Abstract
A growing number of emerging SARS-CoV-2 variants is being identified worldwide, potentially impacting the effectiveness of current vaccines. We report the data obtained in several Italian regions involved in the SARS-CoV-2 variant monitoring from the beginning of the epidemic and spanning the period from October 2020 to March 2021.
Keywords: SARS-CoV-2 virus, Complete genome sequencing, COVID-19 RT-PCR testing, Spike protein, Viral variants
Background
Starting from April 2020 when the Cluster 5 [1] variant was first described, multiple SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2) variants have emerged in different parts of the world. The Alpha variant (formerly known as B.1.1.7) was first identified in the UK from a sample obtained in late September 2020 [2, 3] (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/#:~:text=Naming%20SARS%2DCoV%2D2%20variants,scientists%20and%20in%20scientific%20research), Beta variant (B.1.351) was identified in October 2020 in South Africa [4], and Gamma variant (P.1) was identified in Brazil in December 2020 [5]. These variant strains show multiple changes (deletions and substitutions) in the spike protein (9 in B.1.1.7, 10 in B.1.351, and 12 in P.1) compared with the reference genome Wuhan-Hu-1 sequence (EPI_ISL_406800), some of which affect the receptor-binding domain (RBD) region. The major issue with these variants is their potential to outcompete and rapidly replace formerly prevalent lineages, first in the areas where they likely emerged and subsequently spread in many other countries [6–8]. Increased SARS-CoV-2 diversity raises the concern of escape from pre-existing immunity, elicited either by previous infection or by vaccination.
Main text
Here, we report data from several Italian centers located in Campania, Lazio, Lombardy, Liguria, Marche, Piedmont, Tuscany, Sicily and Umbria, involved in the SARS-CoV-2 variant monitoring from the beginning of the epidemic and spanning the period from October 2020 to March 2021.
Globally, we analysed data from 3744 samples obtained by different techniques: RT-PCR variant screening assays (n = 2095), spike Sanger or Next Generation Sequencing (n = 649) and Whole Genome Sequencing (WGS; n = 1000) (Table 1). Data for the analyses were obtained from each center as a part of routine test of variant monitoring or for research purpose for those not involved in national surveillance but included in the SCIRE (SARS-CoV-2 Italian Research Enterprise) collaborative group. SARS-CoV-2 RNA positive swabs were collected from the respiratory tract of individuals who were either hospitalized or tested within screening programs. SARS-CoV-2 RNA was extracted using different commercial kits such as the Kit QIAsymphony DSP Virus/Pathogen Midi kit on the QIAsymphony automated platform (QIAGEN, Hilden, Germany), the NucleoMag 96 Virus (Macherey-Nagel, Dueren, Germany) on automated KingFisherTM ml Magnetic Particle Processors (Thermo Fisher Scientific, Waltham, MA, USA) and manually with QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany). RT-PCR variant screening assays were performed using TaqPath COVID-19 test (Thermo Fisher Scientific, USA), COVID-19 variant Catcher kit (Clonit srl, Milan, Italy), Allplex SARS-CoV-2 Variants (Arrow Diagnostics srl, Genoa, Italy) or multiplexed RT-qPCR developed by English consortium (https://www.protocols.io/view/multiplexed-rt-qpcr-to-screen-for-sars-cov-2-b-1-1-br9vm966?version_warning=no).
Table 1.
