Abstract
Nowadays, pollution of the environment is a huge problem for humans and other organisms' health. Conventional methods of pollutant removal like membrane filtration or ion exchange are not efficient enough to lower the number of pollutants to standard levels. Biological methods, because of their higher efficiency and biocompatibility, are preferred for the remediation of pollutants. These cost-effective and environment-friendly methods of reducing pollutants are called bioremediation. In bioremediation methods, enzymes play the most crucial role. Enzymes can remedy different types of organic and inorganic pollutants, including PAHs, azo dyes, polymers, organocyanides, lead, chromium, and mercury. Different enzymes isolated from various species have been used for the bioremediation of pollutants. Discovering new enzymes and new subtypes with specific physicochemical characteristics would be a promising way to find more efficient and cost-effective tools for the remediation of pollutants.
1. Introduction
The widespread use of chemicals in industries and militaries, inadequate waste disposal, and accidental leakage cause contamination of soil, water, and air. For instance, there are 34,000 contaminated sites just in Europe that need to be treated. These pollutants are hazardous for humans, other living beings, and even the biogeochemical cycle. Pollutants' stability, low solubility, and resistance to various physical, chemical, and biological degradation pathways are the main reasons for their toxicity [1].
Different physical and chemical methods for cleaning up pollutants have been used, such as oxidizing agents, electrochemical treatments, adsorption of pollutants, ion exchange, and membrane filtration [2]. Despite the adequacy of traditional methods for the high concentration of pollutants, they were not enough for lowering the amount of contamination to regulatory limits [3]. Various disadvantages of traditional methods for cleaning up pollutants include high cost, nonspecificity, and probable secondary contamination production; therefore, ecofriendly and biological methods, called bioremediation, gained interest [4].
Bioremediation is defined as processes and products that are cost-effective and practical to minimize pollutants in the source and diminish danger to the environment and human health [5]. Its main ways of degrading and detoxifying pollutants are through intracellular accumulation or enzymatic transformation [4]. Pollutant properties (i.e., chemical structure, hydrophobicity, and polarity), environmental conditions (i.e., temperature, pH, and redox condition), and soil features (i.e., aggregation, thickness, dissolved organic matter, and pollutants aging) affect biological degradation and contaminants availability [6].
Enzymes are the most efficient bioremediation tools and progress all chemical changes on pollutants. Enzymes' specificity is usually broad enough to act on different molecules with similar structures. Moreover, it is possible to engineer the enzymes for enhancing their stability and efficiency for special conditions or particular substrates [7, 8]. Omics technologies have a significant role in these developments [2].
Using enzymes in bioremediation could be either individually that the isolated enzyme used and added to the contaminated area or as a whole cell, e.g., bacteria, fungi, or algae. In a second way, continuous aeration, inoculation, and nutrition are necessary. Besides, environmental conditions should be prepared for microorganisms living, even though there might still be some toxic compounds in the environment that inhibit microorganisms' activity [1, 9]. The use of individual enzymes has some advantages in comparison with microbial whole cell including greater specificity, more straightforward handling and storage, standardizable activity, more mobility as a result of smaller size, being active in the presence of high concentrations of toxic compounds, and biodegradability that inhibits persistence and recalcitrance [1, 10, 11]. This approach is much more efficient for extracellular enzymes and cofactor-independent enzymes [12, 13].
Enzyme production in the natural environment is low, while it is possible to increase the produced enzyme under controlled conditions. On the other hand, recombinant DNA technology and gene engineering provide many opportunities to produce more efficient and more enzymes [14]. Moreover, nanotechnology offers some tools to increase enzymes' stability by decreasing sensitivity to mechanical stress, preserving the third structure of enzymes, and protecting them against proteases [9].
Enzymatic bioremediation could be in situ or ex situ. In in situ methods with the least disturbance in the environment, the free or immobilized enzyme (adsorbed enzymes on mineral supports that minimize the loss of enzymatic activity) is added to the soil. This approach is less expensive because of no need for excavation and transportation of soil. Ex situ methods are feasible for highly contaminated soils with toxic pollutants or when fast action is essential. During this procedure, soil was excavated and treated in different bioreactors in the best condition for enzymes' activity [1] (Figure 1).
Figure 1.
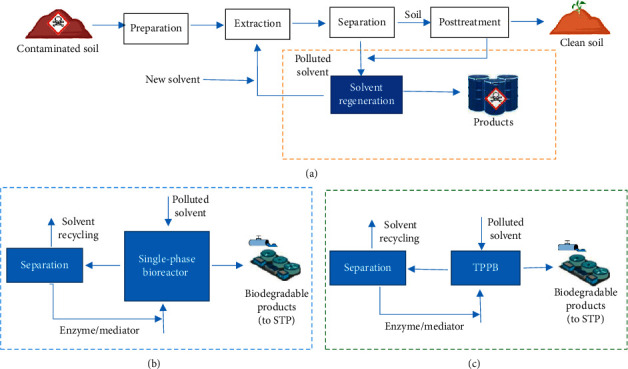
A representative scheme of different methods of soil remediation. (a) Conventional in situ remediation. (b) Using a single-phase bioreactor for solvent extraction. (c) Using a two-phase bioreactor for solvent extraction. Adapted from [1].
Different enzymes like mono- or dioxygenases, halogenases, peroxidases, phosphotriesterases, hydrolases, transferases, and oxidoreductases from various species of bacteria, fungi, algae, and plants have been used for the bioremediation of pollutants [10, 15]. We try to review the most essential enzymes for the bioremediation of pollutants and insight into their mechanism of action.
2. Enzymes for Organic Substrates
Large amounts of organic pollutants, including herbicides, pesticides, dyes, drugs, and plastics, pollute the air, soil, and water every year. Polymers, aromatic molecules, polycyclic aromatic hydrocarbons (PAHs), chlorinated hydrocarbons, steroids, and organocyanides are the most organic compounds that need to be cleaned up worldwide. Their stable structure is the main reason for their toxicity.
2.1. Hydrolases (EC 3)
Esterases, nitrilases, aminohydrolases, lipase, cutinase, and organophosphorus hydrolase are among the hydrolase enzymes used in the bioremediation of different chemicals such as herbicides, pesticides, organophosphorus compounds, nitrile compounds, and polymers [1, 2]. We would review some of them shortly as follows.
2.2. Esterases (3.1)
Esterases catalyze the cleavage of ester bonds in different chemicals like organophosphorus herbicides and pesticides, diethyl glycol adipate, polyurethanes, and aromatic and aliphatic polyesters. Escherichia coli and Pichia pastoris are two bacteria that express and colonize the thermostable kind of enzymes. Moreover, a subgroup of esterases found in E. coli is active in a cold environment and can act on phthalate esters [2].
It is worth noting that the product of esterase reaction with organophosphorus compounds, 3,5,6-trichloro-2-pyridinol (TCP), is metabolized later to less toxic chemicals by aminohydrolase (EC 3.5) [2].
2.3. Nitrilases (EC 3.5.5.1)
Triple bonds between carbon and nitrogen (nitrile group) of herbicides, polymers, and plastics are hydrolyzed stereo-, regio-, or chemoselectively by nitrilases to carboxylic acid and ammonia. Many species can express these enzymes, including Streptomyces sp., Fusarium solani, Rhodococcus rhodochrous, Aspergillus niger, Bacillus pallidus, and Pseudomonas fluorescens. Moreover, an evolution approach on Alcaligenes faecalis tends to isolate a nitrilase that was active in the broader range of pH. Besides, P. fluorescens nitrilase's gene expressed in E. coli is probably the most hopeful nitrilase [16, 17]. Cyanide dihydratase (EC 3.5.5) is one of the nitrilases and degrade cyanide into formate and ammonia. Pseudomonas stutzeri and Bacillus pumilus are two species that express this enzyme. Furthermore, fungal cyanide hydratase (EC 4.2.1.66), isolated from Fusarium lateritium, Neurospora crassa, and Gloeocercospora sorghi, and some other species, is another cyanide-degrading enzyme that metabolizes it to formamide [18]. These enzymes are promising for the bioremediation of wastewaters from coal coking and metal-plating baths [17].
