Abstract
The interior of the cell abounds with reactive species that can accumulate as non-enzymatic covalent modifications (NECMs) on biological macromolecules. These adducts interfere with many cellular processes, for example by altering proteins’ surface topology, enzymatic activity or interactomes. Here, we discuss dynamic NECM modifications on chromatin, which serves as the cellular blueprint. We first outline the chemistry of NECM formation and then focus on the recently identified effects of their accumulation on chromatin structure and transcriptional output. We next describe the known cellular regulatory mechanisms that prevent or reverse NECM formation. Finally, we discuss recently developed chemical biology platforms for probing and manipulating these NECMs in vitro and in vivo.
Keywords: Acylation, glycation, lipidation, chromatin, epigenetics, chemical biology
Prevalent Chemical Reactions in Cells
Many cellular metabolites contain reactive moieties, such as aldehydes, that are key for their cellular function but are also capable of non-specifically modifying macromolecules to form non-enzymatic covalent modifications (NECMs) [1–3]. Some proteins have unique properties that render them susceptible to NECMs: namely a high abundance of accessible reactive functional groups on bases or side chains and comparatively long half-lives. Perhaps unsurprisingly, eukaryotic chromatin (see Glossary) - and in particular the DNA-packaging histone proteins - meet all these criteria and have recently been found to be rich with NECMs [4].
In chromatin, histones interact with DNA via electrostatic interactions between cationic histone tails which are rich in arginine and lysine residues, and the anionic DNA phosphodiester backbone [5]. Modulation of this interaction is key for regulating gene accessibility and is implemented directly or indirectly through various post-translational modifications (PTMs) on the histone tails [6–8]. These modifications, which include acetylation, methylation, and ubiquitination, occur on the highly abundant lysine and arginine residues of the histone tails [4]. The influence of histone PTM patterning on gene expression is the basis of the “Histone Code” hypothesis, wherein combinatorial histone modifications form a code that is interpreted by effector proteins to control gene expression [9].
While many of the modifications on histones are introduced by enzyme “writers”, an increasing number of modifications are being discovered that are derived through non-enzymatic origins [10]. Although, by comparison, many of these PTMs are poorly characterized, and many tend to be deleterious through several mechanisms. Since NECMs react with amine side chains present on lysine and arginine, they can directly disrupt the electrostatic interaction with DNA or block canonical modifications on the same sites. Since epigenetic information needs to be retained accurately across multiple cell divisions, the accumulation of adducts on genome-associated macromolecules may be extremely harmful for the cell (Figure 1) [11].
Figure 1. Effects of histone NECMs on Chromatin.

Non-enzymatic chemical modifications (NECMs) can induce cross-links, abate interactions between DNA and histones, alter histone three-dimensional structure and surface topology, and compete for sites with enzymatic post-translational modifications (PTMs).
Cells have evolved several mechanisms to counter the threat of accumulated NECMs causing damage: 1) metabolite efflux, 2) metabolite scavenging, 3) adduct reversal or editing, and 4) global cellular signaling response [12]. While these mechanisms prevent NECM accumulation and systemic damage, they are undoubtedly vital in reducing the negative effects of histone NECMs in particular. In metabolically aberrant cells, such as cancer cells where anaerobic glycolysis rates are increased (i.e. the Warburg effect), these defense systems are upregulated, most likely to attenuate the toxicity which accompanies their higher metabolic state [13]. Uncovering the mechanisms surrounding this dynamic nature of NECMs could be leveraged to understand the fitness of metabolically dysregulated cells. Here, we describe in detail the mechanism of formation of the three best-characterized classes of histone NECM types: glycation, acylation, and lipidation (Table 1). We then summarize how these are regulated in a cellular context and provide discussion of recent technological advances to track and understand the consequences of these notoriously elusive modifications.
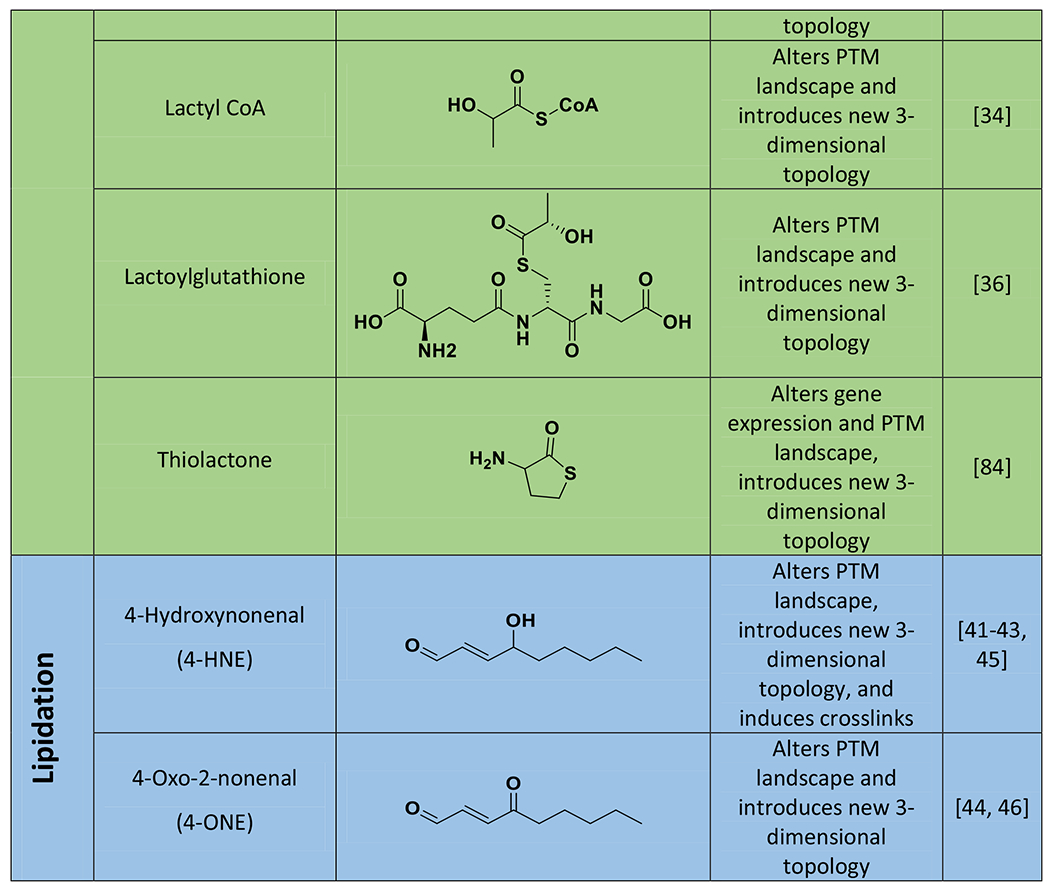
Table 1.
