Abstract
Cryptochromes are blue light photoreceptors that mediate various light responses in plants and mammals. In Arabidopsis (Arabidopsis thaliana), cryptochrome 1 (CRY1) mediates blue light-induced photomorphogenesis, which is characterized by reduced hypocotyl elongation and enhanced anthocyanin production, whereas gibberellin (GA) signaling mediated by the GA receptor GA-INSENSITIVE DWARF1 (GID1) and DELLA proteins promotes hypocotyl elongation and inhibits anthocyanin accumulation. Whether CRY1 control of photomorphogenesis involves regulation of GA signaling is largely unknown. Here, we show that CRY1 signaling involves the inhibition of GA signaling through repression of GA-induced degradation of DELLA proteins. CRY1 physically interacts with DELLA proteins in a blue light-dependent manner, leading to their dissociation from SLEEPY1 (SLY1) and the inhibition of their ubiquitination. Moreover, CRY1 interacts directly with GID1 in a blue light-dependent but GA-independent manner, leading to the inhibition of the interaction between GID1 with DELLA proteins. These findings suggest that CRY1 controls photomorphogenesis through inhibition of GA-induced degradation of DELLA proteins and GA signaling, which is mediated by CRY1 inhibition of the interactions of DELLA proteins with GID1 and SCFSLY1, respectively.
Blue light-dependent interactions of CRY1 with GID1 and DELLA proteins inhibit gibberellin (GA)-induced degradation of DELLA proteins to regulate GA signaling and photomorphogenesis.
Introduction
Light, as both an energy source and an external environmental signal, profoundly influences the entire life span of plants to regulate their growth and development (Fankhauser and Chory, 1997; Deng and Quail, 1999). To monitor the dynamic changes of light in a quantitative and qualitative manner, plants have evolved multiple photoreceptors to perceive light signals. These photoreceptors include the blue/UV-A light receptors cryptochromes (CRYs), phototropins, red/far-red light receptors phytochromes (PHYs, phyA to phyE), and the UV-B receptor UVR8 (Cashmore et al., 1999; Briggs and Christie, 2002; Quail, 2002; Rizzini et al., 2011; Yadav et al., 2020). Among them, CRYs regulate a broad spectrum of physiological processes in plants, including seedling photomorphogenesis, photoperiodic flowering, circadian rhythms, and stomatal opening and development (Ahmad and Cashmore, 1993; Guo et al., 1998; Somers et al., 1998; Toth et al., 2001; Mao et al., 2005; Liu et al., 2008a; Kang et al., 2009). Cryptochromes are present not only in plants, but also in a variety of other organisms from bacteria to humans. In the fruit fly Drosophila melanogaster, cryptochrome serves as photoreceptor to entrain the circadian clock (Emery et al., 1998; Kume et al., 1999), and in mammals, cryptochrome acts as an integral component of the circadian clock. In migratory butterflies and birds, cryptochrome is responsible for sensing the Earth’s magnetic field and providing precise navigation during their long-distance migrations (Gegear et al., 2010).
In Arabidopsis, there are two homologous CRYs: CRY1 and CRY2. CRY1 plays a major role in mediating the blue light promotion of photomorphogenesis, which is characterized by the inhibition of hypocotyl elongation and promotion of anthocyanin accumulation (Ahmad and Cashmore, 1993; Lin et al., 1996), whereas CRY2 acts as the primary blue light photoreceptor that promotes floral initiation under long-day photoperiods (Guo et al., 1998). CRYs possess a photolyase-related N-terminal domain and a distinguishing C-terminal domain. The C-terminal domain of Arabidopsis CRY1 and CRY2 was shown to mediate signaling by its direct interaction with CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1; Wang et al., 2001; Yang et al., 2001), a RING-finger E3 ubiquitin ligase (Deng et al., 1992) that interacts with and targets a set of transcription factors, such as LONG HYPOCOTYL 5 (HY5) and CONSTANS (CO), for degradation (Osterlund et al., 2000; Liu et al., 2008a; Jang et al., 2008) to regulate photomorphogenesis and flowering. Moreover, CRY1 and CRY2 also interact with the COP1 enhancer, SUPPRESSOR OF PHYA-105 1 (SPA1; Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011), which interacts with COP1 to enhance its E3 ligase activity (Seo et al., 2003). The interaction of CRY1/CRY2 with COP1/SPA1 leads to a disruption of the COP1–SPA1 core complex, thus promoting the accumulation of HY5 and CO. The signaling mechanism of CRY1 was also recently demonstrated to involve the inhibition of auxin and brassinosteroid (BR) signaling, which is mediated by blue light-dependent interactions between CRY1 and AUX/IAA and AUXIN RESPONSE FACTOR (ARF) proteins (for auxin), and BRI1-EMS-SUPPRESSOR 1 (BES1)/BES1-INTERACTING MYC-LIKE1 (BIM1; for BRs; Xu et al., 2018; Wang et al., 2018a; Mao et al., 2020).
Gibberellins (GAs) are an essential class of phytohormones that regulate a variety of plant growth and developmental processes, including seed germination, hypocotyl and stem elongation, leaf expansion, and floral initiation (Fleet and Sun, 2005; Pimenta and Lange, 2006). Extensive genetic and biochemical studies have elucidated much of the GA signaling transduction pathway, and identified several key components in GA signaling, which include the GA receptor GA-INSENSITIVE DWARF1 (GID1), the F-box protein SLEEPY (SLY1), and DELLA proteins. GID1 was identified as a soluble GA receptor in rice (Oryza sativa; (Ueguchi-Tanaka et al., 2005). In Arabidopsis, there are three homologous GID1s: GID1a, GID1b, and GID1c, which perform redundant roles in sensing the GA signal (Nakajima et al., 2006). GID1 interacts with DELLA proteins in a GA-dependent manner, which is required for the GA-induced degradation of DELLA proteins (Griffiths et al., 2006; Willige et al., 2007). SLY1 and its close homolog SNEEZY (SNE) are the F-box subunits of a Skp-Cullin-F-box (SCF) E3 ubiquitin ligase complex that interacts with its substrates, the DELLA proteins, to promote their ubiquitination and degradation, and mediate GA responses (Dill et al., 2004; Fu et al., 2004; Ariizumi et al., 2011). The DELLA proteins are the most extensively studied components of the GA signaling cascade. They are nuclear-localized and act as repressors of GA-responsive growth by repressing the expression of GA-responsive genes, and as central regulators to integrate and convey information from multiple developmental pathways. The Arabidopsis genome encodes five DELLA proteins: REPRESSORS OF GA (RGA), GA INSENSITIVE 1 (GAI), RGA-LIKE 1 (RGL1), RGL2, and RGL3. RGA and GAI are the major DELLA proteins that block GA-promoted growth and floral initiation, while RGL2 is the primary DELLA protein suppressing seed germination. In addition, RGA, RGL1, and RGL2 are involved in the repression of floral development (King et al., 2001; Lee, 2002; Tyler et al., 2004; Cheng et al., 2004). Importantly, the GA-triggered degradation of DELLA proteins is a key event in GA signaling, which is dependent on the interactions of DELLA proteins with both GID1 and SLY1/SNE (Silverstone et al., 2001; Dill et al., 2004; Griffiths et al., 2006).
DELLA proteins contain an N-terminal DELLA domain and a C-terminal GRAS domain. The GRAS domain is likely the functional domain, and is presumably responsible for transcriptional regulation. Additionally, the GRAS domain mediates the association of DELLA proteins with SLY1 (Dill et al., 2004). The DELLA domain mediates the interaction between DELLA proteins and GID1, which is essential for the GA-induced degradation of DELLA proteins (Griffiths et al., 2006; Willige et al., 2007). The binding of bioactive GA to GID1 induces a conformational change in GID1, which allows GA–GID1 to interact with DELLA proteins and promote the association of GA–GID1–DELLA with SLY1, thereby targeting the DELLA proteins for degradation via the 26S proteasome (Murase et al., 2008; Shimada et al., 2008; Sun, 2011).
While GA induces hypocotyl elongation, the GA biosynthesis inhibitor paclobutrazol (PAC) represses hypocotyl elongation (Alabadi et al., 2004; Feng et al., 2008). Moreover, the GA biosynthesis-deficient mutant GA requiring 1 (ga1) and loss-of-function mutants in the GA receptor GID1 display shorter hypocotyls when grown under red light (Alabadi et al., 2004; Griffiths et al., 2006; Feng et al., 2008). Notably, the DELLA proteins inhibit hypocotyl elongation in red light (Achard et al., 2007). The direct interaction of DELLA proteins with the basic helix–loop–helix (bHLH) transcription factors PHYTOCHROME INTERACTING 3 (PIF3) and PIF4 was shown to integrate light and GA signaling pathways (de Lucas et al., 2008; Feng et al., 2008), PIF3 and PIF4 are key downstream factors of phytochromes that negatively regulate photomorphogenesis (Ni et al., 1998, 1999; Leivar et al., 2008). These interactions result in the inhibition of the DNA-binding activities of PIF3 and PIF4. Light can also regulate GA biosynthesis through photoreceptors. For example, cryptochromes induce the expression of GIBBERELLIN 2-OXIDASE 1 (GA2ox1), but repress the expression of GA20ox1 or GA3ox1, which leads to a decrease in GA levels and the inhibition of hypocotyl elongation (Zhao et al., 2007). PhyA and phyB mediate the light-induced stabilization of DELLA proteins, which may partially lead to the phytochrome-dependent repression of GA homeostasis (Achard et al., 2007). These studies strongly demonstrate an important role for GA in light-regulated photomorphogenesis.
Our previous study demonstrated that cry1 mutant seedlings show a dramatically enhanced responsiveness to the inhibitor of GA biosynthesis, PAC, and GA in blue light, whereas CRY1-overexpressing seedlings displayed a reduced responsiveness to PAC and GA, indicating that CRY1 mediates blue light suppression of GA responses (Wang et al., 2016). However, whether and how CRY1 regulates GA signaling remains largely unknown. In this study, we show that CRY1 mediates blue light inhibition of GA-induced degradation of DELLA proteins. Through a series of protein–protein interaction studies, we demonstrate that Arabidopsis CRY1 physically interacts with DELLA proteins and the GA receptor GID1 in a blue light-dependent manner. We further demonstrate by a series of protein–protein interaction assays that the blue light-triggered interaction of CRY1 with DELLA proteins and GID1 impairs the interaction of DELLA proteins with SLY1 and GID1. Our findings indicate that GID1 and DELLA proteins act as the direct downstream components of CRY1 in mediating blue light signaling. Photoexcited CRY1 therefore inhibits the association of SCFSLY1 with DELLA proteins to repress their ubiquitination, but also represses the interaction of GID1 with DELLA proteins, which may interfere with the recognition of DELLA proteins by SCFSLY1, and further inhibit their ubiquitination. These two layers of regulatory mechanisms may allow CRY1 to rapidly and efficiently mediate light inhibition of GA signaling, which enables plants to grow and develop in a tightly controlled manner according to the ambient light conditions.
