Abstract
Shoot branching and complex leaf development relies on the establishment of boundaries that precedes the formation of axillary meristems (AMs) and leaflets. The tomato (Solanum lycopersicum) super determinant mutant is compromised in both processes, due to a mutation in Sde1A. Sde1A encodes a protein with a RAWUL domain, which is also present in Polycomb Group Repressive Complex 1 (PRC1) RING finger proteins and WD Repeat Domain 48 proteins. Genetic analysis revealed that Sde1A and Bmi1A cooperate, whereas Bmi1C antagonizes both activities, indicating the existence of functionally opposing PRC1 complexes that interact with Sde1A. Sde1A is expressed at early stages of boundary development in a small group of cells in the center of the leaf-axil boundary, but its activity is required for meristem formation at later stages. This suggests that Sde1A and Bmi1A promote AM formation and complex leaf development by safeguarding a pool of cells in the developing boundary zones. Genetic and protein interaction analyses showed that Sde1A and Lateral suppressor (Ls) are components of the same genetic pathway. In contrast to ls, sde1a mutants are not compromised in inflorescence branching, suggesting that Sde1A is a potential target for breeding tomato cultivars with reduced side-shoot formation during vegetative development.
The RAWUL protein-encoding gene Super determinant1A interacts genetically with the Bmi1 family and plays a crucial role at early steps of axillary meristem formation and complex leaf development in tomato.
Introduction
The remarkable variation in plant architecture is driven primarily by the degree of branching during vegetative and reproductive development. In seed plants, shoot branches develop from established axillary meristems (AMs), an evolutionary innovation that underpinned diversification in shoot architecture (Sussex and Kerk, 2001; Langdale and Harrison, 2008). During the vegetative growth phase, AMs initiate at leaf axils that are located far from the shoot apical meristem (SAM; Greb et al., 2003; Xin et al., 2017; Ponraj and Theres, 2020). This initiation requires the establishment of cells within the leaf-axil boundary domain that have the competence to form new meristems. AMs develop into axillary buds (ABs) and their subsequent development requires the integration of whole-plant signaling, leading to dormant ABs or actively growing side shoots (Domagalska and Leyser, 2011; Martín-Fontecha et al., 2018; Barbier et al., 2019).
The establishment of a boundary domain between the pluripotent stem cells of the SAM and the cells specified for leaf organogenesis precedes the formation of AMs (Wang et al., 2016). Boundary cells show characteristic morphological features, such as small size, infrequent divisions, a parallel arrangement of microtubules, and rigid cell walls (Hussey, 1971; Reddy et al., 2004; Burian et al., 2016). At the molecular level, boundary cells express evolutionarily conserved genes common to pathways that regulate SAM formation (Tian et al., 2014, 2019). For example, central players in boundary establishment and SAM initiation in numerous plant species are orthologs of the NAC-domain transcription factor genes CUP-SHAPED COTYLEDON1 (CUC1), CUC2, and CUC3 in Arabidopsis (Arabidopsis thaliana) and Goblet (Gob) in tomato (Solanum lycopersicum; Aida et al., 1997; Vroemen et al., 2003; Blein et al., 2008; Raman et al., 2008; Berger et al., 2009), which in turn are posttranscriptionally controlled by the conserved miRNA miR164 (Nikovics et al., 2006). The GRAS transcription factor LATERAL SUPPRESSOR (LAS in Arabidopsis and Ls in tomato) functions downstream of CUC/Gob (Schumacher et al., 1999; Greb et al., 2003; Raman et al., 2008; Rossmann et al., 2015). Furthermore, a parallel pathway containing the MYB-domain transcription factors REGULATOR OF AXILLARY MERISTEMS1 (RAX1), RAX2, and RAX3 in Arabidopsis and Blind (Bl) in tomato controls AM initiation (Schmitz et al., 2002; Keller et al., 2006; Müller et al., 2006; Busch et al., 2011).
In species with compound leaves, such as tomato, the regulatory mechanisms that operate during leaf primordium initiation and boundary domain establishment are also recruited for leaflet development. For instance, the leaflet boundary domain is specified by the AM regulators Ls, Gob, and the Bl paralog Potato leaf (Busch et al., 2011). In mature leaves of tomato and other species, these boundaries can form ectopic meristems (EMs) that have the potential to develop into shoots (Rossmann et al., 2015).
The establishment and maintenance of cell pools that are competent to form meristems require the specific activation and repression of transcriptional programs, which involve the spatiotemporal deployment of epigenetic mechanisms that operate on chromatin. One group of enzymes that alters DNA accessibility and exposes cis-regulatory elements to transcription factors is composed of chromatin-remodeling factors. In Arabidopsis, two such factors are the Sucrose Non-Fermentable (SNF2)-type chromatin-remodeling ATPases SPLAYED and BRAHMA. During embryogenesis, both proteins promote the transcription of CUC genes at the cotyledon boundary (Kwon et al., 2006), whereas in the postembryonic phase, SPLAYED promotes the expression of the SAM regulator WUSCHEL (WUS; Kwon et al., 2005). It is currently unknown whether such a mechanism also operates during AM formation. However, a transcriptionally permissive environment at the shoot meristem regulator SHOOT MERISTEMLESS (STM) locus is maintained in young leaf axils (Cao et al., 2020), whereas the WUS locus is characterized by a transcriptionally restrictive environment that gradually becomes transcriptionally permissive during early stages of AM initiation (Wang et al., 2017), indicating the existence of different phases of epigenetic regulation during successive stages of AM formation.
The transcriptional repression of regulatory genes that control the commitment of cells to differentiate is thought to play a major role within the boundary domain, to maintain cellular pluripotency (Wang et al., 2016). Polycomb Group (PcG) proteins are key transcriptional repressors and assemble into two major complexes known as PcG Repressive Complex 1 (PRC1) and PRC2: PRC1 regulates Histone H2A lysine 119 (H2AK119, H2AK121 in Arabidopsis and tomato) monoubiquitylation, while PRC2 regulates Histone H3 lysine 27 (H3K27) trimethylation. PRC1 and PRC2 recruitment and their mechanistic role during the repression of gene expression is an active area of research; however, there remains a profound lack of knowledge concerning their roles, as well as the relative contributions of their individual components during all stages of AM formation. Here, we show that Super determinant 1A (Sde1A) shares an evolutionary origin with the B cell-specific Moloney murine leukemia virus integration site 1 (Bmi1) constituents of PRC1, and that it operates at the early stages of boundary development. Sde1A is required to maintain high expression levels of the shoot meristem regulator genes Tomato Knotted 2 (TKn2, the tomato STM ortholog) and CLAVATA3 (CLV3), as well as the boundary regulators Gob, Ls, and Bl. Together with Bmi1A, Sde1A increases the morphogenetic competence of leaf axils and leaves and promotes the formation of AMs, leaflets, and EMs, indicating the presence of an active epigenetic regulation during the establishment of the leaf-axil boundary domain. In contrast, Bmi1C antagonizes Sde1A and Bmi1A activity, implying that in tomato opposing PRC1 complexes promote or inhibit meristematic competence in combination with Sde1A. Furthermore, protein and genetic interaction analyses indicate that this mechanism requires the activity of the AM regulator Ls.
Results
The sde mutant shows reduced shoot branching and reduced leaf dissection
The sde mutant of tomato is characterized by an altered development of AMs. Under greenhouse conditions, all leaf axils of wild-type (wt, VF36 cultivar) plants developed ABs, whereas ABs failed to develop in sde during early to mid-vegetative development (∼leaf axils 1–6; Figure 1, A–C and E–G). The leaf axils located close to the first inflorescence were not affected and produced ABs (Figure 1G). This trait was fully penetrant in greenhouse and controlled environment conditions, but its expressivity varied from 13% to 90% (Supplemental Figure S1). To discriminate between AM initiation and outgrowth, we scored shoot apices under a stereomicroscope and imaged them by scanning electron microscopy (SEM). During the vegetative phase (2-week-old seedlings), the axils of young leaf primordia (P) 1–5 were barren in wt and sde. In wt, axils of leaf primordia older than P6 developed bulges as the first morphological indication of AM initiation, but these were absent in sde (Figure 1, D and H). During reproductive development (4-week-old plants), wt shoot apices developed ABs at all leaf axils (Supplemental Figure S2, A and B), whereas sde had empty leaf axils (45%), a disorganized mass of cells (11%), ABs that were delayed in development (25%), or fully developed ABs (19%, Supplemental Figure S2, C–F).
Figure 1.
sde is compromised in shoot branching and leaf dissection. A and E, Growth habit of wt (A) and sde (E) plants 8 weeks after sowing. B and F, Close-up of leaf axils from plants shown in A (B) and E (F), respectively. Arrows highlight filled leaf axils in (B) and empty leaf axils in F. C and G, Graphic representation of AB formation. Each column represents a single plant and each square within a column denotes a single leaf axil. Green indicates the presence of an AB or a side shoot, yellow indicates an empty leaf axil. Cotyledons are indicated as ct. D and H, Scanning electron micrographs of vegetative shoot apices (14-d-old seedlings) including the P6 leaf axil. The ovals delimit P6 axils developing an AM in wt (D), and an empty leaf axil in sde (H). I and J, Picture of leaf no. 6 of wt (I) and sde (J) plants showing leaflets separated from the rachis. Examples of first-order (yellow) and second-order (white) leaflets are indicated in I. K and L, Violin plots of the mean number of first-order (K) and second-order (L) leaflets per leaf along the primary shoot (leaves 1–8) in wt and sde plants. Median values are indicated by a black dot. n = 14 in K and L. P-values were determined by two-tailed, two-sample t test. Bars represent 15 cm in (A) and (E), 4 cm in (B) and (F), 50 µm in (D) and (H), and 5 cm in (I) and (J).
