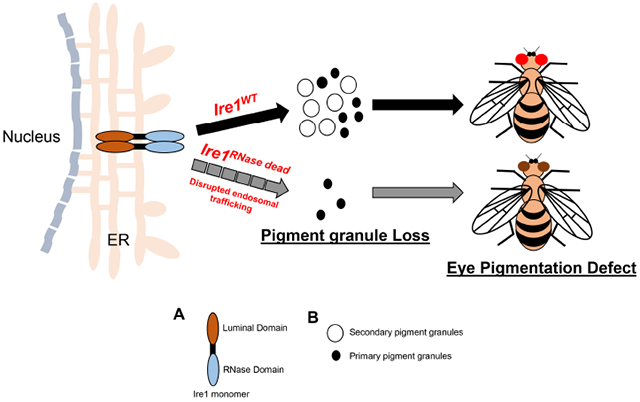
Abstract
Ire1 is an endoplasmic reticulum (ER) transmembrane RNase that cleaves substrate mRNAs to help cells adapt to ER stress. Because there are cell types with physiological ER stress, loss of Ire1 results in metabolic and developmental defects in diverse organisms. In Drosophila, Ire1 mutants show developmental defects at early larval stages and in pupal eye photoreceptor differentiation. These Drosophila studies relied on a single Ire1 loss of function allele with a Piggybac insertion in the coding sequence. Here, we report that an Ire1 allele with a specific impairment in the RNase domain, H890A, unmasks previously unrecognized Ire1 phenotypes in Drosophila eye pigmentation. Specifically, we found that the adult eye pigmentation is altered, and the pigment granules are compromised in Ire1H890A homozygous mosaic eyes. Furthermore, the Ire1H890A mutant eyes had dramatically reduced Rhodopsin-1 protein levels. Drosophila eye pigment granules are most notably associated with late endosome/lysosomal defects. Our results indicate that the loss of Ire1, which would impair ER homeostasis, also results in altered adult eye pigmentation.
Keywords: Ire1, RNase, Drosophila, pigment granules, Rhodopsin-1
Graphical Abstract
Introduction:
Endoplasmic reticulum (ER) is a subcellular organelle where most membrane and secretory proteins undergo synthesis, folding and maturation before being trafficked to their final destination. Failure to properly fold proteins inside this organelle can impose stress broadly throughout the cell. Healthy eukaryotic cells have adaptive responses to those adverse conditions. One of those is the Unfolded Protein Response (UPR), which refers to signaling pathways that regulate gene expression to reduce unfolded/misfolded protein burden [1-3].
In metazoans, the UPR is most notably mediated by the three conserved pathways: Those mediated by Ire1-Xbp1, Perk-Atf4 and Atf6 signaling, respectively [4, 5]. Among these, Ire1 (encoded by Ern1 in mice) is the most conserved throughout phyla, with orthologs present in diverse organisms ranging from yeast to humans [2, 6]. Ire1 is an ER transmembrane protein with a luminal domain (LD) that senses misfolded peptides in the ER, and Kinase and RNase domains on the cytosolic side involved in signaling [6-9]. Upon sensing misfolded proteins inside the ER lumen, Ire1 undergoes dimerization/oligomerization to stimulate Ire1 trans-auto phosphorylation [9-11]. Under these conditions, Ire1’s RNase domain with a conserved Histidine residue gains activity [9, 12-18]. Expressing an RNase dead Ire1 in cultured cells exert dominant negative effects, indicating that Ire1 downstream signaling requires the RNase domains from both of the Ire1 subunits in the active dimer [19].
Upon gaining activity, Ire1’s RNase initiates downstream signaling through the unconventional splicing of the mRNA encoding X-box binding protein −1 (Xbp1). The Xbp1 mRNA has two stem loops that are cleaved by the Ire1 dimer [9]. The resulting spliced Xbp1 isoform encodes a protein that stimulates the transcription of many ER quality control genes, including chaperones, ERAD components, and molecules involved in ER biogenesis, respectively [20-22]. In addition to splicing Xbp1 mRNA, activated Ire1 utilizes its RNase function to cleave a subset of mRNAs by a process termed as regulated Ire1 dependent decay (RIDD) [23-25]. Therefore, impairing the RNase domain of Ire1 is predicted to abolish both Xbp1 splicing and RIDD activation. In addition, recent studies have found that Ire1 also has RNase independent functions in regulating cytoskeletal remodeling [26].
Reflecting the essential role of UPR in normal physiology, loss of Ire1 causes metabolic and development defects across phyla. Examples include lipid metabolism defects in S. cerevisiae [27] and placenta, liver and pancreas dysfunction associated with IRE1α knockouts in mice [5, 28, 29], Drosophila encodes a single Ire1 gene that is essential for early larval development [13, 30]. In addition, mosaic clones of Ire1 during later stages of development revealed Ire1’s crucial roles in developing photoreceptor cells of the Drosophila eye. Xbp1 null mutants do not show phenotypes in the Drosophila eye, indicating that Ire1 regulates photoreceptor homeostasis through an Xbp1-independent mechanism [5, 13, 25]. Notably, these Drosophila Ire1 studies have relied on a single loss-of-function allele, f02170, which has a PiggyBac insertion in Ire1’s coding sequence.
Here, we report previously unrecognized roles of Ire1 based on a CRISPR-Cas9 engineered RNase-dead Ire1 mutant allele. We specifically find that homozygous loss-of-function RNase dead Ire1 eyes exhibit defects in eye pigmentation. We further validate this phenotype through independent genetic methods. In addition, these Ire1 mutant eyes show a more dramatic loss of Rhodopsin-1 (Rh1) protein in photoreceptors. Altogether, our present findings establish a new physiological role of Ire1 in maintaining eye pigmentation dependent on Ire1’s RNase activity.
