Abstract
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has grown into a global pandemic, and no specific antiviral treatments have been approved to date. The angiotensin-converting enzyme 2 (ACE2) plays a fundamental role in SARS-CoV-2 pathogenesis as it allows viral entry into host cells. Here we show that ACE2 nanodecoys derived from human lung spheroid cells (LSCs) can bind and neutralize SARS-CoV-2 and protect the host lung cells from infection. In mice, the nanodecoys were delivered via inhalation therapy and resided in the lungs for over 72 hours post-delivery. Furthermore, inhalation of nanodecoys accelerated clearance of SARS-CoV-2 mimics from the lungs, with no observed toxicity. In cynomolgus macaques challenged with live SARS-CoV-2, four doses of nanodecoys delivered by inhalation promoted viral clearance and reduced lung injury. Our results suggest that LSC-nanodecoys can serve as a potential therapeutic agent for treating COVID-19.
Keywords: Nonhuman primates, SARS-CoV-2, COVID-19, ACE2, Nanodecoy, Lung Spheroid Cells
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen at the center of the current global pandemic, causes coronavirus disease of 2019 (COVID-19)1. Coronaviruses are a common type of virus: alpha (α-) coronaviruses and beta (β-) coronaviruses can infect mammals and often manifest as the common cold or gastrointestinal (GI) discomfort. Rarely, more severe and lethal forms emerge. SARS-CoV-2 is capable of wreaking havoc on the respiratory and immune system by inducing secretion of pro-inflammatory cytokines triggering an increase in alveolar edema, hypoxemia, dyspnea, and systemic inflammatory response syndrome (SIRS)2. Similar to its deadly predecessors, SARS-CoV-1 (the cause of SARS in 2003) and MERS-CoV (the cause of MERS in 2012), SARS-CoV-2 is an enveloped, positive-sense, β-coronavirus with dangerously high human-to-human transmission rates, with the reported R0 (the average number of people that one contagious person will infect) ranging from 2–63,4. Therefore, initial efforts to combat the virus primarily focused on containment to stop the spread and elucidate the pathogenesis of the virus.
It is becoming undeniably evident that in addition to an efficacious vaccine, the development of therapeutics is necessary for completely ending this pandemic and providing a solution to COVID-19 patients who are severely ill. Researchers around the world are in an urgent race to find an effective therapy for COVID-19. According to the published interim results from the World Health Organization’s Solidarity Trial on 15 October 2020, all 4 of the evaluated treatments (remdesivir, hydroxychloroquine, lopinavir/ritonavir, and interferon) had little or no effect on the overall mortality, necessity for mechanical ventilation, and length of hospital stay in hospitalized COVID-19 patients.
Angiotensin-converting enzyme 2 (ACE2), which is present on many cell types and found in almost all tissues, is a carboxypeptidase that plays a pivotal role in host cell viral entry. SARS-CoV-2 specifically attacks ACE2-presenting type II pneumocytes in the lungs and goblet secretory cells in the nasal mucosa by the interaction of their spike protein with ACE25. In this study, we exploit the virus’ cell entry strategy as a Trojan horse to trick the virus. Over the past seven years, our lab has developed Lung Spheroid Cells (LSCs) as a cell therapy to treat lung fibrosis and inflammation from initial rodent studies to an on-going Phase 1 clinical trial (NCT04262167)6–9. LSCs are a natural mixture of resident lung epithelial cells (containing both types I and II pneumocytes) and mesenchymal cells. As resident lung cells, they express ACE2, so we fabricated LSC membrane nanovesicles as ACE2 nanodecoys. Those nanodecoys, acting as cell mimics, are capable of binding to the SARS-CoV-2 Spike (S-) protein and triggering a phagocytic response from macrophages for viral elimination.
Therapeutic antibodies and fusion inhibitors have been developed for targeting the spike protein of SARS-CoV-210. However, more aggressive variants associated with the mutations in the spike protein of SARS-CoV-2 have been discovered11,12. Therefore, antiviral strategies based on the human receptor ACE2, used by the virus to gain host cell entry rather than the viral components, will experience greater interest since no mutations are to be expected on the host cells. Our data support a potential non-invasive therapeutic strategy for neutralizing SARS-CoV-2. Our approach is fundamentally different from the current two strategies: antiviral drugs and vaccines. The LSCs used to fabricate the nanodecoy are generated through a robust, reproducible, and scalable culturing method suitable for producing clinically applicable quantities of cell therapy products. Moreover, this nanodecoy technology is highly translatable as the parental cells are currently in the early clinical trial stage as a potential treatment for pulmonary fibrosis13.
Fabrication of LSC-nanodecoys
The overall rationale of our nanodecoy design is shown in Figure S1. First, LSCs and their parent cells, lung explant-derived cells (EDCs), were screened for ACE2 expression to determine the optimal cell types for nanodecoy fabrication. LSCs and EDCs were analyzed by immunostaining (Figure 1a; Figure S2), immunoblotting, and flow cytometry (Figure 1b–c; Figure S3) for ACE2 expression. In addition, the ACE2 expression levels of HEK293 and human lung fibroblasts were studied using immunoblotting and flow cytometry as controls (Figure S3). LSCs were found to have higher ACE2 expression levels than the other cell types, including their parent cell, EDCs. In comparison, HEK293 and fibroblasts had visibly lower ACE2 expression. Consistent with previous studies, confocal imaging showed that ACE2 was present on the membrane of AQP5+ type I pneumocytes and SFTPC+ type II pneumocytes (Figure 1a), two subpopulations within LSCs5,14. Analysis found ACE2 was co-expressed with other LSC makers such as EpCAM, CD90, and MUC5b (Figure 1d and Figure S4–5). Previous studies have indicated that 83% of ACE2-expressing cells in lung tissue are type II pneumocytes, suggesting that the lungs are the most vulnerable target organs to the SARS-CoV-2 virus15. Thus, our results demonstrated that, as primary resident lung cells, LSCs might serve as an ideal cell type to generate nanodecoys with high levels of ACE2 expression. In contrast, HEK293 cells were used as a control for preparing nanodecoys with a low level of ACE2 expression.
Figure 1. Characterizations of lung spheroid cell-derived nanodecoys.
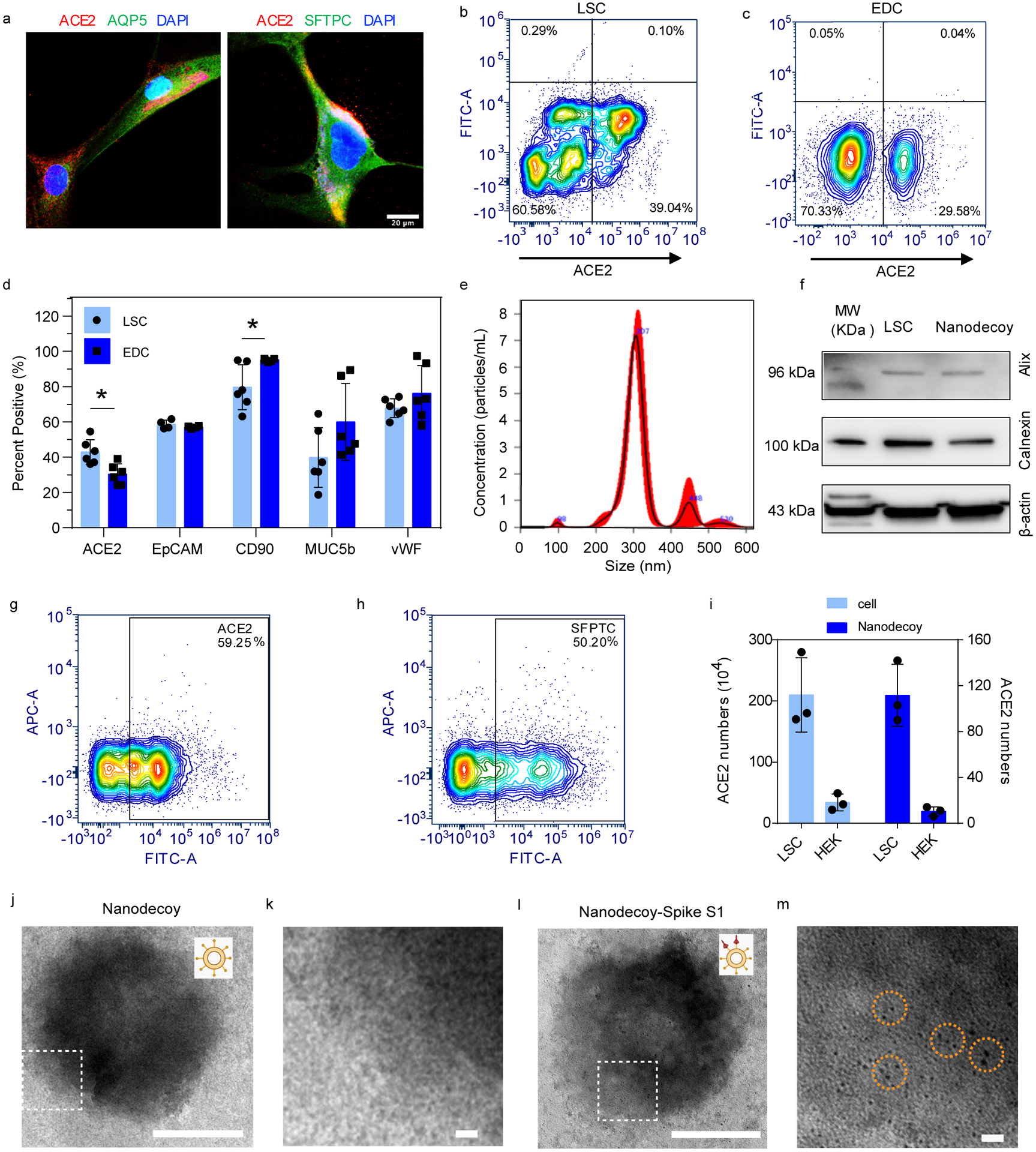
(a) Representative confocal images of LSCs labeled with ACE2, AQP5, and SFTPC antibodies. Three images were taken. Scale bars, 20 μm. (b) Representative flow cytometry analysis of LSCs (b) and EDCs (c) for ACE2 expression and (d) quantitative results of flow cytometry analysis of EDCs and LSCs for ACE2, EpCAM, CD90, MUC5b, and vWF. Data are shown as mean ± SD, n=4 or 6 independent experiments. Statistical analysis was performed by two-way ANOVA with a Tukey post hoc test. See Supplementary Figure S17 for gating strategies. (e) Size measurement of nanodecoys using NTA Nanosight. (f) Western blot of Alix and Calnexin in LSC-nanodecoys and LSCs. Flow cytometry analysis showing the expressions of ACE2 (g) and type II pneumocytes maker SFPTC (h) on LSC-nanodecoys. See Supplementary Figure S17 for gating strategies. (i) Measurement of ACE2 numbers on both cells and nanodecoys. HEK indicates HEK293. Data are shown as mean ± SD, n=3 independent experiments. Transmission electron microscopy (TEM) images showing naked nanodecoys (j) and enlarged figure (k). TEM images showing spike S1-bound nanodecoys (l) and enlarged figure (m). Spike S1 was detected using gold nanoparticle-labeled secondary antibodies with diameters of 10 nm. Cartoon pictures (insets in Figure j and l) were created with BioRender.com.