Tomato plants alternate between vegetative and reproductive growth, which is known as sympodial growth. The primary SAM, which originates during embryogenesis, produces between 8 and 12 leaves before transitioning to reproductive growth as an inflorescence meristem (IM). Vegetative growth subsequently proceeds via sympodial shoots that develop from the uppermost AM. Each sympodial shoot produces three leaves before transitioning to reproductive growth in a continuously reiterated process in indeterminate tomato cultivars. Determinate tomato cultivars harbor a mutation in the Self pruning (Sp) gene and show a progressive reduction in the number of leaves formed by sympodial meristems (Pnueli et al., 1998), which is a desirable trait for tomato field varieties because it facilitates mechanical harvesting. The VF36 cultivar harbors the sp allele and expresses this distinctive sp phenotype. The sde mutation enhanced the sp phenotype, which culminated in sympodial shoots that bypassed vegetative growth and developed as inflorescences, to generate plants with two consecutive inflorescences (Supplemental Figure S3, A–C). This trait showed genotype-by-environment interaction and incomplete penetrance (Supplemental Figure S3, C and D), but the flowering time of the primary shoot and the number of flowers per inflorescence in sde mutants were not affected (Supplemental Figure S3, E and F).
To monitor differences in compound leaf development between sde and wt, we assessed the mean number of first- and second-order leaflets for all leaves of the primary shoot (leaves1–8) in 12-week-old plants. About nine first-order leaflets per leaf developed in wt plants, whereas sde possessed eight (Figure 1, I–K). Similarly, wt plants developed five second-order leaflets on average, whereas sde developed four (Figure 1, I–L), which represented a slight but significant reduction in leaflet number in sde.
Molecular cloning of Sde1A
A backcross F2 population (BCF2) revealed that the sde phenotype segregated as a monogenic recessive trait (Supplemental Figure S4A) and the underlying mutation, referred to as sde1a, was characterized in more detail. Using an F2 population from a cross between sde and the wild tomato species Solanum pennellii, we mapped Sde1A to a region between markers T635 and T725 on chromosome 4 (Figure 2A). Due to low recombination frequency over this interval, the sde1a mutation could not be identified by conventional genetic mapping. Therefore, we followed comparative whole-genome sequencing (WGS) and transcriptome deep sequencing (RNA-seq) approaches. Data from WGS identified 1,977 variants that were evenly distributed throughout the genome of the sde mutant, of which 130 were within the sde1a candidate region (Figure 2B, Supplemental Figure S5A, Supplemental Data Set S1). RNA-seq analysis of vegetative shoot apices revealed no differences in the expression of genes within the target region (Supplemental Data Set S2), indicating that the sde1a mutation does not influence transcript levels, but might impact protein activity. The combination of genome resequencing and RNA-seq analysis uncovered a unique candidate mutation that also fully cosegregated with the sde phenotype in the BCF2 population (Supplemental Figure S4, A and B, Supplemental Data Set S3). A transversion of a guanine to a thymidine nucleotide (251G>T) in the gene Solyc04g049190, referred to as Sde1A hereafter, led to an amino acid exchange of a tryptophan to a leucine residue (W84L) in the encoded protein (Figure 2B, Supplemental Figure S5B, Supplemental Data Set S4). Analysis of the mapped RNA-seq reads in combination with cDNA cloning guided the reannotation of Sde1A, which encodes a ring-finger and WD40-associated ubiquitin-like protein (RAWUL, Supplemental Figure S6). The RAWUL domain is present in the C-terminal region of RING finger proteins from the PRC1 complex (Ring1 and Bmi1) and in the C-terminal region of WD-repeat 48 (Wdr48) proteins (Sanchez-Pulido et al., 2008). The tomato genome contains five genes that encode RING-finger and RAWUL-domain proteins, two of which correspond to Ring1 genes (Ring1A: Solyc02g077890 and Ring1B: Solyc07g053800) and three to Bmi1 genes (Bmi1A: Solyc09g065990, Bmi1B: Solyc06g008600, and Bmi1C: Solyc06g084040). Furthermore, we identified two Wdr48-encoding genes (Wdr48A: Solyc01g098090 and Wdr48B: Solyc08g023570) and an additional Sde1A paralog (Sde1B: Solyc01g009720) in tomato, all of which contain a C-terminal RAWUL domain. However, none of these have been functionally characterized. In the PRC1 complex, Ring1 and Bmi1 interact via the N-terminal RING-finger domain, whereas the RAWUL domain interacts with additional PRC1 core proteins and leads to the formation of functionally distinct PRC1 complexes (Kim, 2017). Recently, the Sde1A homologous genes LAX PANICLE2 (LAX2, Tabuchi et al., 2011) and barren stalk2 (ba2, Yao et al., 2019) from rice (Oryza sativa) and maize (Zea mays), respectively, were shown to regulate shoot branching. However, Basic Local Alignment Search Tool for Protein (BLASTP) searches (Altschul et al., 1997) and protein alignment analysis identified Os12g0479100 and GRMZM2G447297 as the closest Sde1A homologs (Supplemental Figure S7) from rice and maize, respectively, whereas Sde1B is the putative ortholog of LAX2 and ba2. An analysis of Sde1A through Protein Variation Effect Analyzer (PROVEAN, Choi and Chan, 2015) indicated that amino acid W84 located within the RAWUL domain is highly conserved among Sde1 and Bmi1 homologs, suggesting that the W84L amino acid exchange might have a deleterious effect on protein activity.
Figure 2.
Sde1A encodes a RAWUL-domain protein. A, Sde1A maps to a region on chromosome 4 delimited by markers T0635 and T0725. Chromosome 4 from the tomato reference strain Heinz is represented at the top (green). Individual recombinant chromosomes are shown below for the recombinant lines indicated on the left and their corresponding phenotype on the right. White segments denote genomic segments inherited from sde; black segments represent genomic segments inherited from S. pennellii. B, Venn diagrams showing the total number of DNA variants, variants that cause an amino acid change and variants located within the chromosome 4 candidate region, in a comparison between VF36 and sde by whole-genome and RNA-sequencing (WGS and RNA-seq), respectively. C, Schematic representation of the Sde1A locus indicating the genomic region used for complementation and sites targeted by CRISPR-Cas9. The promoter and the terminator are indicated as thick black bars. The conserved (RAWUL) and the nonconserved coding region are indicated in green and white, respectively. sgRNA sequences are represented in red, followed by the adjacent protospacer motif in blue. Partial DNA sequences of individual deletions in the first and fourth exons are shown. The distance between the shown adjacent sequences is denoted by n. The size of the in-frame (green) and frameshift (red) deletions is indicated on the right in bp. D, Violin plots of shoot branching phenotype (% empty leaf axils) of Sde1A CRISPR-Cas9 mutants carrying independent mutations in the VF36 background. E, Violin plots of shoot branching phenotype (% empty leaf axils) of different T1 plants harboring an Sde1A genomic fragment that complements the sde mutant phenotype. Median values are indicated by a black circle. n values in (D) and (E) represents the number of individual plants. P-values were determined by two-tailed, two-sample t tests against wt in (D) and against the sister plants lacking the transgene (null) in (E).
To obtain additional independent sde1a mutant alleles, we edited the Sde1A locus using genome editing via Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated nuclease (Cas9; Belhaj et al., 2013). We designed two pairs of single guide RNAs (sgRNAs) targeting either the first or the fourth exon of Sde1A to alter either the encoded nonconserved region or the conserved RAWUL domain, respectively (Figure 2C;Supplemental Figure S8). Four out of eight independent mutations in the first exon and all five mutations in the last exon recapitulated the sde branching phenotype (Figure 2D;Supplemental Figure S8). In addition, transformation of sde with a Sde1A genomic fragment functionally complemented the sde mutant (Figure 2E). Taken together, three independent approaches validated the sde1a mutation as being causal for the sde phenotype.
To assess the role of Sde1A in the regulation of AM formation in different genetic backgrounds, we crossed sde to the greenhouse cultivar Moneymaker and the field variety M82. Furthermore, we generated additional Sde1A genome-edited alleles in Moneymaker. Compared to the VF36 BCF2 population, F2 populations from crosses between sde and Moneymaker or M82 showed a lower proportion of plants displaying branching defects (∼13% and ∼3%, respectively) that indicated a strong but variable degree of epistasis (Supplemental Figure S4A). Sanger sequencing of the Sde1A locus showed that F2 plants with AM initiation defects comparable to sde are homozygous for sde1a (Supplemental Figure S4A), indicating that the sde phenotype is dependent on sde1a. Moreover, the sde1a allele co-segregated with defects in AM formation in 98% and 93% of all F2 plants that showed AM initiation defects from the crosses between sde and Moneymaker or M82, respectively (Supplemental Figure S4B). In contrast, new sde1a alleles generated in Moneymaker showed a small difference in the pattern of shoot branching compared to that of wt plants (e.g. sde1a-16, Supplemental Figure S8, A and C). However, genetic analysis of sde1a-16 mutant plants in a mixed MM/VF36 genetic background showed a stronger defect on AM formation than the original sde mutant (Supplemental Figure S8, B and C). This result indicated that sde1a-16 is indeed a loss-of-function allele and that sde1a mutants require VF36 modifiers to display the sde phenotype.
Sde1A is specifically expressed at the center of the leaf-axil boundary, whereas Sde1B is broadly expressed
The mRNA of Sde1A predominantly accumulated in the SAM at the vegetative, transition and inflorescence stages, whereas that of Sde1B, the closest Sde1A homolog, was present at low levels in all tissues analyzed (Figure 3A). Furthermore, the analysis of tissue-specific Sde1A expression by RNA in situ hybridization revealed a defined expression domain in the center of the boundary region between the emerging leaf primordium and the SAM, as well as in the presumptive leaflet boundary domain in older leaf primordia (P5, Figure 3, B and C). We observed this expression domain in P1 and P2, whereas Sde1A signal was weak or absent in older leaf axils (Figure 3, B and C; Supplemental Figure S9A). Longitudinal sections revealed that the Sde1A expression domain extends approximately six to seven cell layers in depth, starting from cell layer 2 (Figure 3B). Transverse sections showed that the Sde1A expression domain encompasses about five cell layers along the proximodistal axis (meristem to leaf) and eight cell layers along the lateral axis (Supplemental Figure S9A). This pattern extended through five consecutive sections, creating an ellipsoid-shaped expression domain. We did not detect Sde1A transcripts during the early stages of AM formation, first distinguished as bulges in P6–P7 axils, indicating that Sde1A expression is not required at those stages (Supplemental Figure S9A). To compare the patterns of Sde1A transcript and Sde1A protein localization, we generated a translational fusion between Sde1A and Venus driven by the Sde1A promoter (Sde1A-Venus). Three independent transgenic lines showed that Sde1A-Venus accumulates in a domain highly similar to that of the Sde1A transcript, indicating that the Sde1A protein does not move (Figure 3, D and E). In contrast to Sde1A, RNA in situ hybridization for Sde1B revealed a broad expression in the SAM and leaf primordia (Supplemental Figure S9B).