Materials and Methods:
Generation of CRISPR knock in H890A allele:
H890A knock in allele (referred to as Ire1H890A) was generated through Well Genetics (www.wellgenetics.com) that used a CRISPR/Cas9 – mediated genome editing and homology-dependent repair (HDR) strategy. Specifically, we used a single guide RNA targeting the endogenous locus of Ire1/CG4583 on chromosome 3R (92B1-92B7). To generate the point mutation H890A of Ire1, CAT (Histidine) to GCT (Alanine) mutation was introduced in the targeted genomic position. PBacDsRed was used as a dsDNA plasmid donor knock- in cassette.
Following guide RNA primer pairs were used:
Sense oligo 5’-CTTCGCACTGGCGCCCATATATCCG,
Antisense oligo 5’-AAACCGGATATATGGGCGCCAGTGC
w1118 was used as an injection strain. All the mutant allele containing lines were selected using the DsRed expression marker, and this selection marker was removed using the Cre/Lox strategy from the final balanced fly.
Fly stock maintenance and Constructs:
Drosophila melanogaster strains were maintained on cornmeal/agar molasses fly food in the 25°C incubator. Mutant eye clones were generated by the FLP/FRT technique [31]. where Flipase expression was controlled under the eyeless promoter [32]. To assess the effect of Ire1 mutants on the overall adult eye color, and to assess Rh1 protein levels, we generated adult flies mostly consisting of Ire1 homozygous mutant clones using the FRT, cl-r3 w+ (cell lethal) chromosome as described previously [32].
The following flies lines used for this study were described previously: Ire1f02170 [33], GMR-Gal4 [34], ey-FLP (Bloomington Stock Center, #5576), FRT82B, cl-r3 P[35]90E (Bloomington Stock Center #5620), FRT82B, P{w+}90E (Bloomington Stock Center # 2050), FRT82B, uas-dsRed [25], UAS-xbp1-EGFP [36]. The Ire1 genomic rescue lines P{Ire1 wt genomic rescue} and P{Ire1 H890A genomic rescue} have HA-epitopes fused to the Ire1 protein, and includes the 8.8 kb Ire1 genomic locus as described previously [13].
These lines were used to generate the following stocks for this study:
GMR-Gal4, ey-FLP;; FRT82B, p{w+)90E, cl-r3/TM6B
w1118;; FRT82B, Ire1H890A/TM6B
w1118;; FRT82B, P{w+}90E/TM6B
GMR-Gal4, ey-FLP;UAS-Xbp1-EGFP/Cyo; FRT82B,UAS-DsRed/MKRS
w1118; P{Ire1WT genomic rescue}; FRT82B, Ire1f02170/TM6B
w1118; P{Ire1H890A genomic rescue}; FRT82B, Ire1f02170/TM6B
Imaging of adult fly eye:
Female fly eyes two days after eclosion were imaged under bright field microscopy (Nikon SMZ1500).
For transmission electron microscopy (TEM), the head of the adult female flies were hemi sectioned, eyes were isolated and fixed in 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cadodylate buffer. Samples were processed using the standard core facility service (NYU).
Quantitative estimation of fly eye pigment:
Red (pteridine) and brown (ommochrome) eye pigments were extracted and measured accordingly as described in Ooi et. al. [37]. 20 female fly heads (two days after eclosion) were used for pigment extraction. The absorption spectra were measured by using biospectrometer (eppendorf).
Immunohistochemistry and Western Blot:
Third instar larval eye discs were dissected in 1XPBS and fixed in 4%formaldehyde. After fixation, discs were washed with 1X PBS with 0.2% Triton-X (1X PBT) for 3 times, followed by blocking in 10% normal goat serum for 1 hour at room temperature (RT). Discs were incubated in indicated primary antibodies overnight in 0.2% PBT. After three 10 min washes at RT with 1X PBT, they were incubated with secondary antibodies for 1 hour at room temperature.
Adult fly heads were chopped and lysed in cold lysis buffer (10 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1 mM EDTA; 1% SDS and 1X protease inhibitor). Supernatant was collected upon centrifugation at 13000 RPM for 20 min at 4°C. 10 μl of each sample were loaded onto the 10% SDS-PAGE gel and blotted with the indicated antibodies. Primary antibodies used during the studies are as follows: Rabbit anti-GFP (1:500, Invitrogen # A6455), Rabbit anti-HA (1:1000, Cell signaling, C29F4), Mouse anti-tubulin (1:10000, Biolegend, 903401).
qRT-PCR analysis from adult retina:
For qRT-PCR, 10 pairs of adult retinas were isolated from each indicated genotype and 500 ng of total RNA was extracted using Trizol with random hexamers using Maxima H Reverse transcriptase (Thermo Scientific, cat no: EP0752). Real time PCR was performed on a Biorad CFX96 Touch Real time PCR detection system (Biorad, cat no: 1855196) using Power syber green mix (Life technologies, cat no: 4367659). For mRNA fold change, Rpl15 was used as the normalizing housekeeping gene. Following primers were used during the study:
Ire1 F: GCACTGGCAGCAATGGTA
Ire1 R: AGCACTTCATTTGTGCTGAAGC
Rpl15 F: AGGATGCACTTATGGCAAGC
Rpl15 R: GCGCAATCCAATACGAGTTC
Results:
An RNase dead Ire1 allele shows an eye pigmentation defect and loss of pigment granules in adult fly eyes
Virtually all known Ire1 downstream signaling requires Ire1’s RNase function. Disrupting the RNase domain not only abolishes Ire1 function but also has a dominant negative effect when overexpressed in cultured mammalian cells [19]. To examine how cells respond to the loss of Ire1’s RNase function in vivo, we generated a CRISPR knock-in allele of Ire1 where the essential Histidine residue in the RNase domain was mutated (Materials and Methods). The design of the amino acid substitution in the Drosophila Ire1, Histidine 890 to Alanine (H890A), was based on previous Ire1 structure function studies in yeast and mammals [9, 10](Fig. 1A). Henceforth, we refer to this allele as Ire1H890A.