Results
CRY1 mediates the blue light inhibition of GA-induced degradation of DELLA proteins
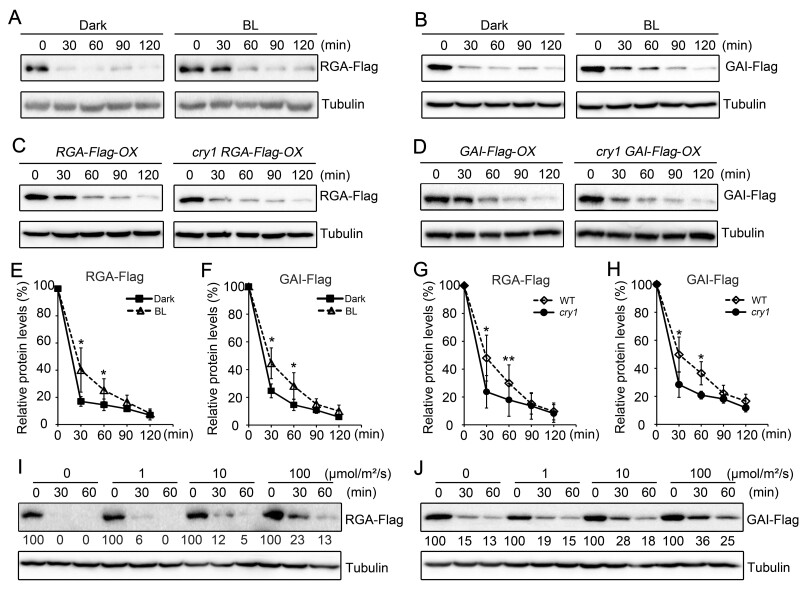
Given our previous demonstration that CRY1 is involved in the repression of GA-induced hypocotyl elongation (Wang et al., 2016) and that CRY1 inhibits auxin signaling by interacting with AUX/IAA proteins to stabilize them (Xu et al., 2018), we explored whether CRY1 might affect GA-triggered degradation of DELLA proteins to regulate GA signaling. To this end, we generated transgenic seedlings overexpressing RGA and GAI tagged with a Flag tag in the wild-type (WT) and cry1 mutant backgrounds (RGA-Flag-OX, GAI-Flag-OX, cry1 RGA-Flag-OX, and cry1 GAI-Flag-OX), to determine whether CRY1 affects GA-induced degradation of DELLA proteins. We used the GA biosynthesis inhibitor PAC to minimize possible interference from endogenous GA potentially resulting from different genetic backgrounds or different light treatments. We performed immunoblot assays using an anti-Flag antibody to detect RGA and GAI proteins levels in GA (GA3)-treated RGA-Flag-OX and GAI-Flag-OX seedlings grown in the dark or illuminated with blue light for 2 h. We observed that both RGA and GAI proteins are degraded much faster in seedlings grown in the dark than in those exposed to blue light (Figure 1, A, B, E, and F). To determine whether CRY1 mediates the blue light inhibition of GA-induced degradation of DELLA proteins, we analyzed RGA and GAI proteins levels in RGA-Flag-OX, cry1 RGA-Flag-OX, GAI-Flag-OX, and cry1 GAI-Flag-OX seedlings adapted in the dark or exposed to blue light for 2 h. GA-induced degradation of both RGA and GAI was faster in the cry1 mutant background than in WT within 1 h of blue light exposure (Figure 1, C, D, G, and H). Moreover, the GA-triggered degradation of RGA and GAI proteins gradually diminished as the blue light fluence rate increased (Figure 1, I and J). We further determined that both red and far-red lights can also inhibit the GA-induced degradation of RGA and GAI (Supplemental Figure S1, A and B).
Figure 1.
CRY1 mediates the blue light inhibition of GA-induced degradation of DELLA proteins. A, B, Blue light inhibits GA-induced degradation of RGA-Flag (A) and GAI-Flag (B) proteins, as shown by immunoblot analysis. RGA-Flag-OX and GAI-Flag-OX seedlings (WT background) were grown on MS medium containing 1-μM PAC in the dark for 5 days, and were then treated with 100-μM GA3 + 1-μM PAC and exposed to blue light (BL, 30 μmol·m–2·s–1) or maintained in the dark for the indicated time. Three independent experiments were performed, and one is shown. C, D, CRY1 mediates the blue light inhibition of GA-induced degradation of RGA-Flag (C) and GAI-Flag (D) proteins, as shown by immunoblot analysis. RGA-Flag-OX and GAI-Flag-OX seedlings (WT and cry1 backgrounds) were grown on MS medium containing 1-μM PAC in the dark for 5 days, and were then treated with 100-μM GA3 + 1-μM PAC and exposed to blue light (30 μmol·m–2·s–1) for the indicated time. Three independent experiments were performed, and one is shown. E–H, Relative protein levels of GA-induced degradation of RGA-Flag and GAI-Flag shown in (A–D) as means ± standard deviation (sd; n = 3; Student’s t test, **P < 0.01, *P < 0.05). I, J, Effects of blue light intensity on GA-induced degradation of RGA-Flag (I) and GAI-Flag (J) proteins, as determined by immunoblot analysis. RGA-Flag-OX and GAI-Flag-OX seedlings (WT background) were grown on MS medium containing 1-μM PAC in the dark for 5 days, and were then treated with 100-μM GA3 + 1-μM PAC and exposed to the indicated light intensities of blue light for the indicated time. Two independent experiments were performed, and one is shown. The immunoblot results were quantified using ImageJ. RGA and GAI protein levels were normalized to the tubulin loading control and the levels at time 0 was set to 100. BL, blue light; DK, dark.
Shade and high temperature conditions induce the COP1-mediated degradation of DELLA proteins, which are stabilized in a light-dependent, but GA-independent fashion (Blanco-Tourinan et al., 2020). To evaluate whether DELLA proteins might undergo degradation in blue light in a COP1-mediated and GA-independent manner, we performed immunoblots with an anti-RGA antibody to analyze endogenous RGA levels in WT and cop1 mutant seedlings treated with PAC and GA that were dark-adapted or exposed to blue light for different lengths of time. GA-induced degradation of RGA was faster in dark-adapted WT and cop1 mutant seedlings compared to those exposed to blue light (Supplemental Figure S2, A–D). However, GA treatment led to a similar progressive decline in RGA protein levels in WT and cop1 mutant seedlings exposed to blue light. reverse transcription quantitative polymerase chain reaction analysis showed that GA treatment induces RGA expression in both WT and cop1 mutant seedlings exposed to blue light (Supplemental Figure S2, E and F). These results indicate that blue light may inhibit the degradation of the DELLA protein, largely in a GA-dependent manner in our conditions.
DELLA proteins act to promote photomorphogenesis in blue light
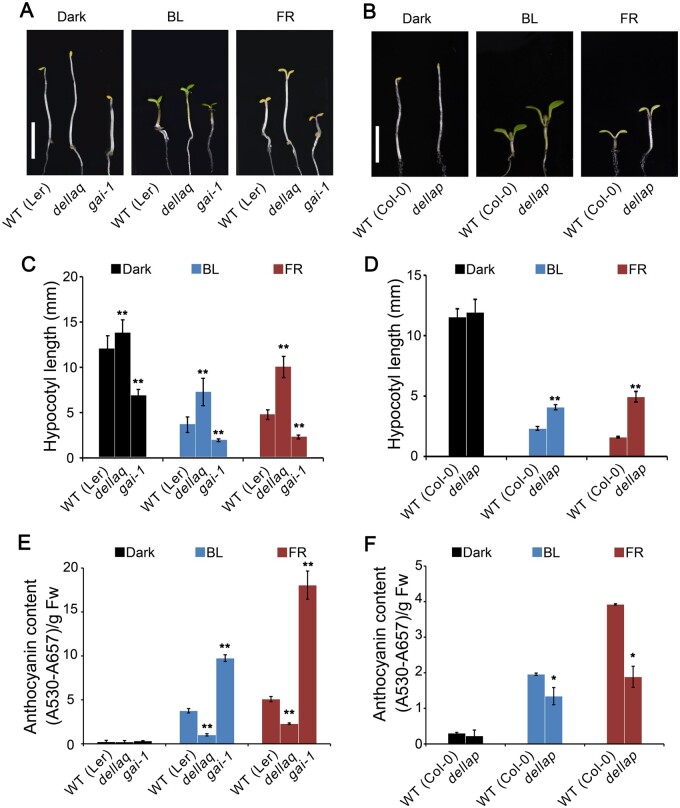
To explore whether DELLA proteins might regulate photomorphogenic development in blue light, we examined the hypocotyl phenotype of DELLA quadruple and pentuple mutants, dellaq and dellap, which are deficient in four and five DELLA proteins, respectively (Achard et al., 2007; Park et al., 2013), and the gain-of-function GAI allele, gai-1 (Peng et al., 1997), under blue light. Both dellaq and dellap seedlings developed taller hypocotyls than WT in blue light, while gai-1 seedlings developed shorter hypocotyls than WT in blue light (Figure 2, A–D). Since DELLA proteins were previously shown to inhibit hypocotyl elongation in red light (Achard et al., 2007; Park et al., 2013), we analyzed the hypocotyl phenotype of dellaq, dellap, and gai-1 mutants in far-red light: compared to the WT, both dellaq and dellap mutants exhibited a dramatically tall hypocotyl phenotype, whereas gai-1 displayed a pronounced short hypocotyl phenotype (Figure 2, A–D). We then analyzed anthocyanin content in dellaq, dellap, and gai-1 mutants grown in blue and far-red lights, and determined that both dellaq and dellap seedlings accumulate less anthocyanin than WT, whereas gai-1 seedlings produced more anthocyanin than WT in blue and far-red lights (Figure 2, E and F). These results suggest that DELLA proteins promote photomorphogenesis in blue and far-red lights.
Figure 2.
DELLA proteins promote photomorphogenesis in blue and far-red lights. A, B, Representative phenotypes of Arabidopsis seedlings of different genotypes grown in continuous darkness (Dark), blue light (BL, 30 μmol·m–2·s–1), or far-red light (FR, 10 μmol·m–2·s–1) for 5 days. Scale bars, 2 mm. C, D, Mean hypocotyl lengths from the seedlings shown in (A) and (B). Data are shown as means ± sd (n = 30; Student’s t test, **P < 0.01). E, F, Analysis of anthocyanin contents from the seedlings shown in (A) and (B). Data are shown as means ± sd (n = 3; Student’s t test, **P < 0.01, *P < 0.05)
CRY1 physically interacts with DELLA proteins in a blue light-specific manner
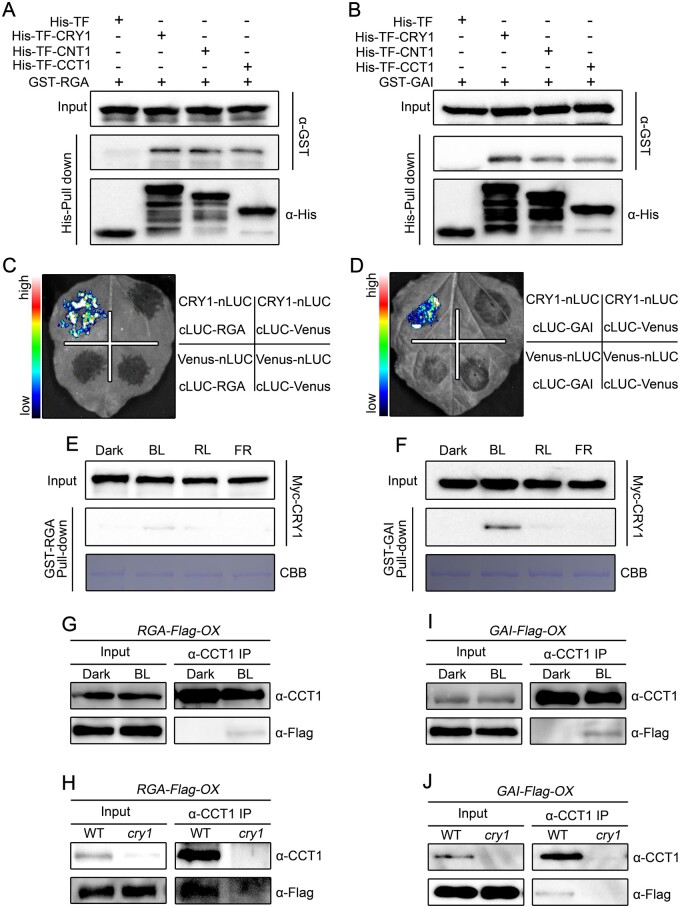
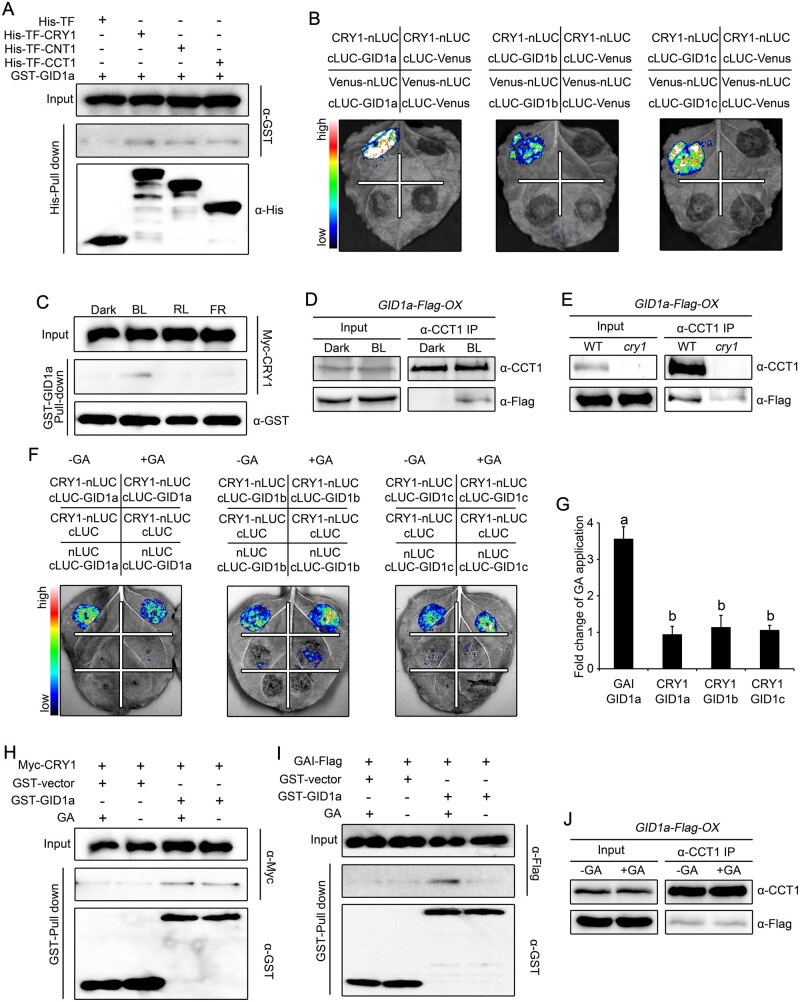
Based on the demonstration that CRY1 inhibits GA-induced degradation of DELLA proteins, and our previous study showing that CRY1 interacts with AUX/IAA proteins to inhibit auxin-induced degradation of these proteins (Xu et al., 2018), we asked whether CRY1 might interact with DELLA proteins. To test this possibility, we first performed pull-down experiments with recombinant His-TF-CRY1 (His-tagged trigger factor-CRY1), His-TF-CNT1 (CRY1 N-terminus), and His-TF-CCT1 (CRY1 C-terminus) as baits and GST–RGA and GST–GAI fusion proteins as prey. As shown in Figure 3, A and B, GST–RGA and GST–GAI were pulled down by His-TFCRY1, His-TF-CNT1, and His-TF-CCT1, but not by the His-tagged Trigger Factor (His-TF) control, indicating that both the N- and C-terminal of CRY1 interact with DELLA proteins. We also performed maltose binding protein (MBP) pull-down experiments with MBP-RGA-Nend and MBP-GAI-Nend, which comprise the N-terminal DELLA domain, and MBP-RGA-M5 and MBP-GAI-M5, which consist of the C-terminal GRAS domain (Supplemental Figure S3, A and B; de Lucas et al., 2008). Recombinant His-TF-CRY1 was pulled down by MBP-RGA-Nend, MBP-RGA-M5, MBP-GAI-Nend, and MBP-GAI-M5 (Supplemental Figure S3, C and D). These results indicate that both the DELLA and GRAS domains mediate the interaction between DELLA proteins and CRY1.