Figure 3.
Sde1A is preferentially expressed in shoot meristematic tissues. A, RT-qPCR analysis of Sde1A and Sde1B relative transcript levels in early vegetative meristems (EVM), late vegetative meristems (LVM), inflorescence meristems (IM), closed flowers, open flowers, roots, internodes, and leaves 1, 4, and 6. The expression level of GAPDH was used as a reference and was set to one. Sde1A transcripts were not detected (nd) in roots, internodes or in the oldest leaf no. 1, which is the leaf that is furthest away from the SAM. B, C, RNA in situ hybridization analysis of longitudinal sections (B) and cross-sections (C) through vegetative shoot apices hybridized with an Sde1A antisense probe. D and E, Confocal images of a line harboring a Sde1A-Venus translational reporter construct. D shows an optical transverse section and E a longitudinal section through a representative shoot apex. Bars in (A) represent the standard error of the mean from three technical replicates. The experiment was repeated twice with similar results. Bars in (B–E) represent 100 µm.
Ectopic expression of Sde1A increases morphogenetic competence in leaves
To test whether Sde1A can promote meristem formation, an activity associated with the boundary domain, we expressed Sde1A under the Arabidopsis UBIQUITIN10 (UBI10) promoter in wt VF36. Out of 15 independent transgenic tomato lines, we selected three with increased Sde1A transcript levels (Supplemental Figure S10, A and B). All three lines showed similar developmental phenotypes (Supplemental Figure S10, C and D). A decrease in plant size due to reduced internode elongation in Sde1A-OExL1 plants correlated with increased Sde1A expression, which is consistent with the longer internodes seen in the sde mutant (Figure 4, A–C). The line Sde1A-OExL1, which showed the highest Sde1A mRNA levels, was selected for detailed characterization.
Figure 4.
Ectopic expression of Sde1A increases morphogenetic competence in tomato leaves. A and B, Growth habit of a wt (A) and a plant overexpressing Sde1A (Sde1A-OExL1, B). C, Violin plot of internode length of wt, sde, and Sde1A-OExL1 plants. D, E, Comparison of leaf complexity in representative leaves (leaf no. 6) of wt (D) and Sde1A-OExL1 (E) plants. F, Ectopic shoot formation in a leaf of an Sde1A-OExL1 plant. G–I, Violin plot of the number of first-order (G) and second-order (H) leaflets and the number of ectopic shoots on leaves (I) of wt, sde, and Sde1A-OExL1 plants. Median values are indicated by a black circle. n values in (C) and (G–I) represent the number of individual plants. P-values were determined by two-tailed, two-sample t tests.
To assess differences in leaf morphogenetic competence, we monitored changes in leaf complexity (leaflet number per leaf) and EM formation in Sde1A-OExL1 plants. On average, wt plants developed between 11 and 5 first- and second-order leaflets per leaf, respectively (Figure 4, D, G, and H). In comparison, the number of first-order leaflets in Sde1A-OExL1 was 41% higher, while it was 20% lower in sde. Similarly, Sde1A-OExL1 plants had almost four-fold more second-order leaflets than wt, whereas sde1 plants had 46% fewer (Figure 4, D, E, G, and H). Typically, tomato leaves form EMs at the distal leaflet boundary (Rossmann et al., 2015) with a mean of approximately one EM per leaf in wt VF36 plants (Figure 4I). In Sde1A-OExL1 plants, about six EMs were present per leaf, whereas we observed none on sde leaves (Figure 4, F and I). Moreover, EMs in some Sde1A-OExL1 plants were not limited to the petiolule–rachis junction, and when the plants had grown to maturity, EMs developed into shoots that covered the entire leaf (Figure 4F;Supplemental Figure S10, E and F).
Sde1 originated in the last common ancestor of embryophytes and shares part of the Bmi1 exon–intron structure, suggesting a common evolutionary history
BLASTP searches identified 138 Sde1A homologs, which were all restricted to embryophytes. The liverwort Marchantia polymorpha and the lycophyte Selaginella moellendorffii each contained a single gene with similarity to Sde1, whereas the genome of the moss Physcomitrium (Physcomitrella) patens contained two genes with similarity to Sde1 (Figure 5A). The genome from the extant relative of flowering plants Amborella trichopoda, the gymnosperm Pinus pinaster, and that of most of the sampled dicots (e.g. Arabidopsis and tomato), contained two Sde1-like genes (Figure 5A;Supplemental Data Set S5). Grasses (e.g. rice and maize), however, usually contained three Sde1-like genes (Figure 5A;Supplemental Data Set S5). Using a similar approach for proteins containing a RAWUL domain, we identified 260 Bmi1, 143 Ring1, and 102 Wdr48 homologous proteins in embryophytes and animals, but none in chlorophytes or rhodophytes (Supplemental Data Set S5). A phylogenetic reconstruction (Jones et al., 1992) showed that Ring1-like and Wdr48-like proteins from animals and plants form closely related clades (Supplemental Figure S11A). In contrast, Bmi1-like proteins from animals and plants clustered within separate clades (Supplemental Figure S11A). Notably, Sde1-like proteins clustered with plant Bmi1-like proteins, suggesting that they share a common evolutionary origin.
Figure 5.

Exon–intron structures of Sde1 and Bmi1 genes suggest they share a common evolutionary history. A, Phylogenetic relationships among embryophytes and representative members of chlorophytes, liverworts, mosses, lycophytes, gymnosperms, and angiosperms. Chlorophytes are represented in blue, basal plants in black and flowering plants in gray. The copy number of Sde1, Bmi1, and Ring1 genes are indicated above. The tree was redrawn based on the relationship of the respective sequenced genomes deposited in Phytozome. B, Gene models of Sde1- and Bmi1-related genes from tomato and Marchantia polymorpha. Red boxes denote the region encoding the C3HC4 ring-finger domain; green boxes represent the region encoding the RAWUL domain.
The gene models of Sde1 and Bmi1 homologous genes from plants revealed structural conservation over the region encoding the RAWUL domain (Figure 5B). For example, the exon–intron borders and the conserved amino acid stretches were located at similar exonic positions in Sde1-like and Bmi1-like genes (Figure 5B;Supplemental Figure S11B). This conservation in gene structure, together with the protein similarity, suggested functional conservation between the RAWUL domains of Sde1-like and Bmi1-like proteins. To explore this possibility, we tested the interaction between Sde1A and known BMI1-interacting proteins by yeast-two hybrid (Supplemental Figure S12). This analysis showed that Sde1A interacts with Like heterochromatin protein 1 (Lhp1, Solyc01g081500). Notably, the mutant sde1a protein showed reduced interaction with Lhp1 (Supplemental Figure S12). In Nicotiana benthamiana mesophyll cells Lhp1 interacted with Sde1A (Supplemental Figure S12). The Lhp1 protein is a component of various PRC1 complexes (Gao et al., 2012; Forderer et al., 2016), indicating that Sde1A may be involved in the regulation of PRC1 activity by competing with Bmi1 for interacting partners or through the formation of noncanonical PRC1 complexes.
Sde1A and Bmi1A synergistically promote AM formation, compound leaf development and EM formation, whereas Bmi1C antagonizes it
In the VF36 background, sde1a behaved as a recessive mutation; however, several genetic analyses suggested that VF36 is deficient in complementary pathways that regulate AM formation (sensitized background, Figure 7, E and F, Supplemental Figures S4, S8, and S19). Primary candidates that may compensate for the loss of Sde1A are Sde1B and the Bmi1 genes (Bmi1A, Bmi1B, Bmi1C), which are also expressed in shoot apices (Supplemental Figure S13A). In Arabidopsis, complete loss of BMI1 function leads to a significant reduction in the PRC1-associated mark H2A monoubiquitylation (H2AK121Ub), resulting in impaired plant development due to the upregulation of embryonic regulators such as BABY BOOM (BBM), FUSCA3 (FUS3), and LEAFY COLYLEDON1 (LEC1) and the meristem regulators STM, WUS, and WUSCHEL-RELATED HOMEOBOX 5 (Xu and Shen, 2008; Bratzel et al., 2010).
Figure 7.
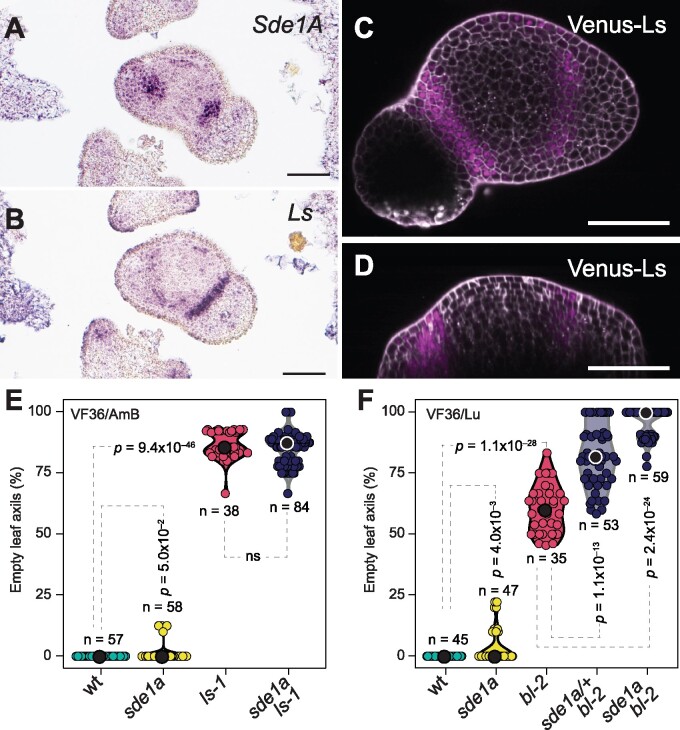
Sde1A and Lateral suppressor are components of a common genetic pathway that regulates AM initiation. A, B, RNA in situ hybridization of consecutive cross-sections of a vegetative shoot apex with an antisense Sde1A (A) or Ls (B) antisense probe. C and D, Confocal images of a line harboring an Ls-Venus translational reporter construct. C shows an optical transverse section and (D) a longitudinal section through a shoot apex. E, Violin plots of the percentage of empty leaf axils in wt, sde1a, ls-1, and sde1a ls-1 plants in a VF36/Antimold B mixed background (VF36/AMB). F, Violin plots to compare the percentage of empty leaf axils in wt, sde1a, bl-2, sde1a/+ bl-2, and sde1a bl-2 plants in a VF36/Lukullus mixed background (VF36/Lu). Median values are indicated by a black or white circle. n values in (E) and (F) represent the number of individual plants. P-values were determined by two-tailed, two-sample t tests. Bars in (A–D) represent 100 µm.