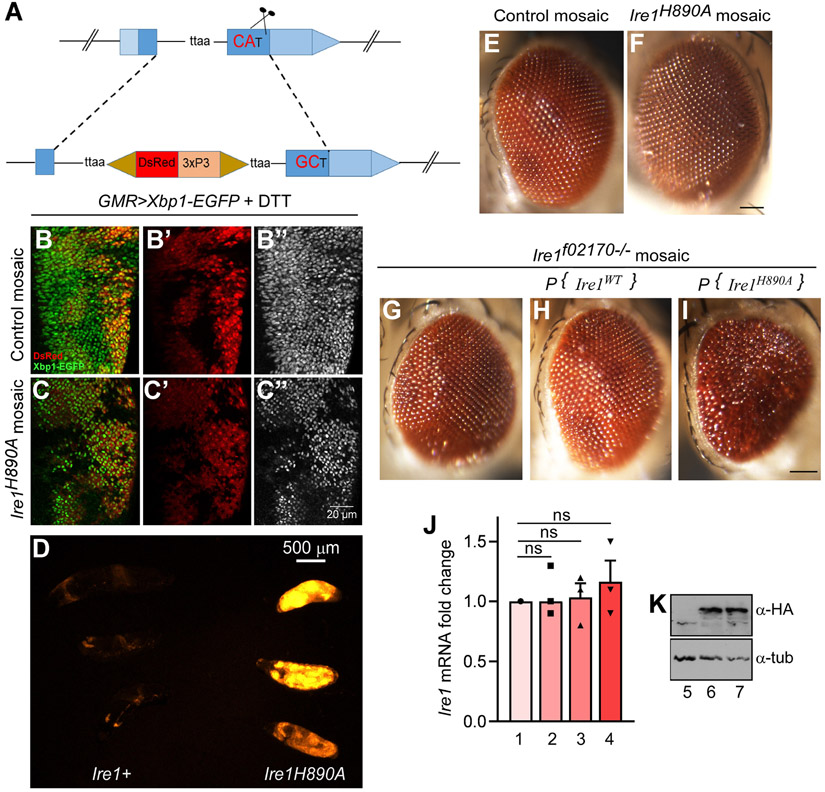
Figure 1: Drosophila eyes with Ire1 RNase dead mosaic clones show an eye pigmentation phenotype.
(A) Schematic representation of Ire1H890A CRISPR knock in mutant allele generation. (B, C) Ire1H890A mosaic clones impair Xbp1 mRNA splicing in response to ER stress. Ire1 RNase activity was assessed through the Ire1 RNase sensor, Xbp1-EGFP (green), expressed through the GMR-Gal4 driver. These eye discs were challenged with DTT treatment. A disc with control wild type mosaic clones (B) and a disc with Ire1H890A homozygous clones that are marked by the absence of DsRed (C). DsRed only channels (B’, C’) and the GFP only channels (B”, C”) are shown. (D) 4E-BPintron-DsRed stress-responsive reporter in the background of Ire1wild type (left) and homozygous Ire1H890A mutant backgrounds (right). Shown are three independent larvae for each genotype, at 24-48 hrs after egg laying. Ire1H890A mutant larvae fail to survive beyond this first instar larval stage. (E and F) Bright field images of the fly eyes bearing either neutral mosaic clones (E) or Ire1H890A homozygous clones (F). To generate eyes mostly consisting of the mutant clones, FRT82, cl (cell lethal) chromosomes were employed (see genotype below). All imaged flies are females. (G, H and I) Fly eyes expressing Ire1 transgenes (Wild type and mutant respectively) with Ire1f02170 mosaic clones. Under these conditions, flies bearing the Ire1H890A rescue transgene shows brown pigmentation not seen in the control flies. (J) qPCR analysis of endogenous Ire1 from the indicated genotypes (see below). Each data was normalized to the qPCR values for Rpl15. n =3 for each genotype. n.s. indicates “not significant.” (K) Adjacent western blot shows the expression of Ire1 transgenes fused with HA-epitopes (upper gel). Anti-tubulin represents the loading control (lower gel). The scale bar in (C”) is for all panels of B and C. There is a separate scale bar for D (approximately 500 μm). Genotypes: (B, B’, B”) GMR-Gal4, ey-FLP/+; UAS-Xbp1-EGFP/+; FRT82B UAS-DsRed/FRT82B. (C, C’, C”) GMR-Gal4, ey-FLP/+: UAS-Xbp1-EGFP/+: FRT82B UAS-DsRed/ FRT82B, Ire1H890A. (D) 4E-BPintron-DsRed/+:+ (left) & 4E-BPintron-DsRed/+:Ire1H890A/H890A (right). (E) GMR-Gal4, ey-FLP/+ ;; FRT82B/ FRT82B, p{w+}90E, cl. (F) GMR-Gal4, ey-FLP/+;;FRT82B, Ire1H890A/ FRT82B, p{w+}90E, cl. (G) GMR-Gal4, ey-FLP/+;;FRT82B , Ire1f02170/ FRT82B, p{w+}90E, cl. (H) GMR-Gal4, ey-FLP/+; P{Ire1WT}/+; FRT82B, Ire1f02170/FRT82B, p{w+}90E, cl. (I) GMR-Gal4, ey-FLP/+; P{Ire1H890A}/+; FRT82B, Ire1f02170/FRT82B, p{w+}90E, cl. (J) In the graph, Bar 1, 2, 3 and 4 represent GMR-Gal4, ey-FLP/+ ;; FRT82B/ FRT82B, p{w+}90E, cl, GMR-Gal4, ey-FLP/+;;FRT82B, Ire1H890A/ FRT82B, p{w+}90E, cl, GMR-Gal4, ey-FLP/+; P{Ire1WT}/+; FRT82B, Ire1f02170/FRT82B, p{w+}90E, cl and GMR-Gal4, ey-FLP/+; P{Ire1H890A}/+; FRT82B, Ire1f02170/FRT82B, p{w+}90E, cl. respectively. (K) Described as lanes 5, 6 and 7, identical to the genotype of Fig G, H and I respectively.
