Abstract
Purpose:
The presence of stage is an important feature to identify in retinal images of infants at risk for retinopathy of prematurity (ROP). The purpose of this study was to implement a convolutional neural network (CNN) for binary detection of stage 1–3 in ROP and evaluate its generalizability across different populations and camera systems.
Design:
Diagnostic validation study of CNN for stage detection.
Subjects, Participants, and/or Controls:
Retinal fundus images obtained from preterm infants during routine ROP screenings.
Methods:
Two datasets were used: 6247 fundus images taken by a RetCam camera from nine North American institutions, and 4647 images taken by a Forus 3nethra camera from four hospitals in Nepal. Images were labeled based on the presence of stage by 1–3 expert graders. Three CNN models were trained using 5-fold cross-validation on datasets from North America alone, Nepal alone, and a combined dataset and evaluated on two held-out test sets consisting of 708 and 247 images from the Nepali and North American datasets respectively.
Main Outcome Measures:
CNN performance was evaluated using area under the receiver operating curve (AUROC) and precision-recall curve (AUPRC), sensitivity, and specificity.
Results:
Both the North American- and Nepali-trained models demonstrated high performance on a test set from the same population: (AUROC/AUPRC) 0.99/0.98 with sensitivity of 94%, and 0.97/0.91 with sensitivity of 73%, respectively. However, the performance of each model decreased to 0.96/0.88 (sensitivity 52%) and 0.62/0.36 (sensitivity 44%) when evaluated on a test set from the other population. Compared to the models trained on individual datasets, the model trained on a combined dataset achieved improved performance on each respective test set: sensitivity improved from 94% to 98% on the North American test set, and from 73% to 82% on the Nepali test set.
Conclusions:
A CNN can accurately identify the presence of ROP stage in retinal images, but performance depends on the similarity between training and testing populations. We demonstrate that internal and external performance can be improved by increasing the heterogeneity of the training dataset features of the training dataset, in this case by combining images from different populations and cameras.
Precis:
The performance and generalizability of deep learning algorithms for detection of stage in retinopathy of prematurity can be improved by training on images from multiple populations and camera systems.
INTRODUCTION
The international classification of retinopathy of prematurity (ICROP)1 has established standard terminology for describing ROP based on retinal findings from an ophthalmoscopic exam, including zone, stage, and plus disease. Using these clinical features, several National Institutes of Health funded studies have determined treatment criteria and follow-up guidelines.2,3 ROP screening has proven to be an effective form of secondary prevention of blindness, yet ROP remains one of the leading causes of childhood blindness particularly in low- and middle-income countries, where there are often too few ROP-trained ophthalmologists to meet the screening burden.4–6 An estimated 50,000 babies develop severe visual impairment or blindness annually in part due to a failure in screening.7
Artificial intelligence (AI) screening systems have the potential to improve the efficiency of disease screening and increase access to care.8 Deep learning (DL) is a subset of AI that includes convolutional neural networks (CNNs), which are a class of neural networks specifically designed for image classification. CNNs have achieved expert-level performance in the diagnosis of gliomas,9,10 melanomas,11 glaucoma,12,13 diabetic retinopathy,14–16 and age-related macular degeneration.17,18 There have also been examples of CNNs developed for all components of ROP classification (zone,19 stage,20–24 and plus disease25,26). However, most of the work has focused on plus disease and used images from the RetCam camera (Natus Medical, Pleasanton, CA), a wide-field fundus camera not widely available in low-income countries due to cost.
It is well-known that CNNs that perform well on one dataset often perform worse on external datasets, a problem known as domain shift.22–25 For example, a CNN trained on images from one population or camera may have significantly decreased performance when evaluated on images taken from another population or camera. There is a gap in knowledge regarding how to successfully translate proof-of-principle efficacy studies into scalable solutions with real-world populations and available medical devices.8 The purpose of this study is to address this gap in knowledge by training a CNN to classify the presence of ROP stage in two different populations and cameras, evaluating each model on the other population, and evaluating the effect of training on both populations to improve external generalizability.
METHODS
This study was approved by the institutional review board (IRB) at the coordinating center (Oregon Health and Science University, Portland) and the other North American study institutions (Columbia University, Cornell University, University of Illinois at Chicago, William Beaumont Hospital, Children’s Hospital Los Angeles, Cedars-Sinai Medical Center, University of Miami, and Asociacion para Evitar la Ceguera en Mexico) as part of the i-ROP consortium. Data was also obtained from an ROP screening program supported by the Helen Keller International at four urban hospitals in Kathmandu, Nepal: Patan Hospital, Kanti Children’s Hospital, Paropakar Maternity and Women’s Hospital, and the Tilganga Institute of Ophthalmology, with local IRB approval. All institutions abided by the Declaration of Helsinki. Informed consent was obtained from the parents of all infants enrolled.
