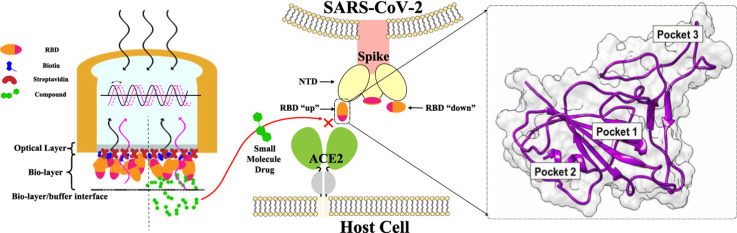
Abstract
Coronavirus Disease 2019 (COVID-19) is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), which enter the host cells through the interaction between its receptor binding domain (RBD) of spike glycoprotein with angiotensin-converting enzyme 2 (ACE2) receptor on the plasma membrane of host cell. Neutralizing antibodies and peptide binders of RBD can block viral infection, however, the concern of accessibility and affordability of viral infection inhibitors has been raised. Here, we report the identification of natural compounds as potential SARS-CoV-2 entry inhibitors using the molecular docking-based virtual screening coupled with bilayer interferometry (BLI). From a library of 1871 natural compounds, epigallocatechin gallate (EGCG), 20(R)-ginsenoside Rg3 (RRg3), 20(S)-ginsenoside Rg3 (SRg3), isobavachalcone (Ibvc), isochlorogenic A (IscA) and bakuchiol (Bkc) effectively inhibited pseudovirus entry at concentrations up to 100 μM. Among these compounds, four compounds, EGCG, Ibvc, salvianolic acid A (SalA), and isoliensinine (Isl), were effective in inhibiting SARS-CoV-2-induced cytopathic effect and plaque formation in Vero E6 cells. The EGCG was further validated with no observable animal toxicity and certain antiviral effect against SARS-CoV-2 pseudovirus mutants (D614G, N501Y, N439K & Y453F). Interestingly, EGCG, Bkc and Ibvc bind to ACE2 receptor in BLI assay, suggesting a dual binding to RBD and ACE2. Current findings shed some insight into identifications and validations of SARS-CoV-2 entry inhibitors from natural compounds.
Keywords: SARS-CoV-2, Bio-layer interferometry, Molecular docking, Natural compounds, Molecular dynamics simulation, Competitive binding
Graphical Abstract
Nomenclature
- ACE2
angiotensin-converting enzyme 2
- Bkc
bakuchiol
- BLI
bio-layer interferometry
- Bvc
bavachin
- Bvcn
bavachinin
- CADD
computer-aided drug design
- CalB
calceolarioside B
- CD
connector domain
- Ceph
cepharanthine
- CH
central helix
- COVID-19
Coronavirus Disease 2019
- CPE
cytopathic effect
- CT
cytoplasmic tail
- CvbD
cyclovirobuxine D
- E
envelope protein
- EGCG
epigallocatechin gallate
- ELISA
Enzyme-linked immunosorbent assay
- FP
fusion peptide
- HR1
heptad repeat 1
- HR2
heptad repeat 2
- Ibvc
isobavachalcone
- ICC
immunocytochemistry
- IscA
isochlorogenic acid A
- Isl
isoliensinine
- M
matrix protein
- MDS
molecular dynamics simulation
- MTT
3-(4,5-dimethylthiazol-2-yl)-3,5-phenytetrazoliumromide
- NTD
N-terminal domain
- RBD
receptor-binding domain
- RBM
receptor-binding motif
- RdRP
RNA-dependent RNA polymerase
- RMSD
Root mean-squared deviation
- RRg3
20(R)-ginsenoside Rg3
- RT
room temperature
- S
spike protein
- SalA
salvianolic acid A
- SARS-CoV-2
Severe acute respiratory syndrome virus 2
- SRg3
20(S)-ginsenoside Rg3
- TimA
timosaponin A III
- TM
transmembrane domain
1. Introduction
Coronaviruses are a group of RNA viruses that can infect a number of hosts, including humans and animals. Severe acute respiratory syndrome virus 2 (SARS-CoV-2) causes an unusual form of viral pneumonia, known as coronavirus diseases 2019 (COVID-19). The high transmission rate of this virus lead to a worldwide pandemic, as announced by the WHO on 11 March 2020 [1]. It is obvious that the urgent need of research is required to understand the structure of virus and mechanism of infection and pathogenic mechanism of SARS-CoV-2 in order to strategize better diagnostic tools and effective therapies against this novel viral infection.
The critical studies on the structure of SARS-CoV-2 reveal that there are two important groups of proteins, structural proteins and non-structural proteins. The structural proteins include the spike protein (S), matrix protein (M), envelope protein (E), whereas the non-structural proteins include proteases and RNA-dependent RNA polymerase (RdRP) [2]. The S protein, which protrudes from the outer surface of all strains of coronaviruses in a homo-trimeric state, is an important recognition site used by the virus for attachment and the subsequent entry into the host cells. The S protein is a complex of two subunits, namely, the S1 and S2 subunits [3]. The S1 subunit, which consists of receptor binding domain (RBD) and N-terminal domain (NTD), binds to the cellular receptors on the host cell membrane. Angiotensin converting enzyme 2 is the classical and the most well-characterized SARS-CoV-2 receptor, whereas CD147 [4] and NRP1 [5] have also been identified as entry receptors for SARS-CoV-2. On the other hand, S2 subunit consists of a cytoplasmic tail (CT), a transmembrane domain (TM), heptad repeat 2 (HR2), connector domain (CD), central helix (CH), heptad repeat 1 (HR1), and fusion peptide (FP). The S1/S2 cleavage site is present at the border between the two subunits, which is cleaved by proteases in the host cells. Such a process is critical for the activation of the S protein leading to the fusion of the viral envelope with the cell membrane of the host cells [6].
The identification of RBD as a crucial structure for viral attachment and entry into the host cells by binding to the ACE2 receptor lead to the development of the potential antiviral drugs, including small-molecules and antibodies targeting the RBD site [6]. Two structural sub-domains are found in the SARS-CoV-2 RBD, the highly conserved core subdomain and the receptor binding motif (RBM). The core subdomain consists of five antiparallel β-strands connected by short helices and loops. Two of the β-strands are connected by a disulfide bond. Four disulfide bonds are found in the RBD, three of which are found in the core subdomain that stabilize the β-sheet. The RBM consisting of the β-5 and β-6 strands as well as loops and alpha helices is present in the RBD [7]. The RBM has a slightly concave surface that binds to the peptidase domain of ACE2. Binding of the S protein to ACE2 destabilizes the pre-fusion conformation in a stable post-fusion structure [8]. The discovery of therapeutic agents that inhibit the specific interaction of RBD with ACE2 would block the attachment and entry of SARS-CoV-2 into human host cells and prevent COVID-19. As the spread of the SARS-CoV-2 mutant viruses globally among different countries, pandemic is getting out of control. Recently, the D614G and N501Y mutants were discovered in Northern Europe and Africa, whereas N439K is commonly found in over 300 countries. The Y453F mutant was originated from mammal mink and has infectivity on humans. Therefore, the search for novel drug candidates for the treatment and prophylaxis against COVID-19, especially for mutant viruses is an urgent task to prevent this pandemic.
Historically, natural products have been known to be critical resources for drug discoveries to treat numerous diseases, including infectious diseases [9]. They are characterized by comprising an enormous diversity of scaffolds as well as by their structural complexity, serving as a reservoir of drug candidates. Furthermore, the long history of their use in traditional medicine provides insights regarding their efficacy and safety [10]. However, the rational manipulation of these vastly diverse chemical and bioactive agents has been a challenge for researchers. Fortunately, the great advances in computational chemistry, chemoinformatics and structural biology lead to development of the field of computer-aided drug design (CADD), which has become a valuable tool in drug discovery and development. Such new advances lead to reduced time and cost, and also provide information that are difficult to obtain using wet laboratory approaches. The computational structure-based approaches in CAAD include molecular docking and molecular dynamics simulation (MDS), which are two indispensable tools used to study molecular recognition in silico. These computational methods have proven to be successful in revealing the potentially active compounds from natural compound libraries. They are used to predict the binding mode and binding affinity of molecular complexes [11]. Despite their great value, the binding affinities predicted by computational techniques are not precise [12] and require further validation using in vitro and in vivo experiments.
Previously, we discovered two natural polyphenol compounds as SARS-CoV-2 entry inhibitors [13], [14]. In the present study, we aimed to discover natural small molecule inhibitors targeting the SARS-CoV-2 RBD from a library of 1871 compounds. We used molecular docking and dynamics coupled with biolayer interferometry (BLI), a real-time detection method for biomolecular interactions. Active compounds were further evaluated using enzyme linked immunosorbent assay, immunocytochemistry (ICC) and live SARS-CoV-2 virus experiments.
2. Material and methods
2.1. Natural small-molecule sources
The compound library including 1871 natural compounds was purchased from Push Biotechnology (Chengdu, China). All compounds were dissolved in DMSO at a concentration of 10 mM and stored at − 20 °C. For further validation, compounds were purchased from the same company.