To confirm whether our engineered allele impairs Ire1’s RNase function, we examined the Ire1 activity sensor Xbp1-EGFP, where EGFP becomes expressed in frame with Xbp1 dependent on Ire1-mediated Xbp1 mRNA splicing [36, 38]. Eye discs expressing Xbp1-EGFP were challenged with 2 mM of the ER stress inducer Dithiothreitol (DTT) for 4 hours at room temperature in S2 media. Confocal microscopy imaging showed that such conditions activated the Xbp1-EGFP reporter in Ire1+ cells of the imaginal discs. However, we found a significant reduction in the Xbp1-EGFP signal from the clones homozygous for the Ire1H890A allele, which were marked by the absence of DsRed (Fig. 1B, C, 1B” and 1C” respectively).
We further examined whether the newly developed Ire1H890A allele is essential for larval development. The well-characterized loss of function allele, Ire1f01270, fails to develop beyond the first instar larval stage (24 – 48 hours after egg laying (AEL)), and shows signs of excessive physiological stress as evidenced by the induction of 4E-BPintron-DsRed reporter [13]. We found that Ire1H890A/H890A homozygous larvae also reach the first instar larval stage, but fail to survive beyond this stage. The surviving Ire1H890A/H890A homozygous larvae also showed intense activation of 4E-BPintron-DsRed reporter (Fig. 1D). These results show that the newly engineered allele (Ire1H890A) shows phenotypes similar to that reported for Ire1f01270 during larval development [13].
We further examined the adult eyes of flies. To generate fly eyes consisting of mostly Ire1H890A homozygous mosaic clones, we used a cell-lethal mutation-bearing FRT chromosome that eliminates most Ire1 wild type cells [32]. We found that these adult eyes made up of Ire1H890A homozygous clones had an eye pigment color different from wild type controls (Fig.1E and 1F, respectively). To corroborate whether the eye pigment phenotype was due to the Ire1H890A allele, we employed an independent method to generate eyes that only expressed the H890A allele. Specifically, we generated eyes made up of primarily Ire1f02170 loss-of-function clones, and in this background, introduced an Ire1H890A genomic rescue transgene (P {Ire1H890A genomic rescue}) that we had reported previously [13]. We compared the eye phenotype with those rescued with an equivalent wild type Ire1 genomic rescue transgene. The eye colors of Ire1f02170 mosaic eyes rescued with P {Ire1H890A genomic rescue} were distinct from that of other control lines, validating our earlier observation that Ire1H890A mutants affect eye pigmentation (Fig.1G, 1H, and 1I, respectively). We performed q-RT-PCR in these fly eyes and confirmed that there were no statistically significant differences in Ire1 transcript levels between the H890A flies and other control conditions (Fig. 1J). These results indicate that the pigmentation phenotype is not a result of any transgene over-expression.
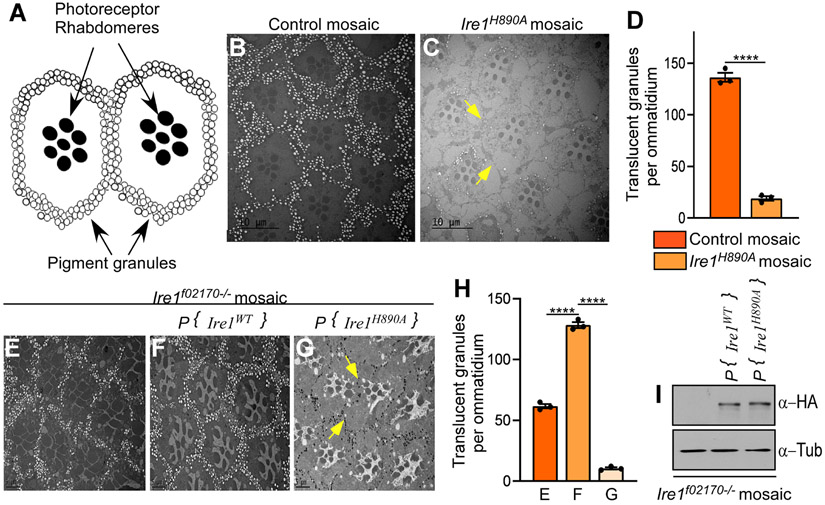
To examine the adult eye at a higher resolution, we further employed Transmission Electron Microscopy (TEM). Adult eyes bearing CRISPR knock-in Ire1H890A homozygous clones (Fig. 2B and 2C respectively), showed loss of secondary pigment cell granules (indicated by the yellow arrow) as compared to control fly eyes (Quantified in Fig. 2D). We independently examined secondary pigment granules in Ire1f02170 homozygous mosaic eyes. Those rescued with P{Ire1H890A genomic rescue} showed a dramatic loss of secondary pigment granules compared to those rescued with Ire1 wild type (Figure 2F, G, H). Upon a closer inspection, Ire1f02170 mosaic eyes without any rescue transgenes also showed a mild reduction of pigment granules, although not as striking as in the Ire1H890A eyes (Figure 2E - H). Together, these results indicate that the Ire1H890A RNase dead mutant allele reveals a previously unrecognized Ire1 role in fly eye pigmentation.