Figure 3.
CRY1 interacts with RGA and GAI in a blue light-dependent manner. A, B, Pull-down assays showing the interactions of CRY1, CNT1, and CCT1 with RGA (A) and GAI (B). His-TF-CRY1, His-TF-CNT1, and His-TF-CCT1 served as baits and were detected with anti-His antibody. GST–RGA and GST–GAI served as preys and were detected with anti-GST antibody. Two independent experiments were performed, and one is shown. C, D, Split-luciferase complementation imaging assays showing the interaction of CRY1 with RGA (C) and GAI (D). Venus-nLUC and cLUC-Venus served as negative controls. Two independent experiments were performed, and one is shown. E, F, Cell-free GST pull-down assays showing the blue light-specific interaction of CRY1 with RGA (E) and GAI (F). GST–RGA and GST–GAI served as baits. Preys were protein extracts prepared from Myc-CRY1-OX seedlings that were dark-adapted and exposed to blue light (BL, 30 μmol·m–2·s–1), red light (RL, 50 μmol·m–2·s–1), or far-red light (FR, 10 μmol·m–2·s–1) for 1 h. Two independent experiments were performed, and one is shown. G–J, co-IP assays showing the blue light-dependent interaction of CRY1 with RGA and GAI. Dark-adapted seedlings of RGA-Flag-OX in WT (G) or cry1 (H), and GAI-Flag-OX in WT (I) or cry1 (J) were maintained in the dark or exposed to blue light (50 μmol/m2/s) for 1 h, followed by immunoprecipitation with an anti-CCT1 antibody. The IP (CRY1) and co-IP signals (RGA and GAI) were detected in immunoblots probed with anti-CCT1 and anti-Flag antibodies, respectively. Two independent experiments were performed, and one is shown.
We then performed split luciferase complementation (split-LUC) assays in Nicotiana benthamiana leaves transiently co-expressing CRY1, CNT1, or CCT1 with RGA or GAI fused to the N- and C-terminal halves of firefly luciferase (CRY1-nLUC, CCT1-nLUC, CNT1-nLUC, and cLUC-RGA or cLUC-GAI). We reconstituted luciferase when CRY1-nLUC, CCT1-nLUC, or CNT1-nLUC was co-expressed with cLUC-RGA or cLUC-GAI, but not with cLUC-Venus, and when cLUC-RGA or cLUC-GAI was co-expressed with Venus-nLUC (Figure 3, C and D; Supplemental Figure S4, A and B). These results confirmed that both CNT1 and CCT1 can mediate the interaction of CRY1 with DELLA proteins. We also performed split-LUC assays in N. benthamiana leaves co-expressing CRY1-nLUC and cLUC-RGA-Nend, cLUC-RGA-M5, cLUC-GAI-Nend, or cLUC-GAI-M5. The deletion of either the entire N-terminal DELLA-containing domain or the C-terminal GRAS-containing domain did not affect the interaction of RGA and GAI with CRY1 (Supplemental Figure S4, C and D). Further co-immunoprecipitation (co-IP) assays with N. benthamiana leaf samples confirmed that both the N-terminal DELLA and C-terminal GRAS domains mediate the interaction of RGA and GAI with CRY1 (Supplemental Figure S4, E and F).
To investigate the effects of light quality on the interaction of CRY1 with DELLA proteins, we performed pull-down experiments with recombinant GST–RGA and GST–GAI proteins incubated with protein extracts prepared from transgenic Arabidopsis seedlings overexpressing Myc-CRY1-OX that were dark-adapted before exposure to blue, red, or far-red light. RGA and GAI successfully pulled down CRY1 from the extracts prepared from seedlings exposed to blue light (Figure 3, E and F), but not from dark-adapted seedlings or seedlings exposed to red or far-red light. These results suggest that CRY1 interacts with RGA and GAI in a blue light-specific manner. To further confirm that CRY1 interacts with DELLA proteins in Arabidopsis, we performed co-IP assays with RGA-Flag-OX, GAI-Flag-OX, cry1 RGA-Flag-OX, and cry1 GAI-Flag-OX seedlings that were dark-adapted and then exposed to blue light for 1 h. As shown in Figure 3, G–J, an IP of endogenous CRY1 also co-immunoprecipitated RGA and GAI in the extracts from RGA-Flag-OX and GAI-Flag-OX seedlings exposed to blue light, but not in the extracts of dark-adapted controls or cry1 RGA-Flag-OX or cry1 GAI-Flag-OX seedlings illuminated with blue light. These results demonstrate that CRY1 interacts with DELLA proteins in a blue light-dependent manner in Arabidopsis.
Given the previous demonstration that DELLA proteins interact with photomorphogenesis-related transcription factors such as PIFs (de Lucas et al., 2008; Feng et al., 2008), we explored whether the interaction between CRY1 and DELLA proteins might influence the interactions of DELLA proteins with PIFs. To test this possibility, we first performed cell-free MBP-PIF3 pull-down experiments with the protein extracts prepared from RGA-Flag-OX and Myc-CRY1-OX seedlings. MBP-PIF3 pulled down similar amounts of RGA-Flag protein in the presence of protein extracts from Myc-CRY1-OX seedlings, whether they were dark-adapted or exposed to blue light (Supplemental Figure S5, A and B). We then performed split-LUC assays to determine whether the interaction of CRY1 with RGA and GAI would interfere with the interaction of RGA and GAI with PIF3 and PIF4 in N. benthamiana cells. However, RGA and GAI showed the same strength of interaction with PIF3 and PIF4 in the presence of CRY1 or the GUS control protein, respectively (Supplemental Figure S5, C–H). These results therefore indicate that the interaction of CRY1 with DELLA proteins may not affect the interaction of DELLA proteins with PIFs.
CRY1 inhibits the interaction of SLY1 with DELLA proteins to inhibit their ubiquitination
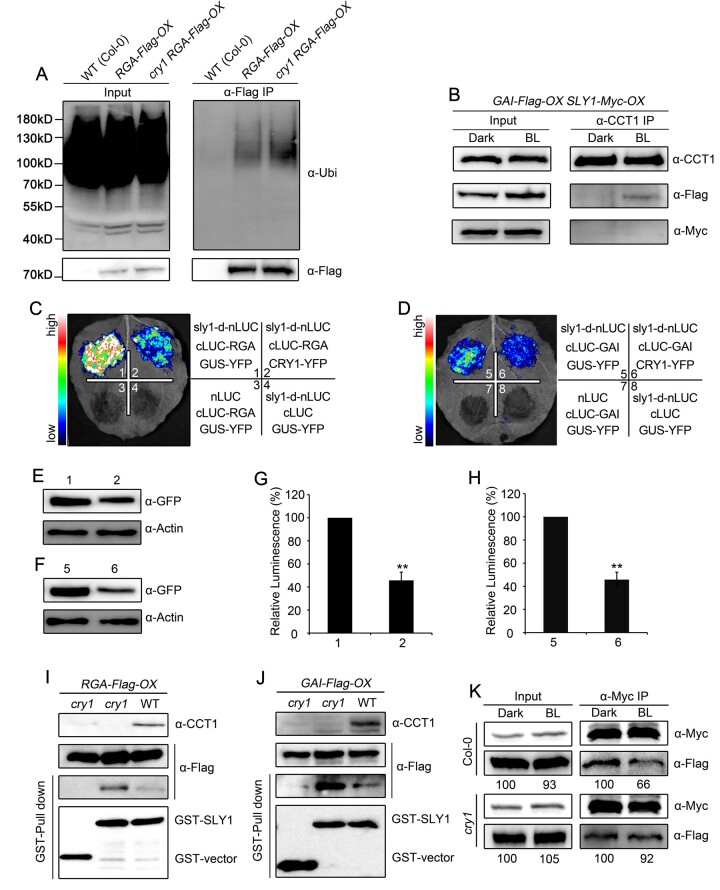
Given that CRY1 interacts with DELLA proteins and inhibits their degradation induced by GA, and that SLY1 is a subunit of the SCFSLY1 E3 ubiquitin ligase that interacts with and ubiquitinates DELLA proteins (Dill et al., 2004; Fu et al., 2004), we asked whether CRY1 might affect the ubiquitination of DELLA proteins. To test this possibility, we first performed IP assays to detect ubiquitinated RGA using an anti-ubiquitin antibody with RGA-Flag-OX and cry1 RGA-Flag-OX seedlings exposed to blue light and then treated with GA and the 26S proteasome inhibitor MG132. At equal RGA levels, much more RGA was ubiquitinated in the cry1 mutant background than in the WT background (Figure 4A). To examine whether CRY1 might inhibit RGA ubiquitination by interacting with SLY1 and its close homolog SNE, we performed split-LUC assays in N. benthamiana leaves co-expressing CRY1-nLUC and cLUC-SLY or cLUC-SNE. However, CRY1 did not interact with either SLY1 or SNE, whereas SLY1 and SNE each interacted with GAI (Supplemental Figure S6, A and B). We further performed co-IP assays to confirm whether CRY1 might interact with SLY1 in Arabidopsis with double transgenic seedlings co-overexpressing SLY1-Myc and GAI-Flag (SLY1-Myc-OX GAI-Flag-OX), which we generated by crossing independent lines overexpressing SLY1-Myc-OX or GAI-Flag-OX, an IP of endogenous CRY1 co-immunoprecipitated GAI, but not SLY1 (Figure 4B). These results demonstrate that CRY1 does not interact with SLY1 in Arabidopsis.
Figure 4.