2.4. Organophosphorus Hydrolase (EC 3.1.8.2)
Organophosphate compounds were developed and used as pesticides and in warfare and even as a drug since 1937. They are neurotoxic, and after a while, they were more than that soil microbiota could remedy all of them. Organophosphorus hydrolase (also known as phosphotriesterase) is one of the enzymes that can serve for organophosphorus compounds bioremediation. It is mostly isolated from Pseudomonas diminuta, although its fungal form is expressed in Aspergillus niger and Penicillium lilacinum. It can act on P-S, P-O, and P-F bonds. This enzyme has Zn2+ as a cofactor in its native form, while assays showed that substitution of Co2+ provides the most potent activity against paraoxon [19]. This enzyme has the fastest catalytic rate and is the most promising enzyme for engineering activity against organophosphates [20].
2.4.1. Peroxidases
(1) Ligninolytic Peroxidases. Ligninolytic enzymes are a family of enzymes with broad applications in bioremediation. This group of enzymes produced by white-rot fungi (WRF) is in the condition of nutrient limitation known as “ligninolytic.” Also, lignocellulosic materials can be an inducer for the production of these enzymes [21]. Due to the high nonspecificity and high nonstereoselectivity of these enzymes, they can degrade a wide range of recalcitrant compounds [22]. They degrade chemicals by pseudo-first-order kinetic via a free-radical-based chain reaction using H2O2 and molecular oxygen [21–24].
Ligninolytic enzymes can be categorized into four main enzymes, including laccase (LAC), lignin peroxidase (LiP), manganese peroxidase (MnP), and versatile peroxidase (VP).
(2) Laccase. For oxidizing phenolic compounds, PAHs, dyes, and pesticides benzenediol: oxygen oxidoreductase, known as laccase, is a suitable enzyme. As an oxidase, laccase substrates go through one of the following pathways: (1) cleavage of aromatic rings, (2) polymerization, and (3) degradation of covalent bonds between monomers. Four atoms of copper are the principal part of the reaction, and oxygen is the last electron receptor [2, 25]. The mechanism of the reaction is shown in Figure 2.
Figure 2.
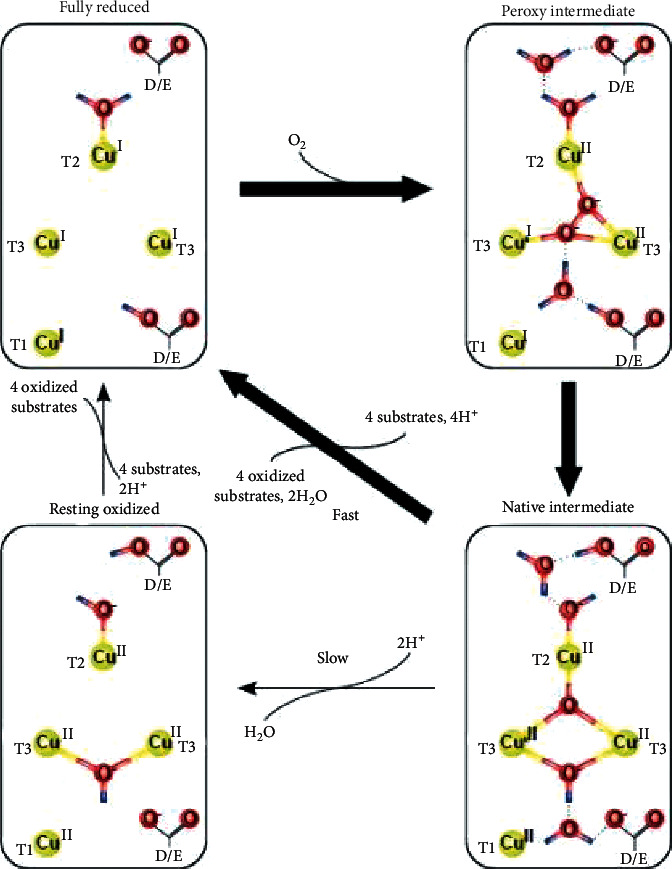
General reaction mechanism of bacterial laccases. Adapted from [26].
Laccase is first discovered in different fungi species like Panus conchatus and Polyporus sp. Later on, laccase was found in Azospirillum lipoferum, as the first bacteria species. Laccase is produced in different Gram-positive bacteria, including Bacillus, Geobacillus, Aquisalibacillus, Lysinibacillus, Staphylococcus, and Streptomyces. Many bacteria produce laccase extracellularly, while some others are unable to secrete the enzyme. Bacterial laccase is more resistant to extreme temperature and pH conditions [1, 25].
There are two kinds of laccase, white and blue. The main difference between these is that blue laccase is dependent on a “mediator” for the degradation of nonphenolic substrates. “Mediator” is an intermediator that laccase oxidizes and turns into oxidized radicals that react with high redox potential or bulky substrates. ABTS (2,20-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid)) and N-heterocycles with N-OH such as violuric acid, N-hydroxybenzotriazole, and N-hydroxy-N-phenylacetamide have been used as effective mediators [25].
Every year, approximately 7 × 104 − 1 × 107 tons of dyes penetrate the environment [25]. Laccase is used for dyes remediation. As an example, a Bacillus licheniformis LS40-derived laccase can decolorize azo, indigo, and anthraquinone dyes by 80% within one hour in the presence of acetosyringone as a mediator [27].
PAHs are xenobiotic pollutants because of their low solubility and degradation rate. Laccase can convert PAHs to their less toxic quinine form and CO2. There are some examples in Table 1. Notably, laccase can degrade some drugs such as diclofenac and mefenamic acid in acidic pH [25].
Table 1.
Examples of laccase's pollutant bioremediation.
(3) Lignin Peroxidase. Lignin peroxidases (LiPs) are a group of heme-containing monomeric enzymes. Their weight ranges between 38 and 43 kDa [30, 31] with iron in the ferric state [21, 32]. LiPs with their high redox potential [32] are capable of breaking alpha and beta carbon bonds, catalyzing the degradation of phenolic and nonphenolic compounds, demethylation, and opening aromatic ring of dyes [33]. LiPs have a high redox potential for oxidizing nonphenolic structures [31].
LiP activity increases in the presence of H2O2 as an electron acceptor. However, high concentrations of H2O2 could damage the LiPs [32]. In the first step of the reaction, Fe3+ binds to H2O2 and oxo-ferryl intermediate named compound I forms. Then, compound I, by a donation of one electron from the substrate, reduces to compound II, finally; by another electron donation from the substrate, iron in heme returns to its ferric resting state, and the enzyme renews to its initial form [31, 34]. In this three-step reaction, the reduction of compound II is the rate-limiting step (Figure 3) [36]. Due to this slow reduction rate, compound I is available for reaction with H2O2 and the formation of a complex between LiP and superoxide (compound III) inactive enzyme [36].
Figure 3.
