Abstract
Cleavage of nascent transcripts is a fundamental process for eukaryotic messenger RNA (mRNA) maturation and for production of different mRNA isoforms. In eukaryotes, cleavage of mRNA precursors by the highly conserved endonuclease CPSF73 is critical for mRNA stability, export from the nucleus, and translation. As an essential enzyme in the cell, CPSF73 surprisingly shows promise as a drug target for specific cancers and for protozoan parasites. In this review, we cover our current understanding of CPSF73 in cleavage and polyadenylation, histone pre-mRNA processing, and transcription termination. We discuss the potential of CPSF73 as a target for novel therapeutics and highlight further research into the regulation of CPSF73 that will be critical to understanding its role in cancer and other diseases.
Keywords: CPSF73/CPSF3, polyadenylation, mRNA 3’ end processing, transcription termination, gene expression, cancer
Cleavage of Nascent Transcripts is a Fundamental Process for Eukaryotic mRNA Maturation and Production of Different mRNA Isoforms
Pre-messenger RNAs (mRNAs) transcribed by eukaryotic RNA polymerase II (Pol II) undergo several modifications, including capping, splicing, adenosine methylation, and cotranscriptional cleavage to form the mature mRNA 3’ end. For most mRNAs, a poly(A) tail is added to the cleaved 3’ end, while mRNAs encoding replication-dependent histones are cleaved but not polyadenylated. This processing step is critical for mRNA stability, export from the nucleus, and translation. Altering the position of the cleavage site, also called the poly(A) site (PAS), in a process called alternative polyadenylation (see Glossary), is well-recognized as a mechanism to alter the protein-coding capacity of mRNAs or to include regulatory sequences in the mRNA’s 3’ untranslated region (UTR) that affect the level of the encoded protein [1–3].
Structural and biochemical studies, including many in the last few years, have elucidated the machinery responsible for 3’ end processing, and have provided a more complete understanding of this important biological process (for reviews, see [1–6]). A critical component of the processing complex is the essential endonuclease CPSF73, which is the focus of this review. Without CPSF73’s cleavage activity, mRNAs cannot be polyadenylated and released from the site of transcription for export to the cytoplasm. Cleavage by CPSF73 is also necessary for the termination of transcription that defines gene boundaries, and thus inhibits transcriptional interference at downstream genes. CPSF73 has been linked to several cancers [7–11], but it was surprising that some cancers show striking susceptibility to inhibition of a protein with such essential roles in gene expression [11]. Furthermore, even though this nuclease is highly conserved in sequence and function across eukaryotes, and even found in archaea (Box 1), inter-species differences in CPSF73 are sufficient to allow development of drugs that specifically inhibit the nuclease of parasitic protozoans [12–15]. However, our understanding of the regulation of mRNA 3’ end processing machinery, including the CPSF73 subunit reviewed here, is still at an early stage, and this deficit impedes potential translational research and applications. In the present review, we summarize our knowledge of the structures of CPSF73, its function in mRNA 3’ end processing and transcription termination, its role in cancer, and its regulation.
Box 1. The structure and function of CPSF73 is conserved from prokaryotes to eukaryotes.
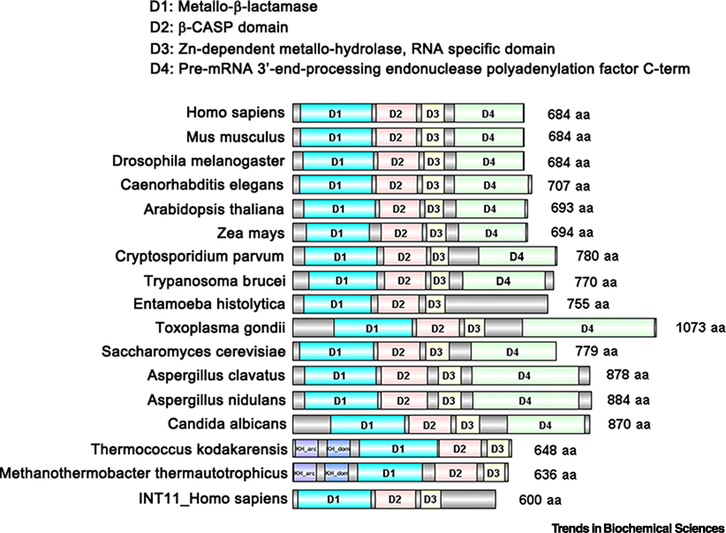
The three-dimensional structure and the amino acid sequence of CPSF73 reveal four functional domains [17]. These domains are a metallo-β-lactamase domain (human CPSF73 amino acids 23–234), a β-CASP domain (amino acids 246–367), a Zn-dependent metallo-hydrolase and RNA specificity domain (amino acid 382–448), and a C-terminal domain (amino acid 477–683) (Figure 1A and Figure I). The first three domains place CPSF73 solidly in a family of nucleases with a metallo-β-lactamase fold. The β -CASP domain is positioned C-terminal to the β-lactamase domain, and the active site of CPSF73 with two zinc ions is located at the interface of these two domains [17]. A hydroxide ion bridging the two zinc ions initiates the cleavage reaction [19]. The Zn-dependent metallo-hydrolase, RNA specificity domain adds essential structural elements to the β -CASP domain and is unique to RNA/DNA-processing nucleases [17, 76, 77]. The C-terminal domain facilitates interactions with other components of the complex such as CPSF100 and symplekin. CPSF100 shares sequence conservation and a similar domain architecture with CPSF73 but has amino acid substitutions in the zinc ligands that make it an inactive nuclease.
The domain architecture of CPSF73 is conserved in plants, fungi, animals, and protozoa, with the greatest conservation being in the N-terminal region containing the endonuclease activity (Figure I). Homologs with similar function in cleavage and transcription termination can even be found in the archaea group of prokaryotes [46, 77–79]. The protozoan and archaea enzymes are most divergent from the CPSF73 of fungi and multicellular eukaryotes, and these differences are sufficient to make CPSF73 homologs in pathogenic parasites good targets for drug treatment (discussed in the section on CPSF73 and diseases). A paralog of CPSF73 within metazoan cells is the endonuclease INT11 (Figure I), which as part of Integrator (INT) complex, forms the mature ends of snRNAs and also participates in termination of enhancer RNA and long non-coding RNA (lncRNA) transcripts [80].
Figure I. Domain organization of CPSF73: comparison from prokaryotes to eukaryotes.
The structural domains and sequence patterns of selected CPSF3 homologs were predicted by InterPro Databases and UniProt Knowledgebase (UniProtKB) using the name of each protein, and were then visually compared with the conserved regions by DOG 2.0 [88]. The Metallo-β-lactamase domain is indicated in blue, the β-CASP domain in red, the Zn-dependent metallo-hydrolase, RNA specificity domain in yellow and the pre-mRNA 3’-end processing endonuclease polyadenylation factor C-term in green. The origin of each protein is at the left; the size in amino acid (aa) residues is at the right. CPSF73 homologs are shown from mammals (Homo sapiens and Mus musculus), other model metazoans (Drosophila melanogaster and C. elegans), plants (Arabidopsis thaliana and Zea mays), parasites (Cryptosporidium parvum, Trypanosoma brucei, Entamoeba histolytica, and Toxoplasma gondii), fungus (Saccharomyces cerevisiae, Aspergillus clavatus, Aspergillus nidulans, and Candida albicans), and Archae bacteria (Thermococcus kodakarensis and Methanothermobacter thermautotrophicus).