Figure 2: Transmission Electron Microscopy (TEM) imaging of adult eyes.
(A) A schematic representation of a Drosophila compound eye unit, ommatidia structure. (B and C) fly eyes with either control wild type mosaic clones (B) or those with Ire1H890A homozygous clones (C). The granules in the secondary pigment cells often appear as translucent, due to the processing artifact during electron microscopy imaging. Here, translucent granules representing secondary pigment granules/ ommatidium from were counted and plotted in the adjacent graph, where **** represents p< 0.0001 (D). (E, F and G) Fly eyes with Irelf02170 homozygous mosaic clones. (E) Mutant clones not rescued with any transgenes. (F) Rescued with an Ire1 wild type transgene. (G) Rescued with the Ire1H890A transgene. Note that eyes with either Ire1 H890A rescue transgene or Ire1H890A CRISPR allele show significant loss of translucent pigment granules as indicated by the yellow arrows. The scale bar represents 10 and 20 μm in the indicated images. Adjacent graph is the quantitative representation of the translucent pigment granules/ommatidium, where **** represents p< 0.0001 (H). (G) Adjacent blot represents the expression of HA-tagged Ire1 transgenes. Here, tubulin is the loading control. Genotypes: (B) GMR-Gal4, ey-FLP/+ ;; FRT82B/ FRT82B, p{w+}90E, cl. (C) GMR-Gal4, ey-FLP/+;; FRT82B, Ire1H890A/FRT82B, p{w+}90E, cl. (E) GMR-Gal4, ey-FLP/+;; FRT82B, Ire1f02170/FRT82B, p{w+}90E, cl. (F) GMR-Gal4, ey-FLP/+;P{ Ire1WT}/+; FRT82B, Ire1f02170/ FRT82B, p{w+}90E, cl. (G) GMR-Gal4, ey-FLP/+; P{Ire1H890A}/+; FRT82B, Ire1f02170/FRT82B, p{w+} [90E] cl. (I) Same genotype as described in Fig E,F and G respectively.
Ire1’s RNase dead mutation causes loss of both red and brown pigments (Pteridine and ommochrome) in the adult fly eye
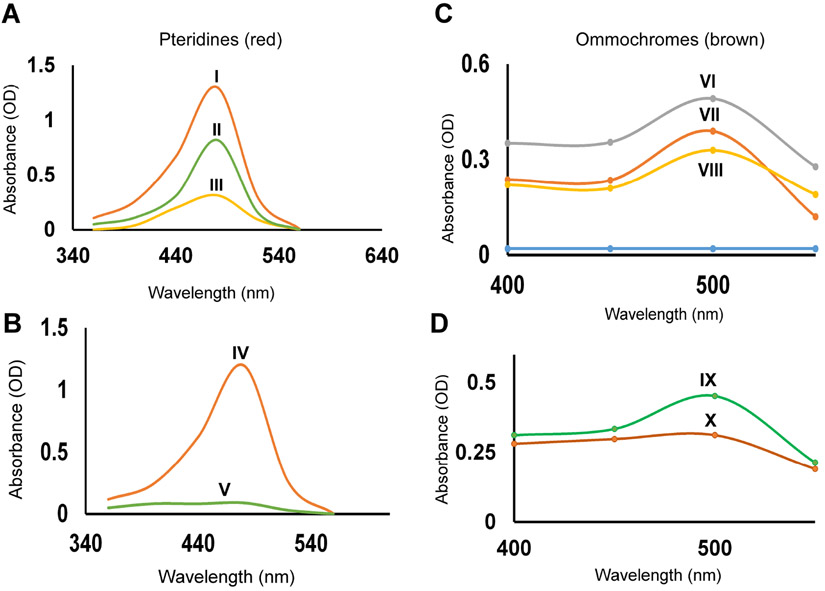
To determine the nature of pigment synthesis affected by the impairment in Ire1’s RNase activity, pigments were extracted and measured from the indicated genotypes. Quantitative estimation shows a significant reduction in both red and brown pigment from both female fly eyes bearing either Ire1H890A transgene or CRISPR knock-in Ire1H890A allele (Fig. 3A, curve III; 3B, curve V; 3C, curve VIII and 3D, curve X respectively). The eyes with the homozygous mutant Ire1f02170 clones also showed a reduction in both pteridine and ommochrome level, but to a more moderate degree (Fig. 3A, curve II and 3C, VII respectively). This observation correlates with our previous finding in Fig. 2 that the mutation in Ire1’s RNase domain results in pigment granule loss, thereby causing pigmentation defect in adult eye.
Figure 3: Quantitative estimation of red (Pteridine) and brown (ommochrome) eye pigments from the female fly eyes.