Structures of molecules that form histone non-enzymatic chemical modifications (NECMs), as well as their detected effects on chromatin physiology.

|
|
AGEs: Advanced Glycation End products are the resulting diverse group of products in a glycation chemical reaction cascade.
Alters PTM landscape refers to affecting the patterning of enzymatically introduced PTMs by competitive or other means.
NECMs that Affect Chromatin Physiology
Glycation
Sugars, their derivatives, and metabolic by-products can often exist within the cell for extended periods of time and at substantial concentrations, facilitating their chemical reaction with proteins [14]. The metabolite’s carbonyl is sufficiently electrophilic to react with and modify nucleophiles in the cell, including small molecules, protein side chains (e.g. lysine, arginine), or nucleobases, a process termed glycation. The initial Schiff base adduct can undergo rapid rearrangement to form more stable “Amadori” ketoamine intermediates which can thereafter react with other species, degrade, or rearrange into a plethora of modifications called advanced glycation end-products (AGEs) [15]. Glycation has been shown to interfere with a number of metabolic and epigenetic processes through the modification of key amino acid side chains on enzymes and histones, as well as the direct modification of DNA [16].
Not all metabolites are equally reactive in glycating histones. It is thought that the rate at which a glycation agent is capable of modifying its substrate is dependent on the amount of time the reacting aldehyde is available [17]. Therefore, glycolysis byproducts such as glyoxal or methylglyoxal (MGO) are extremely reactive, whereas cyclized sugars such as glucose, which primarily exists as a hemi-acetal, are more inert (Figure 2A). Indeed, MGO was recently shown to modify arginine and lysine on histones at key regulatory positions at levels comparable to the endogenous, enzymatically installed modifications, such as methylation and acetylation [18]. The modifications were detected on all four core histones at critical residues involved in both nucleosome stability and “reader” domain binding. RNA-seq data revealed that MGO can alter gene transcription, most notably in cells lacking the major detoxifying enzyme, glyoxalase 1 (GLO1), which normally eliminates MGO. In the same cells, MGO treatment resulted in marked disruption of H2B acetylation and ubiquitination without affecting H2A, H3, and H4 modifications [18]. Altogether, histone MGO modification was found to have global ramifications on histone enzymatic PTMs, the assembly and stability of nucleosomes, and chromatin architecture. The resulting impacts on chromatin structure affect gene accessibility via a two-step mechanism. As lower levels of MGO modifications accumulate, through low concentration or short exposure, chromatin decompacts and gene accessibility is increased due to the disruption of the electrostatic interactions between DNA and histones. By contrast, increased levels of MGO adduct accumulation, through high concentration or long exposure, results in cross-links and hyper compaction of chromatin, leading to a decrease in gene accessibility [19].
Figure 2. Chemical nature of histone glycation, acylation and lipidation NECMs.
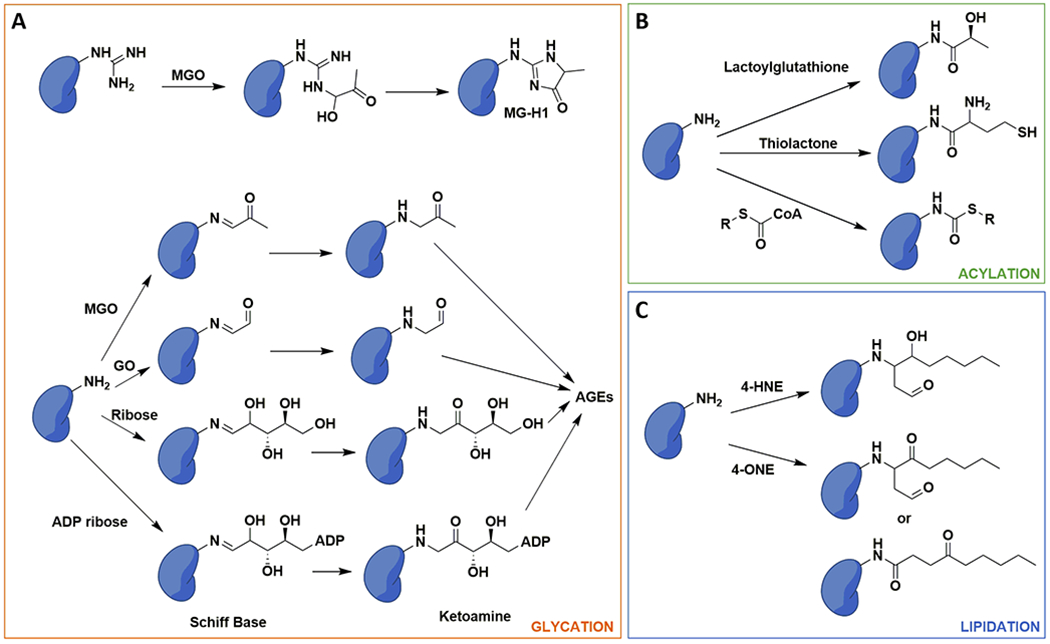
A) Glycation adducts go through stabilized intermediates but can eventually form advanced glycation end products (AGEs). MGO: methylglyoxal; MG-H1: methylglyoxal hydroimidazolone 1; GO: glyoxal. B) Activated acyl groups such as the glyoxalase 1 (GLO1) product lactoylgluathione or metabolic cofactors can modify histone lysine ε-amines. C) Products of lipid peroxidation, including 4-hydroxynonenal (4-HNE) and 4-oxo-2-nonenal (4-ONE) can react with and modify histone amines.
While glucose glycation in general is slow, it does occur in low quantities in the cell to form Maillard products [17]. Subsequent glucose, and more broadly cyclic sugar glycation adducts, can degrade into ketohexones such as 3-deoxyglucosone (3-DG) which are much more reactive and have been demonstrated to modify histones [20]. However, the relatively more labile ribose, as well as its derivates such as the ubiquitous ADP-ribose, have been shown to glycate all four core histones, as well as the linker histone H1 [21, 22]. In solution, the core histones have been shown to react faster with ribose than the linker histone H1; however, in a chromatin context, where H1 is loosely bound to the periphery of the nucleosome and is more accessible, the reverse trend is observed as ribose modifies the linker histone faster than the core histones. In endothelial cells, high concentrations of ribose can generate protein crosslinks and ultimately cause apoptotic cell death. As with MGO, ribose disrupts chromatin PTM patterning and specifically reduces H3 and H4 acetylation; however, without tracking the specific sites and adducts on the site, it remains unclear if this is a direct competition or the result of a more convoluted mechanism [23]. Despite impressive innovations in the field and the discovery of broad epigenetic phenotypes, uncovering a link between site-specific histone glycation and an epigenetic effect remains elusive.