Incorporation of CPSF73 into the mRNA 3’ End Processing Machinery
CPSF73 belongs to the metallo-β-lactamase family of nucleases (Box 1). It has four distinct domains: the metallo-β-lactamase domain, the β -CASP domain, and a Zn-dependent metallo-hydrolase and RNA specificity domain form the nuclease core whose structure has been determined (Figure 1A), and a C-terminal domain which helps recruit CPSF73 into processing machinery. CPSF73 is currently known to reside in two unique complexes depending on its target. It is a core component of the 7-subunit Cleavage and Polyadenylation Specificity Factor (CPSF) needed to produce cleaved and polyadenylated mRNAs [4, 16–18], and it is also a subunit of a simpler 4-subunit factor, the histone pre-mRNA cleavage complex (HCC), that cleaves histone mRNA precursors [19, 20].
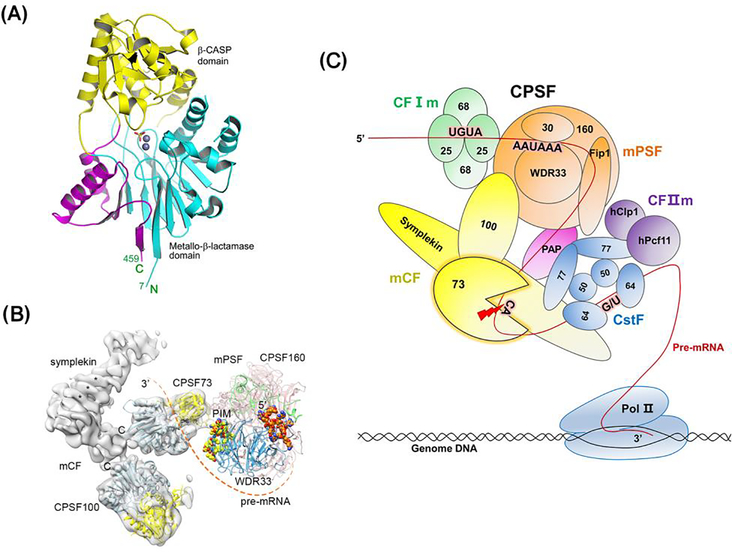
Figure 1. The role of CPSF73 in processing polyadenylated mRNAs.
(A) Crystal structure of the first 459 amino acids of human CPSF73 (PDB id 2i7v) [17]. The active site complexed with zinc ions (grey spheres) and a sulphate ion (stick model) that might mimic the phosphate group of the RNA substrate, is located at the interface of the metallo-β-lactamase domain (blue) and the β-CASP domain (yellow). Additional sequence that is not found in canonical metallo-β-lactamases, called the Zn-dependent metallo-hydrolase and RNA specificity domain, is critical for formation of the active site and is shown in pink. (B) Structure of the Cleavage and Polyadenylation Specificity Factor (CPSF). CPSF73 resides in CPSF, which can be separated into two functional modules, mammalian Polyadenylation Specificity Factor (mPSF) and mammalian Cleavage Factor (mCF). A possible path of the pre-mRNA from the polyadenylation site (PAS) to the CPSF73 active site is indicated with the dashed line. Reproduced from with permission from [16]. (C) The complete polyadenylated mRNA 3’ end processing machinery. The processing machinery is comprised of the CPSF, yellow and orange, Cleavage stimulatory Factor (CstF; blue), Cleavage Factor Im (CFIm; green), CFIIm (purple), and poly(A) polymerase (PAP; pink). In the processing of polyadenylated mRNA transcripts, these factors assemble onto the RNA after RNA Polymerase II (Pol II) moves through the PAS and its immediate flanking sequences. CPSF73 cleaves at the PAS (red arrow), and PAP adds adenosines at the end of the pre-mRNA to form a poly(A) tail.
CPSF and the processing of polyadenylated mRNA
Most eukaryotic mRNAs are cleaved by CPSF, specifically by CPSF73, followed by a non-templated addition of adenosines to the new 3’ end. This biochemical reaction has been best characterized in human cells, but the conservation of the proteins comprising the processing complex suggests the mechanism is similar in all metazoans [21]. CPSF consists of two functionally distinct modules: a cleavage factor (mCF) and a polyadenylation specificity factor (mPSF) (Figure 1B and Box 2) [4, 16]. mCF provides the cleavage activity and contains CPSF100, symplekin, and CPSF73, while mPSF contains CPSF160, CPSF30, Fip1, and WDR33. mPSF is the organizational core of the processing machinery, interacting with mCF, Cleavage stimulatory Factor (CstF), poly(A) polymerase (PAP), and the AAUAAA RNA signal (see below). Recent cryo-electron microscopy (cryo-EM) studies showed that human mCF forms a highly flexible, trilobal structure with the three subunits linked together by interactions of their C-terminal domains (CTDs; Figure 1B [16]).
Box 2. The cleavage and polyadenylation machinery.
Complexes assembled on pre-mRNA in vitro from mammalian extract contain over 80 proteins [22], but fifteen comprise the core machinery that processes polyadenylated mRNAs [4, 5] (see Figure 1C). These separate biochemically into the poly(A) polymerase (PAP) and four complexes: Cleavage and Polyadenylation Specificity Factor (CPSF), Cleavage Stimulatory Factor (CstF), and Cleavage Factors I and II (CFIm and CFIIm). Together, these factors recognize signal sequences around the polyadenylation site (PAS), cleave, and add the poly(A) tail. Structures have been determined for portions of the machinery and these have been summarized in a recent review by Sun, et al [4].
CPSF
CPSF contains seven subunits that are functionally organized into two submodules: the polyadenylation specificity factor (mPSF) and the cleavage factor (mCF) [4, 5, 16]. The mPSF subunits WDR33 and CPSF30 (also known as CPSF4) work together to recognize the AAUAAA hexamer or one of its variants (AAUAAA, A(U/G)UAAA, or UAUAAA) located upstream of the PAS, while CPSF160 (CPSF1) coordinates assembly of the other CPSF components to facilitate AAUAAA binding and recruit mCF [37, 81–84]. The Fip1 subunit of mPSF recruits PAP to the processing complex and can also bind to U-rich sequence upstream of the AAUAAA hexamer to further modulate PAS recognition [85]. mCF provides the cleavage activity and contains CPSF100 (CPSF2), CPSF73 (CPSF3) and symplekin. Symplekin provides scaffolding function. CPSF100 and CPSF73 form an obligate heterodimer with the endonuclease activity provided by CPSF73.
CstF
CstF consists of three subunits present as dimers: CstF50 (CSTF1), either CstF64 (CSTF2) or CstF64tau, and CstF77 (CSTF3). CstF directly binds to the downstream U/GU rich element through the CstF64 subunit and helps assemble a stable processing complex at a bonafide PAS [4–6].