LSC and HEK293 membrane nanovesicles (nanodecoys) were generated by serial extrusion of LSCs or HEK293 cells through polycarbonate membranes with pore sizes of 5 μm, 1 μm, and finally, 0.4 μm with a commercial extruder. The obtained LSC-nanodecoys were characterized by Nanoparticle Tracking Analysis (NTA), showing a homogeneous nanoparticle population with an average size distribution of 320 nm and an average quantity of 5.51×1010 particles/mL produced from 5×106 cells (Figure 1e). In other words, on average, one LSC generated 11,020 nanodecoys. Because whole cells were used to prepare the nanodecoys, we suspected that the nanodecoys were not exclusively generated from the plasma membranes but also from intracellular membranes. To confirm our hypothesis, we also studied the intracellular component of the nanodecoys by testing for Alix (a phylogenetically conserved cytosolic scaffold protein) and Calnexin (a marker of endoplasmic reticulum). Results showed that these two intracellular markers were detected, which supported our claims (Figure 1f). Flow cytometry analysis confirmed the preservation of ACE2 on the surface of the nanodecoys (Figure 1g) as well as type II pneumocyte marker SFTPC (Figure 1h). Moreover, we studied the quantity of ACE2 on both LSCs and HEK293 cells and their nanodecoys by ELISA analysis. The frequency of ACE2 was determined to be 2.1×106 receptors per LSC and 112 receptors per LSC-nanodecoy. In stark contrast, 3.4×105 and 10 ACE2 receptors were found to be present on each HEK293 cell and HEK-nanodecoy, respectively (Figure 1i). Furthermore, transmission electron microscope (TEM) images revealed the spherical morphology of nanodecoys (Figure 1j and 1k).
Nanodecoys bind and neutralize spike S1 in vitro
Having demonstrated the presence of ACE2 on the nanodecoys, we then tested their binding ability to the SARS-CoV-2 S-protein. Spike S1 of the spike protein contains a receptor-binding domain (RBD) that specifically recognizes ACE2. Therefore, we first confirmed that spike S1 could bind to the nanodecoys by TEM with immunogold labeling (Figure 1l and 1m). In a dose-responsive manner, 50% of spike S1 (6.5 ×1010) was captured and bound by 109 LSC-nanodecoys, whereas nanodecoys derived from HEK293 cells failed to bind to spike S1 (Figure 2a). We then examined the binding potency of LSC- and HEK293-nanodecoys using lung cell-based assays (Figure 2b). Spike S1 was found to bind to lung cells after 4 hours of incubation (Figure 2c). DiD-labeled LSC-nanodecoys co-localized with spike S1, while HEK293-nanodecoys did not, suggesting that the LSC-nanodecoys could recognize and competitively bind to spike S1. Additionally, macrophages had a greater internalization efficiency of the nanodecoys than the lung cells did (Figure 2d–2g), indicating the potential clearance of nanodecoys and their neutralized SARS-CoV-2 by macrophages and/or other immune cells, which was confirmed by flow cytometry analysis (Figure 2h–2l). Furthermore, both peripheral blood and alveolar macrophages had the same internalization rate of LSC-nanodecoys (Figure S6).
Figure 2. Neutralization of spike S1 by nanodecoys.
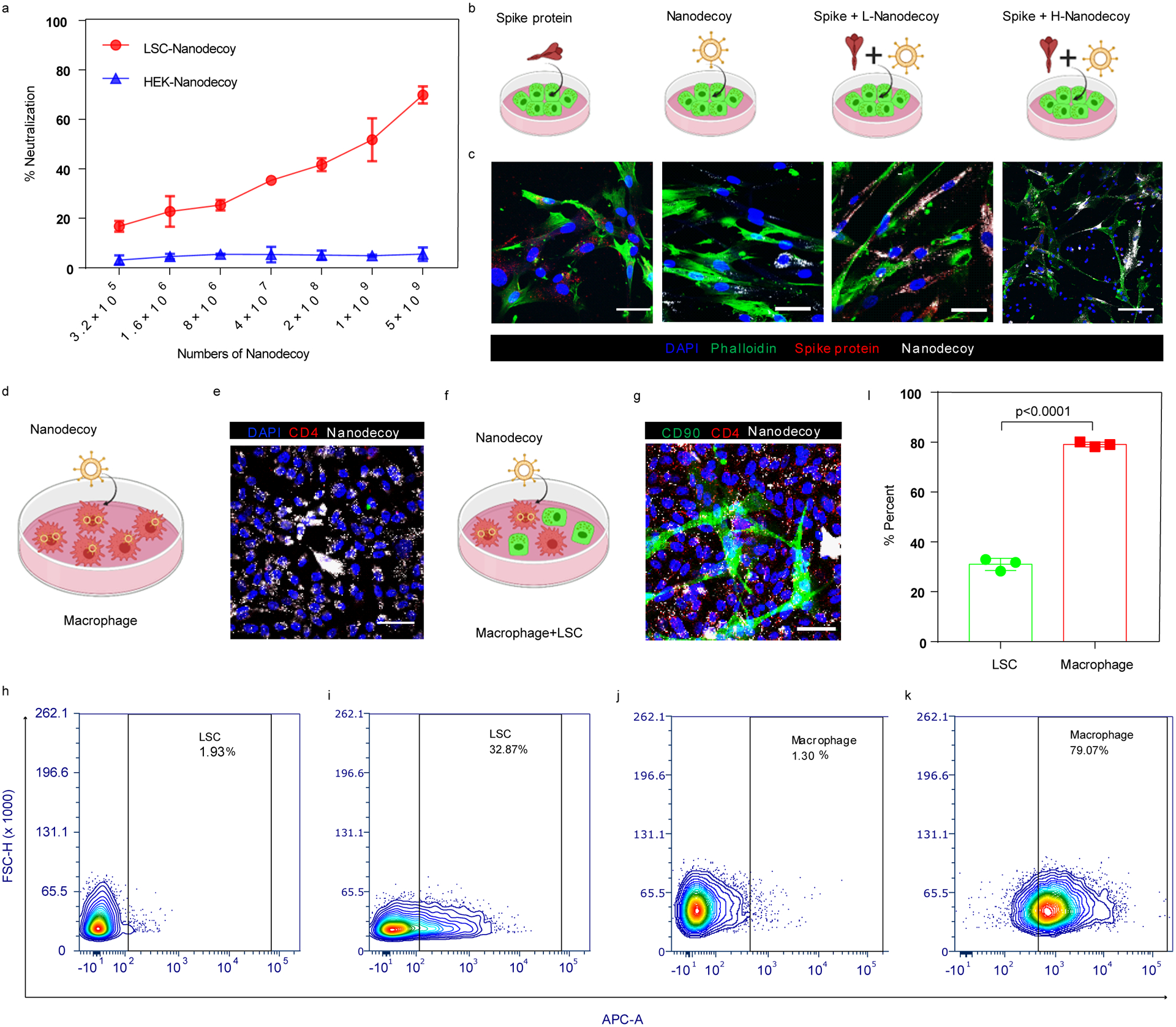
(a) Dose-dependent neutralization of spike S1 by LSC-nanodecoys or HEK-nanodecoys. Data are shown as mean ± SD, n=3 independent experiments. (b) Schematic illustrating the experimental design. (c) Interaction of spike S1 (red) and nanodecoys (white) when co-cultured with lung cells (green). (d) Schematic illustrating the experimental design. (e) Representative confocal images showing internalization of nanodecoys by macrophages (CD4, red). Four images were taken. (f) Schematic illustrating the co-culture experiment and (g) confocal images of the internalization of nanodecoys by macrophages co-cultured with lung cells (CD90, green). Flow cytometry analysis showing internalization of DiD-labeled nanodecoys by LSCs (i) and macrophages (k) and (l) its corresponding quantitation. PBS was used as control group for LSCs (h) and macrophages (j). See Supplementary Figure S18a for gating strategies. Data are shown as mean ± SD, n=3 independent experiments. Statistical analysis was performed with two-tailed Student t-test. Scale bars, 50 μm for Figure 2c, 2e and 2g. Cartoon pictures were created with BioRender.com.