2.2. Recombinant proteins
Mouse Fc-tagged RBD (Catalog: 40592-V05H) and His-tagged human ACE2 (Cat: 10108-H08B) were both purchased from Sino Biological (Beijing, China). RBD was covalently biotinylated with a biotinylation kit (Cat: 1828 M, Genemore, Shanghai, China) as per manufacturer’s protocol. All proteins were stored at − 80 °C.
2.3. Cell culture
Normal human airway epithelial cell line, BEAS-2B, normal human embryonic liver cell line, LO2, and human embryonic kidney cell line, HEK293, were supplied by American Type Culture Collection (Rockville, MD). Cells were cultured in Gibco DMEM (Cat: 12100046) supplemented with 10% Gibco fetal bovine serum and 1% Gibco penicillin-streptomycin-glutamine (Thermo Fisher Scientific, Waltham, USA). Cells were cultured at 37 °C in a humidified incubator containing 5% CO2.
2.4. Molecular docking-based virtual screening
For virtual screening, the crystal structure of RBD of the spike protein SARS-CoV-2 was obtained from PDB (6M17, Chain E). All non-standard residues (water, N-acetyl glucosamine and zinc) were removed in UCSF Chimera. For further processing, the model was prepared in Flare version 3.0 (Cresset, Litlington, UK) by adding missing hydrogens, assignment of optimal ionization states of residues, optimization of spatial positions of polar hydrogens to maximize hydrogen bonding and to minimize steric strain, and reconstruction of unresolved side chains. The compound library was downloaded from Pubchem. The structures were converted into a single file in sdf format using Data warrior software. Energies of all compounds were minimized. Each compound was docked with the prepared RBD of the spike protein SARS-CoV-2 using Flare (Cresset) with default settings using the “fast but accurate” mode. The grid box included the whole domain. Compounds were ordered according to the VS scores of the best binding poses.
2.5. Bio-layer interferometry (BLI) binding kinetics assay
All BLI assays were conducted on an Octet RED96 (FortéBio, Shanghai, China) instrument. A shake speed of 1000 rpm and plate temperature of 30 °C applied to all runs. Phosphate buffer solution (PBS) was used as kinetics buffer. To prepare RBD-bound test probes, Super Streptavidin (SSA) optic fiber probes were run at baseline in PBS for 60 s, loaded in 200 μL of biotinylated RBD solution at 125 μg/mL for 600 s, run at baseline again in PBS for 60 s, and stored at 4 °C dipped in PBS. Ni-NTA probes and 40 μg/mL ACE2 were used to prepare ACE2 probes following the same procedure. For binding kinetics assays, a serial dilution of six concentrations of up to seven drugs dissolved in PBS were added to a black polypropylene 96-well microplate (Greiner Bio-one, Frickenhausen, Germany) with PBS filling the rest of the wells. One row was left as PBS-only negative control. Each well contains a total volume of 200 μL. An assay cycle consists of 120 s of baseline incubation in PBS followed by 120–180 s of association in compound solution followed by 120–180 s of dissociation in PBS, and it was repeated for every concentration and with both an RBD-loaded and a blank probe. Analysis of BLI results was undertaken using FortéBio Data Analysis software version 9.0. The curves were aligned to dissociation, Y axis was aligned to the last 5 s of baseline steps, and the last 5 s of the association step were considered the steady state. Specific binding to RBD was subtracted from blank probe control and PBS negative control by selecting the “Double References” mode. A 1:1 binding model was assumed in binding kinetics analysis. KD, Kon, Koff and R2 values were reported.
2.6. Enzyme-linked immunosorbent assay (ELISA)
The ACE2: SARS-CoV-2 Spike Inhibitor Screening Assay Kit (Cat: 79936) was purchased from BPS Biosciences (San Diego, US). Briefly, Fc-RBD was added to ACE2-His-coated test wells in the presence of 25–200 μM compounds, and to negative control wells containing no compound. Blank wells were left compound-free and mFc-RBD-free. Anti-Fc-horseradish peroxidase substrate was added to each well followed by mixed ELISA ECL Substrate A and B. Chemiluminescence was read with a SpectraMax iD5 Microplate Reader (Molecular Devices, San Jose, US).
2.7. 3-(4,5-dimethylthiazol-2-yl)-3,5-phenytetrazoliumromide (MTT) assay
96-well plates were seeded at a density of 1 × 104 cells/well. Cells were treated with a serial dilution of compounds from 0.78 to 100 μM (200 μM for HEK293). Drug-free wells were used as negative controls. After 72 h (24 h for HEK293) of exposure, 10 μL of 5 mg/mL MTT solution was added to each well. After 4 h of incubation, spent medium was aspirated and 100 μL DMSO was added to each well to dissolve the formazan crystal. Absorbance (A) was read with a SpectraMax Paradigm Microplate Reader (Molecular Devices, San Jose, US) at the wavelength of 570 nm. Cell viability of each treated well was calculated as (Atreated−Apositive control)/(Anegative control−Apositive control)×100%. Cytotoxicity of compounds were indicated by median inhibitory concentration IC50.
2.8. Western blotting
Protein was extracted from lysate of HEK293 cells transfected with plasmid (Vector Builder, Guangzhou, China) expressing human ACE2 with EGFP (pLV[Exp]-Puro-EF1A>hACE2[nm_02184.3]/EGFP) fused to its C-terminus or co-expressing mCherry (pLV[Exp]-mCherry/Neo-EF1A>hACE2 [nm_02184.3] for 48 h after transfection. Primary antibody against ACE2 (sc-390851, Santa Cruz Biotechnology, Dallas, USA) was used at a dilution of 1:1000. Primary antibody against β-actin (sc-47778, Santa Cruz Biotechnology) was used for detection of internal reference at a dilution of 1:1000. HRP-conjugated goat anti-mouse IgG secondary antibody was used at a dilution of 1:5000 (Cat: 32430, Invitrogen, Waltham, USA).
2.9. Pseudovirus assay and immunocytochemistry (ICC)
HEK293 cells in a 10 cm dish were transiently transfected at 50% confluency of cells with 6 μg of plasmid expressing human ACE2 with EGFP fused to its C-terminus or co-expressing mCherry. Transfection efficiency was confirmed under a florescence microscope after overnight incubation. HEK293 cells transiently overexpressing ACE2-EGFP were seeded on glass coverslips in 24-well plates at a density of 2 × 104 cells/well. For pseudovirus assay, wild-type and 4 mutants (D614G, N501Y, N439K & Y453F) SARS-CoV-2 Spike RBD pseudovirus (VectorBuilder) were added at final concentration of 4.66 × 106 TU/mL to each well containing 0, 25, 50 or 100 μM compounds and 5 μg/mL polybrene, respectively. For visualization of RBD binding, 3 μg RBD protein, the mixtures with or without compounds at given concentrations were added to each well followed by 40 min incubation. Cells were rinsed three times for 5 min each time, with PBS containing 0.1% Tween 20 (PBST), fixed with 4% (v/v) paraformaldehyde (PFA) solution, and briefly rinsed with PBST. Goat anti-mouse IgG Fc red fluorescent secondary TRITC-conjugated antibody (Cat: A16089, Invitrogen, Waltham, USA) at a dilution of 1:500 was added. After 2 h of incubation, cells were briefly rinsed with PBST. The coverslips were left to dry and then placed on glass slides and had its edges sealed. Fluorescence intensity and RBD-ACE2 colocalization were visualized under a Leica TCS SP8 Laser Scanning Microscope (Leica, Guangzhou, China). Image J was used for the semi-quantitative immunofluorescence intensity.
2.10. Authentic SARS-CoV-2 viral infection tests
The African green monkey kidney epithelial (Vero E6) cells were purchased from American Type Culture Collection (ATCC, Manassas, USA) and cultured in Dulbecco’s Modified Eagle’s Medium (Gibco, Grand Island, USA) supplemented with 10% fetal bovine serum (FBS) at 37 °C. The clinical isolate of SARS-CoV-2 (Genbank accession no. MT123290.1) was propagated in Vero E6 cells and titrated by the median tissue culture infective dose (TCID50) assay. Experiments involving SARS-CoV-2 were conducted in a biosafety level-3 (BSL-3) laboratory at KingMed Virology Diagnostic & Translational Center and following the protocol approved by the ethics committee of State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, the First Affiliated Hospital of Guangzhou Medical University, Guangzhou Medical University, Guangzhou. Two-fold serially diluted test compounds were incubated with 50 TCID50 of SARS-CoV-2 at room temperature (RT) for 2 h, and the mixture was then added to Vero E6 to allow infection for 2 h. After the inoculum was removed, the cells were further cultured with fresh medium. The infected cells displayed 100% CPE at 72 hpi (hours post-infection). The percentage of cytopathic effect (CPE) in infected cells treated with virus–compound mixture was determined by inspection under an inverted microscope. All active compounds were tested two additional times, and mean CPE percentage of three independent experiments was used to calculate the median effective concentration EC50 and median cytotoxic concentration CC50.