Summary of analyzed data
| Month | WGS (n = 1000) | Spike (n = 649) | Real time (n = 2095) | Total (n = 3744) |
|---|---|---|---|---|
| October 2020 | 137 | 20 | 1 | 158 |
| November 2020 | 168 | 31 | 0 | 199 |
| December 2020 | 139 | 34 | 0 | 173 |
| January 2021 | 291 | 86 | 388 | 765 |
| February 2021 | 193 | 214 | 827 | 1234 |
| March 2021 | 72 | 264 | 879 | 1215 |
WGS whole genome sequencing
Spike sequences were obtained using home-made protocols. Full genome sequences were obtained with different protocols, by a modified version of Artic Protocol (https://artic.network/ncov-2019) using Illumina DNA Prep and IDT ILMN DNA/RNA Index kit (Illumina), by Ion AmpliSeq SARS-CoV-2 Research Panel (Thermo Fisher Scientific, Waltham, Massachusetts, USA) or by CleanPlex® SARS-CoV-2 Panel (Paragon Genomics Inc, Hayward, CA, USA). Sequencing was performed on Illumina Miseq or Nextseq platforms for all samples except for those from Marche that were sequenced with Ion GeneStudioTMS5 System instrument. The results were mapped and aligned to the reference genome obtained from GISAID (https://www.gisaid.org/, accession ID: EPI_ISL_406800) using Geneious Prime software v. 9.1.5 (Biomatters, Auckland, New Zealand) (http://www.geneious.com) or Torrent Suite v. 5.10.1 (Euformatics Oy, Espoo, Finland) or BWA-mem and rescued using Samtools alignment/Map (Hinxton, UK) (v. 1.9).
SARS-CoV-2 sequences were classified using the Pangolin COVID-19 Lineage Assigner tool v. 3.1.5 (https://pangolin.cog-uk.io/) and Nextclade v. 1.4.0 (https://clades.nextstrain.org/). Mutations were identified using Nextclade.
The table shows the analyzed data stratified according to different methodologies used for the variant monitoring and months of sampling collection.
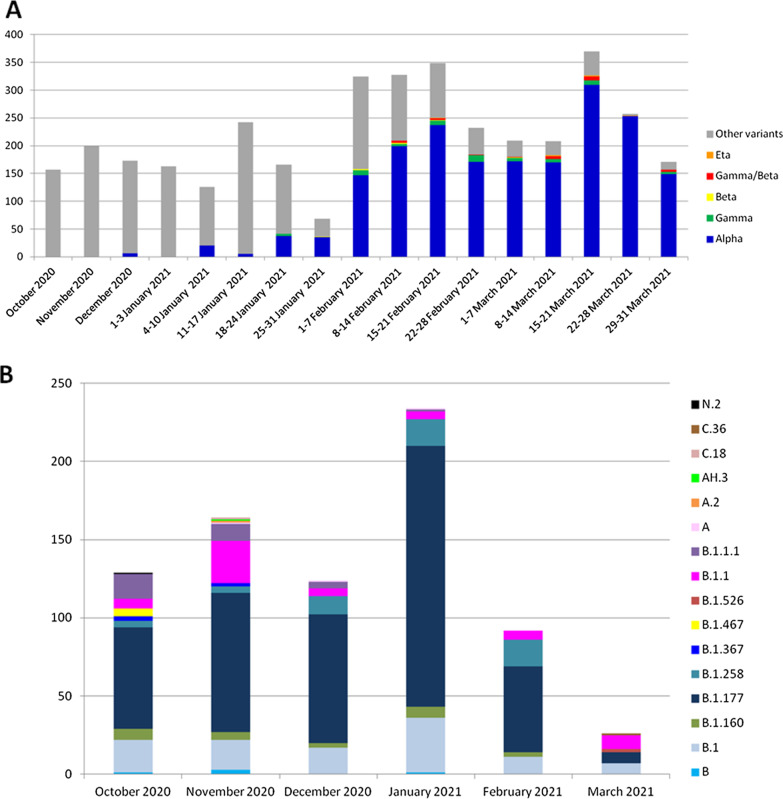
The number of samples considered for variant monitoring increased from 4.2% (158/3744) in October to 32% (1215/3744) in March, due to the increasing interest in viral variants at global level. In the study period, the Alpha variant (B.1.1.7) significantly increased, growing from 3.5% (6/173) in December, to 86.7% (1054/1215) in March (p < 0.001). Particularly, from the second half of December this lineage was already present in Liguria (n = 1 on the 18th), Campania (n = 3, on the 28th) and Marche (n = 2, on the 31st) (EPI_ISL_778869, EPI_ISL_778868), with only one known case related to return from UK. The most important increase in its prevalence was observed from the second week of January (20/126, 16.0%) to the end of the same month (35/68, 51.5%) reaching 73.7% (171/232) at the end of February. In parallel, the prevalence of previously circulating variants significantly decreased from 100% (158/158) to 9.4% (114/1215). The Gamma variant (P.1) was first detected in the second half of January in Campania (n = 1), Lombardy (n = 1) and Umbria (n = 2), subsequently setting at around 2% in February (33/1234) and March (24/1215). Only few Beta and Eta variants (B.1.351 and B.1.525 respectively) cases were identified, never reaching 1% of the total samples analysed, being firstly detected in Liguria at the end of January and in Campania in mid-February, respectively. The B.1.258 lineage (also known as Scottish variant) was firstly observed in Campania, Marche