Acylation
While carboxylate groups alone at physiological pH are poor electrophiles, they can be enzymatically converted into thioesters or even more labile anhydrides, capable of acylation [24]. Advances in mass spectrometry (MS) have revealed a variety of such metabolites including lipids, amino acids, CoA, acetate, malonate, and lactate that can be readily activated and subsequently react with cellular components to form easily hydrolysable esters and thioesters or stable amides (Figure 2B) [25–27]. The simplest forward acylation (acetylation) can be performed enzymatically by histone acetyltransferases, which specifically deposit the mark, or non-enzymatically, resulting in non-directed localization [26]. Conversely, the reverse deacylation process does not occur spontaneously and requires enzymatic regulation, primarily by histone deacetylases (HDACs) or members of the Sirtuin family [28, 29].
In the decades since the discovery of non-enzymatic histone acetylation by acetyl-CoAs, several other histone acylations NECMs have been identified [27, 30]. Generally, acylation occurs on lysine side chains and removes the charge on their substrate amine. The resulting loss of electrostatic binding energy between DNA and the histone tail leads to chromatin decompaction and gene expression [31]. Due to the breadth of reactive molecules capable of non-enzymatic acylation, including crotonylation, butyrylation, or succinylation, the degree to which acylation NECMs are expected to alter histone structure or function varies greatly depending on the chemical structure of the acyl-donor [27]. For example, histone benzoylation, the first described modification by an aromatic acyl group, was identified on 22 lysine residues in cancer cells. The mark was stimulated using the common food preservative and FDA-approved drug sodium benzoate, which is converted into active benzoyl-CoA intracellularly. Notably, ChIP-seq and RNA-seq studies of this mark uncovered substantial physiological consequences of the mark compared to acetylation [32]. Similarly, increased cellular concentrations of lactate in M1 macrophages due to increased glycolysis following hypoxia or bacterial challenge stimulates histone lactylation (also referred to as lactoylation) via a later detected CoA intermediate [33]. Lactylation was identified at 28 sites on core histones in human and mouse cells, and displayed different temporal dynamics from acetylation. Ultimately, in late phase of M1 macrophage polarization it induced homeostatic genes such as Arg1, which is involved in wound healing [34].
The discovery of histone lactylation suggests a potential crosstalk between glycation and acylation, as the latter can also occur in response to cellular upkeep and maintenance through the glyoxalase system. Dicarbonyls such as MGO and glyoxal react with glutathione to form S-D-lactoylglutathione (LGSH), a reaction catalyzed by GLO1. This intermediate can then be hydrolyzed with the assistance of GLO2 to complete the cycle by regenerating glutathione and forming lactate. Alternatively, it can spontaneously react with nearby lysine residues to form lactylLys. The activity of lactylLys depends on GLO2 amounts, as GLO2 knockdown leads to marked increases in LGSH and the subsequent increases in lactylLys levels [34, 35]. Additionally, through the use of an alkyne-tagged MGO analog, these modifications were shown to be enriched on glycolytic enzymes and cause a global decrease in glycolytic output [36].
Whereas the acylations discussed above rely on an acyl-donor being activated by common cellular cofactors like glutathione or CoA, other mechanisms of acylation have also been identified [10, 37]. For example, during error editing in protein biosynthesis, methionyl-tRNA synthetases generate homocysteine thiolactone. The labile thiolactone ring is hydrolyzed and opened by lysine side chains, generating a novel thiol on the surface of the protein, a process termed homocysteinylation (hcy) [38]. Increased hey levels have been associated with suppressed DNA damage repair as well as neural tube defects. Specifically, 39 histone hey sites have been identified in samples from human embryonic brain tissues. Additionally, ChIP-Seq and RNA-seq experiments have demonstrated that an increase in H3K79hcy levels down regulates the expression of selected neural tube closure genes and induces the onset of neural tube defects [39]. However, as with most acylations, it remains a challenge to uncover the extent to which these modifications are enzymatic or non-enzymatic in nature without knowledge of existing “writer” enzymes.
Lipidation
Lipid peroxidation in cells occurs in response to oxidative stress and the breakdown of peroxides leads to the formation of diverse aldehydes (Figure 2C) [40]. Two of these aldehydes, 4-hydroxynonenal (4-HNE) and 4-oxo-2-nonenal (4-ONE) have been extensively studied and found to chemically modify a number of proteins and DNA [41, 42]. Mildly cross-linked HNE-modified proteins are preferentially degraded by the proteasome but extensive cross-linking results in protein aggregation and proteasome inhibition. This is thought to be a defense mechanism to oxidative stress as mild damage can be reversed but extensive damage requires full cellular turnover [43]. Furthermore, stable 4-HNE and 4-ONE adducts on histones have established a potential link between oxidative stress and epigenetics [44]. 4-HNE was initially shown to react with histones using electron paramagnetic resonance in conjunction with protein spin label experiments [45]. While both 4-HNE and 4-ONE have since been shown to form adducts on histones, 4-ONE only attaches to lysine residues while 4-HNE also targets cysteines. Thus, cysteine-deprived histones are more susceptible to 4-ONE over 4-HNE lipidation as the latter will additionally be consumed by the cysteine proteome [44, 46]. The resulting lipid NECMs are much larger than most small molecule PTMs, such as methyl or acetyl marks. This, combined with the fact that both 4-ONE and 4-HNE have been shown to also modify DNA directly, reduce histone-DNA binding as well as a number of other chromatin effector proteins [44, 46]. Together, these may define an additional mechanism behind the observed epigenetic effect of lipidation.
Cellular Defense Mechanisms Against NECMs
Due to the deleterious nature of most NECMs, cells must maintain firm control on the extent of adduct accumulation. This is especially true for metabolically challenged cells, like in the cases of cancer or diabetes [13, 47]. Cells are equipped with a 3-pronged NECM regulatory network that consists of 1) scavenging or deactivating reactive metabolites to prevent the NECM reaction, 2) reversing the adducts or converting them into more tolerable forms, and 3) detecting the damage and activating signaling responses for either repair or induction of cell death. Below we discuss the first two mechanisms as this review centers around histone NECMs, but for detailed reviews on cellular recognition of AGEs and the subsequent cellular response, see [48–50].
Discouraging NECM Formation
Although histones are translated in the cytosol, they are captured by chaperones as they emerge from ribosomes and are immediately escorted to the nucleus where they are deposited into chromatin. Their fleeting cytosolic half-lives, together with the fact that they are mostly masked, reduces the possibility of them accumulating NECMs in the cytosol. Indeed, histones spend most of their lifespan within the nuclear compartment where reactive small molecules, via the large nuclear pores, can freely access and ultimately modify them as well as DNA, and other chromatin-effector proteins.