Datasets and Reference Standards
We used two datasets: 1) nasal, temporal, and posterior pole retinal fundus images in the North American dataset taken by a RetCam camera, and 2) images from all fields of view in the Nepali dataset taken by a 3nethra camera (Forus Health Incorporated, Bengaluru, India).
Images from examinations in the North American dataset were assigned a reference standard diagnosis (RSD) where stage was determined based on consensus from image-based diagnoses by three experts and a clinical exam.27 Since stage was not necessarily present in all 3 views, exams were manually reviewed to verify that individual images reflected the clinical RSD. Images in examinations from the Nepali dataset were labeled with a RSD based on manual review for the presence of stage in an image and validated by one expert grader who also graded the North American dataset. Disagreement was resolved by discussion among both graders. In both datasets, images were excluded if there was evidence of prior treatment, such as laser photocoagulation and scars, if retinal detachment was present (stage≥4), or if artifacts obscured >50% of the image. 50% was an arbitrary cutoff to ensure that image labels accurately corresponded to enough image features to reasonably enable a CNN to learn the features of stage.
Each image was assigned a binary label. Images with stage≥1 visible were labeled “1” and images without visible stage were labeled “0”. Because there was a disproportionately large number of images with no ROP in both datasets, the total number of images with no stage present in the North American and Nepali datasets were randomly downsampled by 50% and 66% respectively to decrease dataset imbalance and to normalize both datasets to around 5000–6000 images.
Image Preprocessing
Prior to CNN input, all images underwent preprocessing using the opencv28 and scipy29 packages in Python (version 3.7).30 Image contrast was first enhanced using contrast limited adaptive histogram equalization (CLAHE) to emphasize pigment differences between the avascular and vascular retina. The green channel was extracted from the image to increase visibility of darker, distinct structures such as vessels and demarcation lines,31 and to decrease the image input size. A Wiener filter was then applied to the image to de-noise aberrations created by CLAHE.32 Prior to CNN training, all images were resized to 224-by-224 pixels, the dimension of the images in the ImageNet dataset used to train ResNet.33 An example of these preprocessing steps is shown in Figure 1.
Figure 1. Image preprocessing.
Images underwent a 3-stage preprocessing step before input to the CNN. The original image (A) was adjusted using contrast limited adaptive histogram equalization (CLAHE) (B), green channel extraction (C), and a Wiener filter for denoising (D). These steps were performed to make features of stage more visible.32
Model Setup and Evaluation
Three types of CNN models were trained using three datasets: images from North America alone, Nepal alone, and a combination of both datasets. Each dataset was split such that 90% of the images were allocated to the training set and 10% were allocated to the test set. The datasets were stratified to ensure similar ratios of images with stage and without stage between the training and test sets, and were also stratified on a patient level such that the training and test sets contained distinctly different patients. Each training set was further split in a 4:1 ratio with patient-level and stage-level stratification into 5 training and validation splits for 5-fold cross validation.
We used a ResNet-15233 architecture pre-trained on images from the ImageNet dataset and Pytorch,34 an open-source DL framework. ResNet is a residual neural network architecture that utilizes “skip” connections, which skip over certain layers in the CNN, and results in more robust backpropagation and learning representations.33 For each cross-validation split, a CNN was trained on the training split augmented with images that randomly underwent rotations (from 0–180°), flips (mirroring images horizontally and vertically), and color jittering (adjusting brightness and contrast). The cross-entropy loss function was minimized using stochastic gradient descent (SGD) for 100 epochs, with a learning rate of 0.001 and batch size of 32 images. Early stopping based on validation loss was implemented to prevent model overfitting if no improvement occurred after 15 epochs. To account for data imbalance, a weighted random sampler was used to equalize the probabilities that images with and without stage would be selected for a given batch. Each trained model was subsequently evaluated on the validation split using the area under the receiver operating characteristics curve (AUROC) and area under the precision-recall curve (AUPRC) scores to assess its ability to accurately label stage on unseen images from different patients. We considered the cross-validated model with the highest AUPRC score the best performing model, since AUPRC scores account for dataset imbalance.
Test Set
The best performing model on cross-validation each dataset was further evaluated against two held-out test sets of unseen images, consisting of 708 images from the North American cohort and 247 images from Nepali dataset. These test sets contained images from unseen patients and were filtered to maintain a similar ratio of non-visible to visible stage as in the training sets. Sensitivity, specificity, AUROC, and AUPRC are reported for each test set.
