Abstract
Schistosomes cause one of the most devastating neglected tropical diseases, schistosomiasis. Their transmission is accomplished through a complex life cycle with two obligate hosts and requires multiple radically different body plans specialized for infecting and reproducing in each host. Recent single-cell transcriptomic studies on several schistosome body plans provide a comprehensive map of their cell types, which include stem cells and their differentiated progeny along an intricate developmental hierarchy. This progress not only extends our understanding of the basic biology of schistosome life cycle but can also inform new therapeutic and preventive strategies against the disease, as blocking the development of specific cell types through genetic manipulations has shown promise in inhibiting parasite survival, growth, and reproduction.
Keywords: Schistosome, life cycle, single-cell sequencing, cell type atlas, stem cells, development
Single-cell analyses open a new era in understanding schistosome biology
Schistosomes are parasitic flatworms that infect hundreds of millions of people worldwide and cause schistosomiasis, a devastating neglected tropical disease [1,2]. Despite the disease’s impact, only a partially effective drug, praziquantel, is available for treatment [3–6]. However, epidemiological studies have shown that the infection burden often rebounds after treatment due to permissive re-infection [7–10]. It has become widely accepted that a major target for disease eradication should be parasite transmission, but a better understanding of the basic biology that enables the life cycle of these parasites is needed in order to develop new therapeutic and preventive strategies.
Schistosomes are members of Trematoda flatworms, one of the largest groups of metazoan endoparasites including tens of thousands of species [11]. Their transmission typically requires passage through multiple hosts, at minimum a mollusk as the intermediate host and a definitive vertebrate host [12–14]. These parasites must build distinct specialized body plans to survive within each host and alternate between asexual and sexual reproduction, maximizing their rate of multiplication [13]. While previous characterizations have documented specific morphological changes of these parasites across different life cycle stages, the cellular and molecular events underlying these changes are only now being understood. In this review, we focus on the recent work that has advanced our understanding of the life cycle and the developmental biology of Schistosoma mansoni, one of the most widespread trematodes. These studies (Table 1) applied single-cell RNA sequencing (scRNAseq) to interrogate the major stages of the schistosome life cycle and provided opportunities to address a series of important questions. Is there a specialized cell population producing new tissues throughout the multi-staged schistosome development? What cell types are schistosomes composed of? Does the cell type composition change as schistosomes progress through life cycle stages? What roles do these changes play in the adaptation of the parasites to various environments? What is the developmental origin of the schistosome germline that enables sexual reproduction? How are the parasite cell types related to those of free-living flatworms that share a common evolutionary ancestor with schistosomes? Just as the completion of the schistosome genome around 2010 transformed the field by establishing the needed framework for molecular work [15–17], the recent progress in characterizing schistosome cell types [18–22] represents a major leap forward by providing the cellular context needed to better understand schistosome biology and develop new therapeutic treatments against schistosomiasis.
Table 1.
Single-cell RNAseq studies on various S. mansoni life cycle stages to date.
| Life cycle stage | Sequenced tissue | scRNAseq method | Reference |
|---|---|---|---|
| mother | G2/M cells | Fluidigm C1 | Wang, et al. 2018 [18] |
| sporocyst | |||
| schistosomulum | whole animal | 10x Chromium | Diaz Soria, et al. 2020 [19] |
| juvenile | G2/M cells | SmartSeq2 | Tarashansky, et al. 2019 [20] |
| juvenile | whole animal | SmartSeq2 | Li, et al. 2021 [21] |
| adults | whole animal | 10x Chromium | Wendt, et al. 2020 [22] |
Stem cells drive the complex life cycle of Schistosoma mansoni
The schistosome life cycle involves parasitic stages in two hosts, with short-lived, free-swimming stages in between (Figure 1 and Box 1). To complete this life cycle, schistosomes develop multiple radically different body plans. Besides embryonic development from the zygote, they modify their body plans post-embryonically three times: from miracidia to sporocysts triggered by the entry into the intermediate molluscan host, from sporocysts to cercariae through multiple rounds of embryogenesis-like development inside the snail host, and finally from cercariae to adult parasites within the mammalian host. In contrast to the sporocysts, which are essentially sacs of cells with no defined body size and shape, adult schistosomes have elaborate organ systems and are unique among the trematodes in that they are dioecious with sexual dimorphism [14]. What changes at the cellular level enable such massive body plan modifications has puzzled scientists for many years.
Figure 1. Schistosomes remodel their body plans multiple times throughout their life cycle.
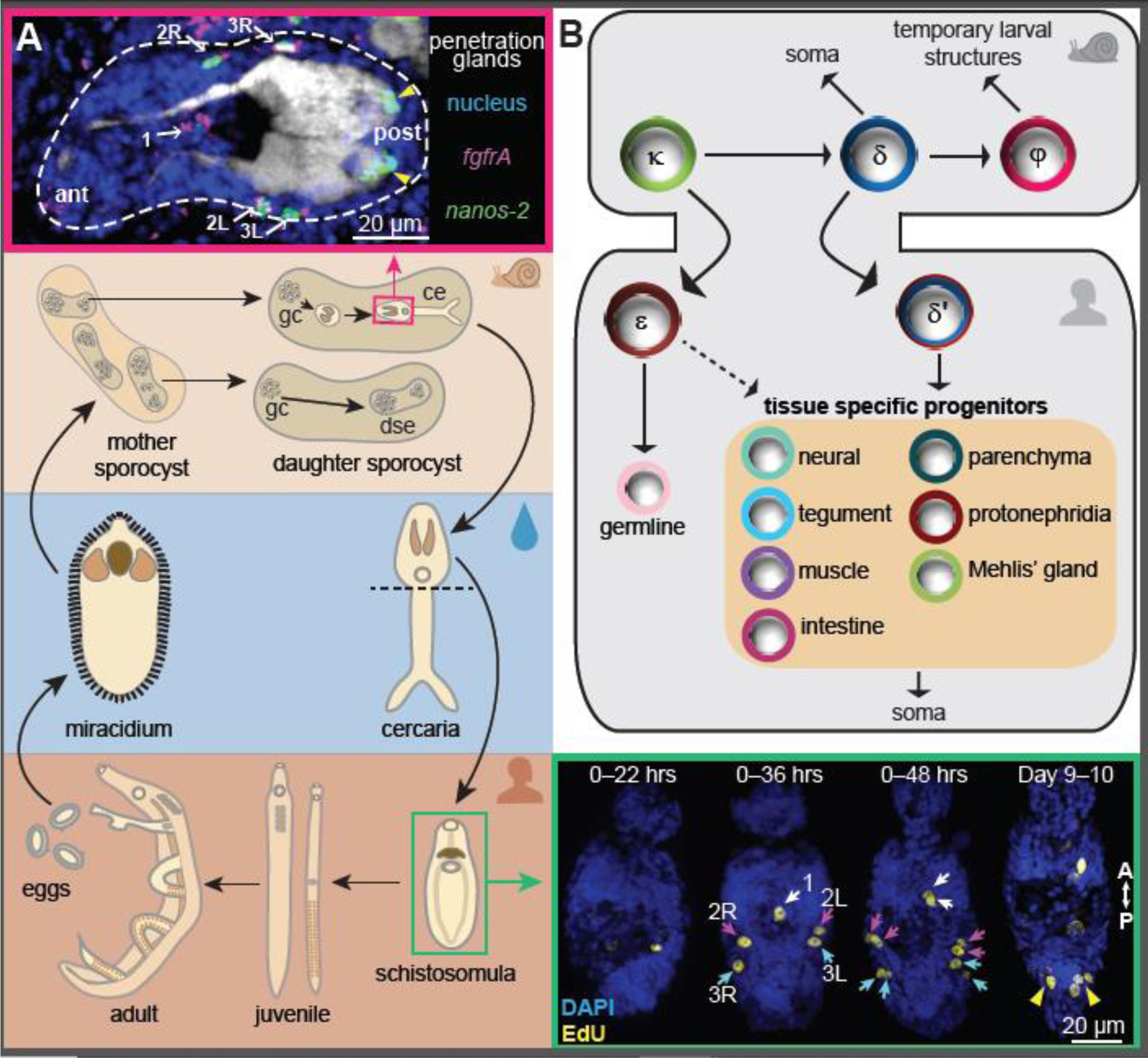
(A) Schematics showing the schistosome life cycle as detail in Box 1. dse.: daughter sporocyst embryo; gc: germinal cells; ce: cercarial embryo. Insets: microscopy images showing that germinal cells are transmitted through the cercarial heads to initiate the intramammalian development. Images correspond to boxed stages in the schematics, and are adapted from ref. 18. Top: a small number of germinal cells are packed at specific locations around the cercarial penetration glands. Arrows: germinal cells that are double positive for fgrfA and nanos-2 (δ-cells); arrowheads: nanos-2+ cells (κ-cells). Bottom right: these cells re-enter the cell cycle and proliferate in schistosomula to expand the stem cell pool. The five anterior stem cells (arrows) first enter the cell cycle and divide synchronously, whereas the two posterior stem cell clusters (arrowheads) begin to proliferate at a later time point. EdU labels newly synthesized DNA. (B) A model for the developmental hierarchy of schistosome germinal/stem cells across life cycle stages, which is adapted from ref. 18. Population specific genes are listed in Table 2.