To unravel the impact of Sde1 and Bmi1 on target loci, we compared the expression of known Bmi1 targets and sde1a misexpressed genes in the tomato sde1 and bmi1 mutants. These experiments showed that individual Sde1 and Bmi1 genes collaborate in synergistic or antagonistic mode depending on the target locus. For instance, TKn2 was upregulated in bmi1b and bmi1c mutants, whereas it was downregulated in sde and sde1b. Wus showed downregulation in sde1b, bmi1a and bmi1b mutants, whereas Clv3 was downregulated in sde and bmi1c (Supplemental Figure S13B). Similarly, the AM regulators Ls, Bl, and Gob showed a coordinated mode of expression. For instance, Ls was upregulated in bmi1a and bmi1b mutants but downregulated in sde. Gob was upregulated in bmi1b and bmi1c mutants, whereas it was downregulated in sde, sde1b, and bmi1a and Bl was downregulated in all sde1 and bmi1 mutants (Supplemental Figure S13B). The sde RNA-seq analysis revealed misexpression of a gene encoding a proline-rich protein (Solyc10g007800) with similarity to PROTODERMAL FACTOR 1 (PDF1) from Arabidopsis, of the gene Solyc08g059700 and the genes Solyc04g081700 and Solyc09g064750 (Supplemental Data Set S2). Solyc08g059700 encodes a protein with similarity to KINASE-INDUCIBLE DOMAIN INTERACTING8 (KIX8) and KIX9 from Arabidopsis and the genes Solyc04g081700 and Solyc09g064750 encode two highly similar long noncoding RNAs. The gene homologous to PDF1 was upregulated only in sde, whereas the lncRNAs were upregulated in sde, sde1b, and bmi1a mutants, and the KIX8/KIX9 homolog was upregulated in all sde1 and bmi1 mutants (Supplemental Figure S13B).
To test for genetic compensation or antagonism between Sde1A, Sde1B, Bmi1A, Bmi1B, and Bmi1C in the processes of meristem formation and complex leaf development, all genes were individually genome-edited using CRISPR-Cas9 technology in the VF36 and Moneymaker backgrounds (Supplemental Figures S14A, S15, A–C). This approach aimed to circumvent potential pleiotropic effects associated with a complete loss of PRC1 activity and consequently, an associated loss of transcriptional regulation at all stages of plant development. First, we analyzed the loss of function of Sde1B, the closest Sde1A homolog, in single and double mutant combinations. Similar to wt plants, sde1b mutants formed AMs. In addition, double mutant combinations showed no increase in the proportion of empty leaf axils during vegetative development compared to that in sde1a mutants, which suggests that these two genes do not function redundantly in AM formation (Supplemental Figure S14B). On the contrary, loss of function of individual Bmi1 genes led to various developmental phenotypes, such as an increase in leaf complexity in bmi1c mutants and an increase in the number of EMs in bmi1b and bmi1c single mutants (Figure 6, A, D, E–G). In contrast, bmi1a mutants showed suppression of EM formation and a modest reduction in leaf complexity (Figure 6, A, B, E–G; Supplemental Figure S16, A–C). To test for genetic interactions between Sde1A and each Bmi1 paralog, we generated double mutants by combining mutant alleles in the sensitized VF36 and the nonsensitized Moneymaker backgrounds. In VF36, loss of Bmi1C function partially suppressed the AM formation defects of one of the strongest CRISPR-Cas9 Sde1A loss-of-function alleles (sde1a-10; Figure 6H), whereas in Moneymaker, loss of Bmi1A function enhanced the weak sde1a-16 defects in AM formation (Figure 6I). However, the increased leaf complexity and EM formation shown in the VF36 bmi1c mutant was suppressed in sde1a bmi1c (Supplemental Figure S16, A–C). Furthermore, although leaf complexity was not strongly reduced in bmi1a mutants, the bmi1a bmi1c double mutant also suppressed the increased leaf complexity and EM formation of bmi1c, indicating that both Sde1A and Bmi1A antagonize Bmi1C (Supplemental Figure S16, A–C). In accordance with this hypothesis, analysis of the sde1a bmi1a double mutant did not show a further decrease in leaf complexity compared to sde1a, indicating that Sde1A and Bmi1A may act in the same pathway to promote leaf complexity and meristem formation (Supplemental Figure S16, A–C). Taken together, this analysis indicates that Sde1A promotes various morphogenetic processes, whereas different Bmi1 paralogs can cooperate to promote or inhibit them.
Figure 6.
Sde1A and Bmi1 genes show synergistic and antagonistic interactions. A–D, Comparison of phenotypes of representative leaf no. 6 of wt (A), bmi1a-2 (B), bmi1b-4 (C), and bmi1c-1 (D) plants. E, F, Violin plots of the number of first-order (E) and second-order (F) leaflets of wt, bmi1a-2, bmi1b-4, and bmi1c-1 plants. G, Violin plots of the mean number of ectopic shoots in wt, bmi1a-2, bmi1b-2, and bmi1c-1 plants (G). H and I, Violin plots of the number of empty leaf axils (%) in sde1a in combination with bmi1a, bmi1b, or bmi1c mutant plants in a sensitized (VF36, H) and a nonsensitized (Moneymaker, I) sde1a background. Median values are indicated by a black or white circle. n values in (E–I) represent the number of individual plants. P-values were determined by two-tailed, two-sample t tests.
Sde1A physically interacts with lateral suppressor
Lateral suppressor (Ls) and Blind (Bl) show overlapping expression domains with Sde1A in the leaf-axil boundary (Busch et al., 2011). Consecutive cross-sections revealed more precisely that this overlap is in the center of the boundary domain (Figure 7, A and B), suggesting that Ls and Bl might physically interact with Sde1A. To compare their accumulation patterns at the protein level, we generated a translational reporter whereby a Venus-Ls fusion is driven by the Ls promoter. Venus-Ls protein and Ls transcript accumulation strongly correlated with each other (Figure 7, B–D). In addition, Sde1A and Ls expression domains overlapped at the mRNA and protein levels (compare Figure 3, B–E with Figure 7, B–D). In yeast, Sde1A interacted with Ls and Bl but not with TKn2 (Supplemental Figure S17); however, in N. benthamiana mesophyll cells, Sde1A interacted only with Ls (Supplemental Figure S17). Ls interaction with the mutant sde1a protein was reduced, as shown by the limited growth of yeast cells in selective medium. Furthermore, deletion of a segment, including the N-terminal 17 amino acids of the RAWUL domain, or deletion of 39 amino acids of the RAWUL C-terminal domain, abolished Sde1A interaction with Ls, indicating that the RAWUL domain is required for the interaction with Ls (Supplemental Figure S17). In addition, Ls was able to interact with the RAWUL domain proteins Sde1B and Bmi1B, suggesting that different RAWUL domain proteins share similar interaction properties.
Genetic analysis showed that sde1a ls-1 double mutants do no enhance the ls-1 mutant phenotype (Figure 7E). In contrast, the bl-2 AM formation defects were strongly enhanced in sde1a bl-2 mutants in vegetative and reproductive stages and in a sde1a dose-dependent manner (Figure 7F). In 61% of sde1a bl-2 double-mutant plants, AMs failed to form at any leaf axil of the primary shoot, including the uppermost leaf axils, which usually develop the sympodial shoot (Supplemental Figure S18); therefore, vegetative growth ceased and the life cycle of those plants terminated in a solitary flower.
Recently, the LAX2 and BA2 proteins in rice and maize were shown to interact with the basic Helix Loop Helix (bHLH) transcription factors LAX1 and BA1, respectively. In tomato, the orthologous bHLH gene Uniflora (Uf: Solyc09g005070) is a major regulator of inflorescence branching (Busch, 2011; Pier, 2015). However, during vegetative development, Uf mRNA was barely detectable (Supplemental Data Set S2) and uf-1 mutants show no branching defects during the vegetative phase. Furthermore, Uf mutations did not enhance the sde1a defects in AM formation (Supplemental Figure S19). Taken together, these results indicate that Sde1A and Ls participate in a common genetic pathway that is independent of Uf, and that may function in parallel to the Bl pathway.