2.11. Plaque formation assay
The virus was incubated with test compounds and then infected Vero E6 cells in the same manner as described in the CPE reduction assay. After removing the inoculum, the cell monolayers were covered with agar overlay (final concentration: 0.6% agar, 2% FBS) and incubated for 48 h at 37 °C with 5% CO2. Following the removal of overlays, the cell monolayer was fixed with 10% formalin and stained with 1% crystal violet. The plaques were then counted and photographed.
2.12. Computational prediction of compound binding sites
The crystallographic structure of the RBD was analyzed by the Fpocket software in order to predict possible druggable binding sites. The PDB file of the RBD was submitted to Fpocket software. The outputs were analyzed using UCSF Chimera software. In order to predict the binding pockets on the RBD, blind docking was performed using web-based program SwissDock. All active compounds were docked on the prepared RBD. SwissDock generated all possible binding modes for each ligand. And the most favorable binding modes at a given pocket were clustered. All ligand clusters were visualized with UCSF Chemiera. A cluster is a predicted binding pocket on the target protein and each cluster for every ligand was inspected for amino acids interacting with the ligand. All of the interacting amino acids with the target receptor were noted for each cluster and compare with the predicted binding pockets obtained from Fpocket. SDF files of the selected compounds were loaded on Flare software (Cresset) and processed using default settings. For each compound, the grid was selected to include the predicted binding pocket identified from the blind docking experiment. Molecular docking was performed using Flare. The best binding pose of each compound was selected for further analysis. The top binding poses of drug-like molecules with SARS-CoV2 RBD from the molecular docking results were subjected to MD simulation using GROMACS package version 5.0.6. The topology of SARS-CoV2 RBD protein was generated by the CAHRMM36 force field. Ligands were converted to Mol2 file format using Avogadro software. Ligand topologies were generated using the CGenFF server. Protein-ligand complexes were generated and solvated in a dodecahedral water box by an explicit SPC water model. The system was neutralized by adding appropriated counter ions. To minimize the energy, the system was allowed to converge at the tolerance of 1000 kJ mol−1 nm−1 with 500 steps of steepest descent. The system was then equilibrated in two phases. The first phase was implemented for NVT equilibration at 300 K. The second phase was implemented for NPT equilibration at 1 bar pressure. The MD simulation was performed to 10 ns time for all the molecules. Root mean-squared deviation (RMSD) and interaction energy were stored in the trajectory for every 2 ps and were analyzed using Grace software.
2.13. BLI cross-competitive binding assay for small-molecules
A test cycle is sequentially consisted of the following BLI machine instructions. All steps were run for 300 s. Concentrations of 200 and 400 μM were used for first compounds. Concentrations of 25, 50 and 100 μM were used for second compounds. One row was left as PBS negative control. All other instrument settings were identical to those of BLI binding kinetics assay. Signals were subtracted from PBS negative control and aligned to dissociation in FortéBio Data Analysis software version 9.0 and exported as raw data tables. Means and standard deviations of the signals elicited by first compound alone (x), second compound alone (y) and second compound after first compound pre-occupation (z) were extracted from the last five seconds of corresponding steps. A test cycle is considered valid when the means of both x and y are above ten times of their respective standard deviations (10 × limit of quantification) which indicate detectable binding. To construct a cross-competition matrix, different initial states of the matrix were generated with every compound being on the first column or first row. A Pearson’s correlation reordering algorithm to reorder the rows these initial state matrices in such a way that most correlated rows or columns are adjacent to each other. Meanwhile, the order of columns is kept identical to that of rows, and vice versa. Thus, for a cross-competition matrix of 8 compounds, 15 reordered matrices corresponding to 15 unique initial states (7 with each of the rest of the 7 compounds moved to the first row, 7 with each of the rest 7 compounds moved to the first column and one at the original state) were obtained. To select an optimal matrix with cells representing competitive interactions aggregated along the diagonal line as much as possible, the total “energy” of the matrix was calculated. Energy of a cell is calculated as (100 – cell value) × distance to the diagonal line. Total energy is sum of the energy of all cells. The one with lowest total energy was selected.
2.14. Maximum tolerated dose
The C57BL/6 mice at the age of 6–8 weeks were obtained from SPF (Beijing) Biotechnology Co., Ltd. All experiments were carried out in accordance with the “Institutional Animal Care and User Committee guidelines” of the Macau University of Science and Technology. Mice were fed ab libitum. Mice (n = 6) were randomly assigned to receive PBS, and two acute high doses of 200 mg/kg/day EGCG or 300 mg/kg/day EGCG by oral administration. Mice were monitored daily for survival and body weight with the above acute lethal dosages for one week. Mice were euthanized on day 8 and the liver, kidney, lung, spleen, and thymus were harvested for weighing and evaluating pathological signs due to toxicity.
2.15. Data analysis and visualization
GraphPad Prism 8.0 “non-linear regression (curve-fit) [Inhibitor] vs. normalized response--Variable slope” was used to calculate IC50 in MTT assay, EC50 in pseudovirus and authentic SARS-CoV-2 virus assay, and CC50 in authentic SARS-CoV-2 virus assay. GraphPad Prism 8 One-way ANOVA was used to process ELISA and animal experiment results. In-house python scripts were used to process BLI cross-competitive assay results.
3. Results
3.1. Identification of natural small-molecule inhibitors against Spike-RBD of SARS-CoV-2
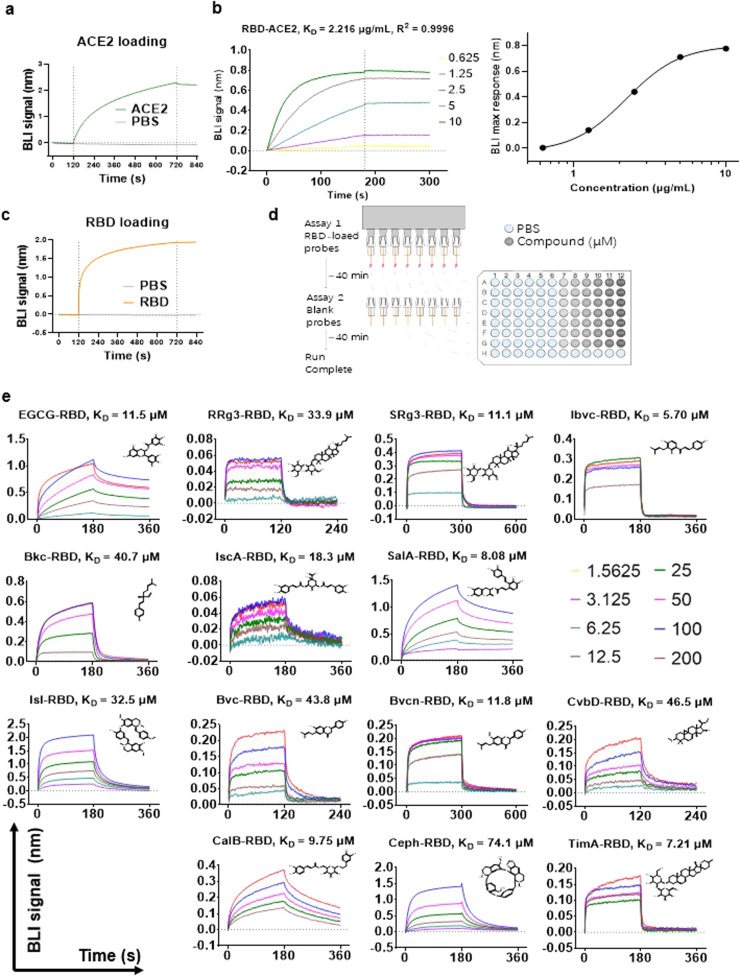
Small molecule inhibitors targeting the interaction between RBD of SARS-CoV-2 and host ACE2 receptor have the potential to work as interventions for prophylaxis and treatment against COVID-19. Here, we report the discovery of natural compounds as potential SARS-CoV-2 entry inhibitors using the molecular docking-based virtual screening coupled with BLI, the latter being a real-time detection method for bio-molecular interaction to compensate for the limited predictive power of the former [15]. Libraries of 1871 natural compounds were virtually docked to the entirety of a SARS-CoV-2 RBD model (PDB: 6M17). We selected 540 compounds mostly with docking scores under − 7.0 kcal/mol for BLI binding affinity assays with the Spike-RBD protein (Table S1). We first validated the mouse Fc-tagged RBD for its binding affinity with His-tagged human recombinant ACE2 immobilized onto nickel-nitrilotriacetic acid (Ni-NTA) probe ( Fig. 1a) with a KD of 2.216 μg/mL (Fig. 1b), which is in agreement with previous reports1. Subsequently, RBD was biotinylated and immobilized on super-streptavidin (SSA) probes (Fig. 1c). RBD-bound SSA probes were sequentially dipped in two-fold serial dilutions of test compounds with concentrations ranging from 200 to 1.56 μM (Fig. 1d). After the BLI screening, 69 compounds were identified to show affinity with RBD (Table S1). Among these compounds, we selected 24 compounds for further validation based on their strong binding affinity and absence of known systemic toxicity or cytotoxicity. To address inter-batch variability, a separate batch of these compounds was purchased for assessment of reproducibility. Only 14 could reproducibly bind at high affinity with Spike-RBD in BLI (Fig. 1e, Table S2). Among these, bavachin (Bvc), isoliensinine (Isl) and cepharanthine (Ceph) were previously found to be effective against authentic SARS-CoV-2 [16].