CNN Feature Analysis
Following training, image features for all images in the North American and Nepali training sets were extracted as high-dimensional vectors from the average pooling layer of the CNN trained on both datasets. The average pooling layer aggregates all features computed by a CNN into 1024 dimensions and was reduced to two dimensions for visualization using uniform manifold approximation and projection (UMAP), a technique that aims to cluster similar features.35 A UMAP scatterplot was generated using the features from the best performing model, with each point representing the feature space of an image. Each point was also labeled by the presence of stage and its dataset of origin.
RESULTS
Dataset
Our final datasets contained 5943 images from 711 patients in the North American dataset and 5009 images from 541 patients from the Nepali dataset. Visible stage was present in 1693 (28.5%) of images in the North American dataset and 252 (5.0%) of images in the Nepali dataset, which contained more images per patient as part of routine screening, including low-risk babies. The mean gestational age for infants in the North American and Nepali cohorts was 26.6 ± 2.2 weeks and 32.6 ± 2.8 weeks, respectively. The mean birth weight for infants in both cohorts was 856.2 ± 293.7 grams and 1949.6 ± 495.8 grams, respectively. The distribution of the validation splits and test sets are shown in Table 1.
Table 1. Training and Test Sets.
All included images from the North American, Nepali, and combined datasets were split such that 90% of the images were distributed into the training set (A) and 10% of the images were distributed into the test set (B). Each dataset was stratified to maintain similar ratios of images with and without stage in each training and test set, as well as separated on a patient-level to ensure all training and test sets contained no overlapping patients. Each training set (A) was further split into 5 cross validation splits, retaining the underlying distribution of stage. Models were created using the North American dataset alone, the Nepali dataset alone, or both datasets combined. A) Training Set, divided into 5 splits for 5-fold Cross Validation
| No. in Training Split | No. In Validation Split | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Total | North American Dataset | Nepali Dataset | Total | North American Dataset | Nepal Dataset | |||||||
|
| ||||||||||||
| Splits | Images | Patients | No Stage | Stage | No Stage | Stage | Images | Patients | No Stage | Stage | No Stage | Stage |
| 1 | 7899 | 895 | 3018 | 1102 | 3599 | 180 | 2138 | 231 | 735 | 380 | 995 | 28 |
| 2 | 7941 | 882 | 2901 | 1161 | 3694 | 185 | 2096 | 244 | 852 | 321 | 900 | 23 |
| 3 | 7977 | 899 | 2998 | 1191 | 3651 | 137 | 2060 | 227 | 755 | 291 | 943 | 71 |
| 4 | 8149 | 909 | 3056 | 1263 | 3661 | 169 | 1888 | 217 | 697 | 219 | 933 | 39 |
| 5 | 8033 | 893 | 2974 | 1145 | 3762 | 152 | 2004 | 233 | 779 | 337 | 832 | 56 |
Cross validation performance
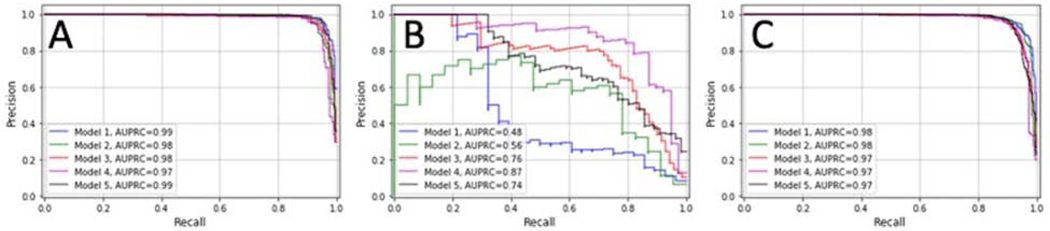
Models trained on North American data alone achieved AUROC scores ranging from 0.98–0.99 (mean±SD, 0.99±0.01) and AUPRC scores ranging from 0.97–0.99 (mean±SD, 0.98±0.01). Models trained on Nepali data alone achieved AUROC scores ranging from 0.95–0.97 (mean±SD, 0.96 ±0.01) and AUPRC scores ranging from 0.48–0.87 (mean±SD, 0.68±0.14). Models trained on images from both North American and Nepal achieved mean ±SD AUROC scores of 0.99±0.01, and mean±SD AUPRC scores of 0.98±0.01 on cross-validation. AUPRC curves are shown in Figure 2 for all three experiments. The models with the highest AUPRC scores were selected for final evaluation on held-out test sets.
Figure 2. Precision-Recall Scores for Cross-Validation of Each Model.