Nanodecoys bind and neutralize SARS-CoV-2 mimics
Next, we fabricated a spike S1 virus to mimic SARS-CoV-2 by modifying a lentivirus without spike S1 to express spike S1 on its surface. Lentiviruses were first modified with Ni nitrilotriacetate (Ni-NTA) (Figure 3a), and then His-tagged spike S1 was conjugated onto the lentivirus through the interaction of Ni with His tag to generate this SARS-CoV-2 mimic (Figure 3b). Immunogold labeling was used to confirm spike S1 on the SARS-CoV-2 mimics. TEM imaging visualized the bare lentivirus (Figure 3c), SARS-CoV-2 mimic (Figure 3d), and the nanodecoy bound SARS-CoV-2 mimics, shown by the presence of spike S1 on the surface of the modified lentivirus together with the nanodecoy (Figure 3e), indicating the SARS-CoV-2 mimics were fabricated successfully. Our examination indicated that there were approximately 6,900 spike S1 per SARS-CoV-2 mimic virus. We found that 2.16×105 LSC-nanodecoys could bind 5×105 SARS-CoV-2 mimics (2.31 SARS-CoV-2 mimics per nanodecoy) while HEK293-nanodecoys showed a lower binding efficiency to SARS-CoV-2 mimics, which we owed to the corresponding low ACE2 level (Figure 3f). This binding interaction is specific since the control lentivirus (without spike S1) had low affinity to LSC-nanodecoys. We co-cultured macrophages and LSCs (Figure 3g) and found SARS-CoV-2 mimics were recognized by LSC-nanodecoys and internalized by macrophages after 4 hours in co-culture (Figure 3h). We then studied the intracellular distribution of the mimics, and confocal imaging showed some of the mimics within the lysosomes while others resided in the cytoplasm (Figure S7). In addition, lentiviruses before and after modification had a slight difference in internalization by LSCs (Figure S8). We next studied the inhibiting internalization effect of nanodecoys by LSCs. Immunocytochemistry (Figure 3i–3l) and flow cytometry (Figure 3m and 3n) confirmed that LSC-nanodecoys could block the entry of SARS-CoV-2 mimics in host cells, but HEK293-nanodecoys could not. Naïve lentiviruses were not efficient in entering lung cells (14.8% infection rate) (Figure 3i). However, spike S1 modified lentiviruses (SARS-CoV-2 mimics) promoted entry into host cells efficiently (73.8% infection rate) (Figure 3j), whereas compared with HEK-nanodecoys, LSC-nanodecoys significantly decreased the internalization of SARS-CoV-2 mimics (from 73.8% to 28.8%) (Figure 3k and 3l). In addition, we studied the dose-dependent blocking effect by LSC-nanodecoys. Flow cytometry analysis showed that increasing doses of LSC-nanodecoys blocked more virus entry into lung cells in a dose-dependent manner (Figure S9). Together these results suggest our nanodecoys could protect the host cells from infection by SARS-CoV-2 mimics.
Figure 3. Neutralization of SARS-CoV-2 mimicking viruses by nanodecoys.
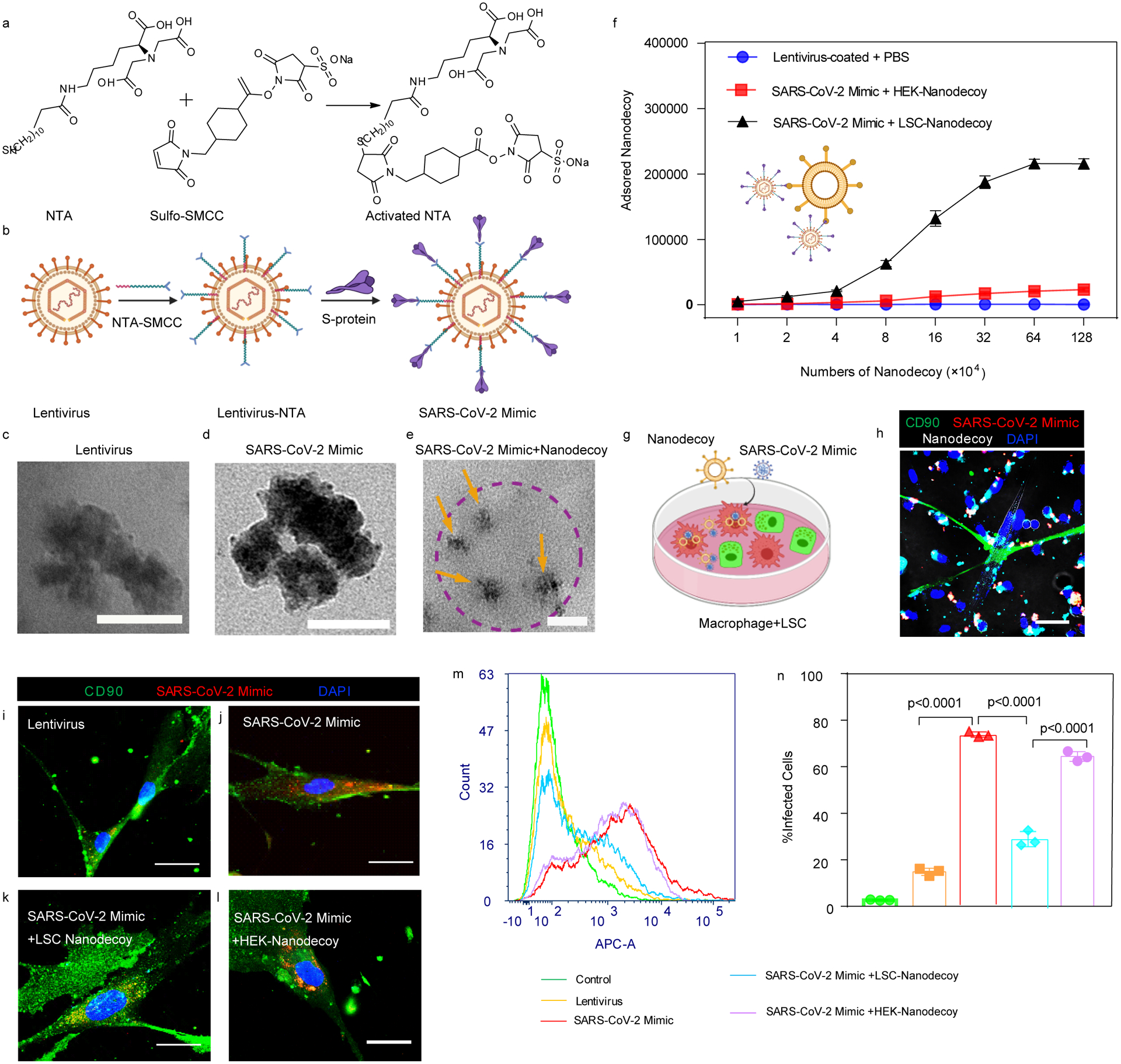
(a) The synthesis of activated NTA and the chemical structure. (b) Schematic illustrating the modification of lentivirus with spike S1 to generate a SARS-CoV-2 mimic. TEM images showing lentivirus (c), SARS-CoV-2 mimic (d), and spike S1 on lentivirus using gold nanoparticle-labeled secondary antibodies with diameters of 10 nm (e), in which SARS-CoV-2 mimicking viruses (yellow arrows) were attached to a nanodecoy (dotted circle). Scale bars, 100 nm for Figure 3c–e. (f) Neutralization assay of SARS-CoV-2 mimics by nanodecoys. Data are shown as mean ± SD, n=4 independent experiments. (g) Schematic showing the experimental design. (h) Nanodecoys (white) neutralize SARS-CoV-2 mimics (red) in a co-culture with lung cells (green) and macrophages. Scale bar, 50 μm. (i-l) Representative confocal images (at least three images were taken per animal) and (m) flow cytometry analysis showing nanodecoys inhibit the SARS-CoV-2 mimic virus (red) entry into the lung cells (green). Scale bars, 50 μm. (n) Corresponding quantitation from (m). See Supplementary Figure S18b for gating strategies. Data are shown as mean ± SD, n=3 independent experiments. Statistical analysis was performed by one-way ANOVA with a Tukey post hoc test. Cartoon pictures were created with BioRender.com.
