Abstract
Beyond its originally discovered role tethering replicated sister chromatids, cohesin has emerged as a master regulator of gene expression. Recent advances in chromatin topology resolution and single cell studies have revealed that cohesin plays a pivotal role regulating highly dynamic chromatin interactions linked to transcription control. The dynamic association of cohesin with chromatin and its capacity to perform loop-extrusion contributes to the heterogeneity of chromatin contacts. Additionally, different cohesin subcomplexes, with specific properties and regulation, controls gene expression across the cell cycle and during developmental cell commitment. Here we discuss the most recent literature in the field to highlight the role of cohesin in gene expression regulation during transcriptional shifts and its relation with human diseases.
Keywords: cohesin, DNA loops, transcription, cell cycle, cell identity, CdLS
Cohesin and gene expression: more than 20 years of relationship
Cohesin is a highly conserved, ring-shaped complex with the ability of encircling chromatin [1]. It comprises two SMC proteins (See Glossary), the kleisin subunit Rad21 and one copy of either STAG1 or STAG2 (Figure 1A). Cohesin loading onto DNA is positively regulated by Nipbl/Mau2 while Wapl and PDS5, in contrast, promote its removal (Figure 1A) [1]. The originally discovered function of cohesin is holding sister chromatids together following DNA replication until chromosomes segregate in mitosis, but it is not the only one. Cohesin also plays a fundamental role in DNA repair, chromatin organization, and gene expression regulation [2,3]. A role for cohesin in gene expression was first inferred in 1999 by connecting Drosophila Nipped-B, a cohesin-associated factor contributing to its DNA loading and loop extrusion activity [4-6], with enhancer-promoter interactions [7]. Further studies have revealed two different-interconnected mechanisms of cohesin action on gene expression. First, at transcriptionally active genes cohesin locally influences RNA Pol II-mediated transcription initiation, elongation and termination [8-11]. Second, in association with CTCF, cohesin has an architectural role in folding chromatin, generating CTCF loop domains and bringing cis-acting elements like enhancers to the vicinity of gene promoters [12]. A complete picture of how cohesin exerts both local and architectural effects on gene expression is not fully understood. Moreover, how cohesin coordinates its dynamic, cell-cycle-regulated association with chromosomes with the role on transcription during cell cycle and fate transitions in development is not completely known. In this review we evaluate our current understanding of these fascinating aspects of cohesin biology and the connection with human disease.
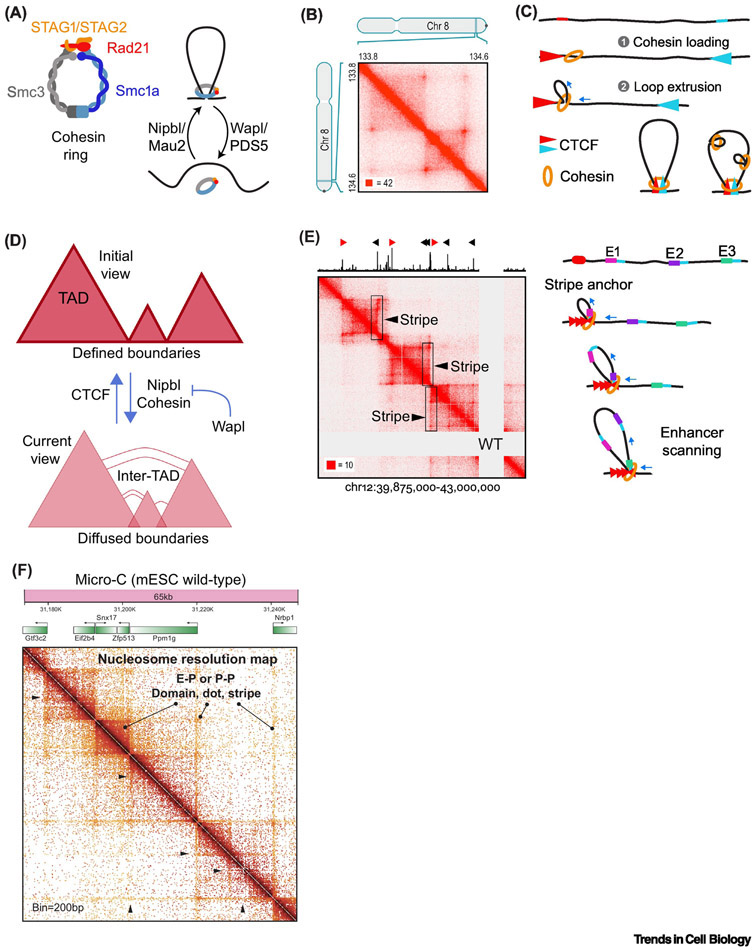
Figure 1. Cohesin and transcription underlies a highly dynamic genome topology.
(A) Schematic representation of a cohesin ring including the main components and the factors involved in its loading or removal from DNA. (B) Genome wide analysis of chromatin interactions evidenced the existence of loop domains termed as CTCF loops (Hi-C map picture from Rao et al. 2017 [12]). (C) Schematic model representing the coordinated action of cohesin loop extrusion activity and oppositely oriented CTCF (red and blue) in the generation of loop domains. (D) Current and previous view of loop domains. Our current vision shows highly diffuse boundaries and inter-CTCF loop interactions prompted by cohesin loading and limited by cohesin retention on chromatin by CTCF or by Wapl-mediated release. (E) In addition to CTCF loops, asymmetrical loading of cohesin mediates the formation of stripe domains highlighted with arrows and rectangles (Hi-C map picture from Vian et al, 2018 [31]) that might represent an enhancer scanning mechanism prompted by cohesin-mediated loop extrusion. (F) Micro-C development provides a nucleosome resolution map that reveals a high complexity of chromatin interactions and the existence of domains, dots and stripes associated with transcription factors and co-activators (Hi-C map picture from Hsieh et al, 2020 [40]).
A CLOSER LOOK INTO LOOP DOMAINS
The 3D organization of eukaryotic genomes is a highly regulated process linked to several functional aspects of DNA metabolism, such as replication and transcription. Eukaryotic genomes are organized on multiple scales. Two compartments are defined based on the transcriptional status. Compartment “A” represents areas with high-gene density and transcriptionally active chromatin, while compartment “B” is defined by transcriptionally repressed regions of heterochromatin [13,14]. Compartments can vary in size and tend to self-associate (i.e. A with A, B with B). In parallel, chromatin is folded and organized into topological associated domains (TADs) spanning hundreds of kilobases [12,15,16] (Figure 1B). Their presence in different species suggests a conserved role for TADs in genome organization and their potential role in evolution has been thoroughly discussed in a recent review [17]. In vertebrates, TAD boundaries preferentially form at convergent CTCF sites where CTCF stalls cohesin to prompt TAD boundary formation by loop extrusion [18,19] (Figure 1C). It is worth noting that TADs can form in the absence of cohesin and CTCF: Drosophila has cohesin-independent TADs, and C. elegans, which lacks CTCF, also forms TADs [17]. Our review will focus on loop domains defined by CTCF and cohesin in vertebrates, which we will refer to as CTCF loop domains. Consistent with this model, deletion of CTCF sites, CTCF, or cohesin subunits weakens CTCF loop boundaries [12,15,16,20-22]. Since the acute elimination of CTCF loops through cohesin or CTCF depletion results in very modest changes in transcript levels [12,20,23], their role and relevance on gene expression regulation remain unclear. Whether CTCF loops have a prominent role in gene expression regulation during cell stage transitions or during cell fate determination events in development requires further investigation. An immediate consequence of CTCF loop formation is the isolation of the DNA contained inside the loop by preventing inter-loop interactions, a concept referred as loop (TAD) insulation. In contrast, contacts between internal sites of the loop are favored. These two topological features of chromatin loops can be visualized in Hi-C maps, a proximity-based ligation method designed to reveal chromatin contacts by DNA sequencing [13] (Figure 1B). Although widely considered stable domains conserved across cell types and species in cell population-based studies [15,16], emerging work have challenged this idea and define CTCF loops as surprisingly dynamic entities. In fact, super resolution imaging methods and single cell approaches are revealing large cell to cell variation that was initially buried in Hi-C studies using cell populations [24-28].