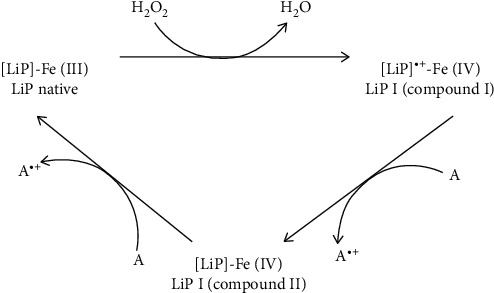
Lignin peroxides catalytic reaction. Adapted from [35].
Veratryl alcohol is a secondary metabolite that can play essential roles in this oxidizing reaction. Veratryl alcohol can be the mediator in the electron transfer reaction; it can play a role in the catalytic cycle of LiP by an oxidizing terminal substrate. Vertaryl alcohol can also prevent the formation of compound III and, if compound III is established, reduce it to its native form [36].
Many WRFs produce LiPs such as Phanerochaete chrysosporium, Trametes versicolor, Bjerkandera adusta, Phlebia radiate, and Ganoderma lucidum [22, 31].
Many technologies have been applied to enhance activity and increase catalytic characteristics of LiPs, such as LiP entrapment in calcium beads [37].
(4) Manganese Peroxidase. Manganese peroxidases (MnPs) are heme-containing glycol proteins with weight ranging from 32 to 62.5 kDa [38]. Like other ligninolytic peroxidases, MnP uses H2O2. By using H2O2, MnP can oxidase Mn2+ to Mn3+. The first step of the reaction is binding an oxygen atom of H2O2 to Fe3+ of heme. Then, by two-electron transfer from Fe3+ to peroxide Fe4+ oxo-porphyrin, compound I radical forms. Then, compound I binds to monochelated Mn2+ and Mn3+ and compound II forms. Finally, by oxidizing another Mn2+ to Mn3+, compound II reduces and the enzyme with Fe3+ reforms (Figure 4) [32, 36].
Figure 4.
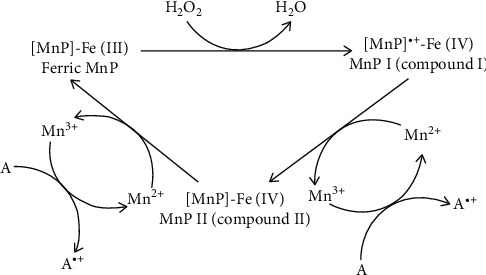
Manganese peroxidase catalytic reaction. Adapted from [35].
Aliphatic organic acids such as lactate and oxalate can induce Mn2+ oxidation rate, and Mn3+-acid chelates have a higher redox potential. MnP activity increases in the presence of glutathione and unsaturated fatty acids, such as tween 80. Many techniques have been utilized to immobilize and enhance the efficacy of bioremediation with MnP, such as making calcium alginate beads and carbon nanotubes [39–41].
MnP can remediate PAHs and nitroaromatic compounds [36, 42], azo dyes [43], and endocrine-disrupting chemicals such as bisphenol A and alkylphenols [44, 45]; moreover, with the contribution of mediators such as lipid and thiyl radicals, MnP is capable of oxidizing nonphenolic structures [34].
Many species of fungi are able to produce MnP, such as Phanerochaete chrysosporium, Trametes versicolor, Irpex lacteus, Dichomitus squalens, and Ganoderma lucidum [45, 46].
(5) Versatile Peroxidase. Versatile peroxidase (VP) is a heme-containing ligninolytic enzyme considered as a hybrid between LiP and MnP. VP has two active sites; therefore, it can oxidize both Mn2+ and veratryl alcohol by a similar mechanism to MnP and LiP, respectively [32, 47].
VP can oxidize both low and high redox potential compounds, polycyclic aromatic hydrocarbons, azo dyes, high molecular weight aromatics, and both phenolic and nonphenolic compounds and environmental pollutants [32, 47, 48].
VP production is less common in WRFs than MnP and LiP, but it can be found in some species such as Pleurotus spp. and Bjerkandera spp. [32].
2.4.2. Horseradish Peroxidase
Horseradish peroxidase (HRP) is an enzyme traditionally extracted and isolated from the root of horseradish (Armoracia rusticana). The most abundant isoenzyme found in the root of horseradish is C isoenzyme (HRPC). HPRC is 44 kDa heme-containing glycopeptide with 308 amino acids, an iron atom in the ferric state in protoporphyrin IX, and two calcium atoms in the central zone [49–51]. HRP catalyzes oxidative reaction using H2O2. In the presence of H2O2, the intermediate compound formed via two-electron oxidation. Then by an oxidable substrate, compound I reduces to compound II. Radical formation occurs via these reactions, and finally, the initial enzyme can be renewed by the reaction of compound II with another substrate molecule. In comparison with LiP compound, I and II are more electronegative in HRP (Figure 5) [52].
Figure 5.
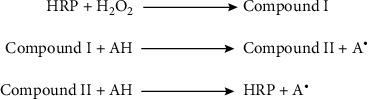
Horseradish peroxidase catalytic cycle. Adapted from [52].
HRP is applicable for removing and remediating phenols, substrate phenols, and alkylphenols, aromatic amines [53, 54], azo dyes [55, 56], endocrine-disrupting compounds [54], and many other environmental pollutants.
Many techniques have been utilized to immobilize and enhance the efficacy of enzyme by nanotechnology [57–59]. Using horseradish root is a standard method; using fertile soil for horseradish cultivation to feed the population has raised concern in recent years [60]. To solve this problem and enhance the efficacy of the enzyme, many biotechnological methods have been experienced, such as recombinant production of HRP in E. coli, yeast, plants, and insect systems [61].
2.5. Cytochrome p450 Monooxygenase (EC 1.14.14.1)
Cytochrome p450 monooxygenases (CYP) are a family of heme-containing enzymes that catalyze different reactions such as N-hydroxylation, N-dealkylation, O-dealkylation, oxidative dehalogenation, and hydroxylation of C-H bonds. CYP derives essential electrons for reactions from NADPH-cytochrome p450 reductase, and the latter enzyme derives electrons from atmospheric oxygen. So, the presence of a reducing agent like NAD (P) H or FAD is necessary [62]. The reaction cycle of CYP450 is shown in Figure 6.
Figure 6.
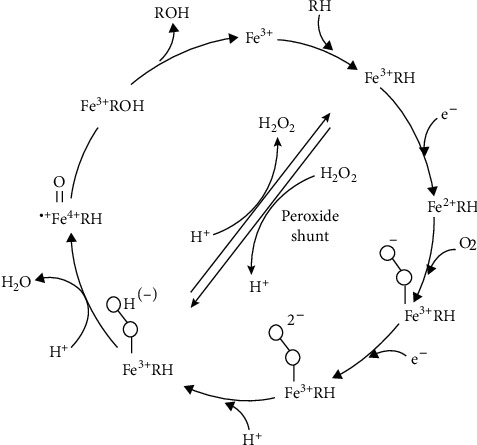
Cytochrome p450 reaction cycle. RH: substrate; ROH: product. Adapted from [63].
CYPs are versatile enzymes expressed in various species of bacteria, fungi, plants, and animals. About 7000 different CYPs have been discovered till now. Saccharomyces, Streptomyces, Basidiomycete, Dehalococcoides, Rhodococcus, Bacillus, Escherichia, and Salmonella are among the genera that their CYPs are used for bioremediation [64, 65].