We next studied the retention and biodistribution of LSC-nanodecoys in mice after inhalation. DiD-labeled nanodecoys were administered to mice by inhalation using a commercially available portable nebulizer for clinical relevance at a dose of 1×1010 nanodecoys per kg of body weight (Figure 4a). As shown in Figure 4b–4c and Figure S10, nanodecoys could still be found in the lungs 72 hours post a single inhalation treatment. In addition to the lungs, the nanodecoys were also detected in the liver, kidney, and spleen, indicating clearance via the reticuloendothelial system as well as the metabolization of the nanodecoys through the body. Moreover, inhalation of nanodecoys had no significant effect on CD68+ macrophage infiltration, indicating their biosafety (Figure S11). Even though some nanodecoys co-localized with APQ5+ (type I) and SFTPC+ (type II) cells (Figure 4d and Figure S12), the majority of nanodecoys were co-localized in macrophages (Figure 4e) after 24 hours in vivo.
Figure 4. Biodistribution of nanodecoys after inhalation.
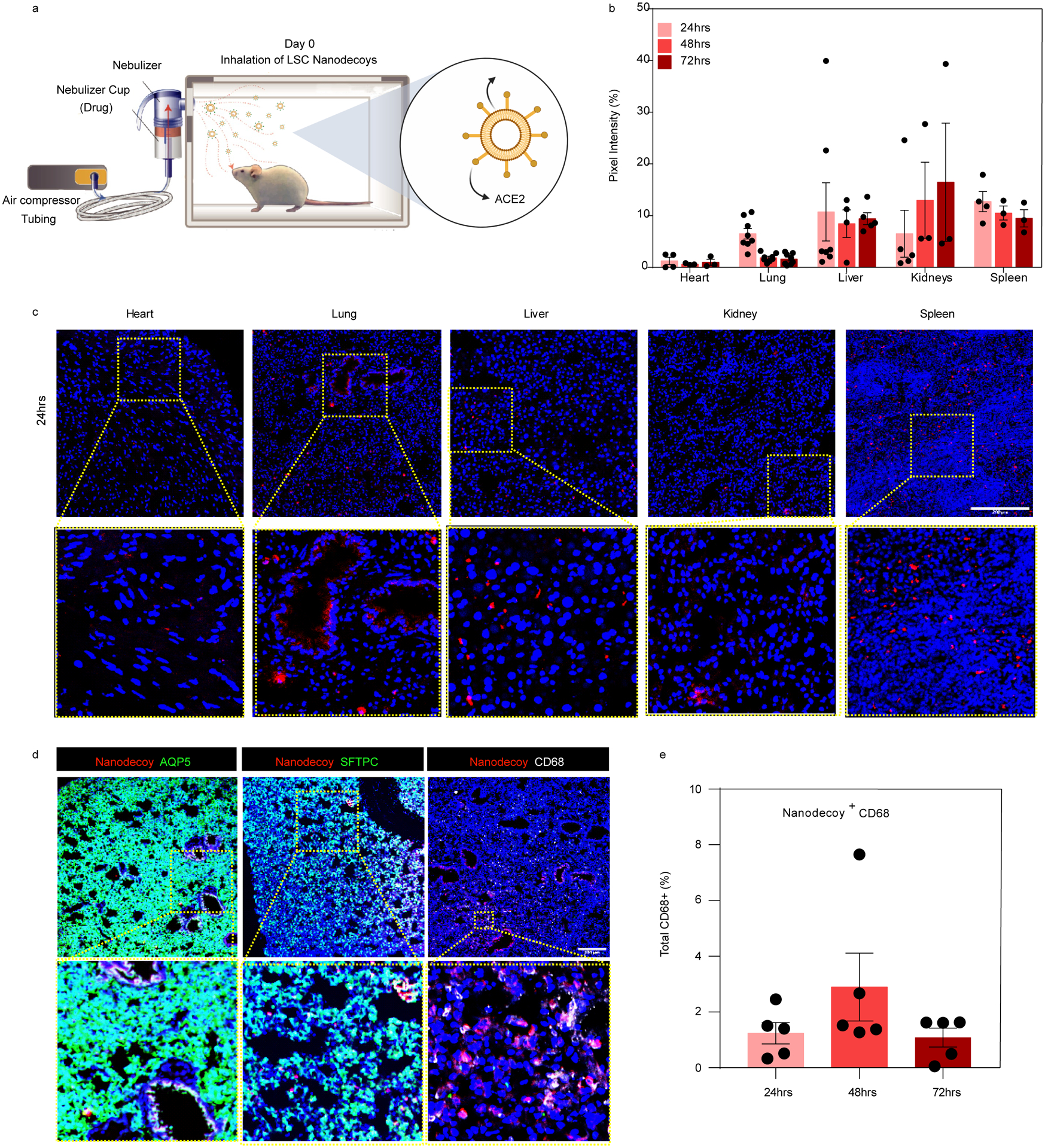
(a) Schematic showing study design of nanodecoy inhalation in CD1 mice. Created with BioRender.com. (b) Corresponding quantitative results from (c) of DiD-labeled nanodecoys in heart, lung, liver, kidney, and spleen tissues. Data are shown as mean ± SD, n=3 animals. At least three images were taken. (c) Representative confocal images of DiD-labeled nanodecoys (red) in tissue sections. (d) Representative confocal images showing nanodecoys in lung tissues co-localizing with lung cells (AQP5, SFTPC) and macrophages (CD68) 24 hrs post-inhalation. Five images were taken. (e) Quantification of the percent of nanodecoy-positive macrophages. Data are shown as mean ± SD, n=3 animals. Scale bars, 200 μm for Figure 4c–d.
We tested whether inhaled LSC-nanodecoys could accelerate the clearance of SARS-CoV-2 mimics in a mouse model (Figure 5a). To mimic infection in human patients, we allowed the mice to receive the SARS-CoV-2 mimics before initiating administration of the therapeutic nanodecoys. Since treatment started 24 hours post viral exposure, not all of the SARS-CoV-2 mimics were intracellular; therefore, nanodecoys could block the viral mimics from entering the cells further. As for the intracellular SARS-CoV-2 mimics, the nanodecoys that were internalized by cells could capture them, avoiding further infection. Ex vivo imaging (Figure 5b and 5c) indicated that the amounts of SARS-CoV-2 mimics were significantly reduced following inhalation of LSC-nanodecoys. Inhalation of the freeform of rACE2 and HEK293-nanodecoy were found to be ineffective. Confocal microscopy confirmed that inhalation of LSC-nanodecoys accelerated the clearance of SARS-CoV-2 mimics (Figure 5d and 5e). Cytokine array analysis (Figure 5f–j) suggested that nanodecoy inhalation did not elevate pro-inflammatory cytokines as compared to the control group. Furthermore, H&E staining of all major organs, hematology, and biochemical parameters indicated no apparent abnormality or adverse effects with LSC or HEK293 nanodecoy inhalation (Figure S13 and S14).
Figure 5. Nanodecoy inhalation accelerates the clearance of the SARS-CoV-2 mimic viruses in a mouse model.
