), NF-YC3 (AT1G54830), NF-YC4 (AT5G63470), NF-YC9 (AT1G08970), IAA19 (AT3G15540), PRE1 (AT5G39860), PRE5 (AT3G28857), SAUR-AC1(AT4G38850), ACS5 (AT5G65800), XTH18 (AT4G30280), ATEXPA8 (AT2G40610), PP2A (AT1G69960), ACT2 (AT3G18780), DET2 (AT2G38050), CPD (AT5G05690), DWF4 (AT3G50660), ROT3 (AT4G36380), CYP90D1(AT3G13730), BR6ox1 (AT5G38970), BR6ox2 (AT3G30180), BIN2 (AT4G18710), BIL1 (AT2G30980), BIL2 (AT1G06390), HY5 (AT5G11260), and BZR1(AT1G75080).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. An overview of the BR biosynthesis pathway.
Supplemental Figure S2. RT-qPCR analysis of BR-responsive gene expression in Col-0 seedlings grown in the light and the dark for 3 d.
Supplemental Figure S3. BR contents of bri1-301 seedlings grown in the light.
Supplemental Figure S4. Hypocotyl phenotypes of various NF-YC mutants treated with BL, PIC, and GA3 in the dark.
Supplemental Figure S5. Hypocotyl lengths of various NF-YC mutants treated with BL, PIC, and GA3 in the light.
Supplemental Figure S6. nf-ycQ plants exhibit significantly lower sensitivity to BL during petiole elongation compared with Col-0.
Supplemental Figure S7. Hypocotyl phenotypes of Col-0 and nf-ycQ seedlings treated with BRZ.
Supplemental Figure S8. BR signaling is activated in light-grown nf-ycQ seedlings.
Supplemental Figure S9. NF-YCs are not involved in regulating the BR pathway in the dark.
Supplemental Figure S10. NF-YCs interact with BIN2, but not BZR1, in yeast two-hybrid assays.
Supplemental Figure S11. In vitro pull-down assay showing the direct interactions between His-NF-YC1/3/4 and GST-BIN2 recombinant proteins.
Supplemental Figure S12. Co-IP assay showing that BR does not mediate the interaction of NF-YC9 and BIN2.
Supplemental Figure S13. Hypocotyl phenotypes of seedlings with various genetic backgrounds grown in the dark.
Supplemental Figure S14. RT-qPCR analysis of BIN2, BIL1, and BIL2 expression.
Supplemental Figure S15. BIN2 levels in 35Spro:BIN2-6MYC and nf-ycQ/35Spro:BIN2-6MYC seedlings grown in the light or the dark for 3 d.
Supplemental Figure S16. Immunoblot analysis of ubiquitinated BIN2.
Supplemental Figure S17. In vitro kinase assay showing that NF-YCs enhance the autophosphorylation activity of BIN2.
Supplemental Figure S18. Detection of de-phosphorylated BZR1 in planta.
Supplemental Figure S19. HDA15 does not regulate BR6ox2 expression.
Supplemental Figure S20. Cell-free degradation assay showing that NF-YC and HY5 regulate BIN2 stabilization in a partially independent manner.
Supplemental Figure S21. Coomassie blue staining of purified recombinant protein used for the in vitro kinase assays.
Supplemental Table S1. Sources of the plant materials used in this study.
Supplemental Table S2. Selected reaction monitoring conditions for protonated or deprotonated plant hormones.
Supplemental Table S3. List of primers used in this study.
Supplemental File S1. Chromatograms and fragmented ions ratio of BR measurements.
Supplemental Data Set S1. Statistical analysis.
Supplementary Material
Acknowledgments
We thank Dr Shuhua Yang for providing 35Spro:BIN2-6MYC seeds, Dr Jun-Xian He for bin2-1 seeds, Dr Jianming Li for anti-BZR1 antibody, and Dr Ziyin Yang for providing technical support.
Funding
This work was supported by a grant from the “Strategic Priority Research Program” of the Chinese Academy of Sciences (No. XDA24010000).
Conflict of interest statement: The authors declare that they have no conflict of interests.