Fig. 1.
Identified natural compounds directly interact with RBD protein by BLI binding kinetics assay. (A) Immobilization of His-tagged ACE2 to Ni-NTA probe. (B) Binding affinity of RBD to ACE2. KD is calculated by fitting of maximum BLI response at different concentrations to a sigmoidal curve. (C) Loading of RBD to SSA probe in association conditions. (D) Schematic illustration of the BLI binding kinetics assay. (E) BLI sensorgrams structures of 14 compounds showing their binding to RBD. Each compound was examined in BLI screening from compound library and a new batch for validation of reproducibility. For each compound, sensorgram of the test with better result is shown. Chemical structures of compounds are displayed.
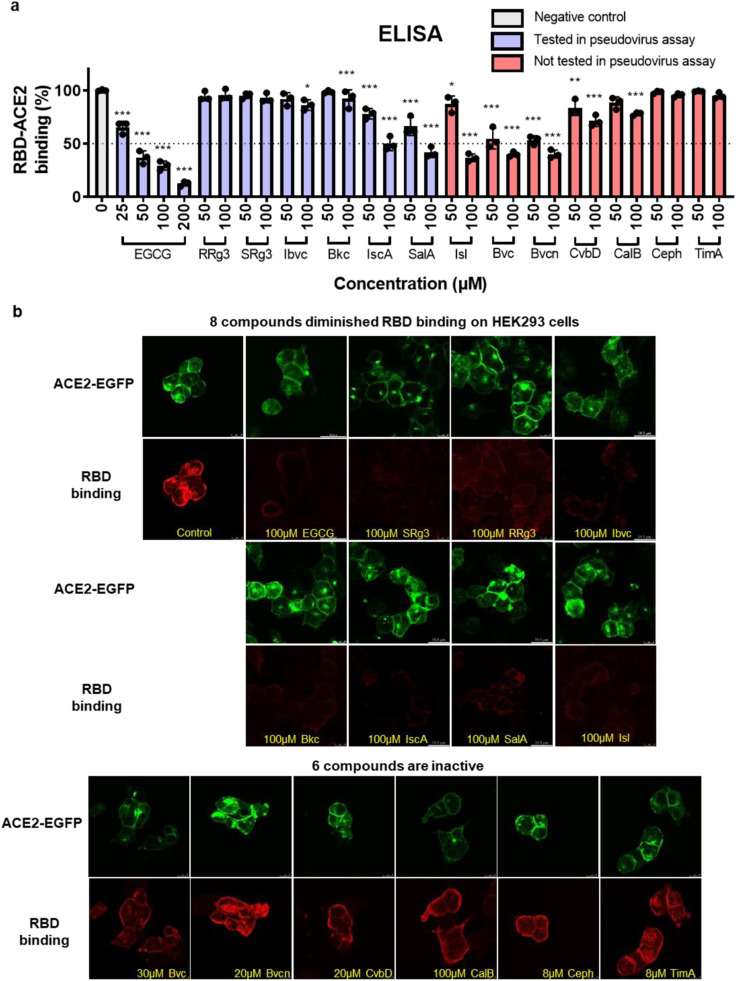
3.2. Eight natural small-molecules suppress RBD-ACE2 interaction via direct inhibition of RBD-ACE2 binding
We substantiated the ability of the 14 compounds to block RBD-ACE2 binding with an ELISA-based kit. Only 6 of these compounds inhibited RBD-ACE2 binding over 50% at 100 μM ( Fig. 2a). In addition, we visualized the inhibitory effects of these compounds on the binding of RBD to ACE2 using HEK293 cells with transient overexpression of human ACE2 fused with EGFP (Fig. S1a, b) followed by immunocytochemistry analysis with a tetramethylrhodamine (TRITC) antibody (Fig. S1c). As shown in Fig. 2b, the RBD binding to cell membrane was blocked by EGCG, 20(S)-ginsenoside Rg3 (SRg3), 20(R)-ginsenoside Rg3 (RRg3), isobavachalcone (Ibvc), bakuchiol (Bkc), isochlorogenic acid A (IscA), salvianolic acid A (SalA) and isoliensinine (Isl), as indicated by reduced fluorescent immunoreactive signal of RBD on the cell surface. On the other hand, Bvc, bavachinin (Bvcn), cyclovirobuxine D (CvbD), calceolarioside B (CalB), Ceph and timosaponin A III (TimA) (all but CalB were used at lower concentrations to circumvent cytotoxicity), were not effective in blocking the RBD-ACE2 binding (Fig. 2b).
Fig. 2.
The natural small-molecules inhibit RBD protein binding to hACE2 by ELISA and ICC assays. (A) ELISA results of 14 compounds. EGCG, IscA, SalA, Isl, Bvc and Bvcn had high activities. In addition, Ibvc, Bkc, CvbD and CalB were weakly active. Ceph, TimA, SRg3 and RRg3 were almost inactive. Data are presented as mean ± SD. Data are analysed with one-way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.005). (B) Visualization of Spike-RBD binding on human ACE2 receptor in HEK293 cells. Representative image of RBD red fluorescence on HEK293 cells in the presence of compounds.
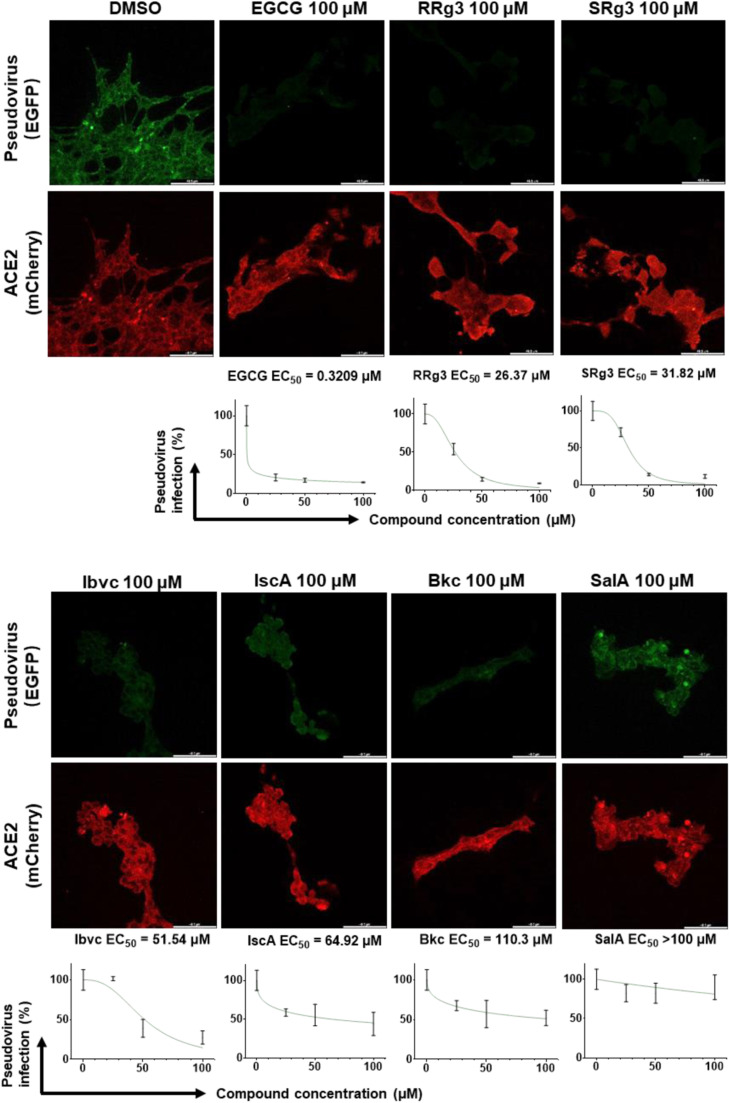
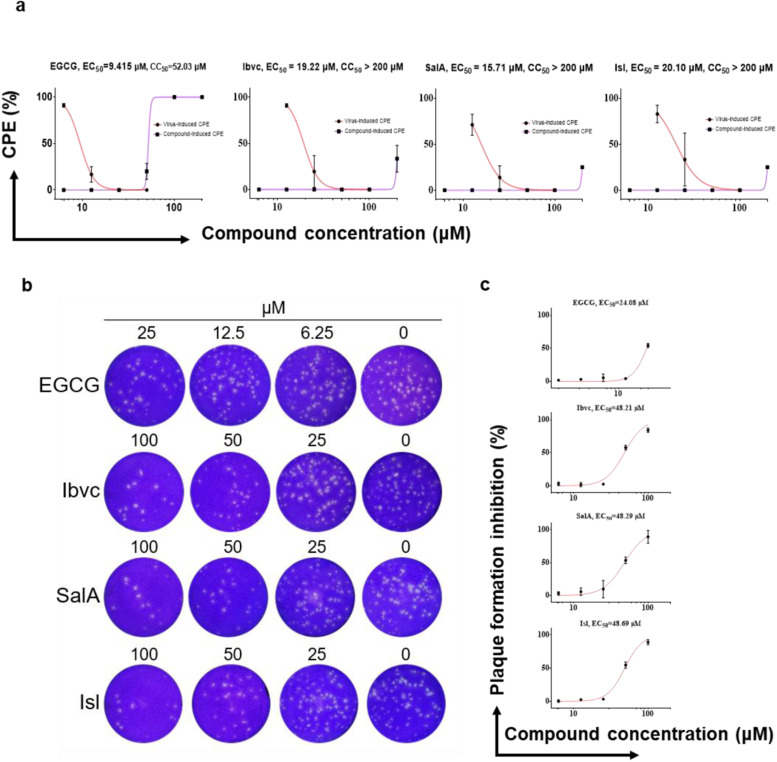
3.3. The natural compounds EGCG, Ibvc and SalA effectively suppress viral infection of SARS-CoV-2-pseudotyped lentivirus and live SARS-CoV-2 virus in hACE2 expressing cells
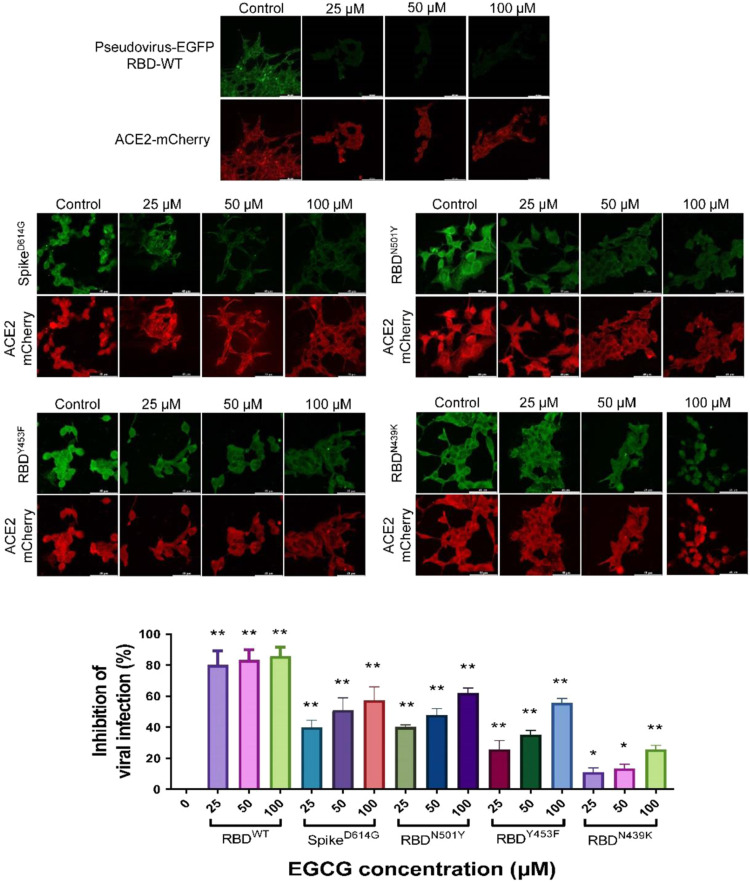
Furthermore, we tested the efficacy of the compounds in protecting HEK293 cells transiently co-overexpressing human ACE2 and mCherry against infection by a SARS-CoV-2-pseudotyped lentivirus encoding EGFP reporter gene (Fig. S2). However, cytotoxicity of 7 compounds in three normal human cell lines, BEAS-2B, LO2 and HEK293, hindered further validation of these compounds in cell-based assay (Fig. S3). 6 of the 7 compounds, EGCG, RRg3, SRg3, Ibvc, IscA and Bkc, effectively inhibited pseudovirus entry at concentrations up to 100 μM, whereas SalA only weakly inhibited pseudovirus entry ( Fig. 3 and Fig. S4). The data from the pseudovirus assay correlated well with those of ICC, suggesting that the compounds’ inhibitory effect on the interaction between RBD and ACE2 is crucial for their antiviral infection ability. Among those 7 compounds effective in inhibiting the pseudoviral entry, 3 of these compounds, EGCG, Ibvc, SalA, were found effective in inhibition of SARS-CoV-2-induced cytopathic effect and plaque formation in Vero E6 cells ( Fig. 4 and Table S3). Consistent with CPE inhibition, these compounds, together with Isl which was excluded from pseudovirus assay due to toxicity on HEK293 cells, also inhibited plaque formation of SARS-CoV-2 in Vero E6 cells, suggesting that these