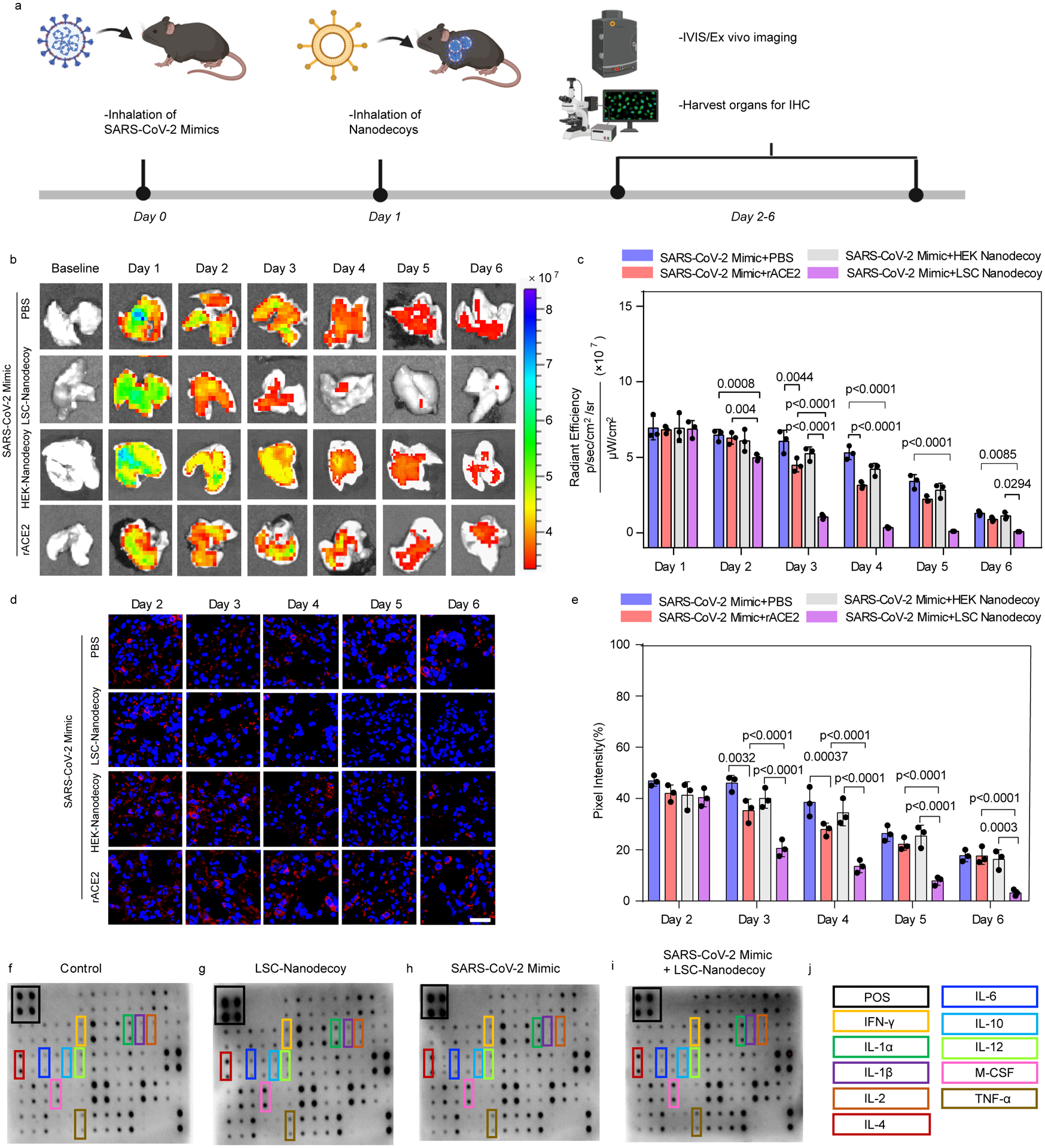
(a) Schematic showing the animal study design. Created with BioRender.com. (b) Representative ex vivo IVIS imaging of lung tissues from mice with various treatments. n=3 animals per group. (c) Quantification of fluorescence intensities of SARS-CoV-2 mimics from the imaging data in (b). Data are shown as mean ± SD, n=3 animals per group. Statistical analysis was performed by two-way ANOVA with a Tukey post hoc test for multiple comparisons. (d) Representative confocal images of AF647-labeled SARS-CoV-2 mimics (red) in lung sections. Scale bar, 50 μm. n=3 animals per group and three images were taken for each animal. (e) Corresponding semi-quantitative analysis of AF647-labeled SARS-CoV-2 mimics in lung tissues. Data are shown as mean ± SD, n=3 animals per group. Statistical analysis was performed two-way ANOVA with a Tukey post hoc test for multiple comparisons. (f-j) Cytokine array analysis of various inflammatory cytokines in the serum 3-days after treatment.
Nanodecoy therapy in SARS-CoV-2 infected nonhuman primates
We then performed a pilot nonhuman primate study to evaluate the safety and preliminary therapeutic efficacy of LSC-nanodecoys. The macaque model can recapitulate many clinical symptoms of SARS-CoV-2 infection and shows a robust viral replication in the upper and lower respiratory tracts. Six cynomolgus macaques were challenged with SARS-CoV-2 by intranasal and intratracheal routes (Figure 6a). Following challenge, the animals were randomized into two treatment arms: inhalation of PBS or LSC-nanodecoys (at a dose of 1010 particles per kg of body weight) at days 2, 3, 4, and 5 post-challenge. Viral loads in bronchoalveolar lavage (BAL) and nasal swabs (NS) were assessed by RT-PCR specific for viral subgenomic RNA (sgRNA, indicative of virus replication). As a result, high levels of sgRNA were observed in the control animals with a median peak of 6.243 log10 RNA copies/mL in BAL and a median peak of 5.595 log10 RNA copies/swab in NS on day 2 (Figure 6b and 6c). sgRNA levels dramatically decreased in nanodecoy-treated animals, with <1.70 log10 reductions of median peak sgRNA in both BAL and NS on day 8 following the challenge. Although sgRNA levels declined in both control and LSC-nanodecoy groups over time, LSC-nanodecoy treatment induced more rapid virus clearance. Negligible difference was observed between the two groups’ hematology parameters (Figure S15). Interestingly, the temperature and body weight fluctuations in the LSC-nanodecoy group were not as drastic as those in control-treated animals (Figure S16).
Figure 6. LSC-nanodecoy inhalation treats SARS-CoV-2 infection in cynomolgus macaques.
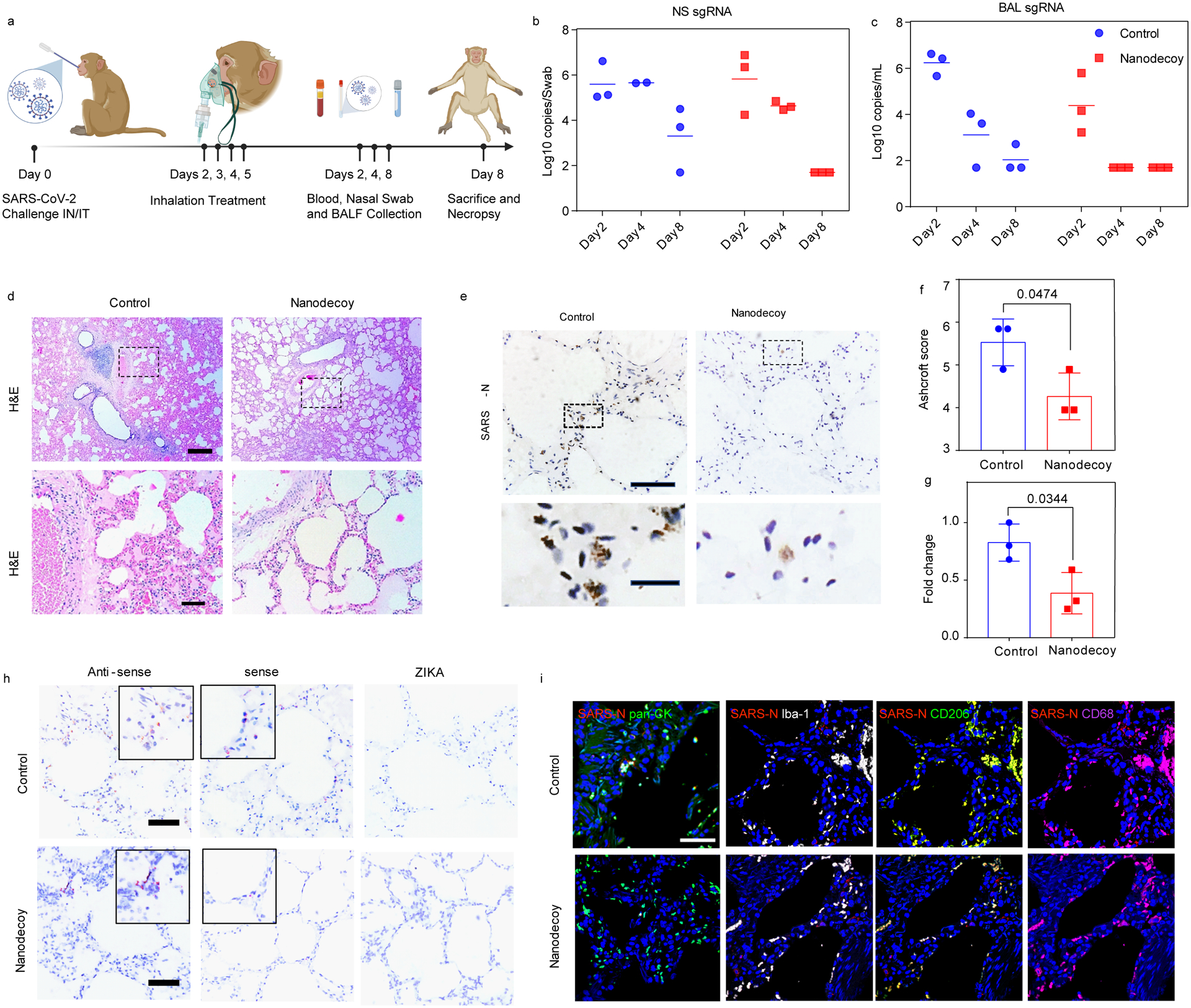
(a) Schematic depicting the cynomolgus macaque study design. Created with BioRender.com. (b and c) Viral subgenomic RNA (sgRNA) copies/Swab in nasal swabs (NS) and bronchoalveolar lavage (BAL) at various timepoints following challenge. Each dot represents data from one animal. n=3 animals per group and three images were taken for each animal. (d) Representative H&E images of fixed lung tissues from SARS-CoV-2 infected cynomolgus macaques and at least three images were taken per animal. Top: scale bar, 500 μm; bottom: scale bar, 100 μm. (e) Representative images of SARS nucleocapsid (SARS-N) immunohistochemistry (IHC) staining in fixed lung tissues from SARS-CoV-2 infected cynomolgus macaques treated with control or LSC-nanodecoys 8-days post-viral challenging. n=3 animals per group and three images were taken for each animal. Top: Scale bar, 100 μm; bottom: scale bar, 20 μm. (f) Quantification of lung fibrosis of infected cynomolgus macaques by Ashcroft scoring; each dot represents data from one animal; data are shown as mean ± SD, n = 3 animals per group. Statistical analysis was performed by two-tailed Student t-test. Ashcroft scoring was performed blindly. (g) Quantitation of positive SARS nucleocapsid numbers in lung tissues of infected cynomolgus macaques. Each dot represents data from one animal; data are shown as mean ± SD, n = 3 animals per group. Statistical analysis was performed with two-tailed Student t-test. (h) Representative images of RNAscope in situ hybridization detection of vRNA in infected cynomolgus macaques. ZIKA as a control probe. n=3 animals per group and three images were taken for each animal. Scale bar, 100 μm. (i) Representative immunofluorescence images of SARS-N (red), pan-CK (green), Ibal-1 (greyscale), CD68 (green) CD206 (magenta) and DAPI (blue). Scale bar, 50 μm. n=3 animals per group and three images were taken for each animal.
At the end of the study, lung tissues of infected cynomolgus macaques were collected and evaluated by histopathology. On day 8 following challenge, multifocal regions of inflammation and evidence of viral pneumonia—including expansion of alveolar septae with mononuclear cell infiltrates, consolidation, and edema—were observed (Figure 6d). Notably, LSC-nanodecoy treatment significantly reduced the numbers of polymorphonuclear cells and neutrophils as compared with the control group. In addition, Ashcroft score analysis revealed that LSC-nanodecoy treatment significantly decreased lung fibrosis (Figure 6f). To detect and visualize the virus in lung tissues, SARS nucleocapsid protein (SARS-N) expression was evaluated by immunohistochemistry (IHC) staining. As shown in Figures 6e and 6g, multifocal positive pneumocytes and alveolar septa were present in control-treated animals. In contrast, the levels of SARS-N protein were decreased substantially with the LSC-nanodecoy treatment. In addition, SARS-CoV-2 viral RNA (vRNA) was evaluated by in situ RNA hybridization (RNAscope). Compared to the control group, the levels of both positive-sense and negative-sense vRNA were diminished after LSC-nanodecoy treatment (Figure 6h), indicative of the reduction of viral replication. The distribution of SARS-CoV-2 in lung tissue was assessed by co-staining SARS-N and pan-cytokeratin (pan-CK, to identify epithelial cells). We found that virus-infected cells greatly overlapped with pan-cytokeratin (pan-CK)-positive cells (Figure 6i), suggesting that they were alveolar epithelial cells. Additionally, foci of virus-infected cells were frequently associated with activated Iba-1+ (ionized calcium binding adaptor as a pan-macrophage marker), CD68+ (monocyte/macrophage marker), and CD206+ (macrophage marker) macrophages (Figure 6i). Consistent with IHC and RNAscope analysis, immunofluorescence results indicated that nanodecoys could decrease virus levels in lung tissues.