Several protein systems have previously been described as being capable of intercepting reactive metabolites and byproducts both in the cytosol and the nucleus before they can reach the histones (Figure 3A). In addition to the glyoxalase cycle mentioned above, aldehyde and lactate dehydrogenases utilize NAD+ as a cofactor and directly oxidize aldehyde and lactate molecules into their respective less deleterious carboxylates [51, 52]. Conversely, aldo-keto reductases use NADPH as a cofactor to reduce aldehydes into alcohols, which are more easily managed [53]. Carnosine synthase 1 (CARNS1) catalyzes the ATP-dependent formation of the dipeptide carnosine from L-histidine and β-alanine, which then scavenges both reactive oxidative and carbonyl species [54, 55].
Figure 3. Cellular housekeeping mechanisms of NECMs.

A) GLO1/GLO2 scavenger system (top). GLO1 can catalyze the reaction between glutathione and methylglyoxal (MGO), forming lactoylglutathione which can then be hydrolyzed by GLO2 to release D-lactate and regenerate glutathione. Small molecule nucleophiles like L-Carnosine (bottom) can scavenge reactive species like MGO by reacting with them and diminishing the pool of reactive small molecule. B) Enzymatic systems can remove the adducts (DJ-1, deacylases, or FN3K) or modify the early intermediates (PAD4) to reduce the likelihood of further damage and downstream consequences.
Amending NECMs
In addition to the preventative measures discussed above, cells have evolved various enzymatic mechanisms to remove NECMs from proteins and DNA at both early and late stages (Figure 3B). One example is the deglycase DJ-1/Park7, which hydrolyzes early MGO glycation adducts from both proteins and DNA to release lactate [18, 19, 56]. In another case, fructosamine 3-kinase (FN3K) catalyzes the removal of glycation adducts by phosphorylating the C-3 hydroxyl on the sugar, leading to the subsequent spontaneous degradation of the adduct, producing a deoxyketone byproduct as well as the regenerated amine [57]. Alternatively, protein arginine deiminase 4 (PAD4) has been shown to convert arginine side chains, including those that have been glycated with MGO, into citrulline. Though it does not regenerate the unmodified substrate, it does stall the glycation cascade to prevent the formation of AGEs as well as protect the arginine from being a glycation substrate again [58].
Several deacylases have been implicated in the repair of non-specific acylation including HDACs and SIRT3 for acetylation, SIRT5 that can remove succinyl acyl groups (see references [59, 60] for summary of the epigenetic roles of Sirtuins), and HDAC11 which acts as a general fatty acid deacylase [32, 61–65]. In addition, SIRT2 wash shown to remove both acetylation as well as histone 4-ONE and lactyllys modifications, which were themselves recently shown to also be removed by HDACs 1-3 [66]. The general low selectivity of these enzymes permits the accommodation of an array of adducts and substrates of differing sizes and shapes. This attribute may be evolved as a fundamental cellular mechanism to prevent the accumulation of non-enzymatic and non-specific acylation.
The activities of the repair enzymes are notably restricted to their cellular localization. So, while FN3K can deglycate histones and attenuate their glycation damage during and immediately after translation, the kinase has no effect on histone chronic NECM accumulation. Conversely, pervasive DJ-1 and PAD4 can affect the accumulated nuclear histone NECM damage as they are present in the nucleus. Interestingly, several of the above enzymes (DJ1, FN3K, and PAD4) are overexpressed in many cancers and have been associated with cancer cell proliferation [67–69]. Consequently, small molecule campaigns have been conducted against DJ-1 and PADI4 to leverage their deglycation activity as a new therapeutic avenue and the resulting molecules may be promising starting points as therapeutic agents [70–72].
Novel Tools Developed for Tracking NECMs
Traditional methods to track NECMs have relied on biophysical approaches including spectroscopic, colorimetric, and chromatographic methods. These methods focused on physical characteristics that changed as NECMs formed, for example the formation of aromatic AGEs, such as pentosidine, which allowed for visualization of glycation on proteins. Additionally, accumulation of many adducts cause physical distortions of protein structure which were detected through spectroscopic analyses. Furthermore, global changes in protein structure were detectable via circular dichroism and various chromatographies. Finally, modifications resulting in cross-links were identified through protein laddering after separation by gel electrophoresis [15]. While effective, these approaches have low sensitivity and thus require extensive accumulation of adducts under extremely harsh and non-physiological conditions, and the scope of identifiable modifications is extremely narrow. There was thus a need for the development of more sensitive and broadly applicable strategies.
Immunoassays
Antibodies were and remain very powerful tools utilized to uncover and study specific protein adducts. Indeed, they have been extensively used in the context of NECMs, particularly in glycation discoveries. However, antibodies have substantial limitations in studying protein glycation. Primarily, glycation adducts are unstable and frequently rearrange to form an array of chemical structures, while antibodies require stable modifications on peptides inoculated in animals for weeks. Using a mixture of adducts as an antigen results in the generation of a less specific antibody [15]. Despite these limitations, powerful and specific antibodies have been commercially developed for certain species, such as pan antibodies against MGO, carboxymethyl lysine, or carboxyethyl lysine. Moreover, chemical synthesis of peptides with the relatively stable MGO arginine modifications, MG-Hs, were used to produce effective antibodies. The success of this pipeline suggests that additional antibodies against other stable NECMs can be similarly generated [73].
Mass Spectrometry
Much of the pioneering work in understanding the diversity of enzymatic histone PTMs came through innovations in MS sample preparations, physical instrumentation, and data analysis including top-down and PTM scanning approaches among others [74–76]. For instance, trypsin digestion limitations on histones, due to an overabundance of lysine and arginine on histone tails, was overcome through the chemical propionylation of these residues prior to digestion [77]. MS analysis of NECMs on histones presents even greater challenges. For example, the acid-based histone extraction and propionylation could alter the structures of NECMs generating additional complexity that is difficult to account for. Additionally, some adducts are constitutional isomers and cannot be distinguished by mass spectrometry. Moreover, many glycation pathways pass through or result in identical structures, thus preventing identification of the initial glycating reagent. Several NECMs also form on lysine and arginine side chains, occluding sites of enzymatic cleavage during sample preparation resulting in peptides that are too long for chromatographic separation, and subsequently pass undetected. During the MS analysis itself, NECMs decrease the net charge on these residues, further reducing their likelihood of detection by ion activation methods. Finally, cross-linking adds an almost insurmountable complexity barrier without prior knowledge of the peptides being cross-linked in addition to the added difficulty of generating quality MS/MS data.
Several of these limitations have been overcome through technical and analytical innovations. Firstly, alternate sample preparations, including a high salt isolation strategy, have been described and recently used to analyze MGO and ribose glycation NECMs on histones while avoiding extreme pH reagents [19, 58, 77, 78]. Secondly, a label-free approach for the quantitative analysis and discovery of Lys and Arg modifications (QuARKMod) for complex biological samples, was developed (Figure 4A). The method involves digestion of protein samples down to individual amino acids followed by collision induced dissociation (CID) MS to determine the absolute concentrations of each amino acid species, with or without modification [79]. This method has since been successfully applied in the study of novel histone NECMs such as lactoylysine and riboLys, as well as MGO adducts [18, 23, 36, 44]. While MS-based methods to study histone NECMs face limitations, the continued development of such approaches will undoubtedly yield further important discoveries in the future.