CRY1 impairs the interaction of DELLA proteins with SLY1. A, In vivo ubiquitination assays showing CRY1-mediated inhibition of RGA ubiquitination in blue light. Seedlings for WT(Col-0), RGA-Flag-OX in WT and cry1 were grown in blue light (50 μmol·m–2·s–1) for 5 days, and then treated with 100-μM GA3 and 50-μM MG132 in liquid MS medium for 2 h, followed by immunoprecipitation with anti-Flag beads. The IP signals (RGA-Flag) were detected in immunoblots probed with anti-ubiquitin, and input signals were detected with anti-Flag antibodies. Two independent experiments were performed, and one is shown. B, Co-IP assays showing no interaction of CRY1 with SLY1 in Arabidopsis. Dark-adapted double transgenic GAI-Flag-OX/SLY1-Myc-OX seedlings were kept in darkness or exposed to blue light (BL, 50 μmol·m–2·s–1) for 1 h, followed by immunoprecipitation with anti-Myc beads. The IP (CRY1) and co-IP signals (SLY1 and GAI) were detected in immunoblots probed with anti-Myc and anti-Flag antibodies, respectively. Two independent experiments were performed, and one is shown. C–H, Split-luciferase complementation imaging assays indicating that CRY1 prevents the interaction between SLY1 with RGA (C) and GAI (D) in N. benthamiana leaves. The quantification of luciferase activity for the samples in (C) and (D) is shown in (G) and (H), respectively. Data are shown as means of biological triplicates ± sd (n = 4; Student’s t test, **P < 0.01). The accumulation of CRY1-YFP and GUS-YFP (negative control) was detected with an anti-GFP antibody in (E) and (F). I, J, Cell-free GST pull-down assays showing that CRY1 prevents the interaction between SLY1 and RGA (I) or GAI (J). GST–SLY1 served as bait. Preys were protein extracts prepared from dark-adapted RGA-Flag-OX GAI-Flag-OX (WT and cry1 backgrounds) seedlings exposed to blue light (50 μmol·m–2·s–1) for 1 h. Two independent experiments were performed, and one is shown. K, Co-IP assays showing that CRY1 prevents the interaction of SLY1 with GAI in Arabidopsis. Total protein was extracted from dark-adapted double transgenic seedlings co-expressing SLY1-Myc and GAI-Flag in the WT and cry1 backgrounds and exposed to blue light (50 μmol·m–2·s–1) for 1 h, followed by immunoprecipitation with anti-Myc beads. The immunoprecipitates were probed with anti-Flag and anti-Myc antibodies. Relative band intensities were normalized for each panel and are shown below each lane. Two independent experiments were performed, and one is shown.
Next, we explored whether CRY1 influences the interaction between SLY1 and DELLA proteins to inhibit their ubiquitination. To this end, we first performed split-LUC assays in N. benthamiana leaves to confirm that SLY interacts with RGA and GAI in the presence of CRY1, using the unrelated protein β-GLUCURONIDASE (GUS) as a negative control. Given the weak interaction between WT SLY1 and DELLA proteins, we generated a construct whereby nLUC was fused to sly1-d, a mutant form of SLY1 with a single amino acid substitution (E138K) that interacts strongly with DELLA proteins in plants (Dill et al., 2004). sly1-d interacted strongly with both RGA and GAI in the presence of GUS, but the strength of the interaction was severely reduced when GUS was replaced by CRY1 (Figure 4, C–H). These results indicate that CRY1 inhibits the interaction of SLY1 with DELLA proteins. We then performed cell-free pull-down assays to evaluate the effects of CRY1 on SLY1-DELLA interaction using GST-SLY1 as bait, and protein extracts prepared from RGA-Flag-OX, cry1 RGA-Flag-OX, GAI-Flag-OX, and cry1 GAI-Flag-OX seedlings. Both RGA and GAI bound to SLY much more strongly in the absence of CRY1 (in samples prepared from cry1 mutant seedlings) than in the presence of CRY1 (Figure 4, I and J), further indicating a role for CRY1 in repressing the interaction between SLY1 and DELLA proteins. To determine whether CRY1 affects the interaction of SLY1 with DELLA protein in Arabidopsis, we performed co-IP assays using double transgenic seedlings co-expressing SLY1-Myc and GAI-Flag in the WT and cry1 mutant backgrounds that were dark-adapted or exposed to blue light. Upon exposure to blue light, the SLY1–GAI interaction was clearly inhibited in the WT background (Figure 4K, upper), but modestly affected in the cry1 mutant background (Figure 4K, lower). Taken together, these results demonstrate that photoactivated CRY1 impairs the interaction of DELLA proteins with SLY1 in planta.
CRY1 physically interacts with GID1 in a blue light-dependent but GA-independent manner
Based on the previous demonstration that GA promotes the interaction between the GA receptor GID1 and DELLA proteins, leading to the subsequent degradation of DELLA proteins (Griffiths et al., 2006; Willige et al., 2007), and our demonstration that CRY1 is involved in the suppression of GA-induced degradation of DELLA proteins, we asked whether CRY1 also interacts with GID1 to influence GA-induced interactions of GID1 with DELLA proteins and stabilize DELLA proteins. To test this possibility, we first performed pull-down experiments with recombinant His-TF-CRY1, His-TF-CNT1, and His-TF-CCT1 proteins as baits and GST-GID1a as prey. As shown in Figure 5A, GST-GID1a was pulled down by His-TF-CRY1, His-TF-CNT1, and His-TF-CCT1, but not His-TF, indicating that CRY1 interacts with GID1a and that both CNT1 and CCT1 mediate this interaction in vitro. We then performed split-LUC assays in N. benthamiana leaves co-expressing CRY1-nLUC, CNT1-nLUC, or CCT1-nLUC and cLUC-GID1a, cLUC-GID1b or cLUC-GID1c: both CNT1 and CCT1 interacted with GID1a, GID1b, and GID1c (Figure 5B; Supplemental Figure S7A). To investigate the effects of light quality on the CRY1–GID1 interaction, we performed cell-free pull-down assays with recombinant GST–GID1a protein and protein extracts prepared from Myc-CRY1-OX dark-adapted seedlings and exposed to blue, red, or far-red light. GST–GID1a pulled down CRY1 from the extracts prepared seedlings exposed to blue light (Figure 5C), but not from those that were dark-adapted or exposed to red or far-red light, supporting the blue light-specific interaction of CRY1 with GID1a.
Figure 5.
CRY1 interacts with GID1a in a blue light-dependent but GA-independent manner. A, Pull-down assays showing the interaction of CRY1, CNT1, and CCT1 with GID1a. His-TF-CRY1, His-TF-CNT1, and His-TF-CCT1 served as baits and were detected with anti-His antibody. GST–GID1a served as prey and was detected with anti-GST antibody. Two independent experiments were performed, and one is shown. B, Split-luciferase complementation imaging assays showing the interaction of CRY1 with GID1a, GID1b, and GID1c. Venus-nLUC and cLUC-Venus served as negative controls. Two independent experiments were performed, and one is shown. C, Cell-free GST pull-down assays showing the blue light-specific interaction of CRY1 with GID1a. GST–GID1a served as bait. Preys were protein extracts prepared from dark-adapted Myc-CRY1-OX seedlings that were exposed to blue light (BL, 30 μmol·m–2·s–1), red light (RL, 50 μmol·m–2·s–1) or far-red light (FR, 10 μmol/m2/s) for 1 h. Two independent experiments were performed, and one is shown. D, E, Co-IP assays showing the blue light-dependent interaction of CRY1 with GID1a in Arabidopsis. Dark-adapted GID1a-Flag-OX (D) and cry1 GID1a-Flag-OX (E) seedlings were maintained in the dark or exposed to blue light (50 μmol·m–2·s–1) for 1 h, followed by immunoprecipitation with anti-CCT1 antibody. The IP (CRY1) and co-IP signals (GID1a) were detected in immunoblots probed with anti-CCT1 and anti-Flag antibodies, respectively. Two independent experiments were performed, and one is shown. F, G, Split-luciferase complementation imaging assays illustrating the interaction of CRY1 with GID1 in a GA-independent manner in N. benthamiana cells. Application of 100-μM GA3 did not affect the interaction of CRY1 with GID1a, GID1b, or GID1c (F). The quantification of luciferase activity for the samples in (F) is shown in (G). Data are shown as means of biological triplicates ± sd (n = 4). Letters “a” and “b” indicate statistically significant differences for the indicated values, as determined by a one-way analysis of variance (ANOVA), followed by Tukey’s least significant difference (LSD) test (P < 0.05). H, I, Cell-free GST pull-down assays showing the GA-independent interaction of CRY1 with GID1. GST–GID1a served as bait. Preys were protein extracts prepared from dark-adapted Myc-CRY1-OX (H) and GAI-Flag-OX (I) seedlings exposed to blue light (50 μmol·m–2·s–1) for 1 h. Extracts were mixed with GST–GID1a, and then 100-μM GA3 was added (+) or not (−). Two independent experiments were performed, and one is shown. J, Co-IP assays showing the GA-independent interaction of CRY1 with GID1 in Arabidopsis. Dark-adapted GID1a-Flag-OX seedlings were exposed to blue light (50 μmol·m–2·s–1) for 1 h, and followed by immunoprecipitation with an anti-CCT1 antibody without (–) or with (−) 100-μM GA3 added. The IP (CRY1) and co-IP signals (GID1a) were detected in immunoblots probed with anti-CCT1 and anti-Flag antibodies, respectively. Two independent experiments were performed, and one is shown.
Next, we generated transgenic lines overexpressing GID1a-Flag in the WT and cry1 mutant backgrounds (GID1a-Flag-OX and cry1 GID1a-Flag-OX), which we then used to perform co-IP assays to confirm whether CRY1 interacts with GID1a in Arabidopsis with seedlings that were dark-adapted and then exposed to blue light. As shown in Figure 5, D and E, an IP of endogenous CRY1 co-immunoprecipitated GID1a from the extracts of GID1a-Flag-OX seedlings exposed to blue light, but not from the extracts of dark-adapted GID1a-Flag-OX seedlings or cry1 GID1a-Flag-OX seedlings exposed to blue light. These results demonstrate that CRY1 interacts with GID1a in a blue light-dependent manner in Arabidopsis.
GID1 interacts with DELLA proteins in a GA-dependent manner (Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Nakajima et al., 2006). We therefore explored whether GA affects the interaction of CRY1 with GID1 next. Accordingly, we first performed split-LUC assays with N. benthamiana leaves co-expressing CRY1-nLUC and cLUC-GID1a, cLUC-GID1b, or cLUC-GID1c, or the controls GAI-nLUC and cLUC-GID1a and treated with GA. This GA treatment did not affect the interaction of CRY1 with GID1a, GID1b, or GID1c (Figure 5, F and G), but dramatically promoted the interaction of GAI with GID1a (Supplemental Figure S7B). We then performed cell-free pull-down experiments with recombinant GST-GID1a protein and protein extracts prepared from blue light-illuminated Myc-CRY1-OX seedlings; we then added 100-μM GA3 to half of the samples to test the influence of GA on protein interaction. GID1a pulled down similar levels of CRY1 with or without exogenous GA added (Figure 5H). However, when we performed the same cell-free pull-down experiments with recombinant GST–GID1a and protein extracts prepared from GAI-Flag-OX seedlings, we discovered that GID1a pulls down much more GAI from the extracts treated with GA relative to control extracts not containing exogenous GA (Figure 5I). To further validate the GA independence of the CRY1–GID1 interaction in Arabidopsis, we generated transgenic plants overexpressing GID1a-Flag in WT (GID1a-Flag-OX) and performed co-IP assays with GID1a-Flag-OX seedlings that were exposed to blue light, with GA added to the extracts during co-IP. As shown in Figure 5J, an IP of endogenous CRY1 co-immunoprecipitated similar levels of GID1a from the extracts of GID1a-Flag-OX seedlings exposed to blue light, irrespective of the addition of GA. Taken together, these results demonstrate that CRY1 interacts with GID1a in a GA-independent manner in Arabidopsis.