High CTCF loop border plasticity
The intensity of loop insulation, measured by the lack of contacts across the boundaries of neighboring loops, is weaker than initially thought [24-28]. This is supported by recent high-resolution Hi-C data and super-resolution imaging that shows evident overlap between CTCF loop boundaries and stochasticity in loop formation between individual cells (Figure 1D) [24-28]. In fact, the analysis of chromatin contacts in single cells unveiled a higher level of heterogeneity that was initially underappreciated when considering cell populations [24-28]. What is the molecular mechanism behind CTCF loop promiscuity? By single-molecule imaging in live mouse embryonic stem cells (mES), Hansen and col. demonstrate a highly dynamic interaction of CTCF with chromatin and suggest a continuous assembly-disassembly cycle for chromatin loops [29]. A more recent study proposed that Nipbl and cohesin are triggers of inter- CTCF loop contacts that are required for proper RNA Pol II frequency burst and expression of genes located at boundary regions [30]. In contrast, CTCF and Wapl inhibit the formation of inter-loop interactions, suggesting that promiscuous CTCF loop borders depend on the dynamic loading and preservation of cohesin into chromatin [30]. This is consistent with the proposed loop-extrusion model, where cohesin action generates multiple contacts as loops extend until encountering a stable CTCF binding site or until undergoing Wapl-mediated release from chromatin (Figure 1D) (Box 1) [4-6]. Thus, the nature of the loop extrusion process might underly the variation on chromatin contacts behind the CTCF loop border heterogeneity observed in single cells. One of the most remarkable implications of dynamic CTCF loop borders is the formation of stripe domains, a sub-class of architectural domains showing asymmetric contact intensity between its two boundaries leading to a band/linear appearance in Hi-C maps [31,32] (Figure 1E). At the anchored border, ChIP-seq data shows enrichment of CTCF, Nipbl, cohesin and TFs that decreases along the stripes. This asymmetry, likely defined by differential CTCF distribution, might represent an enhancer-scanning mechanism between a strongly anchored CTCF-binding site and weak convergent CTCF sites distributed along the stripe domain in an alternative view of the two fixed borders of canonical CTCF loops [31-33]. Again, the loop extrusion activity of cohesin pulling the chromatin from the anchored border might favor the formation of stripes (Figure 1E). Stripe domains mostly locate at super enhancer regions in several cell types from different species and experience higher levels of reorganization during cell commitment in development compared to canonical CTCF loops [31,32]. Although the precise position of RNA Pol II along stripes is still unexplored, RNAseq data suggest that neither transcription nor its direction influence the polarity of these domains [31]. The disruption of the anchored site impairs gene expression of neighborhood genes by weakening DNA-DNA interactions [31,32], suggesting that the architectural role of cohesin on transcription regulation at stripe domains is prominent.
Box 1. Loop extrusion as a source of dynamic chromatin interactions.
The capacity of extruding DNA by the cohesin complex has been proposed as a mechanism to regulate chromatin structure, gene expression and gene recombination by the generation of DNA loops. In 2019, two independent studies provided evidence of this cohesin activity in vitro [4, 5]. By single-molecule imaging and biochemical reconstitution of the complex, both works evidence a bidirectional capacity of cohesin to extrude naked or nucleosome-organized chromatin, which ended with the generation of DNA loops. In addition to ATP, the process requires the presence of Nipbl and MAU2, two key components contributing to cohesin loading into DNA. Interestingly, the process does not require the opening of the cohesin ring neither the topological entrapment of DNA. In a more recent study, Golfier et al., demonstrated the extrusion capacity of cohesin using cell extracts from Xenopus oocytes [6]. Elucidating whether this activity is conserved in vivo constitute an immediate future direction.
The increase on the chromatin motility induced by loop extrusion allows the dynamic formation of DNA contacts and is secured by Nipbl/MAU2-mediated loading of cohesin. In contrast, extrusion capacity is constrained by the stabilization of cohesin, when encounters CTCF, or by cohesin removal through the action of the Wapl/Psd5 complex. In summary, cohesin loop extrusion activity and its regulation constitute a fundamental aspect of genome organization and a source of chromatin contacts with important implications on gene expression regulation.
Internal organization of chromatin loops
Several recent studies have provided insight into the internal organization of TADs. High resolution Hi-C data and super-resolution microscopy chromatin tracing revealed the existence of intra- or sub-loops that retain the key features of CTCF loops [26,34-36]. These loop structures insulate chromatin interactions, are highly dynamic and especially sensitive to cohesin loss and can bring long-distance enhancers to promoters to regulate gene expression [35,37]. However, simultaneous analysis of promoter-enhancer contacts and transcription activity by 3D-FISH or time-lapse microscopy has challenged the traditional view of stable proximity between these elements [38,39]. Surprisingly, increased separation was observed between promoters and enhancers during transcriptional activation suggesting the existence of alternative modes of action for enhancers to the traditional looping model [38,39]. In addition to sub-CTCF loops, the recently developed Micro-C approach provides a higher, nucleosome-resolution contact map (200bp) and revealed the presence of finer-scale structures named dots and Enhancer-Promoter or Promoter-Promoter stripes inside CTCF loops [40,41] (Figure 1F). TFs and co-activators together with RNA Pol II activity prompted the formation of stripes explaining the link between these fine-scale chromatin structures and active histone marks at compartment “A” [40]. The “stirring model” proposes a role for transcription activity in the encounter of enhancers with its target promoters inside CTCF loops by triggering the mobility of this cis-acting elements [42]. Indeed, studies in Drosophila showed a large reorganization of internal loop contacts upon transcriptional activation [43] and, at a locus resolution, that cohesin and elongating RNA Pol II are key factors in the formation of intragenic loops [44]. Overall, while current advances evidence cohesin and CTCF dynamics as key determinants for CTCF loop morphology and inter-loop connectivity at boundaries, at the intra-CTCF loop level cohesin seems to coordinate its role with TFs and RNA Pol II-mediated transcription likely in a CTCF-independent manner. The simultaneous analysis of DNA topology and RNA Pol II phosphovariants will improve our understanding of the crosstalk between cohesin and transcription at intra-CTCF loops.