compounds are effective in blocking the SARS-CoV-2 viral entry. On the other hand, the other 4 compounds were completely or almost inactive in inhibition of viral cytopathic effects. The most potent compound by EC50 in both pseudovirus and authentic SARS-CoV-2 experiment, EGCG, showed no adverse effect on C57BL/6 mice survival (Fig. S5a, b), body weights (Fig. S5c) or organ weights (Fig. S5d) at doses of 200 and 300 mg/kg. Of note, three mutations on the RBD (N439K, Y453F, N501Y) and one in the S1 region (D614G) of SARS-CoV-2 confers SARS-CoV-2 pseudovirus with greater infectivity on HEK293 cells ( Fig. 5). Nonetheless, EGCG effectively inhibited the infection of all mutants, albeit at lower activity. These findings suggest that EGCG could be a safe and potent inhibitor for anti-SARS-CoV-2 infection.
Fig. 3.
The anti-viral effect of natural small-molecules in hACE2 overexpressing HEK293 cells infected by SARS-CoV-2-pseudotyped lentivirus. Representative fluorescence microscopic images of cells expressing ACE2-mCherry, either treated with EGFP-pseudovirus or treated with EGFP-pseudovirus in the presence of 100 μM compounds, are shown; scale bars represent 48.5 µm (upper). Data of pseudovirus entry inhibition at different concentrations are standardized to the mean of fluorescence intensity at 0 μM concentration and presented as mean ± S.D. of three replicates; EC50 is calculated by fitting a sigmoidal curve in GraphPad Prism 8.0 software (lower).
Fig. 4.
The anti-viral effect of natural small-molecules toward live SARS-CoV-2 virus. (A) Inhibitory effects of compounds on CPE effect, such as confluence, number of cells and morphology, induced by authentic SARS-CoV-2 infection in Vero E6 cell culture and cytotoxicity of compounds. (B, C) Compounds effectively inhibit plaque-forming ability of SARS-CoV-2.
Fig. 5.
EGCG inhibits the infection of 4 mutant S-pseudotyped lentivirus in human ACE2 overexpressing cells. HEK293 cells were transiently transfected with hACE2-mCherry (red). After 24 h, the hACE2 overexpressing cells were infected by RBD wild-type and mutant S-pseudotyped lentivirus (D614G, N501Y, N439K & Y453F) (green) in the presence of 0–100 μM EGCG. The infected cells were then replaced with fresh medium and continually incubated for 48 h. All images were captured by confocal microscopy using a Leica SP8 (×40 oil immersion objective lens). Data of pseudovirus entry inhibition at different concentrations are standardized to the mean of fluorescence intensity (quantified by Image J) at 0 μM concentration and presented as mean ± S.D. of three replicates; *P < 0.05, **P < 0.01, student-t-test analysis.
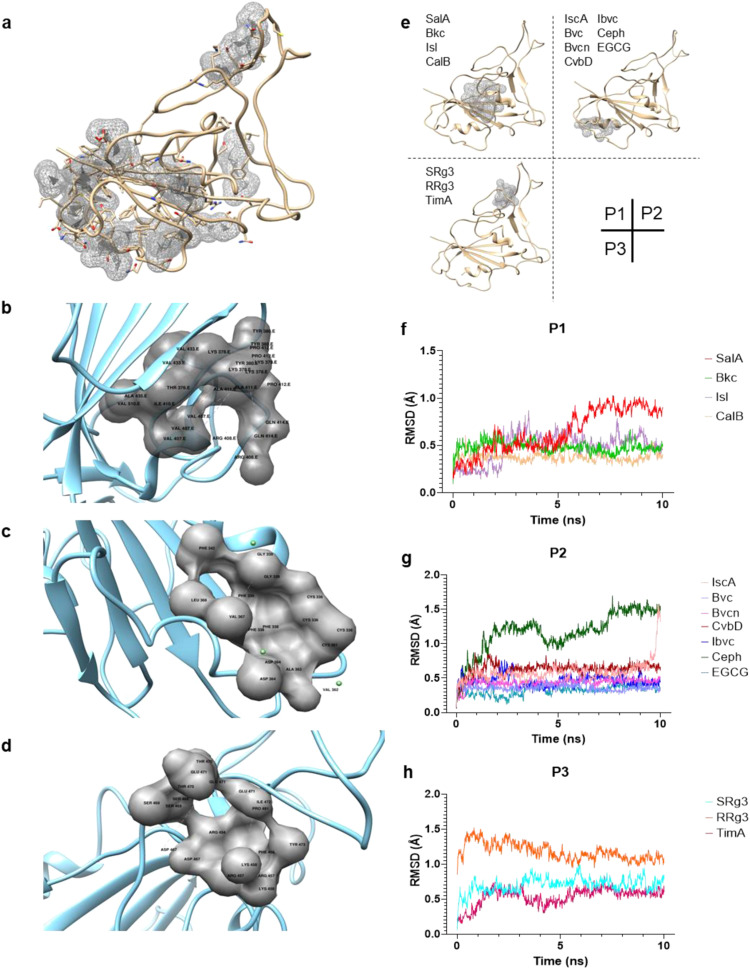
3.4. The identified natural small-molecules target three potential binding pockets of RBD
We noticed some discrepancies between results of ELISA, ICC and pseudovirus assay. For example, SRg3, RRg3, Ibvc and Bkc lacked activity in ELISA, but are active in cell-based assay. We illustrated that these discrepancies may be due to preferential binding of various compounds to their specific binding domains on RBD. Unlike RBD fragments, full-length S protein present on pseudovirus undergoes dynamic conformational changes [17]. Our virtual molecular docking analysis led to prediction of the binding poses of the 14 compounds ( Fig. 6a) and revealed that four, seven and three compounds possibly bind to three potential binding pockets, respectively, hereafter named P1, P2 and P3 (Fig. 6b-e). None of them overlapped with the part of the RBD directly responsible for binding with ACE2, which is a hydrophobic groove in the loop-dominated receptor-binding motif (RBM) [17]. These data suggest that small-molecule inhibitors identified in our study may act allosterically. Notably, the most effective compound EGCG is predicted to bind at P2, which does not harbour any of the mutations D614G, N439K, Y453F, N501Y. This may explain why this compound is still active against mutant pseudoviruses bearing these mutations. All binding poses were confirmed for stability in molecular dynamics simulation, as indicated by equilibration within the first 10 ns (Fig. 6f-h).