CRY1 inhibits the GA-induced interactions of GID1 with DELLA proteins
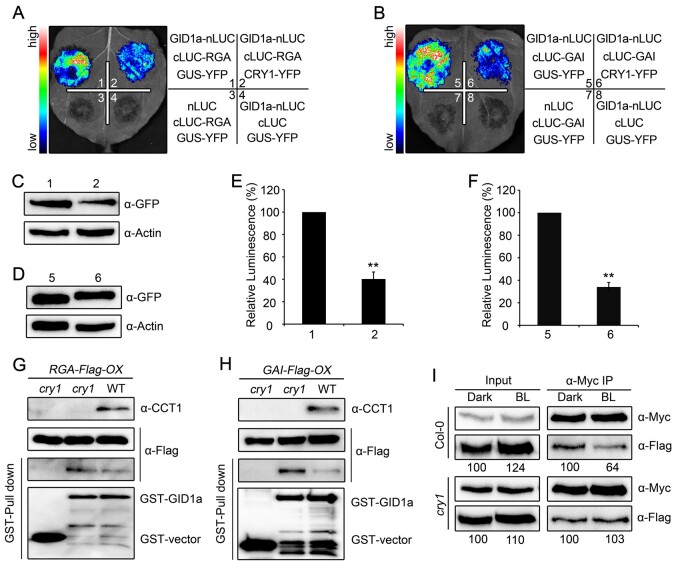
Given the demonstration that the interaction of GID1 with DELLA proteins is essential for the GA-induced degradation of DELLA proteins (Griffiths et al., 2006) and since CRY1 inhibits the GA-induced degradation of DELLA proteins (Figure 1; Supplemental Figure S1), we explored whether the interactions among CRY1, GID1, and DELLA proteins affect GID1–DELLA interactions. We first performed split-LUC assays in N. benthamiana leaves to confirm the interaction between GID1 with RGA in the presence of CRY1 and GUS as an unrelated protein: indeed, GID1a strongly interacted with RGA, but the co-expression of CRY1 reduced the strength of the interaction (Figure 6, A to F). We then performed cell-free pull-down assays with recombinant GST–GID1 protein and protein extracts prepared from dark-adapted RGA-Flag-OX, cry1 RGA-Flag-OX, GAI-Flag-OX, or cry1 GAI-Flag-OX seedlings. Much more RGA-Flag and GAI-Flag proteins were pulled down by GST-GID1a in the absence of CRY1 than in the presence of CRY1 (Figure 6, G and H). Next, we created double transgenic plants co-expressing GAI-Flag and GID1a-Myc in the WT and cry1 mutant backgrounds, and performed co-IP assays to confirm the effects of CRY1 on the interaction between GID1 and GAI with seedlings that were dark-adapted or exposed to blue light. Exposure to blue light led to a dramatic decrease in the GID1a–GAI interaction in the WT background, but had no effect in the cry1 mutant background, as expected (Figure 6I). Taken together, these results demonstrate that photoactivated CRY1 impairs the interaction between DELLA proteins and GID1 in planta.
Figure 6.
CRY1 impairs the interaction of DELLA proteins with GID1. A–F, Split-luciferase complementation imaging assays indicating that CRY1 prevents the interaction of GID1a with RGA (A) and GAI (B) in N. benthamiana leaves. The quantification of luciferase activity for the samples in (A) and (B) is shown in (E) and (F). Data are shown as means of biological triplicates ± sd (n = 4; Student’s t test, **P < 0.01). The accumulation of CRY1-YFP and GUS-YFP was detected with an anti-GFP antibody in (C) and (D). G, H, Cell-free GST pull-down assays showing that CRY1 prevents the interaction of GID1a with RGA (G) and GAI (H). GST-GID1a served as bait. Preys were protein extracts prepared from dark-adapted RGA-Flag-OX and GAI-Flag-OX seedlings in the WT or cry1 backgrounds and exposed to blue light (50 μmol·m–2·s–1) for 1 h. The extracts were mixed with GST-GID1a, in the absence (–) or presence (+) of 100-μM GA3. Two independent experiments were performed, and one is shown. I, Co-IP assays showing that CRY1 prevents the interaction of GID1a with GAI in Arabidopsis. Protein was extracted from 5-day-old dark-adapted double transgenic seedlings GAI-Flag-OX GID1a-Myc-OX in the WT and cry1 backgrounds and exposed to blue light (BL) for 1 h, followed by immunoprecipitation with an anti-Myc beads in the absence (–) or presence (+) of 100-μM GA3. The immunoprecipitates were probed with anti-Flag and anti-Myc antibodies. Relative band intensities were normalized for each panel and are shown below each lane. Two independent experiments were performed, and one is shown.
GID1 and DELLA act genetically downstream of CRY1 to regulate photomorphogenesis in blue light
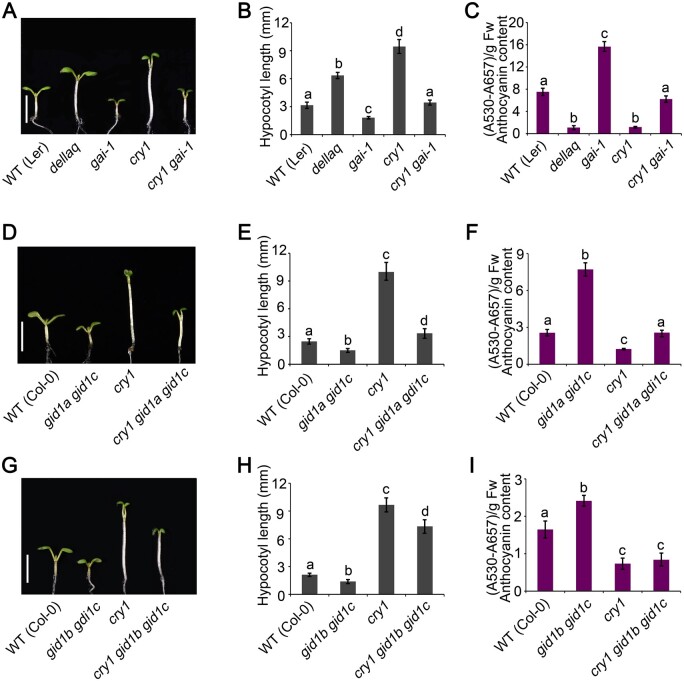
That CRY1 interacts with GID1 in a blue light-dependent manner (Figure 5, C–E) suggested that GID1 is likely involved in CRY1-mediated blue light signaling. To test this hypothesis, we first examined the photomorphogenic phenotype of gid1a gid1c and gid1b gid1c double mutants in blue light. The gid1a gid1c and gid1b gid1c double mutants had shorter hypocotyls and accumulated more anthocyanins than WT in blue light (Supplemental Figure S8). GID1 was previously shown to promote hypocotyl elongation in red light (Griffiths et al., 2006; Feng et al., 2008). We thus also analyzed the photomorphogenic phenotype of gid1a gid1c and gid1b gid1c double mutants in far-red light: compared to WT, the gid1a gid1c and gid1b gid1c double mutants also developed shorter hypocotyls and produced more anthocyanins than the WT in far-red light (Supplemental Figure S8). We then introduced gai-1 into the cry1 mutant background by genetic crossing to generate the cry1 gai-1 double mutant. The cry1 gai-1 mutant developed shorter hypocotyls and accumulated much more anthocyanins than the cry1 mutant when grown in blue light (Figure 7, A–C). However, the cry1 gai-1 double mutant exhibited longer hypocotyls and produced much lower levels of anthocyanins than the gai-1 mutant. These results indicate that GAI acts partially downstream from CRY1 in regulating photomorphogenesis in blue light. Next, we generated the cry1 gid1a gid1c and cry1 gid1b gid1c triple mutants by genetic crossing. Both cry1 gid1a gid1c and cry1 gid1b gid1c triple mutants developed shorter hypocotyls and accumulated much more anthocyanins than the cry1 mutant, but had longer hypocotyls and accumulated much less anthocyanin than gid1a gid1c and gid1b gid1c double mutants (Figure 7, D–I). These results indicate that GID1 may also act partially downstream of CRY1 to regulate photomorphogenesis in blue light. We made several attempts to isolate a gid1a gid1b gid1c triple mutant and a cry1 gid1a gid1b gid1c quadruple mutant, hoping to obtain a stronger photomorphogenic phenotype, but failed, because the gid1a gid1b gid1c mutant was sterile in our hands.
Figure 7.
GID1 and DELLA act genetically downstream from CRY1 to regulate photomorphogenesis. A, Representative phenotypes of gai-1 and cry1 gai-1 mutant seedlings grown in continuous blue light (30 μmol·m–2·s–1) for 5 days. Scale bars, 5 mm. B, C, Mean hypocotyl lengths and anthocyanin contents of the genotypes in (A). D, Representative phenotypes of gid1a gid1c and cry1 gid1a gid1c mutant seedlings grown in continuous blue light (30 μmol·m–2·s–1) for 5 days. Scale bars, 5 mm. E, F, Mean hypocotyl lengths and anthocyanin contents of the genotypes in (D). G, Representative phenotypes of gid1b gid1c and cry1 gid1b gid1c mutant seedlings grown in continuous blue light (30 μmol·m–2·s–1) for 5 days. Scale bars, 5 mm. H, I, Mean hypocotyl lengths and anthocyanin contents of the genotypes in (G). Data in (B), (E), and (H) are means ± sd (n = 30). Data in (C), (F), and (I) are means ± sd (n = 3). Letters “a” to “d” indicate statistically significant differences for the indicated values, as determined by a one-way analysis of variance (ANOVA), followed by Tukey’s least significant difference (LSD) test (P < 0.05).
Discussion
During their lifecycle, the modulation of plant developmental plasticity relies on the perception of external signal such as changes in light quality and quantity, as well as internal signals such as the phytohormones (GA), auxin (IAA), and BRs, to fine-tune the status of growth. Given that light and these phytohormones act to control the same developmental processes, the coordination of their actions is critical to balance plant growth. Significant progress has been made in elucidating the molecular mechanisms underlying this coordination. For example, the signaling crosstalk between GA and light is mediated through the repression of the DNA binding ability of PIF3 and PIF4, pivotal negative regulators of photomorphogenesis (Ni et al., 1998, 1999; Leivar et al, 2008), through the direct interaction of DELLA proteins with PIF3 and PIF4 (de Lucas et al., 2008; Feng et al., 2008). In addition, CRY1- and phyB-mediated light signaling inhibits auxin signaling and hypocotyl cell elongation via the direct interaction of CRY1 and phyB with AUX/IAA proteins to stabilize AUX/IAAs, and with ARFs to inhibit their DNA binding activity (Xu et al., 2018; Mao et al., 2020). CRY1 and UVR8 also mediate the blue and UV-B light repression, respectively, of BR signaling to inhibit hypocotyl elongation through their direct interaction with dephosphorylated BES1 to prevent its DNA binding activity (Liang et al, 2018; Wang et al, 2018a). Our previous study suggested that CRY1 mediates the repression of GA-promoted hypocotyl elongation in blue light through the regulation of GA-responsive genes (Wang et al., 2016). Whether light regulates GA signaling through direct interaction between photoreceptors and key components in GA signaling was unknown.
In the present study, we reveal new connections between regulatory pathways by which Arabidopsis CRY1 mediates the regulation of seedling morphogenesis through inhibition of GA signaling in blue light. Our results suggest that CRY1 may mediate blue light inhibition of GA-induced degradation of DELLA proteins through its blue light-dependent interaction with DELLA proteins (Figures 1 and 3), which leads to inhibition of the interaction between SLY1 and DELLA proteins and suppress their ubiquitination (Figure 4). Furthermore, we show that CRY1 may also mediate blue light inhibition of GA-induced degradation of DELLA proteins through its direct blue light-dependent interaction with the GA receptor GID1 (Figure 5). Such an interaction, together with the blue light-induced interaction of CRY1 with DELLA proteins, may result in the dissociation of DELLA proteins from GID1 (Figure 6). Given that the GA-induced interaction of GID1 with DELLA proteins is essential for their recognition by SCFSLY1 for ubiquitination (Murase et al., 2008; Shimada et al., 2008; Sun, 2011), the repression by photoactivated CRY1 of the GID1-DELLA interaction may interfere with the recognition of DELLA proteins by SCFSLY1, thereby inhibiting their ubiquitination and degradation.