Box 1. Schistosome life cycle.
The schistosome life cycle begins with the zygote undergoing embryogenesis to form the first body plan, a ciliated larva called miracidium enclosed in the eggshell [32,66]. When the egg is excreted from the mammalian host through feces or urine into a body of freshwater, changes in osmolarity and light trigger the hatching of the miracidium, which only survives for a few hours unless it infects a snail host. Once inside the new host, the miracidium transforms its body plan and develops into a mother sporocyst. With most somatic tissues degenerated, the sporocyst forms a sac-like structure and its shape and size are mostly dependent on the space available in the host tissues [18,28]. Each mother sporocyst can asexually produce many daughter sporocysts, which leave the mother and migrate to occupy new areas in the snail host. This process can be repeated multiple rounds, expanding the sporocyst population by the hundred. In some daughter sporocysts, the embryos of the next life cycle stage, cercaria, begin to take shape. Cercariae develop a new body plan consisting of a head and a tail. Mature cercariae burst through the snail tissue and return back to the water where they swim for hours until they find a mammalian host and burrow through its skin. This triggers a series of physiological and morphological changes, initiating their intramammalian development. Cercariae first detach their tails and then remodel their tegument so that it can serve as a nutrient-absorptive and protective layer in the host [44,45]. These highly modified cercarial heads are now called schistosomula. They migrate into the blood vessels and travel through the host body for a couple of weeks, from the lungs to the liver [42,65]. In the course of this journey, cell proliferation is limited, but the remodeling of the body plan and organ systems has already begun [43]. The first set of changes are thought to enable blood feeding. Arrival at the hepatic portal vein marks the beginning of the juvenile stage. Here, influenced by host cues, the parasites undergo massive growth to extend their body posteriorly, increasing their length from a couple hundred micrometers to approximately a centimeter in 2–3 weeks [18]. Of particular significance during this stage is the development of sexual reproductive organs [20]. The reproductive maturation of females requires pairing and mating [22]. Each female can then produce hundreds of eggs daily, from which the cycle restarts.
Early ultrastructural and histological studies recognized a cell population in sporocysts based on their distinct undifferentiated morphology and mitotic activity [23,24]. These cells, historically termed as ‘germinal cells’, were postulated to drive the asexual reproduction of schistosomes at the intramolluscan stages by initiating embryogenesis without fertilization [25,26]. These embryonic stem cell-like cells may have indefinite proliferative capacity, evidenced by the serial transplantation of sporocysts into naïve molluscan hosts that led to continuous propagation of the parasites [27]. However, the molecular and functional properties of these cells remained unresolved for decades after they were first observed. In addition, it was unknown whether germinal cells form a continuous lineage and transit to other life cycle stages.
What is the molecular identity of the germinal cells?
A functional genomic analysis of the germinal cells at the miracidium-to-sporocyst transition provided the first characterization of their molecular identity [28]. The germinal cells were found to abundantly express several post-translational regulators, including homologs of argonaute (i.e., ago2–1) and nanos (i.e., nanos-2), in addition to several members of the vasa/PL10 protein family, which were also shown to be required for the proliferation of these cells [28]. The initial findings were confirmed by a scRNAseq study that sequenced individual germinal cells isolated from in vitro transformed mother sporocysts [18]. This panel of post-transcriptional regulators is reminiscent of a multipotency program in diverse animals [29–31], suggesting that the schistosome germinal cells may exhibit a molecular signature evolutionarily conserved in stem cells.
Do the germinal cells transit to the intramammalian stage?
The identification of highly specific marker genes in the germinal cells has made it possible to label these cells using RNA in situ hybridization and trace their anatomical distributions at various life cycle stages. To transition from the intramolluscan to intramammalian stages, cercariae leave the snail and infect mammalian hosts (Box 1). In the cercarial heads, five germinal cells were found to be dispersed in a regular pattern around the penetration glands, and another two small germinal cell clusters were shown to segregate to the posterior of the glands (Figure 1A) [18]. These two cell populations express slightly different sets of genes and were named δ- and κ-cells, respectively (Figure 1B, Table 2). Upon entering the mammalian host, δ-cells start to proliferate, followed by κ-cells [18]. These observations suggest that the germinal cells transit across life cycle stages and are the only source of new cells at the onset of the intramammalian development. To better distinguish the proliferative cells across the schistosome life cycle, we propose to reserve the use of the historical term ‘germinal cells’ to the intramolluscan stages; accordingly, the progeny of germinal cells in parasites at their intramammalian stages can be generically referred to as ‘stem cells’.
Table 2.