ROLE OF COHESIN ON TOPOLOGY AND GENE EXPRESSION CHANGES DURING CELL DIFFERENTIATION
Analysis of genome organization during cell commitment
In the last few years, the remarkable progress on the methods for chromatin architecture analysis have improved our knowledge about genome topology remodeling during cell differentiation. Compartments show a high degree of plasticity in mammalian cells with a general trend of expansion of the compartment “B” size and an increase in internal chromatin interactions [45,46]. At compartment “A”, both expansion and reduction in size have been observed using different model organisms with a tendency of decreasing the number of DNA contacts inside this compartment in the course of lineage commitment [45,46]. Importantly, switching between compartments frequently occurs and mostly involves transition from “A” to “B” compartment [32]. Comparative analysis of CTCF loops in cell populations from mice and human differentiation models showed low variation of these domains between different cell-types. In contrast, changes in intra-CTCF loop contacts account for most of the cell-specific variation on genome organization [32,45-48]. The same trend was observed in cell populations during somatic mouse cell reprogramming into pluripotent cells with 75% of CTCF loops showing no variation [49]. CTCF loop boundaries show little change in genomic position, but the potential insulation is highly dynamic showing a consistent increase across mouse neural differentiation [46]. In line with this observation, the analysis of cohesin-mediated loops in 24 different human cell lines revealed a significant percentage of shared non-variable loops with higher association to CTCF loop boundaries than variable cell-specific loops [48], reinforcing the idea of boundary strength gaining upon cell differentiation [45,46,48]. Stripe domains represent an exception to this general trend with more stripes forming from loops in the transition of mES to neural stem cells (NSC) than vice versa [32]. The fact that NSC still preserves a high level of pluripotency might explain this particular behavior. In conclusion, the topology of chromatin changes in response to cell differentiation mostly affecting CTCF loop border insulation and intra loop architecture and, widely correlates with gene expression changes underlying cell specification [32,34,45-48,50].
Two rings to coordinate genome topology changes during cell differentiation
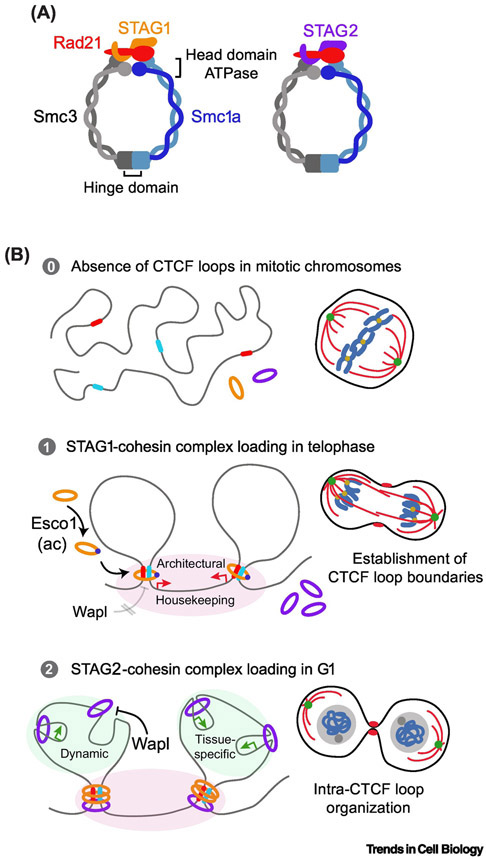
How does cohesin contribute to genome topology dynamics associated with cell differentiation? Cohesin plays a key role in pluripotency maintenance by ensuring a proper network of contacts around loci encoding pluripotency master regulators [51-54]. As a consequence, disruption of cohesin function causes pluripotency loss and premature cell differentiation [51,53-55]. In addition, cohesin is also required to preserve cell-type specific gene expression programs and recent advances have begun to elucidate its role during the differentiation process. A thrilling result is the recent discovery of two distinct cohesin subcomplexes acting differently at CTCF loop borders and at intra-CTCF loop regions [47,56,57] (Figure 2A, Key Figure). STAG1-cohesin preferentially localizes at CTCF loop borders, colocalizes with CTCF, is involved in preserving loop boundaries and shows lower genomic distribution change during differentiation (Figure 2B). Whether it has a prominent contribution to the establishment of anchored borders of stripe domains is unknown. In addition to localizing to CTCF loop borders, STAG2-cohesin mediates short-range interactions at the intra-CTCF loop level influencing TF occupancy and chromatin insulation, promotes the recruitment of PRC1 and the formation of super-enhancers that, collectively, participate in the establishment of cell-type specific expression programs [35,47,56-58] (Figure 2B). In line with this division of functions, specific depletion of STAG2 compromises hematopoietic differentiation and mouse embryonic development but has moderate effects on adult tissues [47,59]. These results anticipate the existence of differential properties and regulation of STAG1- and STAG2-containing complexes in the course of cell differentiation. In fact, STAG1-cohesin shows lower affinity for Wapl, the key factor in cohesin release from chromatin (Figure 2B). Moreover, STAG1-cohesin is specifically stabilized through the acetylation of the Smc3 subunit by Esco1 that may account for the differences between the more stable STAG1-cohesin at CTCF loop borders and the more dynamic STAG2-cohesin enriched at intra-CTCF loop [56,57,60] (Figure 2B). The fact that Wapl and PDS5 protein levels decrease during neural differentiation might stabilize the STAG2-cohesin to ensure cell-type specific programs and suggest a developmental-mediated regulation of cohesin subcomplexes [46]. Recently, a combination of Hi-C and ChIP-seq data from HCT116 cells have revealed a poor correlation of STAG2 specific binding sites with loop anchors, but enrichment on active promoters [61]. In addition, unique STAG2-binding sites mostly match with transcription start sites in human cells [56]. Taken together, these results suggest that STAG1-cohesin mediates cohesin architectural roles while STAG2-cohesin controls local cohesin functions. An exciting future direction will dissect the molecular mechanisms driving the differential assembly and loading of cohesin subcomplexes into chromatin and the determinants of its diverse coordinated function.
Key Figure 2. Two cohesin subcomplexes link chromosome topology and gene expression regulation during cell differentiation and across the cell cycle.
(A) Schematic representation of STAG1- and STAG2- cohesin showing different properties and regulations. (B) Proposed model for the coordinated action of STAG1- and STAG2-cohesin complexes. In the telophase-G1 transition, STAG1-cohesin is loaded into chromatin leading to the formation of CTCF loop boundaries and the expression of housekeeping genes. The Esco1-mediated acetylation of Smc3 and a higher tolerance to Wapl removal are two independent mechanisms underlaying the strength of STAG1-cohesin pool binding to chromatin. STAG2-cohesin, more abundant and dynamic, are sequentially loaded preferentially at intra-CTCF loop regions to regulate internal topology and tissue-specific gene expression.