Shown are quantification values of the eye pigments, pteridines (A, B) and ommochromes (C, D). (A & C) The absorbance (OD) of extracted eye pigments from those of Ire1f02170 homozygous mosaic clones that were rescued with either the wild type Ire1 transgene (I, VI), not rescued with any transgene (II & VII), or rescued with the Ire1 RNase dead mutant (III & VIII) transgene. (B) The absorbance of extracted eye pigments from adult eyes with wild type control mosaic clones (IV & IX), or with the Ire1H890A CRISPR allele clones (V & X). Comparative analysis of eye pigment levels showed a striking loss of both red and brown pigment in presence of Ire1 RNase dead mutation. Genotypes: (I & VI) GMR-Gal4, ey-FLP/+;P{Ire1WT}/+; FRT82B, Ire1f02170/ FRT82B, p{w+}90E, cl. (II & VII) GMR-Gal4, ey-FLP/+;; FRT82B , Ire1f02170/ FRT82B, p{w+}90E, cl. (III & VIII) GMR-Gal4, ey-FLP/+; P{Ire1H890A}/+; FRT82B, Ire1f02170/ FRT82B, p{w+} [90E] cl. (IV & IX) GMR-Gal4, ey-FLP/+;; FRT82B/ FRT82B, p{w+}90E, cl. (V & X) GMR-Gal4, ey-FLP/+;; FRT82B, Ire1H890A/ FRT82B, p{w+}90E, cl. The blue line in Fig. C represents w1118 as a control.
Ire1’s RNase activity is required to maintain Rh1 levels in adult fly eye
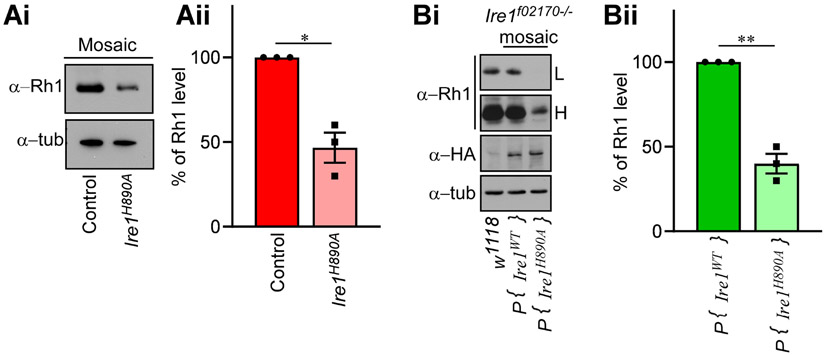
Rhodopsin-1 (Rh1) is a light-detecting GPCR protein in Drosophila photoreceptors that undergoes trafficking and maturation through the secretory pathway [39, 40]. The levels of Rh1 could be influenced by the adjacent pigment cells [41]. Therefore, we examined whether the Ire1H890A allele affects Rh1 levels in adult eyes. Western blot analysis of Rh1 from both eyes with mosaic Ire1H890A clones and those with Ire1f02170 homozygous clones rescued with P{Ire1 H890A genomic rescue} show a significant reduction in the Rh1 levels, as compared to control flies (Fig. 4A and 4B respectively). This observation is consistent with the previous findings where trafficking defect of Rh1 to rhabdomere eventually leads to the reduction of Rh1 [25]. These results support the idea that the Ire1H890A acts as a loss-of-function allele that can be rescued by a wild type allele.
Figure 4: RNase dead mutation of Ire1 causes a severe reduction in Rhodopsin-1 (Rh1) levels.
Shown are western blots from adult head extracts. (Ai & Aii) Samples from eyes with Ire1 H890A mosaic clones have reduced Rh1 levels. (Bi & Bii) Rh1 levels from samples with or without Iref02170 mosaic clones expressing the indicated HA-tagged Ire1 rescue transgenes. Lower gel shows tubulin blots as a loading control. L and H indicate low and high exposures, respectively. For each blot, the intensities of Rh1 bands were normalized to that of tubulin and plotted in the adjacent graph (Aii & Bii respectively). For graph (Bii), high exposure (H) Rh1 blots were used for quantification and Rh1 level was plotted using WT transgene expressing fly as a control. Each data is the representation of three independent experiments. Here, * is p< 0.01 and ** is p<0.001. Genotypes: (A) (lane 1) GMR-Gal4, ey-FLP/+; FRT82B/FRT82B, p {w+} 90E, cl. (lane 2) GMR-Gal4, ey-FLP/+; FRT82B, Ire1 H890A/ FRT82B, p {w+} 90E, cl. (B) (lane 1) w1118. (Lane 2) GMR-Gal4, ey-FLP/+; P {Ire1WT rescue}/+; FRT82B, Ire1f02170/ FRT82B, p {w+} 90E, cl. (lane 3) GMR-Gal4, ey-FLP/+; p{Ire1H890A rescue}/+; FRT82B, Ire1f02170/FRT82B, p{w+}90E, cl.
Discussion:
Previous studies had shown that Ire1 signaling is physiologically essential in various aspects in fly physiology and development including photoreceptor differentiation, rhabdomere morphogenesis and larval development [5, 13, 25]. The indispensable role for Ire1 in metazoan development has also been documented in Arabidopsis and mice placenta development [42, 43]. Our present study reveals a new physiological role of Ire1 in fly eye pigmentation.
Fly eye color is determined by pigment granules, which contain either of the two different types of pigments, the brown ommochromes and the red pteridines [44]. These pigment granules are specialized forms of late endosomes and lysosomes, and pigment granule biogenesis depends on genes that help traffic proteins to the lysosomes [37, 44-48]. A previous study had shown that Ire1 loss disrupts ER structure in photoreceptor cells (PR) and thereby impairs the trafficking dynamics of Rhodopsin-1 (Rh1) [49]. Intriguingly, Ire1 activity was also detected in the pigment cells at the final stage of fly eye development, but no loss-of-function phenotypes were reported in those cells [25] . Since many lysosomal proteins and trafficking factors are initially synthesized and folded in the ER, we interpret that Ire1 loss gives rise to a pigment phenotype by impairing proper protein trafficking to the pigment granules. Our results indicate that Drosophila eye pigment cells have physiological ER stress during fly eye development that requires Ire1 for maintaining the integrity of the ER and the pigment granules.