Fig. 6.
Computational modeling experiment to predict binding sites of natural compounds. (A) Possible binding sites identified by F-pocket. (B-D) Active compounds clusters into three binding sites (B) P1; (C) P2 and (D) P3. (E) Predicted binding pockets. P1 & P2 are located at the core domain of RBD; P3 is located at the distal loop of the RBM. (F-H) Equilibration in 10 ns of all compounds in their respective pockets confirms binding stability.
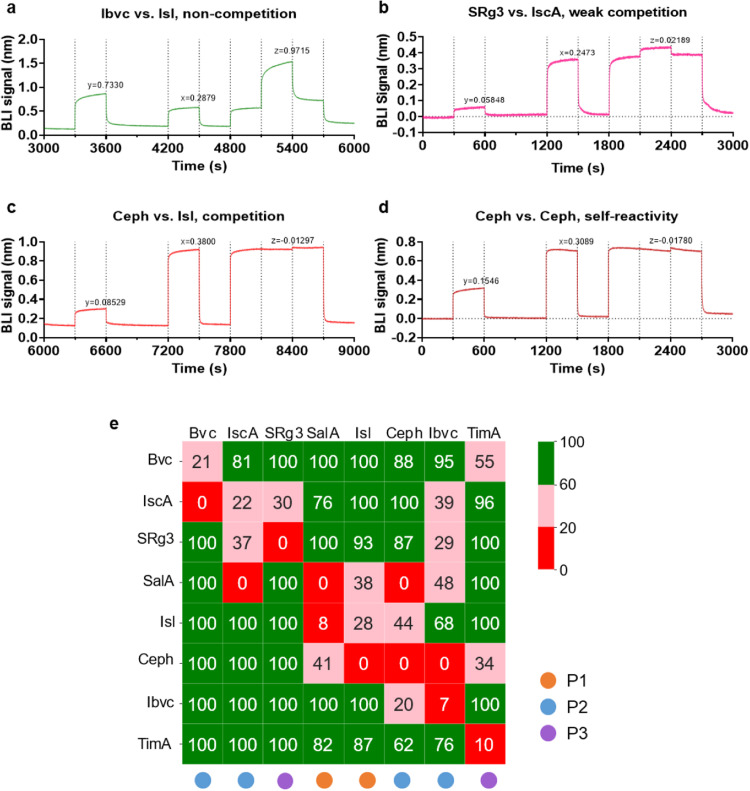
3.5. BLI-based competitive binding assay validates the three potential binding pockets of RBD
We then set out to substantiate the predicted binding poses of these small molecule inhibitors by conducting a BLI-based competitive binding assay, following previously reported methods for epitope binning of antibodies [18]. We assume that saturated binding of RBD by a compound will prevent others that share its binding site from eliciting a signal in BLI, whereas signals of compounds that bind at different sites are independent from each other ( Fig. 7a-d). Here, we were able to obtain reciprocal interactions between 8 compounds (Fig. 7e, Table S4). The rest were excluded from further analysis because the saturating concentration could not be reached even at 800 μM. Surprisingly, results of the competitive binding experiment did not match well with predicted binding sites, possibly due to multiple binding sites or allosteric effects of some compounds. Also, we found that EGCG, Bkc and Ibvc bound to ACE2 receptor in BLI assay (Fig. S6). Thus, it is possible that they work by engaging both RBD and ACE2, which may account for the disagreement between computational prediction and cross-competitive binding assay.
Fig. 7.
Validation of RBD binding pockets with BLI competitive binding assay. (A) Non-competitive interaction. (B) Weakly competitive interaction. (C) Competitive interaction between primary and second compounds in BLI cross-competitive binding assay. (D) Example of a compound blocking its own binding at high concentration. (E) Heatmap matrix representing the results of BLI-based competitive binding experiment of 8 compounds as indicated. Value in each cell represents the percent reduction of the signal of the second compound in the presence of high concentration of the primary compound. It is calculated as signal of mixed compound/signal of second compound alone ×100%, maximizing at 100% and minimizing at 0. Non-competition rate of an interaction was calculated as 100%×|z/y| with a maximum of 100% and minimum of 0. Non-competition rate of ≥ 60% indicates that binding of second compound is not significantly diminished by previous first compound binding; Non-competition rate of ≤ 20% is considered to constitute competition. 0.2 <non-competition rate < 0.6 indicates weak competition. Cut-off values were selected arbitrarily. Among the 8 compounds, IscA and SRg3 diminished binding signals of one another, although they were allocated to P2 and P3, respectively. SalA, Isl and Ceph all reciprocally competed for binding with RBD, although Ceph was allocated to P2 instead of P1 which SalA and Isl were predicted to bind. Ibvc, which was predicted to bind at P2, did mutually compete with Ceph. Interestingly, binding of Ibvc to RBD was weakly inhibited by IscA, SRg3 and SalA, but did not diminish the binding signal of these three compounds. Bvc and TimA, predicted to bind to P2 and P3, respectively, did not have reciprocal interaction with any other compounds.
4. Discussion
Since the outbreak of the new coronavirus in December 2019, it has caused a serious health threat worldwide at a rapid rate. The morbidity and mortality are increasing due to the lack of SARS-CoV-2 specific drugs and currently, remdesivir is the only medication approved by the FDA to treat coronavirus disease 2019 (COVID-19) [19]. Currently, several vaccines have been authorized in globally, however, the side effects, safety and efficacy of these vaccines remained to be fully elucidated. Many European countries, such as Denmark, Norway and Iceland, suspended vaccination of Astra Zeneca vaccine [20] due to its side effects, including blood clots after being vaccinated. Moreover, the reported mutations of SARS-CoV-2 virus are also the major challenge affecting the effectiveness of the vaccines [21]. Under this circumstance, the discovery small-molecules RBD inhibitors maybe another critical approaches to protect the public from viral infection instead of using vaccines. The active ingredients such as EGCG in these products neutralize the virus in vitro, and have less side effects on human body, so they are more easily accepted by people, and can be considered as an effective measure to fight against COVID-19.
Anti-viral small molecule drugs are more effective for the first line of protection against the virus during a pandemic outbreak, especially in the early stage for external use, which makes it more acceptable for the public to apply it to prevent the viral infection. Meanwhile, these small molecule compounds could also be developed to treat COVID-19 patients. SARS-CoV-2 entry inhibitors are mechanistically similar to neutralizing monoclonal antibodies that disrupts RBD binding to ACE2. Clinical trials of this class of drug shows that they are effective on mild and moderate COVID-19 patients. Therefore, it is reasonable to assume that future clinical trials of small-molecule entry inhibitors should primarily include patients with mild and moderate disease.
Our study identified 8 natural small molecules that inhibit the entry of pseudovirus with low cytotoxicity. Some of these compounds have been previously characterized as anti-SARS-CoV-2 agents. Bvc, Isl and Ceph were previously found to be effective against authentic SARS-CoV-2 in high-throughput screening experiments [22], [23]. Interestingly, SalC, a structural relative to SalA identified in our study, is reported to be a fusion inhibitor of SARS-CoV-2 by interfering with the formation of 6-helix bundle [24] but weak binding activity to RBD as measured by BLI. This is in contrast to our data, as SalC is identified in our BLI screening as having strong affinity with SARS-CoV-2 RBD with a KD of 6.2 μM, though it did not proceed to further validation. The discrepancy can be explained by the fact that the other study employed streptavidin probes, whereas the super streptavidin probe we used allows much more sensitive detection of small molecule binding events. Thus, SalC may also bind with RBD in addition to S2 domain of SARS-CoV-2 Spike. Our results warrant further investigation of SalC as a multi-function against SARS-CoV-2 infection by disrupting both RBD-ACE2 interaction and membrane fusion.