CFIm and CFIIm
CFIm contains CFIm25 (also known as NUDT21), which forms a homodimer that further heterodimerizes with two copies of CFIm68 or CFIm59 [28]. CFIm directly binds to the UGUA motifs located upstream of AAUAAA [28]. CFIIm has the hPcf11 and hClp1 subunits. Pcf11 binds G-rich auxillary elements downstream of the PAS and also contains a Pol II C-terminal domain (CTD)-interacting domain (CID) that helps recruit CFIIm to the elongating transcriptional complex.
PABPN1
PABPN1 is a nuclear poly(A) binding protein. It is not needed for the cleavage and polyadenylation steps but acts to limit tail length to 200–300 adenosines [86].
Regulatory factors
Many genes have multiple polyadenylation sites and the choice of which one is used, called alternative polyadenylation, is determined by the level of the core factors and adherence of the PAS to consensus signal sequences, the presence of enhancer or repressor RNA-binding proteins and their recognition sites, the speed of Pol II transcription, and chromatin modifications [1, 87].
Beyond CPSF, processing of pre-mRNA also requires PAP and three additional complexes, summing up to 15 proteins in total (Figure 1C and Box 2): CstF and Cleavage Factors I and II (CFIm and CFIIm) [5, 16, 22]. While several auxiliary proteins have also been identified, the five factors listed above are sufficient for constitutive processing and are described herein as the polyadenylated mRNA processing machinery. This machinery identifies a PAS on pre-mRNA by interacting with specific conserved sequence elements within the transcripts. Specifically, CPSF recognizes one of the main signal motifs AAUAAA or one of its variants located 20–30 nt upstream from the PAS [23–25], CFIm recognizes a UGUA motif located further upstream [26–28], and CstF recognizes a G/U rich element downstream of the PAS [29, 30] (Figure 1C). Protein/protein interactions between the factors stabilize the machinery at the PAS, leading to the cleavage and poly(A) addition reactions.
HCC and the histone mRNA processing machinery
Histone mRNAs are tightly regulated and present in high levels only in S-phase to provide the histone proteins necessary for packaging newly replicated DNA [31]. In plants and budding yeast, all histone mRNAs are polyadenylated by the machinery described above. However, metazoans have evolved a unique processing mechanism in which replication-dependent histone mRNAs are cleaved by HCC, specifically by its CPSF73 subunit, but not polyadenylated (Figure 2A) [31]. Like mCF, HCC contains CPSF73, CPSF100, and symplekin, but also includes CstF64 [32–35]. Interactions of the FLASH protein with the U7 small nuclear ribonucleoprotein (snRNP) and the Stem Loop Binding Protein (SLBP) recruit HCC to the cleavage site [19, 33–35]. Together, these four factors comprise the histone mRNA processing machinery. The cleavage site is defined by two conserved sequences: a stem-loop upstream of the cleavage site that is bound by SLBP, and the histone downstream element (HDE) which base-pairs with U7 snRNP downstream of the cleavage site.
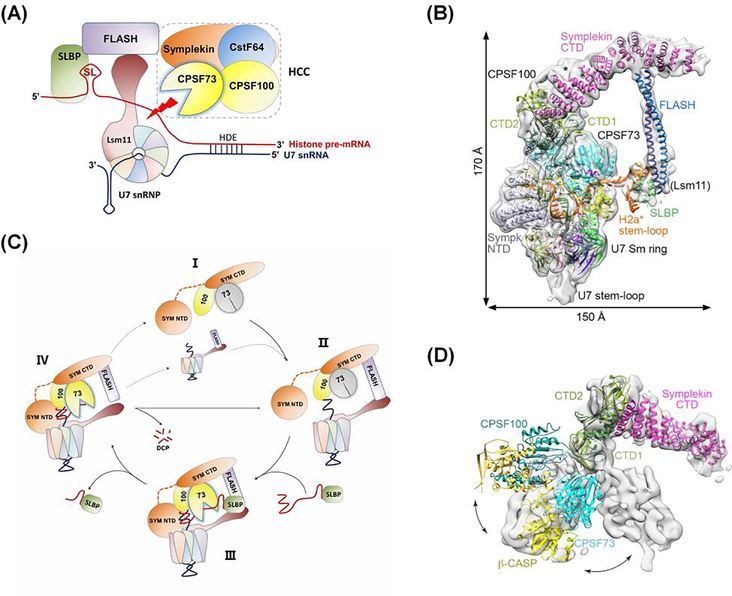
Figure 2. The role of CPSF73 in processing unpolyadenylated histone mRNAs.
(A) The histone mRNA 3’ end processing machinery. The cleavage site is defined by two conserved elements: a stem-loop (SL) upstream of the cleavage site that is bound by stem-loop binding protein (SLBP), and the histone downstream element (HDE) which base-pairs with U7 small nuclear ribonucleoprotein (snRNP) downstream of the cleavage site. The histone cleavage complex (HCC) contains CPSF73, CPSF100, and symplekin, and also CstF64, the subunit of CstF that recognizes the downstream G/U-rich element of polyadenylated mRNA precursors. U7 snRNP interacts with the FLASH protein to recruit HCC to the cleavage site (red arrow). (B) Structure of the histone processing machinery assembled on RNA containing the histone H2A processing site, as determined by cryo-electron microscopy (reproduced from [19]). The positions of the N-Terminal Domain (NTD) and C-terminal Domain (CTD) of symplekin and the two interacting subdomains of the CTDs of CPSF73 and CPSF100 (CTD1 and CTD2) are indicated. CstF64 was not observed in the EM density and is not required for cleavage in vitro. (C) Proposed model for histone pre-mRNA 3’ end processing cycle (adapted from [19]). The U7 snRNP, FLASH, and HCC (State I) come together to form the processing complex (State II), followed by the recognition of the pre-mRNA bound to SLBP, activation of CPSF73 and pre-mRNA cleavage (State III; see also Figure 2B). After cleavage, the upstream product bound to SLBP is released, the downstream product is degraded from the 5’ end (state IV), and the machinery can be recycled.
(D) Conformational differences in the positions of CPSF73, CPSF100, and symplekin relative to each other in the inactive state of mCF (gray surface) compared to that of active HCC (reproduced from [19]).
CPSF73 Endonuclease Activity is Activated by Incorporation into the Processing Machinery
Once the components of the two 3’ end processing machineries were identified, more recent studies have focused on decoding how the individual subunits work together as a whole to execute 3’ end cleavage. A critical prerequisite of this processing is that cleavage does not occur until the processing machinery is fully assembled at a bonafide cleavage site. One property that helps with this is the very weak cleavage activity of CPSF73 on its own [17]. This low activity is due to CPSF73 residing in a closed state in which the catalytic site is not accessible to RNA substrate, as revealed by the structure of human CPSF73 [17] (Figure 1A) and the yeast homolog Ysh1 [36]. Further, CPSF73 on its own does not bind RNA with sequence specificity, and another prerequisite for processing is that the RNA-recognition components of the processing machineries direct CPSF73 to the correct cleavage site. How assembly of the full processing machinery creates the conformational rearrangements needed to position and activate CPSF73 represent a major gap in our understanding of the processing of polyadenylated mRNAs.