Previous studies have indicated that ACE2 is the host receptor for the novel coronavirus (SARS-CoV-2) and that viral entry of SARS-CoV-2 depends on the binding of the viral spike S1 to ACE2 on the host cells16. Therefore, inhibiting the binding of spike S1 to ACE2 is a possible treatment strategy to combat COVID-19. Inspired by this discovery, some prior works have focused on blocking SARS-CoV-2 entry by using recombinant ACE2 (rACE2) protein, such as rACE2 alone or rACE2 fused with an Fc fragment (rACE2-Fc)17,18. However, those protein-based neutralization strategies are limited by their overall short half-life after administration. Furthermore, undesired dosage and distribution of extracellular ACE2 could cause unknown toxicity effects on the body17. In addition, except for ACE2, other components on cell membranes also play roles in virus docking; therefore, targeting ACE2 alone may not be enough.
Previous works have shown several anti-microbial applications by utilizing cell membrane-based nanodecoys19–21. For example, nanodecoys from Aedes albopictus (C6/36) cell membrane-coated gelatin nanoparticles have been developed to trap Zika virus for preventing viral infection22. Also, T-cell-membrane-coated nanoparticles were used as decoys for HIV neutralization owing to the presence of T-cell surface antigens for HIV binding23. In addition to cellular-membrane-based nanodecoys, engineered liposomes have also been fabricated as decoy targets to sequester bacterial toxins produced during active infection in vivo24. Presented here, our strategy represents a potential nanodecoy treatment for COVID-19 (Figure S1). The nanodecoys could be generated from human lung cells in a large scale using commercially available extrusion devices. They not only express natural human ACE2 but also represent a mimic of human lung cells, which are the main targets of SARS-CoV-2.
One concern of drug development is the potential off-target effects and undesired biodistribution. Here, we present a simple and clinically relevant method of nanodecoy delivery via inhalation using a nebulizer instead of traditional intravenous (IV) injection (Figure 4a). Inhalation of nanodecoys resulted in the direct accumulation of the therapeutic particles in the lungs, which is one of the primary sites of SARS-CoV-2 infections and replication. From just one single inhalation treatment, DiD-labeled nanodecoys can still be found in the lungs after 72 hours (Figure 4b). We also detected nanodecoys in the liver, kidney, and spleen throughout the 72 hours, which can be attributed to the metabolization of the nanodecoys, potentially by macrophages. However, additional studies will be required to elucidate the exact clearance mechanism of the nanodecoys. Many factors such as particle size, particle characteristics, breathing pattern, and treatment duration can affect the particles’ biodistribution and which mechanism is utilized for clearance. For instance, particles deposited in the upper airways are often cleared by mucociliary transport; on the other hand, particles that penetrate to the nonciliated respiratory epithelium are often cleared or transported via the alveolar macrophages or enter the bloodstream through the alveolar-capillary barrier. Given that the size of the nanodecoys is approximately 300 nm or 0.3μm, literature has shown that similar nano-sized particles administered via I.V. injection into the bloodstream are easily cleared by the reticuloendothelial system (RES), in which macrophage also played an important role25,26. The ability of the nanodecoys to be recognized and captured by macrophages would contribute to the clearance of SARS-CoV-2 from circulation. Furthermore, we demonstrated that inhalation of nanodecoys could accelerate the removal of SARS-CoV-2 mimics from lungs of infected mice. Mounting evidence indicates that there are increases in inflammatory cytokines in patients with COVID-19, suggesting the existence of cytokine storm in critically ill patients. It has also been demonstrated that peripheral inflammatory monocytes and pathogenic T-cells may induce cytokine storms in severe COVID-19 patients27–29. Thus, excessive macrophage infiltration may cause undesirable effects.
Recently, nanotechnological tools have been used for the treatment of COVID-1930,31 and some recent perspectives and research papers hint at the potential of “nanodecoys” or “nanosponges” for treating SARS-CoV-2 with some basic in vitro or in vivo data. For example, Zhang et al. reported cellular nanosponges as a potential countermeasure to the SARS-CoV-2 virus using an everlasting human lung epithelial type II cell membrane coated PLGA nanoparticles, where in vitro viral neutralization was shown32. Rao et al. generated fused cell membrane nanovesicles derived from genetically engineered cells (which stably express SARS-CoV-2 receptor ACE2) and human monocytes (which display abundant cytokine receptors for adsorbing viruses and inflammatory cytokines). Rao and colleagues employed a mouse model of LPS-induced acute lung inflammation to test their nanovesicles33. To the best of our knowledge, no previous studies have tested nanodecoys in any animal models of live SARS-CoV-2 infection.