While bacterial CYPs are attractive because of their solubility, easy and low-cost production, and self-efficiency (their electron transfer reductases, e.g., FMN, FAD, and p450 monooxygenase, are on a single peptide), mammalian CYPs are membrane-bounded, dependent on a redox partner (e.g., NADPH) and have expansive applications [65]. Bacterial and eukaryotic CYPs can oxidize aliphatic hydrocarbons with 5–16 and 10–16 carbon lengths, respectively [66]. Notably, eukaryotic CYPs need modification at N-terminal, but prokaryotic ones are active in the native form [64].
Dioxins, PCBs (polychlorinated biphenyls), PCDDs (polychlorinated dibenzo-p-dioxins), PCDFs (polychlorinated dibenzofurans), PAHs, aliphatic hydrocarbons, and even Cr (VI) are pollutants that can be degraded and bioremedied by CYPs [1, 14, 65, 67]. In Table 2, the list of different CYPs and their substrates are shown. Immobilizing CYPs can improve their activity even to 10-folds higher than free enzyme. Besides, transgenic plants that can produce special CYPs are a way toward herbicide-resistant plants [65].
Table 2.
Cytochrome p450 subtypes and their substrate for bioremediation.
| Enzyme | Substrate | Reference | |
|---|---|---|---|
| 1 | Rat CYP1A1 | Dibenzo-p-dioxin (DD) and mono-, di-, and trichloro-DD | [65, 67] |
| 2 | F240A | 2,3,7,3,8-Tetrachloro-DD | [65] |
| 3 | CYP101, CYP102, CYP1A1, CYP1A2, CYP1B1 | PAHs | [65] |
| 4 | Dog CYP2B11 | PCBs | [65] |
| 5 | CYP1A1, CYP1A2, CYP1B1 | Low-chlorinated PCDDs | [68] |
| 6 | CYP5145A3 | 1-MCDD, 2-MCDD, 2,3-DCDD | [68] |
| 7 | CYP XplA | RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine) | [69] |
CYPs are interesting enzymes for bioremediation because of their wide range of substrates and diverse oxidative reactions. Among the limitations of using CYPs are their dependency on expensive cofactors, low stability, and low activity [68].
3. Enzymes for Inorganic Substrates
In the presence of toxic heavy metals, most of the microorganisms produce metal-binding peptides such as phytochelatins and metallothioneins, which reduce their toxicity via sequestration [70]. For example, phytochelatin synthase is the enzyme responsible for the production of phytochelatin that, in cooperation with GSH, accumulates heavy metals [71]. Among the limitations of these metal-binding proteins is their nonselectivity. To solve this problem, many microorganisms developed specific pathways for resistance against heavy metals [4]. Obviously, enzymes are the most critical part of these pathways; we would review some of these metal-specific enzymes as follows.
3.1. Arsenic
Arsenic is a heavy metal that exists in nature in organic and inorganic forms. The inorganic forms (As3+ (arsenite) and As5+ (arsenate)) are toxic and may cause enzyme inactivation, carcinoma, hemolysis, keratosis, gangrene, and neurological and cardiovascular diseases [72, 73]. Arsenate and arsenite convert to each other by arsenate reductase and arsenite oxidase through redox reactions. As3+ is more mobile and toxic. As5+ is the terminal electron acceptor in the absence of oxygen and reduces to As3+ [63]. Ferredoxin or glutathione would be the electron source [74]. This process enhances the solubility of As and eases leaching from soil [73]. The final As3+ is excreted through efflux pumps, ArsB and Acr3 [74]. Arsenite oxidase converts As3+ to less toxic As5+ to be used either for a supplemental energy source or as an electron donor for CO2 fixation [74]. The final arsenate is immobile and would be retained by sediments [73].
The methylated form of arsenic is volatile and would be lost from the soil [73]. Interestingly, in methanogenic bacteria, As methylation is coupled with methane biosynthesis and can detoxify soil through this mechanism. Coenzyme M is the biocatalyst of this detoxification process [63].
Many species can remedy As in different ways. The bacterial ones include Acinetobacter sp., Pseudomonas sp., and Sporosarcina ginsengisoli [75]. E. coli, Bacillus idriensis, and Sphingomonas desiccabilis are engineered species for As bioremediation [72]. Some fungi, including Rhizobium sp., Rhizopus sp., Trichoderma sp., Aspergillus flavus, and Penicillium canescens, are As bioremediators too [63, 73]. Moreover, some yeasts like Saccharomyces cerevisiae can reduce arsenate by ArsC (Figure 7), a protein that has As reductase activity. Algae genera, like Hydrodictyon, Oedogonium, Rhizoclonium, and even a plant, Pteris vittata from Pteridaceae, have the potential to be used for bioremediation [75].
Figure 7.
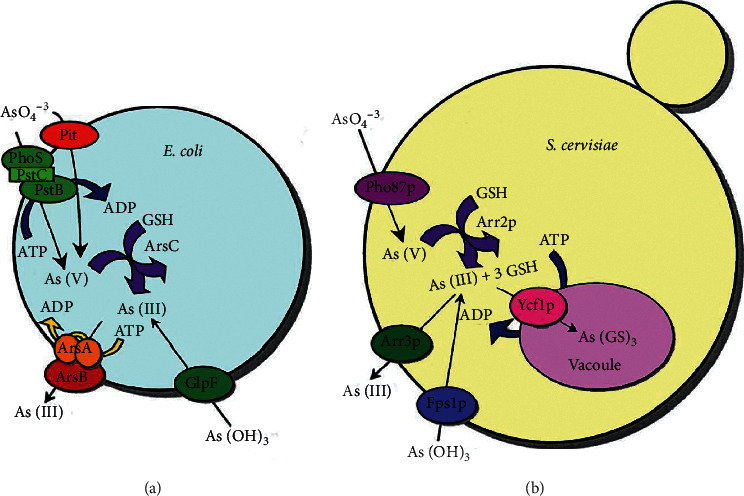
A strategy used for arsenic detoxification using E. coli and S. cerevisiae and their enzymes. Adapted from [63].
3.2. Lead
Lead was found in a small amount in nature before industrialization. However, now, through gasoline burning, different Pb salts originate in and contaminate water, soil, and air [72]. Lead toxicity may cause anemia and appetite loss and gastrointestinal, neurological, and reproductive disorders [73]. Organoleads, especially tetraethyl lead and tetramethyl lead used in gasoline, are toxic forms of lead. They are sensitive to photolysis and volatilization and degrade to dialkyl species. Though, some bacteria can degrade organoleads through bioremediation processes [76].
Cupriavidus metallidurans can remove Pb2+ ions with p-type ATPase and produce inorganic phosphate to sequester Pb2+ in the periplasm [76]. Staphylococcus epidermidis can biomineralize Pb2+ by carbonate. Urease enzymes form different carbonate crystalline Pb2+. It can be mineralized as oxalate and pyromorphite, too [77]. Agaricus bisporus, Rhizopus nigricans, Penicillium canescens, Penicillium chrysogenum, Saccharomyces cerevisiae, Aspergillus niger, and Aspergillus terreus are among biotransforming organisms [72, 73]. Moreover, it is reported that Arthrobacter and Phaeolus schweinitzii can degrade trimethyl lead cations [78].
3.3. Mercury
Mercury is a heavy metal that is toxic in both organic and inorganic forms, although the organic form is more toxic. Hg toxicity would cause neurotoxicity, nephrotoxicity, allergies, and inability to speak [73, 79]. Hg is a rare element in Earth crust, but it spreads and pollutes soil and water because of different humic activities like gold mining, various measurement tools (barometer, thermometer, manometer, etc.), lamps, mercurial fungicides, paper manufacturing industry, and battery cells [72]. Its environmental cycle is shown in Figure 8.