Previous studies have reported that photoreceptor-mediated light signaling regulates the stability of key transcriptional regulators in phytohormone signaling via direct interaction. For example, phyB mediates the red light-induced degradation of ETHYLENE INSENSITIVE 3 (EIN3) by enhancing the binding of EIN3-BINDING F BOX PROTEIN 1 (EBF1) and EBF2, components of an E3 ubiquitin ligase complex, to EIN3 to regulate ethylene responses (Shi et al., 2016). Blue light-triggered interaction of CRY1 with AUX/IAA proteins inhibits their association with TRANSPORT INHIBITOR RESPONSE 1 (TIR1) and thus represses their auxin-induced degradation (Xu et al., 2018). However, whether light regulates the stability of these transcriptional regulators in phytohormone signaling via direct interaction between the photoreceptor and the phytohormone receptor has not been reported. The present study reveals an additional layer in the regulation of the stability of DELLA proteins and GA signaling by light, which can be mediated by the direct interaction of the photoreceptor CRY1 with the phytohormone receptor GID1.
The biological significance of the blue light-dependent interaction of CRY1 with DELLA proteins and GID1 is supported by physiological and genetic analyses. Since CRY1 is the major blue light photoreceptor mediating blue light-induced photomorphogenesis (Ahmad and Cashmore, 1993; Lin et al., 1996), we analyzed the photomorphogenic phenotype of both the loss-of-function and gain-of-function mutants of DELLA proteins and the loss-of-function mutants of GID1a and GID1c or GID1b and GID1c in blue light. Our results demonstrate that DELLA proteins and GID1 promote and inhibit photomorphogenesis in blue light, respectively (Figure 2; Supplemental Figure S7). A genetic interaction study further suggests that DELLA proteins and GID1 act partially downstream from CRY1 to regulate photomorphogenesis in blue light (Figure 7). This conclusion is consistent with previous studies showing that many additional components act directly but separately downstream of CRY1 to regulate photomorphogenesis, such as COP1/SPA1, PIFs, AUX/IAAs, ARFs, BES1/BIM1, ARABIDOPSIS G-PROTEIN BETA 1 (AGB1), and HOMOLOG OF BEE2 INTERACTING WITH IBH 1 (HBI1; Lian et al., 2011, 2018; Liu et al., 2011; Ma et al., 2016; Pedmale et al., 2016; Xu et al., 2018; Wang et al, 2018a, 2018b; Mao et al., 2020).
It is of note that most cryptochrome-interacting proteins also interact with phytochromes and that some phytochrome-interacting proteins also interact with cryptochromes. These include COP1/SPAs, AUX/IAAs, ARFs, PIFs, BES1/BIM1, and AGB1 (Wang et al., 2001, 2018a; Yang et al., 2001; Huq and Quail, 2002; Lian et al., 2011, 2018; Liu et al., 2011; Zuo et al., 2011; Lu et al., 2015; Ma et al., 2016; Pedmale et al., 2016; Wu et al., 2018; Xu et al., 2018, 2019; Mao et al., 2020; Dong et al., 2020). Given previous studies showing that DELLA proteins inhibit hypocotyl elongation in red light (Achard et al., 2007), and our results showing that DELLA proteins also inhibit hypocotyl elongation and promote anthocyanin accumulation in far-red light (Figure 2) and that GID1 promotes hypocotyl elongation and inhibits anthocyanin accumulation in far-red light (Supplemental Figure S8), it is possible that phytochrome signaling may also proceed, at least in part, through an inhibition of GA signaling. Based on these studies and our demonstration that CRY1 physically interacts with GID1 and DELLA proteins to stabilize DELLA proteins and inhibit GA signaling, we speculate that phytochromes may also interact with GID1 and DELLA proteins to regulate GA-induced degradation of DELLA proteins and GA signaling. This possibility will be worth exploring in the future.
A growing body of evidence has shown that cryptochromes physically interact with transcriptional factors, such as CRYPTOCHROME-INTERACTING BASIC-HELIX–LOOP–HELIX 1 (CIB1) and related proteins, PIFs, ARFs, BES1, HBI1, and TARGET OF EARLY ACTIVATION TAGGED 1 (TOE) and TOE2, to directly regulate the transcription of genes involved in photomorphogenesis, thermosensory responses, and flowering (Liu et al., 2008b; Ma et al., 2016; Pedmale et al., 2016; Wang et al., 2018a, 2018b; Mao et al., 2020; Du et al., 2020). Multiple studies have demonstrated that DELLA proteins may act as a central protein that integrates the components from many pathways to coordinate the regulation of plant development. For example, light and GA signaling are integrated through direct interaction of DELLA proteins with PIF3 and PIF4, which leads to the inhibition of their DNA binding activity (de Lucas et al., 2008; Feng et al., 2008). DELLA proteins also interact with ARFs and BRASSINAZOLE-RESISTANT 1 (BZR1) to regulate their DNA binding activity and target gene expression to regulate auxin and BR signaling, respectively (Bai et al., 2012; Oh et al., 2014). Based on these studies and our demonstration that CRY1 inhibits GA-induced degradation of DELLA proteins in blue light, it is reasonable to speculate that CRY1 may indirectly regulate the transcriptional activity of PIFs, ARFs, and BES1 through the inhibition of GA-induced degradation of DELLA proteins. The direct and indirect regulation of transcription by CRY1 may allow plants to respond to changes in light signals rapidly and accurately, thereby fine-tuning their growth and development.
In this study, we establish that the signaling mechanism of Arabidopsis CRY1 involves the inhibition of GA signaling. Based on previous studies and this work, we propose a model describing how CRY1-mediated light signaling regulates GA signaling and photomorphogenesis (Figure 8). In the dark, CRY1 is inactive, and thus cannot interact with GID1 or DELLA proteins. Hence, GA can trigger the formation of the GA–GID1–DELLA complex and promote SCFSLY1–DELLA interaction and the subsequent active degradation of DELLA proteins through the 26S proteasome. PIFs are then released from the repression imposed by DELLA proteins, and can bind to and promote the expression of target genes to promote skotomorphogeneis by enhancing hypocotyl elongation and inhibiting anthocyanin accumulation (Figure 8A); Upon illumination, CRY1 becomes activated and can then interact with GID1 and DELLA proteins to inhibit the GA-induced degradation of DELLA proteins by interfering with their association with GID1 and SCFSLY1 (Figure 8B). Moreover, CRYs can mediate blue light inhibition of GA biosynthesis to stabilize DELLA proteins (Zhao et al., 2007). DELLA proteins thus accumulate and interact with PIFs to repress their DNA binding activity, thereby repressing the expression of their target genes inhibiting photomorphogenesis. The interaction between GID1 and DELLA proteins was previously shown to be induced by GA (Griffiths et al., 2006; Willige et al., 2007), and now we establish that the interaction between CRY1 and DELLA proteins is triggered by blue light. Therefore, this model provides a molecular framework for the antagonistic regulation of the stability of DELLA proteins and promotion of photomorphogenesis by blue light and GA.
Figure 8.
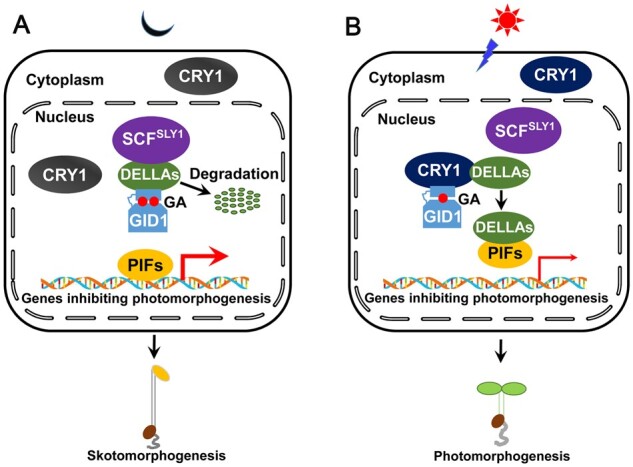
A model illustrating how CRY1 inhibits GA-induced degradation of DELLA proteins and GA signaling. A, B, In the dark, CRY1 is inactive and unable to interact with GID1 or DELLA proteins. GA triggers the interaction of GID1 with DELLA proteins (DELLAs), which are then targeted by SCFSLY1 for ubiquitination and degradation through the 26S proteasome. PIFs are released from the repression of DELLA proteins, and bind to the promoters of their target genes to inhibit photomorphogenesis, and promote their expression and skotomorphogenesis (A); upon blue light illumination, CRY1 is activated and regulates the expression of GA metabolism genes to reduce GA levels. At the same time, CRY1 interacts with GID1 and DELLAs to impair the interaction of DELLAs with SLY1 and GID1. DELLA proteins accumulate and interact with PIFs to inhibit their DNA-binding activity, leading to the inhibition of the expression of genes repressing photomorphogenesis and the promotion of photomorphogenesis (B). Thick and thin red arrows denote gene expression being induced and repressed, respectively. Two (in the dark) and one (in the light) red solid circles denote more and less GA levels, respectively.
Why does CRY1 inhibit GA signaling by interacting with both GID1 and DELLAs? Cryptochrome was the first photoreceptor to evolve about 1.9 billion years ago (Han et al., 2019). Indeed, most red algae lived in deep waters, where blue light served as both their main source of energy and signals for their growth and development. DELLAs originated in plants when they colonized land. The CRY1–DELLAs module may have preceded the acquisition of the GA/GID1 system, because this system appeared in lycophytes (Bowman et al., 2017), and was initially utilized to inhibit COP1-mediated degradation of DELLAs at least in the shade and at high temperatures (Blanco-Tourinan et al., 2020). During evolution, a phytohormone-sensitive lipase evolved into the GA receptor GID1 to regulate GA-induced degradation of DELLAs. To efficiently mediate light regulation of GA signaling, the CRY1–GID1 module evolved to inhibit GA-induced degradation of DELLAs. Given the demonstration that DELLAs are substrates of COP1 and stabilized in a light-dependent, but GA-independent fashion (Blanco-Tourinan et al., 2020), and that CRYs interact with COP1 (Wang et al, 2001; Yang et al, 2001) and regulate plant growth in response to shade and warm temperatures (Ma et al., 2016; Pedmale et al., 2016), it is possible that CRY1 may affect the interaction of COP1 with DELLAs to inhibit their COP1-dependent degradation in the shade and at warm temperatures.
Previous studies have demonstrated that GA binds strongly to GID1, leading to the formation of a hydrophobic surface at its N terminus that facilitates its interaction with DELLA proteins (Murase et al., 2008; Shimada et al., 2008). Given our demonstration that blue light triggers the interaction between CRY1 and GID1 (Figure 5), it will be interesting to explore whether CRY1 affects the binding of GA to GID1 and/or interfere with GA-triggered GID1 conformational change to block its interaction with DELLA proteins in future studies.
Materials and methods
Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana) accessions Columbia (Col-0) and Landsberg erecta (Ler) were used as the WT. The cry1-104 (Col-0), cry1/hy4-2.23N (Ler), dellaq (Ler), dellap (Col-0), gai-1 (Ler), and gid1b gid1c (Col-0) mutants, and transgenic line overexpressing Myc-tagged CRY1 (Myc-CRY1-OX), were described previously (Ahmad and Cashmore, 1993; Yang et al., 2000, 2001; Mao et al., 2005; Sang et al., 2005; Achard et al., 2007; Feng et al., 2008; Park et al., 2013). The plant expression vector pHB-3×Flag was described previously (Wang et al., 2018a). The cDNAs encoding GID1, RGA, and GAI were cloned into pHB-3×Flag to generate vectors overexpressing GID1-Flag, RGA-Flag, and GAI-Flag (pHB-35S:GID1-Flag, pHB-35S:RGA-Flag, and pHB-35S:GAI-Flag), respectively. The DNA fragments encoding the SLY1-Myc and GID1a-Myc fusion proteins were cloned into the plant expression vector pKYL71 (Schardl et al., 1987) to generate pKYL71-35S:SLY1-Myc and pKYL71-35S:GID1a-Myc. All constructs used were confirmed by DNA sequencing. All the primers used for cloning are listed in Supplemental Table S1.