Major germinal/stem cell populations, all expressing the ubiquitous stem cell marker ago2–1 (Smp_179320) and cell cycle genes besides the listed population specific genes1.
| Stem cell population | Population markers | Putative functions |
|---|---|---|
| Intramolluscan stages | ||
| κ |
klf (Smp_172480) nanos-2 (Smp_051920) |
embryonic stem cells |
| δ |
fgfrA (Smp_175590) fgfrB (Smp_157300) nanos-2 (Smp_051920) p53 (Smp_139530) zfp-1 (Smp_145470) |
somatic stem cells |
| φ |
fgfrA (Smp_175590) fgfrB (Smp_157300) hesl (Smp 024860) p53 (Smp_139530) zfp-1 (Smp 145470) |
progenitors of larval structures, including mother and daughter sporocyst epidermis, and cercarial tail |
| Intramammalian stages | ||
| ε |
eled (Smp_041540) klf (Smp_172480) astf (Smp_142120, specific to εα cells) bhlh (Smp_341460, specific to εβ cells) |
pluripotent stem cells giving rise to both soma and germline |
| δ’ |
fgfrA (Smp_175590) fgfrB (Smp_157300) hesl (Smp_024860) klf (Smp_172480) p53 (Smp_139530) nanos-2 (Smp_051920) zfp-1 (Smp_145470) |
somatic stem cells |
| μ |
cabp (Smp_340130) myoD (Smp_167400) |
early muscle progenitors |
| troponin+ (μ’) |
cabp (Smp_340130) myoD (Smp_167400) tcf15 (Smp_015670) troponin (Smp_018250) |
late muscle progenitors |
| tsp2 + | tsp2 (Smp_335630) | tegumental progenitors |
| cpx + |
7b2 (Smp_073270) cpx (Smp_050220) |
neural progenitors |
| cb2 + | cb2 (Smp_141610) | intestinal/parenchymal progenitors |
| hnf4 + |
hnf4 (Smp_174700) prom2 (Smp_179660) |
intestinal progenitors |
| igsf9b + |
igsf9b (Smp_035040) sialidase (Smp_335600) |
flame cell progenitors |
| vwa + | vwa (Smp_157690) | Mehlis’ gland progenitors |
| Germline stem cell (GSC) |
boule (Smp_144860) eled (Smp_041540) nanos-1 (Smp_055740) oc-1 (Smp_196950, specific to male GSCs) |
germline stem cells for both sexes |
| GSC progeny |
eled (Smp_041540) horm2 (Smp_169930) meiob (Smp_333540) nanos-1 (Smp_055740) nuob (Smp_328620) |
putative meiotic GSC progeny |
| S1 |
eled (Smp_041540) nanos-1 (Smp_055740) nr/vf1 (Smp_248100) |
vitellocyte progenitors |
Marker genes listed are by no means exhaustive and selected based on three criteria: (1) they have consistent expression in specific cell types, (2) their cell type specific expression has been confirmed experimentally through in situ hybridization, and (3) if the specific cell type is present in more than one dataset/life cycle stage, their expression is detected across datasets using different single-cell sequencing pipelines.
How do the germinal/stem cells produce differentiated cells during development?
Production of differentiated tissues may be accomplished by either a homogenous pluripotent stem cell pool that directly generates all differentiated cell types or a heterogeneous population that contains stem cells with differential potency and separate differentiation fates. In recent years, several single-cell studies have revealed significant heterogeneity among the schistosome stem cells (Table 2) [18,20,21] and thereby favor the second model.
At the onset of the intramolluscan development, a subset of germinal cells in mother sporocysts are reserved for continuous self-renewal while the rest of the population differentiates to either form embryos of daughter sporocysts or produce somatic structures that enlarge the sporocyst body and support embryogenesis [32]. This process is replicated in some daughter sporocysts to generate more daughters, while others produce cercariae, also through the germinal cells (Figure 1A). Consistent with this functional hierarchy, transcriptomic heterogeneity was identified among the germinal cells [18]. By integrating single-cell gene expression data and anatomical characterizations, Wang et al. proposed the model that germinal cells are composed of three major populations, κ-, δ-, and φ-cells (Figure 1B) [18]. κ-cells express klf, a homolog of KLF4, an important transcriptional regulator governing pluripotency [33], and may represent the most undifferentiated state since they can specify δ-cells that in turn generate embryonic tissues. In contrast, φ-cells may be the progeny of δ-cells, as they are excluded from the embryos and contribute to the somatic structures (e.g., sporocyst epidermis and developing cercarial tails) that are useful only at specific life-cycle stages but not transmitted across.
After the parasite enters the mammalian host and arrives at the host’s portal vein, massive tissue growth is initiated, driven by extensive cell proliferation concentrated towards the posterior of the parasite’s body [18]. A series of single-cell transcriptomic studies focused on this life cycle stage, the juvenile parasites, and observed heterogeneity among the stem cells [18,20,21]. These cells include populations that resemble δ- and κ-cells form the intramolluscan stages with modified gene expression programs and are called δ’- and ε-cells, respectively (Figure 1B, Table 2). The latter can be further divided into two populations based on their transcriptional profiles: εα and εβ, with εα-cells appearing more abundantly during early juvenile development [20,21]. The other identified populations express not only stem cell markers but also genes associated with major differentiated tissue types, including muscle, tegument (the parasites’ epidermis), neural, parenchymal, and intestinal tissues [21,22]. In particular, two distinct stem cell populations express a common set of canonical myogenesis regulators (e.g., myoD), but one lacks functional genes of differentiated muscle such as troponin, suggesting that they may represent sequential stages along the muscle lineage [21]. In situ hybridization experiments confirmed that all these presumptive progenitor populations are in spatial proximity with the corresponding differentiated tissue types and are mitotically active [20,21]. These observations suggest that the schistosome stem cells may consist of both undifferentiated populations and tissue-specific progenitors, potentially one for every differentiated tissue type. Figure 1B summarizes the proposed developmental hierarchy of the schistosome germinal and stem cell populations.
What is the cellular source of the schistosome germline?
Schistosomes multiply asexually inside of the molluscan host with their whole body serving as a brood chamber, but they switch to sexual reproduction at the intramammalian stages by specifying the germline and developing their gonads de novo. Sexually mature male and female worms mate and can produce hundreds of eggs daily [14,34], which are excreted from the host to initiate the next life cycle. This prolific reproductive output can be sustained for many years before the fecundity declines with worm age [7]. Unlike most animals, which segregate their germline during embryonic development [35], the schistosome germline is not specified until the juvenile stage, at which point the parasites have already completed embryogenesis and gone through two additional post-embryonic body plan remodeling processes.