Figure 8.
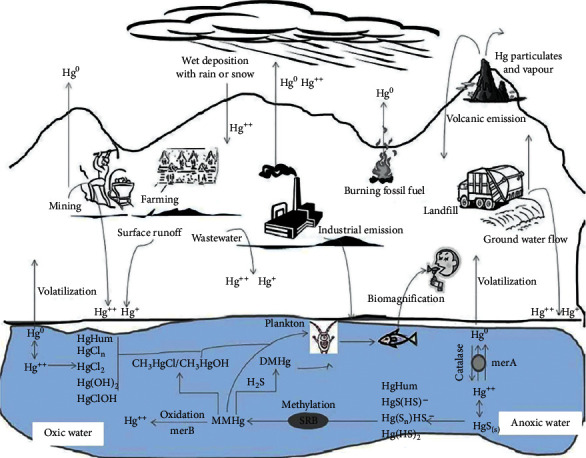
Mercury cycle in the environment. Adapted from [80].
Mercury exists in three forms: metallic mercury (Hg0), mercurous (Hg+1), and mercuric (Hg2+) forms. The most toxic form of Hg is mercuric chloride. Organic mercury can accumulate in living organisms and has an affinity for proteins' sulfhydryl groups. Inorganic mercury has the lowest toxicity because of its low solubility and high vapor pressure. Mercury-resistant bacteria (such as Pseudomonas, Aeromonas, Staphylococcus, Escherichia, Citrobacter, Bacillus, and Rhodococcus) can reduce toxic organic forms of Hg to less toxic metallic Hg. Mercuric reductase is the main enzyme that reduces Hg. The mer operon is the collection of mercury-resistance genes activated in the presence of an inducible concentration of Hg. Mercuric reductase in cooperation with FAD and NADPH, as electron sources, reduces Hg2+ to Hg0. The final metallic mercury is volatile and spreads to the atmosphere [80, 81]. Also, dimethylmercury is volatile and biomethylation can serve as a strategy for Hg bioremediation [73]. The mer operon-independent volatilization of mercury has been discovered, too, in Shewanella oneidensis [82].
Another enzyme that plays a role in mercury bioremediation is organomercurial lyase that breaks the carbon-mercury bonds in organo-Hg compounds [80, 81].
Various microorganisms such as Rhizopus arrhizus, Penicillium canescens, Geobacter sulfurreducens, Pseudomonas putida, Acinetobacter calcoaceticus, Staphylococcus aureus, and Shigella flexneri can remedy mercury [72, 80]. Enterobacter, Pseudomonas, and Bacillus are the most used genera for this purpose [83].
3.4. Chromium
Cr (VI) is the most toxic heavy metal because of its high oxidative potential causing cell damage and mutagenic, carcinogenic, and teratogenic effects [84]. The wide use of chromium and its compounds and mining exerts this pollutant to waters and soils. Bioremediation of hexavalent chromium is through reduction to trivalent species. Pseudomonas, Bacillus, Escherichia, Shewanella, Enterobacter, and Thermus are some genera that are resistant to Cr (VI) and can reduce it. The reduction of hexavalent chromium may occur through aerobic or anaerobic pathways [14]. In the anaerobic process, soluble cytoplasmic enzymes are involved and reduce hexavalent chromium in two steps. In the aerobic reduction of chromium, usually, Cr (VI) is a terminal electron acceptor, while in different species, NADPH, NADH, or formate serves as an electron donor. Chromate reductase, Ni-Fe dehydrogenase, and cytochrome c3 are among the enzymes reported to have hexavalent chromium-reducing activity [85]. Also, Fe2+ and S2- produced in some bacteria can reduce Cr6+ even faster than chromate-reducing bacteria. The detailed mechanism of chromate resistance in bacteria is shown in Figure 9.
Figure 9.
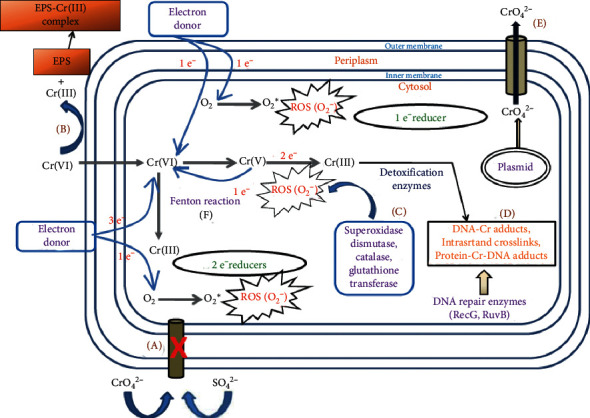
Chromate resistance mechanism in bacteria. (A) Mutation in sulfate uptake transporters. (B) Extracellular reduction of Cr6+ to Cr3+. (C) Intracellular reduction of Cr6+ to Cr3+ by chromate reductase. (D) Reducing oxidative stress and activation of repairing systems. (E) Outflowing of chromate from the cytoplasm. (F) Decreasing oxidative stress by activation of ROS scavenging enzyme. Adapted from [14].
Nitroreductase, iron reductase, flavin reductases, and quinone reductases are bacterial enzymes that reduce Cr6+ [86, 87]. Mammals reduce this pollutant, too, by CYP, aldehyde oxidase, and DT-diaphorase. Some of these bacterial enzymes are extracellular, including nitrate reductases, flavin reductases, and ferrireductases [14].
4. Conclusion
In this review, we aim to provide an insight into the role of the enzyme in the bioremediation of pollutants. While many physical and chemical methods of treating contaminated soil and water are not efficient enough, bioremediation opens a new way to clean up toxic pollutants. Enzymes as practical tools of living organisms are an ecofriendly and bio-based strategy for bioremediation. Microorganisms exposed to contaminated sites and specific pollutants are fascinating sources for the isolation of active enzymes against those pollutants. Interestingly, we may find some enzymes in completely irrelevant places to pollutant sources. Discovering TCP-degrading enzymes and chlorpyrifos-degrading enzymes in the cow rumen microbiome is an instance for this claim [2].
Overall, using enzymes for pollutant bioremediation seems to be a cost-effective, efficient, and practical approach. Although there are still many ways to go, further studies and experiments on enzyme activity and mechanism of action and isolating new enzymes would be a promising way to reduce pollutants and make a healthier environment for humans and all other species.
Contributor Information
Ahmad Gholami, Email: gholami@sums.ac.ir.
Chin Wei Lai, Email: cwlai@um.edu.my.
Wei-Hung Chiang, Email: whchiang@mail.ntust.edu.tw.