A recent structural study on histone pre-mRNA processing broke the ice on this issue and gave significant insight into how interactions within the processing machinery activate CPSF73 [19]. As shown in Figure 2C, HCC is highly dynamic during the histone pre-mRNA 3’ end processing cycle, with CPSF73 in a closed conformation before it is assembled into the histone mRNA processing machinery (State I). In State II, HCC associates with U7 snRNP through interaction of FLASH and the symplekin C-terminal domain (SYM CTD), but CPSF73 remains inactive. Next, SLBP brings the histone pre-mRNA to the machinery in State III, with U7 snRNP binding the HDE, and the architecture of HCC undergoes an extensive change that opens the active site of CPSF73 to receive and cut its RNA substrate. After cleavage, SLBP with cleaved mRNA is released, the downstream cleavage product is degraded by the 5’−3’ exonuclease activity of CPSF73 [18] in State IV, and CPSF73 returns to the inactive state.
However, the comparable dynamic interactions of CPSF73 in CPSF or the 15-protein mRNA processing machinery are not known. As described above, cryo-EM analysis of mCF revealed a highly dynamic trilobal structure held together by interactions of the CTDs of CPSF73, CPSF100, and symplekin [16]. However, in the active histone mRNA processing machinery, the structure of the equivalent HCC is more compact with greatly increased contact between CPSF73 and CPSF100 [19] (Figure 2B, D). The formation of the duplex between U7 snRNA and the HDE, as well as contacts between the CPSF73 β-CASP domain and the Lsm10 subunit of U7 snRNP, between the symplekin CTD and FLASH, and between the symplekin NTD and the HDE-U7 duplex, likely help correctly position CPSF73 and trigger its activation.
In the cryo-EM structure of CPSF (Figure 1B), the position of the cleavage module mCF relative to mPSF is highly variable, with only a small interaction interface detected between CPSF100 and mPSF [16]. Similar structural flexibility was also observed upon reconstitution of the core of the yeast Cleavage/Polyadenylation Factor from recombinant proteins [36]. The analysis of the HCC indicates that the organization and flexibility of mCF must also change dramatically upon assembly of the polyadenylated mRNA processing machinery on its RNA substrate, but the comparable protein/protein interactions that replace those provided by U7 snRNP and FLASH are not known. An added complication in the mechanics of this machinery compared to histone mRNA processing one is that a further reorganization is needed to allow PAP to access the 3’ end formed by CPSF73 without disengaging subunits necessary for polyadenylation from the cleaved RNA. The composition of a post-cleavage complex is not known, but mPSF and PAP are sufficient to reconstitute AAUAAA-dependent polyadenylation in vitro [37]. Determining the activated cleavage and the post-cleavage structures and how transitions are made will be an urgent but challenging goal of future research.
CPSF73 is Needed for Efficient Termination of Transcription.
Transcription termination occurs when the polymerase and nascent RNA are released from the DNA template and is important in defining gene boundaries, recycling RNA Pol II for subsequent rounds of transcription, and controlling gene expression and preventing pervasive transcription (Figure 3). Termination downstream of polyadenylated mRNA genes utilizes a combination of two mechanisms. One results from allosteric effects, involving changes in RNA Pol II processivity upon passage through the PAS and probably mediated by loss of elongation factors and acquisition of termination factors [38, 39]. The second mechanism, known as the torpedo model, depends heavily on cleavage by CPSF73 at the PAS to provide entry for the 5’−3’ exonuclease Xrn2 to digest the nascent RNA; upon reaching RNA Pol II, Xrn2 releases it from the DNA template [38]. CPSF73 also promotes termination of non-coding RNAs (ncRNAs), such as PROMPTs [40].
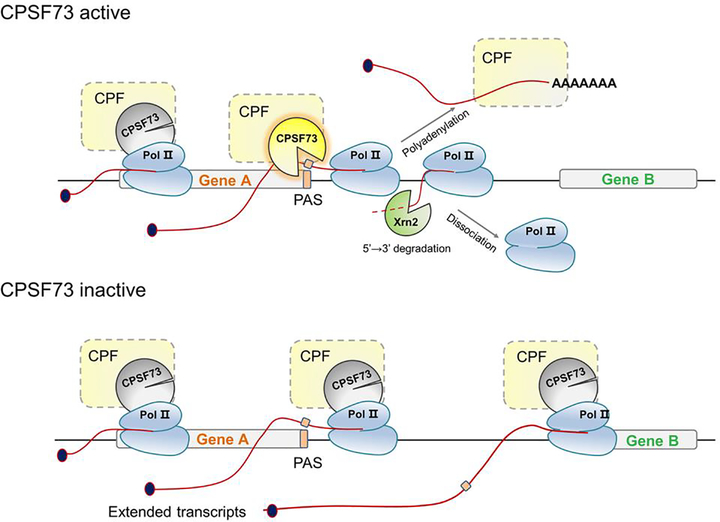
Figure 3. The role of CPSF73 in transcription termination.
During the process of transcription, CPSF73 as part of CPSF is recruited to the elongation complex along with the other cleavage/polyadenylation factors (CPF) that comprise the polyadenylated mRNA processing machinery. Once the polyadenlyation site (PAS) is transcribed, the full processing machinery assembles on the nascent pre-mRNA and active CPSF73 cleaves the transcript at the PAS, followed by polyadenylation and release of the mRNA. Xrn2 degrades the nascent transcript, leading to dissociation of Pol II. When the CPSF73 level is limited or it is inactive due to inhibitors or mutations, cleavage at the PAS is defective, which causes Pol II to continue transcribing and thus generate extended transcripts that can even reach downstream genes.
CPSF73 depletion causes significant readthrough transcription downstream of PASs and histone mRNA cleavage sites (Figure 3) in metazoan cells [11, 20, 41–43], yeast [44, 45], and even archaea [46], and increases the use of distal PASs [8]. Various stress conditions, such as heat shock, osmotic stress, and oxidative stress, as well as diseases such as HSV-1 infection and renal carcinoma, also lead to widespread readthrough transcription [47], but whether this change is through specifically regulating CPSF73 expression and function or through a general down-regulation of the cleavage/polyadenylation machinery, is unknown.
Several biological consequences of readthrough transcription have been identified. Extension into and beyond intergenic regions can either suppress or enhance activity of downstream genes through transcriptional interference [48–50]. Transcription into downstream genes can have unexpected outcomes, such as an increase in circular RNA due to a greater potential for back-splicing [43]. Moreover, addition of 3’ UTR sequence downstream of the preferred PAS can titrate microRNAs (miRNAs), which in turn affects expression of targets of these miRNAs [51], and this, too, would be an expected consequence of a decrease in CPSF73. In summary, inefficient 3’ end processing caused by inhibition of CPSF73 can affect gene expression by directly decreasing mRNA production as well as indirectly through increased 3’UTR length and readthrough transcription into downstream genes.