Discussion
Sde1A modulates AM initiation in tomato
Our findings demonstrate that Sde1A and Bmi1 together shape plant development and provide a layer of regulation that operates at very early stages of AM formation and leaf development. During tomato vegetative development, AM formation manifests morphologically as bulges at P6–P7 leaf axils (Figure 1D), which correlates with the activation of the cell division marker Histone H4 in early P6 leaf axils (Rossmann, 2013) and indicates that AMs probably initiate in P5–P6 leaf axils. The sde mutant, which harbors a hypomorphic sde1a allele, is strongly compromised in AM formation (Figure 1; Supplemental Figures S2, S4, A and B). Sde1A accumulates in the center of the boundary domain in P1–P2 leaf axils, but not in the developing AMs (Figure 3; Supplemental Figure S9A), which indicate that Sde1A activity is required at early stages of boundary development to enable AM initiation at later developmental stages. Notably, sde can form a small number of functional AMs, implying that initiated AMs that develop beyond a critical point can continue normal development (Figure 1; Supplemental Figure S1). In Arabidopsis, the RAX1 (Müller et al., 2006) and the REGULATOR OF AXILLARY MERISTEM FORMATION (Yang et al., 2012) genes show transient mRNA accumulation that ceases before AMs are initiated, similar to Sde1A expression in tomato. By contrast, the expression of the key AM formation regulators Ls/LAS, and Gob/CUC starts during early boundary development in tomato (Busch et al., 2011; Rossmann et al., 2015) and Arabidopsis (Greb et al., 2003; Raman et al., 2008) and is maintained until AMs initiate. The tomato sde mutant shows reduced mRNA levels of these boundary genes as well as of the meristem regulators TKn2 and CLV3, suggesting that Sde1A activates AM formation in part by promoting high expression levels of these genes. In Arabidopsis leaf axils, a transcriptionally permissive epigenetic environment safeguards basal STM expression to allow for STM upregulation during AM initiation (Greb et al., 2003; Shi et al., 2016; Cao et al., 2020). The involvement of Bmi1 genes, either cooperating or antagonizing Sde1A activity, may stabilize a mechanism that safeguards a pool of competent cells at the early stages of the boundary development. Together, these observations indicate the existence of at least three phases of AM development: first, the establishment of a cell pool that is competent to form AMs (P1–P2); second, the maintenance of such cells in a cellular environment of differentiation (P3–P4); and third, the activation of the AM initiation pathway (P5–P6). This view is consistent with the detached meristem hypothesis, which proposes that a few pluripotent cells detach from the primary SAM and associate with the developing leaf axils (Steeves and Sussex, 1989).
Ectopic expression of Sde1A enhances leaf dissection and the formation of EMs at the distal leaflet boundaries (Figure 4, D–I). In contrast, sde mutants show reduced leaf dissection and do not form EMs, indicating that Sde1A promotes these morphogenetic activities. These processes are restricted to tissues that share several features with the leaf axil (Wang et al., 2016), suggesting that Sde1A requires a specific cellular environment and/or the availability of specific interaction partners to exert its function. One such candidate, which is expressed in the leaf axil and the leaflet boundary, is the AM regulator Ls that can interact with Sde1A.
In contrast to the role of Sde1A in AM initiation, rice lax2-1, and maize ba2-3112 mutants show defects in AM maintenance during the vegetative phase. During reproductive development, lax2-1 and ba2-3112 mutants show AM formation defects that were not observed in sde1a mutants. These phenotypes correlate with the accumulation of LAX2 mRNA in vegetative SAMs and developing AMs, and of LAX2 and ba2 mRNA in IM and reproductive AMs. The closest LAX2/ba2 homolog in tomato, Sde1B (Supplemental Figure S7A), is expressed in the SAM, similar to LAX2/ba2; however, no AM initiation or maintenance defects are present in the sde1b mutant, suggesting the existence of redundant functions by homologous genes or differences in the mode of AM regulation in tomato compared to rice and maize. However, it is important to note that the sde1b mutant shows a similar pattern of misexpressed genes as in the sde1a mutant, and both Sde1A and Sde1B interact with Lhp1 and Ls. Besides the differences in expression patterns between the related genes in rice, maize, and tomato, the differences in phenotypic output may indicate different requirements for Sde1 activity at the SAM and in the boundary region: High levels of Sde1 may be required in the leaf axil prior to the establishment of new meristems, whereas low levels of Sde1 may be needed for SAM maintenance. The closest Sde1A homologs in rice and maize are Os12g0479100 and GRMZM2G447297 (Supplemental Figure S7A), which might potentially perform activities similar to Sde1A during AM initiation.
Sde1 probably originated from a Bmi1 gene that lost its RING finger-encoding domain in the last common ancestor of embryophytes
An Sde1-like gene first appeared in embryophytes, with Marchantia polymorpha containing a single-copy gene. The existence of paralogs in several plant species (Figure 5A) suggests that Sde1 then underwent successive rounds of gene duplications followed by retention, probably due to whole-genome duplications in the ancestors of mosses, spermatophytes, and monocotyledons (Paterson et al., 2004; Rensing et al., 2007; Jiao et al., 2011; Vanneste et al., 2014). A previous phylogenetic examination of proteins with RAWUL domains did not detect Sde1 proteins (Sanchez-Pulido et al., 2008). The phylogenetic reconstruction analysis presented here shows that the Sde1 clade clusters with plant Bmi1 proteins in a group that is distinct from the animal Bmi1 clade and the Ring1 and Wdr48 clades of animals and plants. Sde1 proteins lack the N-terminal RING-finger domain present in Bmi1 proteins. However, the phylogenetic relationships and structural gene conservation in the RAWUL domain-encoding region (Figure 5B;Supplemental Figure S11B) suggest that Sde1 is derived from a Bmi1 gene that lost its C3HC4 RING finger-coding region in the last common ancestor of embryophytes, but not in the last common ancestor of seed plants, as postulated by Yao et al. (2019).
Similar to animal and plant BMI1 paralogs, those of Sde1 show low sequence conservation in the RAWUL domain. Because the RAWUL domain is implicated in protein–protein interactions (Kim, 2017), this may imply that variation exists in the interacting partners recruited by Sde1-like proteins, as observed for Bmi1 proteins from humans and mouse (Gao et al., 2012; Fursova et al., 2019; Scelfo et al., 2019).
Sde1A cooperates with BMI1 proteins
In nonplant model systems, PRC1 activity maintains cellular pluripotency and drives differentiation in a tissue-specific manner by the formation of various PRC1 complexes with different enzymatic activities (Aloia et al., 2013; Loubiere et al., 2019). In tomato, the Bmi1-related gene Sde1A is required to maintain high expression levels of the meristem regulators TKn2 and CLV3 and the boundary genes Ls, Bl, and Gob (Supplemental Figure S13B). Furthermore, 74% of the misexpressed genes in sde are upregulated (≥two‐fold, P ≤ 0.05, Supplemental Data Set S2), suggesting a repressive function for Sde1A. In the sensitized (VF36) tomato background, Sde1A functions redundantly with Bmi1A to promote leaf dissection and EM formation, whereas in the nonsensitized (Moneymaker) background, they cooperate to promote AM formation. By contrast, Bmi1C counteracted Sde1A and Bmi1A activity in the VF36 background. This result demonstrated that in tomato, various Bmi1 genes possess opposing roles for the regulation of morphogenetic processes, likely as a result of their differential recruitment to target loci (Figure 6; Supplemental Figure S13). Moreover, Sde1A interacts with the PRC1 component Lhp1 (Supplemental Figure S12), but not with RING1 proteins, suggesting that Sde1A may compete for Lhp1 binding with Bmi1 or that it may form noncanonical PRC1 (ncPRC1) complexes, whose activity is independent of ubiquitylation. In mouse embryonic stem cells, Bmi1 paralogs affect target genes in a manner that can be dependent or independent of RING1 and H2A ubiquitylation (Fursova et al., 2019; Scelfo et al., 2019). In addition, canonical PRC1 (cPRC1) and ncPRC1 complexes cooperatively repress target genes (Fursova et al., 2019; Scelfo et al., 2019). Similarly, the interplay between different BMI1 genes and Sde1A in tomato indicates that cPRC1 and ncPRC1 complexes may also exist in plants. These complexes may promote an epigenetic environment in the center of boundary domains that favors the maintenance of cells with morphogenetic competence, to allow AM formation, leaflet formation, and EM formation (Figure 7G).
Sde1A and Ls are components of a common genetic pathway
Recruitment of PRC1 complexes to target genes is achieved via several mechanisms, one of which involves transcription factors (Merini and Calonje, 2015). In tomato, the transcription factor gene Ls is expressed in a band-shaped domain in leaf axils (Busch et al., 2011), which overlaps with that of Sde1A (Figures 3, D and E, 7, A–D). In addition to the pronounced phenotypic similarity between sde and ls mutants, sde1a does not enhance the ls-1 phenotype. Furthermore, Sde1A and Ls proteins interact in yeast and in planta (Figure 7, C and D), suggesting that Sde1A and Ls participate in a common pathway, in which Ls might guide Sde1A to target genes. The expression patterns and phenotypic features of individual mutants indicate that both genes also possess independent activities. For example, Ls promotes organ separation (Greb et al., 2003; Rossmann, 2013), whereas Sde1A modulates the reproductive transition of sympodial shoots (Supplemental Figures S3 and S18).
The expression domain of Bl, which encodes another regulator of AM initiation, overlaps with that of Ls and Sde1A; however, sde1a and bl-2 mutants show an additive genetic interaction in a Sde1A dose-dependent manner. Double sde1a bl-2 mutants are strongly defective in AM formation, with 61% of plants producing no side shoots. This observation demonstrates that Sde1A and Bl take part in independent pathways that modulate AM initiation during vegetative as well as reproductive development. In rice and maize, the AM regulators LAX1 and BA1 interact with the closest Sde1B homologs LAX2 and BA2 (Supplemental Figure S7). In contrast to LAX1/BA1, the tomato LAX1/ba1 putative ortholog Uf neither regulates axillary branching in the vegetative phase nor interacts genetically with Sde1A (Supplemental Figure S19). Taken together, these results suggest that in tomato, the Uf pathway is not a major regulator of shoot branching. This hypothesis is in contrast to the functions of LAX2/BA2 in rice and maize, respectively, and indicates that the recruitment of Sde1 and Uf homologous gene modules that regulate shoot branching occurs in diverse ways in different plant species.
In contrast to sde, but similar to lax2-1 and ba2-3112, tomato mutants compromised in AM formation during vegetative development also show branching defects during reproductive growth. For instance, ls-1 and bl-2 possess fewer vegetative AMs and fewer flowers per inflorescence (Pier, 2015). These pleiotropic effects have excluded the use of the reduced shoot branching trait in elite greenhouse tomato lines, which normally require pruning to concentrate resources in a limited number of fruits along a single axis. The specific defects in sde1a in vegetative AM formation might represent a valuable tool for breeding tomato cultivars with reduced side-shoot formation.