ROLE OF COHESIN ON GENOME ARCHITECTURAL CHANGES ACROSS THE CELL CYCLE
Architectural genome reorganization during the cell cycle
The genome architecture experiences a massive reorganization as the cell cycle progress, with the most striking example represented by the chromosome compaction in mitosis [62]. In the last few years, the application of novel technologies has improved our understanding of how genomes reorganize during different stages of the cycle. Single-cell Hi-C in mice found that different levels of chromosome organization follow distinct cell-cycle dynamics [63]. Compartments “A” and “B” are absent in compacted mitotic chromosomes, faint in G1, and rise in parallel with DNA replication in S phase with a maximum in G2 before dissociating again upon mitotic entry [63,64]. CTCF loop border positions are invariable, but the strength of insulation varies across the different stages of the cycle [26, 63]. CTCF loop insulation is weakest in mitotic chromosomes but peaks early in G1 before decreasing in S phase during DNA replication [63]. Studies with synchronous HeLa cells reported similar dynamics in humans with a rapid formation of CTCF loops early in G1 [65,66] when Esco1 mediates STAG1-cohesin stablilization [60,67] (Figure 2B). Supporting this conclusion, 4D imaging of protein dynamics upon mitotic entry and exit found a rapid nuclear re-entry of CTCF, STAG1 and Rad21 proteins in late telophase/early G1. In contrast, Wapl and STAG2 show slower nuclear re-entry speed, suggesting that CTCF/STAG1-mediated establishment of CTCF loop boundaries that precede the formation of intra-loop cell-type specific loops dominated by STAG2 complexes [68] (Figure 2B). Whether STAG1- or STAG2-containing complexes have a prominent role in transcription re-start in G1 requires further investigation.
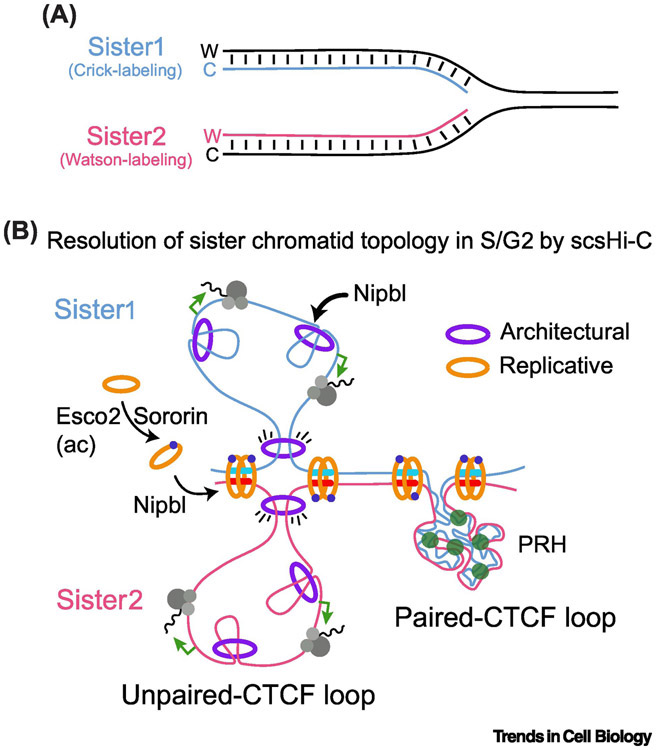
During interphase cohesin contributes to two distinct levels of genome organization: 1) cohesin forms chromatin loops in chromosomes in cis and 2) following DNA replication cohesin complexes link newly replicated sister chromatids until chromosome segregation in anaphase. Separating cis and trans chromosome interactions in the S-G2 stage of the cell cycle has been a significant challenge. Stanyte and col. addressed this question using dCAS9-directed labelling of several loci and live-cell imaging in human cells [69]. Results indicate sister loci separate immediately after replication, which is influenced by the nuclear position and transcriptional state [69]. More recently, a sister-chromatid sensitive Hi-C technology provided a higher resolution analysis of the entire replicated genome topology [70]. The use of nascent DNA-labelling allows the differentiation between cis (intra-chromatid) and trans (inter-chromatid) interactions in Hi-C maps from synchronized HeLa cells that reveal largely separated chromatids in G2 linked at CTCF loop boundaries and the existence of two classes of replicated loop domains: paired-CTCF loops, fully covered by trans interactions, and unpaired-CTCF loops, lacking these types of contacts (Figure 3). While paired CTCF loops mostly correlate with heterochromatin genomic regions of polycomb-repressed genes, the maintenance of unpaired CTCF loops requires dynamic cohesin loading and/or continuous DNA-loop extrusion [70] (Figure 3B). Whether active transcription is also required for replicated CTCF loop unpairing is unknown. Remarkably, two distinct pools of cohesin mediate sister chromatid organization in G2. Sororin-/Esco2-stabilized cohesin pool ensures sister chromatid cohesion while a second dynamic pool mediates loop extrusion and sister chromatid separation [67,70] (Figure 3B). Whether the presence of either STAG1 or STAG2 subunits in the complex define these two G2-associated cohesin pools is not known.
Figure 3. The topology of replicated genome.
(A) By the specific labelling of Watson (W) or Crick(C) strands during DNA replication in S phase, sister-chromatid-sensitive Hi-C approach (scsHi-C) allowed the resolution of chromatin contacts between sister chromatids at S and G2 stages of the cell cycle. (B) Two classes of CTCF loops emerged. Paired CTCF loops, with high level of trans interactions between sister chromatids that correspond to polycomb-repressed heterochromatin (PRH). In contrast, unpaired CTCF loops show abundant cis interactions and poor inter-chromatid connection that correlate with active transcription. In both cases, Sororin and Esco2 stabilizes replicative cohesin showing enrichment at CTCF loop boundaries.
The diversity of cohesin complexes and functions during the cell cycle opens a plethora of questions about the composition, stoichiometry and regulation of the different subcomplexes. Recent studies have estimated the absolute number of cohesin complexes per cell in mESCs and in HeLa cells at different stages of the cell cycle [71,72]. Data revealed a stable 1:1:1:1 stoichiometry of cohesin subunits across the cell cycle (Rad21:Smc1a:Smc3:STAG1/STAG2) and a higher abundance of STAG2- versus STAG1-containing complexes. The maximum difference between STAG2- and STAG1-cohesin pools peaks in G2 when, surprisingly, the dynamic cohesin fraction is lower than in G1 [72]. This presumably unexpected result might be a consequence of the special topology of replicated genomes where cohesive cohesin could constrain the dynamic pool of looping cohesin [70]. A relevant and controversial open question is whether cohesin functions as a single ring as a dimer/oligomer. The differential labelling of a specific cohesin subunit followed by protein-protein interaction analysis have revealed the existence of a moderate but significant percentage of cohesin dimers/oligomers in yeast, mouse and human cells [71,72]. Although the functional relevance of these conformations remains unknown, the increase in cohesin dimer/oligomer levels during DNA replication in S phase suggests a cohesive role on holding sister chromatids in yeast [73]. Future studies will help to elucidate the participation of cohesin dimers/oligomers on the variety of cohesin functions in mammalian cells.