CPSF73 is Regulated at Multiple Levels
Only a few studies have documented regulation of the expression and activity of CPSF73. For example, the HIV-1 Tat protein increases CPSF73 mRNA and protein levels without affecting CPSF100 and CPSF160 mRNAs, and may represent a strategy to regulate both viral and cellular mRNA processing [52]. CPSF73 activity could also be regulated by limiting its recruitment to the 3’ ends of transcribed genes. For example, this recruitment decreases after heat shock [53], possibly due to depletion of the heat-labile symplekin subunit of mCF [54]. Additionally, regulation could be accomplished by blocking the nuclear localization of CPSF73, and indeed, interaction with the cell stress response protein CSR1 keeps CPSF73 in the cytoplasm, resulting in inhibition of 3’ end processing [9]. Unexpectedly, the eukaryotic translation initiation factor eIF4E was found to promote the processing and nuclear export of mRNAs of CPSF73 and other CPSF subunits, and consequently increase their protein level. It physically interacts with the unprocessed transcripts of these genes, as well as with CPSF, providing a mechanism to drive their 3’ end cleavage [55].
Addition of ubiquitin or the ubiquitin-like modifier SUMO can regulate protein stability, subcellular localization, and interaction [56, 57]. Indeed, CPSF73 is a target for SUMO modification (sumoylation), and the major sites of SUMO modification in CPSF73 are highly conserved. De-sumoylation of proteins in extract or depletion of the SUMO-conjugating enzyme Ubc9 inhibited 3’ end processing in vitro, but it is not known if the modifications on CPSF73 were the ones needed for efficient processing [58].
Ysh1, the yeast CPSF73 homolog, is a target for ubiquitin-mediated proteasomal degradation. Ubiquitination of Ysh1 is modulated by the presence of Ipa1, an essential protein that physically interacts with Ysh1 and Mpe1, another cleavage/polyadenylation complex subunit [36, 59]. Mutation of Ipa1 causes degradation of Ysh1 by the proteasome, defects in mRNA 3′ end processing in vitro and in vivo, inefficient transcription termination downstream of PAS and at snoRNA genes, and poor recruitment of the symplekin homolog Pta1 to the 3′ end of an mRNA gene [45, 59]. In the absence of Ipa1, Ysh1 is ubiquitinated by the Ubiquitin-conjugating enzyme Ubc4 and Mpe1, which is a RING ubiquitin ligase in addition to its role in cleavage and polyadenylation [60]. Interestingly, two independent quantitative proteomics screens identified the human Ipa1 homolog, UBE3D, as a CPSF73 interactor [61, 62], and CPSF73 was found to be part of the ubiquitome [63], indicating the mechanism by which Ipa1 regulates polyadenylation may be conserved in humans.
A recent intriguing finding is that CPSF73 is capable of cleaving single-stranded DNA substrates in vitro [18], raising the possibility of CPSF73 playing a role in some aspect of DNA metabolism. This would require redirection of complexes in which CPSF73 is known to reside to non-RNA targets or assembly of CPSF73 into a DNA-specific complex. In this regard, it is interesting that the DNA-activated protein kinase complex (DNA-PK/Ku70/Ku86), which functions in DNA-damage repair, is associated with the polyadenylated mRNA processing machinery that is assembled on exogenous pre-mRNA in crude extract [22].
Alterations in CPSF73 Levels are Associated with Cancer Phenotypes
The studies described below show a correlation of CPSF73 with different cancers (Figure 4, right) and support the idea of CPSF73 as a biomarker for prognosis, as well as a target for cancer treatment.
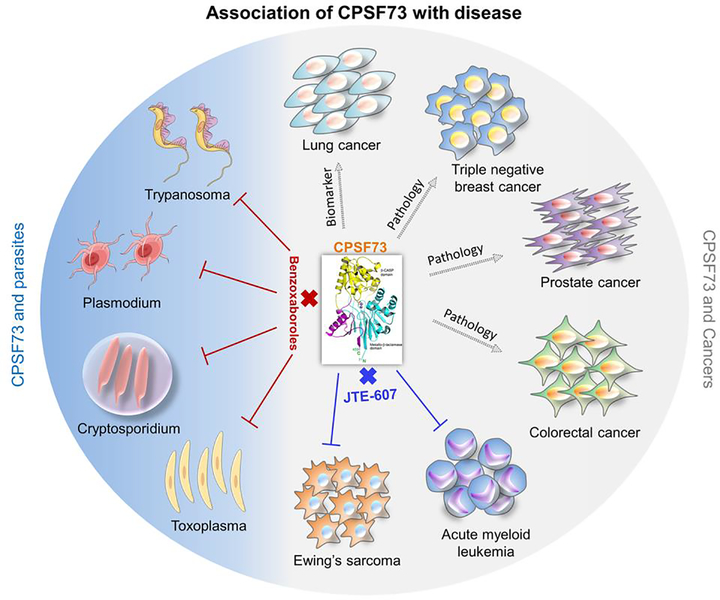
Figure 4. Association of CPSF73 with diseases.
CPSF73 has been implicated in cancers (gray area of the circle) as a biomarker in lung cancer, as a contributor to pathology of prostate, colorectal, and triple-negative breast cancers, and as a new therapeutic intervention for acute myeloid leukemia (AML) and Ewing’s sarcoma (ES) through the CPSF73-specific inhibitor JTE-607. The blue portion of the circle indicates the use of benzoxaboroles as drugs that specifically inhibit the CPSF73 of protozoan parasites, including Trypanosma, Plasmodium, Cryptosporidium, and Toxoplasma, without affecting mammalian CPSF73.
Non-small cell lung cancer
Analysis of overall survival and recurrence-free survival of this disease, which is responsible for 85% of all lung cancer cases [64], showed that CPSF73 over-expression was associated with prognosis and recurrence of the lung adenocarcinoma subtype [7]. According to the Cancer Genome Atlasi, high expression of CPSF73 is also associated with unfavorable prognosis in liver and renal cancers.
Triple negative breast cancer
CPSF73 knock-down significantly reduced migration of triple negative breast cancer cells [8], a highly aggressive breast cancer subtype [65]. In fibroblasts, reduction of CPSF73 levels also slowed fibroblast migration and caused the use of distal PASs in genes with multiple PASs [8]. Triple negative breast cancer cells rely on usage of proximal PASs for oncogene expression [66–68], and expression of shorter 3’ UTRs was an important predictor of patient outcome even beyond established clinical attributes [67]. Thus, CPSF73 levels could affect breast cancer cell properties by altering expression of oncogenes and genes critical for cell movement.
Prostate cancer
CSR1, discussed earlier as a regulator of CPSF73 nuclear localization, is a known tumor suppressor and is significantly down-regulated in prostate cancer [9], a leading cause of cancer-related deaths for men in the United States [69]. Knocking down CPSF73 generated a pattern of cell death in prostate cancer cells similar to that caused by CSR1 induction [9].
Colorectal cancer (CRC)
CRC is the third most frequent cancer and the second leading cause of cancer-related death worldwide [69, 70]. Increasing evidence indicates that deregulation of long non-coding RNAs (lncRNAs) contributes to tumor initiation and progression. The CASC9 lncRNA is frequently up-regulated in CRC, with higher CASC9 levels associated with poor patient outcomes [10].