Materials and methods
Plant material, growth conditions, and phenotyping
Seeds of S. lycopersicum cv VF36 (LA0490) and the sde mutant (in the VF36 background) were a gift from John Yoder (University of California at Davis). Seeds of S. lycopersicum cv Moneymaker (LA2706), cv M82 (LA3475), cv Antimold-B (LA3244), cv Lukullus (LA0534), cv Ailsa Craig (LA2838A), ls-1 mutant (Antimold-B LA0329), bl-2 mutant (Lukullus LA0980), and the wild species S. pennellii (LA0716) were obtained from the Tomato Genetics Resource Center at the University of California at Davis, USA. The uf-1 mutant (Ailsa Craig near-isogenic line MLE567) was kindly provided by Yuval Eshed (The Weizmann Institute of Science, Israel).
Seeds were treated with saturated trisodium phosphate (Na3PO4) for 15 min. After washing with water, seeds were kept in water for 3 d in the dark and then sown on pots containing 50% of coco-peat and 50% perlite. Plants were grown under long-day conditions (16-h light/8-h dark) in a greenhouse under natural light supplemented with artificial light from a combination of high-pressure sodium lamps and high-pressure halogen lamps (∼910 µmol m−2 s−1), a relative humidity of 25%–55%, and day/night temperatures of 30°C/17°C. Controlled long-day conditions (16-h light/8-h dark) were provided in a Bronson walk-in chamber (Bronson Incubator Services B.V.) with Sylvania T12 IRS fluorescent lamps (F65W/33-640 IRS, ∼230 µmol m−2 s−1), a relative humidity of 45%–55% and day/night temperatures of 25°C/20°C. Plants were drip-irrigated in the greenhouse and manually irrigated in the Bronson walk-in chamber, and were fertilized under standard regimes with an electrical conductivity of 2.2 dS m−2, pH= 5.6, NH4 to a total N ratio of 0.05, N/K ratio of 1.8, and P/K+Ca ratio of 0.055.
The number of side shoots was recorded for all leaf axils of the primary shoot. The number of leaves on the primary shoot was used as a proxy for flowering time. Inflorescence branching was calculated as the number of flowers on the primary inflorescence. Leaf complexity was represented by the mean number of first- and second-order leaflets from leaf 1 to leaf 8. Internode length represented the mean length of internodes from leaves one to five. Statistical calculations were carried out by two-tailed, two-sample t test.
For stereomicroscopy and SEM, plants were grown for 2 weeks (vegetative phase) or 4 weeks (reproductive phase) in a Bronson chamber. Scoring of side shoots and internode length measurements were performed for 8-week-old plants grown in a Bronson chamber or in the greenhouse. For leaf complexity phenotyping, 4-week-old plants were transplanted to 5-L pots with standard soil and were grown in the greenhouse for 8 additional weeks. Ectopic shoots on leaves were counted in plants grown in the greenhouse for 12 weeks.
Interspecific mapping population
An interspecific F2 population derived from a cross between S. lycopersicum sde and S. pennellii was generated. Plants grown under long-day conditions were phenotyped for the presence of side shoots. One out of 80 plants displayed the sde phenotype. Selected F2 progeny were backcrossed to sde and a BCF2 population was evaluated with markers described in Tanksley et al. (1992).
DNA sequencing
Total genomic DNA was extracted from leaf tissue using the DNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. Libraries were prepared according to the Illumina TruSeq DNA (PCR-free) protocol and sequenced on an Illumina HiSeq 2500 platform (Illumina) at the Genome Centre of the Max Planck Institute for Plant Breeding Research, Cologne, Germany. In total, 289,329,010 and 281,617,026 paired-end 100-bp reads were obtained for wt and sde samples, respectively. Reads were aligned to the S. lycopersicum reference sequence SL4.0 using the QIAGEN CLC genomics server 11.0 (parameters: No masking, match score = 1, mismatch cost = 2, cost of insertions and deletions = Linear gap cost, insertion cost = 3, deletion cost = 3, length fraction = 0.5, similarity fraction = 0.8, global alignment = No, autodetect paired distances = Yes, nonspecific match handling = Map randomly). Out of all reads obtained, 95% were pair-mapped to the reference genome. Variants were called using the fixed ploidy variant detection module (parameters: ploidy = 2, required variant probability (%) = 98.0, ignore positions with coverage above = 100, Ignore broken pairs = Yes, ignore nonspecific matches = Regions, minimum coverage = 20, minimum count = 2, minimum frequency (%) = 98.0, base quality filter = Yes, neighborhood radius = 5, minimum central quality = 20, minimum neighborhood quality = 15, significance (%) = 1.0, remove pyro-error variants = Yes, in homopolymer regions with minimum length = 3, with frequency below = 0.8). Unique sde variants were identified by removing sde variants that were shared with VF36 (Supplemental Table S1). Variants that changed the amino acid sequence were extracted as candidates that had functional consequences on protein activity (Supplemental Table S4).
RNA-seq
Shoot apices of vegetative plants including four leaf primordia (10-d-old) were harvested using forceps and a stereomicroscope (Leica MZ-16FA). Forty apices per genotype and four biological replicates were sampled. Total RNA was extracted using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions, followed by removal of DNA with DNase I (Roche). Libraries were prepared according to the Illumina TruSeq stranded RNA protocol and sequenced on an Illumina HiSeq 2500 platform (Illumina) at the Genome Centre of the Max Planck Institute for Plant Breeding Research, Cologne, Germany. In total, 73,206,185 and 76,254,799 single-end 100-bp reads were obtained for wt and sde samples, respectively. Reads were aligned to the S. lycopersicum reference sequence SL4.0 using the QIAGEN CLC genomics server 11.0 (parameters: mismatch cost = 2, insertion cost = 3, deletion cost = 3, length fraction = 0.8, similarity fraction = 0.8, global alignment = No, strand-specific = both, and maximum number of hits for a read = 10). Out of all reads obtained, 99% aligned to the reference genome. Changes in gene expression were calculated using the differential expression for RNAreq 2.1 tool of the QIAGEN CLC genomics server 11.0 and using the test of Baggerly et al. (2003). P-values were corrected using false discovery rate (Supplemental Table S2). Differentially expressed genes between wt and sde were identified. Variants were called using the fixed ploidy variant detection module of QIAGEN CLC genomics server 11.0 (parameters: ploidy = 2, required variant probability (%) = 98.0, ignore positions with coverage above = 100,000, ignore nonspecific matches = Reads, minimum coverage = 10, minimum count = 20, minimum frequency (%) = 80.0, base quality filter = Yes, neighborhood radius = 5, minimum central quality = 20, minimum neighborhood quality = 15, significance (%) = 1.0, remove pyro-error variants = Yes, in homopolymer regions with minimum length = 3, with frequency below = 0.8). Unique sde variants were extracted by removing sde variants that were shared with VF36 (Supplemental Tables S1, S3). Variants that altered the amino acid sequence were extracted as candidates that had functional consequences on protein activity (Supplemental Table S4).
Tissue collection, RNA extraction, and RT-qPCR
For RT-qPCR analyses, SAMs including leaf primordia one to four were collected. Twenty apices per genotype and three biological replicates were sampled. Total RNA was extracted using the RNeasy Micro Kit (Qiagen) and digested with DNase I (Roche). One microgram of total RNA was used for cDNA synthesis using the SuperScript III First-Strand Synthesis System (Invitrogen). RT-qPCR was carried out using SYBR Green PCR Master Mix (Applied Biosystems) and a CFX 384 Touch Real-Time PCR Detection System (Bio-Rad). The tomato GAPDH (Solyc05g014470) gene was used as an internal control (Expósito-Rodríguez et al., 2008). All primer sequences can be found in Supplemental Table S6.
Complementation
Plasmid constructs were designed as described by Lampropoulos et al. (2013). A genomic DNA fragment including the presumptive Sde1A promoter was PCR-amplified with CloneAmp HiFi (TakaRa) from the Bacterial Artificial Chromosome (BAC) clone Sly-B-Hba 129H16 (INRA, France) with Sde1A promoter primers and cloned into pGGA000 (Addgene # 48856) using NEBuilder HiFi DNA Assembly (New England Biolabs). The Sde1A coding region was amplified in two fragments using VF36 genomic DNA as a template to synonymously mutate a BsaI site. The first fragment was amplified with Sde1A genomic 1 primers and the second fragment with Sde1A genomic 2 primers. Both PCR fragments were seamlessly assembled into pGGC000. The DNA sequence downstream of the stop codon was amplified with Sde1A terminator primers and cloned into pGGE000. The binary vector pAGM4723 was modified to make it compatible with the Greengate cloning system. To this end, a fragment containing the ccdB+ gene was amplified from pGGZ003 using ccdB+ cassette primers and seamlessly assembled into the BbsI-cut pAGM4723 vector. The modified vector was named pAGM4723GG and deposited at Addgene (#136960). The final complementing construct was assembled using GoldenGate cloning (BsaI) by combining the promoter, genomic and terminator modules with the pGGB003, pGGD002, pGGF008, and pAGM4723GG modules. The Sde1A-Venus translational reporter construct was assembled by combining the Sde1A promoter, genomic, and terminator modules with the pGGB003, pGGD_RT_li_Venus (Addgene # 136973), pGGF008, and pAGM4723GG modules. The Lateral suppressor promoter was amplified from Moneymaker genomic DNA using Ls promoter primers and cloned into pGGA000 using NEBuilder HiFi DNA Assembly (New England Biolabs). The Ls coding region was amplified using genomic Ls primers with Moneymaker genomic DNA as a template, and the PCR fragment was seamlessly inserted into pGGC000. The DNA sequence downstream of the stop codon was amplified in two fragments using Moneymaker genomic DNA as template to synonymously mutate a BsaI site. The first fragment was amplified with Ls terminator 1 primers and the second fragment with Ls terminator 2 primers. Both PCR fragments were seamlessly assembled into pGGE000. The final Venus-Ls construct was assembled as described above by combining the Ls promoter, genomic and terminator modules with the pGGB_RT_Venus_li (Addgene # 136973), pGGD002, pGGF008, and pAGM4723GG modules. The corresponding construct was introduced into Agrobacterium (Agrobacterium tumefaciens) strain GV3101 and used for Agrobacterium-mediated transformation of the sde mutant using young leaf explants (Knapp et al., 1994). The Ls-Venus construct was introduced into the ls-1 mutant by Agrobacterium-mediated transformation. Transgenic plants were tested for presence of the transgene using primers T-DNA Sde1A. All primer sequences can be found in Supplemental Table S6.