Data Availability
The data used to support this study are available upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Eibes G., Arca-Ramos A., Feijoo G., Lema J. M., Moreira M. T. Enzymatic technologies for remediation of hydrophobic organic pollutants in soil. Applied Microbiology and Biotechnology. 2015;99(21):8815–8829. doi: 10.1007/s00253-015-6872-y. [DOI] [PubMed] [Google Scholar]
- 2.Ufarté L., Laville É., Duquesne S., Potocki-Veronese G. Metagenomics for the discovery of pollutant degrading enzymes. Biotechnology Advances. 2015;33(8):1845–1854. doi: 10.1016/j.biotechadv.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Malik A. Metal bioremediation through growing cells. Environment International. 2004;30(2):261–278. doi: 10.1016/j.envint.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Singh S., Kang S. H., Mulchandani A., Chen W. Bioremediation: environmental clean-up through pathway engineering. Current Opinion in Biotechnology. 2008;19(5):437–444. doi: 10.1016/j.copbio.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Kirchhoff M. M. Promoting green engineering through green chemistry. Environmental Science & Technology. 2003;37(23):5349–5353. doi: 10.1021/es0346072. [DOI] [PubMed] [Google Scholar]
- 6.Rao M. A., Scelza R., Acevedo F., Diez M. C., Gianfreda L. Enzymes as useful tools for environmental purposes. Chemosphere. 2014;107:145–162. doi: 10.1016/j.chemosphere.2013.12.059. [DOI] [PubMed] [Google Scholar]
- 7.Theerachat M., Emond S., Cambon E., et al. Engineering and production of laccase from Trametes versicolor in the yeast Yarrowia lipolytica. Bioresource Technology. 2012;125:267–274. doi: 10.1016/j.biortech.2012.07.117. [DOI] [PubMed] [Google Scholar]
- 8.Festa G., Autore F., Fraternali F., Giardina P., Sannia G. Development of new laccases by directed evolution: functional and computational analyses. Proteins: Structure, Function, and Bioinformatics. 2008;72(1):25–34. doi: 10.1002/prot.21889. [DOI] [PubMed] [Google Scholar]
- 9.Rayu S., Karpouzas D. G., Singh B. K. Emerging technologies in bioremediation: constraints and opportunities. Biodegradation. 2012;23(6):917–926. doi: 10.1007/s10532-012-9576-3. [DOI] [PubMed] [Google Scholar]
- 10.Rao M. Role of enzymes in the remediation of polluted environments. Journal of Soil Science and Plant Nutrition. 2010;10(3):333–353. doi: 10.4067/s0718-95162010000100008. [DOI] [Google Scholar]
- 11.Gianfreda L., Rao M. A. Potential of extra cellular enzymes in remediation of polluted soils: a review. Enzyme and Microbial Technology. 2004;35(4):339–354. doi: 10.1016/j.enzmictec.2004.05.006. [DOI] [Google Scholar]
- 12.Scott C., Pandey G., Hartley C. J., et al. The enzymatic basis for pesticide bioremediation. Indian Journal of Microbiology. 2008;48(1):65–79. doi: 10.1007/s12088-008-0007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutherland T., Horne I., Weir K., et al. Enzymatic bioremediation: from enzyme discovery to applications. Clinical and Experimental Pharmacology and Physiology. 2004;31(11):817–821. doi: 10.1111/j.1440-1681.2004.04088.x. [DOI] [PubMed] [Google Scholar]
- 14.Thatoi H., Das S., Mishra J., Rath B. P., Das N. Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: a review. Journal of Environmental Management. 2014;146:383–399. doi: 10.1016/j.jenvman.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Pieper D. H., Martins dos Santos V. A., Golyshin P. N. Genomic and mechanistic insights into the biodegradation of organic pollutants. Current Opinion in Biotechnology. 2004;15(3):215–224. doi: 10.1016/j.copbio.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Nigam V. K., Arfi T., Kumar V., Shukla P. Bioengineering of nitrilases towards its use as green catalyst: applications and perspectives. Indian Journal of Microbiology. 2017;57(2):131–138. doi: 10.1007/s12088-017-0645-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínková L., Rucká L., Nešvera J., Pátek M. Recent advances and challenges in the heterologous production of microbial nitrilases for biocatalytic applications. World Journal of Microbiology and Biotechnology. 2017;33(1):p. 8. doi: 10.1007/s11274-016-2173-6. [DOI] [PubMed] [Google Scholar]
- 18.Park J. M., Trevor Sewell B., Benedik M. J. Cyanide bioremediation: the potential of engineered nitrilases. Applied Microbiology and Biotechnology. 2017;101(8):3029–3042. doi: 10.1007/s00253-017-8204-x. [DOI] [PubMed] [Google Scholar]
- 19.Singh B. K., Walker A. Microbial degradation of organophosphorus compounds. FEMS Microbiology Reviews. 2006;30(3):428–471. doi: 10.1111/j.1574-6976.2006.00018.x. [DOI] [PubMed] [Google Scholar]
- 20.Thakur M., Medintz I. L., Walper S. A. Enzymatic bioremediation of organophosphate compounds-progress and remaining challenges. Frontiers in Bioengineering and Biotechnology. 2019;7:p. 289. doi: 10.3389/fbioe.2019.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barr D. P., Aust S. D. Pollutant degradation by white rot fungi. Reviews of Environmental Contamination and Toxicology. 1994;138:49–72. doi: 10.1007/978-1-4612-2672-7_3. [DOI] [PubMed] [Google Scholar]
- 22.Kaur H., Kapoor S., Kaur G. Application of ligninolytic potentials of a white-rot fungus Ganoderma lucidum for degradation of lindane. Environmental Monitoring and Assessment. 2016;188(10):p. 588. doi: 10.1007/s10661-016-5606-7. [DOI] [PubMed] [Google Scholar]
- 23.Reddy C. A., Mathew Z. British Mycological Society Symposium Series. Amsterdam, Netherlands: Elsevier; 2001. Bioremediation potential of white rot fungi. [Google Scholar]
- 24.Pointing S. B. Feasibility of bioremediation by white-rot fungi. Applied Microbiology and Biotechnology. 2001;57(1-2):20–33. doi: 10.1007/s002530100745. [DOI] [PubMed] [Google Scholar]
- 25.Chauhan P. S., Goradia B., Saxena A. Bacterial laccase: recent update on production, properties and industrial applications. 3 Biotech. 2017;7(5):p. 323. doi: 10.1007/s13205-017-0955-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solomon E. I., Augustine A. J., Yoon J. O2 reduction to H2O by the multicopper oxidases. Dalton Transactions. 2008;30(30):3921–3932. doi: 10.1039/b800799c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu L., Zhao M., Wang T.-N., et al. Characterization and dye decolorization ability of an alkaline resistant and organic solvents tolerant laccase from Bacillus licheniformis LS04. Bioresource Technology. 2012;115:35–40. doi: 10.1016/j.biortech.2011.07.111. [DOI] [PubMed] [Google Scholar]
- 28.Madhavi V., Lele S. Laccase: properties and applications. Bio Resources. 2009;4(4):1694–1717. [Google Scholar]
- 29.Rajeswari M., Bhuvaneswari V. Production of extracellular laccase from the newly isolated Bacillus sp. PK4. African Journal of Biotechnology. 2016;15(34):1813–1826. [Google Scholar]