The key observation to unraveling the cellular origin of the schistosome germline is that a subset of ε-cells activate a germline-specific nanos homolog (nanos-1) in juveniles exceeding a critical body length (~400 μm), indicating that these cells are in the process of committing to the germline fate [18]. These primordial germ cells (PGCs) multiply to form clusters and eventually give rise to gonads, which contain germline stem cells (GSCs) and differentiated germ cells [18,21]. nanos-1 expression persists in GSCs and is required to maintain their molecular identity [18,21,36]. Additional GSC-specific genes have been identified in a recent scRNAseq study that succeeded in capturing GSCs from juvenile parasites [21]. RNAi experiments showed that a genetic program, regulated by onecut homeobox transcription factor and boule mRNA binding protein, balances the proliferation and differentiation of the GSCs [21]. While these studies have proposed a stem cell lineage that begins from ε-cells, converts to PGCs, and ultimately gives rise to GSCs, the molecular events that direct this unusual crossover from the somatic to the germline fate remain unknown and to be addressed by future research (see Outstanding Questions).
Outstanding Questions.
What are the differences between the zygote, which is a totipotent stem cell, and the germinal/stem cells?
What is the developmental lineage of germinal cells during embryonic development from the zygote?
What triggers the germinal cells to enter embryogenesis without fertilization? What determine their in producing daughter sporocyst vs. cercaria? How many germinal cells are needed to initiate the embryogenesis?
Besides germinal cells, what other cell types are required for the asexual reproduction in sporocysts? Are they unique to the intramolluscan life cycle stages?
How do the stem cells convert from somatic to germline fates?
Do schistosomes have pluripotent stem cells that can produce all cell types throughout the life cycle? If transplanted across life cycle stages, can the germinal/stem cells adopt new fates?
What molecular programs regulate the specification of tissue-specific progenitors and the differentiation of each cell type at each life cycle stage?
How do cell types adapt to drastically different environments between hosts?
How do schistosomes adjust their development in response to host cues? What cell types sense and interpret the environment and send signals to other cell types to coordinate the development?
How and when does male vs. female development diverge? What are the key regulators of the sexually dimorphic development?
Do muscle cells provide patterning cues for instructing stem cell differentiation and setting up the body plans?
What cell types respond to praziquantel treatment? What cell types provide praziquantel resistance at the juvenile stage?
How do schistosomes repair tissue damage? What cell types are essential for this process?
Are there cell type specific differences between schistosome species (i.e., S. mansoni, S. japonicum, and S. haematobium)?
How are cell types in other parasitic flatworms (e.g., tapeworms) conserved compared to schistosomes?
Are schistosome stem cells maintained post development?
Adult parasites can survive in their mammalian hosts for decades [7]. This requires repairing aged and damaged tissues over time. Collins et al. proposed that schistosomes achieve this feat using adult stem cells, which exhibit broad gene expression similarities with the germinal cells in sporocysts and stem cells in juveniles [37]. Pulse-chase experiments using a thymidine analog have shown that these cells are responsible for continuously replenishing differentiated tissues, including muscle, intestine, and tegument [37–39].
Given their diverse fates, are the schistosome adult stem cells also a heterogeneous population? Recently, Wendt et al. sequenced ~40,000 cells from adult parasites and constructed a cell type ‘atlas’ of sexually mature males, females, and virgin females, as sexual maturation of the female reproductive organs requires mating [22]. Among the sequenced cells was an abundant population of cells expressing ago2–1, a highly specific and ubiquitous stem cell marker. We performed clustering analysis on these ago2–1+ cells and identified various progenitor populations consistent with those observed in juvenile parasites, including previously characterized tegumental (tsp2+) [38] and intestinal progenitors (hnf4+) [22] (Figure 2A–B and Table 2). The presence of abundant tissue specific progenitors in adult parasites suggest their tissues undergo constant homeostatic turnover, which is surprising especially for tissue types with expected long lifetimes (e.g., nervous system). The ago2-1+ cells also include progenitor populations of sexual reproductive organs, including GSC progeny (meiob+horm2+nuob+) and Mehlis’ gland cells (vwa+). Besides the presumptive tissue progenitor populations, we also noticed a few populations with gene expression signatures characteristic of δ’-cells. However, we failed to identify εα/εβ-cells in the adult dataset, though some of their marker genes (e.g., eled) were found to be expressed more diffusively across multiple stem cell populations. These results suggest that the adult stem cell have a population structure different from that of juveniles (Figure 2C).
Figure 2. Stem cell and esophageal subpopulations at the intramammalian stages.
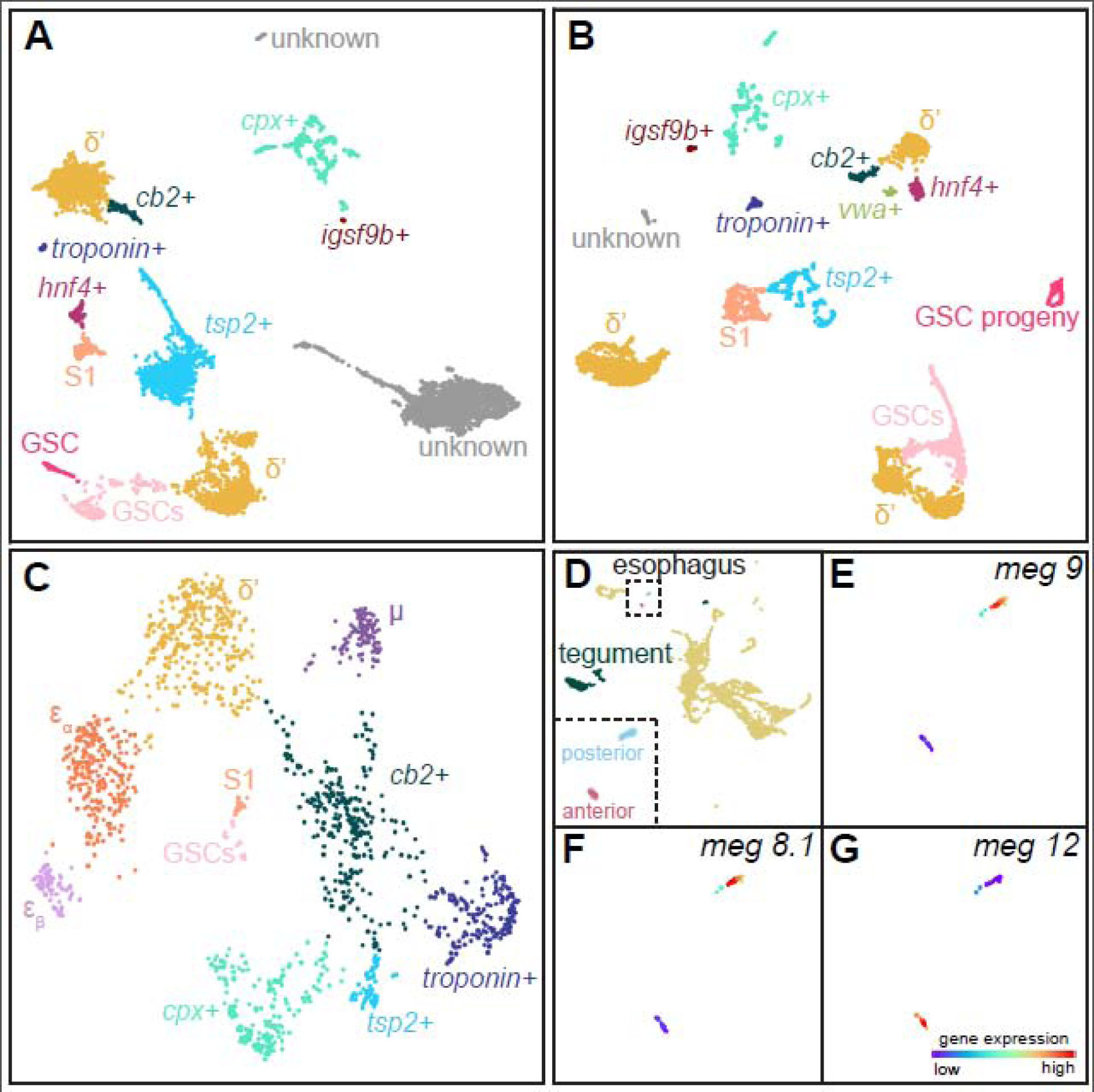