Confocal microscopy
The shoot apices were imaged by confocal microscopy at one time point as described by Burian et al. (2016). Briefly, microscopic observations were carried out using an upright confocal laser-scanning microscope (Leica TCS SP8) with long-working distance water immersion objectives 20×. The cell wall was stained with 0.1% propidium iodide (PI; Sigma-Aldrich) for 3–10 min. Excitation was at 488 nm at 20% full power (and 30% laser output). The image collection was at 505–545 nm for Venus and 600–656 nm for PI.
Yeast two-hybrid and bimolecular fluorescence complementation
To fuse the Gal4 DNA binding domain or activation domain to the test proteins, Sde1A, sde1a, Sde1A N-terminal deletion, Sde1A C-terminal deletion, Sde1B, Bmi1A, Bmi1B, Bmi1C, Ring1A, Ring1B, Lhp1, Ls, and Bl were amplified from cDNA and seamlessly cloned into pGBKT7 (Clontech) digested with NcoI and BamHI or into pGADT7 (Clontech) digested with EcoRI and BamHI. All primer sequences are listed in Supplemental Data Set S6. Yeast strain AH109 was used for cotransformation with pGBKT7 and pGADT7 constructs encoding the candidate interacting proteins. Yeast colonies were selected on medium lacking leucine and tryptophan. Positive clones were tested further by growing three independent yeast colonies in medium lacking Leu, Trp, and His or lacking Leu, Trp, His, and Ade.
DNA fragments for Sde1A, Lhp1, Ls, Bl, and TKn2 were generated by amplification from cDNA and seamlessly cloned into pDEST-VYCE(R)GW digested with XbaI or into pDEST-VYNE(R)GW digested with XbaI. PCR primers used for cloning are listed in Supplemental Table S6. The resulting plasmids were introduced into Agrobacterium strain GV3101 by electroporation and the bacteria used for Agrobacterium-mediated coinfiltration. Briefly, an overnight culture was used to inoculate a fresh culture that was allowed to grow to an OD600 ∼ 0.8. Then, 2 mL of cells was pelleted at 1,000g for 10 min. The cell pellets were resuspended in infiltration medium (5 g L−1d-glucose, 50 mM MES (Sigma), 2 mM Na3PO4, 0.1 mM acetosyringone). This step was repeated twice; cells were then resuspended in 1 mL of infiltration medium. Before coinfiltration, 5–6 weeks old N. benthamiana plants were exposed to white light for one h to allow for stomata opening. For co-infiltration, equal volumes of Agrobacterium were mixed and used to infiltrate N. benthamiana leaves. Plants were then left overnight in the dark and returned to long day conditions; 5-mm2 segments of the infiltrated zone were mounted onto a glass slide covered with water and a coverslip. The interactions were analyzed at 24–48 h post infiltration under a Zeiss LSM 700 confocal microscope.
CRISPR-Cas9 mutagenesis
Plasmid constructs were designed as described by Belhaj et al. (2013) using the Golden Gate cloning system (Engler et al., 2009). Briefly, two CRISPR RNAs (crRNAs) directed towards the coding sequence of the target gene were designed using the CRISPR-P 2.0 tool (Liu et al., 2017). Single guide RNAs (sgRNAs) were developed by PCR by combining the crRNA and the trans-activating crRNA (tracrRNA) using the pICH86966 plasmid as template. Each pair of sgRNA was placed downstream of the Arabidopsis U6 promoter in the Level 1 acceptors pICH47751 (sgRNA1) and pICH47761 (sgRNA2) plasmids. Primers containing the crRNA target sequences, which were used to clone sgRNA1 and sgRNA2, are listed in Supplemental Table S6. The Level 1 constructs pICH47731-NOSpro:NPTII, pICH47742-35S:Cas9, pICH47751-AtU6pro:SP5G-sgRNA-1, and pICH47761-AtU6:SP5G-sgRNA-2 were assembled into the binary Level 2 plasmid pAGM4723. The corresponding constructs were introduced into Agrobacterium strain GV3101 and used for Agrobacterium-mediated transformation (Knapp et al., 1994). Transgenic plants were tested for the presence of the transgene using Cas9 primers. Mutations in the target sequence were analyzed using the primers listed in Supplemental Table S6 and by Sanger DNA sequencing with the same primers. Homozygous plants carrying mutations and lacking the transgene were selected for phenotypic analysis. Mutants were backcrossed to the corresponding wt and analyzed in the BCF2 for co-segregation of the mutant phenotypes. All primer sequences can be found in Supplemental Table S6.
RNA in situ hybridization
In-situ hybridization was performed as described by Coen et al. (1990) with slight modifications. Antisense probes were synthesized from PCR products that used cDNA as a template. The T7 promoter was included within the reverse primer and the DIG RNA Labeling Mix from Roche was used to label RNA with digoxigenin-UTP by in vitro transcription with T7 RNA polymerase (Roche). Primer sequences are listed in Supplemental Table S6.
Shoot apices were fixed with 4% paraformaldehyde and 0.03% Tween-20. Plant material was dehydrated using a series of increasing ethanol concentrations without NaCl. Fixed material was embedded in Paraplast (Kendall) in an ASP300 tissue processor (Leica). Probes were not hydrolyzed. Embedded tissue was sectioned (8 μm) and hybridized according to Coen et al. (1990). After the color reaction, slides were mounted in 30% glycerol and photographed using differential interference contrast microscopy.
Phylogenetic reconstruction
BLASTP searches (Altschul et al., 1997) from public genome and transcriptome databases were performed to retrieve homologous proteins. The databases searched were Phytozome 12 (Goodstein et al., 2011), Metazome 3 (https://metazome.jgi.doe.gov), Gramene (Gupta et al., 2016), Mycocosm (Grigoriev et al., 2013), and NCBI (Johnson et al., 2008). Initially, the RAWUL domains of the proteins Sde1A, Bmi1A, Ring1A, and Wdr48 were used as a query to identify homologs. After the identification of homologs, their RAWUL domains were used to search for related paralogs. To address the relationship between the different RAWUL domains, multiple sequence alignment (Edgar, 2004) followed by a maximum likelihood phylogenetic reconstruction (Felsenstein, 1973) was performed with pairwise distance UPGMA (Michener and Sokal, 1957), hierarchical clustering and a JTT substitution model (Jones et al., 1992). To infer the reliability of the tree, a bootstrap analysis (Felsenstein, 1985) with 1,000 replicas was performed.
Accession numbers
Sequence data from this article can be found in the GenBank data libraries under the BioProject accession number PRJNA643909. The associated sequence for the sde1a mutant allele can be found under accession number MT767035.
Supplemental data
Supplemental Figure S1. sde shows variable expressivity in AM formation.
Supplemental Figure S2. sde shows defects in AM initiation.
Supplemental Figure S3. Sympodial shoots in sde undergo faster developmental transition.
Supplemental Figure S4. Genetic analysis of sde1a in three tomato backgrounds.
Supplemental Figure S5. sde variant distribution per chromosome.
Supplemental Figure S6. Sde1A coding region and protein sequences.
Supplemental Figure S7. Similarity and identity matrix for Sde1-like proteins from tomato, rice, and maize.
Supplemental Figure S8. Sde1A alleles generated by CRISPR-Cas9 in the VF36 and Moneymaker tomato backgrounds.
Supplemental Figure S9. Sde1A and Sde1B in situ hybridization.
Supplemental Figure S10. Ectopic expression of Sde1A in VF36.
Supplemental Figure S11. RAWUL phylogenetic reconstruction.
Supplemental Figure S12. Sde1A interacts with Lhp1.
Supplemental Figure S13. Quantitative expression analysis of Sde1, Bmi1, and shoot meristem regulation genes.
Supplemental Figure S14. Genetic interaction between sde1a and sde1b.
Supplemental Figure S15. Bmi1 alleles generated by CRISPR-Cas9 technology.
Supplemental Figure S16. Genetic interaction between sde1a, bmi1a, and bmi1c.
Supplemental Figure S17. Ls interact with Sde1A, Sde1B, and Bmi1B proteins.
Supplemental Figure S18. sde1a and bl-2 mutually enhance each other’s branching defects.
Supplemental Figure S19. Genetic interaction between sde1a and uf-1.
Supplemental Table S1. Mutant and transgenic line list.
Supplemental Data Set S1. SNPs in sde.
Supplemental Data Set S2. Differentially expressed genes in sde.
Supplemental Data Set S3. SNPs in expressed genes in sde.
Supplemental Data Set S4. SNPs that change amino acid sequence.
Supplemental Data Set S5. Sde1 and Bmi1 copy number in different species.
Supplemental Data Set S6. List of primers used.
Supplemental Data Set S7. Summary of statistical analyses.
Supplemental File S1. Protein alignment of RAWUL proteins shown in Supplemental Figure S11A.
Supplemental File S2. Newick format of the phylogenetic tree shown in Supplemental Figure S11A.
Supplementary Material
Acknowledgments
The authors thank Professor John I. Yoder (UC Davis) for providing the VF36 and sde seeds; Alexandra Kalde, Ursula Pfordt, and Christine Sänger for excellent technical assistance, David Wilson-Sanchez for technical assistance in confocal microscopy, the French Plant Genomic Resource Center (INRA-CNRGV) for providing the sequence and the BAC clone Sly-Hba-129H16. We thank Udhaya Ponraj and John Chandler for critical reading of the manuscript.