Cohesin regulation of gene expression changes during the cell cycle
Gene expression changes across the distinct stages of cell cycle. At low resolution, transcription is mostly silenced in mitotic chromosomes, re-starts early in G1 and continues until the end of G2. This pattern correlates with the dynamic association of RNA Pol II with chromatin. An immediate question arises: are cohesin dynamics and its different subcomplexes involved in cell-cycle associated gene expression changes? The first wave of cohesin loading onto chromatin occurs at the end of mitosis, in telophase, and correlates with RNA Pol II reassociation with chromatin [65,66]. This process might be favored by the rapid nuclear re-entry of the cohesin subunits Rad21 and STAG1 [68] that coordinate the rapid formation of CTCF loops even before cells enter into G1 [65,66]. In a potential model, cohesin-STAG1 might regulate early G1 gene expression by defining CTCF loop borders, that associate with active transcription and genes with ubiquitous expression (Figure 2B) [35,57,60,74,75]. STAG2-cohesin would next establish cell specific gene expression patterns acting at intra-loop regions (Figure 2B). Remarkably, reduced Rad21 levels compromise nascent RNA expression early in G1 linking cohesin with RNA Pol II activity [10]. Elucidating whether cohesin primarily influences RNA Pol II by its architectural or/and local effect on transcription and the specific contribution of the different subcomplexes in the G1 expression program constitute an important future direction.
A second window of massive cohesin loading into chromatin occurs in S phase following DNA replication. Interestingly, cohesin binds to open chromatin near the origins of DNA replication [76,77]. Recent studies in Drosophila demonstrated cohesin binding to both replication origins and active enhancers, raising the possibility that active enhancers determine where in the genome replication starts and/or that replication might contribute to the establishment of promoter-enhancer interactions [78].
Early in mitosis, as chromatin condenses, transcription is silenced except at centromere regions where actively elongating RNA Pol II persists [79]. This pattern resembles the removal of cohesin from chromatin by the prophase pathway, where Wapl mediates the release of cohesin from chromosome arms but not from centromeres. Depletion of Wapl in Xenopus egg extracts and HeLa cells caused chromatin retention of cohesin and actively elongating RNA Pol II, which in turns impairs gene expression reprograming in the transit through mitosis [10]. In conclusion, large-scale cohesin loading and removal play a key role on gene expression changes across the cell cycle with special relevance as cells transition through mitosis.
A NEW VIEW OF CORNELIA DE LANGE SYNDROME ETIOLOGY
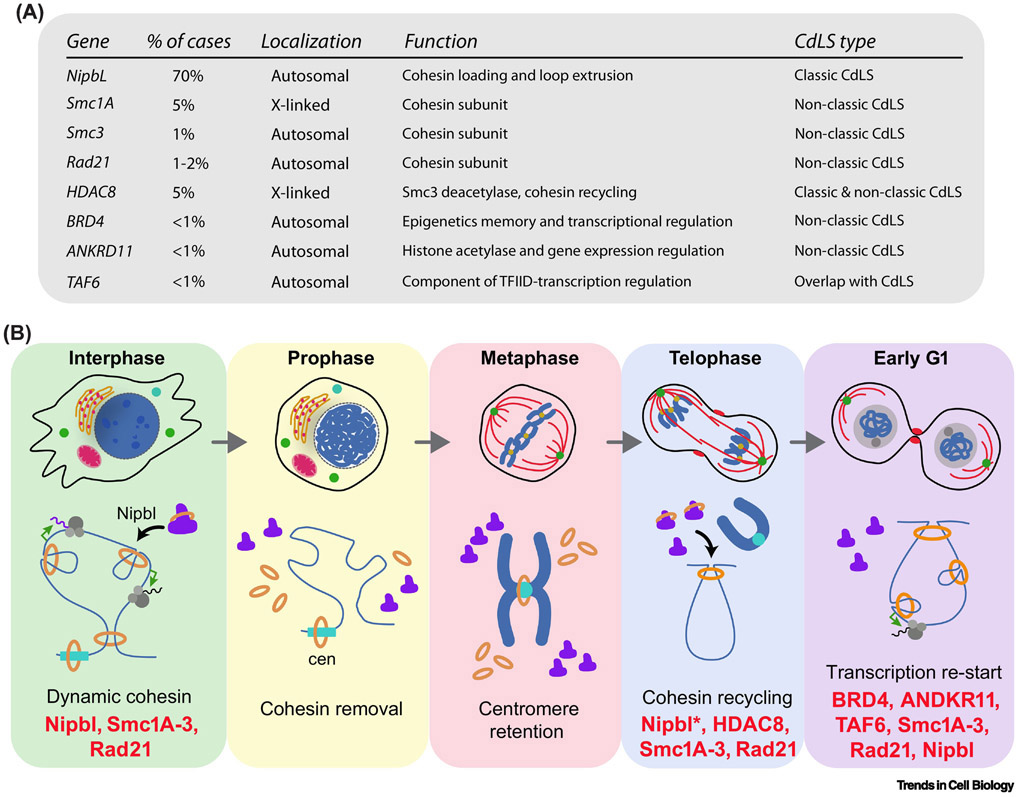
Cohesinopathies represent a family of developmental diseases caused by mutations in cohesin subunits or regulatory-associated factors. The most well-characterized example is Cornelia de Lange Syndrome (CdLS), a malformation disorder with an estimated prevalence of 1 in 10,000 where heterozygous mutations in Nipbl accounts for around 70% of cases [80]. Mutations in genes encoding cohesin subunits as Smc1a, Smc3 or Rad21, or regulators such as HDAC8, have also been linked to CdLS but only in a low proportion of patients [80] (Figure 4A). Interestingly, while chromatid cohesion is not affected, CdLS-associated mutations cause aberrant gene expression patterns associated with developmental defects, indicating the specific impairment of the cohesin role on gene expression regulation [81-84]. In support of this idea BRD4 and ANKRD11, two factors with a direct role in transcription, have also been associated with the CdLS etiology [85,86]. However, a holistic view of how all these factors operate in transcription regulation during development is missing. In light of recent discoveries, we propose a hypothesis that could explain the role of cohesin mutations in CdLS (Figure 4B). The fact that hemizygous mutations in Nipbl cause most CdLS cases indicates that the role of cohesin regulating gene expression depends on precise control of cohesin loading and loop extrusion . Studies with mouse cells derived from Nipbl +/− mice showed a global decrease in cohesin binding linked to gene expression defects [87]. In addition, quantification of Nipbl transcripts in cells derived from CdLS patients evidenced a correlation between the level of reduction and the severity of the symptoms [88]. Proper Nipbl levels may be particularly critical during the two main waves of cohesin loading in the cell cycle: during mitotic exit and following DNA replication in S phase. It is tempting to speculate that Nipbl might be a limiting factor in late telophase since daughter cells receive only half of recycled protein levels inherited from the mother cell. The same principle would apply for the recycling of cohesin subunits showing nuclear re-entry as mitosis ends [68]. In line with this idea, defects on HDAC8 impairs the recycling of cohesin after its removal from chromatin through the prophase pathway [82]. In fact, cohesin retention on metaphase chromosomes due to Wapl depletion showed reduced levels of chromatin-associated cohesin in G1 and aberrant gene expression [10]. During mitosis, Separase degrades ectopic, chromatin-retained cohesin, thus reducing cohesin recycling [89]. BRD4 has a critical role in bookmarking and transcriptional reactivation in G1 by promoting the recruitment of P-TEFb to chromatin [90-93]. Mutations in BRD4 associated with CdLS disrupt its binding to chromatin regions containing H3K27ac marks but have no effect on the interaction with Nipbl [86], which in turn may sequester this factor and compromise cohesin loading early in G1. ANKRD11 mediates histone acetylation required for gene expression during neural development [94]. Although ANKRD11 levels peak in mitosis [95], whether its activity is required for the acetylation of H3K27 is so far unknown. More recently, mutations in TAF6, component of the TFIID complex, have been associated with CdLS [80], indicating that disruption of general regulators of the transcriptional machinery is linked to the origin of this disease. Mutations in EP300, a transcriptional co-activator, and AFF4, subunit of the P-TEFb complex, has been identified in distinct syndromes with overlapping features to CdLS [9,96]. In conclusion, we propose that cohesin loading late in mitosis represent a key step for cohesin function on gene expression regulation. However, other models could also explain the role of cohesin mutations in CdLS. As many developmental transitions require passage through mitosis, the proposed role might be critical during cell differentiation in embryogenesis and a potential molecular framework of the etiology of CdLS and other related syndromes (Figure 4B).