W.Z. and X.H. designed the research. W.Z., Y.T., Y.Y., J.C., Y.H., and H.L. performed the research. W.Z., Y.H., Y.T., C.Z., X.L., and X.H. analyzed data; W.Z. and X.H. wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Xingliang Hou (houxl@scib.ac.cn)
References
- Bernardo-Garcia S, de Lucas M, Martinez C, Espinosa-Ruiz A, Daviere JM, Prat S (2014) BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev 28: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ (2012) Shade avoidance. Arabidopsis Book 10: e0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA (1991) Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee MO, Yoshida S, Feldmann KA, Tax FE (2002) Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3beta-like kinase. Plant Physiol 130: 1506–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Choe S (2013) The regulation of brassinosteroid biosynthesis in Arabidopsis. Crit Rev Plant Sci 32: 396–410 [Google Scholar]
- Clouse SD (2001) Integration of light and brassinosteroid signals in etiolated seedling growth. Trends Plant Sci 6: 1360–1385 [DOI] [PubMed] [Google Scholar]
- Cole A, Frame S, Cohen P (2004) Further evidence that the tyrosine phosphorylation of glycogen synthase kinase-3 (GSK3) in mammalian cells is an autophosphorylation event. Biochem J 377: 249–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M, Galvao VC, Fankhauser C (2016) Light-mediated hormonal regulation of plant growth and development. Annu Rev Plant Biol 67: 513–537 [DOI] [PubMed] [Google Scholar]
- Ding J, Mao LJ, Wang ST, Yuan FB, Feng YQ (2013) Determination of endogenous brassinosteroids in plant tissues using solid-phase extraction with double layered cartridge followed by high-performance liquid chromatography– tandem mass spectrometry. Phytochem Anal 24: 386–394 [DOI] [PubMed] [Google Scholar]
- Dong HX, Liu J, He GH, Liu P, Sun JP (2019) Photoexcited phytochrome B interacts with brassinazoleresistant 1 to repress brassinosteroid signaling in Arabidopsis. J Integr Plant Biol 5: 652–667 [DOI] [PubMed] [Google Scholar]
- Gnesutta N, Kumimote RW, Swain S, Chiara M, Siriwardana C, Horner DS, Holt 3d BF, Mantovani R (2017) CONSTANS imparts DNA sequence specificity to the histone fold NF-YB/NF-YC dimer. Plant Cell 29: 1516–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Liu J, Dong H, Sun J (2019) The blue-light receptor CRY1 interacts with BZR1 and BIN2 to modulate the phosphorylation and nuclear function of BZR1 in repressing BR signaling in Arabidopsis. Mol Plant 12: 689–703 [DOI] [PubMed] [Google Scholar]
- Hou X, Zhou J, Liu C, Liu L, Shen L, Yu H (2014) Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat Commun 5: 4601. [DOI] [PubMed] [Google Scholar]
- Hwang K, Susila H, Nasim Z, Jung JY, Ahn JH (2019) Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol Plant 4: 489–505 [DOI] [PubMed] [Google Scholar]
- Hughes K, Nikolakaki E, Plyte SE, Totty NF, Woodgett JR (1993) Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. EMBO J 12: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Burlingame AL, Wang ZY (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, Wang ZY (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuka T, Kobayashi J, Horiguchi G, Demura T, Sakakibara H, Tsukaya H, Nagatani A (2010) Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol 153:1608–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ye K, Shi Y, Cheng J, Zhang X, Yang S (2017a) BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol Plant 10: 545–559 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J (2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Terzaghi W, Gong Y, Li C, Ling JJ, Fan Y, Qin N, Gong X, Zhu D, Deng XW (2020) Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat Commun 11: 1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QF, He JX (2016) BZR1 interacts with HY5 to mediate brassinosteroid- and light-regulated cotyledon opening in Arabidopsis in darkness. Mol Plant 9: 113–125 [DOI] [PubMed] [Google Scholar]
- Li QF, Hung LC, Wei K, Yu JW, Zhang CH Q, Liu QQ (2017b) Light involved regulation of BZR1 stability and phosphorylation status to coordinate plant growth in Arabidopsis. Biosci Rep 37: BSR20170069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JJ, Li J, Zhu D, Deng XW (2017) Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc Natl Acad Sci USA 114: 3539–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Mei S, Shi C, Yang Y, Peng Y, Ma L, Wang F, Li X, Huang X, Yin Y,. et al. (2018) UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev Cell 44: 512–523 [DOI] [PubMed] [Google Scholar]