Area under the precision-recall curves (AUPRCs) were generated for each convolutional neural network (CNN) trained on images from the North American dataset (A), the Nepali dataset (B), and a combination of both datasets (C). The mean AUPRC score for the models were 0.98, 0.68, and 0.98 respectively.
Test Set Performance
Table 2 shows AUROC and AUPRC, sensitivities, and specificities of the best performing models evaluated on two independent test sets containing unseen images from the North American and Nepali datasets. The best performing model trained on the North American dataset achieved the following performance: AUROC=0.99, AUPRC=0.98, sensitivity=94%, and specificity=96% on the North American test set and achieved lower performance on the Nepali test set (AUROC=0.96, AUPRC=0.88, sensitivity=52%, specificity=99%). The best performing model trained on the Nepali dataset achieved the following performance: AUROC=0.97, AUPRC=0.91, sensitivity=73%, and specificity=99% when evaluated on the Nepali test set, and achieved lower performance when evaluated on the North American test set (AUROC=0.62, AUPRC=0.36, sensitivity=44%, specificity=69%).
Table 2.
Area Under the Receiver Operating Characteristics Curve (AUROC), Area Under the Precision-Recall Curve (AUPRC), Sensitivity, and Specificity of CNNs on test sets from North American and Nepal. The best performing models trained on images from North American, Nepal, and combined datasets were evaluated on two independent test sets from North America and Nepal.
| Model Training Set | Test set | AUROC | AUPRC | Sensitivity | Specificity |
|---|---|---|---|---|---|
|
| |||||
| North America | North America | 0.99 | 0.98 | 94% | 96% |
| North America | Nepal | 0.96 | 0.88 | 52% | 99% |
| Nepal | North America | 0.62 | 0.36 | 44% | 69% |
| Nepal | Nepal | 0.97 | 0.91 | 73% | 99% |
| North America + Nepal | North America | 0.99 | 0.98 | 98% | 96% |
| North America + Nepal | Nepal | 0.98 | 0.92 | 82% | 99% |
AUROC, AUPRC, sensitivity, and specificity of all 3 models evaluated on the North American and Nepali test sets are reported. The model trained on both datasets resulted in slightly increased performance compared to models trained on individual datasets, most notably seen in sensitivity. For example, sensitivity increased from 94% when trained on North American data alone to 98% and from 73% when trained on Nepali data alone to 82%.
Performance on both test sets was slightly increased when applying the model trained on both the North American and Nepali datasets. While AUROC and AUPRC remained approximately the same for both test sets, sensitivity increased from 94% to 98% and from 73% to 82% respectively.
CNN Feature Analysis
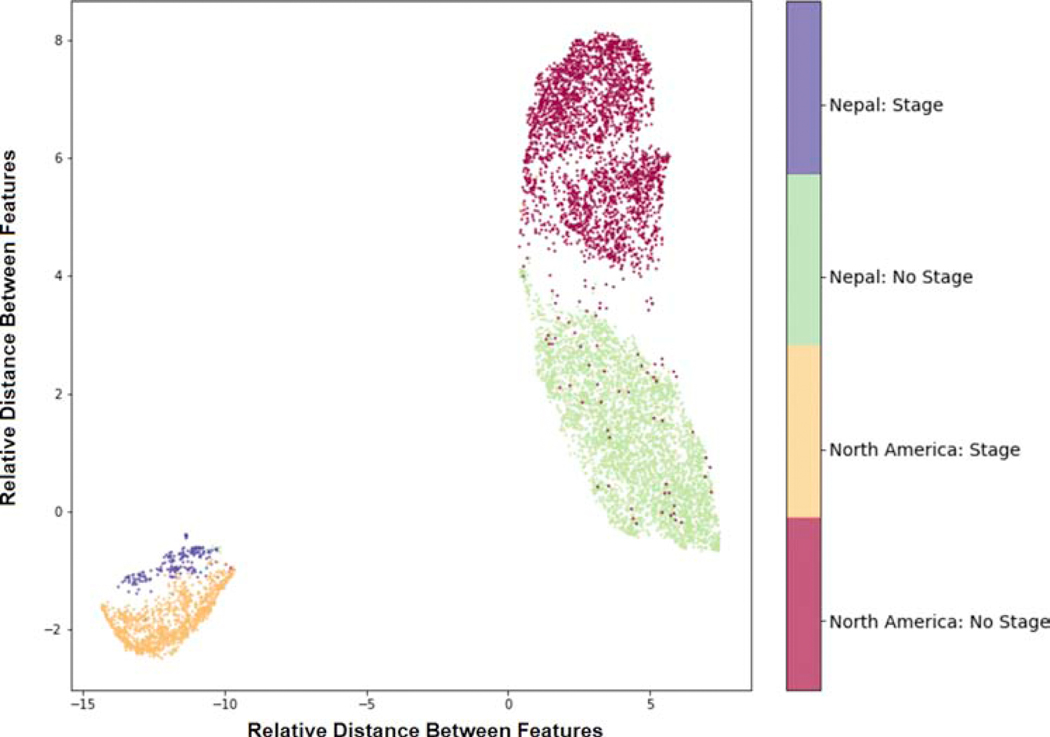
Figure 3 shows a uniform manifold approximation and projection (UMAP) diagram of the average pooling layer from the best performing model trained on images from the combined North American and Nepali datasets. The colored labels represent the presence of stage and dataset of origin.