Figure 4. New strategies to uncover and study histone NECMs.
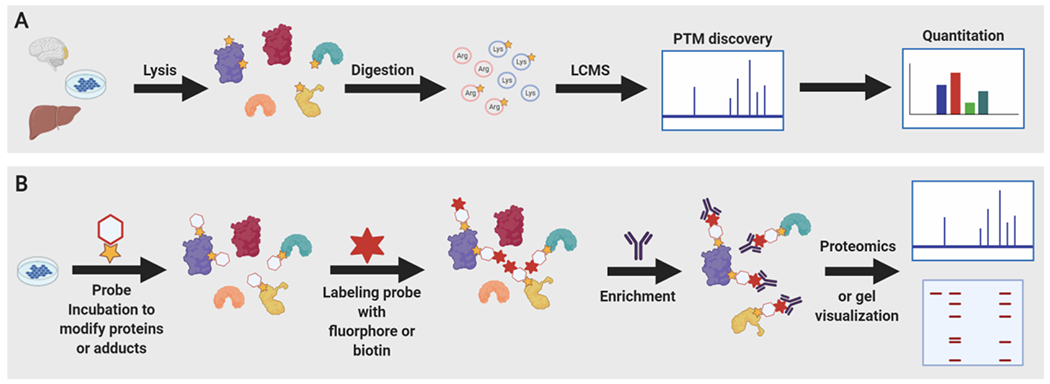
A) Quantitative Analysis and Discovery of Arginine and Lysine Modifications (QuARKMod) is a label-free method to discover and quantitate new modifications on lysine and arginine residues from live cells. Relevant cells or tissues are lysed, proteins are isolated, digested, and analyzed by liquid chromatography mass spectrometry (LCMS), before predicted adduct masses are searched and quantitated. B) General strategy for chemical probe application to reveal or characterize non-enzymatic chemical modifications (NECM) targets. Probes either modify existing NECMs or introduce trackable NECM mimics in cells. The introduced probe can itself be labeled with flurophore or biotin which are used for detection and enrichment for western blot or proteomic analysis.
Chemical Probes
Chemical biology approaches have offered a powerful alternative strategy through the development of chemical analogs of the desired reactive molecules (Figure 4B, Table 2). The analogs contain appended enrichment moieties and have been successfully applied toward the study of a broad range of histone NECMs. For example, two alkynyl-modified MGO probes have been synthesized and used for visualization and enrichment of MGO adducts. The longer of the two was used to characterize the blood MGO proteome, whereas the shorter was used to visualize and enrich histone MGO-glycation NECMs [78, 80, 81]. Similarly, an azidoribose probe was recently synthesized and applied to track histone ribose glycation. Its enrichment handle was utilized to isolate the ribose glycation nuclear proteome, revealing that in addition to histones, several other key nuclear systems, such as those involved in DNA repair, are susceptible to ribose glycation [23].
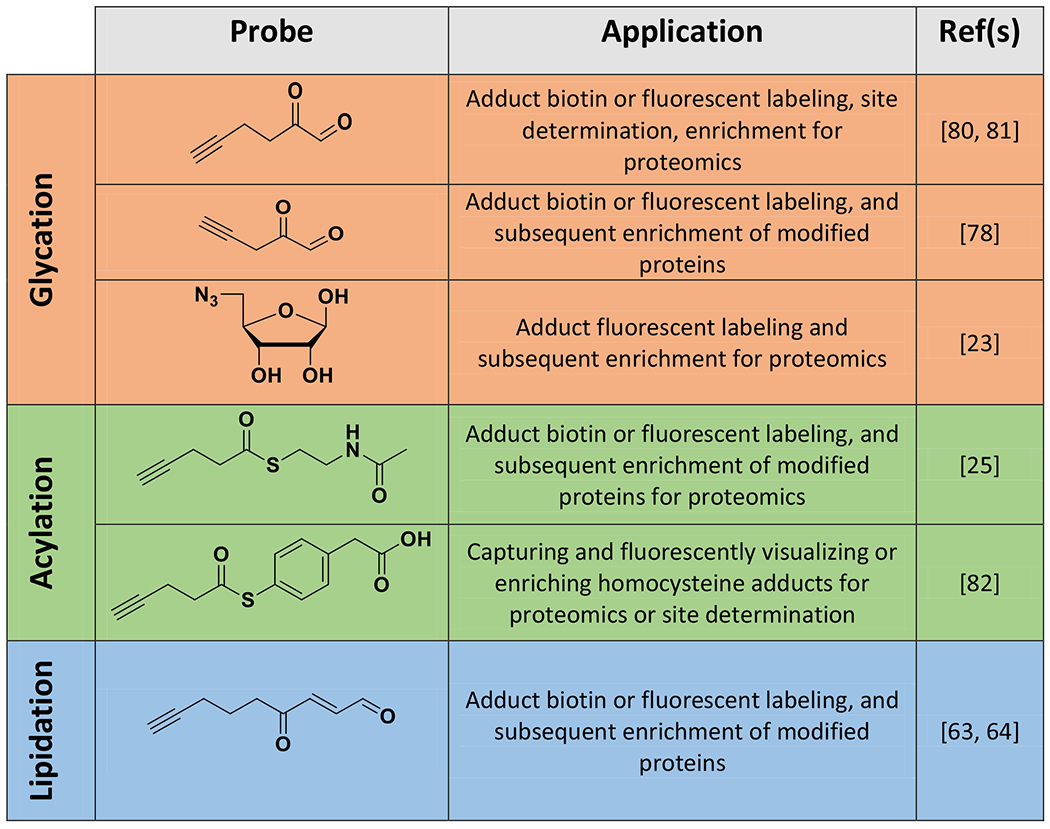
Table 2.
Structures of chemical probes used to characterize and study histone non-enzymatic chemical modifications (NECMs) as well as their reported uses.
|
Additionally, a biotinylated non-specific thioester probe was developed and used in a competition assay with acyl-CoAs to characterize non-enzymatic acylation targets. Applying the method to cancer cell lines uncovered numerous candidate targets of non-enzymatic acylation, including several enzymes in glycolysis [25]. 4-ONE lipidation histone NECMs have also been tracked using an alkynyl analog of the peroxide by-product and this probe was instrumental in the discovery of SIRT2 as an “eraser” of this modification [63, 64]. Although they provide temporal control and useful enrichment capabilities, these probes are limited in that they are by definition non-endogenous and their distinction from the native molecules, though minor, could be crucial. Alternatively, homocysteinylation adducts can be labeled by an alkynyl-fused thioester probe via a biotin switch assay. This probe targets the thiol of the homocysteine, then an S-to-N acyl shift yields the alkynyl modified adduct. By labeling the adduct rather than mimicking it, this probe does not face the same limitations as the chemical analogs discussed above [82]. The development of such technologies will facilitate the advancement of histone NECM research from searching for modifications, into understanding the mechanisms of action linking these NECMs, cellular physiology, and pathology.