Importantly, our data shows that these compounds remain effective against pseudoviruses carrying four mutations. Circulating variants of SARS-CoV-2 carries mutations in the spike protein that promote immune escape and increased infectivity. They may compromise the effectiveness of small molecule entry inhibitors. To assess the activity of our drug candidates on these mutants, we generated mutant pseudoviruses corresponding to mutations found on circulating variants. These include N439K (B.1.141 “Scottish lineage”), Y453F (B.1.1.298 “mink variant”), N501Y (B.1.1.7, B.1.351 and P1) and D614G (found in most variants). This shows that these compounds can be of real-world relevance when used as anti-SARS-CoV-2 drugs.
In addition, our data suggest that these molecules bind to non-overlapping pockets on RBD, analogous to how RBD-targeting antibodies recognize distinct epitopes [25]. It is known that circulating variants of SARS-CoV-2 with mutations on the RBD hampers neutralizing activity of vaccine-induced or convalescent patient-derived antibodies [26]. Antibodies that recognize epitopes distant from ACE2 binding domain remain effective against mutations that are present in circulating variants. And combination of antibodies with non-overlapping epitopes effectively neutralizes escaping variants. However, much of the areas of the RBD are inaccessible to antibody binding due to steric hindrance, limiting the options available to combination antibody therapeutics. Unlike antibodies, small molecule can be designed to targets areas on RBD that are otherwise inaccessible to biomacromolecules. Another advantage of our compounds is that they avoid frequently the mutated RBD region, as exemplified by EGCG. We show that EGCG remains effective against pseudoviruses bearing some of the most prevalent mutations. A possible explanation is that none of the mutations are located on the putative binding site of EGCG. Though we did not test other compounds on mutant pseudoviruses, it is reasonable to assume that other compounds should be still active as long as the mutations do not involve the binding sites of these compounds.
From CPE and plaque reduction assays using of SARS-CoV-2 virus, we found that EGCG, SalA, Ibvc and Isl had significant inhibitory effect on SARS-CoV-2 virus infection. Under the pandemic situation, how to balance the effectiveness, safety and economy of drugs is also an important part for drugs development. In terms of drug efficacy, extensive studies have demonstrated that EGCG exhibits a potent inhibitory effect in H1N1 influenza virus with EC50: 22–28 μM [27], zika virus with EC50: 21.4 µM [28], Ebola virus with EC50: 50 μM [29], and HIV-1 protease kinetics with IC50: 50–100 μM [30], suggesting that EGCG shows a broad spectrum of antiviral effect. As for SalA, this small-molecule exhibits potential pharmacological effects including vasoconstriction [31], anti-inflammation [32], anti-cancer [33]. Recent studies further demonstrated that salvianolic acid A, B and C can inhibit SARS-CoV-2 spike pseudovirus viropexis by binding to both viral RBD and host cells ACE2 receptor [34]. Together with our findings using real SARS-CoV-2 virus, SalA was shown to be effective in prevention of viral infection of COVID-19. Apparently, our CPE and plaque reduction assays indicated that EGCG exhibited a better inhibitory effect on SARS-CoV-2 virus infection at a lower concentration of 25 μM in comparison to SalA and Ibvc. Additionally, in our experiments, EGCG was preincubated with SARS-CoV-2 before infection of cells, ruling out its effects on non-structural proteins. EGCG was recently demonstrated to inhibit SARS-CoV-2 3CL-protease and coronavirus replication by inhibition of 3CL protease [35], suggesting the all-round inhibitory potency of EGCG on SARS-CoV-2. Notably, we also observed that EGCG could inhibit the infection of 4 kinds of mutant S-pseudotyped lentivirus in human ACE2 overexpressing cells, indicating that ECGC has a good prevention prospect for the mutated SARS-COV-2. In terms of drug safety, EGCG has been widely used in foods additive for many years, and less safety problems have been reported under the consumption of recommended dosage. From the pharmacoeconomics aspects, EGCG is extracted from tea, which is widely distributed and cheap in price [36]; whereas SalA is one of the major components in herb Salvia miltiorrhiza [37], which is affordable for medical application. In comparison with these two compounds, Ibvc is derived from the seeds of Psoralea corylifolia and Isl is derived from the embryos of Nelumbo nucifera, which are relative higher in price [38], [39]. Collectively, EGCG is a suitable candidate to be developed as an anti-SARS-CoV-2 virus agent based on its drug efficacy, safety and economic benefits.
5. Conclusion
Overall, our study identified 4 natural small molecules that inhibit the SARS-CoV-2 infection in vitro with low cytotoxicity and no adverse effect on the in vivo system. Among these, EGCG and Ibvc may inhibit the SARS-CoV-2 infection through a dual action by targeting RBD and/or ACE2. Based on the in vitro and in vivo data, we identified EGCG as the most suitable candidate as an anti-SARS-CoV-2 virus agent with a better drug efficacy and safety in animals. In fact, EGCG is also known to inhibit SARS-CoV-2 3CL-protease and coronavirus replication by inhibition of 3CL protease35, suggesting the all-round inhibitory potency of EGCG on SARS-CoV-2. Taken together, our current findings provide an insight into structure-based targeting of RBD inhibition and SARS-CoV-2 infection by the natural small molecule, such as EGCG.
Author contributions
Conception: V.K.W.W., L.L. and S.K.C.; Methodology: V.K.W.W. and D.Q.Z.; BLI experiments: D.Q.Z., C.K.Y. R.H.C and L.J.Y.; Cellular and biochemical experiments: R.H.C, D.Q.Z, Y.Q.Q and L.J.Y.; Authentic SARS-CoV-2 experiments: R.F.L and H.M.J.; Manuscript: D.Q.Z.; Manuscript revision: V.K.W.W and S.K.C; Supervision: V.K.K.W, L.L., S.K.C., Z.F.Y.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Faculty Research Grant from Macau University of Science and Technology (Project code: FRG-20-001), the granted project of Dr. Neher’s Biophysics Laboratory for Innovative Drug Discovery from the Macao Science and Technology Development Fund (Project code: 0033/2019/AFJ; 001/2020/ALC), and Foshan Medicine Dengfeng Project of China (2019–2021).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.phrs.2021.105820.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Data availability
All data used to support the claims of the study can be found in the main text and supplemental materials. Raw data is available from the corresponding author upon reasonable request. Code for analysis of BLI-based competitive binding assay is available from https://github.com/DQ-Zhang/BLICompetitiveBindingAssay.