It is intriguing that this pigment phenotype had evaded detection in previous studies based on the Ire1f02170 mutant allele. That allele has a Piggybac element inserted within the coding sequence, predicted to abolish Ire1 function completely. Our observations with the H890A allele now force us to revisit the assumption that f02170 completely abolishes Ire1 function in mosaic clones. One possible explanation is that the mutant mosaic clones have residual Ire1 mRNA or protein originating from the heterozygotic mother cells. Such perdurance is not unprecedented, as demonstrated in the case of Gal80, which persists when generating MARCM clones [50]. If this is the case, we interpret that Ire1H890A reveals masked phenotypes by acting as a dominant negative against the remaining wild type Ire1 protein within the mosaic clones.
Alternatively, one could argue that Ire1H890A exhibits a neomorphic property, and the pigment phenotype seen in Ire1H890A mutants is not a loss-of-function phenotype. However, we consider this interpretation unlikely because any gain-of-function Ire1H890A phenotype should manifest even when the Ire1H890A allele is present together with the wild type Ire1 allele, which we do not see: The Ire1H890A allele maintained as a heterozygote do not exhibit an eye pigment phenotype. Furthermore, the P {Ire1 H890A genomic rescue} transgene maintained in the Ire1 wild type background has eye pigmentation indistinguishable from control flies.
Rh1 levels that decrease in Ire1H890A correlate with this allele’s effect on pigment cells. While such reduction in Rh1 could be due to Irel’s autonomous role in photoreceptors as had been previously reported [25], it is notable that pigment cells affect Rh1 protein levels[35]. Specifically, Rh1 levels could decrease when there are insufficient 11-cis-3-hydroxyretinal chromophores available in photoreceptor [51-53]. The continuous recycling of these chromophores from the adjacent pigment cell layer is essential for Rh1 maturation [41]. These observations leave open the possibility that Ire1H890A mutants affect Rh1 levels, at least in part, by impacting pigment cells.
In conclusion, our results show that the Ire1H890A mutant allele uncovers phenotypes that were not clear in the Ire1f02170 allele. While eye pigmentation phenotypes are mostly associated with the loss of late endosomal/lysosomal trafficking factors, the current study demonstrates that the phenotype could also manifest by the loss of an ER quality control gene.
Highlights.
Ire1 plays a previously unrecognized role in Drosophila eye pigmentation
The eye pigmentation phenotype is more pronounced in RNase dead mutants
Loss of Ire1 RNase in the eye reduces Rhodopsin-1 protein levels
Acknowledgements
We thank Dr. Jessica Treisman for providing fly stocks, Dr. Mindong Ren for technical advice, and Dr. Justin Kumar for comments on the manuscript. We thank NYU Langone Health DART Microscopy Lab Alice Liang, Chris Petzold and Kristen Dancel-Manning for their consultation and assistance with TEM work. We thank Huai-Wei Huang for advice regarding the adult retina isolation. This core is partially funded by NYU Cancer Center Support Grant NIH/NCI P30CA016087. The overall project was supported by the NIH grant R01 EY020866 to H.D.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: The authors declare no competing interests.
References
- 1.Han J and Kaufman RJ, Physiological/pathological ramifications of transcription factors in the unfolded protein response. Genes Dev, 2017. 31(14): p. 1417–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walter P and Ron D, The unfolded protein response: from stress pathway to homeostatic regulation. Science, 2011. 334(6059): p. 1081–6. [DOI] [PubMed] [Google Scholar]
- 3.Hetz C and Papa FR, The Unfolded Protein Response and Cell Fate Control. Mol Cell, 2018. 69(2): p. 169–181. [DOI] [PubMed] [Google Scholar]
- 4.Ryoo HD, Drosophila as a model for unfolded protein response research. BMB Rep, 2015. 48(8): p. 445–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitra S and Ryoo HD, The unfolded protein response in metazoan development. J Cell Sci, 2019. 132(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox JS, Shamu CE, and Walter P, Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell, 1993. 73(6): p. 1197–206. [DOI] [PubMed] [Google Scholar]
- 7.Cox JS and Walter P, A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell, 1996. 87(3): p. 391–404. [DOI] [PubMed] [Google Scholar]
- 8.Mori K, et al. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell, 1993. 74(4): p. 743–56. [DOI] [PubMed] [Google Scholar]
- 9.Lee KP, et al. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell, 2008. 132(1): p. 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korennykh AV, et al. The unfolded protein response signals through high-order assembly of Ire1. Nature, 2009. 457(7230): p. 687–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, et al. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc Natl Acad Sci U S A, 2010. 107(37): p. 16113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shamu CE and Walter P, Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J, 1996. 15(12): p. 3028–39. [PMC free article] [PubMed] [Google Scholar]
- 13.Huang HW, et al. The requirement of IRE1 and XBP1 in resolving physiological stress during Drosophila development. J Cell Sci, 2017. 130(18): p. 3040–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aragon T, et al. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature, 2009. 457(7230): p. 736–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Credle JJ, et al. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci U S A, 2005. 102(52): p. 18773–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner BM and Walter P, Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science, 2011. 333(6051): p. 1891–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, et al. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci U S A, 2006. 103(39): p. 14343–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korennykh AV, et al. Structural and functional basis for RNA cleavage by Ire1. BMC Biol, 2011. 9: p. 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang K, et al. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest, 2005. 115(2): p. 268–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature, 2002. 415(6867): p. 92–6. [DOI] [PubMed] [Google Scholar]