Funding
The research was co-supported by the Deutsche Forschungsgemeinschaft through the German-Israeli Project Cooperation (FE552/12-1, OR309/1-1) and by the Max Planck Society.
Conflict of interest statement. There are no conflicts of interest.
H.L. and K.T. conceived and designed the research; H.L., G.S., and R.T. performed research; H.L., G.S., and K.T. analyzed the data; H.L. and K.T. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Hernán López (lopez@mpipz.mpg.de).
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia L, Di Stefano B, Di Croce L (2013) Polycomb complexes in stem cells and embryonic development. Development 140: 2525–2534 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggerly KA, Deng L, Morris JS, Aldaz CM (2003) Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics 19: 1477–1483 [DOI] [PubMed] [Google Scholar]
- Barbier FF, Dun EA, Kerr SC, Chabikwa TG, Beveridge CA (2019) An update on the signals controlling shoot branching. Trends Plant Sci 24: 220–236 [DOI] [PubMed] [Google Scholar]
- Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Y, Harpaz-Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, Zinder M, Samach A, Eshed Y, Ori N (2009) The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136: 823–832 [DOI] [PubMed] [Google Scholar]
- Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P (2008) A conserved molecular framework for compound leaf development. Science 322: 1835–1839 [DOI] [PubMed] [Google Scholar]
- Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M (2010) Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol 20: 1853–1859 [DOI] [PubMed] [Google Scholar]
- Burian A, de Reuille PB, Kuhlemeier C (2016) Patterns of stem cell divisions contribute to plant longevity. Curr Biol 26: 1385–1394 [DOI] [PubMed] [Google Scholar]
- Busch BL (2011) Genetic and molecular analysis of aerial plant architecture in tomato. PhD thesis. Universität zu Köln, Germany.
- Busch BL, Schmitz G, Rossmann S, Piron F, Ding J, Bendahmane A, Theres K (2011) Shoot branching and leaf dissection in tomato are regulated by homologous gene modules. Plant Cell 23: 3595–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Wang J, Xiong Y, Yang H, Yang M, Ye P, Bencivenga S, Sablowski R, Jiao Y (2020) A self-activation loop maintains meristematic cell fate for branching. Curr Biol 30: 1893–1904 [DOI] [PubMed] [Google Scholar]
- Choi Y, Chan AP (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31: 2745–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Romero J, Doyle S, Elliott R, Murphy G, Carpenter R (1990) Floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O (2011) Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol 12: 211–221 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, Marillonnet S (2009) Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PloS One 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1973) Maximum likelihood and minimum-steps methods for estimating evolutionary trees from data on discrete characters. Syst Zool 22: 240–249 [Google Scholar]
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Forderer A, Zhou Y, Turck F (2016) The age of multiplexity: recruitment and interactions of Polycomb complexes in plants. Curr Opin Plant Biol 29: 169–178 [DOI] [PubMed] [Google Scholar]
- Fursova NA, Blackledge NP, Nakayama M, Ito S, Koseki Y, Farcas AM, King HW, Koseki H, Klose RJ (2019) Synergy between variant PRC1 complexes defines polycomb-mediated gene repression. Mol Cell 74: 1020–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D (2012) PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45: 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N (2011). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Clarenz O, Schäfer E, Müller D, Herrero R, Schmitz G, Theres K (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F (2013) MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42: D699–D704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Naithani S, Tello-Ruiz MK, Chougule K, D’Eustachio P, Fabregat A, Jiao Y, Keays M, Lee YK, Kumari S. et al. (2016) Gramene Database: navigating plant comparative genomics resources. Curr Plant Biol 7: 10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey G (1971) In vitro growth of vegetative tomato shoot apices. J Exp Bot 22: 688–701 [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97. [DOI] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36; W5–W9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Keller T, Abbott J, Moritz T, Doerner P (2006) Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18: 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CA (2017) The role of RAWUL and SAM in polycomb repression complex 1 assembly and function. Transl Epigenet Ser 5–31 [Google Scholar]
- Knapp S, Larondelle Y, Rossberg M, Furtek D, Theres K (1994) Transgenic tomato lines containing Ds elements at defined genomic—positions as tools for targeted transposon tagging. Mol Gen Genet 243: 666–673 [DOI] [PubMed] [Google Scholar]
- Kwon CS, Chen C, Wagner D (2005) WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev 19: 992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CS, Hibara KI, Pfluger J, Bezhani S, Metha H, Aida M, Tasaka M, Wagner D (2006) A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133: 3223–3230 [DOI] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J (2013) GreenGate-a novel, versatile, and efficient cloning system for plant transgenesis. PloS One 8: e83043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale J A, Harrison CJ (2008) Developmental transitions during the evolution of plant form. InMinelli A, Fusco G, eds, Evolving Pathways: Key Themes in Evolutionary Developmental Biology. Cambridge University Press, Cambridge, UK, pp 299–316 [Google Scholar]
- Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen L-L (2017) CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol Plant 10: 530–532 [DOI] [PubMed] [Google Scholar]
- Loubiere V, Martinez AM, Cavalli G (2019) Cell fate and developmental regulation dynamics by polycomb proteins and 3D genome architecture. BioEssays 41: 1800222 [DOI] [PubMed] [Google Scholar]
- Martín-Fontecha ES, Tarancon C, Cubas P (2018) To grow or not to grow, a power-saving program induced in dormant buds. Curr Opin Plant Biol 41: 102–109 [DOI] [PubMed] [Google Scholar]
- Merini W, Calonje M (2015) PRC1 is taking the lead in PcG repression. Plant J 83: 110–120 [DOI] [PubMed] [Google Scholar]
- Michener CD, Sokal RR (1957) A quantitative approach to a problem in classification. Evolution 11: 130–162 [Google Scholar]
- Müller D, Schmitz G, Theres K (2006) Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A, Bowers J, Chapman B (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101: 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pier AM (2015) Genetic and Evolutionary Analysis of MYB and bHLH Transcription Factors in Secondary Meristem Initiation. Universität zu Köln, Germany [Google Scholar]
- Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E (1998) The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Ponraj U, Theres K (2020) Keep a distance to be different: axillary buds initiating at a distance from the shoot apical meristem are crucial for the perennial life style of Arabis alpina. New Phytol 227: 116–131 [DOI] [PubMed] [Google Scholar]
- Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K (2008) Interplay of miR164, CUP‐SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55: 65–76 [DOI] [PubMed] [Google Scholar]
- Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM (2004) Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development 131: 4225–4237 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Ick J, Fawcett JA, Lang D, Zimmer A, Van de Peer Y, Reski R (2007) An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evol Biol 7: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann S, Kohlen W, Hasson A, Theres K (2015) Lateral suppressor and Goblet act in hierarchical order to regulate ectopic meristem formation at the base of tomato leaflets. Plant J 81: 837–848 [DOI] [PubMed] [Google Scholar]
- Rossmann SE (2013) Analysis of the Lateral Suppressor Pathway in Tomato Axillary Meristem Formation. Universität zu Köln, Germany [Google Scholar]
- Sanchez-Pulido L, Devos D, Sung ZR, Calonje M (2008) RAWUL: a new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics 9: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scelfo A, Fernandez-Perez D, Tamburri S, Zanotti M, Lavarone E, Soldi M, Bonaldi T, Ferrari KJ, Pasini D (2019) Functional landscape of PCGF proteins reveals both RING1A/B-dependent-and RING1A/B-independent-specific activities. Mol Cell 74: 1037–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G, Tillmann E, Carriero F, Fiore C, Cellini F, Theres K (2002) The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc Natl Acad Sci USA 99: 1064–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci USA 96: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Zhang C, Tian C, Wang J, Wang Q, Xu T, Xu Y, Ohno C, Sablowski R, Heisler MG. et al. (2016). Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis. PLoS Genet 12: e1006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM (1989) Patterns in Plant Development. Cambridge University Press, Cambridge, UK [Google Scholar]
- Sussex IM, Kerk NM (2001) The evolution of plant architecture. Curr Opin Plant Biol 4: 33–37 [DOI] [PubMed] [Google Scholar]
- Tabuchi H, Zhang Y, Hattori S, Omae M, Shimizu-Sato S, Oikawa T, Qian Q, Nishimura M, Kitano H, Xie H (2011) LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23: 3276–3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, Devicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB. et al. (1992) High-density molecular linkage maps of the tomato and potato genomes. Genetics 132: 1141–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Zhang X, He J, Yu H, Wang Y, Shi B, Han Y, Wang G, Feng X, Zhang C (2014) An organ boundary‐enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. J Mol Syst Biol 10: 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Wang Y, Yu H, He J, Wang J, Shi B, Du Q, Provart NJ, Meyerowitz EM, Jiao Y (2019) A gene expression map of shoot domains reveals regulatory mechanisms. Nat Commun 10: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K, Baele G, Maere S, Van de Peer Y (2014) Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous–Paleogene boundary. Genome Res 24: 1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian C, Zhang C, Cao X, Zhang T-Q, Wang J-W, Jiao Y (2017) Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell 29: 1373–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hasson A, Rossmann S, Theres K (2016) Divide et impera: boundaries shape the plant body and initiate new meristems. New Phytol 209: 485–498 [DOI] [PubMed] [Google Scholar]
- Xin W, Wang ZC, Liang Y, Wang YH, Hu YX (2017) Dynamic expression reveals a two-step patterning of WUS and CLV3 during axillary shoot meristem formation in Arabidopsis. J Plant Physiol 214: 1–6 [DOI] [PubMed] [Google Scholar]
- Xu L, Shen W-H (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18: 1966–1971 [DOI] [PubMed] [Google Scholar]
- Yang F, Wang Q, Schmitz G, Muller D, Theres K (2012) The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J 71: 61–70 [DOI] [PubMed] [Google Scholar]
- Yao H, Skirpan A, Wardell B, Matthes MS, Best NB, McCubbin T, Durbak A, Smith T, Malcomber S, McSteen P (2019) The barren stalk2 gene is required for axillary meristem development in maize. Mol Plant 12: 374–389 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.