and Piedmont in mid-October, 4 cases were reported in November in Campania (n = 2) and Piedmont (n = 2), thereafter it was only reported in Campania until February reaching a proportion of 18.5% (17/92) of total cases of other variants, including its descendant lineages (B.1.258.3, B.1.258.14). In half of March, two cases of the Iota variant (formally known as New York lineage B.1.526) were identified in Marche. Figure 1 shows the main viral variants observed overtime. Concerning other variants, the main circulating lineage was B.1.177 that represented, together with its descendents (B.1.177.4, B.1.177.8, B.1.177.14, B.1.177.15, B.1.177.52, B.1.177.53, B.1.177.75), more than half of cases (50.4%, 54.3%, 66.1%, 71.4% and 59.8% from October to February). Despite limited data regarding lineage assignment in March (n = 28), we observed a 27% prevalence of B.1 and B.1.177 lineages (Fig. 1, Panel B). Clade assignment was available for a larger number of strains compared to lineage because these data was obtained also by spike sequencing highlighting the clear predominance of 20E(EU1) ranging from 43.2% (68/157) to 67.7% (189/279). Some noteworthy mutations were observed in different lineages: N439K in one lineage A and B.1, E484K in three B.1.177 and A222V + E484K + N501Y in clade 20E.
Fig. 1.
SARS-CoV-2 viral variants observed overtime in Italy. A highlights the variants of concern; B shows all other circulating lineages
Conclusion
This study provides insights into the rapid change in the prevalence of SARS-CoV-2 variants over six months in Italy. Interestingly, we observed the first entry of Alpha variant in Lombardy in late December, in line with the identification of first retrospective data in UK on 20 September [9–11] and those reported by ISS-Istituto Superiore di Sanità (https://www.iss.it/documents/20126/0/11-6-2021+SECONDO+BOLLETTINO+Varianti+SARS-CoV-2.pdf/7febfab3-c32b-7505-cc1e-2176fc12cdf6?t=1623430432808). The introduction of Alpha variant, notoriously associated to higher transmissibility compared to other variants and to an increased rate of mortality reported in some studies, corresponds to the second relevant increase of infections registered in Italy during the second epidemic wave [12–14].
In addition to known variants of concern, we documented other minor variants that could be early warnings of upcoming changes. Continuous monitoring of all variants is mandatory to comprehensively investigate and keep track of virus evolution, particularly along with expanding vaccination still based on original strains that have been largely substituted by novel variants.
Acknowledgements
Members of the collaborative group SCIRE: Claudia Balotta, Maria Gori, Patrizia Bagnarelli, Andreina Baj, Federica Novazzi, Andrea Orsi, Patrizia Caligiuri, Simona Boccotti, Maria Concetta Bellocchi, Loredana Sarmati, Massimo Andreoni, Nicasio Mancini, Elena Criscuolo, Rosa Gallitelli, Sophie Testa, Filippo Dragoni, Maurizio Zazzi. We thank the members of the NGS and bioinformatics core facilities of the Laboratory of Molecular Medicine and Genomics for WGS data generation and analysis and Gianluigi Franci, Massimiliano Galdiero, Pasquale Pagliano, Giuseppe Portella and Teresa Rocco for their scientific contribution.