All constructs were introduced into Agrobacterium (Agrobacterium tumefaciens) strain GV3101, and then transformed into WT Arabidopsis by the floral dip method (Clough and Bent, 1998) to generate transgenic lines overexpressing GID1-Flag, RGA-Flag, and GAI-Flag in WT (Col-0). Double transgenic lines co-overexpressing GAI-Flag and SLY1-Myc or GID1a-Myc (GAI-Flag-OX SLY1-Myc-OX or GAI-Flag-OX GID1a-Myc-OX) was obtained by transforming GAI-Flag-OX plants with GV3101 strains harboring pKYL71-35S:SLY1-Myc or pKYL71-35S:GID1a-Myc constructs, respectively. The transgenic lines were screened on Murashige and Skoog (MS) medium plates containing either 100 mg·mL–1 kanamycin or 50 mg·mL–1 hygromycin. The gai-1 mutant was introgressed into the cry1 (Ler) mutant background to generate the cry1 gai-1 double mutant. GID1-Flag-OX, RGA-Flag-OX, and GAI-Flag-OX and gid1b gid1c mutant plants were crossed with cry1 (Col-0) to generate cry1 GID1-Flag-OX, cry1 RGA-Flag-OX, cry1 GAI-Flag-OX, and cry1 gid1b gid1c triple mutant plants, respectively. The gid1a gid1c and cry1 gid1a gid1c mutants were generated by crossing gid1a with gid1b gid1c and cry1 gid1b gid1c mutants, respectively. All plants were confirmed by phenotypic analyses and/or immunoblot analysis.
Hydrated surface-sterilized seeds with 20% bleach were kept at 4°C for 3 days and then sown on MS medium containing 1% sucrose and 0.8% agar before release at 22°C in continuous white light (100 μmol m−2 s−1, from cool-white fluorescent lamps) for 24 h. Experiments involving blue, light and far-red light illuminations were described previously (Wang et al., 2018a). Light spectra and intensity were measured with a handheld spectroradiometer (ASD) and a Li250 quantum photometer (Li-Cor).
Measurements of hypocotyl length and anthocyanin content
Surface-sterilized seeds of various genotypes were sown on MS medium with 1% sucrose and kept at 4°C for 3 days, before transfer into different light conditions. Seedlings were allowed to grow for 5 days and then photographed with a digital camera (Nikon). Hypocotyl length was determined with Image J software (http://rsbweb.nih.gov/ij/). Anthocyanin content was measured by previously described methods (Li et al., 2014).
DELLA protein degradation assay in Arabidopsis
To analyze the influence of blue light exposure time on GA-induced DELLA protein degradation, immunoblotting assays were performed with anti-Flag antibody (Sigma, F3165) using RGA-Flag-OX and GAI-Flag-OX seedlings or anti-RGA antibody (Agrisera, AS111630) using WT and cop1-4 seedlings grown on MS medium containing 1-μM PAC in the dark for 5 days, and then transferred into liquid MS medium with 100-μM GA3 and 1-μM PAC in the dark or continuous blue light (30 μmol·m–2·s–1) for different lengths of time (0, 15, 30, and 45 min). To analyze the effects of light quality and blue light intensity on GA-induced degradation of DELLA proteins, RGA-Flag-OX and GAI-Flag-OX seedlings grown on MS medium containing 1-μM PAC in the dark, and with 5-day-old etiolated seedlings transferred to liquid MS medium with 100-μM GA3 and 1-μM PAC in the dark or exposed to blue (30 μmol·m–2·s–1), red (50 μmol·m–2·s–1) or far-red light (30 μmol·m–2·s–1) for different lengths of time (30 and 60 min) or continuous blue light at different fluence rates (1, 10, 100 μmol·m–2·s–1). To analyze the effects of CRY1 on GA-induced degradation of DELLA proteins, RGA-Flag-OX, cry1 RGA-Flag-OX, GAI-Flag-OX, and cry1 GAI-Flag-OX seedlings grown on MS medium containing 1-μM PAC in the dark for 5 days were transferred into liquid MS medium containing 100-μM GA3 and 1-μM PAC and kept in the dark or continuous blue light (30 μmol·m–2·s–1) for different lengths of time (30, 60, 90, and 120 min).
Total protein was extracted with lysis buffer (50-mM Tris–HCl, pH 7.5, 150-mM NaCl, 0.2% Triton-X-100) containing 1-mM Pefabloc, 1×EDTA-free Protease Inhibitor Cocktail (Roche), and 50-μM MG132. After centrifugation at 12,000g for 15 min at 4°C, total protein concentration of the supernatant was determined by Bradford assay (Bio-Rad); the supernatant was then mixed with 5× sodium dodecyl (SDS) loading buffer and boiled for 5 min. RGA-Flag and GAI-Flag proteins were detected with anti-Flag antibody (Sigma, F3165, 1:2,000). Immunoblot signals were detected by a Tanon 5200 chemiluminescence/fluorescence image analysis system.
In vitro pull-down assays with proteins produced in Escherichia coli
Pull-down assays were performed as described previously with minor modifications (Xu et al., 2016). The construction of pCold-TF-CRY1, pCold-TF-CNT1, and pCold-TF-CCT1 was described previously (Du et al., 2020). The DNA fragments of GID1a, RGA, GAI, RGA-Nend, RGA-M5, GAI-Nend, and GAI-M5 were cloned into pMAL-c2X (New England Biolabs, NEB) or pGEX-4T-1 (GE Healthcare Life Sciences). His-TF-CRY1, His-TF-CNT1, His-TF-CCT1, GST-GID1a, GST-RGA, GST-GAI, MBP-RGA-Nend, MBP-RGA-M5, MBP-GAI-Nend, and MBP-GAI-M5 recombinant proteins were produced in E. coli (Rosetta). For His and MBP pull-down assays, Ni-NTA Agarose (Qiagen, 30210) and Amylose Magnetic Beads (NEB, E8035S) were used. The protein extracts were separated by 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane (Millipore, ISEQ00010). The membrane was blocked with 5% skimmed milk in PBST solution. After probed with primary and secondary antibodies, the membrane was incubated in ECL and detected by a luminescent imaging workstation (Tanon 5200). Prey proteins were detected by anti-His (GenScript, A00186, 1:2,000) or anti-GST (Genscript, A00865-100, 1:2,000) or anti-MBP antibody (NEB, E8032S, 1:2,000), and bait proteins were visualized by Coomassie Brilliant Blue staining or detected with anti-His antibody.
Split-luciferase assays
For split-LUC assays to detect protein-protein interactions, the cDNA fragments encoding GID1a, GID1b, GID1c, SLY1, sly1-d, SNE, RGA, GAI, RGA-Nend, RGA-M5, GAI-Nend, and GAI-M5 were cloned into pCambia1300-nLUC and pCambia1300-cLUC (Mao et al., 2020). These constructs and those expressing Venus-nLUC, cLUC-Venus, CRY1-nLUC, CNT1-nLUC, and CCT1-nLUC (Du et al., 2020; Wang et al., 2021) were introduced individually into Agrobacterium strain GV3101. The resulting colonies harboring the indicated constructs expressing nLUC or cLUC fusions were grown in LB medium overnight, collected by centrifugation and resuspended in infiltration buffer (liquid MS medium). Bacterial suspensions were then mixed in a 1:1 ratio and infiltrated into N. benthamiana leaves. For split-LUC assays to determine the effects of GA on the interaction of CRY1 with GID1, GV3101 colonies harboring constructs expressing CRY1-nLUC and cLUC-GID1a or cLUC-GID1b or cLUC-GID1c were mixed in a 1:1 ratio and infiltrated into N. benthamiana leaves. Bacterial suspensions bearing GAI-nLUC or SLY1-nLUC and cLUC-GID1a were used as positive and negative controls, respectively. After 2–3 days, N. benthamiana leaves were infiltrated with 1-mM D-luciferin sodium salt substrate (Yeasen) and 100-μM GA3 or an equivalent amount of absolute ethanol (Mock), and kept in the dark for 10 min. LUC signal was collected on a luminescent imaging workstation (Tanon 5200 Chemiluminescence imaging system). For split-LUC assays to determine the effects of CRY1 on the interaction of GID1 or SLY1 with DELLA proteins, GV3101 colonies harboring the constructs expressing cLUC-GID1a or cLUC-sly1-d, RGA-nLUC or GAI-nLUC, and CRY1-YFP or GUS-NLS-YFP (negative control; Wang et al., 2018a) were mixed in a 1:1 ratio and infiltrated into N. benthamiana leaves. After 2–3 days, N. benthamiana leaves were infiltrated with 1-mM D-luciferin sodium salt substrate and kept in the dark for 10 min. LUC signal was collected with a luminescent imaging workstation.
Cell-free pull-down assays with Arabidopsis protein extracts
For assays of blue light-specific CRY1–GID1 and CRY1–DELLA interactions, recombinant GST–GID1, GST–GAI, and GST–RGA bait proteins were first incubated with 10 μL MagneGST glutathione particles (Promega, V8611) and then washed three times with lysis buffer (50-mM Tris–HCl, pH 7.5, 150-mM NaCl, 0.2% Triton X-100). Protein extracts from Myc-CRY1-OX seedlings were used as preys, which were adapted to darkness for 5 days and then remained in the dark for 1 h or exposed to blue (30 μmol·m–2·s–1), red (50 μmol·m–2·s–1), or far-red light (10 μmol·m–2·s–1) for 1 h. All seedlings were homogenized in lysis buffer plus 1-mM Pefabloc, 1×EDTA-free Protease Inhibitor Cocktail (Roche), and 50-μM MG132. After centrifugation at 12,000g for 15 min at 4°C, the supernatant was incubated with bait proteins bound to glutathione particles for 1 h and washed four to five times with 1-mL lysis buffer each time. The precipitates were then eluted with 20-μL SDS 1×loading buffer and subjected to immunoblot analysis with anti-Myc antibody (Millipore, 05-724, 1:2,000).
For assays of CRY1 effects on RGA–PIF3 interactions, we purified recombinant MBP–PIF3 as described previously (Xu et al., 2018). Prey samples were protein extracts prepared from dark-grown RGA-Flag-OX and Myc-CRY1-OX seedlings and maintained in the dark or exposed to blue light (50 μmol·m–2·s–1) for 2 h. The subsequent steps were as described above.
For assays of CRY1 effects on SLY1–DELLA and GID1–DELLA interactions, prey samples were protein extracts prepared from RGA-Flag-OX, GAI-Flag-OX, cry1 RGA-Flag-OX, cry1 GAI-Flag-OX, and Myc-CRY1-OX seedlings first grown in continuous white light for 4–5 days, and dark-adapted for 3 days before being exposed to blue light (30 μmol·m–2·s–1) for 1 h. During the EZview Red Anti-c-Myc Affinity Gel (Sigma, E6654) used for incubation, 100-μM GA3 or ethanol (negative control) was added to the supernatant to explore the influence of GA3 on SLY1–DELLA and GID1–DELLA interactions. The beads were washed three times with lysis buffer containing 100-μM GA3 after incubation. Prey and bait proteins were detected using anti-Flag and anti-GST antibodies, respectively.
Co-IP assays
For co-IP assays of blue light-dependent CRY1–DELLA, CRY1–GID1, and CRY1–SLY1 interactions, RGA-Flag-OX, cry1 RGA-Flag-OX, GAI-Flag-OX, cry1 GAI-Flag-OX, GID1a-Flag-OX, cry1 GID1a-Flag-OX, GAI-Flag-OX/SLY1-Myc-OX, and GAI-Flag-OX GID1a-Myc-OX seedlings were grown in continuous white light for 4–5 days, and then dark-adapted for 3 days. One half of the dark-adapted seedlings was kept in the dark for 1 h, while the other half was exposed to blue light (50 μmol·m–2·s–1) for 1 h. All seedlings were then harvested under a dim green safe light and homogenized in lysis buffer (50-mM Tris–HCl, pH 7.5, 150-mM NaCl, 0.2% Triton X-100) containing 1-mM Pefabloc, 1×EDTA-free Protease Inhibitor Cocktail (Roche), and 50-μM MG132. Total protein concentration was determined by Bradford assay and equal amounts of total protein was incubated in 1 mL of lysis buffer with anti-CCT1 antiserum (Sang et al., 2005) attached to Dynabeads protein G beads (GE healthcare) at 4°C for 2 h. For co-IP assays of the effects of GA3 on the interaction of CRY1 with GID1, protein extracts prepared from GID1a-Flag-OX seedlings were incubated with 100-μM GA3 for 1 h with protein G beads combined with anti-CCT1 antiserum. For co-IP assays of the effects of CRY1 on the interaction of DELLA proteins with SLY1 and GID1a, 5-day-old etiolated double transgenic GAI-Flag-OX SLY1-Myc-OX and GAI-Flag-OX GID1a-Myc-OX seedlings in the WT and cry1 mutant backgrounds were used. For co-IP assays with N. benthamiana leaves, the DNA fragments encoding RGA-Nend-Flag, RGA-M5-Flag, GAI-Nend-Flag, and GAI-M5-Flag were cloned individually into the pHB vector. N. benthamiana leaves were infiltrated with GV3101 colonies harboring these constructs and those expressing Myc-CRY1 and cLUC-NLS-Flag (Sang et al., 2005; Wang et al., 2018a). For IP with anti-Flag antibody, Flag-tagged RGA-Nend, RGA-M5, GAI-Nend, or GAI-M5 constructs were co-infiltrated with Myc-CRY1 in N. benthamiana leaves. Equal amounts of total protein in 1 mL of lysis buffer from N. benthamiana leaves co-infiltrated with Myc-CRY1 and RGA-Nend or RGA-M5 or GAI-Nend or GAI-M5 were incubated with 10-μL anti-Flag beads (Smart-Lifesciences, SA042001) at 4°C for 2 h. All immunoprecipitates were washed three to four times with lysis buffer, and then eluted with 20-μL SDS 1×loading buffer and subjected to immunoblot analysis with anti-CCT1 or anti-Flag or anti-Myc antibody.