Figure 4. Disruption of cohesin-driven re-start of transcription in G1 as a potential mechanism contributing to CdLS etiology.
(A) Several cohesin subunits and related regulatory factors underlies the etiology of CdLS. The disease shows a spectrum in the phenotype of patients including classic (or Typical) cases characterized by specific craniofacial appearance and growth pattern together with defects on limb formation, and a non-classic phenotype. (B) Schematic representation of cohesin dynamic events as cell transition through mitosis showing when each CdLS-associated factor might have a potential role. The telophase/G1 transition emerges as a critical window where cohesin recycling might influence transcription re-start with important implications in cell differentiation during development.
Concluding remarks
Our summary highlights the key role of cohesin regulation of gene expression as cells transition through the cell cycle and during the course of developmental programs. Although significant progress has been made, there are still many unresolved questions (see Outstanding Questions).
Outstanding question box.
Which are the functional consequences of the cell-to-cell differences observed in CTCF loop formation and properties? Are they linked to differences during the cell cycle?
Is cohesin loop extrusion activity operating in vivo? Is this capacity modulated across the cell cycle and/or during cell commitment in development?
How are different RNA Pol II states/phosphovariants distributed along the stripe domains? How do these domains change during the course of the cell cycle?
Are cohesin dimers/oligomers functionally relevant? How is regulated the assembly-disassembly of cohesin dimers/oligomers?
Which are the molecular determinants defining the differential chromatin loading and genome distribution of cohesin STAG1- and STAG2-containing complexes? How are these complexes assembled and why are STAG2-complexes more abundant?
Is recycled cohesin functionally linked to transcription re-start in G1? Is cohesin recycling connected to bookmarking and transcriptional memory? Are STAG1- or STAG2-complexes preferentially recycled and loaded upon mitotic exit?
Is cohesin loading especially sensitive to Nipbl levels during the telophase/G1 transition? Is cohesin misfunction during this timeframe of the cell cycle relevant to CdLS etiology? How is the genome organization and its dynamic affected in a CdLS context?
The analysis of genome topology at the nucleosome and single cell resolution level have uncovered a high dynamism of chromatin contacts. For example, recent single cell studies revealed the surprisingly cell-to-cell variability of CTCF loops, border plasticity and the existence of an intricate network of inter- and intra-CTCF loop connections. Although the dynamic of chromatin contacts is functionally linked to transcription by its influence on RNA Pol II activity, the removal of CTCF loops causes minimal disruption on gene expression. Simultaneous analysis of CTCF loops and gene expression changes in single cells transitioning through different stages of the cell cycle and during cell fate determination events would help to define the precise contribution of these domains to gene expression. A significant fraction of the variability of chromatin contacts rely on the capacity of cohesin to extrude DNA, its dynamic chromatin loading and removal and the existence of distinct subcomplexes showing differential regulation and chromatin binding properties. Although the cohesin loop-extrusion capacity has been thoroughly demonstrated in vitro, the characterization of this process and the determinants of its regulation in vivo constitutes an immediate direction required to advance the field.
Cell differentiation relies on the adjustments of gene expression program and correlates with changes in chromosome organization. Cohesin plays a pivotal role ensuring both, pluripotent and differentiated transcriptional programs and mediates this function though two subcomplexes defined by the presence of STAG1 or STAG2 subunits. These complexes show different dynamics and regulation, genome distribution, chromatin binding properties and, more importantly, distinct roles in gene expression regulation. Deciphering the molecular determinants of the differential loading, genomic distribution and function of STAG1-and STAG2-cohesin will provide a better understanding of cohesin role during development.
Many developmental transitions require the passage of cells through mitosis, a stage defined by the general silencing of gene expression and by the absence of cohesin on chromatin. Whether cohesin is recycled after mitosis and its potential contribution in development by promoting transcription re-start in G1 is unknown. Shedding light on this process may ultimately help to understand the etiology of CdLS, a cohesinopathy associated to aberrant gene expression.
Highlights.
Analysis of chromatin contacts in single cells and at nucleosome resolution evidence a highly dynamic genome topology
Cohesin is key for genome dynamism through the capacity of extruding DNA, its dynamic chromatin loading and removal, and the existence of distinct subcomplexes with differential properties and regulation
STAG1- and STAG2-containing complexes connect cohesin functions in chromosome organization and gene expression regulation in the frame of cell differentiation
Massive cohesin removal and loading as cell transition through mitosis influence gene expression reprogramming across the cell cycle
Cohesin recycling during the telophase/G1 transition might participate in transcription re-start and contribute to cell fate determination during development
Acknowledgements:
This work was supported by a grant from NIH (RO1 GM122893).
Glossary
- AFF4
AF4/FMR2 family member 4, key component of the Super Elongation Complex (SEC) involved in RNA Pol II release from a promoter-paused state.
- ANKRD11
Ankyrin Repeat Domain 11, involved in histone acetylation and gene expression.
- BRD4
bromodomain-containing 4, epigenetic reader of acetylated lysine residues.
- CTCF
CCCTC-binding factor with a key role on chromatin architecture and insulation.
- EP300
E1A-associated cellular p300 transcriptional co-activator protein. Histone acetyltransferase involved in chromatin remodeling and transcription activation.
- Esco1/Esco2
Establishment of cohesin 1 and 2; N-acetyltransferase enzymes with a role in the stabilization of chromatin-bound cohesin.
- HDAC8
Histone deacetylase 8, involved in transcriptional regulation and cell cycle progression.
- Hi-C
an extension method of 3C (chromosome conformation capture) developed to study the 3D architecture of the genomes.