- 30.Akbar M. T., Habib A. M. An insight into the lignin peroxidase of Macrophomina phaseolina. Bioinformation. 2013;9(14):730–735. doi: 10.6026/97320630009730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falade A. O., Nwodo U. U, Iweriebor B. C, Green E, Mabinya L. V, Okoh A. I. Lignin peroxidase functionalities and prospective applications. Microbiology Open. 2017;6 doi: 10.1002/mbo3.394.e00394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Yao B., Su X. Linking enzymatic oxidative degradation of lignin to organics detoxification. International Journal of Molecular Sciences. 2018;19(11):p. 3373. doi: 10.3390/ijms19113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christian V. Mediator role of veratryl alcohol in the lignin peroxidase-catalyzed oxidative decolorization of Remazol Brilliant Blue R. Enzyme and Microbial Technology. 2005;36(2-3):327–332. doi: 10.1016/j.enzmictec.2004.09.006. [DOI] [Google Scholar]
- 34.Abdel-Hamid A., Solbiati J. O., Cann I. K. O. Insights into lignin degradation and its potential industrial applications. Advances in Applied Microbiology. 2013;82:p. 1. doi: 10.1016/B978-0-12-407679-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 35.Abdel-Hamid A. M., Solbiati J. O., Cann I. K. O. Insights into lignin degradation and its potential industrial applications. Advances in Applied Microbiology. 2013;82:1–28. doi: 10.1016/b978-0-12-407679-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 36.Cerniglia C. E. Biodegradation of polycyclic aromatic hydrocarbons. Current Opinion in Biotechnology. 1993;4(3):331–338. doi: 10.1016/0958-1669(93)90104-5. [DOI] [Google Scholar]
- 37.Shaheen R., Asgher M., Hussain F., Bhatti H. N. Immobilized lignin peroxidase from Ganoderma lucidum IBL-05 with improved dye decolorization and cytotoxicity reduction properties. International Journal of Biological Macromolecules. 2017;103:57–64. doi: 10.1016/j.ijbiomac.2017.04.040. [DOI] [PubMed] [Google Scholar]
- 38.Gold M. H., Alic M. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiological Reviews. 1993;57(3):605–622. doi: 10.1128/mmbr.57.3.605-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bilal M., Asgher M. Dye decolorization and detoxification potential of Ca-alginate beads immobilized manganese peroxidase. BMC Biotechnology. 2015;15(1):p. 111. doi: 10.1186/s12896-015-0227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bilal M., Asgher M., Iqbal M., Hu H., Zhang X. Chitosan beads immobilized manganese peroxidase catalytic potential for detoxification and decolorization of textile effluent. International Journal of Biological Macromolecules. 2016;89:181–189. doi: 10.1016/j.ijbiomac.2016.04.075. [DOI] [PubMed] [Google Scholar]
- 41.Chen M., Zeng G., Lai C., et al. Interactions of carbon nanotubes and/or graphene with manganese peroxidase during biodegradation of endocrine disruptors and triclosan. Chemosphere. 2017;184:127–136. doi: 10.1016/j.chemosphere.2017.05.162. [DOI] [PubMed] [Google Scholar]
- 42.Qin X., Zhang J., Zhang X., Yang Y. Induction, purification and characterization of a novel manganese peroxidase from Irpex lacteus CD2 and its application in the decolorization of different types of dye. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0113282.e113282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mielgo I., Lopez C., Moreira M. T., Feijoo G., Lema J. M. Oxidative degradation of azo dyes by manganese peroxidase under optimized conditions. Biotechnology Progress. 2003;19(2):325–331. doi: 10.1021/bp020136w. [DOI] [PubMed] [Google Scholar]
- 44.Hirano T., Honda Y., Watanabe T., Kuwahara M. Degradation of bisphenol A by the lignin-degrading enzyme, manganese peroxidase, produced by the white-rot Basidiomycete, Pleurotus ostreatus. Bioscience, Biotechnology, and Biochemistry. 2000;64(9):1958–1962. doi: 10.1271/bbb.64.1958. [DOI] [PubMed] [Google Scholar]
- 45.Moon D.-S., Song H.-G. Degradation of alkylphenols by white rot fungus Irpex lacteus and its manganese peroxidase. Applied Biochemistry and Biotechnology. 2012;168(3):542–549. doi: 10.1007/s12010-012-9795-4. [DOI] [PubMed] [Google Scholar]
- 46.Xu H., Guo M. Y., Gao Y. H., Bai X. H., Zhou X. W. Expression and characteristics of manganese peroxidase from Ganoderma lucidum in Pichia pastoris and its application in the degradation of four dyes and phenol. BMC Biotechnology. 2017;17(1):19–12. doi: 10.1186/s12896-017-0338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asgher M., Bhatti H. N., Ashraf M., Legge R. L. Recent developments in biodegradation of industrial pollutants by white rot fungi and their enzyme system. Biodegradation. 2008;19(6):771–783. doi: 10.1007/s10532-008-9185-3. [DOI] [PubMed] [Google Scholar]
- 48.Knop D., Levinson D., Makovitzki A., et al. Limits of versatility of versatile peroxidase. Applied and Environmental Microbiology. 2016;82(14):4070–4080. doi: 10.1128/aem.00743-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao X., Huang S., Dong P., et al. Horseradish peroxidase (HRP): a tool for catalyzing the formation of novel bicoumarins. Catalysis Science & Technology. 2016;6(10):3585–3593. doi: 10.1039/c5cy01682g. [DOI] [Google Scholar]
- 50.Bretz R. R., de Castro A. A., Lara Ferreira I. F., Ramalho T. C., Silva M. C. Experimental and theoretical affinity and catalysis studies between halogenated phenols and peroxidases: understanding the bioremediation potential. Ecotoxicology and Environmental Safety. 2020;202 doi: 10.1016/j.ecoenv.2020.110895.110895 [DOI] [PubMed] [Google Scholar]
- 51.Veitch N. C. Horseradish peroxidase: a modern view of a classic enzyme. Phytochemistry. 2004;65(3):249–259. doi: 10.1016/j.phytochem.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 52.Köller G., Möder M., Czihal K. Peroxidative degradation of selected PCB: a mechanistic study. Chemosphere. 2000;41(12):1827–1834. doi: 10.1016/s0045-6535(00)00132-6. [DOI] [PubMed] [Google Scholar]
- 53.Tonegawa M., Dec J., Bollag J.-M. Use of additives to enhance the removal of phenols from water treated with horseradish and hydrogen peroxide. Journal of Environment Quality. 2003;32(4):1222–1227. doi: 10.2134/jeq2003.1222. [DOI] [PubMed] [Google Scholar]
- 54.Zheng W., Colosi L. M. Peroxidase-mediated removal of endocrine disrupting compound mixtures from water. Chemosphere. 2011;85(4):553–557. doi: 10.1016/j.chemosphere.2011.06.064. [DOI] [PubMed] [Google Scholar]
- 55.Mohan S. V., Prasad K. K., Rao N. C., Sarma P. N. Acid azo dye degradation by free and immobilized horseradish peroxidase (HRP) catalyzed process. Chemosphere. 2005;58:1097–1105. doi: 10.1016/j.chemosphere.2004.09.070. [DOI] [PubMed] [Google Scholar]
- 56.Onder S., Celebi M., Altikatoglu M., Hatipoglu A., Kuzu H. Decolorization of naphthol blue black using the horseradish peroxidase. Applied Biochemistry and Biotechnology. 2011;163(3):433–443. doi: 10.1007/s12010-010-9051-8. [DOI] [PubMed] [Google Scholar]
- 57.Li J., Chen X., Xu D., Pan K. Immobilization of horseradish peroxidase on electrospun magnetic nanofibers for phenol removal. Ecotoxicology and Environmental Safety. 2019;170:716–721. doi: 10.1016/j.ecoenv.2018.12.043. [DOI] [PubMed] [Google Scholar]
- 58.Alshawafi W. M., Aldhahri M., Almulaiky Y. Q., et al. Immobilization of horseradish peroxidase on PMMA nanofibers incorporated with nanodiamond. Artificial Cells, Nanomedicine, and Biotechnology. 2018;46(3):S973–S981. doi: 10.1080/21691401.2018.1522321. [DOI] [PubMed] [Google Scholar]
- 59.Šekuljica N. Ž. Immobilization of horseradish peroxidase onto kaolin. Bioprocess and Biosystems Engineering. 2016;39(3):461–472. doi: 10.1007/s00449-015-1529-x. [DOI] [PubMed] [Google Scholar]