Figure 3. Uniform Manifold Approximation and Projection (UMAP) of Feature Maps of Images.
A UMAP was created using the average pooling layer in the model trained on images from both North America and Nepal. Each feature map was reduced to 2 (unitless) dimensions and labeled based on stage and the dataset of origin. The clustering of labeled images demonstrates that the model learned not only the image features associated with the presence of stage, but also with the dataset of origin.
DISCUSSION
This study presents the results of three separate CNNs trained to identify stage 1–3 ROP and has implications for improving the generalizability of AI-diagnostic systems to real-world populations. The key findings from this study are: 1) CNNs can be trained to screen for stage 1–3 ROP in retinal images with high accuracy in images from the same population and camera, 2) the performance of trained CNNs suffers when tested on external samples that differ in features compared to the original training dataset, and 3) both internal and external performance can be improved by combining populations for training.
The first key finding is that CNNs can screen for the presence of stage 1–3 ROP in retinal fundus images with high accuracy from the same population and camera. We trained these CNNs using both the North American and Nepali datasets. The demographic differences of the Nepalese cohort are typical for populations screened in lower-income countries, where fewer extremely premature babies survive, and less premature babies remain at risk for ROP due to differences in neonatal care.36 Of note, variability in the cross-validation performances of the models trained on Nepali data was observed despite the use of equal sampling and may be attributed to the dataset’s significantly smaller number of images with stage. While there exists prior work using CNNs for both binary22,23 and multi-class20,21,24 classification of stage, it is difficult to directly compare performance between these systems due to differences in reported performance metrics as well as demographics and disease prevalence between populations. Since the majority of infants in ROP screening show no or mild disease,37–39 AI-based screening has the potential to increase clinical efficiency by providing real-time diagnosis and reducing time requirements for clinicians to grade eye exams for infants without clinically significant disease. Screening for ROP could be accomplished using the presence of stage, the presence of pre-plus or worse disease,40 or both. Since each eye exam consists of multiple retinal views, each of which can be an independent screening test for stage, the likelihood of missing stage in a given eye is low if it is present in the image set. Future work will need to evaluate the role of AI-based screening and how effective these systems perform in a typical ROP screening population.
Relatedly, our second key finding is that the performance of trained CNNs suffers when tested on external samples that differ in features compared to the original training dataset. This is consistent with previous work, representing a broad problem not only in ophthalmic applications of AI,41 but also throughout medicine.42–45 Factors contributing to dataset differences include demographic features such as gender46 and race,47 disease prevalence, imaging modality, and collection methods.8 Ensuring consistent generalizability prior to clinical deployment is a necessary but unmet need for AI algorithms. In ROP, differences in camera are a particularly important barrier to generalizability since most, if not all, CNNs have been trained using images from the RetCam camera, which is not widely available in low- and middle-income countries.
Therefore, there are opportunities to create CNNs on data from lower cost cameras in populations which may experience the greatest benefit from screening. To our knowledge, this is the first CNN optimized to identify stage using data from the Forus camera. The current precedent for AI screening devices approved by the Food and Drug Administration (FDA) is currently for a single camera system.16 While the FDA has proposed measures that may address generalizability by allowing iterative improvements to AI algorithms based on real-world performance,48 these will require independent regulatory approvals for each system since performance on one system does not necessarily translate to another. Beyond camera systems, rigorous evaluation of a model’s generalizability ought to be assessed across as many variables as possible to ensure that the efficacy seen in research translates to consistent real-world performance.