- 21.Shen X, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell, 2001. 107(7): p. 893–903. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida H, et al. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell, 2001. 107(7): p. 881–91. [DOI] [PubMed] [Google Scholar]
- 23.Hollien J and Weissman JS, Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science, 2006. 313(5783): p. 104–7. [DOI] [PubMed] [Google Scholar]
- 24.Hollien J, et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol, 2009. 186(3): p. 323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coelho DS, et al. Xbp1-independent Ire1 signaling is required for photoreceptor differentiation and rhabdomere morphogenesis in Drosophila. Cell Rep, 2013. 5(3): p. 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urra H, et al. IRE1alpha governs cytoskeleton remodelling and cell migration through a direct interaction with filamin A. Nat Cell Biol, 2018. 20(8): p. 942–953. [DOI] [PubMed] [Google Scholar]
- 27.Nikawa J and Yamashita S, IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol Microbiol, 1992. 6(11): p. 1441–6. [DOI] [PubMed] [Google Scholar]
- 28.Zhang K, et al. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. EMBO J, 2011. 30(7): p. 1357–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwawaki T, Akai R, and Kohno K, IRE1alpha disruption causes histological abnormality of exocrine tissues, increase of blood glucose level, and decrease of serum immunoglobulin level. PLoS One, 2010. 5(9): p. e13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryoo HD, Li J, and Kang MJ, Drosophila XBP1 expression reporter marks cells under endoplasmic reticulum stress and with high protein secretory load. PLoS One, 2013. 8(9): p. e75774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golic KG, Site-specific recombination between homologous chromosomes in Drosophila. Science, 1991. 252(5008): p. 958–61. [DOI] [PubMed] [Google Scholar]
- 32.Newsome TP, Asling B, and Dickson BJ, Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development, 2000. 127(4): p. 851–60. [DOI] [PubMed] [Google Scholar]
- 33.Kang MJ, Chung J, and Ryoo HD, CDK5 and MEKK1 mediate pro-apoptotic signalling following endoplasmic reticulum stress in an autosomal dominant retinitis pigmentosa model. Nat Cell Biol, 2012. 14(4): p. 409–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freeman M, Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell, 1996. 87(4): p. 651–60. [DOI] [PubMed] [Google Scholar]
- 35.Dourlen P, et al. Drosophila fatty acid transport protein regulates rhodopsin-1 metabolism and is required for photoreceptor neuron survival. PLoS Genet, 2012. 8(7): p. e1002833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryoo HD, et al. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J, 2007. 26(1): p. 242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ooi CE, et al. Altered expression of a novel adaptin leads to defective pigment granule biogenesis in the Drosophila eye color mutant garnet. EMBO J, 1997. 16(15): p. 4508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sone M, et al. A modified UPR stress sensing system reveals a novel tissue distribution of IRE1/XBP1 activity during normal Drosophila development. Cell Stress Chaperones, 2013. 18(3): p. 307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinal N and Pichaud F, Dynamin- and Rab5-dependent endocytosis is required to prevent Drosophila photoreceptor degeneration. J Cell Sci, 2011. 124(Pt 9): p. 1564–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang HY and Ready DF, Rescue of photoreceptor degeneration in rhodopsin-null Drosophila mutants by activated Rac1. Science, 2000. 290(5498): p. 1978–80. [DOI] [PubMed] [Google Scholar]
- 41.Tian Y, et al. Neurexin regulates visual function via mediating retinoid transport to promote rhodopsin maturation. Neuron, 2013. 77(2): p. 311–22. [DOI] [PubMed] [Google Scholar]
- 42.Iwawaki T, et al. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci U S A, 2009. 106(39): p. 16657–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mishiba KI, et al. Unfolded protein-independent IRE1 activation contributes to multifaceted developmental processes in Arabidopsis. Life Sci Alliance, 2019. 2(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lloyd V, Ramaswami M, and Kramer H, Not just pretty eyes: Drosophila eye-colour mutations and lysosomal delivery. Trends Cell Biol, 1998. 8(7): p. 257–9. [DOI] [PubMed] [Google Scholar]
- 45.Odorizzi G, Cowles CR, and Emr SD, The AP-3 complex: a coat of many colours. Trends Cell Biol, 1998. 8(7): p. 282–8. [DOI] [PubMed] [Google Scholar]
- 46.Simpson F, et al. Characterization of the adaptor-related protein complex, AP-3. J Cell Biol, 1997. 137(4): p. 835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sevrioukov EA, et al. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol Cell, 1999. 4(4): p. 479–86. [DOI] [PubMed] [Google Scholar]
- 48.Ma J, et al. Lightoid and Claret: a rab GTPase and its putative guanine nucleotide exchange factor in biogenesis of Drosophila eye pigment granules. Proc Natl Acad Sci U S A, 2004. 101(32): p. 11652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Z, et al. Ire1 supports normal ER differentiation in developing Drosophila photoreceptors. J Cell Sci, 2016. 129(5): p. 921–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bohm RA, et al. A genetic mosaic approach for neural circuit mapping in Drosophila. Proc Natl Acad Sci U S A, 2010. 107(37): p. 16378–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu G, et al. Drosophila ninaB and ninaD act outside of retina to produce rhodopsin chromophore. J Biol Chem, 2004. 279(18): p. 18608–13. [DOI] [PubMed] [Google Scholar]
- 52.Ahmad ST, et al. The role of Drosophila ninaG oxidoreductase in visual pigment chromophore biogenesis. J Biol Chem, 2006. 281(14): p. 9205–9. [DOI] [PubMed] [Google Scholar]
- 53.Wang T, Jiao Y, and Montell C, Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J Cell Biol, 2007. 177(2): p. 305–16. [DOI] [PMC free article] [PubMed] [Google Scholar]