- Liu X, Hu P, Huang M, Tang Y, Li Y, Li L, Hou X (2016a) The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat Commun 7: 12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yang C, Li L (2016b) Shade-induced stem elongation in rice seedlings: implication of tissue-specific phytohormone regulation. J Integr Plant Biol 58: 614–617 [DOI] [PubMed] [Google Scholar]
- Luo XM, Lin WH, Zhu S, Zhu JY, Sun Y, Fan XY, Cheng M, Hao Y, Oh E, Tian M. et al. (2010) Integration of light- and brassinosteroid-signaling pathways by a GATA -transcription factor in Arabidopsis. Dev Cell 19: 872–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez C, Ruíz AE, de Lucas M, García SB, Zorrilla JM, Prat S (2018) PIF4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J 37: e99552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers ZA, Holt BF 3rd (2018) NUCLEAR FACTOR-Y: still complex after all these years? Curr Opin Plant Biol 45: 96–102 [DOI] [PubMed] [Google Scholar]
- Myers ZA, Kumimoto RW, Siriwardana CL, Gayler KK, Risinger JR, Pezzetta D, Holt 3rd BF (2016) NUCLEAR FACTOR Y, subunit C (NF-YC) transcription factors are positive regulators of photomorphogenesis in Arabidopsis thaliana. PLoS Genet 12: e1006333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami A, Takahashi K, Inoue SI, Tada Y, Kinoshita T (2019) Brassinosteroid induces phosphorylation of the plasma membrane H+- ATPase during hypocotyl elongation in Arabidopsis thaliana. Plant Cell Physiol 60: 935–944 [DOI] [PubMed] [Google Scholar]
- Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, Vonrhein C, Moras D, Romier C, Bolognesi M, et al. (2013) Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 152: 132–143 [DOI] [PubMed] [Google Scholar]
- Neff MM, Street IH, Turk EM, Ward JM (2005) Interaction of light and hormone signaling to mediate photomorphogenesis. Photomorphogenesis in Plants and Bacteria. Springer Science & Business Media, Berlin, Germany, pp 441–445 [Google Scholar]
- Nolan TM, Vukasinovic N, Liu D, Russinova E, Yin Y (2020) Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32: 295–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF 3rd, Mantovani R (2012) The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24: 4777–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi Y, Hearn TJ, Coates JC (2012) Function and evolution of ‘green’ GSK3/Shaggy-like kinases. Trends Plant Sci 17: 39–46 [DOI] [PubMed] [Google Scholar]
- Siefers N, Dang KK, Kumimoto RW, Bynum W, Tayrose G, Holt BF 3rd (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149: 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SHY, Wang T, Wang LL, Li XM, Jia YC, Liu CH, Huang XH, Xie WB, Wang XL (2018) Natural selection of a GSK3 determines rice mesocotyl domestication by coordinating strigolactone and brassinosteroid signaling. Nat Commun 9: 2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C, et al. (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Tang Y, Liu X, Liu X, Li Y, Wu K, Hou X (2016) Arabidopsis NF-YCs mediate the light-controlled hypocotyl elongation via modulating histone acetylation. Mol Plant 10: 260–273 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY (2012) Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 46: 701–724 [DOI] [PubMed] [Google Scholar]
- Wang W, Lu X, Li L, Lian H, Mao Z, Xu P, Guo T, Xu F, Du S, Cao X, et al. (2018) Photoexcited CRYPTOCHROME1 interacts with dephosphorylated BES1 to regulate brassinosteroid signaling and photomorphogenesis in Arabidopsis. Plant Cell 30: 1989–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei ZY, Yuan T, Tarkowska D, Kim J, Nam HG, Novak O, He K, Gou X, Li J (2017) Brassinosteroid biosynthesis is modulated via a transcription Factor cascade of COG1, PIF4, and PIF5. Plant Physiol 174: 1260–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to -regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yoo MJ, Albert VA, Soltis PS, Soltis DE (2006) Phylogenetic diversification of glycogen synthase kinase 3/SHAGGY- like kinase genes in plants. BMC Plant Biol 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Liu ZJ, Wang JF, Chen YB, He JX (2015) Brassinosteroid is required for sugar promotion of hypocotyl elongation in Arabidopsis in darkness. Planta 242: 881–893 [DOI] [PubMed] [Google Scholar]
- Zhao B, Li J (2012) Regulation of brassinosteroid biosynthesis and inactivation. J Integr Plant Biol 54: 746–759 [DOI] [PubMed] [Google Scholar]
- Zhu JY, Li YY, Cao DM, Yang HG, Oh E, Bi Y, Zhu SW, Wang ZY (2017) The F-box protein KIB1 mediates brassinosteroid- induced inactivation and degradation of GSK3-like kinases in Arabidopsis. Mol Cell 66: 648–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.