References
- 1.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020:2531–6745. doi: 10.23750/abm.v91i1.9397. (Electronic) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang M.-Y., Zhao R., Gao L.-J., Gao X.-F., Wang D.-P., Cao J.-M. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brian D.A., Baric R.S. In: Coronavirus Replication and Reverse Genetics. Enjuanes L., editor. Springer; Berlin Heidelberg: 2005. Coronavirus genome structure and replication; pp. 1–30. [Google Scholar]
- 4.Wang K., Chen W., Zhang Z., Deng Y., Lian J.-Q., Du P., Wei D., Zhang Y., Sun X.-X., Gong L., Yang X., He L., Zhang L., Yang Z., Geng J.-J., Chen R., Zhang H., Wang B., Zhu Y.-M., Nan G., Jiang J.-L., Li L., Wu J., Lin P., Huang W., Xie L., Zheng Z.-H., Zhang K., Miao J.-L., Cui H.-Y., Huang M., Zhang J., Fu L., Yang X.-M., Zhao Z., Sun S., Gu H., Wang Z., Wang C.-F., Lu Y., Liu Y.-Y., Wang Q.-Y., Bian H., Zhu P., Chen Z.-N. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020;5(1):283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Österlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan X., Cao D., Kong L., Zhang X. Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nat. Commun. 2020;11(1):3618. doi: 10.1038/s41467-020-17371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lachance H., Wetzel S., Waldmann H. Role of natural products in drug discovery. Lead. Gener. Approaches Drug Discov. 2010:187–229. [Google Scholar]
- 10.Atanasov A.G., Zotchev S.B., Dirsch V.M., Orhan I.E., Banach M., Rollinger J.M., Barreca D., Weckwerth W., Bauer R., Bayer E.A., Majeed M., Bishayee A., Bochkov V., Bonn G.K., Braidy N., Bucar F., Cifuentes A., D’Onofrio G., Bodkin M., Diederich M., Dinkova-Kostova A.T., Efferth T., El Bairi K., Arkells N., Fan T.-P., Fiebich B.L., Freissmuth M., Georgiev M.I., Gibbons S., Godfrey K.M., Gruber C.W., Heer J., Huber L.A., Ibanez E., Kijjoa A., Kiss A.K., Lu A., Macias F.A., Miller M.J.S., Mocan A., Müller R., Nicoletti F., Perry G., Pittalà V., Rastrelli L., Ristow M., Russo G.L., Silva A.S., Schuster D., Sheridan H., Skalicka-Woźniak K., Skaltsounis L., Sobarzo-Sánchez E., Bredt D.S., Stuppner H., Sureda A., Tzvetkov N.T., Vacca R.A., Aggarwal B.B., Battino M., Giampieri F., Wink M., Wolfender J.-L., Xiao J., Yeung A.W.K., Lizard G., Popp M.A., Heinrich M., Berindan-Neagoe I., Stadler M., Daglia M., Verpoorte R., Supuran C.T. The International Natural Product Sciences T. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discov. 2021;20(3):200–216. doi: 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodrigo Santos Aquino de A., Edeildo Ferreira da S.-J., Thiago Mendonça de A., Marcus Tullius S., Hamilton M.I., Luciana S., Francisco Jaime Bezerra M.-J. Computer-aided drug design applied to secondary metabolites as anticancer agents. Curr. Top. Med. Chem. 2020;20(19):1677–1703. doi: 10.2174/1568026620666200607191838. [DOI] [PubMed] [Google Scholar]
- 12.Xuan-Yu M., Hong-Xing Z., Mihaly M., Meng C. Molecular docking: a powerful approach for structure-based drug discovery. Curr. Comput. -Aided Drug Des. 2011;7(2):146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L.J., Chen R.H., Hamdoun S., Coghi P., Ng J.P.L., Zhang D.W., Guo X., Xia C., Law B.Y.K., Wong V.K.W. Corilagin prevents SARS-CoV-2 infection by targeting RBD-ACE2 binding. Phytomedicine. 2021;87 doi: 10.1016/j.phymed.2021.153591. 153591-153591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen R.H., Yang L.J., Hamdoun S., Chung S.K., Lam C.W.-K., Zhang K.X., Guo X., Xia C., Law B.Y.K., Wong V.K.W. 1,2,3,4,6-pentagalloyl glucose, a RBD-ACE2 binding inhibitor to prevent SARS-CoV-2 infection. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.634176. 634176-634176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R., Lu Y., Wang S. Comparative evaluation of 11 scoring functions for molecular docking. J. Med. Chem. 2003;46(12):2287–2303. doi: 10.1021/jm0203783. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z., Zhang Y., Li X., Zhang H., Xiao S., Deng F., Yuan Z., Ye H., Zhang B. A cell-based large-scale screening of natural compounds for inhibitors of SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5(1):218. doi: 10.1038/s41392-020-00343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao-Chan S., Zachwieja J., Gomez S., Duey D., Lippincott J., Theunissen J.-W. Monoclonal antibody binding-site diversity assessment with a cell-based clustering assay. J. Immunol. Methods. 2014;405:1–14. doi: 10.1016/j.jim.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Lamb Y.N. Remdesivir: first approval. Drugs. 2020;80(13):1355–1363. doi: 10.1007/s40265-020-01378-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wise J. Covid-19: European countries suspend use of Oxford-AstraZeneca vaccine after reports of blood clots. BMJ. 2021:1756–1833. doi: 10.1136/bmj.n699. (Electronic) [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Beltran W.F., Lam E.C., St., Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., Sigal A., Schmidt A.G., Iafrate A.J., Naranbhai V., Balazs A.B. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184(9):2372–2383. doi: 10.1016/j.cell.2021.03.013. e2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z.-R., Zhang Y.-N., Li X.-D., Zhang H.-Q., Xiao S.-Q., Deng F., Yuan Z.-M., Ye H.-Q., Zhang B. A cell-based large-scale screening of natural compounds for inhibitors of SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5(1):218. doi: 10.1038/s41392-020-00343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He C.-L., Huang L.-Y., Wang K., Gu C.-J., Hu J., Zhang G.-J., Xu W., Xie Y.-H., Tang N., Huang A.-L. Identification of bis-benzylisoquinoline alkaloids as SARS-CoV-2 entry inhibitors from a library of natural products. Signal Transduct. Target. Ther. 2021;6(1):131. doi: 10.1038/s41392-021-00531-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang C., Pan X., Xu X., Cheng C., Huang Y., Li L., Jiang S., Xu W., Xiao G., Liu S. Salvianolic acid C potently inhibits SARS-CoV-2 infection by blocking the formation of six-helix bundle core of spike protein. Signal Transduct. Target. Ther. 2020;5(1):220. doi: 10.1038/s41392-020-00325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., Oliveira T.Y., Yang Z., Abernathy M.E., Huey-Tubman K.E., Hurley A., Turroja M., West K.A., Gordon K., Millard K.G., Ramos V., Da Silva J., Xu J., Colbert R.A., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Gazumyan A., Caskey M., Bjorkman P.J., Casellas R., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592(7855):616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., Dingens A.S., Nargi R.S., Sutton R.E., Suryadevara N., Rothlauf P.W., Liu Z., Whelan S.P.J., Carnahan R.H., Crowe J.E., Jr., Bloom J.D. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44–57. doi: 10.1016/j.chom.2020.11.007. e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song J.-M., Lee K.-H., Seong B.-L. Antiviral effect of catechins in green tea on influenza virus. Antivir. Res. 2005;68(2):66–74. doi: 10.1016/j.antiviral.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Carneiro B.M., Batista M.N., Braga A.C.S., Nogueira M.L., Rahal P. The green tea molecule EGCG inhibits Zika virus entry. Virology. 2016;496:215–218. doi: 10.1016/j.virol.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 29.Patrick Reid S., Shurtleff A.C., Costantino J.A., Tritsch S.R., Retterer C., Spurgers K.B., Bavari S. HSPA5 is an essential host factor for Ebola virus infection. Antivir. Res. 2014;109:171–174. doi: 10.1016/j.antiviral.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi K., Honda M., Ikigai H., Hara Y., Shimamura T. Inhibitory effects of (−)-epigallocatechin gallate on the life cycle of human immunodeficiency virus type 1 (HIV-1) Antivir. Res. 2002;53(1):19–34. doi: 10.1016/s0166-3542(01)00189-9. [DOI] [PubMed] [Google Scholar]
- 31.Zhong W., Sun B., Gao W., Qin Y., Zhang H., Huai L., Tang Y., Liang Y., He L., Zhang X., Tao H., Chen S., Yang W., Yang L., Liu Y., Liu H., Zhou H., Sun T., Yang C. Salvianolic acid A targeting the transgelin-actin complex to enhance vasoconstriction. EBioMedicine. 2018;37:246–258. doi: 10.1016/j.ebiom.2018.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng S., Cong H., Ji L., Salvianolic, Acid A. Exhibits anti-inflammatory and antiarthritic effects via inhibiting NF-κB and p38/MAPK pathways. Drug Des. Dev. Ther. 2020;14:1771–1778. doi: 10.2147/DDDT.S235857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin T., Rasul A., Sarfraz A., Sarfraz I., Hussain G., Anwar H., Riaz A., Liu S., Wei W., Li J., Li X. Salvianolic acid A & B: potential cytotoxic polyphenols in battle against cancer via targeting multiple signaling pathways. Int. J. Biol. Sci. 2019;15(10):2256–2264. doi: 10.7150/ijbs.37467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu S., Wang J., Zhang Y., Bai H., Wang C., Wang N., He L. Three salvianolic acids inhibit 2019-nCoV spike pseudovirus viropexis by binding to both its RBD and receptor ACE2. J. Med. Virol. 2021;93(5):3143–3151. doi: 10.1002/jmv.26874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang M., Park R., Park Y.-I., Cha Y.-E., Yamamoto A., Lee J.I., Park J.E.G.C.G. A green tea polyphenol, inhibits human coronavirus replication in vitro. Biochem. Biophys. Res. Commun. 2021;547:23–28. doi: 10.1016/j.bbrc.2021.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinmann J., Buer J., Pietschmann T., Steinmann E. Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea. Br. J. Pharmacol. 2013;168(5):1059–1073. doi: 10.1111/bph.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Y., Li Y., Xue L., Severino R.P., Gao S., Niu J., Qin L.-P., Zhang D., Brömme D. Salvia miltiorrhiza: an ancient Chinese herbal medicine as a source for anti-osteoporotic drugs. J. Ethnopharmacol. 2014;155(3):1401–1416. doi: 10.1016/j.jep.2014.07.058. [DOI] [PubMed] [Google Scholar]
- 38.Kim D.W., Seo K.H., Curtis-Long M.J., Oh K.Y., Oh J.-W., Cho J.K., Lee K.H., Park K.H. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzym. Inhib. Med. Chem. 2014;29(1):59–63. doi: 10.3109/14756366.2012.753591. [DOI] [PubMed] [Google Scholar]
- 39.Lee T., Kwon J., Lee D., Mar W. Effects of Cudrania tricuspidata fruit extract and its active compound, 5,7,3′,4′-tetrahydroxy-6,8-diprenylisoflavone, on the high-affinity IgE receptor-mediated activation of Syk in mast cells. J. Agric. Food Chem. 2015;63(22):5459–5467. doi: 10.1021/acs.jafc.5b00903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Data Availability Statement
All data used to support the claims of the study can be found in the main text and supplemental materials. Raw data is available from the corresponding author upon reasonable request. Code for analysis of BLI-based competitive binding assay is available from https://github.com/DQ-Zhang/BLICompetitiveBindingAssay.