Conclusions
In summary, we provide the first evidence in a nonhuman primate model of live SARS-CoV-2 infection that cell-derived and cell-mimicking nanodecoys can protect lung cells from the infections and damages from SARS-CoV-2. The cynomolgus macaque model recapitulates many clinical features of human patients with COVID-19. Four doses of nanodecoy inhalation led to a reduction of viral load in both BAL and NS 8 days following SARS-CoV-2 challenge. Adverse events such as weight loss, fever, or mortality were not observed. Histopathology, immunohistochemistry, RNAscope, and immunofluorescence analyses of lung tissues demonstrated that the nanodecoys was not only effective in alleviating inflammatory cell infiltration and decreasing pulmonary fibrosis but more importantly, was capable of reducing the levels of SARS nucleocapsid protein (SARS-N) and viral RNA. To those ends, our results suggest that LSC-nanodecoys can serve as a potential therapeutic agent for treating COVID-19.
Methods
Generation of nanodecoys
Nanodecoys were derived from LSCs or HEK293 cells (ATCC® CRL-1573™) by an extruder (AVESTIN LIPOSOFAST LF-50, AVESTIN, Inc). Cells were collected and suspended in PBS at a concentration of 5 × 106 cells/mL. A large volume of cells could be extruded immediately or stored at −80°C until ready. The cells were passed through the extruder twice through 5 μm, 1 μm, and 400 nm pore-sized polycarbonate membrane filters (Avanti Polar Lipids, Inc.) sequentially. The resulting nanodecoys were purified and concentrated using an ultrafiltration centrifuge tube (100 kDa MWCO; Millipore) and centrifuged at 4,500 g for 10 min and washed with PBS. The size and concentration of nanodecoys were measured using Nanoparticle Tracking Analysis system (Nanosight, Malvern). Nanodecoys were stored at 4°C for one week or placed in long-term storage at −80°C. The ACE2 receptors on the nanodecoys were detected using immunoblot, immunostaining, flow cytometry, and transmission electron microscopy (TEM) with immunogold labeling.
ACE2 analysis using ELISA
5×106 of LSC and HEK293 cells were collected and 1010 of LSC-nanodecoy and HEK293-nanodecoy were prepared. They were analyzed with an ACE2 ELISA kit (Abcam, ab235649) according to the manufacturer’s instructions.
In vitro internalization experiments of nanodecoys
Human macrophage primary cells and LSCs (104 cells/mL) were seeded in 4-well culture chamber slides (Thermo Fisher Scientific). Nanodecoys (1 × 106 cells/mL) were then labeled by DiD and incubated with macrophages or LSCs alone, as well as a co-culture of both (1:1) to mimic the in vivo microenvironment. After 4 hours of incubation, free nanodecoys were removed by 3 washes with 1X PBS. Cells were fixed using 4% PFA prior to immunocytochemistry staining with makers for macrophage (CD4; 12-0041-82, Invitrogen) and LSC (CD90; 11-0909-42, Invitrogen) and imaged with an Olympus FLUOVIEW confocal microscope. In addition, to quantify the internalization rate of nanodecoys by the different cell types, cells and nanodecoys were cultured in a T75 flask as previously described and collected for flow cytometry analysis (CytoFlex; Beckman Coulter).
In vitro spike S1 neutralization experiments of nanodecoys
Recombinant spike S1 (Sino Biological 40591-V08H, 10 ng/mL, MW = 76.5 kDa) was added to nanodecoys at different concentrations (5×109, 1×109, 2×108, 4×107, 8×106, 1.6×106, and 3.2×105) and incubated for three hours. After that, the unbound spike S1 was removed by ultracentrifugation (100 kDa). Spike S1 before and after binding to nanodecoys was determined using an ELISA kit (Sino Biological SARS-CoV-2 SPIKE ELISA KIT, Sino Biological) according to manufacturer’s protocol. To study the neutralization of spike S1 with nanodecoys in primary lung derived cells (LSCs), spike S1 was first labeled using NHS-Rhodamine (46406, Thermo Fisher Scientific) according to the manufacturer’s instructions. The RhB-spike S1 (100 ng) was first incubated with LSCs (2×104) in 4-well slides for 1 h and washed with PBS for three times. After that, DiD labeled nanodecoys (2×107) were added and incubated for another 4 h. Cells were washed and fixed using 4% PFA prior to stain with Alexa Fluor™ 488 Phalloidin (Invitrogen™ A12379). Cells were imaged using an Olympus FLUOVIEW confocal microscope.
Generation of SARS-CoV-2 mimicking virus
Spike S1 (40591-V08H; Sino Biological) was conjugated to lentivirus (Cellomics Technology LLC) to create a SARS-CoV-2 mimic. His-tagged spike S1 was linked to Ni nitrilotriacetate (Ni-NTA) through the chemical interaction. NTA with mercapto group (N-[Nα,Nα-Bis(carboxymethyl)-L-lysine]-12-mercaptododecanamide) was first reacted with 4-(N-Maleimidomethyl)cyclohexane-1-carboxylic acid 3-sulfo-N-hydroxysuccinimide ester sodium salt (Sulfo-SMCC) to give NTA-SMCC and then was added to the lentivirus. The NTA groups were conjugated to the lentivirus through the −NH2 group on lentivirus and N-hydroxysuccinimide ester on NTA-SMCC. The free NTA-SMCC was removed by centrifugation using an ultrafiltration tube (100 kDa MWCO; Millipore) to give SARS-CoV-2 mimicking virus (spike S1-lentivirus). The successfully conjugated spike S1 on lentivirus was confirmed using TEM. Briefly, SARS-CoV-2 mimics were incubated with anti-Spike S1 antibodies overnight at 4°C. Free antibodies were removed using an ultrafiltration tube (100 kDa MWCO; Millipore) and washed with PBS three times. Spike S1 on the SARS-CoV-2 mimics was labeled with immunogold (10 nm) antibodies and negatively stained for TEM visualization. The conjugation efficiency of spike S1 on lentivirus was determined using ELISA (Sino Biological SARS-COV-2 SPIKE ELISA KIT, Sino Biological) according to manufacturer’s protocol.
SARS-CoV-2 mimicking virus in cells
LSC cells (104 cells/mL) were seeded in 8-well culture chamber slides (Thermo Fisher Scientific) and allowed to adhere for 24 hrs. SARS-CoV-2 mimics (104 TU/mL) were added into the 8-well slides and incubated for 4 hrs. After that, LSC cells were washed with PBS twice to remove non-internalized nanodecoys and stained with 100 μM Lyso Dye (Invitrogen, green) at 37 °C for 30 min. Subsequently, slides were mounted with ProLong Gold Antifade Mountant with 4,6-diamidino-2-phenylindole (DAPI, Invitrogen, Waltham, MA, USA) and imaged on the Olympus FLUOVIEW CLSM (Olympus; FV3000, Shinjuku, Tokyo, Japan) with an Olympus UPlanSAPO 60x objective (Olympus; 1-U2B832, Shinjuku, Tokyo, Japan).
SARS-CoV-2 mimic neutralization experiment
Nanodecoys were first labeled using DiI. After that, 200 μL of SARS-CoV-2 mimic (5×105) in pH 9.6 coating buffer was added to each well of 96-well plates and incubated at 4°C overnight for coating. In addition, lentiviruses without spike S1 were also coated to the plates as a control. Following the incubation, the protein solution was removed, and the wells were washed with 1×PBS. To study binding, plates were incubated with DiI-labeled nanodecoys at concentrations of 1×104, 2×104, 4×104, 8×104, 1.6×105, 3.2×105, 6.4×105, 1.28×106 for two hours at room temperature. After that, the plates were rinsed with 1× PBS for three times, and fluorescent intensities were determined using a microplate reader (Molecular Devices).
Interaction of SARS-CoV-2 mimic with LSCs was assessed by ICC and flow cytometry. RhB-NHS was first reactivated with NTA-tagged lentivirus and then modified with S1 protein to synthesize RhB-labeled SARS-CoV-2 mimics. LSCs (104 cells/mL) were seeded in 4-well culture chamber slides. RhB-labeled Lentivirus (104 TU/mL), RhB-labeled SARS-CoV-2 mimic (104 TU/mL), RhB-labeled SARS-CoV-2 mimic (104 TU/mL) + LSC nanodecoys (105), RhB-labeled SARS-CoV-2 mimic (104 TU/mL) + HEK nanodecoys (105) were incubated with LSCs, respectively. After four hours of incubation, free SARS-CoV-2 mimics were removed and washed using PBS for three times. Cells were fixed with 4% PFA, stained for LSC markers (FITC-CD90; 11-0909-42, Invitrogen), and imaged with an Olympus FLUOVIEW confocal microscope. The internalization of SARS-CoV-2 mimics by cells was examined by flow cytometry analysis (CytoFlex; Beckman Coulter).