Concluding Remarks
Due to the diverse natures of the cellular metabolome, there are a wide range of NECMs that can affect macromolecules within the proteome. Through their roles in chromatin regulation, the long-lived and arginine- and lysine-rich histone proteins are extremely susceptible to accumulate these NECMs. The resulting modifications can alter their stability, three-dimensional structure, and their functional interactions. These disruptions can lead to several cellular pathophysiological outcomes, including aging and death. As such, multiple layers of regulation including protein scavenging, enzymatic attenuation, and cellular recognition respond to these stresses. These forces of modification and regulation compete, and the resultant has a powerful influence on the cell’s fate and lifespan.
Further development of tools to track, predict and perturb histone NECMs and the systems that surveil them will continue to broaden our knowledge of their effect on cellular function. Although tremendous progress has already been made within the field, much work remains to be done (see Outstanding Questions). We envision that as a new family of modifications, NECMs on histones continue to emerge and further regulatory systems and mechanisms linking them to chromatin dynamics will be established. Finally, the continued development of chemical and biological tools will prompt exciting opportunities for discovery in the field.
Outstanding Questions.
The cellular metabolome is extremely dense and diverse, consisting of thousands of different reactive molecules. Relative to the surfeit of species, only a handful of non-enzymatic covalent modifications (NECMs) have been identified on histones and their effect on chromatin structure and function remain murky. This apparent contradiction raises the question: are there other uncharacterized NECMs that exist on histones and can they play a role in linking metabolism and cell fate through epigenetic mechanisms?
Are all NECMs enzymatically reversible? What other mechanisms exist to regulate NECMs broadly in the cell and more specifically on histones?
What combination of chemical, proteomics, and genetics tools can be utilized to uncover and characterize new NECMs on histones as well as the cellular mechanisms that regulate them?
What chromatin-associated events do NECMs regulate? Are certain histone NECMs recognized and “read” by chromatin associated proteins and are they part of the so-called “histone code”? Furthermore, are some histone NECMs associated with a specific epigenetic state? Can they induce the genetic expression/repression of specific genes resulting in a cellular phenotypic outcome?
Are putative NECM surveillance factors and oncoproteins (including DJ-1, FN3K, PADI4, and GLO1/2) new therapeutic targets in cancer?
Can the chemical composition of NECMs be linked and then calibrated as biomarkers for cell state, age, or disease?
Highlights.
Post-translational modifications (PTMs) of proteins are introduced either by the specific activity of “writer” enzymes or the spontaneous chemical reaction with reactive small molecules forming non-enzymatic covalent modifications (NECMs).
Histones are particularly susceptible to accumulate NECMs due to their long half-lives, unstructured tails, and abundance of accessible reactive arginine and lysine residues that are key sites for regulatory enzymatic PTMs.
NECMs on histones compete with enzymatic PTMs as well as alter chromatin structure, subsequently affecting tightly regulated genetic processes, and ultimately resulting in global cellular phenotype changes.
NECMs can be enzymatically “erased” as well as prevented by scavenger systems including both small molecules and proteins.
Advances in biophysical methods, immunoassays, mass spectrometry, chemical probe development, and chemoproteomics have established a platform which can be leveraged to predict, track, manipulate, and study histone NECMs.
Acknowledgements
We thank Dr. James J. Galligan, Dr. Rasmus Pihl, and Nicholas A. Prescott for their assistance and insight while editing this review. Work in the David lab is supported by the Josie Robertson Foundation, the Pershing Square Sohn Cancer Research Alliance, the NIH (CCSG core grant P30 CA008748, MSK SPORE P50 CA192937, R21 DA044767 and R35 GM138386), the Parker Institute for Cancer Immunotherapy (PICI), and the Anna Fuller Trust. In addition, the David lab is supported by Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and the Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center. Elements of figures were created with BioRender.com.
Glossary
- Acylation
the spontaneous adduction of a small molecule onto the lysine ε-amine or N-terminal amine via an acyl linkage.
- Advanced glycation end products (AGEs)
the resulting diverse group of products in a glycation chemical reaction cascade.
- ChIP-seq
Chromatin Immunoprecipitation-sequencing; the process of identifying the regions where a specific mark is located in terms of genomic position. Involves the cross-linking of DNA and proteins, shearing chromatin, antibody enrichment, reverse cross-linking, digestion, and next-generation sequencing of the isolated DNA fragments.
- Chromatin
the physiologically relevant protein-DNA complex for storage and access of genetic information in eukaryotes. The fundamental unit of this complex is a nucleosome, which is comprised of about 150 DNA base pairs wrapped approximately 1.65 times around a histone octamer. The histone octamer itself contains two copies of each of the four core histones H2A, H2B, H3, and H4.
- Cross-links
covalent bonds linking one polymer chain to another.
- Dicarbonyls
molecules containing two carbonyl groups.
- Electrophiles
chemical species that accept electron pairs to form bonds with nucleophiles.
- Enrichment moieties
functional groups that confer specific purification advantages.
- Epigenetics
the study of phenotypic changes caused by alterations in gene expression rather than the genetic code.
- Glycation
the non-enzymatic modification of biological macromolecules by a reducing sugar or aldehyde-containing sugar derivative via the Mailiard reaction.
- Histone Code
the hypothesis that transcription is partly regulated by chemical modifications, primarily on unstructured histone tails.
- Lactylation
the incorporation of a lactyl moiety onto an ε-amine or N-terminal amine via an acyl linkage.
- Lipidation
the incorporation of a fatty chain as a modification on a protein.
- Glycolysis
the metabolic pathway that describes the breakdown of glucose by enzymes into pyruvate, releasing energy in the process.
- Maillard products
The plethora of species formed from a chemical reaction between amino acids and reducing sugars giving browned food its distinctive color and flavor.
- Propionylation
the reaction of propionyl groups onto the ε-amine or N-terminal amine via an acyl linkage that can be used to remove the ability of trypsin to recognize lysine and arginine residues in mass spectrometry sample preparation.
- Post-translational modifications (PTMs)
the covalent modification of proteins following their biosynthesis.
- RNA-seq
the characterization of the genetic sequences of the RNA in a sample, cell, or tissue.
- Thiolactone
a cyclic chemical compound with a characteristic sulfur adjacent to a carbonyl.