In vivo ubiquitination assays
For assays of the effects of CRY1 on the ubiquitination of DELLA proteins, WT, RGA-Flag-OX, and cry1 RGA-Flag-OX seedlings were grown in blue light (30 μmol·m–2·s–1) for 5 days, and then treated with 100-μM GA3 and 50-μM MG132 in liquid MS medium for 2 h. Total protein was extracted and protein concentration was determined by Bradford assay; equal amounts of total protein in 1 mL of lysis buffer (50-mM Tris–HCl, pH 7.5, 150-mM NaCl, 0.2% Triton X-100) containing 1-mM Pefabloc, 1×EDTA-free Protease Inhibitor Cocktail (Roche), and 50-μM MG132, were incubated with anti-Flag beads at 4°C for 2 h. The immunoprecipitates were washed three to four times with lysis buffer, and then eluted with 20-μL SDS 1×loading buffer and subjected to immunoblot analysis with anti-ubiquitin (CST, 3936S) and anti-Flag antibodies.
Accession numbers
Sequence data from this article can be found in the EMBL/GenBank database or the Arabidopsis Genome Initiative database under the following accession numbers: CRY1 (At4g08920), RGA (At2g01570), GAI (At1g14920), SLY1 (At4g24210), SNE (At5g48170), GID1a (At3g05120), GID1b (At3g63010), and GID1c (At5g27320).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Blue, red and far-red light treatments inhibit GA-induced degradation of DELLA proteins.
Supplemental Figure S2. Blue light inhibits the degradation of DELLA proteins largely in a GA-dependent but COP1-independent manner.
Supplemental Figure S3. Both the N- and C-terminal domains of DELLA proteins mediate the interaction with CRY1.
Supplemental Figure S4. Mapping of domains within CRY1 and DELLA proteins that mediate the interaction of CRY1 and DELLA proteins in N. benthamiana leaves.
Supplemental Figure S5. CRY1 very weakly affects the interaction of DELLA proteins with PIFs.
Supplemental Figure S6. CRY1 does not interact with SLY1 or SNE in N. benthamiana leaves.
Supplemental Figure S7. Both the N- and C-terminal domains of CRY1 mediate the interaction of CRY1 with GID1a in N. benthamiana leaves.
Supplemental Figure S8. GID1 inhibits photomorphogenesis in blue and far-red lights.
Supplemental Table S1. Primers used in this study.
Supplemental File S1. Summary of statistical analyses.
Supplementary Material
Acknowledgments
We thank Drs Suiwen Hou, Xin Zhou, and Xiaoying Zhao for materials assistance, and Dr Xiaoying Zhao for helpful discussions.
Funding
This work was supported by The National Key Research and Development Program of China grant (2017YFA0503802), The National Natural Science Foundation of China grants (31530085, 31900609, 31900207, 32000183), and The Science and Technology Commission of Shanghai Municipality grant (18DZ2260500).
Conflict of interest statement. The authors declare no conflict of interest regarding the publication of this study.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Wenxiu Wang (wangwenxiu85@shnu.edu.cn).
H.-Q.Y., P.X., and W.W. conceived the project; H.-Q.Y., P.X., and W.W. designed the research plan; P.X., H.C., T.L., X.C., L.M., S.D., J.Z., S.Z., and G.Y. carried out the experiments; P.X., W.W., and Z.M. analyzed the data; P.X., W.W., and H.-Q.Y. wrote the manuscript.
References
- Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, Harberd NP (2007) DELLAs contribute to plant photomorphogenesis. Plant Physiol 143: 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Alabadi D, Gil J, Blazquez MA, Garcia-Martinez JL (2004) Gibberellins repress photomorphogenesis in darkness. Plant Physiol 134: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Lawrence PK, Steber CM (2011) The role of two F-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol 155: 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Shang J, Oh E, Fan M, Bai Y, Zentella R, Sun T, Wang Z (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Tourinan N, Legris M, Minguet EG, Costigliolo-Rojas C, Nohales MA, Iniesto E, Garcia-Leomicronn M, Pacin M, Heucken N, Blomeier T, et al. (2020) COP1 destabilizes DELLA proteins in Arabidopsis. Proc Natl Acad Sci U S A 117: 13792–13799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F, et al. (2017) Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171: 287–304 [DOI] [PubMed] [Google Scholar]
- Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7: 204–210 [DOI] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284: 760–765 [DOI] [PubMed] [Google Scholar]
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Deng XW, Quail PH (1999) Signalling in light-controlled development. Semin Cell Dev Biol 10: 121–129 [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH (1992) COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801 [DOI] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun T (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Liu J, He G, Liu P, Sun J (2020) Photoexcited phytochrome B interacts with brassinazole resistant 1 to repress brassinosteroid signaling in Arabidopsis. J Integr Plant Biol 62: 652–667 [DOI] [PubMed] [Google Scholar]
- Du S, Li L, Li L, Wei X, Xu F, Xu P, Wang W, Xu P, Cao X, Miao L, et al. (2020) Photoexcited cryptochrome2 interacts directly with TOE1 and TOE2 in flowering regulation. Plant Physiol 184: 487–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M (1998) CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95: 669–679 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chory J (1997) Light control of plant development. Annu Rev Cell Dev Biol 13: 203–229 [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet CM, Sun TP (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8: 77–85 [DOI] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegear RJ, Foley LE, Casselman A, Reppert SM (2010) Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature 463: 804–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Han X, Chang X, Zhang Z, Chen H, He H, Zhong B, Deng XW (2019) Origin and evolution of core components responsible for monitoring light environment changes during plant terrestrialization. Mol Plant 12: 847–862 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. Embo J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ (2009) Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21: 2624–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KE, Moritz T, Harberd NP (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205 [DOI] [PubMed] [Google Scholar]
- Lee S (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Gene Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Jia KP, Lian HL, Yang X, Li L, Yang HQ (2014) Jasmonic acid enhancement of anthocyanin accumulation is dependent on phytochrome A signaling pathway under far-red light in Arabidopsis. Biochem Biophys Res Commun 454: 78–83 [DOI] [PubMed] [Google Scholar]
- Lian H, Xu P, He S, Wu J, Pan J, Wang W, Xu F, Wang S, Pan J, Huang J,et al. (2018) Photoexcited CRYPTOCHROME 1 interacts directly with G-protein beta subunit AGB1 to regulate the DNA-binding activity of HY5 and photomorphogenesis in Arabidopsis. Mol Plant 11: 1248–1263 [DOI] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Mei S, Shi C, Yang Y, Peng Y, Ma L, Wang F, Li X, Huang X, Yin Y, et al. (2018) UVR8 Interacts with BES1 and BIM1 to Regulate Transcription and Photomorphogenesis in Arabidopsis. Dev Cell 44: 512–523 [DOI] [PubMed] [Google Scholar]
- Lin C, Ahmad M, Cashmore AR (1996) Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J 10: 893–902 [DOI] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C (2008b) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008a) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XD, Zhou CM, Xu PB, Luo Q, Lian HL, Yang HQ (2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8: 467–478 [DOI] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA 113: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Zhang YC, Sang Y, Li QH, Yang HQ (2005) From the cover: A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci USA 102: 12270–12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, He S, Xu F, Wei X, Jiang L, Liu Y, Wang W, Li T, Xu P, Du S, et al. (2020) Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis. New Phytol 225: 848–865 [DOI] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1999) Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400: 781–784 [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Park J, Nguyen KT, Park E, Jeon JS, Choi G (2013) DELLA proteins and their interacting RING Finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell 25: 927–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Huang SS, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PA, Sridevi P, Nito K, Nery JR, et al. (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta LM, Lange T (2006) Gibberellin biosynthesis and the regulation of plant development. Plant Biol (Stuttg) 8: 281–290 [DOI] [PubMed] [Google Scholar]
- Quail PH (2002) PHYTOCHROME PHOTOSENSORY SIGNALLING NETWORKS. Nat Rev Mol Cell Bio 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O'Hara A, Kaiserli E, Baumeister R, Schafer E, Nagy F, Jenkins GI, et al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW, Yang HQ (2005) N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17: 1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl CL, Byrd AD, Benzion G, Altschuler MA, Hildebrand DF, Hunt AG (1987) Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61: 1–11 [DOI] [PubMed] [Google Scholar]
- Seo HS, Yang J, Ishikawa M, Bolle C, Ballesteros ML, Chua N (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Shi H, Shen X, Liu R, Xue C, Wei N, Deng XW, Zhong S (2016) The red light receptor phytochrome B directly enhances substrate-E3 ligase interactions to attenuate ethylene responses. Dev Cell 39: 597–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M (2008) Structural basis for gibberellin recognition by its receptor GID1. Nature 456: 520–523 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y (2001) Repressing a repressor gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Sun TP (2011) The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 21: R338–R345 [DOI] [PubMed] [Google Scholar]
- Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognar L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP (2004) Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Wang C, Dai S, Zhang ZL, Lao W, Wang R, Meng X, Zhou X (2021) Ethylene and salicylic acid synergistically accelerate leaf senescence in Arabidopsis. J Integr Plant Biol. DOI: 10.1111/jipb.13075. [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wang S, Li L, Xu P, Lian H, Wang W, Xu F, Mao Z, Zhang T, Yang H (2018b) CRY1 interacts directly with HBI1 to regulate its transcriptional activity and photomorphogenesis in Arabidopsis. J Exp Bot 69: 3867–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lian H, Zhang L, Mao Z, Li X, Xu F, Li L, Yang H (2016) Transcriptome analyses reveal the involvement of both C and N termini of Cryptochrome 1 in its regulation of phytohormone-responsive gene expression in Arabidopsis. Front Plant Sci 7: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lu X, Li L, Lian H, Mao Z, Xu P, Guo T, Xu F, Du S, Cao X, et al. (2018a) Photoexcited CRYPTOCHROME1 interacts with dephosphorylated BES1 to regulate brassinosteroid signaling and photomorphogenesis in Arabidopsis. Plant Cell 30: 1989–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang W, Xu P, Pan J, Zhang T, Li Y, Li G, Yang H, Lian H (2018) phyB interacts with BES1 to regulate brassinosteroid signaling in Arabidopsis. Plant Cell Physiol 60: 353–366 [DOI] [PubMed] [Google Scholar]
- Xu F, Li T, Xu PB, Li L, Du SS, Lian HL, Yang HQ (2016) DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. Febs Lett 590: 541–549 [DOI] [PubMed] [Google Scholar]
- Xu F, He S, Zhang J, Mao Z, Wang W, Li T, Hua J, Du S, Xu P, Li L, et al. (2018) Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol Plant 11: 523–541 [DOI] [PubMed] [Google Scholar]
- Xu P, Lian H, Xu F, Zhang T, Wang S, Wang W, Du S, Huang J, Yang HQ (2019) Phytochrome B and AGB1 coordinately regulate photomorphogenesis by antagonistically modulating PIF3 stability in Arabidopsis. Mol Plant 12: 229–247 [DOI] [PubMed] [Google Scholar]
- Yadav A, Singh D, Lingwan M, Yadukrishnan P, Masakapalli SK, Datta S (2020) Light signaling and UV-B-mediated plant growth regulation. J Integr Plant Biol 62: 1270–1292 [DOI] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR (2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Zhao X, Yu X, Foo E, Symons GM, Lopez J, Bendehakkalu KT, Xiang J, Weller JL, Liu X, Reid JB, et al. (2007) A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiol 145: 106–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.