Subclustering of ago2–1+ cells from adult male [22] (A) and virgin female [22] (B) parasites. Panel (C) shows juvenile ago2–1+ cells, adapted from ref. 21. Subpopulations of stem cells are annotated using gene expression signatures summarized in Table 2, and labeled by colors. We detected S1 cells in the adult male dataset, which are likely experimental contaminations, as S1-specific genes were shown to be absent in males [67]. (D) Analysis of the schistosomulum cell atlas [19] identifies a cluster containing the two tegumental progeny populations and fully differentiated tegumental cells (Table 3), and two distinct populations of esophageal cells corresponding to the anterior cell mass and posterior gland cells, respectively. Inset: a magnified view of the two esophageal clusters. Similar to many other known gland marker genes [49], the expression of posterior gland markers, meg-9 (E) and meg-8.1 (F), and anterior cell mass marker, meg-12 (G), is highly specific to each cluster. The manifold reconstruction was performed using Self-Assembling Manifolds (SAM) [20]. The SAM algorithm (version 0.7.5) was run using parameters ‘weight_PCs = False’ and ‘preprocessing = StandardScaler’. The SAM source code, tutorials, documentation, stem cell gene expression data, and subpopulation annotations are available through Github (https://github.com/atarashansky/self-assembling-manifold). While ago2–1+ cells form a distinct cluster in juveniles [21], they are distributed in multiple clusters in adults and separated based on their detected ago2–1 expression (>1 unique transcripts). Schistosomulum ago2–1+ cells lack elaborate subpopulation structures, and therefore are not shown.
Single-cell transcriptomic atlases classify cell types across the intramammalian life-cycle stages
Recent single-cell atlases encompass all major stages of the schistosome intramammalian development, schistosomulum [19], juvenile [21], and adult [22], and thereby enable the comparison of cell types across life cycle stages. The intramammalian portion of the schistosome life cycle is of special interest because it is when the parasites cause disease. Understanding whether, when, and what new cell types emerge during these stages can help predict whether therapeutic targets that affect one of these life cycle stages might also be effective on others.
In adults, ten major differentiated tissue types were annotated besides stem cells: muscle, neural, tegument, gut, parenchyma, esophagus, protonephridia, vitellaria, Mehlis’ gland, and the germline (Table 3) [22]. These tissue types can be further divided into ~60 clusters of distinct transcriptomic signatures, with neurons exhibiting the largest diversity with 31 clusters, followed by muscle cells separating into 8 clusters. Whether these molecularly defined clusters correspond to cell types or cell-to-cell variations reflecting regional and functional heterogeneity within individual cell types remains an open question. Similar cell type diversity was also observed in juvenile parasites (Table 3), with the exception of vitellocytes, Mehlis’ gland cells, and gametes as these cell types are expected to emerge only during sexual maturation and yet to be developed at this stage [21].
Table 3.
Major differentiated cell types at the intramammalian life cycle stages, with gametes, vitellarial cells, and Mehlis’ gland cells being unique to adult parasites.
| Cell type | Marker genes1 |
|---|---|
| Muscle | myosin (Smp_045220); troponin (Smp_018250); tpm2 (Smp_031770) |
| Neurons | |
| non-ciliated neurons | 7b2 (Smp_073720) |
| ciliated neurons | 7b2 (Smp_073720); p25α (Smp_097490) |
| kk7+ neurons | kk7 (Smp_194830) |
| Parenchymal | cb2 (Smp_141610); lap (Smp_030000); serpin (Smp_090080); tgfbi (Smp_212710) |
| Intestinal | cb1.1 (Smp_103610); cb1.2 (Smp_067060); ctsl (Smp_343260); hmgbs (Smp_075800) |
| Tegumental | |
| syncytial | annexin b2 (Smp_077720); calpain (Smp_214190); gtp-4 (Smp_105410); npp-5 (Smp_153390); sm25 (Smp_346900); tal (Smp_045200); tsp2 (Smp_335630) |
| progeny 1 | tsp2 (Smp_335630); zfp1-1(Smp_049580) |
| progeny 2 | onzin (Smp_101970); sm13 (Smp_195190); sm25 (Smp_346900); tsp2 (Smp_335630) |
| Protonephdridia | igsf9b (Smp_035040); sialidase (Smp_335600) |
| Esophagus | |
| anterior cell mass | meg-12 (Smp_152630); meg-17 (Smp_180620); pla2 (Smp_031190) |
| posterior gland | foxA (Smp_331700); meg-4.1 (Smp_307220); meg-4.2 (Smp_085840); meg-8.1 (Smp_171190); meg-8.2 (Smp_172180); meg-9 (Smp_125320); meg-11 (Smp_176020); meg-14 (Smp_124000); meg-15 (Smp_010550) |
| Gametes | |
| male | cep162 (Smp_147750); mad1 (Smp_139380) |
| female | bmpg (Smp_078720); clec (Smp_246770) |
| Vitellaria | ataxin-2 (Smp_167830); p48 (Smp_241610) |
| Mehlis’ gland | vwa (Smp_157690) |
Cell type markers are selected based on the same set of criteria used in Table 2.
Diaz Soria et al. made an important observation that a similar set of cell types is present in two-day old schistosomula, which are composed of merely ~900 cells per individual [19]. This study identified and confirmed through in situ hybridization 13 different cell types exhibiting molecular signatures consistent with those in juvenile and adult parasites (Table 3): 3 muscle populations, 4 neural populations, 2 tegument populations, 2 parenchymal/primordial gut populations, an esophageal gland population, and stem cells. The missing cell types are protonephridial cells, though previous anatomical characterizations have noted these cells at this stage [40,41], as well as cell types associated with sexual reproduction (the germline and somatic reproductive organs).