- Warburg effect
a modified form of cellular metabolism found in cancer cells whereby a specialized mixture of mostly anaerobic glycolysis is preferred to aerobic glycolysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harmel R and Fiedler D (2018) Features and regulation of non-enzymatic post-translational modifications. Nat Chem Biol 14, 244–252 [DOI] [PubMed] [Google Scholar]
- 2.DeBerardinis RJ and Thompson CB (2012) Cellular metabolism and disease: what do metabolic outliers teach us? Cell 148, 1132–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cloos PA and Christgau S (2002) Non-enzymatic covalent modifications of proteins: mechanisms, physiological consequences and clinical applications. Matrix Biol 21, 39–52 [DOI] [PubMed] [Google Scholar]
- 4.Erler J, et al. (2014) The role of histone tails in the nucleosome: a computational study. Biophys J 107, 2911–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luger K, et al. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 6.Bannister AJ and Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allfrey VG, et al. (1964) Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci U S A 51, 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips DM (1963) The presence of acetyl groups of histones. Biochem J 87, 258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenuwein T and Allis CD (2001) Translating the histone code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 10.Chan JC and Maze I (2020) Nothing Is Yet Set in (Hi)stone: Novel Post-Translational Modifications Regulating Chromatin Function. Trends Biochem Sci 45, 829–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Probst AV, et al. (2009) Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10, 192–206 [DOI] [PubMed] [Google Scholar]
- 12.Zheng Q, et al. (2020) Non-enzymatic covalent modifications: a new link between metabolism and epigenetics. Protein Cell 11, 401–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liberti MV and Locasale JW (2016) The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci 41, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad S, et al. (2018) Oxidation, glycation and glycoxidation-The vicious cycle and lung cancer. Semin Cancer Biol 49, 29–36 [DOI] [PubMed] [Google Scholar]
- 15.Hellwig M and Henle T (2014) Baking, ageing, diabetes: a short history of the Maillard reaction. Angew Chem Int Ed Engl 53, 10316–10329 [DOI] [PubMed] [Google Scholar]
- 16.Thornalley PJ (2008) Protein and nucleotide damage by glyoxal and methylglyoxal in physiological systems--role in ageing and disease. Drug Metabol Drug Interact 23, 125–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tessier FJ (2010) The Maillard reaction in the human body. The main discoveries and factors that affect glycation. Pathol Biol (Paris) 58, 214–219 [DOI] [PubMed] [Google Scholar]
- 18.Galligan JJ, et al. (2018) Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks. Proc Natl Acad Sci U S A 115, 9228–9233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Q, et al. (2019) Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat Commun 10, 1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashraf JM, et al. (2015) Glycation of H1 Histone by 3-Deoxyglucosone: Effects on Protein Structure and Generation of Different Advanced Glycation End Products. PLoS One 10, e0130630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cervantes-Laurean D, et al. (1996) Glycation and glycoxidation of histones by ADP-ribose. J Biol Chem 271, 10461–10469 [DOI] [PubMed] [Google Scholar]
- 22.Talasz H, et al. (2002) Nonenzymatic glycation of histones in vitro and in vivo. J Cell Biochem 85, 24–34 [PubMed] [Google Scholar]
- 23.Maksimovic I, et al. (2020) An Azidoribose Probe to Track Ketoamine Adducts in Histone Ribose Glycation. J Am Chem Soc 142, 9999–10007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li LO, et al. (2010) Acyl-CoA synthesis, lipid metabolism and lipotoxicity. Biochim Biophys Acta 1801, 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulkarni RA, et al. (2017) Discovering Targets of Non-enzymatic Acylation by Thioester Reactivity Profiling. Cell Chem Biol 24, 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olia AS, et al. (2015) Nonenzymatic Protein Acetylation Detected by NAPPA Protein Arrays. ACS Chem Biol 10, 2034–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simithy J, et al. (2017) Characterization of histone acylations links chromatin modifications with metabolism. Nat Commun 8, 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cress WD and Seto E (2000) Histone deacetylases, transcriptional control, and cancer. J Cell Physiol 184, 1–16 [DOI] [PubMed] [Google Scholar]
- 29.Sauve AA, et al. (2006) The biochemistry of sirtuins. Annu Rev Biochem 75, 435–465 [DOI] [PubMed] [Google Scholar]
- 30.Paik WK, et al. (1970) Nonenzymatic acetylation of histones with acetyl-CoA. Biochim Biophys Acta 213, 513–522 [DOI] [PubMed] [Google Scholar]
- 31.Chang L and Takada S (2016) Histone acetylation dependent energy landscapes in tri-nucleosome revealed by residue-resolved molecular simulations. Sci Rep 6, 34441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H, et al. (2018) Lysine benzoylation is a histone mark regulated by SIRT2. Nat Commun 9, 3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varner EL, et al. (2020) Quantification of lactoyl-CoA (lactyl-CoA) by liquid chromatography mass spectrometry in mammalian cells and tissues. Open Biol 10, 200187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang D, et al. (2019) Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabbani N, et al. (2014) Activity, regulation, copy number and function in the glyoxalase system. Biochem Soc Trans 42, 419–424 [DOI] [PubMed] [Google Scholar]
- 36.Gaffney DO, et al. (2020) Non-enzymatic Lysine Lactoylation of Glycolytic Enzymes. Cell Chem Biol 27, 206–213 e206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diehl KL and Muir TW (2020) Chromatin as a key consumer in the metabolite economy. Nat Chem Biol 16, 620–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakubowski H (1999) Protein homocysteinylation: possible mechanism underlying pathological consequences of elevated homocysteine levels. FASEB J 13, 2277–2283 [PubMed] [Google Scholar]
- 39.Zhang Q, et al. (2018) Elevated H3K79 homocysteinylation causes abnormal gene expression during neural development and subsequent neural tube defects. Nat Commun 9, 3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skrzydlewska E, et al. (2005) Lipid peroxidation and antioxidant status in colorectal cancer. World J Gastroenterol 11, 403–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esterbauer H, et al. (1991) Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med 11, 81–128 [DOI] [PubMed] [Google Scholar]
- 42.West JD and Marnett LJ (2005) Alterations in gene expression induced by the lipid peroxidation product, 4-hydroxy-2-nonenal. Chem Res Toxicol 18, 1642–1653 [DOI] [PubMed] [Google Scholar]
- 43.Castro JP, et al. (2017) 4-Hydroxynonenal (HNE) modified proteins in metabolic diseases. Free Radic Biol Med 111, 309–315 [DOI] [PubMed] [Google Scholar]