Nanodecoys protect lung cells from SARS-CoV-2 mimicking viruses
We studied whether nanodecoys could neutralize SARS-CoV-2 mimic viruses and shelter lung cells from being infected. Macrophages and LSCs (1:1) were co-cultured in 4-well culture chamber slides, and RhB-labeled lentivirus-spike (104 TU/mL) and DiD-labeled nanodecoys (105) were added. After two hours of incubation, free RhB-labeled lentivirus-spike and DiD-labeled nanodecoys were removed and the samples were washed using PBS three times. Cells were fixed with 4% PFA, stained with LSC (FITC-CD90; 11-0909-42, Invitrogen) or macrophages (CD4) markers, and imaged with an Olympus FLUOVIEW confocal microscope. Flow cytometry analysis was performed to confirm the microscopy data.
Biodistribution of nanodecoys in mice
All animal procedures were approved by the Institute Animal Care and Use Committee (IACUC) of North Carolina State University (protocol # 19-806-B). Male CD1 mice (7 weeks) were obtained from Charles River Laboratory (Massachusetts, USA). DiD-labeled nanodecoys (1× 1010 particles per kg of body weight) were delivered to the CD1 mice via inhalation treatment using a nebulizer (Pari Trek S Portable Compressor Nebulizer Aerosol System; 047F45-LCS). Mice were euthanized at 24, 48, and 72 hours. All major organs were collected and were cryo-sectioned for further immunofluorescence analysis of the nanodecoys in vivo biodistribution post-inhalation.
In vivo clearance of the SARS-CoV-2 mimicking virus by nanodecoys in mice
Prior to performing clearance assay, the levels of ACE2 on the nanodecoys were quantified by an ELISA analysis (ab235649, Abcam) and was determined to be 112 ACE2 per nanodecoy. AF647-labeled SARS-CoV-2 mimics (5× 106 per kg of body weight) were first delivered to the Male CD1 mice (7 weeks) via inhalation treatment using a nebulizer (Pari Trek S Portable Compressor Nebulizer Aerosol System; 047F45-LCS). 24 hours later, nanodecoys (1× 1010 particles per kg of body weight) or free rACE2 with the same amount of ACE2 on the nanodecoys were inhaled, respectively. PBS treatment was used as control. Lungs were collected and imaged 1, 2, 3, 4, 5, and 6 days after treatment using Xenogen Live Imager (IVIS). Additionally, lung tissues were cryo-sectioned for further analysis of SARS-CoV-2 mimics biodistribution in vivo post-inhalation. Blood samples were collected for cytokine array analysis (Mouse Cytokine Array C1000, Raybiotech) according to the manufacturer’s instructions.
Toxicity studies in mice
Male CD1 mice (7 weeks) were treated with PBS, LSC- or HEK-nanodecoys (1× 1010 particles per kg of body weight) via inhalation. After 14-day treatment, the blood (blood test) and major organs (H&E) were collected for toxicity evaluation.
Nonhuman primate studies
All animal studies were conducted in compliance with all relevant local, state, and federal regulations and were approved by the Bioqual Institutional Animal Care and Use Committee (IACUC) under approved IACUC # 20-090P. Six Cynomolgus Macaques (three females, three males) were allocated by a counterbalance randomization. All animals were housed at Bioqual, Inc. (Rockville, MD). The macaques were challenged with SARS-CoV-2 using the intranasal and intratracheal routes. The viral inoculum (0.5 mL) will be administered drop-wise into each nostril and 1.0 mL of viral inoculum will be delivered intratracheally using a French rubber catheter/feeding tube, size 10, sterile (cut 4”−6” in length). Macaques were inoculated with a total dose of 1.1×105 PFU SARS-CoV-2. PBS or the LSC-nanodecoys were administered by inhalation using a nebulization and fitted mask daily from days 2–5 following challenge. Bronchoalveolar lavage (BAL), nasal swabs (NS), blood, body weight, and body temperature were monitored or collected throughout the study. Macaques were necropsied on day 8 post-challenge. All immunologic and virologic assays were performed blinded.
Statistics and Reproducibility
All experiments were performed at least three times independently. Results are shown as means ± SD. Comparisons between any two groups were performed using the two-tailed, unpaired Student’s t-test. For multiple group comparison, one-way ANOVA and two-way ANOVA was used with Bonferroni post-correction. A P value less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (HL123920, HL137093, HL144002, HL146153, HL147357, and HL149940 to K.C.) and the American Heart Association (18TPA34230092 and 19EIA34660286 to K.C.). We thank the Analytical Instrumentation Facility at North Carolina State University for TEM analysis (supported by the State of North Carolina and the National Science Foundation ECCS-1542015 and DMR-1726294).
Footnotes
Competing interests
K.C. is a co-founder and equity holder of BreStem Therapeutics Inc. P.U.C.D. reports compensation for consulting from BreStem Therapeutics Inc. The remaining authors declare no competing interests.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Wang C, Horby PW, Hayden FG & Gao GF A novel coronavirus outbreak of global health concern. Lancet (London, England) 395, 470–473 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 323, 1061–1069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao S et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int. J. Infect. Dis 92, 214–217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan J, Li M, Lv G & Lu ZK Monitoring transmissibility and mortality of COVID-19 in Europe. Int. J. Infect. Dis 95, 311–315 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziegler CGK et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181, 1016–1035.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry E et al. Adult Lung Spheroid Cells Contain Progenitor Cells and Mediate Regeneration in Rodents With Bleomycin-Induced Pulmonary Fibrosis. Stem Cells Transl. Med 4, 1265–1274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinh PC et al. Derivation of therapeutic lung spheroid cells from minimally invasive transbronchial pulmonary biopsies. Respir. Res 18, 132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cores J et al. Safety and Efficacy of Allogeneic Lung Spheroid Cells in a Mismatched Rat Model of Pulmonary Fibrosis. Stem Cells Transl. Med 6, 1905–1916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinh P-UC et al. Inhalation of lung spheroid cell secretome and exosomes promotes lung repair in pulmonary fibrosis. Nat. Commun 11, 1064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang S, Zhang X & Du L Therapeutic antibodies and fusion inhibitors targeting the spike protein of SARS-CoV-2. Expert Opin. Ther. Targets 1–7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 182, 1284–1294.e1289 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun 11, 6013 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cores J et al. A pre-investigational new drug study of lung spheroid cell therapy for treating pulmonary fibrosis. Stem Cells Transl. Med 9, 786–798 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamming I et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol 203, 631–637 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Penninger JM, Li Y, Zhong N & Slutsky AS Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 46, 586–590 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan J et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Lei C et al. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun 11, 2070 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruse RL Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Research 9, 72 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang RH, Luk BT, Hu C-MJ & Zhang L Engineered nanoparticles mimicking cell membranes for toxin neutralization. Adv. Drug Deliv. Rev 90, 69–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao L, Tian R & Chen X Cell-Membrane-Mimicking Nanodecoys against Infectious Diseases. ACS Nano 14, 2569–2574 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q et al. Cellular Nanosponges Inhibit SARS-CoV-2 Infectivity. Nano Lett. 20, 5570–5574 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao L et al. A Biomimetic Nanodecoy Traps Zika Virus To Prevent Viral Infection and Fetal Microcephaly Development. Nano Lett. 19, 2215–2222 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Wei X et al. T-Cell-Mimicking Nanoparticles Can Neutralize HIV Infectivity. Adv. Mater 30, e1802233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry BD et al. Engineered liposomes sequester bacterial exotoxins and protect from severe invasive infections in mice. Nat. Biotechnol 33, 81–88 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Hoegger MJ et al. Assessing mucociliary transport of single particles in vivo shows variable speed and preference for the ventral trachea in newborn pigs. Proc. Natl. Acad. Sci 111, 2355–2360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiser M Update on macrophage clearance of inhaled micro- and nanoparticles. J. Aerosol Med. Pulm. Drug Deliv 23, 207–217 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y et al. Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl. Sci. Rev 7, 998–1002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine 57, 102833 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo C et al. Single-cell analysis of two severe COVID-19 patients reveals a monocyte-associated and tocilizumab-responding cytokine storm. Nat. Commun 11, 3924 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Florindo HF et al. Immune-mediated approaches against COVID-19. Nat. Nanotechnol 15, 630–645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin MD et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol 15, 646–655 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q et al. Cellular Nanosponges Inhibit SARS-CoV-2 Infectivity. Nano Lett. 20, 5570–5574 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao L et al. Decoy nanoparticles protect against COVID-19 by concurrently adsorbing viruses and inflammatory cytokines. Proc. Natl. Acad. Sci 117, 27141–27147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