Abbreviations
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- RBD
Receptor-binding domain
- RT-PCR
Real time polymerase chain reaction
- WGS
Whole genome sequencing
Authors' contributions
Conceptualization, MC and MG; formal analysis, AL, AB, SM, GZ, SC, VG, FR, FM, FC, GG, AW, CT, BB, FCS, NC, AC, FS, DF, EVR., IV.; investigation, AL, AB; resources, SM, GZ, VG, FM, AW, BB, FSC, MC, MG; writing—original draft preparation, AL, AB; writing—review and editing, AL, AB; visualization and supervision, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part by Regione Campania (POR Campania FESR 2014/2020 grants: “Monitoring the spread and genomic variability of the Covid 19 virus in Campania using NGS technology”, CUP B14I20001980006 and azione 1.5 “GENOMAeSALUTE”, CUP: B41C17000080007) and by Ministero della Salute COVID-2020-12371817. The funding sources had no role in the study design, data collection analyses, interpretation or writing of the report.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. All consensus genomes are being submitted at the GISAID database (https://www.gisaid.org).
Declarations
Ethics approval and consent to participate
The study was approved by the Ethical Committee of the Sacco Hospital (Prot N. 0012266).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Alessia Lai and Annalisa Bergna contributed equally to this manuscript.
Contributor Information
Alessia Lai, Email: alessia.lai@unimi.it.
collaborative group SCIRE SARS-CoV-2 Italian Research Enterprise:
Claudia Balotta, Maria Gori, Patrizia Bagnarelli, Andreina Baj, Federica Novazzi, Andrea Orsi, Patrizia Caligiuri, Simona Boccotti, Maria Concetta Bellocchi, Loredana Sarmati, Massimo Andreoni, Nicasio Mancini, Elena Criscuolo, Rosa Gallitelli, Sophie Testa, Filippo Dragoni, and Maurizio Zazzi
References
- 1.Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–9. [DOI] [PubMed]
- 4.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 5.Faria NR, Mellan TA, Whittaker C, Claro IM, Candido D, Mishra S, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funk T, Pharris A, Spiteri G, Bundle N, Melidou A, Carr M, et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7; B.1.351 or P.1: data from seven EU/EEA countries; weeks 38/2020 to 10/2021. Euro Surveill. 2021;26:2100348. doi: 10.2807/1560-7917.ES.2021.26.16.2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.European Centre for Disease Prevention and Control . SARS-CoV-2—increased circulation of variants of concern and vaccine rollout in the EU/EEA; 14th update—15 February 2021. Stockholm: ECDC; 2021. [Google Scholar]
- 8.Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, et al. Emergence of SARS-CoV-2 B.1.1.7 lineage—United States; December 29; 2020-January 12; 2021. MMWR Morb Mortal Wkly Rep. 2021;70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Giallonardo F, Puglia I, Curini V, Cammà C, Mangone I, Calistri P, et al. Emergence and spread of SARS-CoV-2 lineages B.1.1.7 and P.1 in Italy. Viruses. 2021;13:794. doi: 10.3390/v13050794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo Menzo S, Marinello S, Biasin M, Terregino C, Franchin E, Crisanti A, Cattelan A. The first familial cluster of the B.1.1.7 variant of SARS-CoV-2 in the northeast of Italy. Infection. 2021;1–5. [DOI] [PMC free article] [PubMed]
- 11.Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. ARTIC Network. 2020.
- 12.Davies NG, Jarvis CI, CMMID COVID-19 Working Group. Edmunds WJ, Jewell NP, Diaz-Ordaz K, Keogh RH. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grint DJ, Wing K, Williamson E, McDonald HI, Bhaskaran K, Evans D, Evans SJ, et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Euro Surveill. 2021;26:2100256. doi: 10.2807/1560-7917.ES.2021.26.11.2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Challen R, Brooks-Pollock E, Read JM, Dyson L, Tsaneva-Atanasova K, Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. All consensus genomes are being submitted at the GISAID database (https://www.gisaid.org).