- 44.Galligan JJ, et al. (2014) Stable histone adduction by 4-oxo-2-nonenal: a potential link between oxidative stress and epigenetics. J Am Chem Soc 136, 11864–11866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drake J, et al. (2004) 4-Hydroxynonenal oxidatively modifies histones: implications for Alzheimer’s disease. Neurosci Lett 356, 155–158 [DOI] [PubMed] [Google Scholar]
- 46.Sun R, et al. (2017) Chemoproteomics Reveals Chemical Diversity and Dynamics of 4-Oxo-2-nonenal Modifications in Cells. Mol Cell Proteomics 16, 1789–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wellen KE and Thompson CB (2010) Cellular metabolic stress: considering how cells respond to nutrient excess. Mol Cell 40, 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt AM, et al. (2000) The biology of the receptor for advanced glycation end products and its ligands. Biochim Biophys Acta 1498, 99–111 [DOI] [PubMed] [Google Scholar]
- 49.Palanissami G and Paul SFD (2018) RAGE and Its Ligands: Molecular Interplay Between Glycation, Inflammation, and Hallmarks of Cancer-a Review. Horm Cancer 9, 295–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bierhaus A, et al. (2005) Understanding RAGE, the receptor for advanced glycation end products. J Mol Med (Berl) 83, 876–886 [DOI] [PubMed] [Google Scholar]
- 51.Zheng Q, et al. (2019) (De)Toxifying the Epigenetic Code. Chem Res Toxicol 32, 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bommer GT, et al. (2020) Metabolite Repair Enzymes Control Metabolic Damage in Glycolysis. Trends Biochem Sci 45, 228–243 [DOI] [PubMed] [Google Scholar]
- 53.Chang KC and Petrash JM (2018) Aldo-Keto Reductases: Multifunctional Proteins as Therapeutic Targets in Diabetes and Inflammatory Disease. Adv Exp Med Biol 1032, 173–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drozak J, et al. (2010) Molecular identification of carnosine synthase as ATP-grasp domain-containing protein 1 (ATPGD1). J Biol Chem 285, 9346–9356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cripps MJ, et al. (2017) Carnosine scavenging of glucolipotoxic free radicals enhances insulin secretion and glucose uptake. Sci Rep 7, 13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richarme G, et al. (2015) Parkinsonism-associated protein DJ-1/Park7 is a major protein deglycase that repairs methylglyoxal- and glyoxal-glycated cysteine, arginine, and lysine residues. J Biol Chem 290, 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szwergold BS, et al. (2001) Human fructosamine-3-kinase: purification, sequencing, substrate specificity, and evidence of activity in vivo. Diabetes 50, 2139–2147 [DOI] [PubMed] [Google Scholar]
- 58.Zheng Q, et al. (2020) Protein arginine deiminase 4 antagonizes methylglyoxal-induced histone glycation. Nat Commun 11, 3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jing H and Lin H (2015) Sirtuins in epigenetic regulation. Chem Rev 115, 2350–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kosciuk J, et al. (2019) Updates on the epigenetic roles of sirtuins. Curr Opin Chem Biol 51, 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seto E and Yoshida M (2014) Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6, a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kutil Z, et al. (2018) Histone Deacetylase 11 Is a Fatty-Acid Deacylase. ACS Chem Biol 13, 685–693 [DOI] [PubMed] [Google Scholar]
- 63.Jin J, et al. (2016) SIRT2 Reverses 4-Oxononanoyl Lysine Modification on Histones. J Am Chem Soc 138, 12304–12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui Y, et al. (2017) Histone Ketoamide Adduction by 4-Oxo-2-nonenal Is a Reversible Posttranslational Modification Regulated by Sirt2. ACS Chem Biol 12, 47–51 [DOI] [PubMed] [Google Scholar]
- 65.Jennings EQ, et al. (2021) Sirtuin 2 Regulates Protein LactoylLys Modifications. Chembiochem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno-Yruela C, et al. (2021) Class I Histone Deacetylases (HDAC1–3) are Histone Lysine Delactylases. BioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao J, et al. (2015) DJ-1 as a human oncogene and potential therapeutic target. Biochem Pharmacol 93, 241–250 [DOI] [PubMed] [Google Scholar]
- 68.Sanghvi VR, et al. (2019) The Oncogenic Action of NRF2 Depends on De-glycation by Fructosamine-3-Kinase. Cell 178, 807–819 e821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuzhalin AE (2019) Citrullination in Cancer. Cancer Res 79, 1274–1284 [DOI] [PubMed] [Google Scholar]
- 70.Lewis HD, et al. (2015) Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat Chem Biol 11, 189–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drechsel J, et al. (2018) Chemical Probe To Monitor the Parkinsonism-Associated Protein DJ-1 in Live Cells. ACS Chem Biol 13, 2016–2019 [DOI] [PubMed] [Google Scholar]
- 72.Tashiro S, et al. (2018) Discovery and Optimization of Inhibitors of the Parkinson’s Disease Associated Protein DJ-1. ACS Chem Biol 13, 2783–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J, et al. (2015) Generation and characterization of antibodies against arginine-derived advanced glycation endproducts. Bioorg Med Chem Lett 25, 4881–4886 [DOI] [PubMed] [Google Scholar]
- 74.Arnaudo AM and Garcia BA (2013) Proteomic characterization of novel histone post-translational modifications. Epigenetics Chromatin 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brunk E, et al. (2018) Characterizing posttranslational modifications in prokaryotic metabolism using a multiscale workflow. Proc Natl Acad Sci U S A 115, 11096–11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tran JC, et al. (2011) Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature 480, 254–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shechter D, et al. (2007) Extraction, purification and analysis of histones. Nat Protoc 2, 1445–1457 [DOI] [PubMed] [Google Scholar]
- 78.Zheng Q, et al. (2020) Synthesis of an Alkynyl Methylglyoxal Probe to Investigate Nonenzymatic Histone Glycation. J Org Chem 85, 1691–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Galligan JJ, et al. (2017) Quantitative Analysis and Discovery of Lysine and Arginine Modifications. Anal Chem 89, 1299–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sibbersen C, et al. (2018) Profiling of Methylglyoxal Blood Metabolism and Advanced Glycation End-Product Proteome Using a Chemical Probe. ACS Chem Biol 13, 3294–3305 [DOI] [PubMed] [Google Scholar]
- 81.Sibbersen C, et al. (2013) Development of a chemical probe for identifying protein targets of alpha-oxoaldehydes. Chem Commun (Camb) 49, 4012–4014 [DOI] [PubMed] [Google Scholar]
- 82.Chen N, et al. (2018) Chemical proteomic profiling of protein N-homocysteinylation with a thioester probe. Chem Sci 9, 2826–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mir AR, et al. (2014) Methylglyoxal mediated conformational changes in histone H2A-generation of carboxyethylated advanced glycation end products. Int J Biol Macromol 69, 260–266 [DOI] [PubMed] [Google Scholar]
- 84.Arakawa S, et al. (2020) Mass spectrometric quantitation of AGEs and enzymatic crosslinks in human cancellous bone. Sci Rep 10, 18774. [DOI] [PMC free article] [PubMed] [Google Scholar]