Knockdown of CASC9 inhibited growth and promoted apoptosis in CRC cells. CPSF73 interacts with CASC9, and knockdown of CPSF73 mimicked the effects of CASC9 knockdown. Moreover, CPSF73 was significantly up-regulated in CRC tissue compared to normal colon samples [10]. Nevertheless, it is not known how CPSF73 contributes to CRC progression, or how CASC9 regulates its activity.
CPSF73 Shows Promise as a Drug Target for Specific Cancers and For Protozoan Parasites
Acute Myeloid Leukemia (AML) and Ewing’s sarcoma (ES)
A recent study showed that CPSF73-dependent pre-mRNA processing is a druggable node in AML and ES [11]. AML is characterized by clonal proliferation of malignant myeloid blast cells in the marrow and impaired normal hematopoiesis; it is an aggressive disease that, if not eradicated in the first round of chemotherapy, becomes increasingly resistant to treatment [71]. ES represents 10–15% of malignant bone tumors and 40–45% of pediatric malignant bone tumors. The cause of ES is unknown, and most cases appear to occur randomly, which makes targeted treatment difficult [72]. Ross, et al. [11] used phenotypic screening combined with chemical genetics to pinpoint CPSF73 as the target of JTE-607, a small molecule originally identified for anti-inflammatory activity but with unknown targets [73]. A separate study confirmed this conclusion [74]. JTE-607 is an ester pro-drug that binds to the CPSF73 active site when its acid form is generated in cells. JTE-607 blocks the interaction of CPSF73 with mRNA, thus preventing cleavage at the PAS and resulting in read-through transcription and formation of DNA-RNA R-loop structures. Inhibition of CPSF73 by JTE-607 alters expression of known downstream effectors in both AML and ES, up-regulates apoptosis, and causes tumor-selective stasis in mouse xenografts [11] (Figure 4, bottom). Surprisingly, despite its potency towards CPSF73, no toxic phenotype was observed in many other cells treated with JTE-607, and no adverse effects were reported from in vivo studies of JTE-607 in both rodents and humans [73, 75]. These findings indicate that even though CPSF73-null mice are embryonic-lethalii, inhibition of CPSF73 by specific molecules could represent a novel therapeutic strategy in the treatment of specific cancers and inflammatory diseases.
Protozoan parasites
CPSF73 has also been explored as a target to treat diseases due to protozoa such as malaria, toxoplasmosis, sleeping sickness, and the diarrhea caused by Cryptosporidium [12–15] (Figure 4, left). Current therapies are unsatisfactory because of limited efficacy, particularly in higher-risk young children or immunocompromised patients, frequent toxic effects, and the rapid emergence of resistance to available drugs. The benzoxaborole compounds AN3661 and AN11736 target the protozoan CPSF73 without affecting the host enzyme and are potent inhibitors of Toxoplasma, Plasmodium, Trypanosoma, and Cryptosporidium growth in vitro [12–15]. The crystal structure of Cryptosporidium CPSF73 revealed that binding of the oxaborole group of AN3661 at the metal-dependent catalytic center of the protozoan CPSF73 blocks cleavage [15]; it is a change in the orientation of a phenylalanine residue in the Cryptosporidium CPSF73 active site allows this benzoxaborole to fit in the catalytic site, an interaction not sterically possible in the human enzyme. Importantly, oral administration of AN3661 protected mice from toxoplasmosis and malaria with efficacy similar to drugs currently in use and without signs of toxicity [13]. These findings indicate druggable differences between the CPSF73 enzymes of mammals and protozoa.
Concluding Remarks
The endonuclease CPSF73, a protein highly conserved from archaea to single-cell and metazoan eukaryotes, plays an essential and central role in the 3’ end processing of mRNA. Though recent structural analyses of the histone mRNA processing complex HCC have started to unveil the mechanisms of its activity, further studies are required to understand how the dynamic interactions of CPSF73 within HCC and CPSF, as well as within the entirety of the 3’ end processing machineries, regulate its localization on mRNA and the timing of cleavage. It also remains to be fully characterized how CPSF73 is regulated at the levels of transcription, 3’ end processing, stability, translation, post-translational modifications, and protein-protein or protein-RNA interactions, and how these factors contribute to the variable CPSF73 expression in cancers (see Outstanding Questions). By integrating knowledge of the structures, mechanisms, and regulation of CPSF73 from different species, we believe we will see the continued development of small molecules and compounds for pathogenic treatment and cancer therapeutics. As dysregulated gene control is a hallmark of tumorigenesis, and the disruption of pathogenic networks is a major pursuit of cancer pharmacology, targeting the 3’ end mRNA processing machineries, and specifically CPSF73, adds yet another tool to these growing fields.
Outstanding Questions.
What interactions trigger CPSF73 nuclease activity during the 3’ end processing of polyadenylated mRNAs, and how are they different from what was observed in the processing of cleaved, non-adenylated histone mRNAs?
How is the mRNA processing machinery reorganized after cleavage to allow poly(A) polymerase to act at the new 3’ end?
In the histone mRNA processing complex, what is the mechanism that converts CPSF73 from an endonuclease to an exonuclease that degrades the downstream cleavage product?
While CPSF73 can be regulated at multiple levels in different cellular contexts, are there general rules as to when a particular mode of regulation is used?
How does interaction with the long non-coding RNA (lncRNA) CASC9 promote CPSF73 activity, and do other lncRNAs regulate its activity?
CPSF73 is phosphorylated at multiple positions – what are the functional consequences of this modification? Further, what are the significances of modifications such as sumoylation and ubiquitination?
Can the interactome of CPSF73 be mapped to completely decode its function in cells?
How can small molecules be used as a chemical tool to further mechanistic understanding of the CPSF complex in different systems?
What is the likelihood that small molecule inhibitors of CPSF73 would actually move to clinical applications, or for treatment of parasitic infections?
Highlights.
The 3’ end processing of eukaryotic mRNA is required for generation of mature mRNA and generally entails cleavage and polyadenylation.
CPSF73 is the essential endonuclease that cleaves precursor mRNA and resides in at least two unique complexes: Cleavage and Polyadenylation Specificity Factor (CPSF) and Histone Cleavage Complex (HCC); these are further assembled into larger processing machineries.
Recent structures of the histone mRNA processing complex provided mechanistic insights into CPSF73 cleavage activity.
Cleavage by CPSF73 is necessary for efficient transcription termination, and where it cleaves can determine the fate of the mRNA and the type of protein produced.
Valuable information regarding how CPSF73 is regulated at different levels, such as mRNA expression, posttranslational modifications, and protein-protein or protein-RNA interactions, have been recently revealed.
This understanding has led to the development of small molecule inhibitors of CPSF73 which have therapeutic potential for treating cancer, inflammatory diseases, and protozoan infections.
Acknowledgement
We thank members of the Moore lab for comments. The Moore lab is supported by grants from the National Science Foundation (MCB1714603) and the National Institutes of Health (1R01 AI152337-01).