- Nipbl/MAU2
Nipped-B-like protein and MAU2 chromatid cohesion factor homolog; form a heterodimeric complex contributing to the DNA loading and loop extrusion capacity of cohesion.
- P-TEFb
positive transcription elongation factor, multiprotein complex regulating RNA Pol II activity.
- Rad21
α-kleisin subunit of the human cohesin complex, also known as double-strand break repair protein Rad21 homolog.
- Separase
protease required for mitotic progression. At the metaphase-anaphase transition separase degrade cohesive cohesin at centromere regions facilitating sister chromatid segregation.
- Smc1a and Smc3
structural maintenance of chromosomes (SMC); protein subunits that form a heterodimer that constitute an essential part of the cohesin complex.
- Sororin
stabilizes cohesin complex association with chromatin antagonizing the action of Wapl/PDS5 complex.
- STAG1 and STAG2
stromal antigen 1 and 2, subunits of the human cohesin complex.
- TAF6/TFIID
TATA Box Binding Protein (TBP)-Associated Factor 6 (TAF6), component of the transcription factor IID (TFIID) that regulate the initiation of RNA Pol II-mediated transcription.
- Wapl/PDS5
Wings apart-like protein homolog and Precocious dissociation of sisters 5 homolog; form a heterodimer involved in the dissociation of cohesin from DNA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests:
The authors declare no competing interests.
List of References
- 1.Yatskevich S et al. (2019) Organization of Chromosomal DNA by SMC Complexes. Annu. Rev. Genet. 53, 445–482 [DOI] [PubMed] [Google Scholar]
- 2.Peters JM et al. (2008) The cohesin complex and its roles in chromosome biology. Genes Dev. 22, 3089–3114 [DOI] [PubMed] [Google Scholar]
- 3.Nasmyth K and Haering CH (2009) Cohesin: Its Roles and Mechanisms. Annu. Rev. Genet 43, 525–558 [DOI] [PubMed] [Google Scholar]
- 4.Davidson IF, et al. (2019) DNA loop extrusion by human cohesin. Science. 366 (6471), 1338–1345 [DOI] [PubMed] [Google Scholar]
- 5.Kim Y et al. (2019) Human cohesin compacts DNA by loop extrusion. Science. 366 (6471), 1345–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golfier S et al. (2020) Cohesin and condensin extrude DNA loops in a cell cycle-dependent manner. eLife, 9, e53885. DOI: 10.7554/eLife.53885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rollins RA et al. (1999) Nipped-B, a Drosophila Homologue of Chromosomal Adherins, Participates in Activation by Remote Enhancers in the cut and Ultrabithorax Genes. Genetics. 152 (2), 577–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorsett D and Merkenschlager M (2013) Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr. Opin. Cell Biol 25, 327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izumi K et al. (2015) Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat. Genet 47, 338–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perea-Resa C et al. (2020) Cohesin Removal Reprograms Gene Expression upon Mitotic Entry. Mol. Cell. 78, 127–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nanavaty V et al. (2020) DNA Methylation Regulates Alternative Polyadenylation via CTCF and the Cohesin Complex. Mol. Cell 78, 752–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao SSP et al. (2017) Cohesin Loss Eliminates All Loop Domains. Cell. 171, 305–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman-Aiden E et al. (2009) Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 326, 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowley MJ and Corces VG (2018) Organizational principles of 3D genome architecture. Nat. Rev. Genet 19, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon JR et al. (2012) Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature. 485, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao SSP et al. (2014) A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 159, 1665–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szabo Q et al. (2019) Principles of genome folding into topologically associating domains. Sci. Adv 5. DOI: 10.1126/sciadv.aaw1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanborn AL et al. (2015) Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA 112, 6456–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fudenberg G et al. (2016) Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 15, 2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nora EP et al. (2017) Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 169, 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarzer W et al. (2017) Two independent modes of chromatin organization revealed by cohesin removal. Nature. 551, 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wutz G et al. (2017) Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 15;36(24), 3573–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghavi-Helm Y et al. (2019) Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet 51(8), 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flyamer IM et al. (2017) Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 544, 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens TJ et al. (2017) 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 544, 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bintu B et al. (2018) Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science. 362, eaau1783. DOI: 10.1126/science.aau1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finn EH et al. (2019) Extensive Heterogeneity and Intrinsic Variation in Spatial Genome Organization. Cell. 176, 1502–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mateo LJ et al. (2019) Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature. 568, 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen AS et al. (2017) CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife. e25776. DOI: 10.7554/eLife.25776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luppino JM et al. (2020) Cohesin promotes stochastic domain intermingling to ensure proper regulation of boundary-proximal genes. Nat. Genet 52, 840–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vian L et al. (2018) The Energetics and Physiological Impact of Cohesin Extrusion. Cell. 173, 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrington C et al. (2019) Enhancer accessibility and CTCF occupancy underlie asymmetric TAD architecture and cell type specific genome topology. Nat. Commun. 10, 2908. DOI: 10.1038/s41467-019-10725-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen AS et al. (2019) Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Mol. Cell 76, 395–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips-Cremins JE et al. (2013) Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell. 153, 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthews BJ and Waxman DJ (2018) Computational prediction of CTCF/cohesin-based intra-TAD loops that insulate chromatin contacts and gene expression in mouse liver. eLife. e34077. DOI: 10.7554/eLife.34077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norton HK et al. (2018) Detecting hierarchical genome folding with network modularity. Nat. Methods 15, 119–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu W et al. (2017) Identifying topologically associating domains and subdomains by Gaussian Mixture model And Proportion test. Nat. Commun 8 (535). DOI: 10.1038/s41467-017-00478-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexander JM et al. (2019) Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife. 8, e41769. DOI: 10.7554/eLife.41769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benabdallah NS et al. (2019) Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol. Cell 76, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh TS et al. (2020) Resolving the 3D Landscape of Transcription-Linked Mammalian Chromatin Folding. Mol. Cell 78, 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krietenstein N et al. (2020) Ultrastructural Details of Mammalian Chromosome Architecture. Mol. Cell 78, 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu B et al. (2018) Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science. 359, 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cardozo Gizzi AM et al. (2019) Microscopy-Based Chromosome Conformation Capture Enables Simultaneous Visualization of Genome Organization and Transcription in Intact Organisms. Mol. Cell 74, 212–222 [DOI] [PubMed] [Google Scholar]