The third key finding is that both internal and external performance can be improved by combining populations for training. The improved performance of the combined model suggests that some degree of heterogeneity (different image characteristics) in the training data (Figure 3), rather than simply the added amount of training data, contributed to this increased performance. Interestingly, Figure 3 also demonstrates that the CNN retained strong classification performance of stage despite learning features unique to each population and/or camera. This may support the development of two-stage CNNs (i.e. one for each camera, and then one for classification), or may lead to more robust CNNs that perform well across platforms agnostic to which camera took the images. In addition to training using images from multiple camera systems, another method to overcome differences in learned features between camera systems is through the use of adversarial training, which aim to minimize a CNN’s ability to learn variables besides the outcome of interest.49 This would be especially useful if we want to deploy these algorithms in regions where expensive fundus cameras are unavailable, and lower-cost camera systems such as smartphone-based devices are being explored for clinical use. Images from these systems often differ in field of view, magnification, and image quality compared to RetCam images.50 One potential limitation to the approach of combining multiple datasets into a single combined dataset for training is that there are practical, ethical, and regulatory issues with data sharing that limit widespread use of multi-institutional dataset training. Future work may focus adaptively fine-tuning the model across multiple institutions without the data leaving the institution.51
Our study has additional limitations. First, our dataset was filtered for image quality, and thus is not representative of a real-world screening population. It will be important to establish quality thresholds as part of image analysis protocols since other real-world images may be lower quality and result in worse performance. Second, we tested preprocessing for the North American dataset and applied this preprocessing to Nepali images without testing other preprocessing methods on these images. This may in part explain the relatively lower performance on these data. Future studies should focus on preprocessing for different types of datasets as well as methods that standardize images such as CNN pipelines with segmentations.26,51,52 Third, while our UMAP showed that our CNNs learned features consistent with accurately identifying stage, interpreting the CNN features was beyond the scope of our study. To address explainability, there may be opportunities for future studies to elucidate CNN imaging features associated with stages 1–3. Fourth, our CNN only classified stage, one component of the ICROP classification. Future work should focus on achieving high performance on multi-class classification of stage and combining automated classifiers for zone, stage, and plus disease to predict need for treatment.
AI-based screening has the potential to improve the accessibility of expert-level diagnosis for multiple diseases. As we move from proof-of-principle efficacy studies to real-world effectiveness, there is a need to develop and validate generalizable models to different populations and camera systems. Overall, we will need to develop systems of care that integrate and sustain these technologies in health systems that lead to improved patient care, and in the case of ROP, more efficient, effective, and accessible ROP screening.
B) Test Sets
| No. | ||||
|---|---|---|---|---|
|
| ||||
| Dataset | Images | Patients | No Stage | Stage |
|
| ||||
| North America | 708 | 72 | 497 | 211 |
| Nepal | 247 | 54 | 203 | 44 |
Acknowledgments
Financial Support:
This study was supported by grants T15LM007088, R01EY19474, R01EY031331, K12EY027720, and P30EY10572 from the National Institutes of Health (Bethesda, MD); and by unrestricted departmental funding, a Medical Student Research Fellowship (JSC), and a Career Development Award (JPC) from Research to Prevent Blindness (New York, NY). The Nepal screening program was supported by support from the United States Agency for International Development and Helen Keller International.
Conflicts of Interest:
R.V. Paul Chan is on the Scientific Advisory Board for Phoenix Technology Group (Pleasanton, CA), a Consultant for Novartis (Basel, Switzerland), and a Consultant for Alcon (Ft. Worth, TX). Michael F. Chiang is a consultant for Novartis (Basel, Switzerland), and an equity owner of Inteleretina (Honolulu, HI). Michael F. Chiang, J. Peter Campbell, R.V. Paul Chan, and Jayashree Kalpathy-Cramer receive research support from Genentech. R.V. Paul Chan receives research support from Regeneron.