Glossary
- Alternative polyadenylation:
a mechanism by which altering the site of mRNA 3’ end processing yields polyadenylated mRNA isoforms of different lengths that can have different amounts of coding sequence or different regulatory elements in the mRNA’s 3’ UTR that govern mRNA stability, translation or localization; commonly observed during development, differentiation, carcinogenesis, change in growth rate, or in response to stress or external signals.
- Metallo-β-lactamase family:
a family of enzymes that vary in biological functions and substrate specificities but fold into a common 3-dimensional structure epitomized the founding member of this family, a hydrolase that inactivates β-lactam antibiotics.
- Torpedo model:
a model of termination in which cleavage of an elongating RNA Pol II transcript provides an entry site for the human Xrn2 (yeast Rat) 5’−3’ exonuclease to degrade the RNA, thus honing in on its target like a torpedo, and upon impact, dislodging RNA Pol II from the DNA template.
Footnotes
Resources
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gruber AJ and Zavolan M (2019) Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet 20, 599–614 [DOI] [PubMed] [Google Scholar]
- 2.Tian B and Manley JL (2017) Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Bio 18, 18–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren F, et al. (2020) Alternative Polyadenylation: a new frontier in post transcriptional regulation. Biomark Res 8, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun YD, et al. (2020) Recent molecular insights into canonical pre-mRNA 3 ‘-end processing. Transcr-Austin 11, 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y and Manley JL (2015) The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes & development 29, 889–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiang K, et al. (2014) Delineating the structural blueprint of the pre-mRNA 3’-end processing machinery. Mol Cell Biol 34, 1894–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ning Y, et al. (2019) CPSF3 is a promising prognostic biomarker and predicts recurrence of non-small cell lung cancer. Oncol Lett 18, 2835–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitra M, et al. (2018) Alternative polyadenylation factors link cell cycle to migration. Genome Biol 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu ZH, et al. (2009) CSR1 induces cell death through inactivation of CPSF3. Oncogene 28, 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo KL, et al. (2019) LncRNA CASC9 interacts with CPSF3 to regulate TGF- signaling in colorectal cancer. J Exp Clin Canc Res 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross NT, et al. (2020) CPSF3-dependent pre-mRNA processing as a druggable node in AML and Ewing’s sarcoma. Nature chemical biology 16, 50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonoiki E, et al. (2017) A potent antimalarial benzoxaborole targets a Plasmodium falciparum cleavage and polyadenylation specificity factor homologue. Nature communications 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palencia A, et al. (2017) Targeting Toxoplasma gondii CPSF3 as a new approach to control toxoplasmosis. Embo Mol Med 9, 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wall RJ, et al. (2018) Clinical and veterinary trypanocidal benzoxaboroles target CPSF3. Proceedings of the National Academy of Sciences of the United States of America 115, 9616–9621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swale C, et al. (2019) Metal-captured inhibition of pre-mRNA processing activity by CPSF3 controls Cryptosporidium infection. Science translational medicine 11 [DOI] [PubMed] [Google Scholar]
- 16.Zhang YX, et al. (2020) Structural Insights into the Human Pre-mRNA 3 ‘-End Processing Machinery. Molecular cell 77, 800–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandel CR, et al. (2006) Polyadenylation factor CPSF-73 is the pre-mRNA 3 ‘-end-processing endonuclease. Nature 444, 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang XC, et al. (2020) Studies with recombinant U7 snRNP demonstrate that CPSF73 is both an endonuclease and a 5’−3’ exonuclease. Rna 26, 1345–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun YD, et al. (2020) Structure of an active human histone pre-mRNA 3 ‘-end processing machinery. Science 367, 700–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan KD, et al. (2009) A Core Complex of CPSF73, CPSF100, and Symplekin May Form Two Different Cleavage Factors for Processing of Poly(A) and Histone mRNAs. Molecular cell 34, 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neve J, et al. (2017) Cleavage and polyadenylation: Ending the message expands gene regulation. RNA biology 14, 865–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi YS, et al. (2009) Molecular Architecture of the Human Pre-mRNA 3 ‘ Processing Complex. Molecular cell 33, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beaudoing E, et al. (2000) Patterns of variant polyadenylation signal usage in human genes. Genome research 10, 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian B, et al. (2005) A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic acids research 33, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X and Bartel DP (2017) Widespread Influence of 3’-End Structures on Mammalian mRNA Processing and Stability. Cell 169, 905–917 e911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkataraman K, et al. (2005) Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes & development 19, 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu J, et al. (2005) Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. Rna 11, 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Q, et al. (2010) Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3 ‘ processing. Proceedings of the National Academy of Sciences of the United States of America 107, 10062–10067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDonald CC, et al. (1994) The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol 14, 6647–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou ZF, et al. (1994) Sequence and position requirements for uridylate-rich downstream elements of polyadenylation signals. Nucleic acids research 22, 2525–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marzluff WF and Koreski KP (2017) Birth and Death of Histone mRNAs. Trends Genet 33, 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XC, et al. (2009) FLASH, a Proapoptotic Protein Involved in Activation of Caspase-8, Is Essential for 3 ‘ End Processing of Histone Pre-mRNAs. Molecular cell 36, 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang XC, et al. (2013) A Complex Containing the CPSF73 Endonuclease and Other Polyadenylation Factors Associates with U7 snRNP and Is Recruited to Histone Pre-mRNA for 3 ‘-End Processing. Mol Cell Biol 33, 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabath I, et al. (2013) 3 ‘-End processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors. Rna 19, 1726–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skrajna A, et al. (2018) Protein composition of catalytically active U7-dependent processing complexes assembled on histone pre-mRNA containing biotin and a photo-cleavable linker. Nucleic acids research 46, 4752–4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill CH, et al. (2019) Activation of the Endonuclease that Defines mRNA 3 ‘ Ends Requires Incorporation into an 8-Subunit Core Cleavage and Polyadenylation Factor Complex. Molecular cell 73, 1217–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schonemann L, et al. (2014) Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes & development 28, 2381–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eaton JD and West S (2020) Termination of Transcription by RNA Polymerase II: BOOM! Trends Genet 36, 664–675 [DOI] [PubMed] [Google Scholar]
- 39.Cortazar MA, et al. (2019) Control of RNA Pol II Speed by PNUTS-PP1 and Spt5 Dephosphorylation Facilitates Termination by a “Sitting Duck Torpedo” Mechanism. Molecular cell 76, 896–908 e894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ntini E, et al. (2013) Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nature structural & molecular biology 20, 923–928 [DOI] [PubMed] [Google Scholar]