- 44.Rowley MJ et al. (2019) Condensin II Counteracts Cohesin and RNA Polymerase II in the Establishment of 3D Chromatin Organization. Cell Rep. 26, 2890–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dixon JR (2015) Chromatin architecture reorganization during stem cell differentiation. Nature. 518, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonev B et al. (2017) Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 171, 557–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viny AD et al. (2019) Cohesin Members Stag1 and Stag2 Display Distinct Roles in Chromatin Accessibility and Topological Control of HSC Self-Renewal and Differentiation. Cell Stem Cell. 25(5), 682–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grubert F et al. (2020) Landscape of cohesin-mediated chromatin loops in the human genome. Nature. 583, 737–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stadhouders R et al. (2018) Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat. Genet 50, 238–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuartero S et al. (2018) Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat. Immunol 19, 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Apostolou E et al. (2013) Genome-wide Chromatin Interactions of the Nanog Locus in Pluripotency, Differentiation, and Reprogramming. Cell Stem Cell. 12, 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei Z et al. (2013) Klf4 Organizes Long-Range Chromosomal Interactions with the Oct4 Locus in Reprogramming and Pluripotency. Cell Stem Cell. 13, 36–47 [DOI] [PubMed] [Google Scholar]
- 53.Zhang H et al. (2013) Intrachromosomal Looping Is Required for Activation of Endogenous Pluripotency Genes during Reprogramming. Cell Stem Cell. 13, 30–35 [DOI] [PubMed] [Google Scholar]
- 54.Khaminets A et al. (2020) Cohesin controls intestinal stem cell identity by maintaining association of Escargot with target promoters. eLife. 9, e48160. DOI: 10.7554/eLife.48160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu NQ et al. (2021) WAPL maintains a cohesin loading cycle to preserve cell-type-specific distal gene regulation. Nat. Genet 53, 100–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kojic A et al. (2018) Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat. Struct. Mol. Biol 25, 496–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cuadrado A et al. (2019) Specific Contributions of Cohesin-SA1 and Cohesin-SA2 to TADs and Polycomb Domains in Embryonic Stem Cells. Cell Rep. 27, 3500–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cuadrado A and Losada A (2020) Specialized functions of cohesins STAG1 and STAG2 in 3D genome architecture. Curr. Opin. Genet 61, 9–16 [DOI] [PubMed] [Google Scholar]
- 59.De Koninck M et al. (2020) Essential Roles of Cohesin STAG2 in Mouse Embryonic Development and Adult Tissue Homeostasis. Cell Rep. 32(6). DOI: 10.1016/j.celrep.2020.108014 [DOI] [PubMed] [Google Scholar]
- 60.Wutz G et al. (2020) ESCO1 and CTCF enable formation of long chromatin loops by protecting cohesinSTAG1 from WAPL. eLife 9, e52091. DOI: 10.7554/eLife.52091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casa V et al. (2020) Redundant and specific roles of cohesin STAG subunits in chromatin looping and transcriptional control. Genome Res. 30, 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibcus JH et al. (2018) A pathway for mitotic chromosome formation. Science. 359, eaao6135. DOI: 10.1126/science.aao6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nagano T et al. (2017) Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 547, 61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dekker J et al. (2013) Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet 14, 390–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abramo K et al. (2019) A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat. Cell. Biol 21, 1393–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang H et al. (2019) Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature. 576, 158–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alomer RM et al. (2017) Esco1 and Esco2 regulate distinct cohesin functions during cell cycle progression. Proc. Natl. Acad. Sci. USA 114, 9906–9911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cai Y et al. (2018) Experimental and computational framework for a dynamic protein atlas of human cell division. Nature. 561, 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stanyte R et al. (2018) Dynamics of sister chromatid resolution during cell cycle progression. J. Cell Biol 217, 1985–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mitter M et al. (2020) Conformation of sister chromatids in the replicated human genome. Nature. 586, 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cattoglio C et al. (2019) Determining cellular CTCF and cohesin abundances to constrain 3D genome models. eLife 8, e40164. DOI: 10.7554/eLife.40164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holzmann J et al. (2019) Absolute quantification of cohesin, CTCF and their regulators in human cells. eLife, e46269. DOI: 10.7554/eLife.46269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi D et al. (2020) The acetyltransferase Eco1 elicits cohesin dimerization during S phase. J. Biol. Chem 295, 7554–7565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hug CB et al. (2017) Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell. 169, 216–228 [DOI] [PubMed] [Google Scholar]
- 75.Rowley MJ et al. (2017) Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Mol. Cell 67, 837–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guillou E et al. (2010) Cohesin organizes chromatin loops at DNA replication factories. Genes. Dev 24(24), 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.MacAlpine HK et al. (2010) Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 20(2), 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pherson M et al. (2019) Cohesin occupancy and composition at enhancers and promoters are linked to DNA replication origin proximity in Drosophila. Genome Res. 29(4), 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chan FL et al. (2012) Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Nat. Acad. Sci 109, 1979–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kline AD et al. (2018) Diagnosis and management of Cornelia de Lange syndrome: first international consensus statement. Nat. Rev. Genet 19, 649–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu J et al. (2009) Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 7(5):e1000119. DOI: 10.1371/journal.pbio.1000119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deardorff MA et al. (2012) HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 489, 313–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Remeseiro S et al. (2013) Reduction of Nipbl impairs cohesin loading locally and affects transcription but not cohesion-dependent functions in a mouse model of Cornelia de Lange Syndrome. Biochim. Biophys. Acta 12, 2097–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mannini L et al. (2015) Mutant cohesin affects RNA polymerase II regulation in Cornelia de Lange syndrome. Sci. Rep 5:16803. DOI: 10.1038/srep16803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parenti I et al. (2016) Broadening of cohesinopathies: exome sequencing identifies mutations in ANKRD11 in two patients with Cornelia de Lange-overlapping phenotype. Clin. Genet 89, 74–81 [DOI] [PubMed] [Google Scholar]
- 86.Olley G et al. (2018) BRD4 interacts with NIPBL and BRD4 is mutated in a Cornelia de Lange–like syndrome. Nat. Genet 50, 329–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Newkirk DA et al. (2017) The effect of Nipped-B-like (Nipbl) haploinsufficiency on genome-wide cohesin binding and target gene expression: modeling Cornelia de Lange syndrome. Clin. Epigenetics 9:89. DOI: 10.1186/s13148-017-0391-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaur M et al. (2016) NIPBL expression levels in CdLS probands as a predictor of mutation type and phenotypic severity. Am. J. Med. Genet. C. Semin. Med. Genet 172(2),163–70 [DOI] [PubMed] [Google Scholar]
- 89.Tedeschi A et al. (2013) Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature. 501, 564–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang Z et al. (2008) Brd4 Recruits P-TEFb to Chromosomes at Late Mitosis To Promote G1 Gene Expression and Cell Cycle Progression. Moll. Cell Biol 28, 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dey A et al. (2009) Brd4 Marks Select Genes on Mitotic Chromatin and Directs Postmitotic Transcription. Mol. Biol. Cell 20, 4899–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao R et al. (2011) Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol 13, 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Behera V et al. (2019) Interrogating Histone Acetylation and BRD4 as Mitotic Bookmarks of Transcription. Cell Rep. 27, 400–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gallagher D et al. (2015) Ankrd11 Is a Chromatin Regulator Involved in Autism that Is Essential for Neural Development. Dev. Cell 32, 31–42 [DOI] [PubMed] [Google Scholar]
- 95.Walz K et al. (2015) Characterization of ANKRD11 mutations in humans and mice related to KBG syndrome. Hum. Genet 134, 181–190 [DOI] [PubMed] [Google Scholar]
- 96.Woods SA (2013) Exome sequencing identifies a novel EP300 frame shift mutation in a patient with features that overlap Cornelia de Lange syndrome. Am. J. Med. Genet. A 164A(1), 251–258 [DOI] [PubMed] [Google Scholar]