- 60.Singh S., Malhotra S., Mukherjee P., et al. Peroxidases from an invasive Mesquite species for management and restoration of fertility of phenolic-contaminated soil. Journal of Environmental Management. 2020;256 doi: 10.1016/j.jenvman.2019.109908.109908 [DOI] [PubMed] [Google Scholar]
- 61.Krainer F. W., Glieder A. An updated view on horseradish peroxidases: recombinant production and biotechnological applications. Applied Microbiology and Biotechnology. 2015;99(4):1611–1625. doi: 10.1007/s00253-014-6346-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lamb S. B., Lamb D. C., Kelly S. L., Stuckey D. C. Cytochrome P450 immobilisation as a route to bioremediation/biocatalysis. FEBS Letters. 1998;431(3):343–346. doi: 10.1016/s0014-5793(98)00771-6. [DOI] [PubMed] [Google Scholar]
- 63.Sher S., Rehman A. Use of heavy metals resistant bacteria-a strategy for arsenic bioremediation. Applied Microbiology and Biotechnology. 2019;103(15):6007–6021. doi: 10.1007/s00253-019-09933-6. [DOI] [PubMed] [Google Scholar]
- 64.Lamb D. C., Kelly D. E., Masaphy S., Jones G. L., Kelly S. L. Engineering of heterologous cytochrome P450 in Acinetobacter sp.: application for pollutant degradation. Biochemical and Biophysical Research Communications. 2000;276(2):797–802. doi: 10.1006/bbrc.2000.3541. [DOI] [PubMed] [Google Scholar]
- 65.Kumar S. Engineering cytochrome P450 biocatalysts for biotechnology, medicine and bioremediation. Expert Opinion on Drug Metabolism & Toxicology. 2010;6(2):115–131. doi: 10.1517/17425250903431040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pinto É. S. M., Dorn M., Feltes B. C. The tale of a versatile enzyme: alpha-amylase evolution, structure, and potential biotechnological applications for the bioremediation of n-alkanes. Chemosphere. 2020;250 doi: 10.1016/j.chemosphere.2020.126202.126202 [DOI] [PubMed] [Google Scholar]
- 67.Sakaki T., Yamamoto K., Ikushiro S. Possibility of application of cytochrome P450 to bioremediation of dioxins. Biotechnology and Applied Biochemistry. 2013;60(1):65–70. doi: 10.1002/bab.1067. [DOI] [PubMed] [Google Scholar]
- 68.Sakaki T. Practical application of cytochrome P450. Biological and Pharmaceutical Bulletin. 2012;35(6):844–849. doi: 10.1248/bpb.35.844. [DOI] [PubMed] [Google Scholar]
- 69.Rylott E. L., Jackson R. G., Sabbadin F., et al. The explosive-degrading cytochrome P450 XplA: biochemistry, structural features and prospects for bioremediation. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2011;1814(1):230–236. doi: 10.1016/j.bbapap.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 70.Cobbett C., Goldsbrough P. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annual Review of Plant Biology. 2002;53(1):159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]
- 71.Kang S. H., Singh S., Kim J.-Y., Lee W., Mulchandani A., Chen W. Bacteria metabolically engineered for enhanced phytochelatin production and cadmium accumulation. Applied and Environmental Microbiology. 2007;73(19):6317–6320. doi: 10.1128/aem.01237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pratush A., Kumar A., Hu Z. Adverse effect of heavy metals (As, Pb, Hg, and Cr) on health and their bioremediation strategies: a review. International Microbiology. 2018;21(3):97–106. doi: 10.1007/s10123-018-0012-3. [DOI] [PubMed] [Google Scholar]
- 73.Kapahi M., Sachdeva S. Bioremediation options for heavy metal pollution. Journal of Health and Pollution. 2019;9(24) doi: 10.5696/2156-9614-9.24.191203.191203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamamura S., Amachi S. Microbiology of inorganic arsenic: from metabolism to bioremediation. Journal of Bioscience and Bioengineering. 2014;118(1):1–9. doi: 10.1016/j.jbiosc.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 75.Ojuederie O. B., Babalola O. O. Microbial and plant-assisted bioremediation of heavy metal polluted environments: a review. International Journal of Environmental Research and Public Health. 2017;14(12) doi: 10.3390/ijerph14121504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Naik M. M., Dubey S. K. Lead resistant bacteria: lead resistance mechanisms, their applications in lead bioremediation and biomonitoring. Ecotoxicology and Environmental Safety. 2013;98:1–7. doi: 10.1016/j.ecoenv.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 77.Rahman Z., Singh V. P. Bioremediation of toxic heavy metals (THMs) contaminated sites: concepts, applications and challenges. Environmental Science and Pollution Research. 2020;27(22):27563–27581. doi: 10.1007/s11356-020-08903-0. [DOI] [PubMed] [Google Scholar]
- 78.Teeling H., Cypionka H. Microbial degradation of tetraethyl lead in soil monitored by microcalorimetry. Applied Microbiology and Biotechnology. 1997;48(2):275–279. doi: 10.1007/s002530051050. [DOI] [PubMed] [Google Scholar]
- 79.Azimi S., Moghaddam M. S. Effect of mercury pollution on the urban environment and human health. Environment and Ecology Research. 2013;1(1):12–20. doi: 10.13189/eer.2013.010102. [DOI] [Google Scholar]
- 80.Mahbub K. R., Bahar M. M., Labbate M., et al. Bioremediation of mercury: not properly exploited in contaminated soils! Applied Microbiology and Biotechnology. 2017;101(3):963–976. doi: 10.1007/s00253-016-8079-2. [DOI] [PubMed] [Google Scholar]
- 81.Singh S., Kumar V. Mercury detoxification by absorption, mercuric ion reductase, and exopolysaccharides: a comprehensive study. Environmental Science and Pollution Research. 2020;27(22):27181–27201. doi: 10.1007/s11356-019-04974-w. [DOI] [PubMed] [Google Scholar]
- 82.Xu J., Bravo A. G., Lagerkvist A., Bertilsson S., Sjöblom R., Kumpiene J. Sources and remediation techniques for mercury contaminated soil. Environment International. 2015;74:42–53. doi: 10.1016/j.envint.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 83.Velásquez-Riaño M., Benavides-Otaya H. D. Bioremediation techniques applied to aqueous media contaminated with mercury. Critical Reviews in Biotechnology. 2016;36(6):1124–1130. doi: 10.3109/07388551.2015.1100156. [DOI] [PubMed] [Google Scholar]
- 84.Tadishetty Hanumanth Rao S., Papathoti N. K., Gundeboina R., Mohamed Y. K., Mudhole G. R., Bee H. Hexavalent chromium reduction from pollutant samples by achromobacter xylosoxidans SHB 204 and its kinetics study. Indian Journal of Microbiology. 2017;57(3):292–298. doi: 10.1007/s12088-017-0654-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chardin B., Giudici-Orticoni M.-T., De Luca G., Guigliarelli B., Bruschi M. Hydrogenases in sulfate-reducing bacteria function as chromium reductase. Applied Microbiology and Biotechnology. 2003;63(3):315–321. doi: 10.1007/s00253-003-1390-8. [DOI] [PubMed] [Google Scholar]
- 86.Gonzalez C. F., Ackerley D. F., Lynch S. V., Matin A. ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. Journal of Biological Chemistry. 2005;280(24):22590–22595. doi: 10.1074/jbc.m501654200. [DOI] [PubMed] [Google Scholar]
- 87.Ackerley D. F., Gonzalez C. F., Park C. H., Blake R., Keyhan M., Matin A. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Applied and Environmental Microbiology. 2004;70(2):873–882. doi: 10.1128/aem.70.2.873-882.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support this study are available upon request.