Footnotes
Meeting Presentation:
A preliminary version of this work was accepted as an abstract at ARVO, 2020.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Classification of Retinopathy of Prematurity*. The International Classification of Retinopathy of Prematurity Revisited. Arch Ophthalmol. 2005;123(7):991–999. [DOI] [PubMed] [Google Scholar]
- 2.Multicenter Trial of Cryotherapy for Retinopathy of Prematurity: Preliminary Results. Arch Ophthalmol. 1988;106(4):471–479. [DOI] [PubMed] [Google Scholar]
- 3.Revised Indications for the Treatment of Retinopathy of Prematurity: Results of the Early Treatment for Retinopathy of Prematurity Randomized Trial. Arch Ophthalmol. 2003;121(12):1684–1694. [DOI] [PubMed] [Google Scholar]
- 4.Wallace DK. Fellowship training in retinopathy of prematurity. J Am Assoc Pediatr Ophthalmol Strabismus JAAPOS. 2012;16(1):1. [DOI] [PubMed] [Google Scholar]
- 5.Nagiel A, Espiritu MJ, Wong RK, et al. Retinopathy of prematurity residency training. Ophthalmology. 2012;119(12):2644–5.e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong RK, Ventura CV, Espiritu MJ, et al. Training fellows for retinopathy of prematurity care: a Web-based survey. J AAPOS Off Publ Am Assoc Pediatr Ophthalmol Strabismus. 2012;16(2):177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blencowe H, Lawn JE, Vazquez T, Fielder A, Gilbert C. Preterm-associated visual impairment and estimates of retinopathy of prematurity at regional and global levels for 2010. Pediatr Res. 2013;74 Suppl 1(Suppl 1):35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scruggs BA, Chan RVP, Kalpathy-Cramer J, Chiang MF, Campbell JP. Artificial Intelligence in Retinopathy of Prematurity Diagnosis. Transl Vis Sci Technol. 2020;9(2):5–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang P, Grinband J, Weinberg BD, et al. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. Am J Neuroradiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Yan L-F, Zhang X, et al. Glioma Grading on Conventional MR Images: A Deep Learning Study With Transfer Learning. Front Neurosci. 2018;12:804–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Li L, Wormstone IM, et al. Development and Validation of a Deep Learning System to Detect Glaucomatous Optic Neuropathy Using Fundus Photographs. JAMA Ophthalmol. 2019;137(12):1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, He Y, Keel S, Meng W, Chang RT, He M. Efficacy of a Deep Learning System for Detecting Glaucomatous Optic Neuropathy Based on Color Fundus Photographs. Ophthalmology. 2018;125(8):1199–1206. [DOI] [PubMed] [Google Scholar]
- 14.Abràmoff MD, Lavin PT, Birch M, Shah N, Folk JC. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. Npj Digit Med. 2018;1(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gargeya R, Leng T. Automated Identification of Diabetic Retinopathy Using Deep Learning. Ophthalmology. 2017;124(7):962–969. [DOI] [PubMed] [Google Scholar]
- 16.Gulshan V, Peng L, Coram M, et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016;316(22):2402–2410. [DOI] [PubMed] [Google Scholar]
- 17.Burlina PM, Joshi N, Pacheco KD, Freund DE, Kong J, Bressler NM. Use of Deep Learning for Detailed Severity Characterization and Estimation of 5-Year Risk Among Patients With Age-Related Macular Degeneration. JAMA Ophthalmol. 2018;136(12):1359–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grassmann F, Mengelkamp J, Brandl C, et al. A Deep Learning Algorithm for Prediction of Age-Related Eye Disease Study Severity Scale for Age-Related Macular Degeneration from Color Fundus Photography. Ophthalmology. 2018;125(9):1410–1420. [DOI] [PubMed] [Google Scholar]
- 19.Zhao J, Lei B, Wu Z, et al. A Deep Learning Framework for Identifying Zone I in Ret CamImages. IEEE Access. 2019;7:103530–103537. [Google Scholar]
- 20.Ding A, Chen Q, Cao Y, Liu B. Retinopathy of Prematurity Stage Diagnosis Using Object Segmentation and Convolutional Neural Networks. Preprint. Posted April 3, 2020. arXiv:2004.01582. [Google Scholar]
- 21.Chen G, Zhao J, Zhang R, Wang T, Zhang G, Lei B. Automated Stage Analysis of Retinopathy of Prematurity Using Joint Segmentation and Multi-instance Learning. In: Fu H, Garvin MK, MacGillivray T, Xu Y, Zheng Y, eds. Ophthalmic Medical Image Analysis. Springer International Publishing; 2019:173–181. [Google Scholar]
- 22.Mulay Supriti, Ram Keerthi, Sivaprakasam Mohanasankar, Vinekar Anand. Early detection of retinopathy of prematurity stage using deep learning approach. SPIE. Proceedings Vol 10950. 2019. [Google Scholar]
- 23.Hu J, Chen Y, Zhong J, Ju R, Yi Z. Automated Analysis for Retinopathy of Prematurity by Deep Neural Networks. IEEE Trans Med Imaging. 2019;38(1):269–279. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y-P, Basanta H, Kang EY-C, et al. Automated detection of early-stage ROP using a deep convolutional neural network. Br J Ophthalmol. 2020:bjophthalmol-2020–316526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Z, Simkin S, Lai C, Dai S. Deep Learning Algorithm for Automated Diagnosis of Retinopathy of Prematurity Plus Disease. Transl Vis Sci Technol. 2019;8(6):23–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown JM, Campbell JP, Beers A, et al. Automated Diagnosis of Plus Disease in Retinopathy of Prematurity Using Deep Convolutional Neural Networks. JAMA Ophthalmol. 2018;136(7):803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan MC, Ostmo S, Jonas K, et al. Development and Evaluation of Reference Standards for Image-based Telemedicine Diagnosis and Clinical Research Studies in Ophthalmology. AMIA Annu Symp Proc AMIA Symp. 2014;2014:1902–1910. [PMC free article] [PubMed] [Google Scholar]