The finding that schistosomulum cell types are consistent with later developmental stages is unexpected in two ways. First, there is minimal cell proliferation in the schistosomula at this young age: the first five cell divisions occur between 36–48 hours after the transformation from cercaria to schistosomulum (Figure 1) [18]. This indicates that all major tissue types in a two-day old schistosomulum are made during the cercarial development in the snail host. In support of this view, when we reanalyzed the schistosomulum dataset, the tegumental progenitors are the only tissue specific progenitor population that we were able to identify, suggesting that the production of new differentiated cells is mostly absent at this stage. A single-cell analysis of cercarial development should help determine the exact time when these cell types are specified and distinguish cell lineages that build structures either specific to cercaria (e.g., sensory nerve endings and acetabular glands involved in host location and penetration) or shared by cercaria and schistosomulum.
Second, despite previously documented morphological and functional changes of several tissues as schistosomula develop into adults [42,43], the associated cell types seem to be surprisingly consistent. For example, as a key adaptation to the environment in the mammalian host, the cercarial tegument is shed and replaced from below by preformed membrane inclusions [44–46]. This modification is thought to involve multiple cell types as there are two kinds of membrane inclusions in the tegumental syncytium; these inclusions appear to be packaged at different Golgi apparatus, which may originate from different cell bodies [47]. Indeed, we identified four tegumental cell types present at all three intramammalian life cycle stages, including the tegumental progenitors (ago2–1+nanos2+tsp2+) within the stem cell compartment, two distinct progeny populations, tsp2+zfp-1-1+ and tsp2+sm13+sm25+ cells, and fully differentiated tegumental cells expressing many characteristic genes such as annexin b2, calpain, npp-5 and gtp-4 (Table 3). These cell types are consistent with the tegumental lineage previously proposed in adult parasites [38], suggesting that the same differentiation process may underlie the tegumental development and homeostatic turnover. Whether the two progeny populations represent sequential maturation states or parallel differentiation pathways remains an important question to be resolved.
Another example showing consistency in cell type composition between stages is the esophageal tissues, an essential organ required for both blood feeding and lysing ingested host immune cells [48–50]. In adults, the esophagus is divided into the anterior cell mass and the posterior gland cells, which express distinct sets of microexon genes (megs) and potentially play different functional roles [49]. Consistent with characterizations in the adult parasites, in the two-day old schistosomulum dataset we also found two distinct esophageal cell clusters (Figure 2DG): one expressed the anterior cell mass markers, meg-12, −17, whereas the other expressed posterior gland markers, meg-4.1, −4.2, −8.1, −8.2, −9, −11, −14, −15, in addition to a forkhead-box transcription factor foxA. Lee et al. reported recently that knockdown of foxA led to the loss of the posterior esophageal gland while leaving the anterior cells unaffected, supporting the difference between these two cell types [50].
Besides all the commonalities between the three datasets, we also noticed a few important differences. It is immediately evident that the schistosomulum and juvenile datasets lack the cell types that pertain to several sexual reproductive organs. Cell type diversity for some tissue types (e.g., neural and muscle) also varies, with the adult parasites showing the largest diversity. Progenitor cell types may also be modified to address the need of rapidly replenishing certain tissues at specific life cycle stages. For example, Wendt et al. identified an abundant hnf4+ gut progenitor population in adults. RNAi studies showed that hnf4 is required for gut maintenance and blood feeding [22]. This progenitor population is absent in schistosomula. In juveniles, a few hnf4+ stem cells are present but do not form a distinct cluster, instead they mix with the cb2+ progenitor population, which may be associated with both parenchymal and intestinal tissues (Table 2).
Altogether, the cell atlases spanning schistosome intrammamalian development reveal a common set of persisting cell types. In order to generate different structures and develop functions specific to each life cycle stage, cells could either modify their gene expression programs while maintaining their cell type identity or adjust their relative abundance with respect to other cell types. Since distinguishing these two potential mechanisms is challenging using the current datasets, understanding the developmental plasticity of these cells will be an important question to address in the future (see Outstanding Questions).
Schistosome cell types exhibit broad conservation with cell types in free-living flatworms
Previous phylogenomic analyses suggest that parasitic flatworms evolved from free-living ancestors [51–53]. Although the conservation and diversification in life cycle, morphological, and genomic traits between parasitic and free-living flatworms have been extensively studied [51–54], the degree to which cell types are conserved between them has only been examined recently [19,21,55]. Among organisms that have annotated cell type atlases, the planarian, Schmidtea mediterranea, is most closely related to parasitic flatworms and therefore offers a useful comparison [56–58]. While initial comparison between schistosomes and planarians drew parallels between the schistosome germinal/stem cells and the planarian adult stem cells, neoblasts [18,28], it was limited to a handful of stem cell-specific factors.
To systematically compare planarian and schistosome cell types, Diaz Soria et al. trained a random forest model [59] to classify cell types in a planarian single-cell dataset [57], and then used the same model to assign labels to the schistosomulum cell types based on the expression of orthologous genes [19]. This model successfully linked stem cells between species. Of the differentiated cell types, the best match was a Sm-kk7+ neural population in schistosomulum and neurons expressing otoferlin 1 (otf1) in the planarian, but the mapping of other cell types was significantly weaker.
More recently, Tarashansky et al. developed a method (SAMap) for mapping single-cell transcriptomes across evolutionarily distant species [55], and used it to compare the cell types of juvenile schistosome and planarian [21,56]. SAMap revealed broad cell type homology supported by shared gene expression programs, including stem cells, neural, muscle, intestine, tegumental/epidermal, protonephridia, and parenchymal cell types. The shared gene expression signatures include both known cell type specific markers and numerous conserved transcriptional regulators [55].
Consistent with the schistosomulum comparison, SAMap linked Sm-kk7+ cells to the planarian otf1+ neurons [19,55]. Additionally, SAMap found both organisms to have neural compartments composed of ciliated, non-ciliated, dopaminergic, and peptidergic neurons, respectively mapped to their analogues in the other species, though their functions remain to be further characterized. The schistosome and planarian muscle cell types share a common set of transcriptional regulators such as myoD and tcf15, which are core regulators of myocyte specification in diverse animals [60]. In addition to the molecular machinery that is typically associated with contractility, these muscle cells express a common set of wnt homologs, with some showing regional specificity: wnt-2 is concentrated towards the anterior end of the parasite body [19,22] whereas wnt-11 has high expression at the posterior [55]. In planarians, wnt-2 and wnt-11 are also expressed in the anterior and posterior regions respectively, providing the positional cues for setting up the body plan during tissue regeneration [61–63]. The presence of a similar Wnt gradient in schistosome muscles reveals a deep conservation of cell type specific gene expression programs, and raises important questions about muscle functions in instructing stem cells differentiation and guiding body plan patterning (see Outstanding Questions).