- 41.Pettinati I, et al. (2018) Biosynthesis of histone messenger RNA employs a specific 3 ‘ end endonuclease. eLife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eaton JD, et al. (2018) Xrn2 accelerates termination by RNA polymerase II, which is underpinned by CPSF73 activity. Genes & development 32, 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang DM, et al. (2017) The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Molecular cell 68, 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaughency P, et al. (2014) Genome-Wide Mapping of Yeast RNA Polymerase II Termination. Plos Genet 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson EL, et al. (2019) Ipa1 Is an RNA Polymerase II Elongation Factor that Facilitates Termination by Maintaining Levels of the Poly(A) Site Endonuclease Ysh1. Cell reports 26, 1919–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanders TJ, et al. (2020) FttA is a CPSF73 homologue that terminates transcription in Archaea. Nat Microbiol 5, 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vilborg A, et al. (2017) Comparative analysis reveals genomic features of stress-induced transcriptional readthrough. Proceedings of the National Academy of Sciences of the United States of America 114, E8362–E8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shearwin KE, et al. (2005) Transcriptional interference--a crash course. Trends Genet 21, 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crisp PA, et al. (2018) RNA Polymerase II Read-Through Promotes Expression of Neighboring Genes in SAL1-PAP-XRN Retrograde Signaling. Plant Physiol 178, 1614–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gullerova M and Proudfoot NJ (2010) Transcriptional interference and gene orientation in yeast: noncoding RNA connections. Cold Spring Harb Symp Quant Biol 75, 299–311 [DOI] [PubMed] [Google Scholar]
- 51.Park HJ, et al. (2018) 3’ UTR shortening represses tumor-suppressor genes in trans by disrupting ceRNA crosstalk. Nat Genet 50, 783–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calzado MA, et al. (2004) Human immunodeficiency virus type 1 tat increases the expression of cleavage and polyadenylation specificity factor 73-kilodalton subunit modulating cellular and viral expression. J Virol 78, 6846–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cardiello JF, et al. (2018) Heat Shock Causes a Reversible Increase in RNA Polymerase II Occupancy Downstream of mRNA Genes, Consistent with a Global Loss in Transcriptional Termination. Mol Cell Biol 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kolev NG and Steitz JA (2005) Symplekin and multiple other polyadenylation factors participate in 3 ‘-end maturation of histone mRNAs. Genes & development 19, 2583–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis MR, et al. (2019) Nuclear eIF4E Stimulates 3 ‘-End Cleavage of Target RNAs. Cell reports 27, 1397–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sundvall M (2020) Role of Ubiquitin and SUMO in Intracellular Trafficking. Curr Issues Mol Biol 35, 99–108 [DOI] [PubMed] [Google Scholar]
- 57.Hershko A (1983) Ubiquitin - Roles in Protein Modification and Breakdown. Cell 34, 11–12 [DOI] [PubMed] [Google Scholar]
- 58.Vethantham V, et al. (2007) Sumoylation modulates the assembly and activity of the pre-mRNA 3’ processing complex. Mol Cell Biol 27, 8848–8858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costanzo M, et al. (2016) A global genetic interaction network maps a wiring diagram of cellular function. Science 353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee SD, et al. (2020) Regulation of the Ysh1 endonuclease of the mRNA cleavage/polyadenylation complex by ubiquitin-mediated degradation. RNA biology 17, 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hein MY, et al. (2015) A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 [DOI] [PubMed] [Google Scholar]
- 62.Huttlin EL, et al. (2017) Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen T, et al. (2014) mUbiSiDa: A Comprehensive Database for Protein Ubiquitination Sites in Mammals. PloS one 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas A, et al. (2015) Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol 12, 511–526 [DOI] [PubMed] [Google Scholar]
- 65.Rakha EA and Ellis IO (2009) Triple-negative/basal-like breast cancer: review. Pathology 41, 40–47 [DOI] [PubMed] [Google Scholar]
- 66.Mayr C and Bartel DP (2009) Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia Z, et al. (2014) Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3 ‘- UTR landscape across seven tumour types. Nature communications 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erson-Bensan AE and Can T (2016) Alternative Polyadenylation: Another Foe in Cancer. Mol Cancer Res 14, 507–517 [DOI] [PubMed] [Google Scholar]
- 69.Siegel RL, et al. (2018) Cancer Statistics, 2018. Ca-Cancer J Clin 68, 7–30 [DOI] [PubMed] [Google Scholar]
- 70.Bray F, et al. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-Cancer J Clin 68, 394–424 [DOI] [PubMed] [Google Scholar]
- 71.Saif A, et al. (2018) Acute Myeloid Leuemia: Is That All There Is? Cureus 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ben Kridis W, et al. (2017) A Review of Ewing Sarcoma Treatment: Is it Still a Subject of Debate? Rev Recent Clin Tria 12, 19–23 [DOI] [PubMed] [Google Scholar]
- 73.Kakutani M, et al. (1999) JTE-607, a novel inflammatory cytokine synthesis inhibitor without immunosuppression, protects from endotoxin shock in mice. Inflamm Res 48, 461–468 [DOI] [PubMed] [Google Scholar]
- 74.Kakegawa J, et al. (2019) JTE-607, a multiple cytokine production inhibitor, targets CPSF3 and inhibits pre-mRNA processing. Biochem Biophys Res Commun 518, 32–37 [DOI] [PubMed] [Google Scholar]
- 75.Borozdenkova S, et al. (2011) Effects of a cytokine inhibitor, JTE-607, on the response to endotoxin in healthy human volunteers. Int Immunopharmacol 11, 1837–1843 [DOI] [PubMed] [Google Scholar]
- 76.Roux CM, et al. (2011) Characterization of Components of the Staphylococcus aureus mRNA Degradosome Holoenzyme-Like Complex. J Bacteriol 193, 5520–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nishida Y, et al. (2010) Crystal structure of an archaeal cleavage and polyadenylation specificity factor subunit from Pyrococcus horikoshii. Proteins 78, 2395–2398 [DOI] [PubMed] [Google Scholar]
- 78.Silva AP, et al. (2011) Structure and activity of a novel archaeal beta-CASP protein with N-terminal KH domains. Structure 19, 622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phung DK, et al. (2013) Archaeal beta-CASP ribonucleases of the aCPSF1 family are orthologs of the eukaryal CPSF-73 factor. Nucleic acids research 41, 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mendoza-Figueroa MS, et al. (2020) The Integrator Complex in Transcription and Development. Trends Biochem Sci 45, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chan SL, et al. (2014) CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3’ processing. Genes & development 28, 2370–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun Y, et al. (2018) Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc Natl Acad Sci U S A 115, E1419–E1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clerici M, et al. (2018) Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex. Nature structural & molecular biology 25, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Clerici M, et al. (2017) Structural insights in to the assembly and polyA signal recognition mechanism of the human CPSF complex. eLife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lackford B, et al. (2014) Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. The EMBO journal 33, 878–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuhn U, et al. (2009) Poly(A) Tail Length Is Controlled by the Nuclear Poly(A)-binding Protein Regulating the Interaction between Poly(A) Polymerase and the Cleavage and Polyadenylation Specificity Factor. Journal of Biological Chemistry 284, 22803–22814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaczmarek Michaels K, et al. (2020) Regulation of alternative polyadenylation in the yeast Saccharomyces cerevisiae by histone H3K4 and H3K36 methyltransferases. Nucleic acids research 48, 5407–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ren J, et al. (2009) DOG 1.0: illustrator of protein domain structures. Cell Res 19, 271–273 [DOI] [PubMed] [Google Scholar]