- 28.Bradski G The OpenCV Library. Dr Dobb’s Journal of Software Tools, 2000. [Google Scholar]
- 29.Virtanen P, Gommers R, Oliphant TE, et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat Methods. Published online 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Rossum G, Drake FL Jr. Python Reference Manual. Centrum voor Wiskunde en Informatica Amsterdam; 1995. [Google Scholar]
- 31.Raja DSS, Vasuki S. Automatic detection of blood vessels in retinal images for diabetic retinopathy diagnosis. Comput Math Methods Med. 2015;2015:419279–419279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X Image denoising using local Wiener filter and its method noise. Optik. 2016;127(17):6821–6828. [Google Scholar]
- 33.He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition.ArXiv151203385 Cs. Preprint. Posted December 10, 2015. arXiv:1512.03385. [Google Scholar]
- 34.Paszke A, Gross S, Massa F, et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In: Wallach H, Larochelle H, Beygelzimer A, Alché-Buc F textquotesingle, Fox E, Garnett R, eds. Advances in Neural Information Processing Systems 32. Curran Associates, Inc.; 2019:8024–8035. [Google Scholar]
- 35.McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. ArXiv180203426 Cs Stat. Preprint. Posted February 9, 2018. Last Revised December 6, 2018. arXiv:1802.03426. [Google Scholar]
- 36.Quinn GE. Retinopathy of prematurity blindness worldwide: phenotypes in the third epidemic. Eye Brain. 2016;8:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khorshidifar M, Nikkhah H, Ramezani A, et al. Incidence and risk factors of retinopathy of prematurity and utility of the national screening criteria in a tertiary center in Iran. Int J Ophthalmol. 2019;12(8):1330–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lad EM, Hernandez-Boussard T, Morton JM, Moshfeghi DM. Incidence of Retinopathy of Prematurity in the United States: 1997 through 2005. Am J Ophthalmol. 2009;148(3):451–458.e2. [DOI] [PubMed] [Google Scholar]
- 39.Yau GSK, Lee JWY, Tam VTY, et al. Incidence and risk factors for retinopathy of prematurity in multiple gestations: a Chinese population study. Medicine (Baltimore). 2015;94(18):e867–e867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Redd TK, Campbell JP, Brown JM, et al. Evaluation of a deep learning image assessment system for detecting severe retinopathy of prematurity. Br J Ophthalmol. 2019;103(5):580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ting DSW, Cheung CY-L, Lim G, et al. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA. 2017;318(22):2211–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang K, Beers AL, Brink L, et al. Multi-Institutional Assessment and Crowdsourcing Evaluation of Deep Learning for Automated Classification of Breast Density. J Am Coll Radiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Badgeley MA, Zech JR, Oakden-Rayner L, et al. Deep learning predicts hip fracture using confounding patient and healthcare variables. Npj Digit Med. 2019;2(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLOS Med. 2018;15(11):e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.AlBadawy EA, Saha A, Mazurowski MA. Deep learning for segmentation of brain tumors: Impact of cross-institutional training and testing. Med Phys. 2018;45(3):1150–1158. [DOI] [PubMed] [Google Scholar]
- 46.Larrazabal AJ, Nieto N, Peterson V, Milone DH, Ferrante E. Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proc Natl Acad Sci. 2020;117(23):12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adamson AS, Smith A. Machine Learning and Health Care Disparities in Dermatology. JAMA Dermatol. 2018;154(11):1247–1248. doi: 10.1001/jamadermatol.2018.2348 [DOI] [PubMed] [Google Scholar]
- 48.Health C for D and R. Artificial Intelligence and Machine Learning in Software as a Medical Device. FDA. https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device. Published online January 28, 2020. Accessed June 17, 2020. [Google Scholar]
- 49.Janizek JD, Erion G, DeGrave AJ, Lee S-I. An Adversarial Approach for the Robust Classification of Pneumonia from Chest Radiographs. Preprint. Posted on January 13, 2020. arXiv:2001.04051. [Google Scholar]
- 50.Patel TP, Aaberg MT, Paulus YM, et al. Smartphone-based fundus photography for screening of plus-disease retinopathy of prematurity. Graefes Arch Clin Exp Ophthalmol. 2019;257(11):2579–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mehta N, Lee CS, Mendonça LSM, et al. Model-to-Data Approach for Deep Learning in Optical Coherence Tomography Intraretinal Fluid Segmentation. JAMA Ophthalmol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Fauw J, Ledsam JR, Romera-Paredes B, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24(9):1342–1350. [DOI] [PubMed] [Google Scholar]