Most importantly, SAMap linked ε-cells in schistosome juveniles with the planarian pluripotent neoblasts [55,58], and mapped several homologous tissue specific progenitors between the two species through a large number of genes including transcription factors soxP2, myoD, p53, hnf4, and sox2, which are conserved in the pluripotent, muscle, epidermal/tegumental, intestinal, and neural populations, respectively [55]. The conservation of key transcription factors in the tissue progenitors suggests that stem cells may follow similar differentiation pathways between planarians and schistosomes. In contrast, adult schistosomes lack the stem cell population that maps to planarian pluripotent neoblasts, implicating that pluripotent cells may be a transient population during development but not maintained for tissue homeostasis in adult schistosomes. This is consistent with the observation that, in contrast to planarians that can regenerate their entire body from small tissue fragments, schistosome adults have very limited regenerative ability beyond wound healing [64]. Experimentally delineating the potency of schistosome stem cells at different life cycle stages is an important task for future research (see Outstanding Questions).
Concluding Remarks
Schistosomes propagate through a complex life cycle during which their body plans undergo dramatic transformations multiple times. These changes are essential for the parasite’s survival, transmission, and multiplication. This review summarizes the developmental, cellular, and molecular events underlying the schistosome life cycle by synthesizing recent single-cell RNAseq studies on S. mansoni. These studies have identified important heterogeneity among stem cells that drive the schistosome development and reproduction, classified a variety of differentiated cell types across life cycle stages, and enabled the comparison between parasitic and free-living flatworms to explore the evolutionary origin of these cell types. Understanding the molecular and cellular events involved in regulating and sustaining the schistosome life cycle may further our understanding of cell type differentiation and developmental plasticity (see Outstanding Questions).
The analyses of the schistosome cell types have also provided information that may help combat the disease. For example, the differentiation of intestinal progenitor cells in adult parasites has been shown to be essential for the gut maintenance and blood feeding, inhibition of which reduced the parasite size and alleviated the pathology in vivo [22]. Similarly, blocking the development of esophageal glands exposes the parasites to the attack of host immune cells in vivo, likely due to the parasite’s inability to degrade ingested immune cells within the esophagus [50]. A newly discovered pathway that controls GSC differentiation provides routes to disrupt the production of gametes, which in turn should stop the parasite’s sexual reproduction [21], though this effect still needs to be confirmed in vivo. Beyond these examples, meticulous comparisons of cell types across life cycle stages may define new targets to inhibit the parasite development and therefore the disease transmission.
Highlights.
Single-cell transcriptomic studies discover cellular and molecular events that regulate the schistosome life cycle, providing targets for inhibiting parasite survival, growth, and transmission.
Germinal/stem cells exhibit high transcriptomic heterogeneity, consistent with the need to generate a diverse set of differentiated cell types, including the germline, and to build several distinct body plans throughout the life cycle.
Comparison of schistosomulum, juvenile, and adult somatic cell types suggest that they may be specified early on during the cercarial development in the molluscan host.
Cross-species comparisons reveal a broad and deep conservation of cell types between schistosomes and free-living planarian flatworms, and propose that a putative pluripotent stem cell population is restricted to the parasite development and not maintained for tissue homeostasis in adults.
Acknowledgments
We thank J. J. Collins III and C. L. Diaz Soria for sharing their adult and schistosomulum datasets before publication. We also thank J. Lee for numerous discussions and his critical reading of the manuscript. DNS is a Bio-X Bowes Graduate Student Fellow. BW is a Beckman Young Investigator. This work was supported by an NIH grant 1R35GM138061.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Hoffmann KF et al. (2014) Halting harmful helminthes: vaccines and new drugs are needed to combat parasitic worm infections. Science 346, 168–169 [DOI] [PubMed] [Google Scholar]
- 2.Hotez PJ and Fenwick A (2009) Schistosomiasis in Africa: an emerging tragedy in our new global health decade. PLoS Negl. Trop. Dis 3, e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergquist R et al. (2017) Controlling schistosomiasis with praziquantel: how much longer without a viable alternative? Infect Dis Poverty 6, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doenhoff MJ et al. (2009) Praziquantel: Its use in control of schistosomiasis in sub-Saharan Africa and current research needs. Parasitology 136, 1825–1835 [DOI] [PubMed] [Google Scholar]
- 5.Chan JD et al. (2017) The anthelmintic praziquantel is a human serotoninergic G-protein-coupled receptor ligand. Nat. Commun 8, 1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li P et al. (2018) Nanomedicine approaches against parasitic worm infections. Adv. Healthc. Mater 7, 1701494. [DOI] [PubMed] [Google Scholar]
- 7.Buck JC et al. (2020) Concomitant immunity and worm senescence may drive schistosomiasis epidemiological patterns: an eco-evolutionary perspective. Front. Immunol 11,160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sokolow SH et al. (2015) Reduced transmission of human schistosomiasis after restoration of a native river prawn that preys on the snail intermediate host. Proc. Natl. Acad. Sci. USA 112, 9650–9655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guidi A et al. (2010) Praziquantel efficacy and long-term appraisal of schistosomiasis control in Pemba Island. Trop. Med. Int. Heal 15, 614–618 [DOI] [PubMed] [Google Scholar]
- 10.Webster BL et al. (2013) Praziquantel treatment of school children from single and mixed infection foci of intestinal and urogenital schistosomiasis along the Senegal River Basin: monitoring treatment success and re-infection patterns. Acta Trop. 128, 292–302 [DOI] [PubMed] [Google Scholar]
- 11.Loker ES and Hofkin BV (2015) Parasitology: A Conceptual Apporach, Garland Science. [Google Scholar]
- 12.Viney M and Cable J (2011) Macroparasite life histories. Curr. Biol 21, R767–R774 [DOI] [PubMed] [Google Scholar]
- 13.Clark WC (1974) Interpretation of life history pattern in the Digenea. Int. J. Parasitol 4, 115–123 [DOI] [PubMed] [Google Scholar]
- 14.Basch PF (1991) Schistosomes: development, reproduction, and host relations, Oxford University Press. [Google Scholar]
- 15.Berriman M et al. (2009) The genome of the blood fluke Schistosoma mansoni. Nature 460, 352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young ND et al. (2012) Whole-genome sequence of Schistosoma haematobium. Nat. Genet 44, 221–225 [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y et al. (2009) The Schistosoma japonicum genome reveals features of host–parasite interplay. Nature 460, 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B et al. (2018) Stem cell heterogeneity drives the parasitic life cycle of Schistosoma mansoni. eLife 7, e35449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diaz Soria CL et al. (2020) Single-cell atlas of the first intra-mammalian developmental stage of the human parasite Schistosoma mansoni. Nat. Commun 11, 6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarashansky AJ et al. (2019) Self-assembling manifolds in single-cell RNA sequencing data. eLife 8, e48994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li P et al. (2020) Single-cell analysis of Schistosoma mansoni identifies a conserved genetic program controlling germline stem cell fate. Nat. Commun 12, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wendt G et al. (2020) A single-cell RNA-seq atlas of Schistosoma mansoni identifies a key regulator of blood feeding. Science. 369, 1644–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan SCT (1980) The fine structure of the miracidium of Schistosoma mansoni. J. Invertebr. Pathol 36, 307–372 [DOI] [PubMed] [Google Scholar]
- 24.Schutte H (1974) Studies of South African strain of Schistosoma mansoni. S. Afr. J. Sci 70, 327–346 [Google Scholar]
- 25.Cort WW et al. (1954) Germinal development in the sporocysts and rediae of the digenetic trematodes. Exp. Parasitol 3, 185–225 [DOI] [PubMed] [Google Scholar]
- 26.Whitfield PJ and Evans NA (1983) Parthenogenesis and asexual multiplication among parasitic platyhelminths. Parasitology 86, 121–160 [DOI] [PubMed] [Google Scholar]
- 27.Jourdane J and Theron A (1980) Schistosoma mansoni: cloning by microsurgical transplantation of sporocysts. Exp. Parasitol 50, 349–357 [DOI] [PubMed] [Google Scholar]
- 28.Wang B et al. (2013) Functional genomic characterization of neoblast-like stem cells in larval Schistosoma mansoni. eLife 2, e00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solana J (2013) Closing the circle of germline and stem cells: the primordial stem cell hypothesis. Evodevo 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juliano CE et al. (2010) A conserved germline multipotency program. Development 137, 4113–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alié A et al. (2015) The ancestral gene repertoire of animal stem cells. Proc. Natl. Acad. Sci. USA 112, E7093–E7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galaktionov KV and Dobrovolskij AA (2003) The biology and evolution of Trematodes, Springer, Dordrecht [Google Scholar]
- 33.Takahashi K and Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 34.Geyer KK et al. (2011) Cytosine methylation regulates oviposition in the pathogenic blood fluke Schistosoma mansoni. Nat. Commun 2, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whittle CA and Extavour CG (2017) Causes and evolutionary consequences of primordial germ-cell specification mode in metazoans. Proc. Natl. Acad. Sci. USA 114, 5784–5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer H et al. (2016) NF-YB Regulates spermatogonial stem cell self-renewal and proliferation in the planarian Schmidtea mediterranea. PLoS Genet. 12, e1006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins JJ et al. (2013) Adult somatic stem cells in the human parasite Schistosoma mansoni. Nature 494, 476–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wendt GR et al. (2018) Flatworm-specific transcriptional regulators promote the specification of tegumental progenitors in Schistosoma mansoni. eLife 7, e33221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins JJ et al. (2016) Stem cell progeny contribute to the schistosome host-parasite interface. eLife 5, e12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins JJ et al. (2011) An atlas for Schistosoma mansoni organs and life-cycle stages using cell type-specific markers and confocal microscopy. PLoS Negl. Trop. Dis 5, e1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson RA and Webster LA (1974) Protonephridia. Biol. Rev. Camb. Philos. Soc 49. 127–160 [DOI] [PubMed] [Google Scholar]
- 42.Wilson RA (2009) The saga of schistosome migration and attrition. Parasitology 136, 1581–1592 [DOI] [PubMed] [Google Scholar]
- 43.Clegg JA (1965) In vitro cultivation of Schistosoma mansoni. Exp. Parasitol 16, 133–147 [DOI] [PubMed] [Google Scholar]
- 44.Hockley DJ and McLaren DJ (1973) Schistosoma mansoni: changes in the outer membrane of the tegument during development from cercaria to adult worm. Int. J. Parasitol 3, 13–20 [DOI] [PubMed] [Google Scholar]
- 45.Tyler S and Hooge M (2004) Comparative morphology of the body wall in flatworms (Platyhelminthes). Can. J. Zool 82, 194–210 [Google Scholar]
- 46.Skelly P and Shoemaker C (2001) The Schistosoma mansoni host-interactive tegument forms from vesicle eruption of a cyton network. Parasitology 122, 67–73 [DOI] [PubMed] [Google Scholar]
- 47.Wilson RA (1974) The tegument of Schistosoma mansoni: observations on the formation, structure and composition of cytoplasmic inclusions in relation to tegument formation. Parasitology 68, 239–258 [PubMed] [Google Scholar]
- 48.Li XH et al. (2013) The schistosome oesophageal gland: initiator of blood processing. PLoS Negl. Trop. Dis 7, e2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson RA et al. (2015) The schistosome esophagus is a ‘hotspot’ for microexon and lysosomal hydrolase gene expression: implications for blood processing. PLoS Negl. Trop. Dis 9, e0004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J et al. (2020) The esophageal gland mediates host immune evasion by the human parasite Schistosoma mansoni. Proc. Natl. Acad. Sci. USA 117, 19299–19309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laumer CE et al. (2015) Nuclear genomic signals of the “microturbellarian” roots of platyhelminth evolutionary innovation. eLife 4, e05503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Littlewood DTJ and Waeschenbach A (2015) Evolution: a turn up for the worms. Curr. Biol 25, R457–R460 [DOI] [PubMed] [Google Scholar]
- 53.Tsai IJ et al. (2013) The genomes of four tapeworm species reveal adaptations to parasitism. Nature 496, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collins JJ and Newmark PA (2013) It’s no fluke: The planarian as a model for understanding schistosomes. PLoS Pathog. 9, e1003396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarashansky AJ et al. (2020) Mapping single-cell atlases throughout metazoa unravels cell type evolution. bioRxiv DOI: 10.1101/2020.09.28.317784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fincher CT et al. (2018) Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360, eaaq1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plass M et al. (2018) Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360, eaaq1723. [DOI] [PubMed] [Google Scholar]
- 58.Zeng A et al. (2018) Prospectively isolated tetraspanin+ neoblasts are adult pluripotent stem cells underlying planaria regeneration. Cell 173, 1593–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pandey S et al. (2018) Comprehensive identification and spatial mapping of habenular neuronal types using single-cell RNA-seq. Curr. Biol 28, 1052–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brunet T et al. (2016) The evolutionary origin of bilaterian smooth and striated myocytes. eLife 5, e19607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scimone ML et al. (2016) Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. eLife 5, e12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Witchley JN et al. (2013) Muscle cells provide instructions for planarian regeneration. Cell Rep. 4, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reddien PW (2018) The cellular and molecular basis for planarian regeneration. Cell 175, 327–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wendt GR and Collins JJ (2016) Schistosomiasis as a disease of stem cells. Curr. Opin. Genet. Dev 40, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nation CS et al. (2020) Schistosome migration in the definitive host. PLoS Negl. Trop. Dis 14, e0007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jurberg AD et al. (2009) The embryonic development of Schistosoma mansoni eggs: proposal for a new staging system. Dev. Genes Evol 219, 219–234 [DOI] [PubMed] [Google Scholar]
- 67.Wang JP et al. (2019) Systematically improved in vitro culture conditions reveal new insights into the reproductive biology of the human parasite Schistosoma mansoni. PLoS Biol. 17, e3000254. [DOI] [PMC free article] [PubMed] [Google Scholar]


